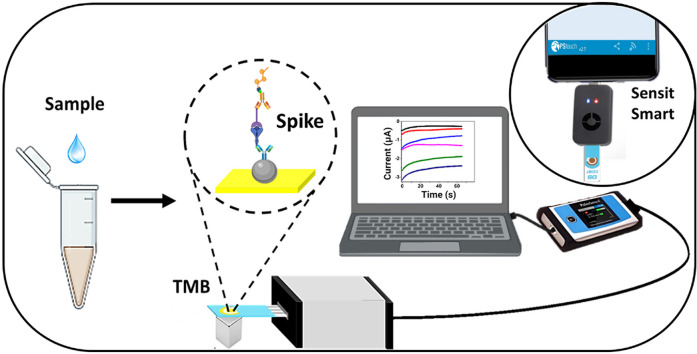
Abstract
Rapid, straightforward, and massive diagnosis of coronavirus disease 2019 (COVID-19) is one of the more important measures to mitigate the current pandemics. This work reports on an immunosensor to rapidly detect the spike protein from the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The immunosensing device entraps the spike protein linked to angiotensin-converting enzyme host receptor (ACE2) protein in a sandwich between carboxylated magnetic beads functionalized with an anti-spike antibody and an anti-ACE2 antibody, further labeled with streptavidin (poly)horseradish peroxidase (HRP) reporter enzyme. The particles were confined at the surface of screen-printed gold electrodes, whose signal resulting from the interaction of the enzyme with a mediator was recorded in a portable potentiostat. The immunosensor showed a sensitivity of 0.83 μA∗mL/μg and a limit of detection of 22.5 ng/mL of spike protein, with high reproducibility. As a proof-of-concept, it detected commercial spike protein-supplemented buffer solutions, pseudovirions, isolated viral particles and ten nasopharyngeal swab samples from infected patients compared to samples from three healthy individuals paving the way to detect the virus closer to the patient.
Keywords: SARS-CoV-2, Spike protein, ACE2, Immunosensor, Coronavirus, Magnetic beads
Graphical abstract
1. Introduction
Coronavirus disease 2019 (COVID-19) is an infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), first reported in Wuhan (China) in December 2019 [1]. The SARS-CoV-2 belongs to the Coronaviridae family and is the seventh virus of this family known to infect humans, causing severe symptoms (mainly respiratory failure) that may lead to death, affecting people worldwide regardless of race or gender, age or social status [2].
The SARS-CoV-2 genome is a single strand + RNA of 29903 nucleotides with approximately 80% homology with SARS-CoV, a coronavirus described in 2002 [3]. The virus structure consists of a cellular-derived bilipid membrane and a set of the structural proteins spike (S), membrane (M), envelope (E) and nucleocapsid (N). The M, E and S proteins are located on the virion's envelope, and the N protein is associated with the viral genome. Protein S is a highly N-glycosylated 180 KDa trimeric class I fusion glycoprotein that consists of two subunits known as S1 and S2 [[4,5]] and is responsible for binding to the ACE2 protein in the cell membrane. The receptor-binding domain (RBD) within the S1 subunit is responsible for interaction with the ACE2 receptor, while the S2 subunit facilitates virus-cell membrane fusion and forms the spike stem [[1,[5], [6], [7]]]. The RBD region of the SARS-CoV-2 binds to the ACE2 receptor with a 10–20 fold higher affinity than SARS-CoV, which facilitate viral entry and provide an explanation for the easiness of virus spread from person to person [[8,9]] thereby generating significant concern throughout the whole world population.
Rapid, simple and specific detection of the virus is paramount to decrease its speed of spread. Standard diagnostics include molecular methods that detect the viral genome (RNA) by reverse transcription-polymerase chain reaction (RT-PCR), immunoassays that detect viral structural proteins, and indirect serological methods that detect anti-SARS-CoV-2 IgG and IgM antibodies [10]. However, these methods are time-consuming and have highly qualified personnel and specialized equipment requirements.
Biosensor-based devices are characterized by high sensitivity, low detection limit and high specificity derived from the combination of material properties and biological recognition systems [11]. They are also amenable for miniaturization and integration into portable analysis systems using minimal volumes of reagents and samples for fast and user-friendly assays [12]. Electrochemical biosensors are the most extended. A complete characterization of the electrochemical process at an electrode surface is achieved readily by electrochemical techniques. Chronoamperometry can be specific by adequately selecting the working potential and enjoys high sensitivity and simplicity [[13,14]; therefore, it is the preferred technique to follow electrochemical reactions once the electrochemical process has been well-characterized. Outstanding features of biosensors have been used in the detection of bacterial pathogens [15], infections [16], cancer biomarkers [[17], [18]], autoimmune diseases [19] and virus identification [[20], [21]], including SARS-CoV-2 immunosensors [[[22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34]]], and genosensors [[[35], [36], [37], [38]]], being up-and-coming detection tools that promise to solve diagnosis limitations in this and coming pandemics [39].
Here, we report an electrochemical immunosensor for the straightforward detection of the spike protein from SARS-CoV-2. This is the first sandwich-type assay based on an anti-spike antibody immobilized on carboxylated magnetic beads (MBs) that capture the spike-ACE2 complex. The immunocomplex is further linked to a biotinylated anti-ACE2 antibody and an enzymatic system based on streptavidin (poly)HRP for amplification and reading. All variables involved in the immunosensor development were rapidly screened by spectrophotometry. The analytical performance was finally interrogated by chronoamperometry in solutions of commercial spike protein, SARS-CoV-2 pseudovirions, SARS-CoV-2 viral particles obtained from cell culture isolation and nasopharyngeal swabs samples from infected patients. Overall, the approach rapidly and straightforwardly detected the spike protein, offering opportunities to detect SARS-CoV-2 closer to the patient in remote settings and minimizing people and samples displacement and thus virus spreading.
2. Experimental
2.1. Reagents and solutions
Dynabeads™ MyOne™ carboxylic acid (Ref. 65011) and Pierce™BCA Protein Assay Kit (Ref. 23225) were obtained from Thermo Fisher Scientific. SARS-CoV-2 (2019-nCoV) spike antibody, Rabbit PAb (Ref. 40591-T62); SARS-CoV-2 (2019-nCoV) spike S1-His Recombinant Protein (Ref. 40591-V08H) and ACE2 Protein, Human, Recombinant (Ref. 10108-H05H) were obtained from Sino Biological. Human ACE2 Biotinylated Antibody (Ref. BAF933) was obtained from R&D systems. Streptavidin-HRP conjugate (Ref. OR3L-200UG) and soluble TMB (Ref. 613544-100 ML) were obtained from Merck. N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) (Ref. E6383-5G), N-Hydroxysuccinimide (NHS) (Ref. 130672-5G) and 2-(N-morpholino) ethanesulfonic acid sodium salt (MES) were purchased from Sigma-Aldrich. Streptavidin-poly-HRP-20 (Ref. 85R-200) and streptavidin-poly-HRP-80 (Ref. 65R-S105PHRP) were obtained from Fitzgerald. SARS-CoV-2 (COVID-19) spike antibody (Ref. GTX135356) was obtained from GeneTex. SARS-CoV-2 spike antibodies (Ref. AM038105, AM002414, AM043105, AM001414 and AM009105) were obtained from Active Motif. Potassium hydrogen phosphate (K2HPO4) and disodium hydrogen phosphate (Na2HPO4) were acquired from PanReac AppliChem. Potassium dihydrogen phosphate (KH2PO4), potassium chloride (KCl) and sodium chloride (NaCl) were obtained from J.T.Baker®. Sulfuric acid (H2SO4) was purchased from Honeywell FlukaTM. RT-qPCR commercial kit (Maxima SyBR Green/Rox qPCR master mix, Thermo Scientific), HEK-293 cell line (Ref. CRL-1573) and Vero-E6 cells were kindly donated by the Immunovirology Group from the University of Antioquia. Miniprep (250 reactions) (Ref. FAPDE 300) was purchased in Favorgen and linear polyethyleneimine (PEI), MW 25000, transfection grade was obtained from Polysciences. 50 mm Tris–HCl (pH 7.4), 100 mm NaCl, 0.1 mm EDTA (TNE buffer) was prepared in-house.
