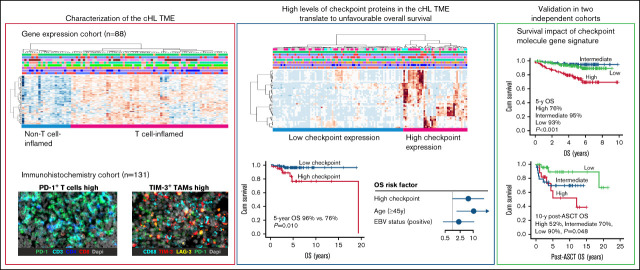
Key Points
Composition of the cHL microenvironment is heterogeneous and differs in the proportions of T cells and macrophages.
High T cell and macrophage-mediated checkpoint protein expression in the cHL microenvironment associates with inferior OS.
Visual Abstract
Abstract
Emerging evidence indicates a major impact for the tumor microenvironment (TME) and immune escape in the pathogenesis and clinical course of classical Hodgkin lymphoma (cHL). We used gene expression profiling (n = 88), CIBERSORT, and multiplex immunohistochemistry (n = 131) to characterize the immunoprofile of cHL TME and correlated the findings with survival. Gene expression analysis divided tumors into subgroups with T cell-inflamed and -noninflamed TME. Several macrophage-related genes were upregulated in samples with the non–T cell-inflamed TME, and based on the immune cell proportions, the samples clustered according to the content of T cells and macrophages. A cluster with high proportions of checkpoint protein (programmed cell death protein 1, PD-1 ligands, indoleamine 2,3 dioxygenase 1, lymphocyte-activation gene 3, and T-cell immunoglobulin and mucin domain containing protein 3) positive immune cells translated to unfavorable overall survival (OS) (5-year OS 76% vs 96%; P = .010) and remained an independent prognostic factor for OS in multivariable analysis (HR, 4.34; 95% CI, 1.05-17.91; P = .043). cHL samples with high proportions of checkpoint proteins overexpressed genes coding for cytolytic factors, proposing paradoxically that they were immunologically active. This checkpoint molecule gene signature translated to inferior survival in a validation cohort of 290 diagnostic cHL samples (P < .001) and in an expansion cohort of 84 cHL relapse samples (P = .048). Our findings demonstrate the impact of T cell- and macrophage-mediated checkpoint system on the survival of patients with cHL.
Introduction
In classical Hodgkin lymphoma (cHL), sparse malignant Hodgkin Reed-Sternberg (HRS) cells are embedded into extensive tumor microenvironment (TME) consisting mostly of benign immune cells, such as T and B lymphocytes, macrophages, eosinophils, plasma cells, and mast cells.1 HRS cells are in close crosstalk with immune cells, and by producing cytokines and chemokines, they recruit immune cells with an aim of creating immunosuppressive, tumor growth–promoting TME.2
Recruitment of inflammatory cells into the TME plays a significant role in the pathogenesis of cHL and survival of HRS cells.1 T lymphocytes composed of CD4+ helper T cells, CD4+ regulatory T cells (Tregs), and CD8+ cytotoxic T cells (CTLs) represent the most numerous cell type of the TME.3,4 cHL TME is especially dominated by CD4+ T cells, which are skewed toward phenotypes of T helper (Th) 2 cells and Tregs.5-7 Tregs inhibit cytotoxic cells and are responsible for sustaining the immunosuppressive state of the tumor.3,7 High CTL content has been shown to translate into poor outcome,8,9 whereas both positive and negative associations of Tregs with survival have been reported.8,10-12 Previous studies have also reported that HRS cells have developed various mechanisms to evade T cell-mediated tumor evasion, one being downregulated or lost HLA expression.13
In addition to T cells, tumor-associated macrophages (TAMs) are an essential component of the cHL TME. They are considered to promote tumor growth and suppress antitumor immunity.14 High TAM content has been demonstrated to translate to poor outcome in cHL,15,16 but there are also studies with contradictory findings.17,18
Immune checkpoint molecules such as programmed cell death protein 1 (PD-1), indoleamine 2,3 dioxygenase 1 (IDO-1), lymphocyte-activation gene 3 (LAG-3), and T-cell immunoglobulin and mucin domain containing protein 3 (TIM-3) restrict immune function, and tumor cells use them to avoid host immune surveillance.13,19 In most cHL cases, HRS cells have a genetic aberration of the CD274/PDCD1LG2 locus of the chromosome 9p24.1, resulting in increased expression of PD-1 ligands (PD-L) 1 and 2 and leading to activation of the PD-1 pathway in T cells.20 Furthermore, increased expression of PD-L1 on HRS cells has been shown to translate to superior progression-free survival after PD-1 blockade,21 whereas high proportion of PD-1+ and PD-L1+ leukocytes in the TME translate to poor event-free survival after standard chemotherapy.22 We have previously shown that the adverse prognostic impact of TAMs on survival depends on PD-L1 and IDO-1 expression.23 In addition, LAG-3+ T cells have recently been identified as important mediators of immune suppression in cHL.24
Altogether, the complex interactions between immune cells and HRS cells have still largely remained uncharacterized. The purpose of our study was to characterize immunological profiles of cHL focusing on checkpoint protein-expressing T cells and TAMs and correlate the findings with clinical characteristics and outcome after standard chemotherapy.
