Key Points
Rab27a-dependent secretion of leukemic EVs promotes leukemia engraftment and immunosuppressive potential of Treg cells in vivo.
Leukemic EVs containing 4-1BBL protein promote eTregs displaying specific signature (CD39, CCR8, CD30, TNFR2, CCR4, TIGIT, and IL21R).
Abstract
Chronic and acute myeloid leukemia evade immune system surveillance and induce immunosuppression by expanding proleukemic Foxp3+ regulatory T cells (Tregs). High levels of immunosuppressive Tregs predict inferior response to chemotherapy, leukemia relapse, and shorter survival. However, mechanisms that promote Tregs in myeloid leukemias remain largely unexplored. Here, we identify leukemic extracellular vesicles (EVs) as drivers of effector proleukemic Tregs. Using mouse model of leukemia-like disease, we found that Rab27a-dependent secretion of leukemic EVs promoted leukemia engraftment, which was associated with higher abundance of activated, immunosuppressive Tregs. Leukemic EVs attenuated mTOR-S6 and activated STAT5 signaling, as well as evoked significant transcriptomic changes in Tregs. We further identified specific effector signature of Tregs promoted by leukemic EVs. Leukemic EVs-driven Tregs were characterized by elevated expression of effector/tumor Treg markers CD39, CCR8, CD30, TNFR2, CCR4, TIGIT, and IL21R and included 2 distinct effector Treg (eTreg) subsets: CD30+CCR8hiTNFR2hi eTreg1 and CD39+TIGIThi eTreg2. Finally, we showed that costimulatory ligand 4-1BBL/CD137L, shuttled by leukemic EVs, promoted suppressive activity and effector phenotype of Tregs by regulating expression of receptors such as CD30 and TNFR2. Collectively, our work highlights the role of leukemic extracellular vesicles in stimulation of immunosuppressive Tregs and leukemia growth. We postulate that targeting of Rab27a-dependent secretion of leukemic EVs may be a viable therapeutic approach in myeloid neoplasms.
Introduction
Myeloid neoplasms, including chronic and acute myeloid leukemia (CML/AML), are characterized by evasion of antileukemic effector immune response and induction of immunosuppression. In CML and AML patients at diagnosis, effector cells of the immune system (CD8+ T cells, NK cells) are exhausted and dysfunctional.1-3 Simultaneously, suppressive immune subsets, including suppressive myeloid cells4 and Foxp3+ regulatory T cells (Tregs), dominate the microenvironment.4 Tregs are increased in peripheral blood5,6 and bone marrow (BM) of leukemic patients.7,8 Importantly, high levels of Tregs in blood and BM of AML patients predict inferior response to chemotherapy, disease relapse, and shorter survival.9,10 Depletion of Tregs in a mouse model of MLL-AF9 AML reduced leukemic burden and increased survival.11 In CML, Tregs were shown to decrease in patients responding to tyrosine kinase inhibitors, and low Treg levels were associated with treatment-free remission.12
Although direct Treg targeting and subsequent elimination would seem like a viable therapeutic strategy, precise depletion of Tregs is difficult to achieve in patients, and it may lead to severe autoimmune adverse events.13 Downregulation of proleukemic Tregs and improved therapeutic outcome could be achieved by targeting factors that drive expansion and activity of Treg subsets in leukemias. Even though some candidates, including coinhibitory PD-1/PD-L1, Gal9/TIM-3 pathways, and IDO enzyme, were implicated in Tregs expansion,3,14-16 Treg-driving mechanism have been poorly investigated in myeloid neoplasms.
Extracellular vesicles (EVs) have recently emerged as important mediators of intercellular communication. EVs are particles released by all types of cells and present in body fluids. They are divided into small exosomes, medium microvesicles, and large apoptotic bodies.17 EVs have been largely implicated in both immune cell activation and immunosuppression.18 In solid tumors, EVs inhibit antitumor activity of T cells19 and are responsible for induction and expansion of Tregs.20,21 In leukemias, including myeloid neoplasms, leukemic EVs promote (in an autocrine manner) growth and drug resistance of leukemic cells,22,23 as well as modify stromal and vascular components of the BM niche.24-26 Until now, immunomodulatory properties toward T cells have only been described for AML-derived EVs, which inhibit effector function of CD8+ cytotoxic T lymphocytes.27
Using mouse ex vivo models, we previously observed involvement of CML-derived leukemic EVs in regulation of suppressive activity of murine Tregs and Foxp3 expression.28 Here, we use ex vivo models with human lymphocytes and EVs to show that leukemic EVs, containing 4-1BBL/CD137L/TNFSF9 protein, promote induction and immunosuppressive polarization of human Tregs by modulating mTOR and STAT5 signaling. We identify specific subsets of effector Tregs (eTreg), characterized by expression of novel tumor Treg markers, such as CD30, CCR8, TNFR2, CD39, and TIGIT. Finally, in vivo in a mouse model of CML-like disease, Rab27a-dependent leukemic EVs promote leukemic engraftment, which was associated with higher abundance of activated, immunosuppressive Tregs. Our results indicate that inhibition of leukemic EVs secretion, to attenuate Tregs, may contribute to improved therapeutic outcome in myeloid leukemias.
Methods
Cell lines
Human cell lines CML-K562 (ATCC, CCL-243) and AML-MOLM-14 (DSMZ, ACC777) were cultured in supplemented Iscove's modified Dulbecco's medium or RPMI 1640 media, respectively. Murine 32D BCR-ABL1+GFP+ cells were obtained from 32D BCR-ABL1+ cells and cultured as previously described.28 To obtain stable knock-out cell lines without expression of Rab27a or 4-1BBL, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) technology was used, using “all-in-1” plasmids encoding guide RNA, Cas9, and red fluorescent protein (RFP) (Merck-Sigma-Aldrich).
Primary human samples from patients
Plasma (source of primary EVs) was obtained from whole blood of 10 leukemic (7 CML and 3 AML; before starting treatment and at diagnosis) patients and 10 healthy donors. Blood was processed as described in supplemental Methods. Material from patients was collected with their informed written consent, under the approval of the Bioethics Committee of the Medical University of Warsaw (KB/107/2018) and Ethics and Bioethics Committee of the Cardinal Stanislaw Wyszynski University in Warsaw (KEiB-19/2017), and in accordance with the Declaration of Helsinki and Polish regulations.
Extracellular vesicles isolation and characterization
EVs released by leukemic cells were isolated from cell culture medium, conditioned for 24 hours by either K562 (CML EVs) or MOLM-14 (AML EVs) cells (seeded in media with EVs-depleted fetal bovine serum). EVs were isolated by differential ultracentrifugation as previously described.17 Pelleted EVs were resuspended in either nonsupplemented AIM V medium (for functional assay with T cells) or phosphate-buffered saline (for EVs characterization). EVs characterization and uptake analysis (supplemental Figure 1A-F) was performed as previously described17 (described in detail in supplemental Methods). For all experiments (unless specified in dose-dependent studies), T cells were treated with either 3 × 109 CML EVs or 5 × 109 AML EVs, equivalent of EVs released by 1 × 108 cells. EVs from plasma were isolated using size-exclusion chromatography (qEV original 35 nm columns, Izon) and fractions 7 to 9 (supplemental Figure 1G-H) were pooled and concentrated using Amicon Ultra-2 10kDa (Merck). Isolation and characterization of EVs was performed according to guidelines of the International Society for Extracellular Vesicles29 and the EV-TRACK consortium30 (EV-TRACK database entry EV210187).
Primary T cell (Treg) isolation and culture
Human lymphocytes were obtained from buffy coats of healthy donors (different donor each experiment) from the Regional Center for Blood Donation and Blood Care in Warsaw, Poland (in accordance with the Declaration of Helsinki and Polish regulations). Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation (Lymphoprep, STEMCELL). T cells, including Tregs, were sorted using BD FACS Aria II (gating strategy: supplemental Figure 2A). Sorted T cells (CD4+CD25hiCD127lo Treg, CD4+CD25− conventional T cells/Tconv) were cultured in AIM V medium (Gibco), stimulated with antibodies anti-CD3 (coated wells, 5 μg/mL; Biolegend) and anti-CD28 (soluble, 1 μg/mL; Biolegend), and supplemented with IL-2 (100 IU/mL for Tconv/Foxp3 induction, 50 IU/mL for Tregs; Peprotech). Cultures were maintained for 6 days to analyze Foxp3 induction in Tconv, 5 days to analyze Tregs, and 18 hours to analyze phosphorylation of signaling molecules in T cells.
In vivo mouse model of CML
To study CML in vivo in immunocompetent animals, murine 32D BCR-ABL1+GFP+ cells, either wild-type (wt) or Rab27a−/−, were injected into genetically-matched male C3H mice, 8 to 10 weeks old (Figure 2A). All experimental procedures were performed according to the guidelines of Poland’s National Ethics Committee for Animal Experimentation and approved by the First Local Ethics Committee for Animal Experimentation in Warsaw (835/2019, 1059/2020). In brief, 1 × 106 32D BCR-ABL1+GFP+ cells were injected intraperitoneally into nonirradiated animals. Control, nonleukemic mice were mock injected with NaCl. Following 2 months, blood, BM, and spleens were isolated and used for subsequent analyses. Development of leukemia-like disease was assessed by analysis of engraftment of GFP+ cells by flow cytometry. Animals that had GFP+ cells engrafted into BM, blood, and spleen (supplemental Figure 5A) were further analyzed.
