Summary
Extracellular vesicles (EVs) are ubiquitously secreted by almost all tissues and carry many cargoes, including proteins, RNAs, and lipids, which are related to various biological processes. EVs are shed from tissues into the blood and expected to be used as biomarkers for diseases. Here, we isolated EVs from EDTA plasma and serum of six healthy subjects by an affinity capture isolation method, and a total of 4,079 proteins were successfully identified by comprehensive EV proteomics. Our reliable and detailed catalog of the differential expression profiles of EV proteins in plasma and serum between healthy individuals could be useful as a reference for biomarker discovery. Furthermore, tissue-specific protein groups co-regulated between blood EVs from healthy individuals were identified. These EV proteins are expected to be used for more specific and sensitive enrichment of tissue-specific EVs and for screening and monitoring of disease without diagnostic imaging in patient blood in the future.
Subject areas: Omics, Proteomics, Biochemical analysis
Graphical abstract

Highlights
-
•
Catalog of EV proteome created by state-of-the-art proteome analysis technologies
-
•
Plasma and serum EV proteome profiles showed a difference in healthy individuals
-
•
Novel standard reference proteins in plasma and serum EVs were identified
-
•
Tissue-specific EV marker candidates were presented by the informatics approach
Omics; Proteomics; Biochemical analysis
Introduction
Extracellular vesicles (EVs) are small lipid bilayer-enclosed particles ubiquitously released by almost every cell type and are present in body fluids, including urea, blood, and cerebrospinal fluid (CSF) (Fang et al., 2009; Ikeda et al., 2021; Muraoka et al., 2020a, 2021; Thery et al., 2018). EVs are classified into exosomes (50–150 nm), which are secreted into the extracellular space after the fusion of multivesicular bodies (MVBs) with the plasma membrane (PM); microvesicles (MVs) (150–1,000 nm), which are shed from the PM; and apoptotic bodies (1,000–5,000 nm), which are produced from apoptotic cells (DeLeo and Ikezu, 2018; Kowal et al., 2016). Several proteins, such as tetraspanin proteins, syntenin-1, TSG101 and Alix, have been reported as markers for exosomes (Al-Nedawi et al., 2008; Taylor et al., 2007). On the other hand, the markers for MVs are glycoprotein 1b, actinin-4, heat shock protein (HSP) 90B1, and myosin light chain (Castellana et al., 2009; Frühbeis et al., 2013). EVs carry nucleic acids, such as microRNA, mRNA, and noncoding RNA (ncRNA), lipids, and proteins, which can transfer to recipient cells for cell-to-cell communication (Delpech et al., 2019; Hoshino et al., 2020; Muraoka et al., 2020b; You et al., 2020). EVs have been extensively investigated for their function and roles in intracellular communication during cancer development and neurodegenerative disease progression (Hoshino et al., 2015; Hosseini et al., 2016; Ma et al., 2021; Patel and Weaver, 2021; Ruan et al., 2020; Sun et al., 2021).
Hoshino et al. reported a database of the EV proteome from 426 human samples, which included tissue explants and bodily fluids from control and cancer samples (Hoshino et al., 2020). The average number of proteins detected in the EVs was 1,482 proteins in 131 tissue explant EVs and 269 proteins in 127 blood EVs. Blood is the most difficult body fluid to isolate EVs and identify EV proteins due to its high concentration of nonvesicular materials such as free proteins and protein aggregates (Tian et al., 2019). Most recently, attention has turned to plasma- and serum-derived EVs as liquid biopsy; this is because various EVs are shed into the blood and contain cargo that is representative of origin tissue. Thus, the detailed catalog of blood EV profiling between healthy individuals is an important resource for disease biomarker studies.
Most EV studies for discovery biomarkers have been performed using bulk EVs isolated from body fluids, including serum and plasma. Bulk EV-based liquid biopsy is excellent in that it can monitor the entire body; however, it has the disadvantage that changes in certain tissues are diluted. Therefore, the ability to enrich tissue-derived EVs is very attractive because it increases the possibility of direct monitoring of tissue-specific changes without the need to collect tissue. Recently, several researchers have reported the feasibility of detecting and isolating EVs shed from the brain, liver, prostate, and other tissues in body fluids (Agliardi et al., 2019; Allelein et al., 2021; Lee et al., 2020; Newman et al., 2021; Rodrigues et al., 2021; Skalnikova et al., 2019). Brain, prostate, and liver-tissue-specific EVs, which were isolated using L1CAM, SNAP25, PSMA, and ASGR1 antibodies from serum samples, were used for disease biomarker studies and drug metabolism studies (Agliardi et al., 2019; Allelein et al., 2021; Lee et al., 2020; Newman et al., 2021; Rodrigues et al., 2021; Skalnikova et al., 2019). However, these proteins are known to be expressed in other tissues, and the specificity and sensitivity need to be carefully confirmed for screening and monitoring of disease (Norman et al., 2021).
Herein, we provide the comprehensive protein composition of EVs separated from the EDTA plasma and serum of six healthy subjects by affinity capture (AC) using high-sensitivity data-independent acquisition (DIA) proteomics. As a novel approach to identify tissue-specific proteins suitable for tissue-derived EV enrichment, in this study, we attempted to identify candidate tissue-derived EV markers by selecting tissue-specific proteins defined by the Human Protein Atlas (HPA) from a highly comprehensive EV proteome dataset, as well as EV proteins that are co-regulated between blood EVs obtained from healthy individuals.
Results
Biochemical and morphological characterization of EVs separated from plasma and serum
Our experimental design for protein profiling of plasma- and serum-derived EVs is outlined in Figure 1A. Blood from six healthy subjects was collected with EDTA for plasma and without anticoagulants for serum. All samples were centrifuged at 1,200 g and passed through 0.45-μm filters, and EVs were separated with an AC isolation kit for proteomics analysis. The NTA results identified the EV population, which included MVs and exosomes with sizes between 100 and 300 nm (Figure 1B). The particle counts were 2.70 × 1011 and 5.70 × 1011 in 150 μL of plasma and serum and between 1.10 × 109 and 1.28 × 109 in 100 μL of separated plasma EV and serum EV fractions, respectively (Table 1). There were 2.37 × 107 particles / μg protein in plasma, 1.31 × 109 particles / μg protein in the separated EV fraction from plasma, 5.00 × 107 particles / μg protein in serum, and 1.50 × 109 particles / μg protein in the separated EV fraction from serum. The results suggest that the AC method can efficiently enrich EV particles and remove highly abundant contaminant proteins from the plasma and serum. The separated EVs were observed by Transmission Electron Microscopy (TEM), which showed exosomal morphology, as commonly seen in separated plasma and serum EVs (Figure 1C). EV markers such as CD9 and FLOT1 (flotillin 1) were clearly represented in both EV fractions, whereas contamination markers such as apolipoprotein B (ApoB) and human serum albumin (HSA) in the MISEV 2018 guidelines (Thery et al., 2018) were completely absent in the serum EV fraction, but ApoB was found at a low level in the plasma EV fraction (Figure 1D). The western blotting results were consistent with the proteomics results (Tables S1 and S2).
Figure 1.
Characterization of EVs separated from human plasma and serum by AC
(A) Workflow used for proteomics analysis of EVs from the plasma and serum of six healthy subjects. EVs were separated from plasma samples and serum samples from six healthy subjects using the AC method (MagCapture Exosome Isolation kit). The separated EVs were analyzed by MS2 on an Orbitrap Fusion Lumos Tribrid Mass Spectrometer.
(B) EVs separated from plasma and serum were examined for size and number by NTA. The black line shows the fitting curve. The red line represents the error bar. Y axis: EV particle counts [/mL], X axis: EV particle size [nm].
(C) TEM image of EVs separated from plasma and serum. Scale bar, 100 nm. Left, plasma EV; right, serum EV.
