Summary
Background
Dyslipidaemia is highly prevalent in individuals with type 2 diabetes mellitus (T2DM). Numerous studies have sought to disentangle the causal relationship between dyslipidaemia and T2DM liability. However, conventional observational studies are vulnerable to confounding. Mendelian Randomization (MR) studies (which address this bias) on lipids and T2DM liability have focused on European ancestry individuals, with none to date having been performed in individuals of African ancestry. We therefore sought to use MR to investigate the causal effect of various lipid traits on T2DM liability in African ancestry individuals.
Methods
Using univariable and multivariable two-sample MR, we leveraged summary-level data for lipid traits and T2DM liability from the African Partnership for Chronic Disease Research (APCDR) (N = 13,612, 36.9% men) and from African ancestry individuals in the Million Veteran Program (Ncases = 23,305 and Ncontrols = 30,140, 87.2% men), respectively. Genetic instruments were thus selected from the APCDR after which they were clumped to obtain independent instruments. We used a random-effects inverse variance weighted method in our primary analysis, complementing this with additional sensitivity analyses robust to the presence of pleiotropy.
Findings
Increased genetically proxied low-density lipoprotein cholesterol (LDL-C) and total cholesterol (TC) levels were associated with increased T2DM liability in African ancestry individuals (odds ratio (OR) [95% confidence interval, P-value] per standard deviation (SD) increase in LDL-C = 1.052 [1.000 to 1.106, P = 0.046] and per SD increase in TC = 1.089 [1.014 to 1.170, P = 0.019]). Conversely, increased genetically proxied high-density lipoprotein cholesterol (HDL-C) was associated with reduced T2DM liability (OR per SD increase in HDL-C = 0.915 [0.843 to 0.993, P = 0.033]). The OR on T2DM per SD increase in genetically proxied triglyceride (TG) levels was 0.884 [0.773 to 1.011, P = 0.072] . With respect to lipid-lowering drug targets, we found that genetically proxied 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) inhibition was associated with increased T2DM liability (OR per SD decrease in genetically proxied LDL-C = 1.68 [1.03-2.72, P = 0.04]) but we did not find evidence of a relationship between genetically proxied proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibition and T2DM liability.
Interpretation
Consistent with MR findings in Europeans, HDL-C exerts a protective effect on T2DM liability and HMGCR inhibition increases T2DM liability in African ancestry individuals. However, in contrast to European ancestry individuals, LDL-C may increase T2DM liability in African ancestry individuals. This raises the possibility of ethnic differences in the metabolic effects of dyslipidaemia in T2DM.
Funding
See the Acknowledgements section for more information.
Keywords: Type 2 diabetes mellitus, Lipid traits, Mendelian Randomization, PCSK9, HMGCR
Research in context.
Evidence before this study
Type 2 Diabetes Mellitus (T2DM) is a metabolic disorder characterized by insulin resistance, hyperglycaemia, and pancreatic β-cell dysfunction. Dyslipidaemia is frequent in people with T2DM and has been linked to the pathogenesis of T2DM. Several research have investigated the link between dyslipidemia and T2DM risk, utilizing either traditional observational or more contemporary Mendelian Randomization approaches. However, there are some significant flaws in the existing corpus of research. Confounding and reverse causality are inherent risks in traditional observational research. Adiposity and a variety of other metabolic variables, for example, are likely to muddle the link between dyslipidemia and T2DM risk.
Added value of this study
In this MR study we used univariable and multivariable two-sample MR, by leveraging on summary-level data for lipid traits and T2DM liability of African ancestry individuals. We highlight the negative causal effect of increased genetically proxied LDL-C, TC and HMGCR inhibition on T2DM liability and, a protective causal effect of HDL-C on T2DM liability in individuals of African ancestry. Our finding that increased genetically proxied HMGCR inhibition is associated with increased T2DM liability in African ancestry individuals is also consistent with MR findings in European ancestry individuals and large-scale randomized data from clinical trials of statin efficacy.
Implications of all the available evidence
Our study was the first to shed light on the causal relationship between lipid traits and T2DM liability in African ancestry individuals including importance of statin therapy for primary and secondary cardiovascular disease prevention in African, adding to existing knowledge in etiology of T2DM.
Alt-text: Unlabelled box
Introduction
Type 2 Diabetes Mellitus (T2DM) is a metabolic disorder characterized by insulin resistance, hyperglycaemia, and pancreatic β-cell dysfunction.1 Dyslipidaemia, which consists of increased LDL-C, increased TG, and reduced HDL-C levels, is common in individuals with T2DM and is implicated in the pathogenesis of T2DM.2 For example, experimental evidence suggests that dyslipidaemia may impair normal pancreatic β-cell function, in part by affecting protein folding and trafficking in β-cell endoplasmic reticulum.1,2 In Africa alone, there are more than 19 million people with T2DM and numbers in sub-Saharan Africa are projected to increase by 47.5% by 2030.2 As the economies of African countries become increasingly urbanized, the clinical burdens of dyslipidaemia and T2DM will grow throughout the continent. Accordingly, there is a clinical imperative to understand the relationship between dyslipidaemia and T2DM in African ancestry individuals.