20 mM MES buffer pH 6.5 was used to activate MBs and conjugate anti-spike antibodies. 0.15 M PBS buffer 1X pH 7.4 (PBS) was used to conjugate the spike-ACE2 complex and anti-ACE2 antibodies, and 0.15 M buffer PBS 1X pH 7.4 with 0.05% Tween 20 (PBST) for each of the washing steps. 0.15 M buffer PBS 1X pH 7.4 with 0.05% Tween 20 and 1% casein (PBST-C) was used to block the particles after anti-spike conjugation and conjugation of the streptavidin-(poly) HRP 80 enzyme complex. 15 M buffer PBS 1X pH 7.4 with 0.05% Tween 20 and 1% BSA (PBST-B) was used for blocking the particles after anti-spike conjugation and for conjugation of the streptavidin-HRP and streptavidin-(poly) HRP 20 enzyme complex. ChemCruz Radioimmunoprecipitation lysis buffer (RIPA, Ref. sc-24948) with 1% phenylmethylsulfonyl fluoride (PMSF), 1% sodium orthovanadate and 2% protease inhibitor cocktail were used for lysing viral particles and samples.
2.2. Apparatus and chips
The electrochemical measurements were performed with a three-electrode cell configuration screen-printed gold electrode (SPAuE, Ref. 220BT, from DropSens) in a PalmSens4 potentiostat with a PS Trace software analyzer and a Sensit Smart smartphone workstation. The chips consist of a 4 mm gold working electrode, a gold counter electrode, and a silver pseudo-reference electrode, respectively, printed on the same strip.
2.3. Production of pseudovirions
Pseudovirions were assembled by co-transfection of HEK-293 cells with three plasmids: murine leukemia virus (MLV)-Gag-Pol encoding the Gag and Pol proteins of MLV, luciferase plasmid encoding the luciferase reporter gene, the packaging signal and the 5'/3′ long repetitive sequences of MLV and SARS-CoV-2 S plasmid encoding the spike protein of SARS-CoV-2, which were donated by Gary Whittaker at Cornell University, USA. HEK-293 cells were seeded in a 6-well plate, 800000 cells per well, and incubated at 37 °C and 5% CO2 overnight. Co-transfection was achieved using 800 ng of SARS 2 S plasmid, 600 ng of MLV-Gag-Pol plasmid and 600 ng of luciferase plasmid per well in the presence of 1 g/L PEI. Then culture medium was added and the cells were incubated at 37 °C and 5% CO2 for 72 h and the supernatant of the transfected cells was collected and stored in aliquots previous centrifugation at 25000 rpm and filtration with a 0.45 μm filter [40]. Following the same protocol, pseudovirions expressing the spike protein of MERS-CoV, SARS-CoV and the glycoprotein of the vesicular stomatitis virus (VSV) were obtained as controls to evaluate the biosensor's specificity using a MERS-CoV S, SARS-CoV S and VSV plasmid, respectively.
2.4. Characterization of pseudovirions
Infectivity of pseudovirions was assessed by Vero-E6 cells transduction using 10-fold serial dilutions of the supernatant obtained from cotransfected HEK-293. Luciferase activity was determined at 72 h after transduction, and expression level was quantified using a commercial kit (Promega) after cell lysis. Luminescence signal was determined by each dilution, evaluated by triplicate, using a Varioskan. In addition, pseudovirions were quantified in copies/ml by RT-qPCR using the primers CTCACTGAGACTACATCAGC and TCCAGATCCACAACCTTCGC and a commercial kit (Maxima SyBR Green/Rox qPCR master mix, Thermo Scientific), previous RNA extraction.
In addition, Spike protein expression was demonstrated in transfected HEK-293 cells by western blot using polyclonal anti-SARS-CoV-2 Spike antibody and HRP-conjugated secondary antibody.
2.5. Assembly of the immunosensor
1 mL of carboxylated MBs was functionalized with an anti-spike antibody by transferring 40 μL of commercial MBs to a 1.5 mL vial, washing twice with 1 mL of MES, placing the vial in a magnetic rack for 2 min and removing the supernatant. Subsequently, the carboxylated MBs were activated with 15.4 mg of EDC (400 mM) and 2.2 mg of NHS (100 mM) dissolved in 200 μL of MES for 30 min at room temperature under 1000 rpm constant stirring. After removing the supernatant, the activated MBs were incubated with 200 μL of 12 μg/mL anti-spike solution in MES for 2 h at room temperature under constant stirring. The antibody-coated MBs were washed thrice with 1 mL of PBST and resuspended in 1 mL PBS. For each sensor, 50 μL of functionalized particles were transferred to a 1.5 mL vial and removed the supernatant. 50 μL of PBST-C was added as blockage agent, incubated for 1 h at 37 °C and later washed with 200 μL of PBST.
Separately, 0.1–2 μg/mL of commercial spike protein and 2 μg/mL ACE2 protein were mixed in a total volume of 50 μL of PBS and incubated at 37 °C for 45 min under stirring. On the other hand, 1 μL of inactivated viral particles at 1 × 1011 copies/mL were diluted in 99 μL RIPA, vortexed for 1.5 min and sonicated in a sonication bath for 1 min. Dilutions of viral particles were prepared from 1 × 107 to 1 × 105 copies/mL. Subsequently, 25 μL of diluted viral particles were mixed with 2 μg/mL ACE2 protein and completed the volume up to 50 μL with PBS. In either case, all the spike-containing solution volume was added to the antibody-coated MBs button and incubated for 30 min at 37 °C under constant stirring, followed by washing twice with 200 μL of PBST. The captured spike-ACE2 complex was incubated with 50 μL of 2 μg/mL anti-ACE2-biotin in PBS for 30 min at 37 °C under stirring and washed thrice with PBST. 50 μL of 50 ng/mL of streptavidin(poly) HRP in PBST-C were later added and incubated for 30 min at 37 °C, followed by five washing steps with PBST and finally, the immunocomplex was resuspended in 10 μL of PBST. The total immunosensor assembly time was approximately 90 min, once MBs have been modified with the anti-spike antibody and blocked, which stock solution is stable for 20 days at 4 °C.
2.6. Characterization of surface coverage
The amount of anti-spike antibodies at the surface of the magnetic particles was calculated indirectly, i.e., estimating the one in the supernatant. For this purpose, 25 μL of the supernatant was added to a 96-well plate, and 200 μL of BCA working reagent was added, incubated at 37 °C for 30 min and after cooling to room temperature, the absorbance was measured at 562 nm. Known concentrations of anti-spike antibodies were assessed to calculate the antibody concentration from a calibration curve.
