1. Introduction
Britz et al. [1] objected to our conclusion that ricefishes of the family Adrianichthyidae dispersed eastward ‘out-of-India’ after the collision of the Indian subcontinent with Eurasia and subsequently diversified in Southeast-East Asia [2], based mainly on the following three points: (i) artefacts involved in ancestral area reconstruction by BioGeoBEARS in RASP [3], (ii) uncertainty of the phylogenetic position of Oryzias setnai because of long-branch attraction (LBA) and (iii) inadequate calibration using the fossil species †Lithopoecilus brouweri. Our replies to these three points are as follows.
2. Artefact in ancestral area reconstruction
First, we were aware that short trees lead to inaccurate maximum-likelihood estimates in BioGeoBEARS (http://phylo.wikidot.com/biogeobears-mistakes-to-avoid#toc2). This was likely the case for the tree used in Yamahira et al. [2] because the estimated parameters and likelihoods were completely identical between DEC and DEC + J models without scaling up tree branches (see electronic supplementary material, table S1a and b). Indeed, likelihoods also did not differ between the two models in most of the analyses in [1]. However, we considered that this may be a bug in RASP (v. 4.2); re-analysis with the original BioGeoBEARS (v. 1.1.2) [4–6] generated reasonable parameter estimates and likelihoods without scaling up branch lengths. This re-analysis (electronic supplementary material, table S1c) supported the DEC + J model (unlike in [1]) but estimated that the most recent common ancestor (MRCA) of Adrianichthyidae was distributed not only in the Indian subcontinent but also in Southeast Asia. We apologize and correct our previous result in [2].
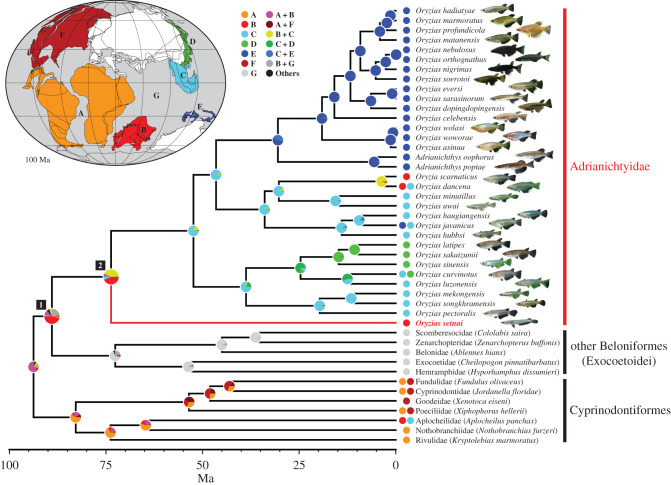
Our interest then shifted to how this intercontinental distribution across the Tethys Sea was shaped. Because the cladogenesis of Cyprinodontiformes—the outgroup of Beloniformes to which Adrianichthyidae belongs—largely reflects the breakup of the supercontinent Gondwana in deep Mesozoic times [7–11], we conducted an additional BioGeoBEARS analysis, expanding the scope to include Cyprinodontiformes (see electronic supplementary material, supplementary material and methods and table S2 for details). The result revealed that an intercontinental distribution of the MRCA of Adrianichthyidae between India and Southeast Asia was not best supported; the most probable distribution area was on the Indian subcontinent only (node 2 in figure 1), and the MRCA of Beloniformes was also distributed only on the Indian subcontinent (node 1 in figure 1). These results strongly support the ‘out-of-India’ dispersal scenario.
Figure 1.
Ancestral areas at each node of the phylogenetic tree of Beloniformes and Cyprinodontiformes reconstructed under the DEC + J model by BioGeoBEARS.
Britz et al. [1] concluded that the fragmentation of a widely distributed coastal ancestral species better explains the historical biogeography of Adrianichthyidae. However, this ‘vicariance’ scenario is a premature conclusion derived from lack of consideration of the ancestral species' origin.