Numerous studies using either conventional observational2, 3, 4, 5, 6 or more recent Mendelian Randomization methods7, 8, 9, 10, 11, 12, 13, 14 have investigated the relationship between dyslipidaemia and T2DM liability. However, the existing body of evidence has some important limitations. Conventional observational studies are inherently vulnerable to confounding. For instance, the relationship between dyslipidaemia and T2DM liability is likely confounded by adiposity and a range of related metabolic factors.2,15 Altered lipid concentrations may also be a consequence, rather than a cause, of insulin resistance.15 By virtue of the random inheritance of genetic variants, their presence in the germline and their non-modifiable nature, MR studies are comparatively less vulnerable to confounding16, 17, 18 and thus, under specific assumptions,18 are capable of isolating and estimating the causal effect of dyslipidaemia on T2DM liability. However, MR studies addressing this question have primarily focused on European ancestry individuals and none to date have been performed in individuals of African ancestry. This is important because the frequency and distribution of genetic variants may differ across populations and so the extent to which findings in European ancestry individuals are applicable to African ancestry individuals is uncertain.
In this study, we used two-sample MR to investigate the effect of genetically proxied levels of circulating lipid traits (LDL-C, HDL-C, TC and TG) and genetically proxied levels of two lipid-lowering drug targets (HMGCR and PCSK9) on T2DM liability in African ancestry individuals.
Methods
Data sources
We obtained genetic association estimates from summary statistics of large-scale genome-wide association studies (GWAS). For the exposures, the genetic associations are based on up to 13,612 individuals (36.9% men) from the African Partnership for Chronic Disease Research (APCDR).19 Genetic association estimates for T2DM liability were obtained from 23,305 cases and 30,140 controls of African American ancestry individuals in the Million Veteran Program (MVP) (87.2% men)16 (Table 1). Participant consent and ethical approval were obtained in the original studies.
Table 1.
Demographic characteristics of the exposure and outcome study cohorts.
| Characteristic | MVP | APCDR |
|---|---|---|
| MVP cohort size, n | 53,445 (23,305 cases; 30,140 controls) | 13,612 |
| Age at enrolment, mean | 61.7 | 44.82 |
| Male gender, n (%) | 87.2 | 36.9 |
| BMI (kg/m2), mean | 30.8 | 26.3 |
| LDL-C (mmol/L) | NA | 2.54 |
| HDL-C (mmol/L) | NA | 1.63 |
| Triglycerides (mmol/L) | NA | 2.85 |
| Dyslipidemia, n | 28,689 | NA |
| T2DM Diagnosis (MVP) | ||
| Case | T2D based on ≥ 2 diagnosis codes for T2D, excluding T1D and other diabetes-inducing conditions. | |
| Control | No T2D, T1D, or diabetes-related conditions | |
*MVP: Million Veteran Program; APCDR: African Partnership for Chronic Disease Research; BMI: body mass index; LDL-C: low-density lipoprotein cholesterol; HDL-C: high-density lipoprotein cholesterol; T2D:Type 2 Diabetes, T1D:Type 1 Diabetes, NA: Non-Applicable
Genetic instruments
The genetic instruments for each exposure were selected as single-nucleotide polymorphisms (SNPs) that are associated with the corresponding exposure at p<10−6 in the APCDR dataset and that were also available in the outcome summary statistics (Supplementary1: ST1-ST4). Such p-value threshold was used to enable a sufficient number of genetic variants for MR sensitivity analyses. For the drug targets, we examined LDL-C associated variants within ±250kb of HMGCR and PCSK9. To identify instruments for HMGCR inhibition, we used a more lenient threshold of p<10−4 (Supplementary1: ST5, ST6).
We harmonized the effect alleles in the exposure and outcome datasets, and excluded palindromic variants with MAF>0.4. The remaining variants were clumped at linkage disequilibrium (LD) r2<0.001 (based on the 1000 Genomes African reference panel) within a 10 Mb window. For the drug targets, the clumping was done at r2<0.1.20 F-statistics was calculated for the individual variants used as instruments to quantify instrument strength using method described Burgess et al.21 The statistical power was evaluated by calculating the approximate minimum detectable odds ratio (OR) for each lipid trait at a power of 0.8, given the sample size of the exposure, total variance explained by the instruments and type 1 error rate of 0.05.22 To examine potential ancestral differences in the variants used as instruments, we compare the effect sizes used here to the effect sizes of these variants in European ancestry GWAS.23
Statistical analysis
In this two-sample MR analysis, the random-effects inverse-variance weighted (IVW) method implemented in the Mendelian Randomization R package24 was used as the main analysis. This method gives a consistent causal estimate if the genetic variants employed as instruments meet the instrumental variable assumptions.17 Horizontal pleiotropy, where the variants affect the outcome independently of the exposure, is a common violation of the assumptions required by the IVW method. Therefore, we used MR-Egger, weighted median and weighted mode methods as sensitivity analyses, as these methods are more robust to potential pleiotropic effects of the variants employed as instrumental variables.18
As the lipid traits are both genetically and phenotypically correlated with each other, we accounted for this by conducting multivariable MR (MVMR)25 to examine the mutually adjusted direct effects of the lipid traits. TC was omitted from the MVMR due to its collinearity with the sum of HDL-C and LDL-C. The considered instruments in MVMR were SNPs that associated with any of HDL-C, LDL-C, or TG at p<10−6 in the APCDR dataset (Supplementary1: ST7), with similar exclusions as in univariable MR described above, and clumping conducted based on the lowest SNP-wise p-value with the considered lipid traits. We calculated the conditional F-statistics for each exposure as described in Sanderson et al.26 with the covariance between the effect of genetic variants on each exposure fixed at 0.