2.7. Electrochemical measurements
For the electrochemical reading, the SPAuE was activated by cyclic voltammetry in 0.1 M sulfuric acid from 1.6 to - 0.2 V at 0.05 V/s scan rate for 11 consecutive cycles (when the peak reached about −1.5 V). A baseline was established with only TMB commercial solution containing H2O2. The immunocomplex was then confined at the working electrode, placing a magnet behind it. After removing the supernatant, 50 μL of TMB was added and the resultant current was measured by chronoamperometry at a potential of −150 mV for 65 s.
2.8. Measurement of pseudovirions and viral particles
Viral particles were isolated after culturing a nasopharyngeal swab sample from an infected patient following established biosafety and ethical standards in a biosafety level (BSL)-3 facility (donated by the Immunovirology Group from the University of Antioquia). The viral particles were inactivated by ultraviolet (UV)-light exposure for 30 min with a 6-W power lamp.
Both pseudovirions and viral particles were diluted to a concentration of 1 × 106 copies/mL in RIPA buffer, then vortexed for 1 min and 30 s and sonicated for 1 min. From this stock solution, the respective concentrations were achieved by diluting in PBS buffer before being tested. It is important to note that it is not necessary to dilute the pseudovirions or viral particles. The starting stock solution was 10.6 × 108 copies/mL and 4.3 × 107 copies/mL, respectively. Following the established protocol in which a previous incubation with the ACE2 protein is performed and maintaining the conditions of standardized volumes, the maximum concentration of 1 × 106 copies/mL was used as a starting point, followed by serial dilutions in PBS buffer to obtain the calibration curve.
2.9. Measurement of patient samples
The Tropical Diseases Study and Control Program (PECET - by its Spanish acronym) laboratory from the Universidad de Antioquia kindly donated Nasopharyngeal swab samples. For analysis by RT-PCR in a BSL-2 facility following established ethical and biosafety standards, they were inactivated by UV-light and the RT-PCR protocol briefly described below was followed.
Samples obtained by nasopharyngeal swabs were immediately deposited in a transport medium, followed by RNA extraction from 200 to 300 μL samples on the same day. The extraction was done by extraction using the kingFisher Flex robot and the MagMAXTM Viral/Pathogen II (MVP II) Nucleic Acid Isolation kit (Thermo Fisher) or using manual extraction with the Quick RNA Viral Kit (Zymo Research). The amplification reaction was done using the Berlin protocol, with some modifications. For this, the Luna® Universal Probe One-Step RT-qPCR Kit (New England BioRad) was used following the manufacturer's instructions. Primers were used to detect the E gene and primers and a probe for the Human RNAsaP gene were also used as control. Some borderline positivity samples were confirmed by amplifying the N gene described in the Berlin protocol or the GeneFinder™ COVID-19 Plus RealAmp Kit (OSANG Healthcare Co., Ltd, Korea). The primers, probes and the respective concentrations used for the Berlin protocol are in the following reference [41]. Additionally, a negative control was included in all runs in which water was added to replace the sample. The RT-PCR reactions were performed in a BioRad, CFX96 thermal cycler using 55 °C for 10 min for reverse transcription, followed by 95 °C for 3 min and then 45 cycles at 95 °C for 15 s and 58 °C for 30 s. Extracts were frozen at −80 °C until use.
For testing the samples with the immunosensor, the nasopharyngeal swabs non-inactivated were progressively thawed and lysed in RIPA buffer at 1/10 dilution by vortexing for 1 min and 30 s, followed by sonication for 1 min. All sample assays were performed in a BSL-2 laboratory following established biosafety and ethical standards.
2.10. Statistical analysis
Statistical analysis on the specificity of results and evaluation of patient samples used R studio software. Data were analyzed by the variance method (ANOVA) and compared the samples using the least significant difference (LSD) and Tukey methods with a confidence level of 95%. ∗∗∗ indicates a significance level p less than 0.001, ∗∗ values between 0.001 and 0.01, ∗ values between 0.01 and 0.05; and - values greater than 0.05.
3. Results and discussion
3.1. Optimization of the immunosensor
Spike protein is repeated 50–200 times on the viral surface [42] and consists of approximately 1273 amino acids [43], of which around 200 correspond to the RBD region (amino acid residues 331–524) located in the S1 subunit of the protein [44]. ACE2 is a protein of 805 amino acids formed by the N-terminal peptidase and the C-terminal choleric domains. The peptidase domain is the one that binds tightly to the RBD fragment of the spike protein [3,40] and this interaction occurs with a 10–20 fold higher affinity compared to SARS-CoV [ [8,45]], making ACE2 a potential biorecognition element for virus detection. Although the spike-ACE2 protein interaction occurs, forming the complex, the spike protein still has several epitopes available to be detected by a polyclonal anti-spike antibody [46]. Therefore, we took advantage of these features to assemble an immunosensor based on the spike-ACE2 protein complex in a sandwich between the corresponding antibodies with high efficiency while minimizing unspecific interactions [47]. In addition, magnetic particles pre-concentrated the spike protein directly from the samples, facilitating washing steps and decreasing the effects of potential interferents present in the complex matrix while confining the immunocomplex at the SPAuEs [48]. In this context, the immunosensor response further interrogated in a portable potentiostat offers a versatile and cost-affordable but robust alternative to detect the virus.
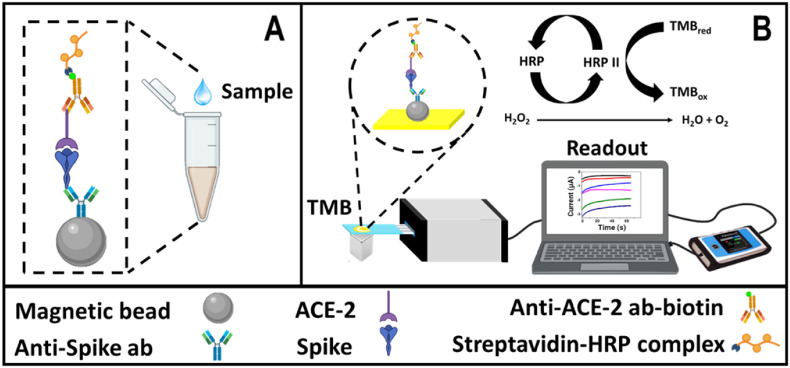
The immunosensor assembly design is described in Scheme 1 A. It consists of carboxylated magnetic particles as a support platform to bind primary amine groups from the anti-spike antibodies by covalent coupling with N-ethyl-N′-(3-(dimethylamino)propyl)carbodiimide (EDC) and N-hydroxysuccinimide (NHS). Some of the anchored antibodies eventually have the Fab region exposed and available to interact with the spike protein epitope from the spike-ACE2 complex with high affinity. This ACE2 protein later interacts with a biotinylated anti-ACE2 antibody and the streptavidin-(HRP) complex reporter. Finally, the immunocomplex is transferred to an SPAuE and, after adding TMB as a substrate of the HRP enzyme in the presence of H2O2, it generates a change in current over time detectable by chronoamperometry that correlates well with changes in the concentration of the spike protein (pseudovirions and viral particles) analyzed as depicted in Scheme 1B.
Scheme 1.