Methods
Patients
The study material consisted of clinical data and formalin-fixed paraffin-embedded diagnostic tumor tissue samples from patients with primary cHL diagnosed between years 1993 and 2012 who were treated or followed at the Helsinki University Hospital (further information provided in supplemental Methods). The patients were divided into 2 cohorts: the first cohort (gene expression cohort) consisted of 88 patients; the second cohort (immunohistochemistry [IHC] cohort) consisted of 131 patients. The study design is shown in supplemental Figure 1. The gene expression cohort consisted of patients who had enough tissue to extract RNA and was designed to be enriched in patients with poor prognosis (elderly patients, patients with relapsed/refractory [R/R] diseases), whereas the IHC cohort consisted of patients from whom enough representative tumor tissue was available for the construction of a tissue microarray (TMA).
The study was approved by the Ethics Committee in Helsinki University Hospital and the Finnish National Authority for Medicolegal Affairs.
Gene expression data
NanoString nCounter digital gene expression profiling with 770-gene PanCancer Immune Profiling Panel (XT-CSO-HIP1-12; NanoString Technologies, Seattle, WA) was used to assess the expression of immune response genes from the 88 tumor samples of the gene expression cohort. The protocol has been described previously.25 After exclusion of internal reference genes and the genes not expressed in >10% of the samples, the final analysis included 706 genes. As validation cohorts, we used gene expression data from 130 diagnostic samples from patients with cHL who were treated at the British Columbia Cancer Agency15 and from 290 diagnostic samples from patients with cHL who were enrolled into E2496 Intergroup trial with available pretreatment biopsies and consent.26 In addition, as an expansion cohort, we used 84 relapse samples from patients with R/R cHL who were treated at the British Columbia Cancer Agency.27
In silico immunophenotyping
To assess the proportions of the distinct immune cells in the TME, we applied CIBERSORT28 for our gene expression cohort and the validation cohort of 130 diagnostic cHL samples.15 CIBERSORT analysis is described in more detail in supplemental Methods.
IHC
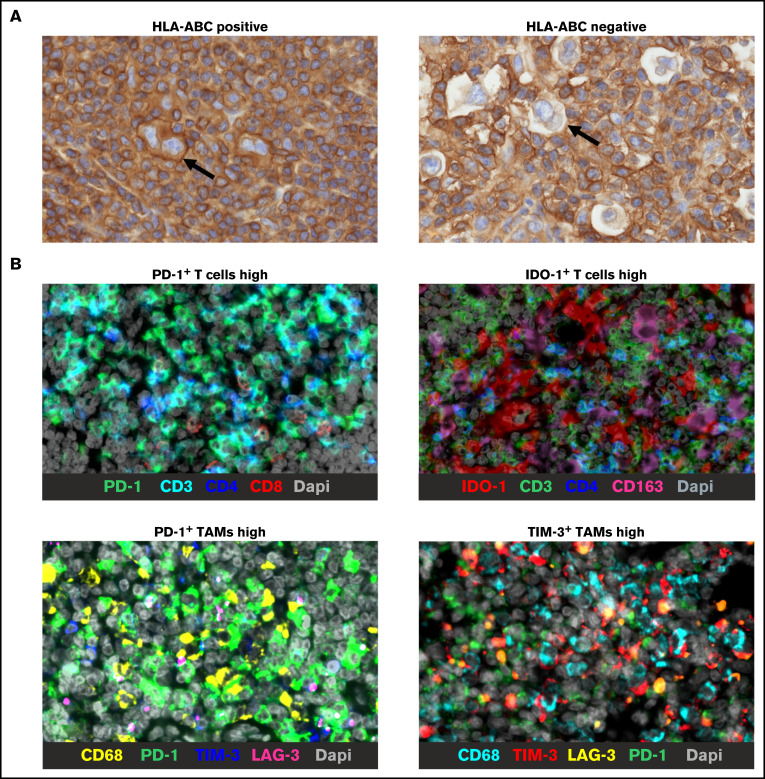
For IHC analysis, the TMAs were stained with primary antibodies for HLA-ABC, β2 microglobulin (B2M), and HLA-DR as previously described.25 Based on the membranous staining of HRS cells, cases were scored either as positive or negative (more detail in supplemental Methods). Examples of representative images of HLA-ABC stainings are shown in Figure 1A.
Figure 1.