(D) Assessment of EV markers (CD9 and FLOT1) and non-EV markers (ApoB and HSA) in plasma, serum, and EV fractions by western blotting. In terms of undiluted solution, 10 μL of EVs (equivalent to 15 μL of plasma and serum) and 0.001 μL of plasma and serum were loaded onto SDS-PAGE gels. “See also Figure S8.”
Table 1.
Enrichment of EVs separated from plasma and serum with an AC kit
| Human plasma | Plasma EV fraction | Human serum | Serum EV fraction | |
|---|---|---|---|---|
| Particle numbera | 2.70 × 1011/150 μL plasma | 1.10 × 109/100 μL | 5.70 × 1011/150 μL serum | 1.28 × 109/100 μL |
| EV protein (μg)b | 11,400 | 0.898 | 11,400 | 0.84 |
| Particles/proteins (μg) | 2.37 × 107 | 1.31 × 109 | 5.00 × 107 | 1.50 × 109 |
The number of particles isolated from EVs was measured by NTA.
The EV protein concentrations were measured by BCA.
Protein profiling of plasma- and serum-derived EVs
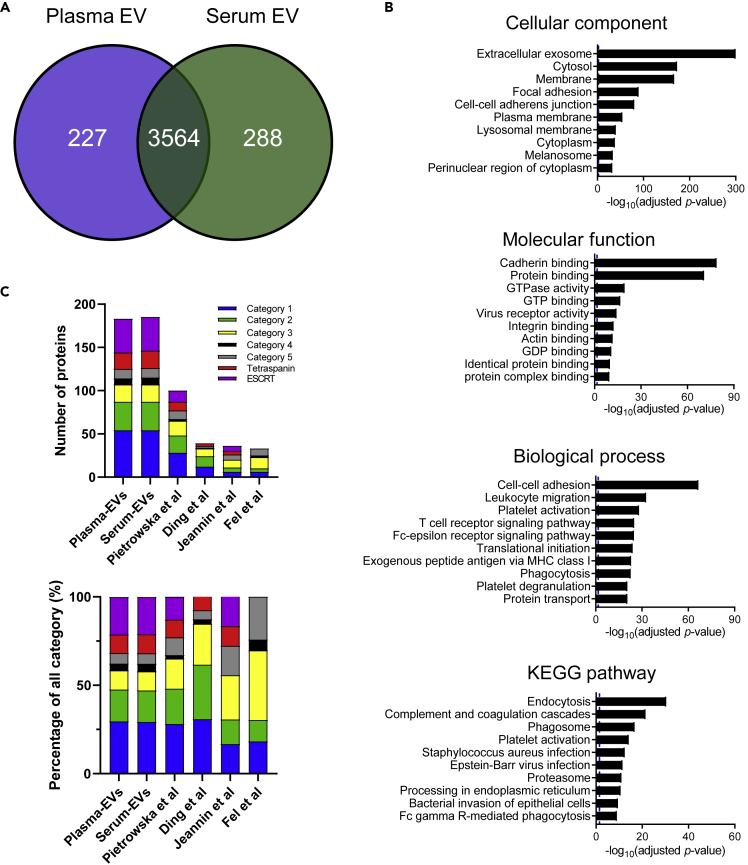
Data-independent acquisition label-free quantitative proteomics analysis of the separated EV fraction identified a total of 4,079 proteins (Table S1). Figure 2A shows a Venn diagram comparing plasma-derived EV proteins with serum EV proteins. We subjected the proteomics dataset to Gene Ontology (GO) analysis with the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Dennis et al., 2003). The identified proteins from plasma and serum EVs showed enrichment of extracellular exosomes in the cellular component category and protein binding in the molecular function category, respectively. The biological process and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway categories showed enrichment of leukocyte migration, the T cell receptor signaling pathway, endocytosis, and phagosomes, which are related to immune functions (Figure 2B). Figure 2C shows the identified number and the percentage of EV proteins and non-EV proteins, which have been reported in the MISEV 2018 guidelines (Thery et al., 2018), in our study and four previous studies (Ding et al., 2020; Fel et al., 2019; Jeannin et al., 2018; Pietrowska et al., 2021) (Table S2). Our study found 54 proteins in both plasma- and serum-derived EVs in category 1 (transmembrane GPI-anchored proteins associated with the PM and/or endosomes), whereas 28, 12, 6, and 6 EV proteins were identified in other studies. The 33 proteins in both plasma- and serum-derived EVs were identified in category 2 (cytosolic proteins recovered in EVs) in our study and 20, 12, 5, and 4 EV proteins in other studies. In the category of tetraspanin and the endosomal sorting complexes required for transport (ESCRT) family, 20 proteins were identified in both plasma- and serum-derived EVs, whereas 17, 9, 9, 13 proteins were identified in other studies. The percentages of proteins belonging to category 3 (major components of non-EV co-isolated structures) were 10.9% and 10.8% in our study and 17.0%, 23.1%, 13.9%, and 12.1% in previous studies, respectively (Figure 2C).
Figure 2.
Protein profiling of plasma- and serum-derived EVs
(A) Venn diagram of the proteins identified in plasma- and serum-derived EVs by label-free proteomics analysis.
(B) GO analysis using DAVID Bioinformatics Resources 6.8. The top 10 enriched GO terms in the biological process, cellular component, molecular function, and KEGG pathway categories are represented.
(C) The bar graph comparing the proteomic data of our study and four previous studies of the EV proteome in the category indicated by the MISEV 2018 guidelines. The top panel shows the number of identified proteins in these categories. The bottom panel shows the percentage of total proteins (all categories). Category 1 (blue): transmembrane GPI-anchored proteins associated with the PM and/or endosomes; category 2 (green): cytosolic proteins recovered in EVs; category 3 (yellow): major components of non-EV co-isolated structures; category 4 (black): transmembrane, lipid-bound, and soluble proteins associated with intracellular compartments other than PM/endosome; and category 5 (gray): secreted proteins recovered with EVs, tetraspanin family proteins (red), ESCRT family proteins (purple). “See also Tables S1 and S2.”
Proteomics comparison of plasma- and serum-derived EV proteins
Principal component analysis (PCA) showed a large separation of the plasma and serum EV proteomes (Figure 3A). Figure 3B shows the scatter plots of the signal intensity for 1,573 common proteins, which were quantified in all subjects at technical triplicates, between plasma EV and serum EV (Figures S1 and S2 and Table S3). The expression levels were positively correlated between plasma EVs and serum EVs (r = 0.7381, p < 0.0001). However, 157 proteins (10.1%) were 5 times different between plasma and serum. Functional enrichment analysis of the 1,547 proteins revealed that the pathways related to complement and coagulation cascades and cholesterol metabolism were overrepresented, and platelet activation was underrepresented in plasma EVs compared with serum EVs (Figure S3 and Table S4). We calculated the coefficient of variation (CV) for each individual protein across all subjects within plasma or serum (Figure 3C). The cytoplasmic protein NCK1 showed a low CV in both plasma (CV = 10.95) and serum (CV = 8.70). Beta-actin-like protein (ACTBL2) (CV = 3.64) in plasma EVs and ADP-ribosylation factor (ARF6) (CV = 4.42) in serum EVs may be good standard EV markers (Figure 3D, Tables 2 and 3). CD9, CD81, and CD63 had higher CVs in both plasma and serum (Figure 3D). Spearman’s correlation coefficient and p value were calculated for the 1,573 proteins, and a heatmap of the protein-protein correlation matrix based on these EV proteins was generated (Figure S4 and Tables S5, S6, S7, and S8). These data show the different expression patterns of EV proteins between plasma EVs and serum EVs. Proteins with low CV values might be suitable for housekeeping proteins for EV protein analysis.