Conceptual schematic of the immunosensor design based on the Spike-ACE-2 complex. A) Shandwich immunosensor assembly on the magnetic bead-based platform and B) enzyme-amplified electrochemical reaction and chronoamperometric signal readout [49].
As preliminary steps, each parameter involved in the immunosensor development was screened rapidly and systematically by spectrophotometry. As indicated above, carboxylated magnetic beads served as a platform to covalently couple the anti-spike antibody using the carbodiimide chemistry [[48,49]], following a slightly modified protocol reported by the manufacturer but expecting a high yield surface coverage [50]. A higher signal-to-noise ratio was evident when incubating the activated particles with the anti-spike antibody in MES buffer pH 6.0 for 2 h compared to 12 h, or PBS buffer pH 7.4 for 12 h (recommended by the manufacturer) or 2 h, respectively (see supporting information (S.I.) Figs. S1–A), as expected from the instability of the acyl urea intermediate of EDC in PBS [52]. The effect of 1% casein and 1% bovine serum albumin (BSA) on the immunosensor response was evaluated based on the signal-to-noise ratio and determined casein as the optimal blocking agent (see Figs. S1–B).
The amount of MBs supporting the immunoassay was tested between 5 and 40 μg, which maximum signal was reached at 20 μg (Figs. S2–A). Subsequently, the MBs' maximum coating capacity was evaluated empirically from 6 to 48 μg/mL anti-spike antibody based on the signal response (see Figs. S2–B). Although the maximum screened signal was obtained with 24 μg/mL, it didn't represent a significant increase concerning 12 μg/mL, which is more cost-efficient; therefore, such last-mentioned concentration was selected as optimal for further steps (see Figs. S2 and B). Surface coverage was estimated by indirectly quantifying the antibody concentration in the supernatant after conjugation to the magnetic particles, using a BCA kit (see Fig. S3). 3 μg/mL was determined as the concentration immobilized at the MBs, which is 75% efficiency in the coupling reaction as compared to the one reported by the manufacturer (4–6 μg/mL) [51], indicating a proper bioconjugation process. Besides, having empirically found the MB-based supporting platform's optimal surface coverage will ensure optimal electrochemical and optical readout. Remarkably, when different references of commercially available anti-spike antibodies were evaluated, it was found that the spike S1-His Recombinant Protein (Ref. 40591-V08H) used herein interacted only with the corresponding antibody of the same commercial company (see Fig. S4). Although the antibodies were kept at the storage conditions recommended by the manufacturer, they did not have the expected stability. Yet, the mentioned protein-antibody marriage was used in the following steps.
The ACE2 protein and anti-ACE2 antibody concentrations were assessed between 0.5 and 3 μg/mL, obtaining 2 μg/mL as optimal in both cases (see Figures S2, C and D). Finally, the concentration of streptavidin-HRP, also crucial for getting the higher signal response while minimizing the noise, was interrogated between 50 and 200 ng/mL. Although the signal seems to reach a plateau at 100 ng/mL (Figs. S2 and E), 50 ng/mL produced a higher signal/noise ratio, so this was selected as optimal condition (Figs. S2 and F).
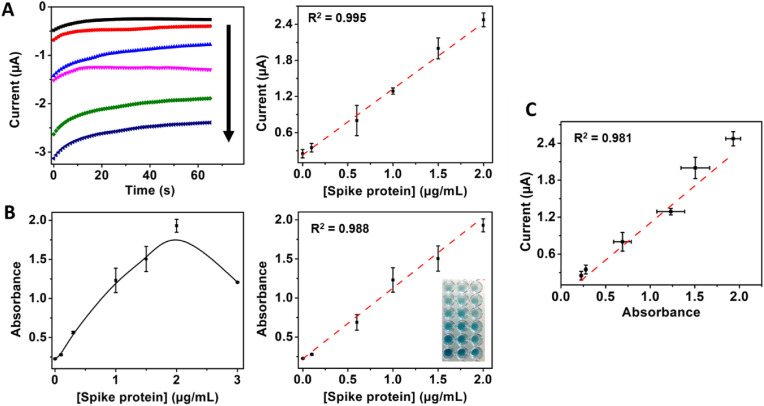
Once the optimal conditions were screened, the MB-based immunoplatform was confined at the SPAuE surface by placing a magnet behind the working electrode to check its performance with an electrochemical readout. It was achieved by chronoamperometry by applying a fixed potential of −150 mV and using a commercial TMB solution, as detailed in the experimental section. Once the performance was confirmed, the immunosensor was challenged with a buffer supplemented with concentrations of commercial spike protein ranging from 0 to 3 μg/mL to construct a calibration curve. Fig. 1 A shows the electrochemical signal response was concentration-dependent in a linear range between 0 and 2 μg/mL, with a sensitivity of 1.13 μA∗mL/μg, a LOD of 100 ng/mL and high linearity (R2 = 0.9948). The optical signal response was linear in the same range, with a sensitivity of 0.86 mL/μg, LOD of 17 ng/mL and R2 of 0.9883 (see Fig. 1B). Although the optical method is slightly more sensitive than the electrochemical one, the signals were well-correlated (R2 = 0.9814), as shown in Fig. 1C, indicating the great potential of the immunosensor not only for spectrophotometric bench laboratory testing but for electrochemical detection closer to the patient and remote settings. Despite being more time-consuming than our immunosensor, the ELISA-like assay permits more samples to be detected simultaneously, but both techniques with high reproducibility.
Fig. 1.
Immunosensor response with increasing concentrations of spike protein amplified with streptavidin-HRP. A) Electrochemical and B) spectrophotometric response with 0, 0.1, 0.6, 1, 1.5 and 2 μg/mL spike protein, C) Correlation between electrochemical and optical reading.
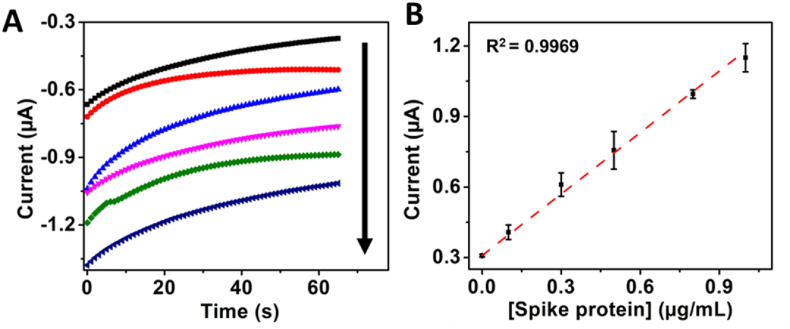
Streptavidin (poly) HRP-20 and streptavidin (poly) HRP-80 bioconjugated enzymatic complexes, with 100 and 400 HRP molecules per streptavidin molecule and the same concentration of streptavidin HRP were coupled to the immunosensor format in an attempt to increase its analytical performance. It was observed that there was a significant improvement in the signal while keeping constant the corresponding background signal as reported for this kind of complexes [53]. Therefore, based on the signal-to-noise ratio, 50 ng/mL streptavidin (poly) HRP-80 complex was defined as optimal (see Fig. S5). With the new enzymatic complex, the enhanced immunosensor format produced a signal that was spike protein concentration-dependent in a linear range between 0 and 1.0 μg/mL, with a sensitivity of 0.83 μA∗mL/μg, a LOD of 22.55 ng/mL and R2 = 0.997 (see Fig. 2 ). By replacing the standard streptavidin HRP with the streptavidin (poly) HRP-80 bioconjugated enzymatic complex, the LOD of the resulting immunosensor was lowered more than 4-fold mainly due to an increased signal without increasing the background. Such a low LOD is comparable to other SARS-CoV-2 spike protein immunosensors reported in the literature [34] and lower than other reports [[28,32]]. Furthermore, it highlights the potential of the immunosensor for the detection of the spike protein in infected patients, considering that the viral loading in samples from infected patients has been reported to be between 104 and 1011 copies/mL, with an average loading of 105 copies/mL [[[54], [55], [56]]]. Therefore, based on approximately 100 copies of spike protein per virion [55] and the 180–200 kDa molecular weight of spike protein [57], we can expect a concentration of approximately 0.3 pg/mL to 3 μg/mL spike protein in samples from patients for the reported viral loading (104 to 1011 copies/mL).
