Summary
While microbial communities inhabit a wide variety of complex natural environments, in vitro culturing enables highly controlled conditions and high-throughput interrogation for generating mechanistic insights. In vitro assemblies of gut commensals have recently been introduced as models for the intestinal microbiota, which plays fundamental roles in host health. However, a protocol for 16S rRNA sequencing and analysis of in vitro samples that optimizes financial cost, time/effort, and accuracy/reproducibility has yet to be established. Here, we systematically identify protocol elements that have significant impact, introduce bias, and/or can be simplified. Our results indicate that community diversity and composition are generally unaffected by substantial protocol streamlining. Additionally, we demonstrate that a strictly aerobic halophile is an effective spike-in for estimating absolute abundances in communities of anaerobic gut commensals. This time- and money-saving protocol should accelerate discovery by increasing 16S rRNA data reliability and comparability and through the incorporation of absolute abundance estimates.
Subject areas: Microbiology, Methodology in biological sciences
Graphical abstract

Highlights
-
•
Systematic optimization of 16S library generation from gut in vitro communities
-
•
Community diversity and composition are generally reproducible after streamlining
-
•
Halomonas elongata is an effective spike-in for estimating absolute abundance
-
•
This time- and money-saving protocol should accelerate discovery
Microbiology; Methodology in biological sciences
Introduction
The gut microbiota plays critical roles in many aspects of health and disease (Manor et al., 2020; Zheng et al., 2020). Sequencing and the development of associated computational analysis tools (Callahan et al., 2016) have enabled quantification of the relative abundance of both culturable and unculturable species within communities of interest, as well as distinctions between community composition in healthy and diseased states (Mayer et al., 2014; Shreiner et al., 2015; Sonnenburg and Backhed, 2016). However, mechanistic insights that lead to predictions about the microbiota response to perturbations and interventions remain elusive. While various animal models have increased our understanding of host-microbe interactions (Becker et al., 2011; Faith et al., 2011; Mark Welch et al., 2017; Reyes et al., 2013; Rezzonico et al., 2011; Turnbaugh et al., 2009), recent studies have employed a complementary approach: multispecies microbial communities assembled in vitro to study function under highly controlled conditions and in high throughput (Aranda-Díaz et al., 2022; Cheng et al., 2021). Such in vitro communities can be highly diverse, stable, and reproducible, and 16S rRNA gene (hereafter referred to to as “16S rRNA”) and metagenomic sequencing of these communities have been used successfully to predict the response of gut commensals to antibiotics (Aranda-Díaz et al., 2022), uncover strain-nutrient interactions (Cheng et al., 2021), and reinforce or disprove ecological hypotheses (Chang et al., 2021; Goldford et al., 2018; Sanchez et al., 2021). While these successes attest to the power of surveying community function in this manner, a protocol for preparing such in vitro samples for sequencing that optimizes cost, time/effort, and accuracy/reproducibility has yet to be comprehensively established.
Typical library preparation steps include (1) genomic DNA (gDNA) extraction, (2) gDNA normalization, (3) 16S rRNA gene amplification by PCR, (4) PCR-product clean-up and quantification, and (5) normalization of PCR products and pooling (Figure 1A). Each of these steps involves choices that could affect sequencing outcomes, and also imposes costs in terms of money and time. After a library is sequenced and sequence files are demultiplexed, data analysis pipelines involve (1) learning of error rates and sample inference to identify amplicon sequence variants (ASVs, a proxy for species) and (2) phylogeny assignment using a taxonomy reference database (Figure 1B). The dependence of relative abundance quantification within in vitro communities on analysis parameters and database selection has yet to be established.
Figure 1.
Schematic of 16S rRNA sequencing protocol optimization
(A) Typical protocol for preparation of 16S rRNA gene libraries for sequencing.
(B) Analysis pipeline using DADA2 (Callahan et al., 2016).
(C) Optimized protocol reduces monetary cost and time/effort while retaining accuracy and reproducibility and adding absolute abundance quantification.
Previous studies aimed at identifying how library preparation methodology affects the biological interpretation of 16S rRNA sequencing data reported that choice of DNA extraction kit (Ariefdjohan et al., 2010; Claassen et al., 2013; Costea et al., 2017; Kennedy et al., 2014), PCR amplification parameters (e.g. enzyme choice, gDNA input concentration, and number of PCR cycles) (Gohl et al., 2016), and reference database when annotating ASVs (Balvociute and Huson, 2017) can all be sources of variability. As these studies focused on stool samples or mock communities with relatively low diversity, we sought to determine a simple, efficient 16S rRNA sample preparation and data analysis protocol appropriate for complex in vitro communities of gut commensals. We systematically investigated each step of a common protocol and determined those with a significant impact that introduce bias, and/or can be omitted. Mimicking previous studies that used a spike-in control to measure the absolute abundance of each ASV (Rao et al., 2021), we also established a straightforward absolute abundance quantification protocol appropriate for anaerobically grown in vitro communities of gut commensals using the halophilic, strictly aerobic Proteobacterium Halomonas elongata. Our results indicate that most conclusions based on sequencing of in vitro communities are not sensitive to alteration of many steps of the library preparation and analysis protocol. Thus, we propose a simplified protocol with reduced monetary cost and experimental effort (Table 1) that still results in highly accurate and reproducible results (Figure 1C).
Table 1.
Time and monetary cost breakdown for standard and optimized 16S rRNA sample preparation protocols
| Step | Option | Standard protocol |
Optimized protocol |
||
|---|---|---|---|---|---|
| Hours per 96 samplesa | Cost per 96 samples | Hours per 96 samplesa | Cost per 96 samples | ||
| DNA extraction | Ultra-Clean | 1 | $282 | 1 | $282 |
| Blood and Tissue | 1 | $350 | 1 | $350 | |
| PowerSoil | 2 | $575 | 2 | $575 | |
| DNA quantificationb | 0.25 | $117 | n.n. | ||
| DNA normalization into PCR | 0.5 | $0 | n.n. | ||
| Polymerase for PCR | Platinum II | 3 | $348 | ||
| 5PRIMEc | 3 | $65 | |||
| AccuStart | 3 | $172 | 3 | $57d | |
| PCR clean-up | 0.5 | $180 | 0.5 | $7 | |
| PCR-product quantificationb | 0.25 | $117 | n.n | ||
| Individual sample PCR pool | 0.5 | $0 | 0.125 | $0 | |
| Total | Ultra-Clean w/AccuStart | 6 | $868 | 4.6 | $346 |
| Blood and Tissue w/AccuStart | 6 | $936 | 4.6 | $414 | |
| PowerSoil w/AccuStart | 7 | $1,161 | 5.6 | $639 | |
Time calculations assume the use of a semi-automatic 96-well pipetting system.
DNA/PCR-product quantification estimates include colorimetric assay reagents and fluorescence 96-well microplates, and assume two replicates per sample. n.n.: not necessary based on the results in this study.
5PRIME polymerase has been discontinued, hence AccuStart becomes the best choice for polymerase based on cost.
For 25-μL PCRs.