For drug targets with MR evidence at P<0.05, we performed colocalization analysis as a sensitivity analysis. Coloc method27 applied here is a Bayesian method which, assuming a maximum of one causal variant per locus, calculates posterior probabilities (PP) for the competing models of (i) no causal variants, (ii) causal variant on the exposure, (iii) causal variant on the outcome, (iv) distinct causal variants, and (v) shared causal variant. A high PP for model (v) (PPshared) gives support to the obtained MR results, while a high PP for model (iv) (PPdistinct) would suggest that the MR results are due to confounding by LD, that is, the observed MR result is not causal, but due to LD between two distinct variants. We set the prior probabilities of any variant in the region associated with exposure, outcome, or both traits, at 10−4. The liberal prior was used as colocalization was done as a sensitivity analysis, after evidence for association in MR.
To evaluate the possibility of reverse causality, we conducted MR analysis treating T2DM liability as the exposure, and each lipid trait as an outcome in turn. The genetic instruments for T2DM liability were selected as variants associated with risk of T2DM at p<5e-8 in the MVP GWAS summary statistics, with harmonisation and clumping done as in the main analysis.
Role of funding source
Funding sources had no role in the conduct or reporting of the research.
Results
Table 2 shows the summary of the instruments for each exposure. The F-statistics for individual variants ranged between 14 and 1050, indicating a low risk of substantial weak instrument bias. The comparison of the effect sizes between African and European ancestries are given in Supplementary 2.
Table 2.
Instrumental variables for the exposures.
| NSNPs | Median F (range) | Approximate minimum detectable Odds Ratio | |
|---|---|---|---|
| Primary exposures | |||
| HDL-C | 18 | 32 (23–270) | >1.10 or <0.91 |
| LDL-C | 23 | 33 (24–1050) | >1.06 or <0.94 |
| Total C | 18 | 33 (14–461) | >1.09 or <0.92 |
| TG | 11 | 28 (22–114) | >1.14 or <0.87 |
| Secondary exposures | |||
| HMGCR inhibition | 1 | 18 | >1.90 or <0.53 |
| PCSK9 inhibition | 8 | 47 (26–86) | >1.14 or <0.87 |
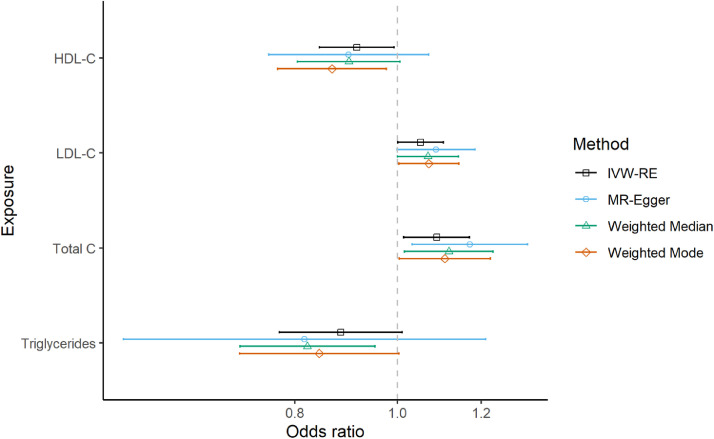
In the main analysis, we found evidence of associations between genetically predicted LDL-C and T2DM( [OR] per 1-standard deviation increase in the genetically proxied exposure 1.05, 95% CI 1.000 to 1.106, P = 0.046). Genetically predicted TC was also associated with increased risk of T2DM (OR = 1.089 (1.014 to 1.170, P = 0.019). Genetically predicted HDL-C (OR = 0.915 (0.843 to 0.993, P = 0.033)) and TG (OR = 0.884 (0.773 to 1.011, P = 0.072)) associated with lower T2DM risk (Figure 1). The results were robust in the sensitivity analyses employing different MR methods more robust to horizontal pleiotropy (Figure 1, Supplementary3:ST1).
Figure 1.
Forest plot of the odds ratios and their 95% confidence intervals between lipid traits and genetically predicted T2DM risk using different MR methods. IVW-RE = inverse-variance weighted random-effects. HDL-C: high-density lipoprotein cholesterol; LDL-C: low-density lipoprotein cholesterol; Total C: total cholesterol.