Fig. 2.
A) Immunosensor electrochemical response with increasing concentrations of 0, 0.1, 0.3, 0.5, 0.8 and 1 μg/mL of the spike protein amplified with streptavidin-HRP (poly) 80 and B) Corresponding calibration curve.
Although other biosensors with slightly lower LODs have been reported in the literature [58,59], our immunosensor has good enough analytical properties for practical applications being advantageous in terms of easiness of sample processing without the need to modify electrodes and use robust and specialized equipment. Remarkable, MBs preconcentrates the spike protein at the surface of the SPAuEs, simplifies washing steps and reduces assays time. The use of such electrodes is expected to be of similar performance concerning those from carbonaceous materials but worst than those modified with nanomaterials such as graphene and Pusian blue, plausible approaches to improve the analytical performance of the resultant biosensors [34].
3.2. Detection of pseudovirions
Developing protocols and devices to detect hazardous pathogens may represent a risk for researchers when evaluating samples with active pathogens, requiring BSL-3 settings. In contrast, pseudovirions expressing the spike protein and heterologous viral proteins reduce the risk, requiring only BSL-2 facilities [40]. Pseudovirions have been proposed to investigate virus-host interaction but herein were used as a good substitute of virions for biosensor characterization to speed up the process while protecting the safety of researchers [60]. Pseudovirions were assembled by co-transfection of three plasmids carrying the genes MLV gal-pol, luciferase and SARS-CoV-2 S into the HEK-293T cell line and characterized by luciferase expression and RT-qPCR.
Pseudoviral particles harboring the SARS-CoV-2 spike glycoprotein were then produced in cell culture following the protocol detailed in the experimental methods section. The viability of pseudovirions was demonstrated by transduction of Vero-E6 cells by luciferase expression after 72 h post-transduction. The assay confirmed the viability of the pseudovirions (see Figs. S6 and A) with high efficiency and reproducible results being higher for SARS-CoV-2 (7.76 × 108 URL/mL) than for MERS-CoV pseudovirions (5.66 × 107 URL/mL) but lower than the VSV (8.74 × 1010 URL/mL) counterparts used as controls. A western blot assay confirmed the presence of SARS-CoV-2 spike protein in the HEK-293 cells cotransfected with MLV Gag-Pol, luciferase and SARS-CoV-2 spike plasmids and control HEK-293 cells (see Figs. S6 and B). Finally, pseudovirions were quantified in copies/mL by a real-time PCR (RT-PCR) assay. Table 1 of the S.I. file shows a 3.7-fold higher concentration of pseudovirions after ultracentrifugation (25.000 rpm 90 min 4 °C TNE in buffer) than untreated pseudovirions. Furthermore, concentrations of 1.1 × 109, 5.2 × 108 and 3.3 × 108 copies/mL were obtained for SARS-CoV-2, SARS-CoV and MERS-CoV pseudovirions, respectively.
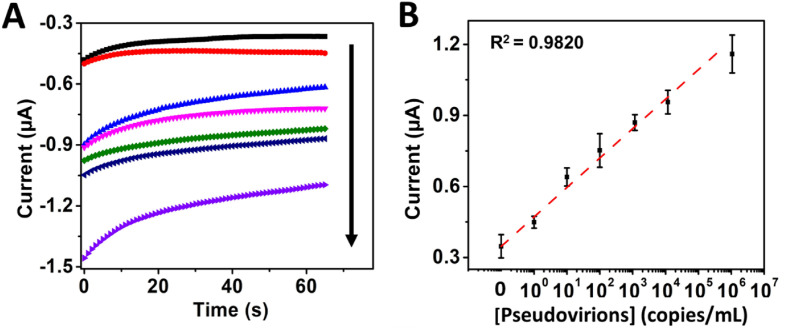
The immunosensor was then tested with 1 to 1 × 106 copies/mL of artificially assembled pseudovirions, which resultant electrochemical signal was viral particle-dependent in this range, with a sensitivity of 1.28 μA∗mL/copies, a LOD of 0.12 copies/mL, and high linearity, i.e., R2 = 0.982 (see Fig. 3 ). However, the immunosensor responded with lower sensitivity 6.0 × 10−7 μA∗mL/copies and worst LOD of 4.17 × 104 copies/mL (R2 = 0.987) in a range between 1 and 5 × 105 copies/mL of viral particles cultured from an infected patient inactivated by UV-light exposure for 30 min and lysed with RIPA buffer before the measurements (see Fig. S7). Inactivation of the viral particles was explained by Refs. [61,62]. They reported that UV-light irradiation could cause damage to genetic material and other viral components, which can potentially decrease the binding capacity of the RBD from the spike protein to the ACE2 protein, in agreement with the decreased sensitivity of our immunosensor in UV-inactivated viral particles. Therefore, inactivation (denaturalization) of viral particles only by lysis would be a better strategy for pretreatment and interrogation of clinical samples. Although such protocol breaks the viral particles membrane, it does not damage the spike protein, thus maintaining its binding capacity to the ACE2 protein.
Fig. 3.
Immunosensor electrochemical response with increasing concentrations of 0, 1, 10, 102, 103, 104, and 106 copies/mL pseudovirions amplified with streptavidin-HRP (poly) 80 and B) Corresponding calibration curve.
3.3. Specificity of the immunosensor
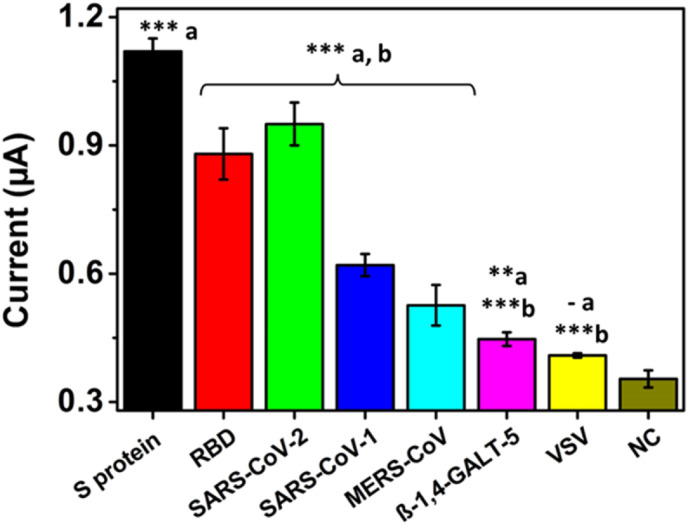
Considering that specificity is a highly desired feature in a biosensor to obtain reliable results, this feature was evaluated later. Fig. 4 shows the 1 μg/mL spike protein signal compared to the commercial RBD at the same concentration and 1 × 105 copies/mL of lysed SARS-CoV-2 pseudovirions. Although the commercial spike protein signal was 1.35-fold and 1.2-fold higher than those of the RBD region and lysed pseudovirions, they had statistically significant differences from the negative control without analyte (∗∗∗p < 0.001). Statistically significant differences were also observed when comparing the signals from the same number of pseudovirions of SARS-CoV-2, SARS-CoV and MERS-CoV (∗∗∗p < 0.001). Since SARS-CoV-2 shares 79.9% homology with SARS-CoV and 40% homology with MERS-CoV, the anti-spike antibody bound to the MBs may present cross interactions, which explain the small current signals obtained. Specificity was also evaluated with another glycoprotein (β-1.4-GALT-5) and VSV pseudovirions as controls. In both cases, there were statistically significant differences (∗∗∗p < 0.001) with commercial spike protein and SARS-CoV-2 pseudovirions and very low significance (∗∗p < 0.01 and - p > 0.05) with the negative control, respectively. These results demonstrated that the immunosensor efficiently detected SARS-CoV-2 spike protein with higher affinity than glycoproteins from other coronaviruses and VSV.