Results
Efficiency of DNA extraction is approximately constant across commercial kits
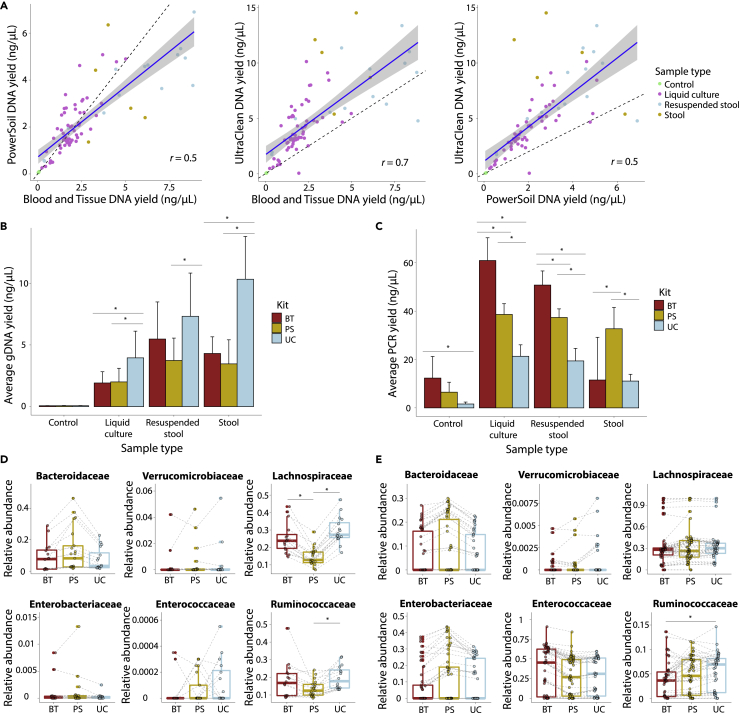
Previous studies have shown that the choice of DNA extraction kit for sample preparation from human stool is a source of variability in 16S rRNA and metagenomic sequencing (Ariefdjohan et al., 2010; Claassen et al., 2013; Costea et al., 2017; Kennedy et al., 2014). However, it is unclear how much of this variability arises from specific properties of stool as opposed to the intrinsic microbial constituents of the community under study. To address this question, we made use of 57 in vitro communities passaged in complex media that are highly diverse and stable (Aranda-Díaz et al., 2022), along with 21 stool samples, either dry or resuspended in PBS and with a ∼100-fold range in biomass input, for comparison. The in vitro communities were derived from feces of humanized mice, hence are composed of human gut commensals, and preserve most of the major families with relative abundances similar to the corresponding mouse feces (Aranda-Díaz et al., 2022). We evaluated the effectiveness of three DNA extraction kits (Ultra-Clean Microbial (UC), Blood and Tissue (BT), and PowerSoil (PS)) based on gDNA yield, subsequent PCR yield, and community composition; these kits were selected based on their common usage for a range of microbiome-related applications.
Across in vitro communities with a range of compositions, gDNA yield was reasonably correlated (R=0.5–0.7) between pairs of kits (Figure 2A), although the UC kit typically resulted in gDNA concentrations ∼2-fold higher than the BT and PS kits (Figure 2B). Higher gDNA yield did not translate to higher yield from PCR amplification of 16S rRNA (STAR Methods), presumably because the number of PCR cycles was sufficient to saturate the reaction, but the UC kit produced consistent PCR yields across all tested sample types and exhibited the lowest levels of amplification from negative controls (PBS or no input) (Figure 2C).
Figure 2.
Different extraction kits yield similar gDNA yield, PCR yield, and community composition
(A) Efficiency of gDNA extraction was constant across the three kits tested for all sample types. A linear fit to the data is shown as a blue line with a 95% CI in gray. Controls are PBS with no biomaterial input. Resuspended stool refers to 30 mg of dry stool resuspended in 10–100 μL of PBS.
(B) The Ultra-Clean (UC) kit resulted in higher gDNA yield than the PowerSoil (PS) and Blood&Tissue (BT) kits, especially for stool samples. Shown are averages (n = 10 controls, 57 liquid cultures, 14 resuspended stool, 7 dry stool) and error bars represent 1 SD. Significant differences between kits are denoted by asterisks (p < 0.05, ANOVA and HSD test).
(C) The UC kit resulted in more consistent PCR-product yield across sample types and lower amplification from control samples than the PS and BT kits. Shown are averages (n = 10 controls, 57 liquid cultures, 14 resuspended stool, 7 dry stool) and error bars represent 1 SD.
(D and E) Relative abundance was similar across kits for all families (r > 0.8), except for Enterococcaceae, Ruminococcaceae, and Lachnospiraceae in stool samples (D) and Ruminococcaceae in in vitro community samples (E). Each sample is depicted by a circle and dashed lines connect the same sample across kits. Cases in which median relative abundance was significantly different between kits are denoted by asterisks (p < 0.05, ANOVA and HSD test).
After sequencing, we used DADA2 (Callahan et al., 2016) to quantify the relative abundance of ASVs in all stool and in vitro community samples. For stool samples, the kits led to similar representation of most major taxonomic families (R > 0.8), except for the Lachnospiraceae and Ruminococcaceae families, whose relative abundances were lower by ∼10%–15% (a ∼50% relative decrease) and by ∼6% (a ∼30% relative decrease), respectively, in samples extracted with the PS kit compared with the BT and UC kits, and the Enterococcaceae, whose extremely low abundance in these samples led to substantial variability between kits (Figure 2D). While to our knowledge no other study has compared these specific kits to each other, lower Lachnospiraceae abundance in stool samples extracted using the PS kit compared to other DNA extraction kits has been previously documented (Kennedy et al., 2014). The families for which we observed significant differences across kits are all Gram-positive. To determine whether Gram-positive taxa generally exhibited variability across kits, we analyzed the relative abundance of genera within the Lachnospiraceae, Ruminococaceae, and Enterococcaceae that were present in ≥5 samples and at ≥1% relative abundance. A few genera exhibited significant differences: Blautia and Dorea (Lachnospiraceae) were overrepresented in samples extracted using the UC kit compared to the BT and PS kits, Ruminococcus_1 (Ruminococcaceae) and Lachnoclostridium (Lachnospiraceae) were overrepresented in samples extracted using the BT kit compared to the PS kit, and Fusicatenibacter (Lachnospiraceae) was underrepresented in samples extracted with the PS kit compared to the UC and BT kits (Figure S1A). Nonetheless, most genera were similarly represented among all three kits. These results suggest that the kit dependence of gDNA extraction efficiency from stool samples is genus-specific, especially in the Lachnospiraceae and Ruminococcaceae families, and that differences are not generalizable across all Gram-positive taxa.
For the in vitro community samples, the only significant difference between kits was in the abundance of the Ruminococcaceae family. Compared with stool samples, these changes were lower in magnitude; Ruminococcaceae relative abundance was generally higher by ∼1%–2% (a ∼15%–30% relative increase) in samples extracted using the UC kit compared with the BT and PS kits (Figure 2E). At the genus level, Blautia (Lachnospiraceae) was overrepresented in samples extracted using the UC kit compared to the BT and PS kits, and Tyzzerella (Lachnospiraceae) was overrepresented in samples extracted with the UC kit compared to the PS kit; all other genera were similarly represented across all kits (Figure S1B).
Taken together, these data suggest that the UC kit is slightly preferable for in vitro community sample preparation in terms of gDNA yield and PCR yield consistency, with the added advantage that it currently costs ∼$1100 for 384 samples, which is at present $250 and $1,150 cheaper than the BT and PS kits, respectively. In addition, the elevated relative abundance of the Ruminococcaceae family while levels of all other families were maintained in both in vitro community and stool samples extracted using the UC kit as compared to the BT and PS kits suggests that the UC kit is more efficient at extracting gDNA from Ruminococcaceae cells, independent of sample type.