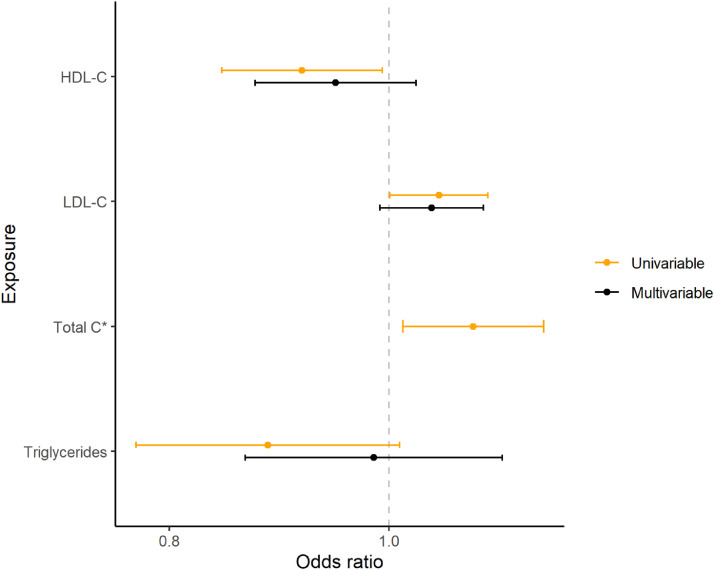
MVMR analyses showed similar estimates for HDL-C and LDL-C as in the univariable MR, while the association between genetically predicted TG and T2DM risk was notably attenuated towards the null (Figure 2, Supplementary3:ST2).
Figure 2.
Forest plot showing the odds ratios and their 95% confidence intervals of univariable and multivariable MR of lipid traits vs T2DM. *Total C omitted from multivariable MR due to collinearity with the sum of HDL-C and LDL-C. HDL-C: high-density lipoprotein cholesterol; LDL-C: low-density lipoprotein cholesterol; Total C: total cholesterol.
In the drug target analyses, we found evidence for genetically predicted HMGCR inhibition being associated with increased T2DM risk (OR per 1-SD decrease in genetically proxied reduction in LDL-C via HMGCR inhibition 1.68, 95% CI =1.03-2.72, P=0.04; Supplementary3:ST3). Colocalization analysis showed some support for shared causal variant (PPshared=0.18; PPshared/(PPshared+PPdistinct) = 0.81), and deemed confounding by LD unlikely (PPdistinct=0.04). There was no evidence for association between genetically predicted PCSK9 inhibition and T2DM risk (OR [95% CI] 0.94 [0.82-1.08], P=0.41).
The reverse MR analyses did not show evidence of genetic liability to T2DM being associated with lipid levels (Supplementary4).
Discussion
Principal findings in context
This study provides genetic evidence in support of a deleterious causal effect of increased genetically proxied LDL-C, TC and HMGCR inhibition on T2DM liability and, conversely, a protective causal effect of HDL-C on T2DM liability in individuals of African ancestry. Although the effect of both LDL-C and HDL-C on T2DM liability were somewhat attenuated after mutual adjustment in our MVMR model, MR estimates were similar in both univariable and multivariable MR. In contrast, the effect of genetically proxied TG on T2DM was inconsistent across univariable and multivariable MR. We did not find evidence to support a causal effect of genetically proxied PCSK9 levels on T2DM liability.
Our univariable MR result of increased genetically proxied HDL-C levels being associated with a decreased risk of T2DM in African ancestry individuals is consistent with previous MR studies in European ancestry individuals.7,9,12,14 Although the effect was slightly attenuated in MVMR such that the 95% CI included the null, the magnitude of the MR estimates was similar in direction and magnitude across both univariable and multivariable MR (see Figure 2). This attenuation therefore likely reflects insufficient statistical power as opposed to measured horizontal pleiotropy being accounted for and identified during MVMR, though repeating the MVMR analysis once more summary-level data are available for African ancestry individuals will be necessary to confirm this. Indeed, the same phenomenon of directionally consistent albeit attenuated MR estimates for HDL-C has been observed in MVMR analyses in European ancestry individuals.9 In addition to MR, genetic support for a causal effect of HDL-C on T2DM liability comes from experimental studies of monogenic disease. The mutations in the ABCA1 gene that characterize Tangier disease cause both low levels of plasma HDL-C and abnormal insulin secretion in humans.28,29 Thus, our study adds weight to the increasing genetic evidence to support the importance of HDL-C as a therapeutic target in treating the diabetogenic side effects of statins and, potentially, in the treatment of T2DM itself.9,13
In contrast, the effect of TG on T2DM liability was significantly attenuated in our MVMR model after adjusting for LDL-C and HDL-C, suggesting that TG does not exert a direct causal effect on T2DM liability independently of LDL-C and HDL-C. Consistent with our study, the same attenuation has been observed in MVMR studies in European ancestry individuals11 and numerous previous MR studies did not detect a causal effect of genetically proxied TG on T2DM liability.7,9,14 It should, however, be noted that targeting TG may yet be of therapeutic value in Type 2 Diabetics with dyslipidaemia in light of data suggesting that hypertriglyceridaemia may contribute to residual cardiovascular risk in statin-optimized patients with T2DM.30
Comparatively, similar associations have not been reported in other non-European populations. In the study by Sobrin et al.,8 there was no evidence of genetically predicted plasma lipid levels being associated with risk of diabetic retinopathy in Chinese ancestry populations. In a bidirectional MR study exploring the causal relationships between lipid and glycemic levels in Indian population by Agarwal et al.,31 the authors report evidence of only genetically predicted TG levels being associated with glycemic traits.