Fig. 4.
Immunosensor specificity when detecting 1 μg/mL spike, RBD and β-1,4-GALT-5 proteins and 1 × 105 copies/mL MERS, SARS-CoV-1, SARS-CoV-2 and VSV pseudovirion supernatants. Statistically significant differences (a) with respect to the negative control and (b) with respect to the commercial spike protein, ∗∗∗(p < 0.001), ∗∗(p < 0.01), ∗(p < 0.05) and – (p > 0,05).
3.4. Detection of SARS-COV-2 spike protein in clinical samples
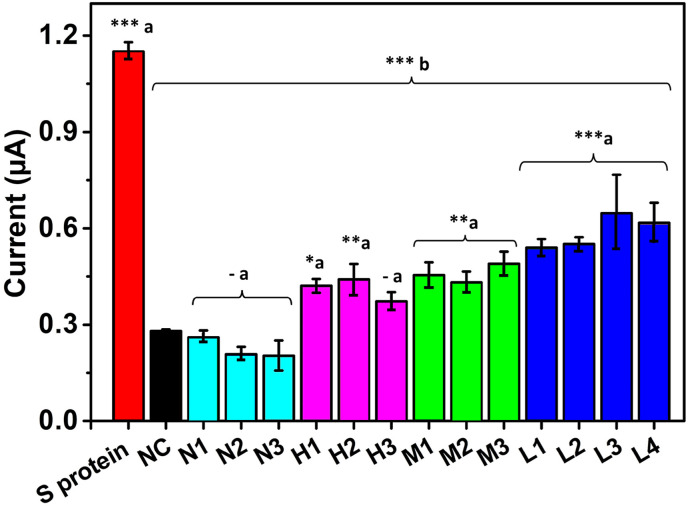
Once the specificity was proved, the immunosensor was challenged by detecting the spike protein in samples obtained from ten patients and results compared to three samples from healthy individuals, positive and negative by RT-PCR, respectively. The samples were obtained by nasopharyngeal swabbing, kindly donated by the PECET group from the University of Antioquia and classified based on the cycle threshold (C.T.) measured values (as higher the C.T., the lower the viral loading, see Table 2 in the S.I. file). The immunosensor signal of 1 μg/mL commercial spike protein was compared with three negative samples, three samples with high, three with medium and four with low C.T. values. The samples were measured at a 1/10 dilution and lysed in RIPA buffer, using only 5 μL of sample for the electrochemical measurement. The 90% of samples positive by RT-PCR had statistically significant differences (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001) concerning the negative control. Similarly, samples negative by RT-PCR had no statistically significant differences from the negative control (p > 0.05). Furthermore, the results were consistent with the C.T. reported for each sample, i.e., all the ten positive samples by RT-PCR and the three negative ones were positive and negative, respectively with our immunosensor, demonstrating the excellent detection capability of the as-developed immunosensor even in samples with high C.T. (see Fig. 5 ). An estimation of the concentration of the spike protein in each sample is in Table 2. Yet, rather than an exact amount of spike protein found, it must be interpreted as negative samples or positive samples with low, middle or high viral loading. To avoid the sample preparation and simplify the assay, samples with low C.T. were evaluated both lysed and raw. It was observed that detection of the spike protein from SARS-CoV-2 could be achieved with statistically significant differences with the negative control (∗p < 0.05) even without sample pretreatment (see Fig. S8). It is important to highlight that the number of samples is low and clinical validation with samples from a cohort of patients positive for SARS-CoV-2 and healthy individuals by the PCR gold standard is required. Yet, this proof-of-concept opens the way to sensitive detection of SARS-CoV-2 using simpler sample handling, i.e., the availability of a portable, easy-to-use device that may offer the possibility of point-of-care implementation.
Fig. 5.
Immunosensor response of different lysed patient samples compared to a commercial spike protein-positive control and a negative control without the target protein. Cyan, purple, green and blue columns correspond to negative samples and high, medium and low C.T positive samples. Statistically significant differences (a) with respect to the negative control and (b) with respect to the commercial spike protein, ∗∗∗(p < 0.001), ∗∗(p< 0.01), ∗(p < 0.05) and – (p> 0,05). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. Detection with Sensit Smart smartphone potentiostat
We compared the signal obtained with the PalmSens4 potentiostat with a Sensit Smart pocket potentiostat to detect the virus closer to the patient. Fig. S9 shows both the positive and negative controls have a similar signal in both potentiostats, without statistically significant differences (p > 0.05), evidencing the possibility of implementing the immunosensor assay in this Sensitive Smart potentiostat maintaining the high detection performance.
4. Conclusion
It was developed a proof of concept immunosensor with the ability to detect the SARS-CoV-2 spike protein by electrochemical reading. The immunosensor takes advantage of the high-affinity interaction between the spike protein and the ACE2 human host protein used herein as bioreceptor-like for the first time, forming a spike-ACE2 protein complex. The immunosensor showed outstanding analytical performance with a sensitivity of 0.83 μA∗mL/μg and a LOD of 22.55 ng/mL, using the streptavidin (poly) HRP-80 enzymatic complex as an amplification system. The immunosensor specifically detected pseudovirions synthetically assembled herein with spike protein expressed on their outermost surface with LOD of 0.12 copies/mL, but with lower sensitivity with UV-light inactivated viral particles. Finally, the high detection capacity of the immunosensor was demonstrated in raw and lysed samples from infected patients and compared with samples from healthy individuals using only 5 μL of the sample. The approach does not need RNA extraction, complex, robust equipment, or specialized personnel compared to RT-PCR, highlighting the device's potential for SARS-CoV-2 detection closer to the patient and minimizing sample volume, displacement and virus spreading.