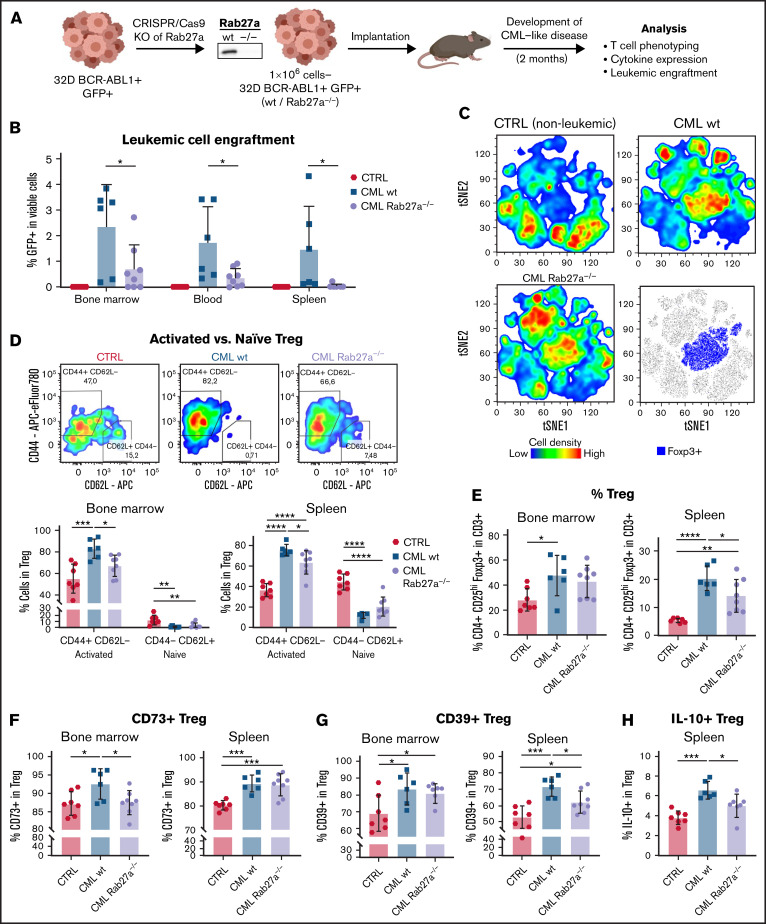
Figure 2.
Rab27a-dependent leukemic EVs promote engraftment of CML cells and activation, amount, and suppressive phenotype of Tregs in vivo in mice. (A) Experimental pipeline of experiments using in vivo model of CML/leukemia-like disease. (B) Leukemic spreading and engraftment of leukemic (GFP+) cells in BM, blood, and spleen of mice with leukemia-like disease. (C) Unsupervised tSNE clustering of CD3+ T cells from spleens of control, healthy mice (CTRL), and animals with leukemia-like disease, induced by wt or Rab27a−/− CML cells. The bottom right graph shows localization of Foxp3+ cells on the tSNE map. Data from 3 to 4 mice (per group) from single experiment were used as representative groups. In each group, 30 000 viable CD3+ T cells were clustered, 7500 to 10 000 from each animal (obtained by downsampling in FlowJo). (D) Distribution of activated (CD44+CD62L−) and naïve (CD44−CD62L+) Treg subsets in BM and spleen of mice bearing leukemia-like disease. Representative density plots showing expression of CD44 and CD62L by Tregs in the BM are shown. Treg amount (E) and expression of CD73 (F), CD39 (G), and IL-10 (H) on Tregs in BM and spleen of mice bearing leukemia-like disease. In each graph, data are presented as mean ± SD, 1-way ANOVA with Tukey’s posttest, *P < .05, **P < .01, ***P < .001, ****P < .0001. N = 6 to 8 animals per group from 3 different experiments (different litters/groups of animals and leukemic cells’ injections). Gating strategy for Treg phenotyping is shown in supplemental Figure 7A-B. SD, standard deviation.
High resolution spectral flow cytometry and data analysis
For 23-color phenotyping of Tregs treated with leukemic EVs, cells from ex vivo cultures were stained with surface antibody cocktail (including viability dye, supplemental Table 1) in Brilliant Stain Buffer Plus (BD) for 30 minutes. Cells were fixed and permeabilized for intracellular staining by eBioscience Foxp3/Transcription Factor Staining Buffer Set (Invitrogen). Intracellular proteins were stained for 30 minutes. Samples were acquired using CYTEK Aurora spectral flow cytometer and analyzed in FlowJo (BD). For manual analysis, cells were gated as shown in supplemental Figure 12A. For computational analyses, each sample was downsampled to obtain 7500 Treg. For tSNE and FlowSOM,31 all samples were concatenated and processed using specific plugins in FlowJo v10. For FlowSOM, cells were clustered into 6 populations (49 nodes), based on 15 parameters (Figure 5B). Generated clustering strategies were then used to cluster and quantify detected populations in individual samples and experimental conditions. Remaining flow cytometry assays and antibody specifics are described in supplemental Methods and Tables.
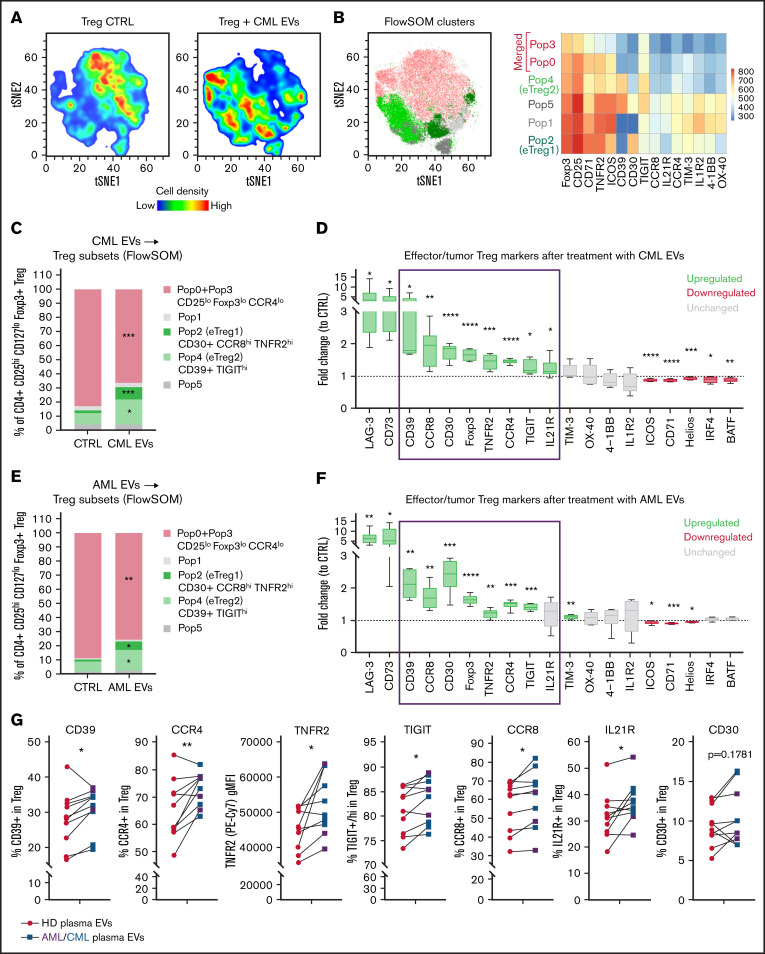
Figure 5.
Human eTreg subsets and effector/tumor Treg signature are driven by leukemic EVs (A) Unsupervised tSNE clustering of CTRL and CML EV (K562-derived)-treated Tregs. (B) Unsupervised FlowSOM clustering of CTRL and CML EVs(K562-derived)-treated Tregs. Identified 5 to 6 FlowSOM clusters/populations were overlaid onto the tSNE map. Heatmap shows relative expression of selected markers by 6 FlowSOM populations. For panels A and B, data from 16 samples (8 per group), 7500 events/sample, were concatenated and used to create tSNE map and FlowSOM clustering scheme. (C) Abundance of Treg subsets (identified by FlowSOM as in panel B) in CTRL and CML EV (K562-derived)-treated Tregs. (D) Expression (fold change to CTRL samples) of tumor Treg markers after treatment of Tregs with CML EVs (K562-derived). For TNFR2, Foxp3, IRF4, and BATF, gMFI was analyzed; for other markers, percent of positive cells. For panels C and D, data are from 4 experiments, n = 8. (E) Abundance of Treg subsets (identified by FlowSOM as in panel B) in CTRL and AML EV (MOLM-14–derived)-treated Tregs. (F) Expression (fold change to CTRL samples) of tumor Treg markers after treatment of Tregs with AML EVs (MOLM-14–derived). For TNFR2, Foxp3, IRF4, and BATF, gMFI was analyzed; for other markers, percent of positive cells. For panels E and F, data are from 4 experiments, n = 6. For panels C through F, statistics were unpaired t tests with Welch’s correction. *P < .05, **P < .01, ***P < .001, ****P < .0001. (G) Expression of selected tumor Treg markers after treatment of Tregs with primary patients’ plasma EVs, compared with healthy donors’ (HD) plasma EVs. For leukemic group, plasma from 3 AML and 7 CML patients was used. For TNFR2, gMFI was analyzed; for other markers, percent of positive cells. N = 10 CML/AML patients and 10 healthy donors. Pairing was done for samples that were used to treat the same batch of (primary) Tregs. Two-tailed paired t test. *P < .05, **P < .01. Gating strategy for Treg phenotyping is shown in supplemental Figure 12A. gMFI, geometric mean fluorescence intensity.