Our finding that increased genetically proxied HMGCR inhibition is associated with increased T2DM liability in African ancestry individuals is also consistent with MR findings in European ancestry individuals9 and large-scale randomized data from clinical trials of statin efficacy.32 Clinically, this implies that it is equally important to counsel African ancestry individuals on the small increase in the risk of T2DM upon commencing statin therapy for primary and secondary cardiovascular disease prevention. Interestingly, mediation analysis has shown that obesity is a mediator of the diabetogenic effect of LDL lowering.33 We could not include obesity in our MVMR model due to insufficient GWAS data on obesity in individuals of African ancestry, but it will be interesting to explore in future studies whether this finding extends to African ancestry individuals, in whom differences in the cardiometabolic effects of adiposity are well-documented.
Strikingly, in contrast to findings in European ancestry individuals, we found that increased genetically proxied LDL-C levels were associated with an increased risk of T2DM in African ancestry individuals9,13,32 an association that was attenuated, albeit similar in magnitude, after adjusting for HDL-C and TG in our MVMR model. In other words, our study suggests that in African ancestry individuals, elevated LDL-C levels increase T2DM liability and yet lowering LDL-C levels via the HMGCR pathway also increase T2DM liability. Taken together, this raises the possibility of diabetogenic, LDL-C-increasing pathways, acting independently of HMGCR, that exist in African ancestry individuals but not in European ancestry individuals. In this regard, it is clinically and biologically plausible that the effect of LDL-C on T2DM liability differs across ancestries. Longitudinal data from the Isfahan cohort study support a positive association between LDL-C and T2DM liability in a non-European population.27 African ancestry individuals have been shown to exhibit insulin resistance in the absence of the typical profile of dyslipidaemia and central adiposity observed in European ancestry individuals and indeed, lipid metrics adept at detecting T2DM liability in European ancestry individuals often fail to do so African ancestry individuals.34, 35, 36 Moreover, despite having a more favourable atherogenic profile, consisting of lower small LDL-C, higher HDL-C and lower TG levels, African Americans have a lower angiographic burden of coronary artery disease37 and a 2-4-fold greater risk of cardiovascular disease outcomes in comparison to white Americans.37,38 Hence, if the atherogenicity of certain lipid subfractions and their corresponding propensity to cause downstream cardiometabolic sequalae differs between African ancestry and European ancestry individuals, so too may the diabetogenicity of LDL-C. If, for example, individuals carrying more LDL-C-increasing alleles are on average more likely to be prescribed statins, which are themselves diabetogenic, then this might induce a spurious association between increased genetically proxied LDL-C levels and T2DM liability.39 However, it is very unlikely that all people with high LDL-C in the African study were treated with statins, these differences could equally be present in MR studies in European ancestry individuals where the opposite effect is seen. We therefore do not believe that such confounding explains this finding, although future GWAS studies incorporating longitudinal data may provide further clarity. In summary, observed differences in the propensity of LDL-C to cause T2DM in African ancestry individuals vs European ancestry individuals may reflect the dissimilarity in the underlying biology of diabetic dyslipidaemia across ancestries but more studies are necessary to explore this further. Another possible explanation for this observed differences across ancestries may be that the pathophysiology of T2DM vary across populations and different socio-economic settings.40 However, it is worthy to note that even if the disease pathophysiology is the same, changing levels and manifestations of risk variables in both populations can also influence these observed differences.
Our findings in examining the link between T2DM and lipid levels do not indicate a causal effect of increased T2DM on lipid levels as reported in observational studies,41,42 which could be attributable to potential confounders in observational studies. Our findings are consistent with those of Vatner et al., where they demonstrated that TG synthesis and levels are unaffected by insulin resistance, insulin action, or insulin levels.43
Strengths
Our study has number of strengths. Under certain assumptions,18 MR is more resistant to confounding and is therefore better able to distinguish causation from correlation in comparison to traditional observational studies.44,45 SNPs that were employed as instruments were all from genetic regions encoding LDL-C, HDL-C, TG, HMGCR and PCSK9 or from proximate genetic loci; thus, a strong biological link between our genetic instruments and exposures of interest strengthens the validity of these variants with respect to the assumption of relevance in the IV framework. Given the propensity of certain genetic variants associated with one lipid fraction to also associate with another, the use of multivariable MR is particularly suited to disentangling the direct causal effect of different lipid fractions on T2DM liability. MVMR enables us to incorporate a degree of measured horizontal pleiotropy into our analysis, which, in comparison to univariable analyses, reduces bias owing to violation of the exclusion-restriction assumption.25 We also interrogated the robustness of our IVW estimates to the presence of pleiotropic variants using a range of sensitivity analyses and our MR estimates were consistent across these different robust methods. Whilst previous MR studies on this question have focused on European ancestry individuals, ours is the first to focus on African ancestry individuals. Given that African ancestry individuals are underrepresented in cardiovascular disease randomized clinical trials and observational studies, there is a risk of inappropriately generalizing findings in European ancestry to African ancestry individuals.46,47 To this end, our study provides valuable genetic evidence that pertains directly to the effect of lipid traits and lipid-lowering therapies on T2DM liability in African ancestry individuals.