Figure 3.
Proteomics comparison of plasma- and serum-derived EV proteins
(A) PCA of plasma and serum EV proteomics data from six pair-samples by Perseus software. Circles and squares show plasma and serum samples, respectively, from six individual healthy subjects.
(B) Scatterplot showing correlations between serum and plasma EV protein expression levels, which were measured using label-free proteomics. The Pearson’s r was 0.7381 (p < 0.0001).
(C) Frequency histogram of the CV for plasma- and serum-derived EV proteome data.
(D) The dot plot of proteins with low CVs for standard proteins in plasma EVs, serum EVs, and both plasma and serum EVs. Data are represented as mean ± SEM “See also Figures S1–S4 and Tables S3, S4, S5, S6, S7, and S8.”
Table 2.
Top 10 stably identified proteins in six plasma-derived EV samples
Table 3.
Top 10 stably identified proteins in six serum-derived EV samples
Enrichment of tissue-specific proteins in plasma and serum EVs
To design the isolation of tissue- and organelle-derived EVs from circulating EVs in blood for clinical biomarker applications, we searched tissue-specific proteins within the plasma and serum EV proteomics data using a combination of RNA sequencing analysis with HPA as a reference and coregulation analysis across individuals (Tables S1, S5, S6, S7, and S8). Protein coregulation analysis based on the correlation coefficient protein expression level is able to capture the interaction of protein-protein associations with high accuracy (Johnson et al., 2020; Lapek et al., 2017). Therefore, if the expression of certain EV proteins is positively correlated, it suggests that these proteins co-localize in EVs secreted from a particular tissue. Proteins with at least 4-fold increased expression in the tissue of interest compared with other tissues were defined as tissue-specific proteins in the HPA dataset (Table S1). Thus, we screened EV proteins with more than 4-fold increased expression in our plasma and serum EV proteomic datasets. Figure 4A shows a histogram of the number of tissue-specific proteins in the HPA dataset and our dataset. A total of 242 proteins were defined as liver tissue-specific proteins in the HPA dataset, and 120 and 112 proteins in plasma and serum EVs, respectively, were annotated as liver tissue-specific proteins (Figure 4A and Table 4). For the liver-tissue-specific proteins in EVs, KEGG pathway analysis showed enrichment of complement and coagulation cascades and the PPAR signaling pathway (Figure S5). Complement proteins and albumin are defined as non-EV proteins in the MISEV 2018 guidelines; however, it is possible that these proteins are liver-tissue-specific EV proteins because they are mainly produced in the liver. Moreover, cytochrome P450 family 4 subfamily F member 3 (CYP4F3), which is involved in drug metabolism, was identified in blood EVs. Comparison of quantitative values showed that liver-tissue-specific proteins were more enriched in plasma EVs than in serum EVs (Table S1). These results indicate that plasma EVs might be useful to assess drug metabolism in blood. Interestingly, 34 and 36 brain-tissue-specific proteins were enriched in plasma and serum EVs, respectively (Table 4). Co-regulation analysis revealed that seven proteins or three proteins in plasma EVs or serum EVs had a strong positive correlation and a p value < 0.001 in plasma EVs and serum EVs (Figures 4B and S6, Tables S9 and S10). The strong positive correlation among these brain-specific protein groups suggests that they were enriched in brain-derived EVs in a highly specific manner. Moreover, four proteins (Syntaxin-binding protein 1 (STXBP1), Neuronal membrane glycoprotein M6-a (GPM6A), Phosphatidylserine decarboxylase proenzyme 2 (PSD2), Rab GDP dissociation inhibitor alpha (GDI1)) and two proteins (Choline transporter-like protein 1 (SLC44A1), 2',3'-cyclic-nucleotide 3'-phosphodiesterase (CNP)) were neuron type-specific and oligodendrocyte-specific proteins, respectively, in brain tissue (Sharma et al., 2015). The correlation coefficients between 13 brain-specific proteins are shown in Figure S7 and Tables S9 and S10. Figure 4C shows the direct and indirect protein-protein interactions, specifically those of neuron-specific (blue) and oligodendrocyte-specific (green) proteins, in brain tissue and the protein-protein correlation values among 13 proteins (with direct interactions) and 10 proteins (with indirect interactions). GPM6A, STXBP1, and GDI1 interact indirectly through amyloid precursor protein (APP), microtubule-associated protein tau (MAPT), presenilin-1 (PSEN1), and huntingtin (HTT), which are related to neurodegenerative diseases, including Alzheimer disease (AD), progressive supranuclear palsy (PSP), and Huntington disease (HD). Interestingly, PSEN1, MAPT, APP, and HTT might be contained in neuron-derived EVs, which express GPM6A, STXBP1, and GDI1, according to protein-protein interaction analysis, and EVs might be released from brain tissue into the blood through the blood-brain barrier. Furthermore, several proteins were defined as intestine (Angiotensin-converting enzyme (ACE) and 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3 (PLCB3)), skeletal muscle (Xin actin-binding repeat-containing protein 2 (XIRP2), Serine/threonine-protein kinase 25 (STK25), Fructose-bisphosphate aldolase A (ALDOA), Pyruvate kinase PKM, Phosphoglucomutase-1 (PGM1), UV excision repair protein RAD23 homolog A (PAD23A), and Charged multivesicular body protein 1b (CHMP1B)), and other tissue-specific proteins in healthy blood EVs by HPA and coregulation analysis (Tables S1, S5, S6, S7, and S8). These data can provide information for the development of high-sensitivity EV-based biomarker assays using blood for the monitoring of disease.
Figure 4.
Enrichment of tissue-specific proteins in plasma and serum EVs
(A) Enrichment of tissue-specific proteins in plasma- and serum-derived EVs. Gray bar: tissue-specific proteins by RNA sequencing of data from the HPA. Blue bar: identified tissue-specific proteins in plasma EVs. Green bar: identified tissue-specific proteins in serum EVs.
(B) Protein networks of seven proteins or three proteins with a strong positive correlation (p < 0.001) between brain tissue-specific proteins in plasma EVs or serum EVs.
(C) Protein-protein interaction networks of 13 proteins (STXBP1, GPM6A, PSD2, GDI1, SLC44A1, CNP, MBP, GNG7, SLC6A5, NRGN, YQHAH, TMEM59L, LLGL1), which were identified as brain-tissue-specific proteins by IPA in plasma and serum EVs. Pale blue symbols represent four neuron-specific proteins (STXBP1, GPM6A, PSD2, GDI1) in brain tissue, whereas green symbols represent oligodendrocyte-specific proteins (SLC44A1, CNP) in brain tissue. Gray symbols show proteins identified in this study. “See also Figures S5–S7 and Tables S1, S5, S6, S7, S8, S9, and S10.”
Table 4.
Enrichment of tissue-specific proteins in EVs separated from plasma and serum
| Source (tissue) | Tissue enrichmenta | Total | Plasma and Serum EVs | Plasma EVs | Serum EVs |
|---|---|---|---|---|---|
| Testis | 950 | 23 | 14 | 21 | 16 |
| Brain | 488 | 39 | 31 | 34 | 36 |
| Liver | 242 | 123 | 109 | 120 | 112 |
| Lymphoid | 123 | 13 | 13 | 13 | 13 |
| Intestine | 122 | 15 | 11 | 14 | 12 |
| Skin | 113 | 11 | 11 | 11 | 11 |
| Skeletal muscle | 111 | 19 | 16 | 18 | 17 |
| Placenta | 91 | 11 | 10 | 11 | 10 |
| Retina | 87 | 4 | 3 | 4 | 3 |
| Pancreas | 64 | 2 | 2 | 2 | 2 |
| Blood | 57 | 14 | 13 | 13 | 14 |
| Kidney | 53 | 4 | 4 | 4 | 4 |
| Salivary gland | 42 | 6 | 2 | 5 | 3 |
| Bone marrow | 29 | 15 | 13 | 15 | 13 |
| Heart muscle | 29 | 3 | 3 | 3 | 3 |
| Parathyroid | 22 | 6 | 5 | 5 | 6 |
| Breast | 19 | 2 | 1 | 2 | 1 |
| Lung | 13 | 2 | 1 | 2 | 1 |
| Thyroid gland | 13 | 1 | 1 | 1 | 1 |
| Tongue | 11 | 3 | 2 | 3 | 2 |
| Adrenal | 9 | 1 | 1 | 1 | 1 |
| Fallopian tube | 6 | 1 | 1 | 1 | 1 |
| Vagina | 3 | 2 | 2 | 2 | 2 |
The number of genes with enriched expression in a particular tissue compared with other tissues.