CRediT authorship contribution statement
Viviana Vásquez: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – original draft, and graphs. Maria-Cristina Navas: Conceptualization, Methodology, discussion, pseudovirions production and editing. Javier A. Jaimes: Methodology, Writing – review & editing, plasmids production, discussion, review and editing. Jahir Orozco: Conceptualization, Methodology, Formal analysis, discussion, Resources, Writing – review & editing, Supervision, and, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge MinCiencias for funding the project Nanobiosensors to detect SARS-CoV-2 rapidly (Cod. 1115101576765). J.O acknowledges financial support from the Minciencias, the University of Antioquia and the Max Planck Society through the Cooperation agreement 566-1, 2014. The authors are thankful to the PECET and the Immunovirology Group from the University of Antioquia for donations of patient samples and viral particles and Vero cells, respectively and CECOLTEC for the Sensit Smart pocket potentiostat donation. The authors also thank the technical assistance of Maria Camila Lopez-Osorio and Melissa Montoya-Guzman of the Gastrohepatology Group from the University of Antioquia; and EPM and Ruta N for hosting the Max Planck Tandem Groups. The authors want to acknowledge Gary Whittaker at Cornell University for providing the plasmids needed for the pseudovirion system.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aca.2022.339718.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Malik Y.A. 2020. Properties of Coronavirus and SARS-CoV-2; pp. 3–11. Malaysia. [PubMed] [Google Scholar]
- 2.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. Mar. 2020;367(6485):1444. doi: 10.1126/SCIENCE.ABB2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabaan A.A., et al. SARS-CoV-2, SARS-CoV, and MERS-CoV: a comparative overview. 2020. https://www.infezmed.it/media/journal/Vol_28_2_2020_7.pdf [Online]. Available: [PubMed]
- 5.Sino Biological Human coronavirus spike. 2020. https://www.sinobiological.com/research/virus/human-coronavirus-spike Nov. 05, 2020.
- 6.Zhang L., Guo H. Biomarkers of COVID-19 and technologies to combat SARS-CoV-2. Adv. Biomark. Sci. Technol. Jan. 2020;2:1–23. doi: 10.1016/j.abst.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo Z., et al. Combating the coronavirus pandemic: early detection, medical treatment, and a concerted effort by the global community. Research. Jun. 2020;2020:1–35. doi: 10.34133/2020/6925296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J., Sun P.D. High affinity binding of SARS-CoV-2 spike protein enhances ACE2 carboxypeptidase activity. J. Biol. Chem. Dec. 2020;295(52):18579–18588. doi: 10.1074/JBC.RA120.015303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J., et al. 2020. Molecular Interaction and Inhibition of SARS-CoV-2 Binding to the ACE2 Receptor; pp. 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kevadiya B.D., et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021 205. Feb. 2021;20(5):593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orozco J., Jiménez-Jorquera C., Fernández-Sánchez C. Gold nanoparticle-modified ultramicroelectrode arrays for biosensing: a comparative assessment. Bioelectrochemistry. Jun. 2009;75(2):176–181. doi: 10.1016/J.BIOELECHEM.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Patel S., Nanda R., Sahoo S., Mohapatra E. Biosensors in health care: the milestones achieved in their development towards lab-on-chip-analysis. Biochem. Res. Int. 2016;2016 doi: 10.1155/2016/3130469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guy O.J., Walker K.A.D. Graphene functionalization for biosensor applications. Silicon Carbide Biotechnol. A Biocompatible Semicond. Adv. Biomed. Devices Appl. Jan. 2016:85–141. doi: 10.1016/B978-0-12-802993-0.00004-6. second ed. [DOI] [Google Scholar]
- 14.Franklin R.K., Martin S.M., Strong T.D., Brown R.B. Chemical and biological systems: chemical sensing systems for liquids. Ref. Modul. Mater. Sci. Mater. Eng. Jan. 2016 doi: 10.1016/B978-0-12-803581-8.00549-X. [DOI] [Google Scholar]
- 15.Vásquez G., Rey A., Rivera C., Iregui C., Orozco J. Amperometric biosensor based on a single antibody of dual function for rapid detection of Streptococcus agalactiae. Biosens. Bioelectron. Jan. 2017;87:453–458. doi: 10.1016/J.BIOS.2016.08.082. [DOI] [PubMed] [Google Scholar]
- 16.Echeverri D., Garg M., Varón Silva D., Orozco J. Phosphoglycan-sensitized platform for specific detection of anti-glycan IgG and IgM antibodies in serum. Talanta. Sep. 2020;217 doi: 10.1016/j.talanta.2020.121117. [DOI] [PubMed] [Google Scholar]
- 17.Kumar S., et al. Electrochemical paper based cancer biosensor using iron oxide nanoparticles decorated PEDOT:PSS. Anal. Chim. Acta. May 2019;1056:135–145. doi: 10.1016/J.ACA.2018.12.053. [DOI] [PubMed] [Google Scholar]
- 18.Campuzano S., Pedrero M., Pingarrón J.M. Non-invasive breast cancer diagnosis through electrochemical biosensing at different molecular levels. Sensors 2017. Aug. 2017;17(9):1993. doi: 10.3390/S17091993. Vol. 17, Page 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghorbani F., Abbaszadeh H., Mehdizadeh A., Ebrahimi-Warkiani M., Rashidi M.R., Yousefi M. Biosensors and nanobiosensors for rapid detection of autoimmune diseases: a review. Microchim. Acta. Dec. 01, 2019;186(12. Springer):1–11. doi: 10.1007/s00604-019-3844-4. [DOI] [PubMed] [Google Scholar]
- 20.Cajigas S., Alzate D., Orozco J. Gold nanoparticle/DNA-based nanobioconjugate for electrochemical detection of Zika virus. Microchim. Acta 2020 18711. Oct. 2020;187(11):1–10. doi: 10.1007/S00604-020-04568-1. [DOI] [PubMed] [Google Scholar]
- 21.Alzate D., Cajigas S., Robledo S., Muskus C., Orozco J. Genosensors for differential detection of Zika virus. Talanta. Apr. 2020;210:120648. doi: 10.1016/j.talanta.2019.120648. [DOI] [PubMed] [Google Scholar]
- 22.Seo G., et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. Apr. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 23.Muñoz J., Pumera M. 3D-Printed COVID-19 immunosensors with electronic readout. Chem. Eng. J. Dec. 2021;425:131433. doi: 10.1016/J.CEJ.2021.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahmati Z., Roushani M., Hosseini H., Choobin H. Electrochemical immunosensor with Cu2O nanocube coating for detection of SARS-CoV-2 spike protein. Mikrochim. Acta. Mar. 2021;188(3):105. doi: 10.1007/s00604-021-04762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karakuş E., Erdemir E., Demirbilek N., Liv L. Colorimetric and electrochemical detection of SARS-CoV-2 spike antigen with a gold nanoparticle-based biosensor. Anal. Chim. Acta. Oct. 2021;1182:338939. doi: 10.1016/J.ACA.2021.338939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehsan M.A., Khan S.A., Rehman A. Screen-printed graphene/carbon electrodes on paper substrates as impedance sensors for detection of coronavirus in nasopharyngeal fluid samples. Diagnostics 2021. Jun. 2021;11(6):1030. doi: 10.3390/DIAGNOSTICS11061030. Vol. 11, Page 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Idili A., Parolo C., Alvarez-Diduk R., Merkoçi A. Rapid and efficient detection of the SARS-CoV-2 spike protein using an electrochemical aptamer-based sensor. ACS Sens. Aug. 2021;6(8):3093–3101. doi: 10.1021/ACSSENSORS.1C01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vadlamani B.S., Uppal T., Verma S.C., Misra M. Functionalized tio2 nanotube-based electrochemical biosensor for rapid detection of sars-cov-2. Sensors. Oct. 02, 2020;20(20. MDPI AG):1–10. doi: 10.3390/s20205871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vezza V.J., et al. An electrochemical SARS-CoV-2 biosensor inspired by glucose test strip manufacturing processes. Chem. Commun. (Camb). Mar. 2021 doi: 10.1039/d1cc00936b. [DOI] [PubMed] [Google Scholar]