PCR-product yield and community composition are largely unaffected by gDNA input or PCR reaction volume
The concentration of gDNA available for PCR amplification can vary across samples due to the total input biomass and the efficiency of gDNA extraction. As a result, some protocols include a normalization step (Figure 1A) to ensure that gDNA input is approximately constant across all samples. Because such normalization is time-consuming, especially for in vitro community experiments that contain hundreds to thousands of samples, we explored whether the total amount of input gDNA would affect in vitro community composition by serial diluting the input gDNA from three samples before performing 16S rRNA amplification and sequencing. For all three gDNA samples, the yield after PCR amplification was similar across a 729-fold range of dilution (Figure 3A), suggesting that a protocol with 35 PCR cycles is sufficient for most samples.
Figure 3.
Concentration of gDNA input does not affect PCR yield or community composition
(A) PCR-product yield remained essentially constant across a wide range of dilutions of gDNA input for the three in vitro community samples tested.
(B and C) Community composition at the family level was conserved up to a 27-fold dilution of gDNA input. For dilutions ≥81-fold (outside the typical range of gDNA input concentrations), the relative abundance of the Ruminococcaceae, Verrucomicrobiaceae, and Lachnospiraceae families exhibited changes of 10%–30%, while the relative abundance of the Enterococcaceae family exhibited larger changes in samples 1 and 2, likely due to its low relative abundance (<0.5%) (C). Verrucomicrobiaceae family members were not detected in Sample 1.
(D and E) Across 84 samples, the relative abundance of all ASVs (D) and number of ASVs detected (E) were similar between 75-μL and 25-μL PCR volumes. Individual dots are data points, and dashed lines are x = y. r-values are Pearson’s correlation coefficients, and p-values are from two-tailed Student’s t-tests.
Despite the insensitivity of PCR yield after clean-up to gDNA input concentration, PCR overcycling or biases such as jackpotting could in principle impact the relative abundance of particular taxa. Nonetheless, the composition of all three communities was broadly conserved throughout dilutions up to 27-fold (Figure 3B), suggesting that overcycling was not a concern. Across samples, relative abundances of the Enterococcaceae, Ruminococcaceae, and Lachnospiraceae families ranged from 0.2% to 2%, 3% to 5%, and 17% to 25%, respectively, and remained essentially constant for a particular sample across gDNA dilutions up to 27-fold (Figure 3C). When gDNA was diluted ≥81-fold, the relative abundances of these families increased to 0.5%–3%, 5%–6%, and 20%–30%. In the two samples in which the relative abundance of the Verrucomicrobiaceae family was >0.1%, Verrucomicrobiaceae abundance shifted slightly from ∼50% to 44% for dilutions ≥81-fold (Figure 3C).
In our 16S rRNA protocol (STAR Methods), each PCR reaction of 75 μL contains 3 μL of extracted gDNA at a typical concentration of ∼1–10 ng/μL. The average molecular weight of a deoxynucleotide monophosphate is ∼300 g/mol and thus a typical five Mbp bacterial genome weighs ∼5 × 10−15 g, hence 3–30 ng of gDNA corresponds to ∼106–107 genomes. For a taxon such as the Ruminococcaceae family that is present at ∼4% relative abundance, a 729-fold dilution reduces the number of genome copies to ∼50–500. Thus, the systematic changes in Ruminococcaceae abundance with lower gDNA input are unlikely to be explained by stochastic jackpot events due to low genome copy numbers. Instead, the PCR likely has a slight bias that favors Ruminococcaceae ASVs; lowering gDNA input 729-fold requires an additional ∼9.5 PCR cycles to reach the same DNA yield, so the observed ∼25% bias in Ruminococcaceae abundance would be explained by a slight amplification bias of 2%–3% per cycle (and the amplification bias for other taxa must be even smaller). However, for ASVs near the limit of detection (0.1%), an 81-fold dilution would result in <10 genome copies in the input gDNA, and thus a high probability of introducing noise into abundance quantification due to jackpotting. The concentration of gDNA extracted from our in vitro communities varied by only ∼10-fold, indicating that the bias introduced by not normalizing gDNA input is likely <10% or even lower but could be more substantial for certain families like the Ruminococcaceae across sets of samples with a broader variation in gDNA.
In sum, the amount of gDNA available for PCR amplification does not affect PCR-product yield, but may affect the resulting relative abundance of certain taxa for samples with very low gDNA concentration. While these differences are important to note, the changes in relative abundance induced by systematic PCR amplification bias are small for the range of cell densities and gDNA concentrations typically obtained from in vitro communities and are much smaller than the intrinsic relative abundance differences across samples (Figure 3B). Therefore, normalization of gDNA prior to PCR can be omitted in most experiments involving in vitro communities. Considering the cost of DNA quantification reagents and materials, omitting this step saves ∼$120 and ∼0.5–1 h of sample preparation time per 96-well plate (Table 1), as well as avoiding additional steps that could introduce contamination.
Because a 75-μL reaction yields more than sufficient DNA for subsequent pooling and sequencing (Figure 3A), we asked whether reducing the PCR reaction volume would enable saving on reagents without sacrificing data quality. We performed 75-μL and 25-μL PCR reactions for each of 84 samples and compared sequencing results. Despite the variation in community diversity and composition across samples, reducing reaction volumes from 75 μL to 25 μL generally preserved community composition (Figure 3D). The largest outliers were a Collinsella aerofaciens ASV in a community derived from the stool of a healthy human (0.035 relative abundance in the 25-μL reaction, not detected in the 75-μL reaction) and a Clostridium citroniae ASV in an in vitro community derived from a humanized mouse after antibiotic treatment (0.012 relative abundance in the 75-μL reaction, not detected in the 25-μL reaction). Nonetheless, when comparing these two ASVs across all samples, the relative abundance of these two ASVs across all samples was not significantly different between 75-μL and 25-μL reactions (p = 0.06 for C. aerofaciens and p = 0.84 for C. citroniae, two-sided Student’s t test). The number of ASVs detected was also highly similar between 75-μL and 25-μL reactions (Figure 3E); the largest difference was in two communities derived from humanized mice fed a polysaccharide-deficient diet for which the 25-μL reaction detected nine more ASVs than the 75-μL reaction in each case. Across all samples, ASVs that were unique to one reaction volume were not biased toward either volume and were present at low relative abundance (0.0028 ± 0.0033, mean ± SD). Thus, reducing PCR volumes to 25 μL can further save on the cost of reagents without introducing systematic biases or sacrificing 16S data quality.
Different DNA polymerases result in similar amplification across taxa
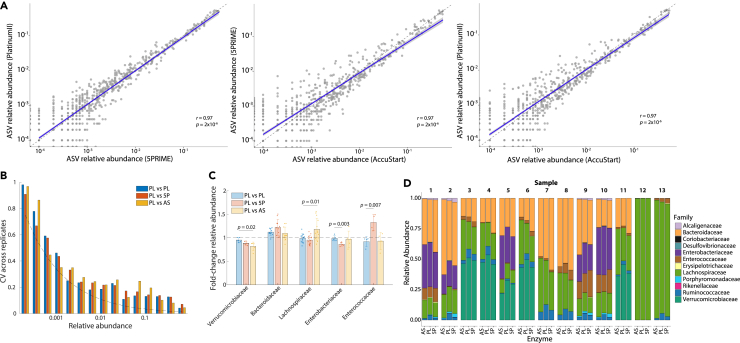
Another factor that could introduce biases during 16S rRNA sample preparation is the choice of DNA polymerase for amplification, which can be dictated by price, availability, or historical practices of a lab. To quantify potential biases, we amplified the 16S rRNA gene of gDNA extracted from 13 in vitro community samples with Platinum II Hot-Start PCR Master Mix (PL), 5PRIME HotMaster Mix (5P), and AccuStart II PCR Super-Mix (AS), three commonly used polymerases for 16S rRNA sample preparation, and compared the relative abundances of all ASVs after sequencing. The resulting relative abundances were highly correlated between pairs of DNA polymerases (Figure 4A). The coefficient of variation (SD/mean) for ASV relative abundance between replicate samples was negatively correlated with relative abundance and was considerably higher for ASVs with relative abundances <0.1% compared with >0.1% (∼0.2 versus >0.5, Figure 4B). These variations were largely comparable across DNA polymerases (Figure 4B), indicating that the choice of DNA polymerase does not impact the resulting community composition.