Statistical analysis
Data were plotted and statistics were performed using GraphPad Prism v9. Statistical tests used are indicated in figure legends. Unless indicated otherwise, statistics were performed in comparison with control (CTRL). Significant differences (P < .05) are marked on graphs with asterisks (*P < .05, **P < .01, ***P < .001, ****P < .0001).
Results
EVs released by myeloid leukemia cells induce Foxp3 and upregulate suppressive phenotype and activity of Tregs
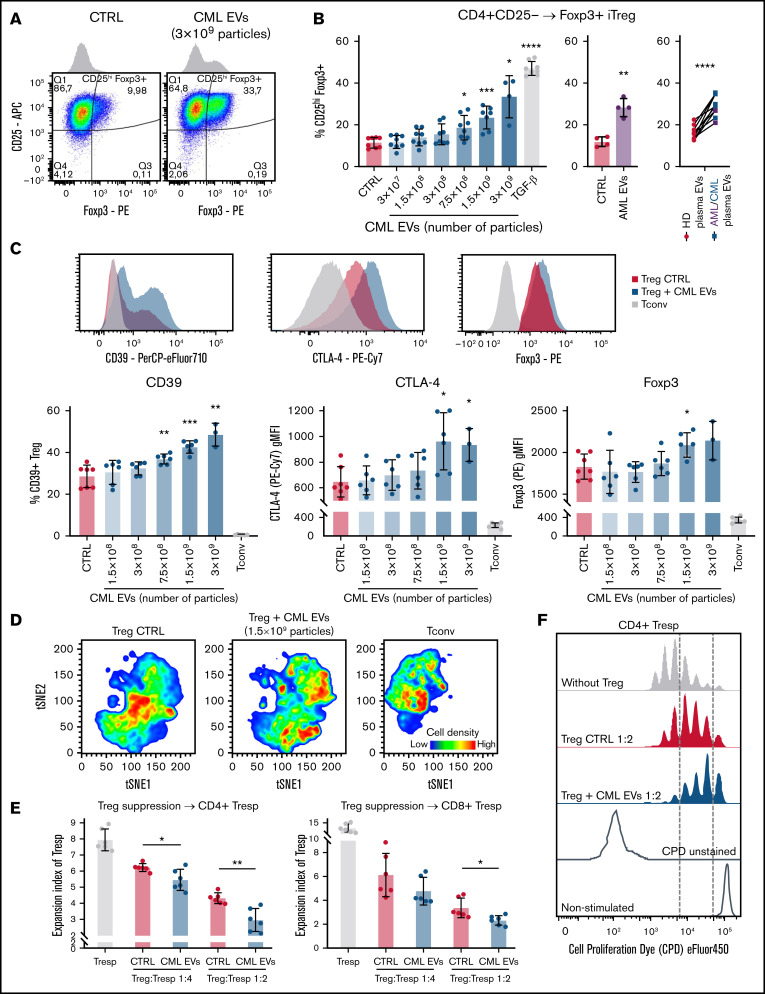
In tumors, immunosuppressive milieu can either expand and drive effector polarization of already differentiated Treg or induce expression of Treg-specific transcription factor Foxp3 in nonregulatory, CD4+CD25− conventional T cells and turn them into CD25hiFoxp3+induced regulatory T cells (iTregs) (CD4+CD25− → Foxp3+ iTreg).32 Therefore, we studied the impact of leukemic EVs on Foxp3 induction, phenotypic changes, and suppressive activity of human Tregs. We performed ex vivo cultures of purified (sorted) human CD4+CD25hiCD127lo Tregs or CD4+CD25− Tconv, together with EVs released by CML-K562 cells (CML EVs) or AML-MOLM-14 cells (AML EVs) (characterized in supplemental Figure 1A-C). Interaction of EVs with human T cells/Tregs was confirmed by tracking fluorescent signal of carboxyfluorescein succinimidyl ester (CFSE)–labeled EVs in a culture with lymphocytes (supplemental Figure 1D-F).
CML EVs induced expression of Foxp3 in CD4+CD25− cells, similarly to TGF-β, a widely recognized Foxp3 inducer (Figure 1A-B). Foxp3 induction was also observed after treatment with AML EVs and primary EVs from plasma of leukemic patients (as compared with healthy donor EVs) (Figure 1B). Induced expression of Foxp3 distinguished a separate population of iTreg (Figure 1A).
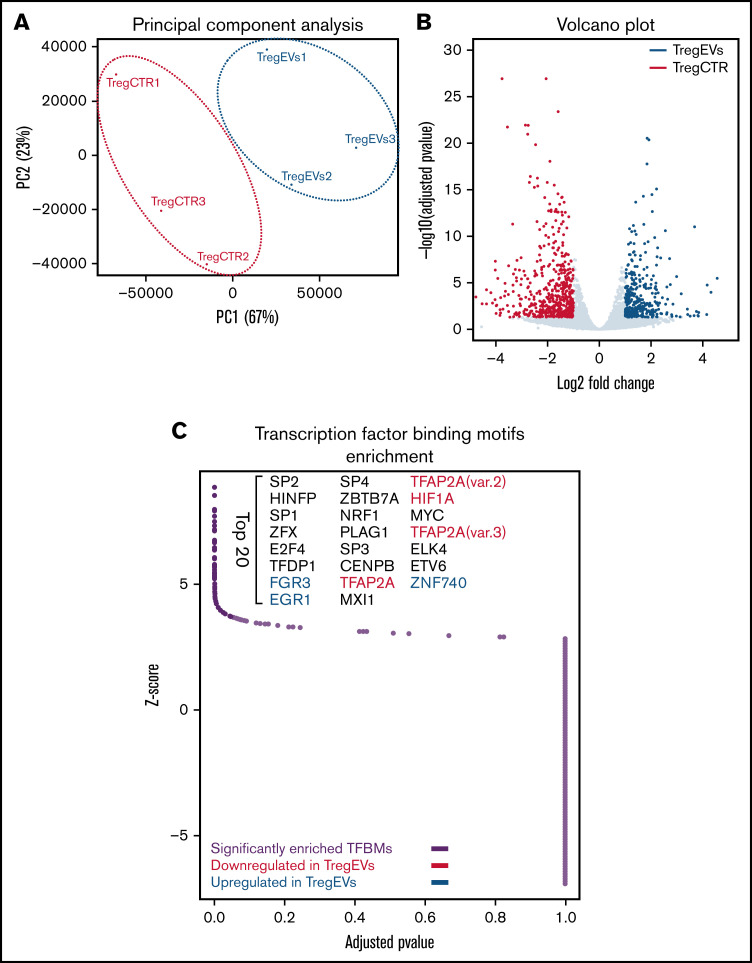
Figure 1.
Leukemic EVs promote induction, suppressive phenotype, and activity of human Tregs. (A) Representative dot plots and histograms of iTregs induced from CD4+CD25− Tconv in control conditions and after treatment with CML EVs (K562-derived). (B) Induction of Foxp3+ cells (iTregs) from CD4+CD25− Tconv cells by leukemic EVs, either released by CML-K562 cells (left panel), AML-MOLM-14 cells (middle panel, 5 × 109 particles), or primary patients’ plasma EVs, compared with healthy donors’ (HD) plasma EVs (right panel). For CML-K562, data are from 4 experiments, n = 8 (except n = 4 for 3 × 109 CML EVs); for AML-MOLM-14 from 4 experiments, n = 4. Mean ± SD is presented, unpaired t test with Welch’s correction, compared with CTRL. For plasma EVs, n = 10 CML/AML patients (3 AML, 7 CML) and 10 healthy donors. Pairing was done for samples that were used to treat the same batch of primary CD4+CD25− Tconv. Two-tailed paired t test. For panels A and B, cells were gated as in supplemental Figure 2B. (C) Influence of leukemic EVs (K562-derived) on key proteins that drive suppressive activity of Tregs: CD39, CTLA-4, and transcription factor Foxp3. Non-Treg CD4+Tconv (CD25−) cells were used as negative controls. Representative histograms are shown (for CD39, 3 × 109 CML EVs; for CTLA-4 and Foxp3, 1.5 × 108 CML EVs). Data are from 3 experiments, n = 6 (n = 3 for 3 × 109 particles). (D) Unsupervised tSNE clustering of Tregs, Treg + CML EVs (K562-derived, 1.5 × 109 particles), and Tconv, based on CD39, CTLA-4, CD25, and Foxp3. In each group, 60 000 cells were clustered, 10 000 from each replicate (obtained by downsampling). For panels C and D, cells were gated as in supplemental Figure 3A. (E) In vitro suppressive activity of control and leukemic EV (K562-derived)-treated Tregs, pronounced as expansion index (EI) of responder cells (Tresp). Lower expansion index corresponds to higher suppressive activity. Data are from 3 experiments, n = 6. (F) Representative proliferation profiles of CD4+ responder cells in an in vitro suppression assay with control and CML EV (K562-derived)-treated Tregs. For panels C and E, mean ± SD is presented, unpaired t test with Welch’s correction, compared with CTRL. *P < .05, **P < .01, ***P < .001, ****P < .0001. SD, standard deviation.