Discussion
Plasma- and serum-derived EVs are considered liquid biopsies for disease-associated changes because EVs are shed into the blood from the tissue of origin. Tissue-derived EVs are thought to reflect tissue changes more directly than bulk EVs; thus, tissue-derived EV analyses have been reported for several tissues. Rodrigues et al. reported that liver-derived EVs were separated from serum samples using a novel two-step protocol that combines a size exclusion chromatography (SEC) column and an anti-ASGR1 immunocapture step for drug metabolism and biomarkers of liver disease (Newman et al., 2021; Rodrigues et al., 2021). Allelein et al. reported that PSMA, which is located on the membrane, isolated prostate-derived EVs from cell culture supernatants with immunomagnetic beads and became a promising candidate for liquid-biopsy-based diagnostics (Allelein et al., 2021). In a brain-tissue-specific EV study, L1CAM was identified as a brain-tissue-specific protein; however, Norman et al. reported that L1CAM was not present in blood-derived EVs and was present as a soluble protein in blood (Lee et al., 2020; Norman et al., 2021). Although the tissue-specific markers used in the aforementioned examples are based on previous knowledge, large-scale tissue-specific expression information such as HPA has recently become publicly available, making it possible to select proteins with higher tissue specificity. We successfully identified tissue-specific EV proteins in healthy plasma and serum using a combination of the HPA database and protein coregulation analysis, which may be a suitable method for selecting tissue-specific proteins in plasma and serum EVs (Tables S1, S5, S6, S7, and S8) (Johnson et al., 2020; Lapek et al., 2017; Rodrigues et al., 2021).
Brain-derived EVs are expected to be a monitoring source for the brain, as brain tissue cannot be harvested alive. In 2021, the Food and Drug Administration (FDA) approved Aduhelm (aducanumab), which reduces soluble and insoluble amyloid beta in the brain, for the treatment of early Alzheimer disease (AD) (Sevigny et al., 2016). Thus, we need to screen patients with early AD from healthy adults for aducanumab therapy and monitor the progression of AD. In this study, we identified 39 brain-tissue-specific proteins in our blood EV proteome data and showed a strong positive correlation across some brain-tissue-specific proteins. STXBP1, GPM6A, PSD2, and GDI1 were found to be neuron-cell-specific proteins in brain tissue, and STXBP1, GPM6A, and GDI1 in particular interact indirectly through APP, MAPT, and HPP, which are related to neurodegenerative disease and may be enriched in the same EVs in the blood (Inoue et al., 2015; Ito et al., 2018; Sharma et al., 2015). The detection of APP and MAPT in brain-tissue-specific neuronal EVs, which are enriched by STXBP1, GPM6A, and GDI1 protein, is expected to screen for early AD and monitor the progression of AD. It has been reported that brain-derived EVs can be detected in the blood (Lee et al., 2020; Ricklefs et al., 2019; Sproviero et al., 2018), but it is not yet known how they are released from brain tissue into the blood through the blood-brain barrier (BBB). These brain-tissue-specific proteins may elucidate the mechanism by which EVs cross the BBB. Furthermore, liver-, intestine-, skeletal muscle-, and other tissue-specific proteins were found to be co-regulated in healthy blood EVs. Future studies on the separation of EVs from plasma and serum by tissue-specific EV proteins would provide new biomarkers for diagnosing and monitoring the progression of tissue-specific disease without diagnostic imaging.
There is no consensus on standard reference proteins for EV protein study normalization in plasma and serum EVs. In the present study, we found several proteins that were detected more commonly among the samples within a 10% CV value than commonly used EV markers, such as CD9, CD81, and CD63. These proteins, mentioned later, may be useful as endogenous housekeeping controls in plasma and serum for EV protein assays. The plasma EV proteins ACTBL2, NPEPPS, and DBNL; the serum EV proteins ARF6, GNAI2, and HSPH1; and the plasma and serum EV proteins NCK1, GNAI2, and SLC39A10 had low CV values among each EV sample (Figure 3D, Tables 4 and S3). GNAI2 was found in the top 100 most abundant EV proteins reported in ExoCarta (Keerthikumar et al., 2016). Therefore, our CV data may serve as information for finding standard reference candidate proteins in plasma and serum EVs for EVs or for selecting serum or plasma for the detection of specific target markers. Our data will be more robust with larger sample validation in the future.
Proteome profiling of plasma and serum EVs has been performed following ultracentrifugation (UC), SEC, precipitation, and AC in downstream proteomics analysis (Ding et al., 2020; Ebrahimkhani et al., 2018; Fel et al., 2019; Jeannin et al., 2018; Pietrowska et al., 2021). Our study showed more than 4,000 proteins in EV proteomics and identified more EV markers than previous studies. Our approach effectively isolated EVs containing EV markers from plasma and serum (Tables 1 and S2). Regarding non-EV proteins such as albumin and apolipoprotein, we identified them at a lower ratio than previous studies (Figure 2C). This AC method captures and enriches phosphatidylserine (PS)+ EVs, including apoptotic bodies from body fluids. Matsumura et al. reported that two subtypes of EVs containing different rates of PS+ EVs could be separated from the culture media of cancer cells by density gradient UC methods (Matsumura et al., 2019). However, it is not known how many PS- EVs exist in biospecimens. Most recently, Matsumoto et al. reported that the PS− EV subpopulation in blood is present for a long time due to its escape from macrophage uptake (Matsumoto et al., 2021). Thus, somatic-cell-derived EVs in the endogenous blood are mainly PS− EVs, which would allow for highly sensitive and specific detection (Matsumoto et al., 2021). Previously, PS+ EVs were shown to be involved in tumor growth and metastasis and are expected to be tumor and neurological disease markers (Birge et al., 2016; Lea et al., 2017; Sharma et al., 2017). Further studies are important to clarify the characteristics of PS+ EVs and PS− EVs in blood and to compare with the results of EVs isolated by other isolation methods, including UC and SEC, which are able to isolate a more comprehensive EV population for biomarker discovery.
There were differences in plasma and serum EV profiles across individuals (Figures 3A and 3B). However, the contribution of Components 1 and 2 in Figure 3A is 74.4% and 6.6%, indicating that the difference between plasma and serum has a greater influence on the PCA than the difference between individuals. In addition, the top five PC1 loading factors included alpha-actinin (ACTN1), coronin-1C (CORO1C), Tubulin alpha-8 (TUBAB8), Filamin-A (FLNA), and Tubulin alpha-4 (TUBA4), whereas the bottom included fibrinogen beta, gamma, Complement 2 (C2), matrix Gla (MGP), and Fibrinogen alpha (FGA). Fibrinogen family proteins, which are synthesized in the liver, are known to be abundant in plasma, and they are enriched in plasma EVs compared with serum EVs (Tennent et al., 2006). The KEGG enrichment analysis revealed that proteins related to complement and coagulation cascades and cholesterol metabolism were enriched in plasma EVs, whereas proteins related to platelet activation were enriched in serum EVs (Figure S2). Gyorgy et al.. and Palviainen et al. showed that serum contains more platelet-derived EVs than acid citrate dextrose (ACD) or EDTA plasma because ACD and EDTA inhibit platelet activation in blood samples after blood collection (György et al., 2014; Palviainen et al., 2020). The HPA data showed higher expression levels of fibrinogen and lipoproteins in the liver than in other tissues (Table S1), and liver-tissue-specific proteins were increased in plasma EVs compared with serum EVs. The results of the differences between serum and plasma EV proteomes are consistent with existing reports as described earlier. The differences in sensitivity and reproducibility between serum and plasma EVs for each protein were also revealed for nearly 4,000 proteins in this study. Our proteomic profiling of plasma EVs and serum EVs would be helpful for determining whether plasma or serum should be used for biomarker discovery of organ-specific diseases.