Representative IHC images. Representative images of HLA-ABC membrane positive and negative HRS cells (arrows indicate HRS cells) (A) and mIHC stainings showing examples of high proportions of checkpoint positive T cells and TAMs (B). Bars in all images represent 20 µm.
For multiplex IHC (mIHC), TMAs were stained with primary antibody panels, including markers to identify different types of T cells (CD3, CD4, CD8, and FoxP3) and TAMs (CD68 and CD163), checkpoint molecules (PD-1, PD-L1, IDO-1, LAG-3, and TIM-3), and CD30 to recognize HRS cells (supplemental Table 1; Figure 1B). The data analysis was performed digitally with the CellProfiler version 2.2.0.23,29 A detailed description of mIHC procedures is in supplemental Methods.
Statistical analysis
Softwares R (version 4.0.0) and IBM SPSS version 25.0 (IBM, Armonk, NY) were used for statistical analyses. Hierarchical clustering was performed with R pheatmap package using Euclidean distance with ward.D linkage. Pathway analyses were performed with DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov)30 using all genes from the PanCancer Immune Profiling Panel as a background. Pearson χ-square and Fisher’s exact tests were used to evaluate the frequencies of distinct clinical characteristics, and Mann-Whitney U test was used to compare 2 groups. Outcome associations were analyzed by Cox univariable and multivariable analyses. Survival rates were estimated using the Kaplan-Meier method with log-rank test. Forty-five years was used as a cutoff for the comparative age group analysis. Further information on statistical methods is provided in supplemental Methods.
Results
Patient characteristics and outcome
Baseline characteristics and survival are presented in Table 1. As expected, the outcome was worse in the gene expression cohort due to enrichment of the patients with adverse prognostic factors. In the IHC cohort, poor freedom from treatment failure (FFTF) was associated with advanced stage (HR, 8.02; P = .001) whereas overall survival (OS) was inferior in patients with higher age (≥45 years) (HR, 7.73; P = .004) or Epstein-Barr virus (EBV) positivity (HR, 4.10; P = .036) (supplemental Table 2).
Table 1.
Patient baseline characteristics and outcome
| Characteristic | Gene expression cohort n = 88 (%) | IHC cohort n = 131 (%) |
|---|---|---|
| Median follow-up time, mo (range) | 42 (12-164) | 55 (7-229) |
| Age (y) | ||
| Median (range) | 32 (16-84) | 29 (16-83) |
| <45 | 55 (62.5) | 100 (76) |
| ≥45 | 33 (37.5) | 31 (24) |
| Gender | ||
| Male | 43 (49) | 60 (46) |
| Female | 45 (51) | 71 (54) |
| Histological subtype | ||
| Nodular sclerosis | 65 (74) | 102 (78) |
| Mixed cellularity | 20 (23) | 22 (17) |
| Lymphocyte-rich | 2 (2) | 6 (4) |
| Unclassified cHL | 1 (1) | 1 (1) |
| Stage (Ann Arbor classification) | ||
| Limited (I-IIA) | 32 (36) | 56 (43) |
| Advanced (IIB-IV) | 55 (63) | 74 (56) |
| NA | 1 (1) | 1 (1) |
| EBV status | ||
| Negative | 46 (52) | 89 (68) |
| Positive | 26 (30) | 34 (26) |
| NA | 16 (18) | 8 (6) |
| Treatment | ||
| ABVD | 74 (84) | 112 (85) |
| BEACOPPesc | 4 (5) | 9 (7) |
| ABVD+BEACOPPesc | 4 (5) | 4 (3) |
| CHOP | 4 (5) | 4 (3) |
| Other | 2 (2) | 2 (2) |
| Radiotherapy* | 44 (50) | 78 (60) |
| Relapses | 25 (28) | 29 (22) |
| Deaths | 10 (11) | 10 (8) |
| cHL-related deaths | 8 (80) | 6 (60) |
| 5-y FFTF | 66% | 80% |
| 5-y OS | 85% | 91% |
NA, not assigned.
Including chemo- and radiotherapy and radiotherapy only.