Figure 4.
Community composition is not affected by choice of DNA polymerase during amplification
(A) Pair-wise correlations of ASV relative abundances quantified after amplification of the same sample with different DNA polymerases indicated that all three enzymes result in similar community compositions (correlation coefficients and p-values were computed using F-tests). A linear fit to the data is shown as a blue line and the 95% CI is shown in gray.
(B) ASV relative abundance coefficient of variation (CV, SD/mean) in two experiments was similar across DNA polymerase enzymes regardless of relative abundance. Dashed line is the theoretical limit of CV due to sample size.
(C) Different polymerase enzymes can systematically affect the fold-change in the relative abundance of certain families relative to a separate run with PL, but in most cases only by <20%. Each sample is depicted by a circle and error bars represent 1 SD. p-values are shown for cases in which the fold-change was statistically significant (t-test).
(D) Variations in community composition between DNA polymerase enzymes were small compared to the intrinsic differences in community composition across samples. n = 1 replicate per community.
Although community composition was largely insensitive to the choice of polymerase, it was possible that each DNA polymerase systematically favors the amplification of certain taxonomic groups. To test this hypothesis, we compared the relative abundances of five families, all of which were present at high relative abundance in many of our in vitro communities and hence biases would be more apparent. To minimize the effects of statistical fluctuations, we only included samples for which the relative abundance of each of the five families was >1%. Compared to results using PL, 5P systematically underestimated the relative abundance of the Enterobacteriaceae family and overestimated that of Enterococcaceae, while AS slightly underestimated the relative abundance of Verrucomicrobiaceae and overestimated that of Lachnospiraceae (Figure 4C). While these variations were statistically significant across samples, the fractional changes in relative abundance were small (<20%) and were typically smaller than the intrinsic differences across samples (Figure 4D).
Thus, the choice of DNA polymerase can be a source of variability for a few families but typically does not affect conclusions based on community composition.
Normalization of PCR product input when pooling can be omitted to reduce preparation time without compromising read distribution
When assembling a pooled library for sequencing, it is often customary to quantify the PCR product concentration and normalize the amount of DNA from all samples that will make up the library. This step is expensive (costs of DNA quantification reagents, PCR-product clean-up kits, and microplates for fluorescence-based assays sum to >$300 per 96 samples) and time consuming (amplified and cleaned DNA from each PCR sample must be manually manipulated and pooled), especially for in vitro community experiments whose potential for high throughput means that they typically involve hundreds to thousands of samples. Because we observed variability in the average DNA yield from PCR reactions of only ∼30% (Figure 2C), we hypothesized that omitting this normalization step would not adversely affect the resulting distribution of reads across samples.
To test this hypothesis, we prepared one library with normalization and two libraries without normalization, in which rather than normalizing input DNA to 200 ng/sample, we simply used 10 μL of PCR product from each sample without PCR clean-up (yielding ∼200 ng of DNA given an average PCR product of 20 ± 8 ng/μL) to construct the pooled library. Although these libraries were prepared from different samples and sample types, the distributions of read number per sample from the two non-normalized libraries were similar to that of the normalized library (Figure 5).
Figure 5.
The distribution of reads per sample obtained from non-normalized libraries is highly similar to that of a normalized library
Regardless of whether normalization of PCR-product yield was performed prior to pooling, sequencing resulted in normal distributions of read counts centered on ∼104 reads, indicating that normalization is not necessary.
Thus, under conditions in which the PCR was saturated, normalization of input PCR product can be omitted from pooled library construction protocol without compromising results.
Similar sequencers at different facilities produce equivalent community compositions
As the number of sequencing facilities expands to accommodate increasing demand, research groups may find it beneficial to employ multiple such facilities, raising the question of whether differential handling or variation in equipment affects the distribution of reads per sample and the resulting composition of communities. To address this question, we sent aliquots of a pooled library prepared from 192 undefined and 192 defined in vitro communities to two facilities for comparison: the Genome Analysis Core at Mayo Clinic and the Chan Zuckerberg BioHub. Illumina MiSeq sequencers were used at both facilities.
The distributions of reads per sample were almost identical (Figure S2), and the number of reads per sample was highly correlated (R=0.99) between the two facilities (Figure 6A). ASV relative abundance across all samples was also highly correlated between facilities (R=0.99 ± 0.001, Figure 6B), indicating that community composition was extremely similar between facilities. Indeed, the ASVs that were present only in the Mayo Clinic run or only in the BioHub run accounted for only 0.015 ± 0.03% and 0.007 ± 0.02% of the communities, respectively. These data indicate that 16S rRNA results generated by the same type of sequencer are likely to be highly comparable across facilities.
Figure 6.
Read count and community composition are almost identical for sequencing of the same samples at different facilities
(A) The reads per sample for sequencing of the same pool of samples were highly correlated (n = 368) between the Mayo Clinic and BioHub facilities. A linear fit to the data is shown as a blue line and the 95% CI is shown in gray. Correlation coefficients were computed using F-tests.
(B) ASV relative abundance was highly correlated between sequencing runs at the two facilities. The scatter plot shows ASV relative abundance for one representative sample. A linear fit to the data is shown as a blue line and the 95% CI is shown in gray. Inset: the distribution of correlation coefficients of ASV relative abundance from sequencing at the two facilities across all samples.
Pooling samples during DADA2 analysis can increase the number of detected ASVs, at the expense of compute time and potential inclusion of contaminants
Previous studies have analyzed differences between OTU-level and ASV-level bioinformatic workflows for 16S rRNA analysis (Prodan et al., 2020). Here, we chose to focus on DADA2 based on its documented sensitivity and its broad use in the field. There are three methods that can be applied when performing sample inference with DADA2 (Callahan). Briefly, sequences from all samples can be analyzed (1) separately before being combined into a final sequence table using the “no pool” option (pool = FALSE, the default option), (2) together using the “true pool” option (pool = TRUE), or (3) separately but using information from all samples that is shared using the “pseudopool” option (pool = pseudo) (for a more detailed explanation, see (Callahan)). The “true pool” option increases sensitivity, allowing for detection of more ASVs as may be preferable when exploring new and complex communities, but it is computationally expensive and may report false positives such as contaminants that are prevalent at very low frequencies (Callahan).
To explore how these sample inference options affect analyses of in vitro communities, we analyzed the 16S rRNA sequencing data from the 192 undefined in vitro communities generated by the two facilities described in the previous section. Sample inference with DADA2 was performed on the same sequencing files using one of the three pooling options. The “true pool” option identified 4-fold more ASVs (∼780) and required a computing time that was ∼5–8 times longer (∼4.5 h for 768 samples with 104 reads on average per sample) than either the “pseudopool” (∼1 h, ∼150 ASVs) or the “no pool” (∼0.6 h, ∼140 ASVs) options (Figure 7A).