Limitations of the study
Comprehensive MS profiling of plasma and serum EV can provide a detailed catalog of the differential expression profiles of EV proteins in plasma and serum among healthy individuals and a list of tissue-specific EV proteins in healthy individuals. The tissue-specific EV proteins were screened by comparison with our plasma/serum EV proteomics dataset and RNA sequencing analysis (HPA dataset). Proteome studies of human tissue have successfully quantified 13,000 proteins (Jiang et al., 2020; Wang et al., 2019), but there were fewer tissue-specific proteins than with HPA dataset. Further investigation is necessary to identify and quantify more proteins in healthy human tissues by comprehensive proteome analysis to compare tissue-specific proteins and plasma/serum EV proteins of human. In addition, as they have only been identified in a small cohort of healthy individuals, it would be preferred to replicate them in a larger sample of healthy individuals and patients with diseases.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Human CD9 (D3H4P) | Cell Signaling Techonology | Cat# 13403; RRID:AB_2732848 |
| Rabbit anti-Human FLOT1 (EPR6041) | Abcam | Cat# ab133497; RRID:AB_11156367 |
| Mouse anti-Human Alix (3A9) | Cell Signaling Techonology | Cat# 2171; RRID:AB_2299455 |
| Mouse anti-Human ApoB (A-6) | Santa Cruz BioteB6:C7chnology | Cat# sc-393636 |
| Mouse anti-Human serum albumin (MAB1455) | R&D Systems | Cat# 188835 |
| Anti-mouse IgG, HRP-linked antibody | Cell Signaling Techonology | Cat# 7076; RRID:AB_330924 |
| Anti-rabbit IgG, HRP-linked antibody | Cell Signaling Techonology | Cat# 7074; RRID:AB_2099233 |
| Biological samples | ||
| Human blood | Healthy adult | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 0.45-μm Spin-X centrifuge tube | Corning | CLS8162 |
| Heparin sodium solution | Fujifilm WAKO Pure Chemical Corporation | 085-00134 |
| Carbon-film grid | Nisshin EM | 2634 |
| Filter paper | ADVANTEC | 21055 |
| 4% to 20% gradient gel | Dream Realization & Communication Co. Ltd | NXV-2761HP20 |
| Immobilon-P membrane, PVDF 0.45-μm | Millipore | IPVH304F0 |
| Blocking One solution | Nacalai Tesque | 03953-95 |
| Tris(2-carboxyethyl)phosphine (TCEP) | WAKO | 209-19861 |
| Iodoacetamide | Nacalai Tesque | 19302-54 |
| L-cysteine | Nacalai Tesque | 1030912 |
| Lys-C | WAKO | 121-02541 |
| Trypsin solution | WAKO | 202-20081 |
| Critical commercial assays | ||
| Detergent compatible (DC) protein assay | Bio-Rad | 500-0116 |
| MagCapture Exosome Isolation Kit PS version 2 | Fujifilm WAKO Pure Chemical Corporation | 290-84103 |
| Deposited data | ||
| Deposition of mass spectrometry proteomics data | jPOST repository | PXD025750 |
| Software and algorithms | ||
| Nanosight NTA 3.3 software | Malvern Panalytical Inc | https://www.malvernpanalytical.com/ |
| DIA-NN software (Ver. 1.7.12) | Demichev et al. (2020) | https://github.com/vdemichev/DiaNN (Demichev et al., 2020) |
| UniProt database | UniProt Consortium | https://www.uniprot.org/ (Consortium et al., 2020) |
| Perseus ver. 1.6.14.0 | Tyanova et al., 2016a, Tyanova et al., 2016b | https://maxquant.org/perseus/ (Tyanova et al., 2016b) |
| GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com |
| Ingenuity Pathway Analysis | QIAGEN | https://digitalinsights.qiagen.com/products-overview/discoveryinsights-portfolio/analysis-andvisualization/qiagen-ip |
| DAVID Bioinformatics Resources 6.8 | Huang et al. (2009) | https://david.ncifcrf.gov/ (Huang et al., 2009) |
| STRING database | Szklarczyk et al. (2017) | https://string-db.org/ (Szklarczyk et al., 2017) |
| IBM SPSS software ver. 27 | IBM | https://www.ibm.com/ |
| R studio (ver. 1.4.1103) | R Studio | https://rstudio.com/ |
| Venny_2.1 | http://bioinfogp.cnb.csic.es/tools/venny/ | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Project reader Jun Adachi (jun_adachi@nibiohn.go.jp)
Materials availability
This study did not generate any new unique reagents.
Experimental model and subject details
The study involved the use of human peripheral blood from six healthy adults. Six healthy subjects (gender; 3 males and 3 females, age; 52.2 ± 10.7) from the Cancer Institute Hospital of the Japanese Foundation for Cancer Research provided EDTA plasma samples and matched serum samples. The Institutional Review Board of the Cancer Institute Hospital of the Japanese Foundation for Cancer Research approved the protocol, and all participants provided informed consent.
Method details
Separation of EVs from human plasma and serum samples
EVs were separated from human plasma and serum using the MagCapture Exosome Isolation Kit PS version 2 (# 290-84103 Fujifilm WAKO Pure Chemical Corporation). Briefly, 150 μl of plasma and serum were added to 350 μl of Tris-buffered saline (TBS) and centrifuged at 1,200 g for 20 min at 4°C. The supernatant was filtered through a 0.45-μm Spin-X centrifuge tube (# CLS8162 Corning), and then 5.0 μl of 1 U/μl heparin sodium solution (# 085-00134 Fujifilm WAKO Pure Chemical Corporation) was added only to the EDTA plasma samples to maintain the anticoagulant effect instead of EDTA. The EVs were eluted with 100 μl of elution buffer, and 80 μl of eluate was used for proteomics analysis.
Nanoparticle tracking analysis (NTA)
All samples were diluted in double-filtered PBS (dfPBS) at least 1:10 or more to obtain particles within the target reading range for the Nanosight 300 machine (Malvern Panalytical Inc), which is 10-100 particles per frame. Using a manual injection system, five 30-sec videos were taken for each sample at 21°C. Analysis of particle counts was carried out with Nanosight NTA 3.3 software (Malvern Panalytical Inc) with a detection threshold of 5.
Transmission electron microscopy (TEM)
EVs separated from plasma and serum were analyzed by TEM. Five microliters of the EV sample was adsorbed for 10 sec on a carbon-film grid (# 2634 Nisshin EM). Excess liquid was removed with filter paper (# 00021055 ADVANTEC), and the grid was then floated briefly on a drop of water (to wash away phosphate or salt), blotted on filter paper, and stained with 2% uranyl acetate (# 22400 EMS) in water for 10 sec. After removing the excess uranyl acetate with filter paper, random fields were photographed using a HITACHI H-7600 electron microscope at 100 kV.