Gene expression analysis identified a gene signature enriched for T-cell signaling-related genes
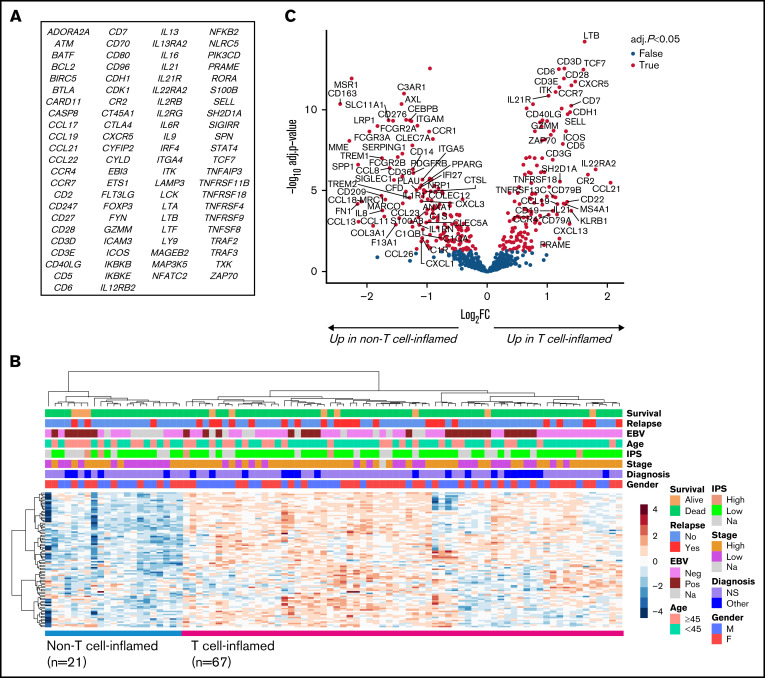
Unsupervised hierarchical clustering of the 706 genes from the NanoString analysis revealed 6 gene signatures with differential expression among cHL samples (supplemental Figure 2). According to pathway analysis, these signatures were enriched for genes related to T-cell signaling, cytokines, viral infections/carcinogenesis, B-cell signaling, focal adhesion/extracellular matrix interactions, and antigen processing and presentation. We chose to focus on the T-cell signaling signature (Figure 2A), consisting of 90 genes, most significantly associated with “co-stimulatory signal during T-cell activation” (P < .001) and “T-cell receptor signaling” (P < .001) pathways (supplemental Table 3). Reclustering the T-cell signature genes according to their expression levels separated patients into 2 groups with high (T cell inflamed; n = 67) and low (non–T cell inflamed; n = 21) expression (Figure 2B). Regarding clinical baseline characteristics, high stage was associated with T cell-inflamed TME (P = .026), whereas gender, age, histological subtype, and EBV status were equally distributed between the 2 subgroups (supplemental Table 4). In addition, the T-cell signature did not translate to FFTF (P = .121) or OS (P = .389; data not shown).
Figure 2.
T-cell signature divides the cHL TME into T cell-inflamed and -noninflamed groups. (A) List of 90 genes included in the T-cell signature. (B) Reclustering the T-cell signature genes separates patients into groups with T cell-inflamed (n = 67) and –noninflamed (n = 21) cHL TME. (C) Volcano plot showing differentially expressed genes between the samples with T cell-inflamed and -noninflamed TME. Named genes represent those with absolute log2 fold change ≥1 and adjusted P < .05.
As expected, differential gene expression analysis between the T cell-inflamed and -noninflamed subgroups revealed genes related to T-cell receptors and function (eg, CD3D/E, ITK, TCF7, CD6, CD28, CD5, and CD7) to be more expressed in the T cell-inflamed subgroup (Figure 2C; supplemental Table 5). Interestingly, genes associated with B cells (MS4A1, CD19, and CD79A/B) were highly expressed in the same group, whereas several genes related to macrophages (CD163, MSR1, MARCO, MRC1, and SIGLEC1) were upregulated in the non–T cell-inflamed subgroup.