Figure 7.
Pooling samples during DADA2 analysis of in vitro communities increases the number of detected ASVs but does not change the final community composition
(A) All three pooling options yielded similar numbers of families and genera, but the “true pool” option yielded ∼4-fold more ASVs.
(B and C) Approximately 25% of the ASVs uniquely detected by the “true pool” option were assigned to the Escherichia/Shigella or the Enterococcus genera (B) and ∼50% of the ASVs uniquely detected by the “pseudopool” option were assigned to the Escherichia/Shigella genus (C). Nonetheless, the relative abundance of all uniquely detected ASVs was low. Error bars represent 1 SD.
(D) All three pooling options yielded almost identical community composition at the genus level, as demonstrated by a representative sample.
Approximately 25% of the ASVs uniquely identified by the “true pool” option had a taxonomic assignment to the Escherichia or Enterococcus genera, with very low and highly variable abundances of 0.06 ± 0.05% and 0.02 ± 0.02%, respectively. All other ASVs mapped to 41 distinct genera at average abundances of <0.01 ± 0.01% (Figure 7B). For the “pseudopool” option, ∼50% of the uniquely identified ASVs mapped to the Escherichia genus, at the slightly higher but still low abundance of 0.2 ± 0.09% compared with the “true pool” option. All other unique ASVs mapped to six other genera at average abundance <0.05% (Figure 7C).
The uneven taxonomic distribution of the ASVs included through pooling suggests that they may be contaminants; regardless, they collectively account for only 0.8% ± 0.1% or 0.1% ± 0.1% of total abundance across communities for the “true pool” and “pseudopool” options, respectively. Consequently, the community compositions obtained with the three pooling options were almost identical even at the genus level (Figures 7D and S3).
Based on these results, we propose that the “no pool” option during sample inference with DADA2 is preferable for analyses of in vitro communities due to its low computing time and comprehensive community composition determination without the potential introduction of noise, with the caveat that the limit of detection is higher.
Sequencing depth affects ASV detection sensitivity in DADA2
To account for natural variations in read count across samples (Figure 5A), sequencing data are often rarefied so that all samples have the same number of ASV reads (Jha et al., 2018). To determine whether rarefaction fully compensates for differences in sequencing depth, we analyzed 73 samples in two ways: (1) we directly processed the fastq files through DADA2, then rarefied the resulting ASV reads to 10,000 for each sample (Figure 8A, left); or (2) all fastq files were first downsampled to 20,000 reads per sample, processed with DADA2, and then rarefied again to obtain 10,000 ASV reads (Figure 8A, right). The downsampling process mimics a sequencing run with lower sequencing depth than the original data, which have median read count ∼100,000.
Figure 8.
Sequencing depth affects DADA2 sensitivity
(A) Two analysis pipelines, with or without downsampling prior to DADA2 processing.
(B) Downsampling prior to DADA2 processing did not affect the overall abundance of most ASVs in the samples tested. Individual dots are the relative abundance of an ASV in one sample, solid black line is x = y, dashed lines denote the limit of detection after rarefying to 10,000 ASV reads. Blue and orange dots highlight instances in which an ASV was detected in one analysis but not the other, including some cases in which the relative abundance was >0.1%. The histograms on the left and bottom are the distributions of the blue and orange dots, respectively.
The relative abundance of all ASVs in the 73 samples was strongly correlated between the two analyses (r = 0.92, p < 10−10, n = 16,060, two-sided Student’s t test, Figure 8B). There were only 28 instances in which an ASV was detected after downsampling but not in the original data (Figure 8B, left histogram). By contrast, there were 920 instances in which an ASV was detected in the original data but not after downsampling (Figure 8B, bottom histogram). In some of these instances, the ASV relative abundance was >0.1%, corresponding to at least ∼100 reads in the raw fastq file; these ASVs were distributed across many major families. Because it is very unlikely (p ∼ 2 × 10−10) that 5-fold downsampling would eliminate all 100 reads corresponding to that of ASV, these data suggest that the sensitivity and accuracy of DADA2 can be dependent on sequencing depth.
Choice of database for taxonomic assignment of ASVs is unlikely to affect conclusions that are based on overall community composition
Taxonomic assignment of ASVs detected via 16S rRNA sequencing can be performed using various databases, with the most widely used databases in the gut microbiome field being SILVA and Greengenes. The SILVA database contains taxonomic information for all three domains of life and is manually curated (Yilmaz et al., 2014). The Greengenes database contains information from the bacterial and archeal domains and is curated based on de novo tree inference (McDonald et al., 2012).
To determine how taxonomic assignment by the two databases impacts analyses of in vitro communities, we examined the percentage of annotated ASVs and the relative abundance of the five major taxonomic families in our dataset involving 192 undefined and 192 defined in vitro communities. The two databases annotated a similar percentage of ASVs at the genus and family levels; Greengenes annotated a slightly higher proportion at the species level (Figure 9A). The number of unique taxa assigned was also similar, with each database outperforming the other for particular samples (Figure 9B). These differences may emerge because the SILVA database undergoes frequent updates, while the last update performed on the Greengenes database was in 2012. When examining overall community composition, the relative abundance of almost all major taxonomic families was similar between the two databases, with the lone exception being the Lachnospiraceae family in undefined communities (Figure 9C). These results indicate that while the two databases may annotate ASVs with different names, taxonomic assignments performed with different databases are likely to be comparable at least at the family level and therefore the conclusions drawn from analyzing changes in composition are unlikely to be affected.
Figure 9.
Community richness and diversity is unaffected by ASV annotation differences between the SILVA and Greengenes (Gg) reference databases
(A and B) The databases annotated similar percentages of ASVs (A) and yielded similar numbers of unique taxa (B) at the family, genus, and species levels for undefined communities (complex communities derived from stool samples, left) and defined communities (synthetic 14- or 15-member communities, right).
(C) Most major taxonomic families exhibited similar relative abundances regardless of annotation database. The only case for which a significant difference was observed was the Lachnospiraceae family in undefined communities (∗: p < 0.05, ANOVA and HSD-test).
H. elongata as a spike-in control enables accurate estimation of absolute abundance using only a small fraction of the reads
A major limitation of 16S rRNA analyses is that results typically are relative abundances, even though the absolute abundances of a given set of microbes may have important biological consequences. Methods to quantify absolute abundance in stool samples have been developed, but they are not widely implemented. These methods include bacterial cell counting via flow cytometry (with or without the inclusion of propidium monoazide to exclude free-floating DNA), qPCR, and the use of external standards such as synthetic chimeric DNA or cells from organisms that are not present in the samples to be analyzed (Galazzo et al., 2020; Rao et al., 2021; Tkacz et al., 2018). Inspired by the multi-kingdom spike-in method (MK-SpikeSeq) (Rao et al., 2021), in which a defined number of microbial cells from the bacteria, fungi, and archaeal kingdoms are added to stool samples before performing amplicon sequencing, we sought to determine whether a single external spike-in control could provide an accurate estimate of absolute abundance across a wide range of in vitro communities. We selected H. elongata ATCC 33173 because it is halophilic and strictly aerobic (Ventosa et al., 1998), and therefore unlikely to be present in human stool samples or anaerobically grown in vitro communities.