Protein concentrations
The detergent compatible (DC) protein assay (# 500-0116 Bio-Rad) was used to determine the protein concentration for each sample. Due to the limited amount of sample, plasma and serum samples were diluted 1:1000, EVs were diluted 1:10 before loading into the assay, and a 1:5:40 ratio of sample to reaction components (A’ and B) was used. All assay mixtures were incubated at 37°C for 15 min before the protein concentration was read on an iMark Microplate Absorbance Reader (# 1681130J1 Bio-Rad) at 750 nm.
Western blotting
EV samples from plasma and serum and plasma and serum samples were run in a 4% to 20% gradient gel (# NXV-2761HP20, Dream Realization & Communication Co. Ltd., Japan) and electrotransferred to an Immobilon-P membrane, PVDF 0.45-μm (# IPVH304F0, Millipore). The membrane was blocked in Blocking One solution (# 03953-95 Nacalai Tesque) before being immunoblotted with specific primary antibodies (CD9 (D3H4P), # 13403 Cell Signaling Technology; Alix (3A9), # 2171 Cell Signaling Technology; flotillin (FLOT1), # EPR6041 Abcam; apolipoprotein B (A-6), # sc-393636 Santa Cruz Biotechnology; human serum albumin (MAB1455), # 188835 R&D Systems). The membrane was incubated with HRP-labeled secondary antibodies (mouse, # 7076S; rabbit, # 7074S; Cell Signaling Technology) and scanned using a LAS4000 multicolor microscope (Fujifilm).
In-solution digestion
Separated EV fractions were lysed with lysis buffer (12 mM sodium deoxycholate, 12 mM sodium lauroyl sarcosinate, 50 mM ammonium bicarbonate (ABC)), and then the mixed samples were vortexed for 5 min at room temperature followed by spin down and boiling for 10 min at 95°C. The samples were reduced with 10 mM tris(2-carboxyethyl)phosphine (TCEP) (# 209-19861 WAKO) for 30 min at 37°C and alkylated with 20 mM iodoacetamide (# 19302-54 Nacalai Tesque) for 30 min at 37°C in the dark. Subsequently, L-cysteine (# 1030912 Nacalai Tesque) was added to the samples to a final concentration of 21 mM, and then the mixed sample was incubated for 10 min at room temperature. The samples were digested with 2 μL of 1 mAU/μL Lys C (#121-02541 WAKO) and 0.5 μg/μL trypsin solution (#202-20081 WAKO) overnight at 37°C. The digested peptides were desalted via the stop-and-go-extraction tip (StageTip) protocol (Rappsilber et al., 2007), dried via vacuum centrifugation, and resuspended in 2% acetonitrile 1% trifluoroacetic acid for liquid chromatography and tandem mass spectrometry (LC-MS/MS) processing.
Mass spectrometry
Nano-LC-MS/MS
Nano-LC-MS/MS analysis was conducted with an LTQ-Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, USA) equipped with an UltiMate 3000 Nano LC system (Thermo Fisher Scientific, Bremen, Germany) and an HTC-PAL autosampler (CTC Analytics, Zwingen, Switzerland). Peptides were separated on an analytical column (75 μm × 50 cm, packed in-house with ReproSil-Pur C18-AQ, 1.9 μm resin, Dr. Maisch, Ammerbuch, Germany), and separation was achieved using a 145-min gradient of 5 to 30% acetonitrile in 0.1% formic acid at a flow rate of 280 nL/min. Data were acquired using data-independent acquisition mode (DIA). An Orbitrap Fusion Lumos mass spectrometer was used for gas-phase fractionation (GPF)-DIA acquisition of a pooled sample for library, and full mass spectra were acquired with the following parameters: a 2-m/z isolation window, a resolution of 120,000, an automatic gain control (AGC) target of 4 × 105 with an injection time of 86 ms, and a normalized collision energy of 30. The five GPF-DIA runs collectively covered 395-805 m/z (i.e., 395-485, 475-565, 555-645, 635-725, and 715-805 m/z). For the individual samples for proteome profiling acquisition, full mass spectra were acquired with the following parameters: an 8-m/z isolation window, a resolution of 50,000, an AGC target of 4 × 105 with an injection time of 86 ms, and a normalized collision energy of 30.
Quantification and statistical analysis
MS data analysis
MS data (raw file) were processed with DIA-NN software (Ver. 1.7.12) (Demichev et al., 2020). Database searching included all entries from the Homo sapiens UniProt database (downloaded in April 2020, taxonomy ID: 9606) and contaminant database (Tyanova et al., 2016a). This database was concatenated with one composed of all protein sequences in the reversed order. The search parameters were as follows: up to two missed cleavage sites, 7-30 peptide length, carbamidomethylation of cysteine residues (+57.021 Da) as static modifications, protein names from FASTA for implicit protein grouping, robust LC (high precision) for quantification strategy, and RT-dependent for cross-run normalization. Precursor ions were adjusted to a 1% false discovery rate (FDR). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the jPOST repository with the dataset identifier PXD025750 (Okuda et al., 2017). Protein quantitation values were exported for further analysis in Microsoft Excel or Prism9.
Statistical analysis
Statistical analysis was conducted using Perseus ver. 1.6.14.0 (Tyanova et al., 2016b), IBM SPSS software ver. 27 and GraphPad Prism9. Bivariate correlation analysis was applied to examine differences between proteins in proteomics data and demographics data by Spearman’s rank and Pearson’s using IBM SPSS software ver.27 and R studio (ver. 1.4.1103) (Corrplot, psych, Hmisc and ggplot2 package). The Gene Ontology (GO) of identified proteins was elucidated with the Database for Annotation, Visualization, and Integrated Discovery (DAVID) Bioinformatics Resources 6.8. Protein networks and enrichment analysis were generated using Ingenuity Pathway Analysis (IPA) (QIAGEN) and the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (version. 11.0). Venn diagram analysis was generated using Venny_2.1 (http://bioinfogp.cnb.csic.es/tools/venny/).
Acknowledgments
The author thanks S. Kawamoto (Hanaichi Ultra-Structure Research Institute, Co., Ltd.) for electron microscopic imaging services and R. Ukekawa and S. Ozaki (Fujifilm Wako Pure Chemical Co.) for experimental support. Graphical abstract was created with BioRender.com.
Funding: This work is in part funded by Cabinet Office of Japan Government for the Public/Private R&D Investment Strategic Expansion PrograM (PRISM).
Author contributions
S.M. and J.A. designed research; S.M., M.H., and J.I. performed research; S.M. and J.A. analyzed data; S.N. provided plasma and serum; S.M. and J.A. wrote the paper; S.M., T.T., and J.A. edited the paper.
Declaration of interests
S.M., T.T., and J.A. are named inventors on invention disclosures and patents involving Blood EV proteome. The other authors declare no commercial or financial conflict of interest.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104012.
Supplemental information
Data and code availability
The datasets generated in this study are available via ProteomeXchange with identifier PXD025750.