In silico immunophenotyping-based hierarchical clustering separated patients according to T-cell and macrophage composition of the TME
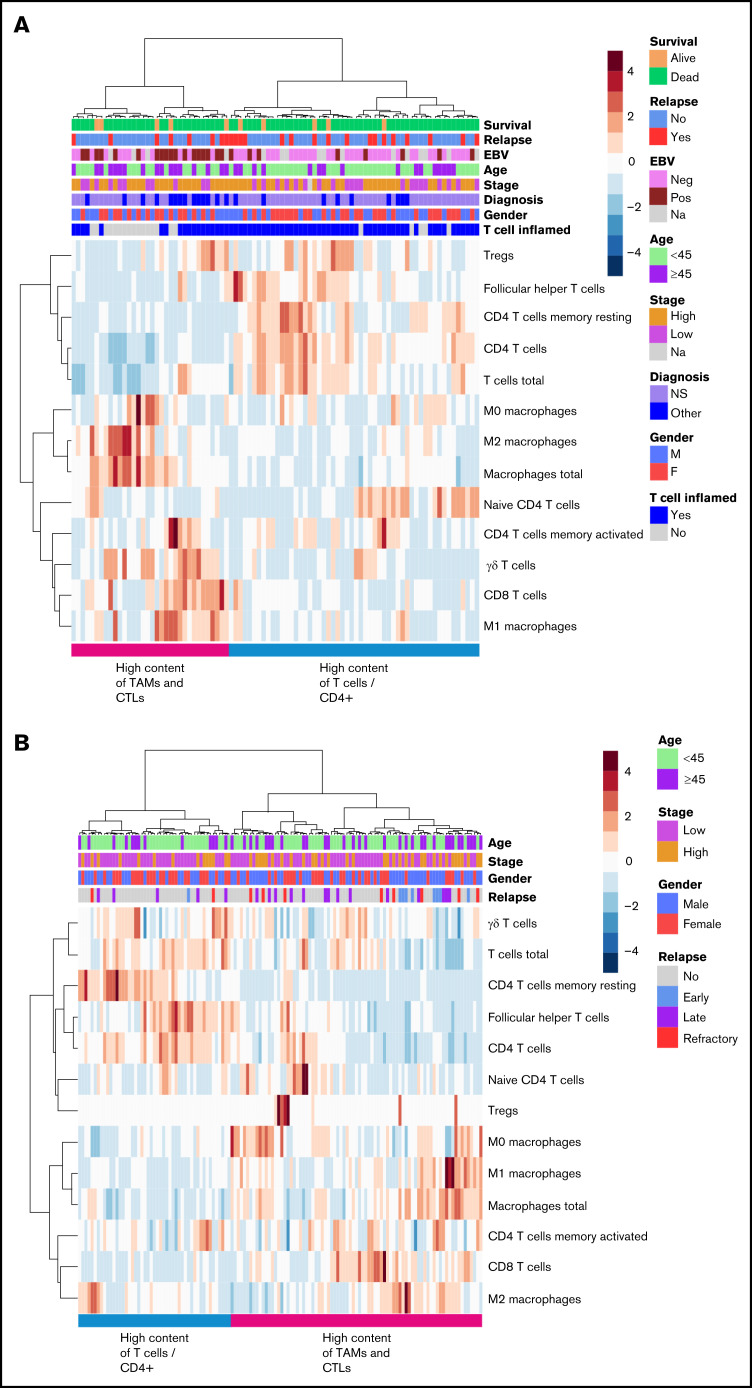
Next, we performed in silico immunophenotyping using CIBERSORT with its built-in signature matrix describing the expression fingerprints of 22 immune cell phenotypes. Hierarchical clustering based on 13 different T-cell and macrophage phenotypes separated patients with cHL into 2 clusters. The first cluster included patients with higher content of TAMs and CTLs in their tumor tissue, and the second cluster encompassed patients with higher content of CD4+ T cells and T cells in general (Figure 3A). In line with this finding, T cells and TAMs (rs = −0.45; P < .001) as well as CD4+ and CD8+ T cells (rs = −0.30; P = .013) correlated inversely with each other. The finding was validated in the cohort of 130 patients with primary cHL (Figure 3B).15
Figure 3.
In silico immunophenotyping of T cells and TAMs. Hierarchical clustering of T-cell and TAM proportions based on in silico immunophenotyping by CIBERSORT using NanoString data of the gene expression cohort (A) and gene expression data from an independent validation cohort (B).
The TME can be divided into four immune signatures based on the contents of T cells and TAMs
To study TME composition in more detail, we performed mIHC with T-cell and macrophage markers. We found a good correlation between gene expression levels and corresponding cell proportions (supplemental Figure 3). In addition, in silico deconvoluted cell proportions correlated well with mIHC data (supplemental Figure 4).
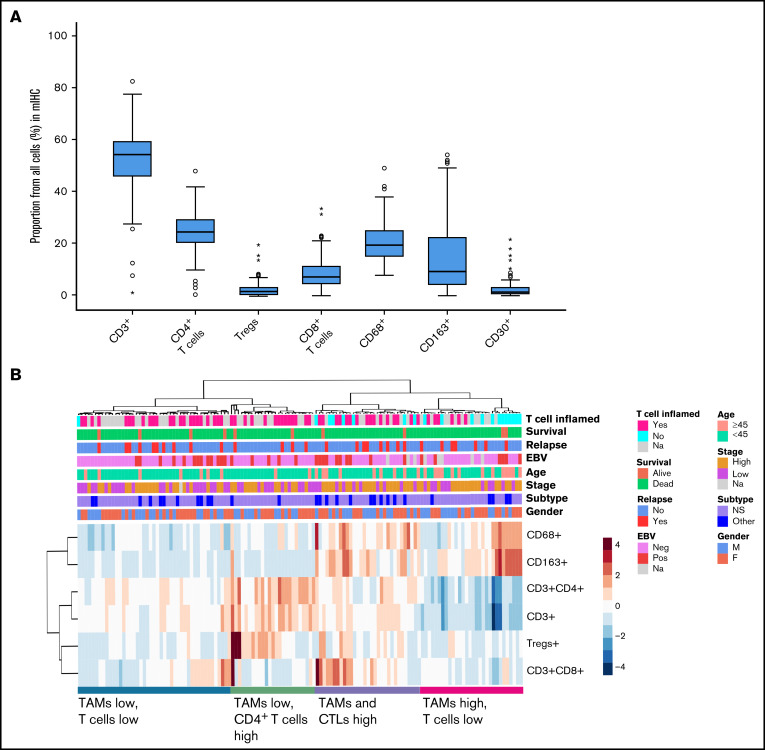
A large proportion of the whole tumor cellularity represented T cells (median 55%, range 1.1% to 83%), the most abundant T-cell type being CD4+ T cells (median 25%, range 0.5% to 48%) (Figure 4A). The median proportion of CTLs was 7.4% (range 0.2% to 34%) and that of Tregs was 1.7% (range 0% to 20%). Furthermore, 20% (range 8.0% to 49%) of the tumor cellularity were TAMs, and approximately half of them were CD163+ M2-like TAMs (median 9.5%, range 0.2% to 54%). The median proportion of CD30+ cells was low (1.5%, range 0.1% to 22%).