Treatment of sorted CD4+CD25hiCD127lo Tregs with CML EVs led to elevated expression of molecules responsible for suppressive activity: CD39, CTLA-4, and Foxp3 (Figure 1C). Unsupervised analyses of flow cytometric data based on these markers and activation molecule CD25 already indicated EVs-mediated polarization of Tregs into heterogenous cell states (Figure 1D; supplemental Figure 3B) and expansion of highly suppressive subsets, as revealed by FlowSOM clustering (supplemental Figure 3C-D). Using “nonconditioned medium” control we confirmed that effects observed in our experiments were specific to CML EVs (supplemental Figure 4A-C). Consequently, a functional in vitro suppression assay confirmed that Tregs treated with CML EVs more potently inhibited proliferation of effector responder CD4+ and CD8+ T cells (Tresp), confirming superior suppressive activity (Figure 1E-F).
Overall, we demonstrate that leukemic EVs directly induce differentiation and promote suppressive phenotype and activity of human Tregs.
Rab27a-dependent secretion of leukemic EVs promotes Treg activity and leukemic engraftment in vivo
Rab27a is a significant regulator of EVs biogenesis, and Rab27a deficiency downregulates secretion of EVs, though it is not the only protein engaged in EVs secretion.33 Due to its major function, Rab27a deficiency is sufficient to downregulate EVs secretion and has been used to study effect of continuous EVs secretion in in vivo models of cancer.19 To evaluate physiological relevance of leukemic EVs, stable Rab27a-deficient (Rab27a−/−) 32D BCR-ABL1+GFP+ cells, which secrete 30% to 40% less EVs (supplemental Figure 5C-D), were used in an in vivo model of CML-like disease (Figure 2A).
Diminished release of EVs by Rab27a−/− BCR-ABL1–expressing cells led to reduced engraftment of leukemic cells into blood, spleen, and BM (Figure 2B), whereas Rab27a deficiency did not affect their clonogenic potential, proliferation, and cell cycle (supplemental Figure 5E). These results suggest that leukemic EVs modulated disease development by affecting other cells in the microenvironment, including Tregs.
We analyzed Tregs in mice, focusing on spleen and BM: tissues where CML develops and which encompass full spectrum of cellular Treg interactions. Representative tSNE visualization of multiparameter flow cytometry data has already confirmed that in Rab27a−/− leukemia–like disease, Foxp3+ Tregs cluster differently (predominantly as cells expressing lower CD25, CD39, CD44), as compared with wt leukemia and healthy animals (Figure 2C; supplemental Figure 6A). Detailed analysis of CD25hiFoxp3+ Tregs has revealed that Rab27a-dependent secretion of EVs significantly promotes expansion of CD44+CD62L−activated Tregs in both BM and spleen, contrary to naive CD44−CD62L+ Tregs (Figure 2D). Moreover, Rab27a deficiency resulted in lower Treg numbers in spleen, as well as diminished expression on Tregs of suppressive CD39, IL-10 in spleen and CD73 in BM (Figure 2E-H). Differences between spleen and BM in some phenotypic features of Treg in Rab27a−/− leukemia may be an outcome of different tissue context, as BM consists primarily of myeloid cells, whereas T and B cells are most abundant in the spleen. This may also be relevant in terms of CD39/CD73 modulation, as these ectoenzymes may be expressed by other cells in BM/spleen niche.34 Overall, significant changes in the amount of activated CD44+CD62L− Tregs in both tissues show that leukemic EVs expand Tregs with immunosuppressive potential in vivo, even though leukemic EVs did not entirely affect Treg phenotype or Treg numbers in the BM.
In our model, Rab27a−/− CML did not significantly influence other immunosuppressive cells, such as Bregs or myeloid suppressive cells (supplemental Figure 8A-D), which additionally exhibited low abundance, compared with mouse models of solid tumors.35,36 This implicated direct modulation of Tregs by Rab27a-dependent leukemic EVs rather than indirectly via B cell-T-cell, or macrophage-T cell interactions.
Altogether, in vivo data strongly support our hypothesis that secretion of leukemic EVs and EVs-mediated upregulation of activated, immunosuppressive Tregs facilitate development of myeloid leukemias.
Furthermore, pharmacological targeting of EVs secretion by inhibition of Rab27a or nSMase2 has attenuated expansion of Tregs in cultures of human PBMCs with myeloid leukemia cell lines K562 and MOLM-14 (supplemental Figure 9). This suggests that effects similar to genetic targeting of Rab27a (described above) may also be achieved by pharmacological inhibition of EVs secretion.
Leukemic EVs drive Tregs by modulating mTOR and STAT5 signaling and remodeling of the transcriptome
Foxp3 and Tregs undergo complex molecular regulation.37,38 To identify molecular drivers involved in modulation of Tregs by leukemic EVs, we analyzed activation of signaling pathways crucial for regulation of Foxp3 and Treg biology (by phospho-specific flow cytometry)38 in both human Tregs and CD4+CD25− Tconv differentiating into iTregs, as well as performed transcriptomic analysis of human Tregs.
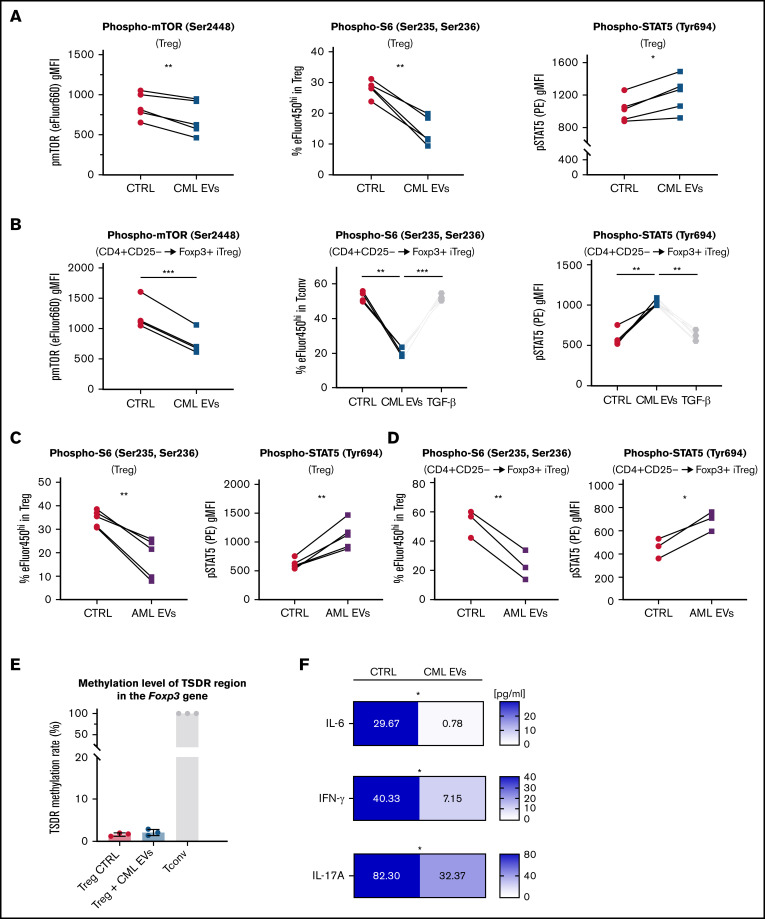
Leukemic (both CML and AML) EVs downregulated phosphorylation of mTOR and its downstream effector protein S6, parallel to upregulation of phosphorylated STAT5, in both Tregs and CD4+CD25− Tconv upon Foxp3 induction by leukemic EVs (Figure 3A-D). Such changes have been described as favorable for Treg differentiation, stability, and suppressive function, as activated STAT5 binds the Foxp3 promoter to drive its transcription, whereas mTOR pathway inhibits Foxp3 expression.38,39 On the other hand, phosphorylation of p38, p65/RelA, and SMAD2/3 were not changed (supplemental Figure 10A-B). To validate whether signaling changes induced by leukemic EVs promote genetic and functional stability of Tregs, we analyzed demethylation of the Treg-specific demethylated region (TSDR) in the Foxp3 gene and secretion of both non–Treg-specific and immunoregulatory cytokines.40 TSDR was demethylated in both CTRL and leukemic EVs-treated Tregs (Figure 3E). However, CML EVs blocked secretion of IL-6, IL-17A, and IFN-γ, preventing polarization into unstable, Th1/Th17-like subsets (Figure 3F), even though secretion of immunoregulatory IL-10 and TGF-β was unaffected (supplemental Figure 10C). These results clearly show that leukemic EVs not only upregulate suppressive features of Tregs but also maintain their stability. Both processes are likely modulated by downregulated mTOR-S6 and upregulated STAT5 signaling.