16S rRNA sequencing of an axenic H. elongata culture demonstrated that there was no mismapping from H. elongata to other species, and H. elongata was not present in any of our anaerobically grown in vitro communities of gut commensals. Therefore H. elongata was an appropriate candidate as a spike-in control (Figure 10A). To test the range over which a spike-in would enable absolute abundance quantification, we serially diluted a saturated H. elongata culture (STAR Methods), mixed it with an in vitro community at various volumetric ratios, and performed gDNA extraction and 16S rRNA sequencing on the mixtures. Mixing with H. elongata was performed prior to gDNA extraction to determine whether variation in extraction efficiency would introduce substantial noise. The amount of spike-in H. elongata culture was linearly correlated with its relative abundance as long as the spike-in ratio was >0.5% of the reads (Figure 10B). For spike-in ratios ≥1%, the coefficient of variation was <0.2 across replicates (Figure 10C). Thus, H. elongata spiked in at 1% can serve as an effective external standard for quantifying the absolute abundance of ASVs in a community.
Figure 10.
Halomonas elongata (H.e.) is an effective external standard for estimating the absolute abundance of ASVs in in vitro communities
(A) Left: virtually all ASVs from sequencing of an H. elongata culture mapped to H. elongata. Right: H. elongata ASVs were practically absent from all of the anaerobically grown in vitro communities tested. Shaded regions are ±0.5% relative abundance around the mean values.
(B) For volumetric ratios of H. elongata to in vitro culture >0.5%, the amount of spike-in was linearly correlated with its resulting relative abundance. Gray data points are replicates (n = 8), orange dots and error bars are mean ± 1 standard deviation (SD), and the orange dashed line is a linear fit of the mean values that are >0.1%. The black dashed line denotes the detection limit of relative abundance.
(C) H. elongata abundance normalized by its mean at each dilution ratio. Gray points are individual replicates (n = 8), orange dots and error bars are mean ± 1 SD, and the black dashed lines denote 20% variation from the mean. For samples with spike-in ratios ≥1%, most replicates exhibited <20% variation.
(D) Optical density (OD) is a reasonable, but imperfect, proxy for in vitro culture biomass (normalized gDNA, estimated using the H. elongata spike-in). Each circle represents an in vitro community sample.
The optical density (OD) of an in vitro community can be used as a proxy for total biomass to estimate the absolute abundance of ASVs. We compared the performances of OD and a spike-in control with 10% (v/v) H. elongata using 96 in vitro communities with a range of community compositions. The total amount of gDNA estimated from the spike-in control was correlated with community final OD (r = 0.5, p = 10−6, F-test) (Figure 10D), suggesting that OD is indeed a reasonable proxy for gDNA.
However, there was a large degree of variation between OD and spike-in metrics. We hypothesized that this variation was due to different DNA-to-biomass ratios across species, such that OD was affected by both total gDNA and the specific ASVs present in a community. To test this hypothesis, we focused on samples with OD between the 25th and 75th percentiles. Among these samples, OD and estimated total gDNA were no longer correlated (r = 0.21, p = 0.16, two-sided Student’s t-test). However, samples with higher total gDNA also had higher abundance of an Escherichia fergusonii ASV (p < 10−14, two-sided Student’s t test), suggesting that the OD metric was biased by community composition, for instance due to variable 16S rRNA copy number across species. Taken together, we conclude that a spike-in control during DNA extraction and 16S rRNA sequencing of mixed communities allows for more accurate estimation of ASV absolute abundance than OD, albeit with the cost of diverting a small fraction of reads from the species in the community to the spike-in.
Discussion
As the gut microbiome research field continues to expand, the ability to compare datasets across labs and studies has become increasingly important. A typical pipeline for 16S rRNA gene sequencing sample preparation and data analysis consists of multiple steps, each with various options that can potentially lead to variable results. Determining whether and how each choice affects biological interpretations and using that knowledge to establish efficient and reproducible protocols should prove valuable for comparing across datasets.
Here, we presented a systematic streamlining of the steps in a typical 16S rRNA protocol that resulted in a simple and efficient strategy appropriate for complex in vitro communities of gut commensals (Figure 1B). Importantly, many steps could be omitted or simplified without affecting community composition, enabling future studies to save time and money (Table 1). For example, the monetary savings of switching from the PowerSoil to the Ultra-Clean DNA extraction kit is ∼$300 per 96-well plate (currently ∼$575 versus ∼$280) and the time expenditure decreases from 2 to 1 h per plate. By omitting the gDNA and PCR-product normalization steps and performing PCR clean-up on the pooled library instead of the individual PCR products, the major cost of sample preparation drops from ∼$700 to ∼$290 per 96-well plate and the time expense drops from ∼3 to ∼1.6 h per 96-well plate.
Because DNA extraction kits and DNA polymerase enzyme mixes are the two major costs of a typical standard 16S rRNA sample preparation protocol (Table 1), costs could be reduced by purchasing or purifying the individual components to make “homemade” kits/mixes. Such a strategy could be economically optimal in the long run if the number of samples to be processed is large enough, but would likely augment sample preparation time and, especially for PCR, establishing which components work best together and with what parameters would require substantial optimization. Fortunately, our data show that the cheapest and most time-efficient commercial kit (Ultra-Clean) resulted in the highest gDNA yields (Figure 2B) and most consistent PCR yields (Figure 2C), and normalization of gDNA or PCR-product yield did not significantly improve the narrowness of the distribution of reads per sample (Figures 3 and 5). Data obtained using different DNA polymerases for amplification (Figure 4) or different sequencing facilities (Figure 6) were largely comparable, so the most affordable and/or convenient choice can be used. Finally, we determined during data analysis that the least computationally demanding sample inference pooling option in DADA2 is likely preferable for most in vitro communities (Figure 7), that detection of ASVs can depend on sequencing depth (Figure 8), and that the choice of database used to annotate ASVs is unlikely to affect biological interpretations regarding community composition (Figure 9). Taken together, implementing this time- and money-saving protocol should accelerate future studies, particularly those involving high-throughput in vitro community experiments.
Adding to the relative simplicity and reliability of this 16S rRNA sequencing protocol, we developed a spike-in method for quantifying absolute abundance (Figure 10) that enables differentiation between changes in relative abundance that are a true representation of fluctuations in the level of a species versus those indirectly caused by expansion or contraction of other species. This step can be kept consistent between experiments using aliquots from one batch of spike-in cells that have been grown to the same state, and we described a simple calibration for determining the appropriate level of spike-in to add to communities with varying biomass. We identified the bacterium H. elongata as a suitable spike-in choice for gut-derived in vitro communities. Although other organisms might be more appropriate for other communities (e.g. soil and marine), our method provides a proof-of-principal method that can be modified and then similarly implemented. Using a culture-based spike-in rather than a standard based on DNA controls for any global source of variations during DNA extraction. Although other factors such as variability in 16S copy number and species-specific DNA extraction efficacy could confound estimates of absolute abundance, our cell spike-in provides a reliable method for comparing the absolute abundance of a given species across many samples. Taken together, the strategies we have described in this study should enhance progress in the gut microbiome field by facilitating more efficient and informative studies of in vitro communities.