Figure 4.
Immunophenotypes of different cell types as determined by mIHC analysis. (A) Median proportions of different cell types in the TME. (B) Hierarchical clustering of T-cell and TAM proportions from all cells (%) divides TME into 4 different immune cell clusters.
Based on hierarchical clustering of the T cells and TAMs, patients were separated into 4 clusters: (1) TAMs high and T cells low, (2) TAMs high and CTLs high, (3) TAMs low and CD4+ T cells high, and (4) TAMs low and T cells low (Figure 4B), further corroborating heterogeneity of cHL TME. Furthermore, the proportion of T cells did not correlate with CD68+ TAMs (rs = −0.11; P = .20) and had an inverse correlation with the proportion of CD163+ TAMs (rs = −0.47; P < .001). Consistent with gene expression data, these clusters did not correlate with survival (FFTF, P = .839; OS, P = .996). Higher age (≥45 years) was associated with higher proportion of TAMs, whereas other clinical characteristics (gender, cHL subtype, stage, EBV status) were not significantly different according to the TAM and T-cell proportions. Instead, a T cell-inflamed TME was more common in patients with low TAM contents (supplemental Table 6).
Association of HRS cells’ membranous HLA complexes with T cells
HLA-ABC and B2M are components of HLA I, and as expected, their expression correlated well with each other (rs = 0.60; P < .001), whereas expression of HLA-DR correlated neither with the expression of HLA-ABC (rs = 0.03; P = .758) nor with the expression of B2M (rs = 0.13; P = .216). HRS cells had lost their membranous HLA-ABC from 76 (70%), B2M from 88 (80%), and HLA-DR from 67 (63%) cases, which agreed with previous findings.31 CTL counts were higher both in HLA-ABC (P = .034) and B2M (P < .001) positive cases as compared with negative ones (supplemental Figure 5A-B), whereas there was no correlation between CD4+ T cells and HLA I complexes. Furthermore, HLA-DR expression did not correlate with CD4+ T cell or CTL counts, and HLA-ABC, B2M, and HLA-DR expression did not correlate with survival (data not shown).
Checkpoint expression in T cells and TAMs
Next, we characterized the immunophenotypes of T cells and TAMs according to their PD-1, PD-L1, IDO-1, LAG-3, and TIM-3 checkpoint molecule expression. We first discovered that most of the studied immunosuppressive molecules were expressed in T cells (supplemental Figure 6). Interestingly, 47% of IDO-1+ cells were T cells, although previously IDO-1 expression has been described mainly in macrophages and dendritic cells.32 Compared with T cells, a smaller proportion of the checkpoint molecule–expressing cells were CD68+ TAMs (supplemental Figure 6).
We also studied the distribution of checkpoint molecule expression between CTLs and CD4+ T cells and found that CTLs expressed more frequently PD-1 (P = .003), IDO-1 (P < .001), and TIM-3 (P = .005) than CD4+ T cells (supplemental Figure 7A). Likewise, the expression of PD-1 and IDO-1 was higher in CD68+ TAMs than in M2-like CD163+ TAMs (PD-1, P = .043; IDO-1, P < .001), whereas no difference was seen in PD-L1, LAG-3, and TIM-3 expression (supplemental Figure 7B).
HLA-ABC–positive cases correlated with higher proportions of IDO-1+ (P = .022) and LAG-3+ (P = .007) CTLs, whereas B2M-positive cases associated with higher proportions of PD-1+ (P < .002), IDO-1+ (P < .001), LAG-3+ (P < .001), and TIM-3+ (P < .001) CTLs. Consistent with previous results,24 HLA-DR–negative cases correlated only with high proportion of LAG-3+CD4+ T cells (P = .039) (supplemental Figure 5A-C).