Figure 3.
Leukemic EVs inhibit mTOR-S6 and upregulate STAT5 signaling in both human Tregs and CD4+CD25− Tconv (differentiating into iTregs) and maintain stability of human Tregs. (A) Analysis of phosphorylation of mTOR, S6, and STAT5 in Tregs after treatment with CML EVs (K562-derived). Data are from 5 experiments, n = 5. Single data points, connected for each experiment, are presented. (B) Analysis of phosphorylation of mTOR, S6, and STAT5 in Tconv differentiating into iTregs (CD4+CD25− → Foxp3+ iTreg as in Figure 1A-B) after treatment with CML EVs (K562-derived). Data are from 4 experiments, n = 4. Single data points, connected for each experiment, are presented. (C) Analysis of phosphorylation of S6 and STAT5 in Tregs after treatment with AML EVs (MOLM-14-derived). Data are from 5 experiments, n = 5. Single data points, connected for each experiment, are presented. For panels A and C, cells were gated as in supplemental Figure 3A, using antibodies conjugated with fluorochromes as in supplemental Table 1. (D) Analysis of phosphorylation of S6 and STAT5 in Tconv differentiating into iTregs (CD4+CD25− → Foxp3+ iTreg as in Figure 1A-B) after treatment with AML EVs (MOLM-14–derived). Data are from 3 experiments, n = 3. Single data points, connected for each experiment, are presented. For panels B and D, cells were gated as in supplemental Figure 2B, using antibodies conjugated with fluorochromes as in supplemental Table 1. (E) Level of methylation of TSDR region in the Foxp3 gene in Tregs treated with leukemic EVs (K562-derived; Tconv served as positive control). Data are from 3 experiments, n = 3. Mean ± SD is presented. (F) Secretion of effector/proinflammatory cytokines (pg/ml, calculated per 1 × 106 cells; final concentration shown on graphs) detected in cultured medium of control Tregs or Tregs cultured with leukemic EVs (K562-derived). Data are from 6 experiments, n = 6. For all experiments in the figure, statistics was 2-tailed paired t test. *P < .05, **P < .01, ***P < .001. SD, standard deviation.
Analysis of Tregs by RNA sequencing revealed significant remodeling of the transcriptome and elevated expression of 356 genes due to treatment with CML EVs (Figure 4A-B), as well as influence on biological processes, such as RNA metabolism (supplemental Figure 11B-C). We analyzed genes described as characteristic for Tregs in cancer41-45 and observed a visible trend of upregulated expression for CCR4, TFRC, TNFRSF1B (encoding TNFR2), ENTPD1 (CD39), TNFRSF8 (CD30), IL1R1, HAVCR2 (TIM-3), and TGFB1 (supplemental Figure 11D). However, in most cases, the difference was not statistically significant, therefore we additionally verified these observations on protein level (Figure 5D,F). Analysis of transcription factor–binding motifs (TFBMs) of differentially expressed genes identified several transcription factors potentially engaged in modulation of Tregs by leukemic EVs, such as EGR1, EGR3, ZBTB7A(LRF), E2F4, or TFDP1 (Figure 4C; supplemental Figure 11E). Overall, RNA sequencing further signified that leukemic EVs affect Treg, by global remodeling of gene expression, including upregulation of genes responsible for immunosuppressive function. Analysis of transcription factor–binding motifs pinpointed a set of transcription factors that modulate these changes in Tregs and may be relevant for immunosuppression in myeloid leukemias.
Figure 4.
Leukemic extracellular vesicles (K562-derived) remodel human Treg transcriptome. (A) Principal component analysis (PCA) of transcriptomes of control (TregCTR) and CML EVs-treated Tregs (TregEVs). (B) Volcano plot comparing differences in gene expression between CTRL and CML EVs-treated Tregs (TregEVs). Treatment with CML EVs led to 356 upregulated (marked in blue) and 566 downregulated (marked in red) differentially expressed genes (DEGs). Genes were considered as differentially expressed when −log10 (adjusted P value) < .05 and |log fold change (LFC)|>1. (C) Analysis of transcription factor binding motifs (TFBMs) enriched in DEGs (|LFC|>1), performed using PSCAN software. Significantly enriched TFBMs are indicated by dark purple dots, and top 20 hits are named. Transcription factors (TFs) that are differentially expressed at mRNA level are additionally indicated in either blue (upregulated by CML EVs) or red (downregulated by CML EVs). RNA sequencing was performed for n = 3 per group. For each replicate, Tregs obtained from a different donor were used. mRNA, messenger RNA.
Tregs driven by leukemic EVs are constituted by heterogenous effector subsets and characterized by upregulated CD39, CCR8, CD30, TNFR2, CCR4, TIGIT, and IL21R
To evaluate changes in human Tregs on single cell protein level, we developed a 23-color panel for spectral flow cytometry to analyze markers of effector/tumor Treg.41-45 tSNE clustering revealed that leukemic EVs promoted expansion of heterogenous Treg subsets (Figure 5A). Using FlowSOM we clustered Tregs into 6 populations (Figure 5B). As 2 of them (Pop0 and Pop3) exhibited very similar expression pattern of analyzed markers and established 1 joint cluster on tSNE, we decided to merge them and analyze together as 1 population (Pop0+Pop3) (Figure 5B, marked in pink). Two identified populations were upregulated (Pop2, Pop4) and 1 was downregulated (Pop0+Pop3), by CML and AML EVs (Figure 5C,E). Both upregulated populations exhibited high expression of functional effector markers (Figure 5B; supplemental Figure 12B), thus we named them effector Treg1 (eTreg1/Pop2) and effector Treg2 (eTreg2/Pop4). eTreg1 (Pop2) could be distinguished by high expression of CD30, CCR8, TNFR2, whereas eTreg2 (Pop4) by high expression of CD39 and TIGIT (Figure 5B; supplemental Figure 12B). As newly described effector subsets exhibited a distinct phenotype, it suggests specialized functions of eTreg1 and eTreg2 in the context of leukemic microenvironment. The EVs-downregulated population (Pop0+Pop3) had lower expression of markers such as CCR4, CD25, Foxp3, ICOS (supplemental Figure 12B), thus it constitutes a less-effector and probably less suppressive population.
Classical, manual gating analysis of the entire Treg population revealed that CML and AML EVs upregulated expression of several effector tumor Treg markers, identifying a specific leukemic EVs-driven Treg signature that includes elevated expression of CD39, CCR8, CD30, Foxp3, TNFR2, CCR4, TIGIT, and IL21R markers (Figure 5D,F, blue frame). Although LAG-3 and CD73 were highly upregulated, their expression was limited to <1% of Tregs. Altogether, these data demonstrate that leukemic EVs mediate development of specific subsets of eTreg cells and promote EVs-dependent signature of Tregs (elevated expression of CD39, CCR8, CD30, Foxp3, TNFR2, CCR4, TIGIT, and IL21R).
Finally, we verified whether primary EVs isolated from plasma of CML/AML patients promote similar effector polarization of human Tregs and the same specific marker signature of Tregs. The treatment of Tregs with primary EVs from the plasma of leukemic patients (compared with healthy donor EVs) led to elevated expression of the signature molecules CD39, CCR8, TNFR2, CCR4, TIGIT, IL21R, and CD30 (Figure 5G). This provides evidence supporting the hypothesis that leukemic EVs, present in the circulation (plasma), can influence immune cells in distant tissues to facilitate immunosuppression outside the BM.
Collectively, leukemic EVs, both released in vitro by CML/AML cells and of primary origin (plasma EVs), upregulate specific signature of Tregs (characterized by high expression of CD39, CCR8, TNFR2, CCR4, TIGIT, IL21R, and CD30) that includes 2 effector immunosuppressive Treg subsets.