References
- Agliardi C., Guerini F.R., Zanzottera M., Bianchi A., Nemni R., Clerici M. SNAP-25 in serum is carried by exosomes of neuronal origin and is a potential biomarker of Alzheimer’s disease. Mol. Neurobiol. 2019;56:5792–5798. doi: 10.1007/s12035-019-1501-x. [DOI] [PubMed] [Google Scholar]
- Allelein S., Medina-Perez P., Lopes A.L.H., Rau S., Hause G., Kölsch A., Kuhlmeier D. Potential and challenges of specifically isolating extracellular vesicles from heterogeneous populations. Sci. Rep. 2021;11:11585. doi: 10.1038/s41598-021-91129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A., Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- Birge R.B., Boeltz S., Kumar S., Carlson J., Wanderley J., Calianese D., Barcinski M., Brekken R.A., Huang X., Hutchins J.T., et al. Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ. 2016;23:962–978. doi: 10.1038/cdd.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana D., Zobairi F., Martinez M.C., Panaro M.A., Mitolo V., Freyssinet J.-M., Kunzelmann C. Membrane microvesicles as actors in the establishment of a favorable prostatic tumoral niche: a role for activated fibroblasts and CX3CL1-CX3CR1 axis. Cancer Res. 2009;69:785–793. doi: 10.1158/0008-5472.can-08-1946. [DOI] [PubMed] [Google Scholar]
- Consortium T.U., Bateman A., Martin M.-J., Orchard S., Magrane M., Agivetova R., Ahmad S., Alpi E., Bowler-Barnett E.H., Britto R., et al. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2020;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo A.M., Ikezu T. Extracellular vesicle biology in Alzheimer’s disease and related tauopathy. J. Neuroimmune Pharmacol. 2018;13:292–308. doi: 10.1007/s11481-017-9768-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpech J.-C., Herron S., Botros M.B., Ikezu T. Neuroimmune crosstalk through extracellular vesicles in health and disease. Trends Neurosci. 2019;42:361–372. doi: 10.1016/j.tins.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demichev V., Messner C.B., Vernardis S.I., Lilley K.S., Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods. 2020;17:41–44. doi: 10.1038/s41592-019-0638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Ding X.-Q., Wang Z.-Y., Xia D., Wang R.-X., Pan X.-R., Tong J.-H. Proteomic profiling of serum exosomes from patients with metastatic gastric cancer. Front. Oncol. 2020;10:1113. doi: 10.3389/fonc.2020.01113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimkhani S., Vafaee F., Hallal S., Wei H., Lee M.Y.T., Young P.E., Satgunaseelan L., Beadnall H., Barnett M.H., Shivalingam B., et al. Deep sequencing of circulating exosomal microRNA allows non-invasive glioblastoma diagnosis. NPJ Precis. Oncol. 2018;2:28–29. doi: 10.1038/s41698-018-0071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Q., Strand A., Law W., Faca V.M., Fitzgibbon M.P., Hamel N., Houle B., Liu X., May D.H., Poschmann G., et al. Brain-specific proteins decline in the cerebrospinal fluid of humans with Huntington disease. Mol. Cell Proteomics. 2009;8:451–466. doi: 10.1074/mcp.m800231-mcp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fel A., Lewandowska A.E., Petrides P.E., Wiśniewski J.R. Comparison of proteome composition of serum enriched in extracellular vesicles isolated from polycythemia vera patients and healthy controls. Proteomes. 2019;7:20. doi: 10.3390/proteomes7020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frühbeis C., Fröhlich D., Kuo W.P., Amphornrat J., Thilemann S., Saab A.S., Kirchhoff F., Möbius W., Goebbels S., Nave K.-A., et al. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013;11:e1001604. doi: 10.1371/journal.pbio.1001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- György B., Pálóczi K., Kovács A., Barabás E., Bekő G., Várnai K., Pállinger É., Szabó-Taylor K., Szabó T.G., Kiss A.A., et al. Improved circulating microparticle analysis in acid-citrate dextrose (ACD) anticoagulant tube. Thromb. Res. 2014;133:285–292. doi: 10.1016/j.thromres.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Hoshino A., Costa-Silva B., Shen T.-L., Rodrigues G., Hashimoto A., Mark M.T., Molina H., Kohsaka S., Giannatale A.D., Ceder S., et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A., Kim H.S., Bojmar L., Gyan K.E., Cioffi M., Hernandez J., Zambirinis C.P., Rodrigues G., Molina H., Heissel S., Mark M.T. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182:1044–1061.e18. doi: 10.1016/j.cell.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini H., Obradović M.M.S., Hoffmann M., Harper K.L., Sosa M.S., Werner-Klein M., Nanduri L.K., Werno C., Ehrl C., Maneck M., et al. Early dissemination seeds metastasis in breast cancer. Nature. 2016;540:552–558. doi: 10.1038/nature20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ikeda A., Nagayama S., Sumazaki M., Konishi M., Fujii R., Saichi N., Muraoka S., Saigusa D., Shimada H., Sakai Y., Ueda K. Colorectal cancer–derived CAT1-positive extracellular vesicles alter nitric oxide metabolism in endothelial cells and promote angiogenesis. Mol. Cancer Res. 2021 doi: 10.1158/1541-7786.mcr-20-0827. [DOI] [PubMed] [Google Scholar]
- Inoue M., Hur J., Kihara T., Teranishi Y., Yamamoto N.G., Ishikawa T., Wiehager B., Winblad B., Tjernberg L.O., Schedin-Weiss S. Human brain proteins showing neuron-specific interactions with γ-secretase. FEBS J. 2015;282:2587–2599. doi: 10.1111/febs.13303. [DOI] [PubMed] [Google Scholar]
- Ito Y., Honda A., Igarashi M. Glycoprotein M6a as a signaling transducer in neuronal lipid rafts. Neurosci. Res. 2018;128:19–24. doi: 10.1016/j.neures.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Jeannin P., Chaze T., Gianetto Q.G., Matondo M., Gout O., Gessain A., Afonso P.V. Proteomic analysis of plasma extracellular vesicles reveals mitochondrial stress upon HTLV-1 infection. Sci. Rep. 2018;8:5170–5177. doi: 10.1038/s41598-018-23505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Wang M., Lin S., Jian R., Li X., Chan J., Dong G., Fang H., Robinson A.E., Consortium Gte., Snyder M.P. A quantitative proteome map of the human body. Cell. 2020;183:269–283.e19. doi: 10.1016/j.cell.2020.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.C.B., Dammer E.B., Duong D.M., Ping L., Zhou M., Yin L., Higginbotham L.A., Guajardo A., White B., Troncoso J.C., et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med. 2020;26:769–780. doi: 10.1038/s41591-020-0815-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerthikumar S., Chisanga D., Ariyaratne D., Saffar H.A., Anand S., Zhao K., Samuel M., Pathan M., Jois M., Chilamkurti N., et al. ExoCarta: a web-based compendium of exosomal cargo. J. Mol. Biol. 2016;428:688–692. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J., Arras G., Colombo M., Jouve M., Morath J.P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Thery C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U S A. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapek J.D., Greninger P., Morris R., Amzallag A., Pruteanu-Malinici I., Benes C.H., Haas W. Detection of dysregulated protein association networks by high-throughput proteomics predicts cancer vulnerabilities. Nat. Biotechnol. 2017;35:983–989. doi: 10.1038/nbt.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea J., Sharma R., Yang F., Zhu H., Ward E.S., Schroit A.J. Detection of phosphatidylserine-positive exosomes as a diagnostic marker for ovarian malignancies: a proof of concept study. Oncotarget. 2017;8:14395–14407. doi: 10.18632/oncotarget.14795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.E., Winston-Gray C., Barlow J.W., Rissman R.A., Jeste D.V. Plasma levels of neuron- and astrocyte-derived exosomal amyloid beta1-42, amyloid beta1-40, and phosphorylated tau levels in schizophrenia patients and non-psychiatric comparison subjects: relationships with cognitive functioning and psychopathology. Front. Psychiatry. 