Limitations of the study
We limited our analyses to the most commonly used commercial DNA kits and PCR enzymes and to data obtained from MiSeq sequencers, with a primary focus on in vitro cultured bacterial communities. It is possible that our conclusions may not generalize to all microbial communities, DNA extraction methods, PCR enzymes, PCR reaction conditions, and/or to data obtained from all sequencers (particularly those that yield greater sequencing depth). Nonetheless, our systematic investigation of a broad range of parameters led to an optimized 16S rRNA sequencing protocol that is a robust, time- and cost-efficient method that can be readily adopted. Our methods and analyses pipelines can also be easily adapted to study the effects of other changes to the protocol.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| Quant-iT dsDNA High Sensitivity Assay kit | Invitrogen | Cat. #Q33120 |
| DNeasy PowerSoil HTP 96-kit | Qiagen | Cat. #12955-4 |
| DNeasy 96 Blood and Tissue Kit | Qiagen | Cat. #69581 |
| DNeasy UltraClean 96 Microbial Kit | Qiagen | Cat. #10196-4 |
| AccuStart II PCR SuperMix | Quantabio | Cat. #95137-100 |
| PlatinumTM II HotStart PCR Master Mix | ThermoFisher | Cat. #14000013 |
| 5PRIME HotMasterMix | Quantabio | Cat. #2200410 |
| Experimental models: Organisms/strains | ||
| Stool-derived in vitro communities | This paper | N/A |
| Deposited data | ||
| Stanford Digital Repository | This paper | https://doi.org/10.25740/ct503zg9433 |
| Software and algorithms | ||
| Stanford Digital Repository | This paper | https://doi.org/10.25740/vh225xq6457 |
Resource availability
Lead contact
Communication regarding this article should be directed to the lead contact, Kerwyn Casey Huang (kchuang@stanford.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human fecal samples or in vitro gut bacterial community cultures were used to conduct analyses in this study as specified in the text. The in vitro communities were derived from the feces of humanized mice (Aranda-Díaz et al., 2022) and cultured anaerobically in Brain Heart Infusion (BHI, BD 211069) medium or M9+10% BHI medium plus one of 23 carbon sources (Figure S3). Halomonas elongata cells were grown aerobically with shaking at 30°C in ATCC medium 1097.
Method details
Baseline protocol for 16S library preparation
DNA from fecal samples or 50 μL of saturated bacterial cultures were extracted using an extraction kit such as the DNeasy UltraClean 96 Microbial Kit (Qiagen, 10196-4). Three microliters of extracted gDNA were used for PCR in 75-μL volumes containing Earth Microbiome Project-recommended 515F/806R primer pairs (0.4 μM final concentration) and a polymerase such as that of the 5PRIME HotMasterMix (Quantabio, 2200410) to generate V4 region 16S rRNA amplicons. The following thermocycler conditions were used: 94°C for 3 min, 35 cycles of [94°C for 45 s, 50°C for 60 s, and 72°C for 90 s], then 72°C for 10 min. PCR products were individually cleaned up and quantified using the UltraClean 96 PCR Cleanup Kit (Qiagen, 12596-4) and the Quant-iT dsDNA High Sensitivity Assay kit (Invitrogen, Q33120) before 200 ng of PCR product for each sample were manually pooled. Pooled libraries were then sequenced with 250- or 300-bp paired-end reads on a MiSeq (Illumina).
DNA extraction
gDNA from ∼30 mg of human fecal samples (dry or resuspended in PBS) or 50 μL of bacterial cultures were extracted using the DNeasy PowerSoil HTP 96-kit (Qiagen, 12955-4), the DNeasy UltraClean 96 Microbial Kit (Qiagen, 10196-4), or the DNeasy 96 Blood and Tissue Kit (Qiagen, 69581) following the manufacturers’ protocols. All other aspects of the standard 16S rRNA sequencing library preparation protocol described in the section above were then followed.
16S rRNA gene amplification
For testing the dependence on gDNA input concentration, the standard 16S rRNA library preparation protocol described above was followed with the following modifications: the extracted DNA was diluted 1- to 729-fold in 3-fold steps before 3 μL were used in the 75-μL PCRs. For testing the dependence on polymerase enzymes, the AccuStart II PCR SuperMix (Quantabio, 95137-100) or the PlatinumTM II HotStart PCR Master Mix (ThermoFisher, 14000013) were used instead of 5PRIME HotMasterMix. Thermocycler conditions were held constant.
Library construction without normalization
gDNA was extracted from 50 μL of bacterial cultures or human gastrointestinal tract samples, and PCR was performed using the standard 16S rRNA library preparation protocol described above. Ten microliters of each PCR product were pooled without quantification or individual clean up in a 50-mL conical vial. The pooled PCR product mix (∼4-7 mL) was cleaned up and concentrated using the Macherey-Nagel NucleoSpin® Gel and PCR Clean-up, Mini Kit (Fisher, 740609). Sequencing was performed as described in the standard protocol above.
16S rRNA sequencing data analysis
Samples were demultiplexed with QIIME2 v. 2021.2 and subsequent processing was performed using DADA2 (Callahan et al., 2016). truncLenF and truncLenR parameters were set to 240 and 180, respectively, and the pooling option parameter was set to “pool=FALSE” unless otherwise indicated. All other parameters were set to the default. The taxonomies of the resulting ASVs were assigned using the assignTaxonomy function and the SILVA reference database as default or the GreenGenes database when testing the dependence on database. Code for data analysis can be found at https://bitbucket.org/kchuanglab/optimization16S/src/master/.
Spike-in for absolute abundance estimation
Halomonas elongata ATCC 33173 was grown aerobically overnight from a colony to saturation in ATCC medium 1097 [80 g/L NaCl, 7.5 g/L casamino acids, 5.0 g/L peptone, 1.0 g/L yeast extract, 3.0 g/L sodium citrate, 20 g/L MgSO4•7H2O, 0.5 g/L K2HPO4, 0.05 g/L Fe(NH4)2(SO4)2•6H2O, pH adjusted to 7.0] at 30°C with shaking. The saturated H. elongata culture was mixed with in vitro communities at various volumetric ratios prior to gDNA extraction. Ideally all comparisons between samples should use the same H. elongata culture preparation.
Quantification and statistical analysis
Data were analyzed in R studio and MATLAB (The MathWorks, Inc). Statistical significance was calculated using ANOVA and HSD-tests or two-tailed Student’s t-tests, as specified in the text and in the figure legends. r-values are Pearson’s correlation coefficients. Values of n used for each analysis are specified in the figure legends.
Acknowledgments
The authors thank members of the Huang and Relman labs for helpful discussions. The authors acknowledge funding from a Howard Hughes Medical Institute International Student Research Fellowship (to A.A.-D.), a Stanford Bio-X Bowes Fellowship (to A.A.-D.), a Siebel Scholarship (to A.A.-D.), an NDSEG Graduate Fellowship (to R.C.), James S. McDonnell Postdoctoral Fellowships (to K.X. and H.S.), the Thomas C. and Joan M. Merigan Endowment at Stanford University (to D.R.), the Stanford Microbiome Therapies Initiative (to A.I.C., D.R., and K.C.H.), NSF grants EF-2125383 and IOS-2032985 (to K.C.H.), NIH R01 AI147023 (to D.R. and K.C.H.), and NIH RM1 Award GM135102 (to K.C.H.). D.R. and K.C.H. are Chan Zuckerberg BioHub Investigators.
Author contributions
A.I.C., A.A.-D., R.C., K.X., H.S., and K.C.H. designed the research; A.I.C., A.A.-D., R.C., and H.S. performed the research; A.I.C., R.C., and H.S. analyzed the data; D.R. and K.C.H. supervised the research; and A.I.C., H.S., and K.C.H. wrote the paper. All authors reviewed the paper before submission.
Declaration of interests
The authors declare no competing interests.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103907.