Leukemic EVs contain 4-1BBL protein, which contributes to Treg activity and effector phenotype
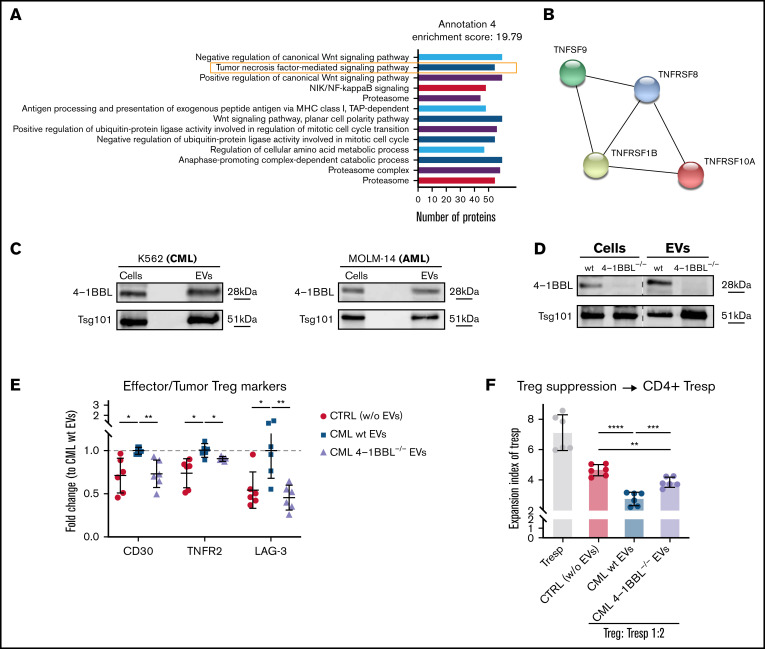
Finally, we aimed to identify specific protein content of EVs that influences human Tregs. Mass spectrometry, followed by functional annotation of detected proteins, identified groups of proteins connected to immune response, such as TNF signaling, including 4-1BBL/TNFSF9/CD137L (Figure 6A-B). Presence of 4-1BBL in CML and AML EVs was confirmed by western blotting (Figure 6C). TNF receptor superfamily was recently implicated in Treg function, though by far mainly in the small intestine, colon, and during colitis.46-48 We hypothesized that it may also contribute to EVs-mediated eTreg polarization in myeloid leukemias. Using CRISPR/Cas9 mutagenesis, we generated 4-1BBL–deficient K562 CML cells, leading to the absence of 4-1BBL in CML cells and EVs (Figure 6D; supplemental Figure 13A). Tregs treated with 4-1BBL–deficient CML EVs no longer upregulated CD30, TNFR2, and LAG-3 (Figure 6E, but not the remaining receptors upregulated by leukemic EVs, such as CD39 or Foxp3; supplemental Figure 13B-C), as well as exhibited weaker suppressive activity in a functional in vitro suppression assay (Figure 6F; supplemental Figure 13D). These data demonstrate the regulatory role of 4-1BBL in promotion of Treg activity and effector phenotype.
Figure 6.
Leukemic EVs contain 4-1BBL/TNFSF9 protein, which contributes to effector phenotype and suppressive activity of human Tregs. (A) Protein groups identified (annotation 4 only, out of 18 statistically significant annotations; details in supplemental Table 4) in mass spectrometric profiling of CML EVs (K562-derived). Number of proteins in each group is indicated. Data were collected from 4 experiments (n = 4). (B) Proteins from TNF (TNFSF9/4-1BBL) and TNF receptor (TNFRSF1B/TNFR2, TNFRSF8/CD30, TNFRSF10A/DR4) superfamilies identified in proteomic analysis of CML EVs (K562-derived). (C) Western blot analysis of 4-1BBL/TNFSF9 protein in K562 (CML) and MOLM-14 (AML) cells and EVs. An equal amount of protein was loaded on gels. Data are representative for 4 experiments (CML EVs) and 2 experiments (AML EVs). (D) Western blot analysis of 4-1BBL/TNFSF9 protein in wt and 4-1BBL−/− K562 (CML) cells and released EVs. An equal amount of protein was loaded on gels. Data from the same gel are presented (marked with dashed line). (E) Expression (fold change to CML wt EVs samples) of CD30, TNFR2, and LAG-3 after treatment of Tregs with CML EVs (K562-derived) from wt and 4-1BBL−/− CML cells. For TNFR2, gMFI was analyzed; for CD30 and LAG-3, percent of positive cells. Data are from 3 experiments, n = 6. (F) In vitro suppressive activity of Tregs treated with either wt or 4-1BBL–deficient CML EVs (K562-derived), toward CD4+ responder T cells, pronounced as expansion index (EI) of responder cells (Tresp). Lower expansion index corresponds to higher suppressive activity. Data are from 3 experiments, n = 6. For panels E and F, mean ± SD is presented, unpaired t-test with Welch’s correction. *P < .05, **P < .01, ***P < .001, ****P < .0001. gMFI, geometric mean fluorescence intensity; SD, standard deviation.
Overall, we postulate that TNF superfamily protein 4-1BBL contributes to effector immunosuppressive polarization of Tregs promoted by leukemic EVs.
Discussion
In the presented study, we report leukemic (CML and AML) extracellular vesicles as novel, significant drivers of human immunosuppressive Foxp3+ Tregs with an effector phenotype, including 2 distinct effector subsets (Figure 7). We show this by complementary approaches, using pure leukemic EVs released by human cell lines and primary EVs isolated from the plasma of leukemic patients, compared with EVs of healthy donors. Using the mouse model of CML-like disease, we showed that leukemic EVs-Tregs interaction facilitates the development of leukemia-like disease, which implicates importance for human leukemia.
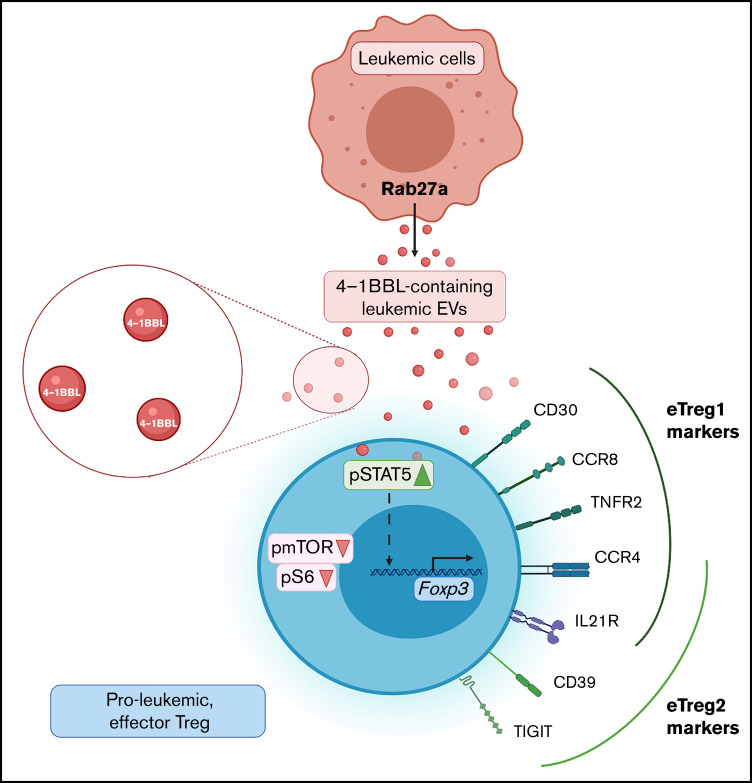
Figure 7.
Proposed model of proleukemic eTreg generation, driven by 4-1BBL containing leukemic extracellular vesicles. Leukemic cells secrete Rab27a-dependent EVs that contain 4-1BBL protein. EVs are internalized by Tregs to upregulate STAT5 and attenuate mTOR-S6 signaling. As a result, Foxp3 expression is driven and Tregs upregulate specific EVs-mediated phenotypic marker signatures (CD39, CCR8, CD30, TNFR2, CCR4, TIGIT, and IL21R), responsible for immunosuppressive, proleukemic function. Leukemic EVs-driven Tregs include 2 distinct eTreg subsets - CD30+CCR8hiTNFR2hi eTreg1 and CD39+TIGIThi eTreg2. CCR4 and IL21R are expressed by both eTreg1 and eTreg2 subsets.
EVs enable intercellular communication between distant tissues and cells. EVs may thus promote the growth of leukemic cells outside the BM by creating leukemia-permissive microenvironment in faraway tissues. It has already been established that Tregs and immunosuppression are critical elements that facilitate such leukemia-supporting conditions.3,9,11 We found that circulating, primary EVs from plasma of CML and AML patients induced Foxp3 and promoted effector signature of Tregs. This supports our hypothesis that leukemic EVs in circulation may drive proleukemic Tregs outside the BM and established a previously undescribed mechanism facilitating spreading of leukemic blasts.
Indeed, one of our critical findings, significant for myeloid neoplasms, is that EVs secretion facilitates leukemia development in vivo in mice, demonstrated in a model of leukemia-like disease induced by Rab27a−/− CML cells with downregulated EVs secretion. Rab27a deficiency and diminished release of EVs in vivo partially reversed expansion of proleukemic, activated Tregs in the BM and spleen. In parallel, significantly lower engraftment of leukemic cells was observed. Importantly, we excluded the role of autocrine influence of EVs on growth and clonogenicity of leukemic cells and excluded the involvement of B cells or immune myeloid cells in the identified effects. These observations strongly suggest direct modulation of Tregs by CML EVs in vivo and the relevance of EVs-Tregs interaction for leukemia progression. Even if leukemic EVs have been shown to modulate other components of the BM niche,24,25 and we cannot entirely conclude that the effect depends solely on Tregs and immunosuppression, our data indicate significant involvement of Tregs. In a translational context, our in vivo discoveries provide rationale for therapeutic targeting of EVs/Rab27a in leukemias. Similar conclusions have been drawn by Poggio et al,19 based on mouse models of Rab27a−/− colorectal and prostate cancers, where targeting EVs also targeted immune checkpoint molecule PD-L1. EVs secretion may be clinically targeted by pharmacological inhibitors of Rab27a49 or other EV-regulatory hubs (nSMase2).50 In our experiments, such treatment indeed attenuates expansion of Tregs in cultures of human PBMCs and leukemic cells. Overall, our discoveries identified leukemic, Rab27a-dependent EVs and EVs-Tregs interaction as potential, previously unrecognized, therapeutic targets in myeloid neoplasms.