3. Phylogenetic position of Oryzias setnai
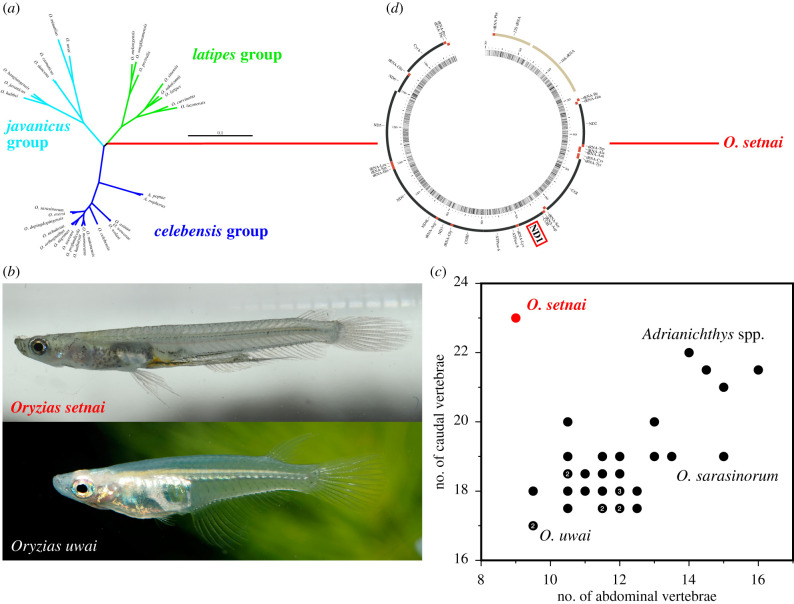
Second, we tested for the presence of LBA for O. setnai by estimating the phylogenetic position of this species after removing all outgroups, as proposed by [12]. We found that the position of O. setnai did not change (i.e. the branch leading to O. setnai split off from the internal branch that separates the latipes group from others; figure 2a), indicating that the effect of LBA, if any, is not substantial. It is also clear that fig. 1 in [1] did not successfully resolve deep divergences among families and orders within Atherinomorpha; for example, the flyingfish Cheilopogon pinnatibarbatus (Beloniformes) and the silverside Menidia menidia (Atheriniformes) were close to each other, implying that the finding of O. setnai nested among other adrianichthyids is unreliable.
Figure 2.
(a) Maximum-likelihood phylogeny of 33 adrianichthyid species based on mitochondrial (11 233 bp) and nuclear (4204 bp) sequences. We followed [1] except that all non-adrianichthyid outgroup species were excluded, and no outgroup was set. (b) Males of Oryzias setnai (photo by V. K. Anoop) and O. uwai (photo by N. Hashimoto). (c) Relationship between the number of abdominal and caudal vertebrae among 34 adrianichthyid species (see electronic supplementary material, table S3 for source references of raw data). Numbers within circles represent the number of species having the same combination of abdominal and caudal vertebral numbers. (d) Draft structure of the O. setnai mitochondrial genome. Note transposition of the ND1 gene downstream of the COI gene.
We were aware that Parenti [13] estimated from morphological comparisons that O. setnai is sister to O. uwai, so we respectfully correct our assertion to ‘no molecular study has investigated its phylogenetic position’. We agree with [13] in terms of the view that O. setnai is a member of adrianichthyids, for which monophyly is supported by 17 synapomorphic characters, such as the lack of vomer and rostral cartilage. However, we disagree that O. setnai is phylogenetically located within other adrianichthyids [1,13], because it is highly autapomorphic. It is the only adrianichthyid species having internal fertilization, with male anterior anal-fin rays modified into an intromittent organ (figure 2b), and a bilaterally asymmetric female body [13]. Moreover, the number of abdominal and caudal vertebrae of O. setnai is disproportionally uncommon compared with other adrianichthyids (figure 2c). We also reported in [2] that mitochondrial genome gene order differs from that typical of vertebrates including other adrianichthyids (figure 2d; see also electronic supplementary material, figure S1 in [2]). These highly autapomorphic traits of O. setnai are consistent with our phylogenetic estimation that this species is sister to all other adrianichthyids.
4. Usage of †Lithopoecilus brouweri as fossil calibration
Third, we included †L. brouweri in our fossil calibration [2] because Parenti [13] had classified this species within Adrianichthyidae, but we realized that this classification was only tentative. We therefore re-estimated the divergence time of O. setnai, excluding †L. brouweri from the calibration. Excluding this fossil species did not greatly affect the divergence time estimation for O. setnai; it was estimated to have diverged about 71 million years ago (Mya) (electronic supplementary material, figure S1), whereas it was 74 Mya in [2]. This outcome indicates that although we still think that †L. brouweri is the common ancestor of O. sarasinorum and O. eversi (extant species endemic to Sulawesi [14]), the divergence time estimation for O. setnai is independent of its authenticity.
5. Conclusion
In summary, the points raised by Britz et al. [1] do not undermine our conclusions about the origin and evolutionary history of Adrianichthyidae (i.e. an eastward ‘out-of-India’ dispersal). We emphasize that ‘earth and life evolve together from something more ancestral’.