2020;11:532624. doi: 10.3389/fpsyt.2020.532624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., McGuire M.H., Mangala L.S., Lee S., Stur E., Hu W., Bayraktar E., Villar-Prados A., Ivan C., Wu S.Y., et al. Gain-of-function p53 protein transferred via small extracellular vesicles promotes conversion of fibroblasts to a cancer-associated phenotype. Cell Rep. 2021;34:108726. doi: 10.1016/j.celrep.2021.108726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A., Takahashi Y., Ogata K., Kitamura S., Nakagawa N., Yamamoto A., Ishihama Y., Takakura Y. Phosphatidylserine-deficient small extracellular vesicle is a major somatic cell-derived sEV subpopulation in blood. iScience. 2021;24:102839. doi: 10.1016/j.isci.2021.102839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura S., Minamisawa T., Suga K., Kishita H., Akagi T., Ichiki T., Ichikawa Y., Shiba K. Subtypes of tumour cell-derived small extracellular vesicles having differently externalized phosphatidylserine. J. Extracell. Vesicles. 2019;8:1579541. doi: 10.1080/20013078.2019.1579541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka S., Jedrychowski M.P., Yanamandra K., Ikezu S., Gygi S.P., Ikezu T. Proteomic profiling of extracellular vesicles derived from cerebrospinal fluid of Alzheimer’s disease patients: a pilot study. Cells. 2020;9:1959. doi: 10.3390/cells9091959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka S., Lin W., Chen M., Hersh S.W., Emili A., Xia W., Ikezu T. Assessment of separation methods for extracellular vesicles from human and mouse brain tissues and human cerebrospinal fluids. Methods. 2020;177:35–49. doi: 10.1016/j.ymeth.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka S., Jedrychowski M.P., Iwahara N., Abdullah M., Onos K.D., Keezer K.J., Hu J., Ikezu S., Howell G.R., Gygi S.P., Ikezu T. Enrichment of neurodegenerative microglia signature in brain-derived extracellular vesicles isolated from Alzheimer’s disease mouse models. J. Proteome Res. 2021;20:1733–1743. doi: 10.1021/acs.jproteome.0c00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman L.A., Fahmy A., Sorich M.J., Best O.G., Rowland A., Useckaite Z. Importance of between and within subject variability in extracellular vesicle abundance and cargo when performing biomarker analyses. Cells. 2021;9:485. doi: 10.3390/cells10030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman M., Ter-Ovanesyan D., Trieu W., Lazarovits R., Kowal E.J.K., Lee J.H., Chen-Plotkin A.S., Regev A., Church G.M., Walt D.R. L1CAM is not associated with extracellular vesicles in human cerebrospinal fluid or plasma. Nat. Methods. 2021;18:631–634. doi: 10.1038/s41592-021-01174-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S., Watanabe Y., Moriya Y., Kawano S., Yamamoto T., Matsumoto M., Takami T., Kobayashi D., Araki N., Yoshizawa A.C., et al. jPOSTrepo: an international standard data repository for proteomes. Nucleic Acids Res. 2017;45:D1107–D1111. doi: 10.1093/nar/gkw1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palviainen M., Saraswat M., Varga Z., Kitka D., Neuvonen M., Puhka M., Joenväärä S., Renkonen R., Nieuwland R., Takatalo M., Siljander P.R.-M. Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo—implications for biomarker discovery. PLoS One. 2020;15:e0236439. doi: 10.1371/journal.pone.0236439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M.R., Weaver A.M. Astrocyte-derived small extracellular vesicles promote synapse formation via fibulin-2-mediated TGF-β signaling. Cell Rep. 2021;34:108829. doi: 10.1016/j.celrep.2021.108829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrowska M., Zebrowska A., Gawin M., Marczak L., Sharma P., Mondal S., Mika J., Polańska J., Ferrone S., Kirkwood J.M., et al. Proteomic profile of melanoma cell-derived small extracellular vesicles in patients’ plasma: a potential correlate of melanoma progression. J. Extracell. Vesicles. 2021;10:e12063. doi: 10.1002/jev2.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Ricklefs F.L., Maire C.L., Reimer R., Dührsen L., Kolbe K., Holz M., Schneider E., Rissiek A., Babayan A., Hille C., et al. Imaging flow cytometry facilitates multiparametric characterization of extracellular vesicles in malignant brain tumours. J. Extracell. Vesicles. 2019;8:1588555. doi: 10.1080/20013078.2019.1588555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A.D., Dyk M.V., Sorich M.J., Fahmy A., Useckaite Z., Newman L.A., Kapetas A.J., Mounzer R., Wood L.S., Johnson J.G., Rowland A. Exploring the use of serum-derived small extracellular vesicles as liquid biopsy to study the induction of hepatic cytochromes P450 and organic anion transporting polypeptides. Clin. Pharmacol. Ther. 2021;110:248–258. doi: 10.1002/cpt.2244. [DOI] [PubMed] [Google Scholar]
- Ruan Z., Pathak D., Kalavai S.V., Yoshii-Kitahara A., Muraoka S., Bhatt N., Takamatsu-Yukawa K., Hu J., Wang Y., Hersh S., et al. Alzheimer’s disease brain-derived extracellular vesicles spread tau pathology in interneurons. Brain. 2020;21:72. doi: 10.1093/brain/awaa376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevigny J., Chiao P., Bussière T., Weinreb P.H., Williams L., Maier M., Dunstan R., Salloway S., Chen T., Ling Y., et al. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature. 2016;537:50–56. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- Sharma K., Schmitt S., Bergner C.G., Tyanova S., Kannaiyan N., Manrique-Hoyos N., Kongi K., Cantuti L., Hanisch U.-K., Philips M.-A., et al. Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 2015;18:1819–1831. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., Huang X., Brekken R.A., Schroit A.J. Detection of phosphatidylserine-positive exosomes for the diagnosis of early-stage malignancies. Br. J. Cancer. 2017;117:545–552. doi: 10.1038/bjc.2017.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalnikova H.K., Bohuslavova B., Turnovcova K., Juhasova J., Juhas S., Rodinova M., Vodicka P. Isolation and characterization of small extracellular vesicles from porcine blood plasma, cerebrospinal fluid, and seminal plasma. Proteomes. 2019;7:17. doi: 10.3390/proteomes7020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproviero D., Salvia S.L., Giannini M., Crippa V., Gagliardi S., Bernuzzi S., Diamanti L., Ceroni M., Pansarasa O., Poletti A., Cereda C. Pathological proteins are transported by extracellular vesicles of sporadic amyotrophic lateral sclerosis patients. Front. Neurosci. 2018;12:12093. doi: 10.3389/fnins.2018.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Lin F., Sun W., Zhu W., Fang D., Luo L., Li S., Zhang W., Jiang L. Exosome-transmitted miRNA-335-5p promotes colorectal cancer invasion and metastasis by facilitating EMT via targeting RASA1. Mol. Ther. Nucleic Acids. 2021;24:164–174. doi: 10.1016/j.omtn.2021.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.R., Robinson M.B., Gifondorwa D.J., Tytell M., Milligan C.E. Regulation of heat shock protein 70 release in astrocytes: role of signaling kinases. Dev. Neurobiol. 2007;67:1815–1829. doi: 10.1002/dneu.20559. [DOI] [PubMed] [Google Scholar]
- Tennent G.A., Brennan S.O., Stangou A.J., O’Grady J., Hawkins P.N., Pepys M.B. Human plasma fibrinogen is synthesized in the liver. Blood. 2006;109:1971–1974. doi: 10.1182/blood-2006-08-040956. [DOI] [PubMed] [Google Scholar]
- Thery C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., Ayre D.C. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Gong M., Hu Y., Liu H., Zhang W., Zhang M., Hu X., Aubert D., Zhu S., Wu L., Yan X. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles. 2019;9:1697028. doi: 10.1080/20013078.2019.1697028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Wang D., Eraslan B., Wieland T., Hallström B., Hopf T., Zolg D.P., Zecha J., Asplund A., Li L.-H., Meng C., et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 2019;15:e8503. doi: 10.15252/msb.20188503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y., Borgmann K., Edara V.V., Stacy S., Ghorpade A., Ikezu T. Activated human astrocyte-derived extracellular vesicles modulate neuronal uptake, differentiation and firing. J. Extracell. Vesicles. 2020;9:1706801. doi: 10.1080/20013078.2019.1706801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated in this study are available via ProteomeXchange with identifier PXD025750.