Recent studies of Tregs in cancer have identified new molecules specific for tumor Tregs, such as CCR8,42 CD30, IL21R,44 and others,51 as well as specific eTreg subsets.41 Remarkably, our findings provide evidence that leukemic EVs contribute to the expansion of highly suppressive effector subsets of Tregs and promote specific effector signatures of Tregs, which has not been previously dissected in the cancer field. The eTreg1 subset we described had high expression of CCR8, CD30, and TNFR2, as well as transcription factor IRF4 (supplemental Figure 12B). It thus resembles an effector, IRF4-driven Treg population described in lung cancer.41 On the other hand, tissue and cellular context for these molecules in myeloid neoplasms remain to be elucidated, such as expression of CCR8 chemokine ligands in the leukemic BM. Presence of TNFR2, which is usually expressed by T cell receptor–activated Tregs,44 implies interaction with antigen-presenting cells, such as tolerogenic dendritic cells.52 The ubiquitous expression of CD39 and TIGIT on the eTreg2 subset we identified suggests different function of this population in the leukemic microenvironment. TIGIT acts as a coinhibitory receptor, capable of inhibiting Th1- and Th17-polarized CD4+ T cells,53 whereas CD39 is an ectoenzyme, converting adenosine triphosphate to adenosine (jointly with CD73), also to inhibit effector T cells.54 Moreover, CD39 on Tregs can support hematopoietic stem cells in the BM,55 which implies possible interaction of eTreg2 with leukemic stem cells. Thus, even though identified populations of Tregs have well-documented immunosuppressive phenotype and function, it would be important to dissect the precise relevance of eTreg1 and eTreg2 in the leukemic microenvironment, both functionally and spatially. The significance of leukemic EVs-Tregs interaction is further highlighted by our discoveries of molecular regulators. We identified transcription factors that may be responsible for driving eTreg or markers of eTregs in myeloid neoplasms. Some of them, such as EGR3,56 E2F4,45 ZBTB7A/LRF,57 or TFDP1,41 have already been implicated in regulation of Tregs, as well as in tumors. We also pinpoint that although leukemic EVs modulate mTOR and STAT5 pathways, they do not engage TGF-β/SMAD signaling, which classically induces de novo Foxp3 expression,58 implicating a new modality of Treg induction and modulation.
Finally, we detected a functional role of TNF superfamily member 4-1BBL/TNFSF9/CD137L in leukemic EVs and propose that its presence contributes to the amplified immunosuppressive potential of Tregs. According to ExoCarta database, 4-1BBL protein was previously not detected in EVs59 but was identified on hematopoietic and progenitor cells in the BM.60 Moreover, 4-1BB receptor is highly expressed on Tregs in tumors.32,41 Our conclusion is also supported by recent findings, which have demonstrated relevance of 4-1BBL/4-1BB signaling in Treg activation, physiological function, and transcriptomic identity.46-48 Our results pinpoint 4-1BBL as a new protein engaged in the upregulation of suppressive activity and eTreg phenotype in leukemia, and we show, to our knowledge for the first time, that 4-1BBL signaling may occur via EVs. Such findings may have a diagnostic and therapeutic value, and 4-1BBL expression in leukemic EVs could be considered in liquid biopsy approaches as an early biomarker of leukemia and immunosuppression in CML/AML. Substantial advances in the extracellular vesicles field have been developed for such approach, in terms of quick and effective phenotyping of EVs in plasma, for instance by EV-cytometry.61
In conclusion, we discovered a proleukemic, immunosuppressive mechanism, dependent on 4-1BBL-containing EVs derived from CML and AML cells. Leukemic EVs act as drivers of effector subsets and highly suppressive phenotype of Tregs. Our findings demonstrate the rationale to target Rab27a-dependent EV secretion, which may lead to prospective therapeutic applications aimed at attenuating immunosuppression in myeloid neoplasms.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors would like to acknowledge Grażyna Mosieniak for granting generous access to the Nanosight NS300 nanoparticle analyzer and Ewa Wąsiewicz for collecting clinical samples from leukemic patients. Figures 2A and 7 and supplemental Figure 5B were created with biorender.com online software. RNA sequencing was performed at Genomics Core Facility (GeneCore) EMBL Heidelberg.
This work was supported by grants from the Polish National Science Centre: 2013/10/E/NZ3/00673 (K.P.), 2018/29/N/NZ3/01754 (J.S.), and 2014/15/B/NZ7/00966 (U.W.). J.S., K.P., and M.B.-O. were supported by the Foundation for Polish Science grant TEAM TECH Core Facility Plus/2017-2/2 (POIR.04.04.00-00- 23C2/17-00, cofinanced by the European Union under the European Regional Development Fund). E.M. and J.M. were funded from the Foundation for Polish Science International Research Agendas Programme financed from the Smart Growth Operational 2014-2020 (Grant Agreement No. MAB/2018/6). S.D.B. has been a Marylou Ingram Scholar of the International Society for Advancement of Cytometry (ISAC) for the period 2016-2020.
Authorship
Contribution: J.S., L.T.-K., M.B.-O., A.K., B.V.L., and W.G.-P. performed ex vivo experiments; J.S., L.T.-K., B.V.L., S.C., P.P., and E.K. performed in vivo experiments and analyses; W.D. isolated RNA and prepared libraries for RNA sequencing; E.M. and J.M. analyzed RNA sequencing data; J.S., S.D.B., and A.C. analyzed and discussed flow cytometry data; D.C. performed proteomics; U.W., W.G.-P., and G.B. provided primary material; J.S. and K.P. conceptualized and supervised the project and experiments; J.S. prepared the figures and manuscript draft; J.S., L.T.-K., S.D.B, S.C., E.K., U.W., J.M., T.S., A.C., and K.P. prepared and reviewed final version of manuscript; and all authors have read and agreed to the published version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interest.
Correspondence: Katarzyna Piwocka, Laboratory of Cytometry, Nencki Institute of Experimental Biology, 3 Pasteur St, 02-093 Warsaw, Poland; e-mail: k.piwocka@nencki.edu.pl.