Limitations
Our study has limitations. We used a more lenient significance threshold of p<10−6 to select SNPs in our genetic instruments, which may lead to inclusion of weak or invalid instruments. Despite this, the F-statistics of the genetic instrument for each outcome were all > 10. Statistical power may have been insufficient to detect a causal effect for genetically proxied TG and PCSK9 levels on T2DM liability in our univariable analysis and for LDL-C and HDL-C in our multivariable model. Genetic association estimates used in this study were derived from meta-analyzing studies with demographically heterogenous populations and so despite adjusting for age, sex and population stratification, such heterogeneity may introduce bias into MR estimates.47 Our MR estimates should not be extrapolated to estimate the effect of changing circulating lipid levels in subgroups of African ancestry individuals with particularly high or low circulating lipid levels.45 Despite finding no evidence of significant pleiotropy in our sensitivity analyses, we cannot definitively exclude the possibility of horizontal pleiotropy whereby variants in our genetic instrument influence T2DM liability via pathways independent of the lipid traits investigated here.44 We also did not have information available on statin use among the participants in MVP, and its potential impact on our results is unclear. In addition, the peculiarities of the MVP, including the use of a single genotyping array for the diverse ethnicities48 may have influenced the genetic association estimates.
The APCDR cohort had only about one-third of men, whereas the MVP had 87.2 percent men, implying that the effect of lipids on T2DM liability was primarily assessed by comparing lipid levels in females to T2DM liability in males. As a result, the results presented in this study should be interpreted in the context of T2DM liability in men.
Conclusion
Overall, we found MR evidence of a protective effect of HDL-C and an adverse effect of HMGCR inhibition on the risk of T2DM. Our finding of higher genetically proxied LDL-C levels increasing T2DM liability in African ancestry individuals raises the possibility of ancestral differences in the metabolic effects of dyslipidaemia in T2DM.
Data sharing statement
All scripts for the analysis are available from the authors upon request.
Contributors
SF, DG and SB conceptualised the study. OS, VK, YH and SF performed the analyses. OS, VK, SF, and DG verified the underlying data. OS, VK, SB, SF and DG wrote the first draft of the manuscript. All authors read and approved the final version of the manuscript.
Declaration of interests
DG is employed part-time by Novo Nordisk and has received consultancy fees from Policy Wisdom.
No potential conflicts of interest relevant to this article were reported by all other authors.
Acknowledgement
The authors thank Million Veteran Program (MVP) staff, researchers, and volunteers, who have contributed to MVP, and especially participants who previously served their country in the military and now generously agreed to enroll in the study. (See https://www.research.va.gov/mvp/ for more details). The citation for MVP is Gaziano, J.M. et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70, 214-23 (2016). This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by the Veterans Administration (VA) Cooperative Studies Program (CSP) award #G002
SF is an international Intermediate Fellow funded by the Wellcome Trust grant (220740/Z/20/Z) at the MRC/UVRI and LSHTM. TC is an international training fellow supported by the Wellcome Trust grant (214205/Z/18/Z). DG was supported by the British Heart Foundation Centre of Research Excellence (RE/18/4/34215) at Imperial College, and a National Institute for Health Research Clinical Lectureship (CL-2020-16-001) at St. George's, University of London. VK is supported by the Academy of Finland Project 312123, and European Union's Horizon 2020 research and innovation programme under Grant Agreement No 848158 (EarlyCause). M. Nakabuye acknowledges the support of Makerere University Non-Communicable Diseases (MacNCD). This research is made possible by the MakNCD Research Training Program: National Institutes of Health (NIH) 1D43TW011401-01 through the Fogarty International Center (FIC). C Soremekun, B. Udosen, and S Fatumo acknowledge H3Africa Bioinformatics Network (H3ABioNet) Node, National Biotechnology Development Agency (NABDA), and the Center for Genomics Research and Innovation (CGRI) Abuja, Nigeria. Dr Mason is funded by the EC-Innovative Medicines Initiative (BigData@Heart). Dr Burgess is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (204623/Z/16/Z). This research was funded by UK Research and Innovation (UKRI) Medical Research Council (MC_UU_00002/7) and supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre (BRC-1215-20014). The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care. The funding sources did not have any role in designing the study, performing analysis, or communicating findings.
Dr. Segun Fatumo is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103953.