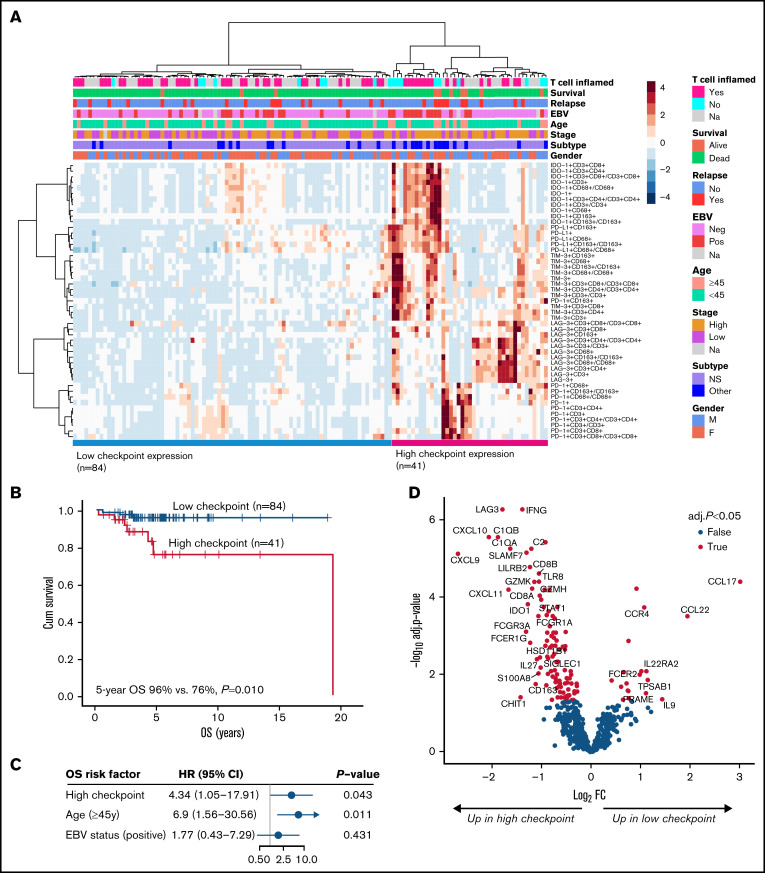
High overall expression of checkpoint molecules predicts inferior OS
When data for the checkpoint molecule–expressing T cells and TAMs were clustered, 2 distinct groups with high and low checkpoint molecule expression were identified (Figure 5A). A subgroup of mainly EBV-positive cHLs had higher expression of IDO-1, PD-L1, and TIM-3, whereas LAG-3 positivity was associated with EBV negativity. A smaller subgroup of cases had high expression of PD-1 but lacked the expression of other checkpoint molecules. Interestingly, high checkpoint expression translated to unfavorable OS (5-year OS 76% vs 96%; P = .010) (Figure 5B), whereas no correlation with FFTF was seen (P = .586, data not shown). High checkpoint expression associated with other than nodular sclerosis (NS) cHL subtype (P = .017) and B2M positivity (P = .009) (supplemental Table 7), but the adverse prognostic impact on OS was independent of the subtype (HR, 4.78; P = .030) and B2M status (HR, 4.79; P = .039) (supplemental Table 8). In multivariable analysis with other prognostic factors for OS (age and EBV status), the adverse prognostic impact of high checkpoint expression on poor OS was sustained (Figure 5C).
Figure 5.
Checkpoint expression in the TME according to mIHC analysis and association with survival. (A) Hierarchical clustering of all distinct checkpoint molecule-expressing cells, including T cells and TAMs (proportions from all cells or from specific cells). (B) OS according to high and low expression of the checkpoint molecule clusters. (C) Forest plot visualizing the impact of checkpoint molecule cluster on OS in multivariable analysis. (D) Volcano plot showing differentially expressed genes between patients with high and low checkpoint molecule cluster. The highlighted 34 genes represent those with absolute log2 fold change ≥1 and adjusted P < .05.
As expected, differential gene expression analysis with the NanoString gene expression cohort revealed high levels of the checkpoint molecule–encoding genes (LAG3, IDO1) in the tumor samples with high number of checkpoint molecule–expressing immune cells (Figure 5D; supplemental Table 9). In addition, they had higher expression of genes related to markers of cytotoxicity (CD8A/B, GZMK/H, CXCL9, CXCL10, and CXCL11) and macrophages (CD163 and SIGLEC1), probably reflecting high checkpoint molecule positivity in T cells (especially in CTLs) and in TAMs, but hypothetically also reflecting the composition of the TME surrounding the checkpoint-expressing cells. Similar to gene expression data, high checkpoint protein levels were associated with higher content of CTLs (P < .001) and CD163+ M2-like TAMs (P = .016) and lower content of Tregs (P = .085). In patients with low checkpoint protein expression, upregulated genes included CCL17 and CCL22, which are highly expressed by HRS cells and recruit Th2 cells and Tregs,33 providing rationale for lower count of Tregs in that patient group. Of note, the proportion of HRS cells was equally distributed between the samples with low and high checkpoint protein expression (data not shown).