- 30.Singh N.K., et al. Hitting the diagnostic sweet spot: point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Biosens. Bioelectron. May 2021;180:113111. doi: 10.1016/j.bios.2021.113111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao W., Shurin M.R., Wheeler S.E., He X., Star A. Rapid detection of SARS-CoV-2 antigens using high-purity semiconducting single-walled carbon nanotube-based field-effect transistors. ACS Appl. Mater. Interfaces. Feb. 2021;13:10321–10327. doi: 10.1021/acsami.0c22589. [DOI] [PubMed] [Google Scholar]
- 32.Mojsoska B., Larsen S., Olsen D.A., Madsen J.S., Brandslund I., Alatraktchi F.A. Rapid SARS-CoV-2 detection using electrochemical immunosensor. Sensors. Jan. 2021;21(2):390. doi: 10.3390/s21020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yousefi H., et al. Detection of SARS-CoV-2 viral particles using direct, reagent-free electrochemical sensing. J. Am. Chem. Soc. Feb. 2021;143:1722–1727. doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
- 34.Fabiani L., et al. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. Jan. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao H., et al. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor. Actuator. B Chem. Jan. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farzin L., Sadjadi S., Sheini A., Mohagheghpour E. A nanoscale genosensor for early detection of COVID-19 by voltammetric determination of RNA-dependent RNA polymerase (RdRP) sequence of SARS-CoV-2 virus. Mikrochim. Acta. Mar. 2021;188(4):121. doi: 10.1007/s00604-021-04773-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. Dec. 2020;14(12):17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng Y., et al. An electrochemical biosensor for sensitive analysis of the SARS-CoV-2 RNA. Biosens. Bioelectron. Aug. 2021;186:113309. doi: 10.1016/J.BIOS.2021.113309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumari V., Rastogi S., Sharma V. Springer; Cham: 2019. Emerging Trends in Nanobiosensor; pp. 419–447. [Google Scholar]
- 40.Millet J.K., et al. Production of pseudotyped particles to study highly pathogenic coronaviruses in a biosafety level 2 setting. JoVE. Mar. 2019;2019(145) doi: 10.3791/59010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corman V., et al. Users looking for a workflow protocol consult the last three pages of this document. 2020. https://virologie-ccm.charite.de/en/ Sep. 25, 2021. [Online]. Available:
- 42.Szunerits S., et al. The role of the surface ligand on the performance of electrochemical SARS-CoV-2 antigen biosensors. Anal. Bioanal. Chem. 2021. Feb. 2021:1–11. doi: 10.1007/S00216-020-03137-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hussain M., et al. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J. Med. Virol. Sep. 2020;92(9):1580–1586. doi: 10.1002/JMV.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basu A., Sarkar A., Maulik U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci. Rep. Oct. 2020;10(1):1–15. doi: 10.1038/s41598-020-74715-4. 2020 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delgado J.M., Duro N., Rogers D.M., Tkatchenko A., Pandit S.A., Varma S. Molecular basis for higher affinity of SARS-CoV-2 spike RBD for human ACE2 receptor. Proteins Struct. Funct. Bioinforma. Sep. 2021;89(9):1134–1144. doi: 10.1002/PROT.26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biological S. Anti-coronavirus spike antibody, 40591-T62 | Sino biological. 2021. https://www.sinobiological.com/antibodies/cov-spike-40591-t62 Sep. 01, 2021.
- 47.Stills H.F. Jan. 2012. Polyclonal Antibody Production,” Lab. Rabbit. Guinea Pig, Hamster, Other Rodents; pp. 259–274. [DOI] [Google Scholar]
- 48.Haghighi A.H., Khorasani M.T., Faghih Z., Farjadian F. Effects of different quantities of antibody conjugated with magnetic nanoparticles on cell separation efficiency. Heliyon. Apr. 2020;6(4):e03677. doi: 10.1016/J.HELIYON.2020.E03677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.BioRender. https://biorender.com/ Nov. 12, 2021.
- 50.Thermo Fisher Scientific DynabeadsTM MyOneTM carboxylic acid. 2021. https://www.thermofisher.com/order/catalog/product/65012#/65012 Sep. 01, 2021.
- 51.Thermo Fisher Scientific Surface-activated dynabeads. 2021. https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fbrochures%2FSurface_Activated_Dynabeads.PDF Sep. 05, 2021.
- 52.Sehgal D., Vijay I.K. A method for the high efficiency of water-soluble carbodiimide-mediated amidation. Anal. Biochem. Apr. 1994;218(1):87–91. doi: 10.1006/ABIO.1994.1144. [DOI] [PubMed] [Google Scholar]
- 53.Ojeda I., Moreno-Guzmán M., González-Cortés A., Yáñez-Sedeño P., Pingarrón J.M. Electrochemical magnetoimmunosensor for the ultrasensitive determination of interleukin-6 in saliva and urine using poly-HRP streptavidin conjugates as labels for signal amplification. Anal. Bioanal. Chem. Aug. 2014;406(25):6363–6371. doi: 10.1007/S00216-014-8055-6. 2014 40625. [DOI] [PubMed] [Google Scholar]
- 54.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. The Lancet Infectious Diseases; 2020. Viral Load of SARS-CoV-2 in Clinical Samples.https://alatorax.org/es/biblioteca/viral-load-of-sars-cov-2-in-clinical-samples Sep. 03, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bar-On Y.M., Flamholz A., Phillips R., Milo R. Sars-cov-2 (Covid-19) by the numbers. Elife. Mar. 2020;9 doi: 10.7554/ELIFE.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liv L., Çoban G., Nakiboğlu N., Kocagöz T. A rapid, ultrasensitive voltammetric biosensor for determining SARS-CoV-2 spike protein in real samples. Biosens. Bioelectron. Nov. 2021;192:113497. doi: 10.1016/J.BIOS.2021.113497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang Y., Yang C., Xu X., Xu W., Liu S. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020 419. Aug. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Funari R., Chu K.Y., Shen A.Q. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens. Bioelectron. Dec. 2020;169:112578. doi: 10.1016/j.bios.2020.112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao J., et al. Magnet-assisted electrochemical immunosensor based on surface-clean Pd-Au nanosheets for sensitive detection of SARS-CoV-2 spike protein. Electrochim. Acta. Feb. 2022;404:139766. doi: 10.1016/J.ELECTACTA.2021.139766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Millet J., Whittaker G. Murine leukemia virus (MLV)-based coronavirus spike-pseudotyped particle production and infection. Bio-Protocol. 2016;6:23. doi: 10.21769/bioprotoc.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patterson E.I., et al. Methods of inactivation of SARS-CoV-2 for downstream biological assays. bioRxiv. May 2020 doi: 10.1101/2020.05.21.108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hadi J., Dunowska M., Wu S., Brightwell G. Control measures for SARS-CoV-2: a review on light-based inactivation of single-stranded RNA viruses. Pathog. 2020. Sep. 2020;9(9):737. doi: 10.3390/PATHOGENS9090737. Vol. 9, Page 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.