Appendix. Supplementary materials
References
- 1.Ashcroft F.M., Rorsman P. Diabetes mellitus and the β cell: the last ten years. Cell. 2012;148:1160–1171. doi: 10.1016/j.cell.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mooradian A.D. Dyslipidemia in type 2 diabetes mellitus. Nat Clin Pract Endocrinol Metab. 2009;5:150–159. doi: 10.1038/ncpendmet1066. [DOI] [PubMed] [Google Scholar]
- 3.Gaillard T., Osei K. Ethnic differences in serum lipids and lipoproteins in overweight/obese African-American and white American women with pre-diabetes: significance of NMR-derived lipoprotein particle concentrations and sizes. BMJ Open Diabetes Res Care. 2016;4 doi: 10.1136/bmjdrc-2016-000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haffner S.M., D'Agostino R., Goff D., et al. LDL size in African Americans, Hispanics, and non-Hispanic whites: the insulin resistance atherosclerosis study. Arterioscler Thromb Vasc Biol. 1999;19:2234–2240. doi: 10.1161/01.atv.19.9.2234. [DOI] [PubMed] [Google Scholar]
- 5.Gaillard T., Schuster D., Osei K. Differential impact of serum glucose, triglycerides, and high-density lipoprotein cholesterol on cardiovascular risk factor burden in nondiabetic, obese African American women: implications for the prevalence of metabolic syndrome. Metabolism. 2010;59:1115–1123. doi: 10.1016/j.metabol.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 6.Burns S.F., Lee S., Arslanian S.A. In vivo insulin sensitivity and lipoprotein particle size and concentration in black and white children. Diabetes Care. 2009;32:2087. doi: 10.2337/dc09-0380. LP –2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fall T., Xie W., Poon W., et al. Using genetic variants to assess the relationship between circulating lipids and type 2 diabetes. Diabetes. 2015;64:2676–2684. doi: 10.2337/db14-1710. [DOI] [PubMed] [Google Scholar]
- 8.Sobrin L., Chong Y.H., Fan Q., et al. Genetically determined plasma lipid levels and risk of diabetic retinopathy: a Mendelian Randomization study. Diabetes. 2017;66:3130–3141. doi: 10.2337/db17-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White J., Swerdlow D.I., Preiss D., et al. Association of lipid fractions with risks for coronary artery disease and diabetes. JAMA Cardiol. 2016;1:692–699. doi: 10.1001/jamacardio.2016.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt A.F., Swerdlow D.I., Holmes M.V., et al. PCSK9 genetic variants and risk of type 2 diabetes: a mendelian randomisation study. Lancet Diabetes Endocrinol. 2017;5:97–105. doi: 10.1016/S2213-8587(16)30396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan W., Sun W., Yang S., et al. LDL-C plays a causal role on T2DM: a Mendelian randomization analysis. Aging (Albany NY) 2020;12:2584–2594. doi: 10.18632/aging.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan S., Larsson S.C. An atlas on risk factors for type 2 diabetes: a wide-angled Mendelian Randomisation study. Diabetologia. 2020;63:2359–2371. doi: 10.1007/s00125-020-05253-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bu S.Y. Genetically mediated lipid metabolism and risk of insulin resistance: insights from Mendelian Randomization studies. J Lipid Atheroscler. 2019;8:132–143. doi: 10.12997/jla.2019.8.2.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi Q., Liang L., Doria A., et al. Genetic predisposition to dyslipidemia and type 2 diabetes risk in two prospective cohorts. Diabetes. 2012;61:745–752. doi: 10.2337/db11-1254. 2012/02/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Athyros V.G., Doumas M., Imprialos K.P., et al. Diabetes and lipid metabolism. Hormones. 2018;17:61–67. doi: 10.1007/s42000-018-0014-8. [DOI] [PubMed] [Google Scholar]
- 16.Vujkovic M., Keaton J.M., Lynch J.A., et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet. 2020;52:680–691. doi: 10.1038/s41588-020-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davey Smith G., Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23:R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slob E.A.W., Burgess S. A comparison of robust Mendelian randomization methods using summary data. Genet Epidemiol. 2020;44:313–329. doi: 10.1002/gepi.22295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurdasani D., Carstensen T., Fatumo S., et al. Uganda genome resource enables insights into population history and genomic discovery in Africa. Cell. 2019;179:984. doi: 10.1016/j.cell.2019.10.004. -1002.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gill D., Georgakis M.K., Walker V.M., et al. Mendelian randomization for studying the effects of perturbing drug targets. Wellcome Open Res. 2021;6:16. doi: 10.12688/wellcomeopenres.16544.2. PMID: 33644404; PMCID: PMC7903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgess S., Davies N.M., Thompson S.G. Bias due to participant overlap in two-sample Mendelian Randomization. Genet Epidemiol. 2016;40:597–608. doi: 10.1002/gepi.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol. 2014;43:922–929. doi: 10.1093/ije/dyu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham S.E., Clarke S.L., Wu K.H.H., et al. The power of genetic diversity in genome-wide association studies of lipids. Nature. 2021;600:675–679. doi: 10.1038/s41586-021-04064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yavorska O.O., Burgess S. Mendelian Randomization: an R package for performing Mendelian Randomization analyses using summarized data. Int J Epidemiol. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess S., Thompson S.G. Multivariable Mendelian Randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251–260. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanderson E., Davey Smith G., Windmeijer F., et al. An examination of multivariable Mendelian Randomization in the single-sample and two-sample summary data settings. Int J Epidemiol. 2019;48:713–727. doi: 10.1093/ije/dyy262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giambartolomei C., Vukcevic D., Schadt E.E., et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLOS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rickels M.R., Goeser E.S., Fuller C., et al. Loss-of-function mutations in ABCA1 and enhanced β-cell secretory capacity in young adults. Diabetes. 2015;64:193–199. doi: 10.2337/db14-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vergeer M., Brunham L.R., Koetsveld J., et al. Carriers of loss-of-function mutations in ABCA1 display pancreatic beta-cell dysfunction. Diabetes Care. 2010;33:869–874. doi: 10.2337/dc09-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexopoulos A.S., Qamar A., Hutchins K., et al. Triglycerides: emerging targets in diabetes care? Review of moderate hypertriglyceridemia in diabetes. Curr Diabetes Rep. 2019;19:13. doi: 10.1007/s11892-019-1136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal T., Lyngdoh T., Dudbridge F., et al. Causal relationships between lipid and glycemic levels in an Indian population: a bidirectional Mendelian randomization approach. PLoS One. 2020;15:1–14. doi: 10.1371/journal.pone.0228269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swerdlow D.I., Preiss D., Kuchenbaecker K.B., et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet. 2015;385:351–361. doi: 10.1016/S0140-6736(14)61183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu P., Moon J.Y., Daghlas I., et al. Obesity partially mediates the diabetogenic effect of lowering LDL cholesterol. Diabetes Care. 2021 doi: 10.2337/dc21-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goff L.M., Ladwa M., Hakim O., et al. Ethnic distinctions in the pathophysiology of type 2 diabetes: a focus on black African-Caribbean populations. Proc Nutr Soc. 2020;79:184–193. doi: 10.1017/S0029665119001034. [DOI] [PubMed] [Google Scholar]
- 35.Zoratti R., Godsland I.F., Chaturvedi N., et al. Relation of plasma lipids to insulin resistance, nonesterified fatty acid levels, and body fat in men from three ethnic groups: relevance to variation in risk of diabetes and coronary disease. Metabolism. 2000;49:245–252. doi: 10.1016/s0026-0495(00)91507-5. [DOI] [PubMed] [Google Scholar]
- 36.Bell R.A., Chen H., Saldana S., et al. Comparison of measures of adiposity and cardiovascular disease risk factors among African American adults: the Jackson heart study. J Racial Ethn Health Disparities. 2018;5:1230–1237. doi: 10.1007/s40615-018-0469-y. 2018/02/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark L.T., Ferdinand K.C., Flack J.M., et al. Coronary heart disease in African Americans. Heart Dis. 2001;3:97–108. doi: 10.1097/00132580-200103000-00007. [DOI] [PubMed] [Google Scholar]
- 38.Gillum R.F., Mussolino M.E., Madans J.H. Diabetes mellitus, coronary heart disease incidence, and death from all causes in African American and European American women: the NHANES I epidemiologic follow-up study. J Clin Epidemiol. 2000;53:511–518. doi: 10.1016/s0895-4356(99)00208-5. [DOI] [PubMed] [Google Scholar]
- 39.Fall T., Xie W., Poon W., et al. Using genetic variants to assess the relationship between circulating lipids and type 2 diabetes. Diabetes. 2015;64:2676–2684. doi: 10.2337/db14-1710. [DOI] [PubMed] [Google Scholar]
- 40.PrayGod G., Filteau S., Range N., et al. β-cell dysfunction and insulin resistance in relation to pre-diabetes and diabetes among adults in north-western Tanzania: a cross-sectional study. Trop Med Int Health. 2021;26:435–443. doi: 10.1111/tmi.13545. [DOI] [PubMed] [Google Scholar]
- 41.Parhofer K.G. Interaction between glucose and lipid metabolism: More than diabetic dyslipidemia. Diabetes Metab J. 2015;39:353–362. doi: 10.4093/dmj.2015.39.5.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu C.P., Vergès B. IntechOpen; Rijeka: 2011. Lipid Disorders in Type 1 Diabetes. p. Ch. 3. [Google Scholar]
- 43.Vatner D.F., Majumdar S.K., Kumashiro N., et al. Insulin-independent regulation of hepatic triglyceride synthesis by fatty acids. Proc Natl Acad Sci USA. 2015;112:1143–1148. doi: 10.1073/pnas.1423952112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith G.D., Ebrahim S. Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol Engl. 2003:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 45.Davies N.M., Holmes M.V., Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prasanna A., Miller H.N., Wu Y., et al. Recruitment of black adults into cardiovascular disease trials. J Am Heart Assoc. 2021;10 doi: 10.1161/JAHA.121.021108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fatumo S., Karhunen V., Chikowore T., et al. Metabolic traits and stroke risk in individuals of African ancestry: Mendelian Randomization analysis. Stroke. 2021;52:2680–2684. doi: 10.1161/STROKEAHA.121.034747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hunter-Zinck H., Shi Y., Li M., et al. Genotyping array design and data quality control in the million veteran program. Am J Hum Genet. 2020;106:535–548. doi: 10.1016/j.ajhg.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.