Contributor Information
Handuo Shi, Email: handuo@stanford.edu.
Kerwyn Casey Huang, Email: kchuang@stanford.edu.
Supplemental information
Data and code availability
Sequencing data and code have been deposited at the Stanford Data Repository and are publicly available as of the date of publication. DOIs are listed in the key resources table. Any additional information required to reanalyze the data in this paper is available from the lead contact upon request.
References
- Aranda-Díaz A., Ng K.M., Thomsen T., Real-Ramírez I., Dahan D., Dittmar S., Gonzalez C.G., Chavez T., Vasquez K.S., Nguyen T.H., et al. Establishment and characterization of stable, diverse, fecal-derived in vitro microbial communities that model the intestinal microbiota. Cell Host Microbe. 2022;30:260–272. doi: 10.1016/j.chom.2021.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariefdjohan M.W., Savaiano D.A., Nakatsu C.H. Comparison of DNA extraction kits for PCR-DGGE analysis of human intestinal microbial communities from fecal specimens. Nutr. J. 2010;9:23. doi: 10.1186/1475-2891-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balvociute M., Huson D.H. SILVA, RDP, Greengenes, NCBI and OTT - how do these taxonomies compare? BMC Genomics. 2017;18:114. doi: 10.1186/s12864-017-3501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker N., Kunath J., Loh G., Blaut M. Human intestinal microbiota: characterization of a simplified and stable gnotobiotic rat model. Gut Microbes. 2011;2:25–33. doi: 10.4161/gmic.2.1.14651. [DOI] [PubMed] [Google Scholar]
- Callahan B. Pooling for sample inference. https://benjjneb.github.io/dada2/pool.html#pooling-for-sample-inference
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.Y., Vila J.C.C., Bender M., Li R., Mankowski M.C., Bassette M., Borden J., Golfier S., Sanchez P.G.L., Waymack R., et al. Engineering complex communities by directed evolution. Nat. Ecol. Evol. 2021;5:1011–1023. doi: 10.1038/s41559-021-01457-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A.G., Aranda-Díaz A., Jain S., Yu F., Iakiviak M., Meng X., Weakley A., Patil A., Shiver A.L., Deutschbauer A., et al. Systematic dissection of a complex gut bacterial community. Preprint at bioRxiv. 2021 doi: 10.1101/2021.06.15.448618. [DOI] [Google Scholar]
- Claassen S., du Toit E., Kaba M., Moodley C., Zar H.J., Nicol M.P. A comparison of the efficiency of five different commercial DNA extraction kits for extraction of DNA from faecal samples. J. Microbiol. Methods. 2013;94:103–110. doi: 10.1016/j.mimet.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costea P.I., Zeller G., Sunagawa S., Pelletier E., Alberti A., Levenez F., Tramontano M., Driessen M., Hercog R., Jung F.E., et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 2017;35:1069–1076. doi: 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- Faith J.J., McNulty N.P., Rey F.E., Gordon J.I. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galazzo G., van Best N., Benedikter B.J., Janssen K., Bervoets L., Driessen C., Oomen M., Lucchesi M., van Eijck P.H., Becker H.E.F., et al. How to count our microbes? The effect of different quantitative microbiome profiling approaches. Front. Cell. Infect. Microbiol. 2020;10:403. doi: 10.3389/fcimb.2020.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl D.M., Vangay P., Garbe J., MacLean A., Hauge A., Becker A., Gould T.J., Clayton J.B., Johnson T.J., Hunter R., et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 2016;34:942–949. doi: 10.1038/nbt.3601. [DOI] [PubMed] [Google Scholar]
- Goldford J.E., Lu N., Bajic D., Estrela S., Tikhonov M., Sanchez-Gorostiaga A., Segre D., Mehta P., Sanchez A. Emergent simplicity in microbial community assembly. Science. 2018;361:469–474. doi: 10.1126/science.aat1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A.R., Davenport E.R., Gautam Y., Bhandari D., Tandukar S., Ng K.M., Fragiadakis G.K., Holmes S., Gautam G.P., Leach J., et al. Gut microbiome transition across a lifestyle gradient in Himalaya. PLoS Biol. 2018;16:e2005396. doi: 10.1371/journal.pbio.2005396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy N.A., Walker A.W., Berry S.H., Duncan S.H., Farquarson F.M., Louis P., Thomson J.M., Consortium U.I.G., Satsangi J., Flint H.J., et al. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS One. 2014;9:e88982. doi: 10.1371/journal.pone.0088982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor O., Dai C.L., Kornilov S.A., Smith B., Price N.D., Lovejoy J.C., Gibbons S.M., Magis A.T. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 2020;11:5206. doi: 10.1038/s41467-020-18871-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch J.L., Hasegawa Y., McNulty N.P., Gordon J.I., Borisy G.G. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl. Acad. Sci. U S A. 2017;114:E9105–E9114. doi: 10.1073/pnas.1711596114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer E.A., Knight R., Mazmanian S.K., Cryan J.F., Tillisch K. Gut microbes and the brain: paradigm shift in neuroscience. J. Neurosci. 2014;34:15490–15496. doi: 10.1523/JNEUROSCI.3299-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Price M.N., Goodrich J., Nawrocki E.P., DeSantis T.Z., Probst A., Andersen G.L., Knight R., Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodan A., Tremaroli V., Brolin H., Zwinderman A.H., Nieuwdorp M., Levin E. Comparing bioinformatic pipelines for microbial 16S rRNA amplicon sequencing. PLoS One. 2020;15:e0227434. doi: 10.1371/journal.pone.0227434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao C., Coyte K.Z., Bainter W., Geha R.S., Martin C.R., Rakoff-Nahoum S. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature. 2021;591:633–638. doi: 10.1038/s41586-021-03241-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A., Wu M., McNulty N.P., Rohwer F.L., Gordon J.I. Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc. Natl. Acad. Sci. U S A. 2013;110:20236–20241. doi: 10.1073/pnas.1319470110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezzonico E., Mestdagh R., Delley M., Combremont S., Dumas M.E., Holmes E., Nicholson J., Bibiloni R. Bacterial adaptation to the gut environment favors successful colonization: microbial and metabonomic characterization of a simplified microbiota mouse model. Gut Microbes. 2011;2:307–318. doi: 10.4161/gmic.18754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A., Vila J.C.C., Chang C.Y., Diaz-Colunga J., Estrela S., Rebolleda-Gomez M. Directed evolution of microbial communities. Annu. Rev. Biophys. 2021;50:323–341. doi: 10.1146/annurev-biophys-101220-072829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreiner A.B., Kao J.Y., Young V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015;31:69–75. doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg J.L., Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacz A., Hortala M., Poole P.S. Absolute quantitation of microbiota abundance in environmental samples. Microbiome. 2018;6:110. doi: 10.1186/s40168-018-0491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ridaura V.K., Faith J.J., Rey F.E., Knight R., Gordon J.I. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventosa A., Nieto J.J., Oren A. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 1998;62:504–544. doi: 10.1128/MMBR.62.2.504-544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz P., Parfrey L.W., Yarza P., Gerken J., Pruesse E., Quast C., Schweer T., Peplies J., Ludwig W., Glockner F.O. The SILVA and "all-species living tree project (LTP)" taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Liwinski T., Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data and code have been deposited at the Stanford Data Repository and are publicly available as of the date of publication. DOIs are listed in the key resources table. Any additional information required to reanalyze the data in this paper is available from the lead contact upon request.