References
- 1.Dufva O, Pölönen P, Brück O, et al. Immunogenomic landscape of hematological malignancies [published correction appears in Cancer Cell. 2020;38(3):424–428]. Cancer Cell. 2020;38(3):380-399.e13. [DOI] [PubMed] [Google Scholar]
- 2.Le Dieu R, Taussig DC, Ramsay AG, et al. Peripheral blood T cells in acute myeloid leukemia (AML) patients at diagnosis have abnormal phenotype and genotype and form defective immune synapses with AML blasts. Blood. 2009;114(18):3909-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swatler J, Turos-Korgul L, Kozlowska E, Piwocka K. Immunosuppressive cell subsets and factors in myeloid leukemias. Cancers (Basel). 2021;13(6):1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pyzer AR, Stroopinsky D, Rajabi H, et al. MUC1-mediated induction of myeloid-derived suppressor cells in patients with acute myeloid leukemia. Blood. 2017;129(13):1791-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes A, Clarson J, Tang C, et al. CML patients with deep molecular responses to TKI have restored immune effectors and decreased PD-1 and immune suppressors. Blood. 2017;129(9):1166-1176. [DOI] [PubMed] [Google Scholar]
- 6.Szczepanski MJ, Szajnik M, Czystowska M, et al. Increased frequency and suppression by regulatory T cells in patients with acute myelogenous leukemia. Clin Cancer Res. 2009;15(10):3325-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brück O, Dufva O, Hohtari H, et al. Immune profiles in acute myeloid leukemia bone marrow associate with patient age, T-cell receptor clonality, and survival. Blood Adv. 2020;4(2):274-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brück O, Blom S, Dufva O, et al. Immune cell contexture in the bone marrow tumor microenvironment impacts therapy response in CML. Leukemia. 2018;32(7):1643-1656. [DOI] [PubMed] [Google Scholar]
- 9.Williams P, Basu S, Garcia-Manero G, et al. The distribution of T-cell subsets and the expression of immune checkpoint receptors and ligands in patients with newly diagnosed and relapsed acute myeloid leukemia. Cancer. 2019;125(9):1470-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamble AJ, Lind EF. Targeting the immune microenvironment in acute myeloid leukemia: a focus on T cell immunity. Front Oncol. 2018;8:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang R, Feng W, Wang H, et al. Blocking migration of regulatory T cells to leukemic hematopoietic microenvironment delays disease progression in mouse leukemia model. Cancer Lett. 2020;469:151-161. [DOI] [PubMed] [Google Scholar]
- 12.Irani YD, Hughes A, Clarson J, et al. Successful treatment-free remission in chronic myeloid leukaemia and its association with reduced immune suppressors and increased natural killer cells. Br J Haematol. 2020;191(3):433-441. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka A, Sakaguchi S. Targeting Treg cells in cancer immunotherapy. Eur J Immunol. 2019;49(8):1140-1146. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Q, Munger ME, Highfill SL, et al. Program death-1 signaling and regulatory T cells collaborate to resist the function of adoptively transferred cytotoxic T lymphocytes in advanced acute myeloid leukemia. Blood. 2010;116(14):2484-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curti A, Pandolfi S, Valzasina B, et al. Modulation of tryptophan catabolism by human leukemic cells results in the conversion of CD25Ϫ into CD25ϩ T regulatory cells. Blood. 2007;109(7):2871-2877. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Q, Munger ME, Veenstra RG, et al. Coexpression of Tim-3 and PD-1 identifies a CD8ϩ T-cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood. 2011;117(17):4501-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swatler J, Dudka W, Piwocka K. Isolation and characterization of extracellular vesicles from cell culture conditioned medium for immunological studies. Curr Protoc Immunol. 2020;129(1):e96. [DOI] [PubMed] [Google Scholar]
- 18.Marar C, Starich B, Wirtz D. Extracellular vesicles in immunomodulation and tumor progression. Nat Immunol. 2021;22(5):560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poggio M, Hu T, Pai C-C, et al. Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell. 2019;177(2):414-427.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin Y, Cai X, Chen X, et al. Tumor-secreted miR-214 induces regulatory T cells: a major link between immune evasion and tumor growth. Cell Res. 2014;24(10):1164-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mrizak D, Martin N, Barjon C, et al. Effect of nasopharyngeal carcinoma-derived exosomes on human regulatory T cells. J Natl Cancer Inst. 2014;107(1):363. [DOI] [PubMed] [Google Scholar]
- 22.Nehrbas J, Butler JT, Chen D-W, Kurre P. Extracellular vesicles and chemotherapy resistance in the AML microenvironment. Front Oncol. 2020;10:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raimondo S, Saieva L, Corrado C, et al. Chronic myeloid leukemia-derived exosomes promote tumor growth through an autocrine mechanism. Cell Commun Signal. 2015;13(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mineo M, Garfield SH, Taverna S, et al. Exosomes released by K562 chronic myeloid leukemia cells promote angiogenesis in a Src-dependent fashion. Angiogenesis. 2012;15(1):33-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huan J, Hornick NI, Goloviznina NA, et al. Coordinate regulation of residual bone marrow function by paracrine trafficking of AML exosomes. Leukemia. 2015;29(12):2285-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dumontet E, Pangault C, Roulois D, et al. Extracellular vesicles shed by follicular lymphoma B cells promote polarization of the bone marrow stromal cell niche. Blood. 2021;138(1):57-70. [DOI] [PubMed] [Google Scholar]
- 27.Szczepanski MJ, Szajnik M, Welsh A, Whiteside TL, Boyiadzis M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-beta1. Haematologica. 2011;96(9):1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swatler J, Dudka W, Bugajski L, Brewinska-Olchowik M, Kozlowska E, Piwocka K. Chronic myeloid leukemia-derived extracellular vesicles increase Foxp3 level and suppressive activity of thymic regulatory T cells. Eur J Immunol. 2020;50(4):606-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Deun J, Mestdagh P, Agostinis P, et al. ; EV-TRACK Consortium . EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods. 2017;14(3):228-232. [DOI] [PubMed] [Google Scholar]
- 31.Van Gassen S, Callebaut B, Van Helden MJ, et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87(7):636-645. [DOI] [PubMed] [Google Scholar]
- 32.Wing JB, Tanaka A, Sakaguchi S. Human FOXP3+ regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity. 2019;50(2):302-316. [DOI] [PubMed] [Google Scholar]
- 33.Ostrowski M, Carmo NB, Krumeich S, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1):19-30, 1-13. [DOI] [PubMed] [Google Scholar]
- 34.Moesta AK, Li X-Y, Smyth MJ. Targeting CD39 in cancer. Nat Rev Immunol. 2020;20(12):739-755. [DOI] [PubMed] [Google Scholar]
- 35.Sosnowska A, Chlebowska-Tuz J, Matryba P, et al. Inhibition of arginase modulates T-cell response in the tumor microenvironment of lung carcinoma. OncoImmunology. 2021;10(1):1956143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilanc P, Wojnicki K, Roura A-J, et al. A Novel oral arginase 1/2 inhibitor enhances the antitumor effect of PD-1 inhibition in murine experimental gliomas by altering the immunosuppressive environment. Front Oncol. 2021;11:703465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haiqi H, Yong Z, Yi L. Transcriptional regulation of Foxp3 in regulatory T cells. Immunobiology. 2011;216(6):678-685. [DOI] [PubMed] [Google Scholar]
- 38.Lu L, Barbi J, Pan F. The regulation of immune tolerance by FOXP3. Nat Rev Immunol. 2017;17(11):703-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delgoffe GM, Kole TP, Zheng Y, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30(6):832-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominguez-Villar M, Hafler DA. Regulatory T cells in autoimmune disease. Nat Immunol. 2018;19(7):665-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvisi G, Brummelman J, Puccio S, et al. IRF4 instructs effector Treg differentiation and immune suppression in human cancer. J Clin Invest. 2020;130(6):3137-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plitas G, Konopacki C, Wu K, et al. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45(5):1122-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azizi E, Carr AJ, Plitas G, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174(5):1293-1308.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Simone M, Arrigoni A, Rossetti G, et al. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity. 2016;45(5):1135-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magnuson AM, Kiner E, Ergun A, et al. Identification and validation of a tumor-infiltrating Treg transcriptional signature conserved across species and tumor types. Proc Natl Acad Sci USA. 2018;115(45):E10672-E10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ronin E, Lubrano di Ricco M, Vallion R, et al. The NF-κB RelA transcription factor is critical for regulatory T cell activation and stability. Front Immunol. 2019;10:2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasanthakumar A, Liao Y, Teh P, et al. The TNF receptor superfamily-NF-κB axis is critical to maintain effector regulatory T cells in lymphoid and non-lymphoid tissues. Cell Rep. 2017;20(12):2906-2920. [DOI] [PubMed] [Google Scholar]
- 48.Lubrano di Ricco M, Ronin E, Collares D, et al. Tumor necrosis factor receptor family costimulation increases regulatory T-cell activation and function via NF-κB. Eur J Immunol. 2020;50(7):972-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson JL, Ramadass M, He J, et al. Identification of neutrophil exocytosis inhibitors (Nexinhibs), small molecule inhibitors of neutrophil exocytosis and inflammation: druggability of the small GTPase Rab27a. J Biol Chem. 2016;291(50):25965-25982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tallon C, Hollinger KR, Pal A, et al. Nipping disease in the bud: nSMase2 inhibitors as therapeutics in extracellular vesicle-mediated diseases. Drug Discov Today. 2021;26(7):1656-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prlic M, Mair F, Erickson J, et al. Extricating human tumor-unique immune alterations from non-malignant tissue inflammation. Nat. Port. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kleijwegt FS, Laban S, Duinkerken G, et al. Critical role for TNF in the induction of human antigen-specific regulatory T cells by tolerogenic dendritic cells. J Immunol. 2010;185(3):1412-1418. [DOI] [PubMed] [Google Scholar]
- 53.Lucca LE, Dominguez-Villar M. Modulation of regulatory T cell function and stability by co-inhibitory receptors. Nat Rev Immunol. 2020;20(11):680-693. [DOI] [PubMed] [Google Scholar]
- 54.Plitas G, Rudensky AY. Regulatory T cells: differentiation and function. Cancer Immunol Res. 2016;4(9):721-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hirata Y, Furuhashi K, Ishii H, et al. CD150high bone marrow Tregs maintain hematopoietic stem cell quiescence and immune privilege via adenosine. Cell Stem Cell. 2018;22(3):445-453.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morita K, Okamura T, Inoue M, et al. Egr2 and Egr3 in regulatory T cells cooperatively control systemic autoimmunity through Ltbp3-mediated TGF-β3 production. Proc Natl Acad Sci USA. 2016;113(50):E8131-E8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carpenter AC, Wohlfert E, Chopp LB, et al. Control of regulatory T cell differentiation by the transcription factors Thpok and LRF. J Immunol. 2017;199(5):1716-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463(7282):808-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.La Trobe University. Bundoora, Victoria. ExoCarta: Exosome protein, RNA and lipid database. http://exocarta.org/index.html. Accessed 27 July 2021.
- 60.Lee S-W, Park Y, So T, et al. Identification of regulatory functions for 4-1BB and 4-1BBL in myelopoiesis and the development of dendritic cells. Nat Immunol. 2008;9(8):917-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rikkert LG, Beekman P, Caro J, et al. Cancer-ID: toward identification of cancer by tumor-derived extracellular vesicles in blood. Front Oncol. 2020;10:608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.