Acknowledgements
We thank Ralf Britz and Lukas Rüber for providing files for their re-analysis. We thank Gabe Yedid, PhD, from Edanz (https://jp.edanz.com) for editing a draft of this manuscript.
Footnotes
The accompanying comment can be viewed at http://doi.org/10.1098/rsbl.2021.0568.
Data accessibility
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.nvx0k6dtx [15].
Authors' contributions
K.Y.: conceptualization, formal analysis and writing—original draft; S.F.: formal analysis and writing—review and editing; Y.T.: formal analysis and writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
We received no funding for this study.
References
- 1.Britz R, Parenti LR, Rüber L. 2022. Earth and life evolve together- a comment on Yamahira et al. Biol. Lett. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamahira K, et al. 2021. Mesozoic origin and ‘out-of-India’ radiation of ricefishes (Adrianichthyidae). Biol. Lett. 17, 20210212. ( 10.1098/rsbl.2021.0212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu Y, Blair C, He XJ. 2020. RASP 4: ancestral state reconstruction tool for multiple genes and characters. Mol. Biol. Evol. 37, 604-606. ( 10.1093/molbev/msz257) [DOI] [PubMed] [Google Scholar]
- 4.Matzke NJ. 2013. Probabilistic historical biogeography: new models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Front. Biogeogr. 5, 242-248. ( 10.21425/F55419694) [DOI] [Google Scholar]
- 5.Matzke NJ. 2014. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst. Biol. 63, 951-970. ( 10.1093/sysbio/syu056) [DOI] [PubMed] [Google Scholar]
- 6.Matzke NJ. 2018. BioGeoBEARS: BioGeography with Bayesian (and likelihood) evolutionary analysis with R scripts. version 1.1.1, published on GitHub on November 6, 2018. See 10.5281/zenodo.1478250. [DOI]
- 7.Murphy WJ, Collier GE.1997. A molecular phylogeny for aplocheiloid fishes (Atherinomorpha, Cyprinodontiformes): the role of vicariance and the origins of annualism. Mol. Biol. Evol. 14, 790-799. ( 10.1093/oxfordjournals.molbev.a025819) [DOI] [PubMed] [Google Scholar]
- 8.Sparks J, Smith W. 2005. Freshwater fishes, dispersal ability, and nonevidence: ‘gondwana life rafts’ to the rescue. Syst. Biol. 54, 158-165. ( 10.1080/10635150590906019) [DOI] [PubMed] [Google Scholar]
- 9.Samonds KA, Godfrey LR, Ali JR, Goodman SM, Vences M, Sutherland MR, Irwin MT, Krause DW.. 2012. Spatial and temporal arrival patterns of Madagascar's vertebrate fauna explained by distance, ocean currents, and ancestor type. Proc. Natl Acad. Sci. USA 109, 5352-5357. ( 10.1073/pnas.1113993109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa WJEM. 2013. Historical biogeography of aplocheiloid killifishes (Teleostei: Cyprinodontiformes). Vert. Zool. 63, 139-154. [Google Scholar]
- 11.Pohl M, Milvertz FC, Meyer A, Vences M. 2015. Multigene phylogeny of cyprinodontiform fishes suggests continental radiations and a rogue taxon position of Pantanodon. Vert. Zool. 65, 37-44. [Google Scholar]
- 12.Siddall ME, Whiting MF. 1999. Long-branch abstractions. Cladistics 15, 9-24. ( 10.1111/j.1096-0031.1999.tb00391.x) [DOI] [Google Scholar]
- 13.Parenti LR. 2008. A phylogenetic analysis and taxonomic revision of ricefishes, Oryzias and relatives (Beloniformes, Adrianichthyidae). Zool. J. Linn. Soc. 154, 494-610. ( 10.1111/j.1096-3642.2008.00417.x) [DOI] [Google Scholar]
- 14.Horoiwa M, et al. 2021. Mitochondrial introgression by ancient admixture between two distant lacustrine fishes in Sulawesi Island. PLoS ONE 16, e0245316. ( 10.1371/journal.pone.0245316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamahira K, Fujimoto S, Takami Y. 2022. Data from: Earth and life evolve together from something ancestral—reply to Britz et al. Dryad Digital Repository. ( 10.5061/dryad.nvx0k6dtx) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Yamahira K, Fujimoto S, Takami Y. 2022. Data from: Earth and life evolve together from something ancestral—reply to Britz et al. Dryad Digital Repository. ( 10.5061/dryad.nvx0k6dtx) [DOI] [PMC free article] [PubMed]
Data Availability Statement
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.nvx0k6dtx [15].