Gene expression levels of the immunosuppressive cytokine IL27 were elevated in the samples with high checkpoint molecule expression, supporting the hypothesis that checkpoint-expressing cells are dysfunctional and exhausted. Controversially, the same group also had increased expression of T-cell activation marker IFNG, which has divergent roles, with both pro- and antitumoral effects.34
The gene expression profile associated with high checkpoint molecule TME predicts survival of patients with cHL
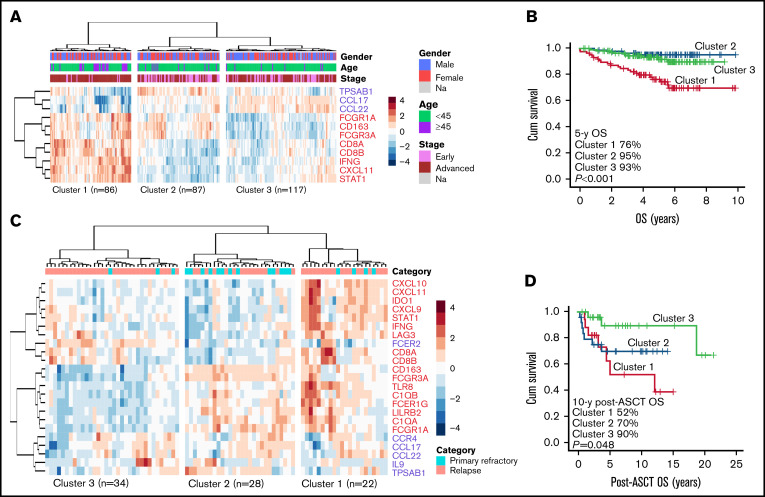
To validate the association of checkpoint molecule phenotype with survival, we used gene expression data from an independent cHL patient cohort consisting of 290 diagnostic samples.26 We used the gene signature, which was differentially expressed based on the checkpoint molecule expression in our IHC cohort (fold change ≥2; adjusted P < .05; supplemental Table 9), and performed hierarchical clustering with 11 genes overlapping in the validation cohort. This resulted in separation of the patients into 3 clusters (Figure 6A). Interestingly, patients with higher expression of the gene signature corresponding to high checkpoint molecule expression (Cluster 1) had worse OS (P < .001) compared with those with lower expression (Figure 6B), verifying the association of high checkpoint molecule proportions with poor outcome. Similarly, in an independent expansion cohort with 84 cHL relapse samples,27 the gene signature (23 overlapping genes) separated the patients into 3 clusters (Figure 6C). Higher expression of the signature corresponding to high checkpoint molecule proportions (Clusters 1 and 2) associated with inferior post-autologous stem cell transplantation (post-ASCT) OS in 69 patients with ASCT as a treatment of R/R disease (P = .048) (Figure 6D).
Figure 6.
The gene expression profile associated with high checkpoint molecule expression in the TME predicts survival of patients with cHL. (A) Hierarchical clustering of diagnostic samples in the cHL validation cohort (n = 290) using the checkpoint molecule gene signature (11 overlapping genes) separated the samples into 3 clusters: Cluster 1 corresponds to higher, Cluster 2 to intermediate, and Cluster 3 to lower checkpoint expression. Early stage was defined as stages I to II and advanced stage as III to IV. (B) Kaplan-Meier estimates of OS according to the checkpoint signature clusters in the validation cohort. (C) Hierarchical clustering of relapse samples in the cHL expansion cohort (n = 84) using the checkpoint molecule gene signature (23 overlapping genes) separated the samples into 3 clusters: Cluster 1 corresponds to higher, Cluster 2 to intermediate, and Cluster 3 to lower checkpoint expression. Primary refractory disease was defined as progression during primary treatment or within 3 months after it ended. (D) In the expansion cohort, 69 of 84 patients received ASCT as a treatment of R/R disease. Kaplan-Meier estimates demonstrating post-ASCT OS according to the checkpoint signature clusters.
Discussion
The TME plays a significant role in supporting tumor growth in cHL, raising interest in exploring new predictive biomarkers from benign TME cells with the aim of developing more effective targeted treatment strategies. Here, we characterized the immune cell composition of the cHL TME and correlated the findings with clinical course and survival of patients.
Based on the gene expression profiling, cHLs could be separated into subgroups with T cell-inflamed or -noninflamed TME, emphasizing the heterogeneity of the tumor-infiltrating T-cell composition. Interestingly, cHLs with non–T cell-inflamed TME overexpressed genes related to macrophages, further demonstrating that TME consists of either high content of T cells or TAMs. Consistent with this finding, in silico and mIHC-based cell immunophenotypes separated the cHL samples into distinct clusters with varying T-cell and TAM contents. Altogether, cell phenotype data strengthen the hypothesis that cHL TME is enriched either in T cells or TAMs. These data also are consistent with a recent whole-slide image analysis showing that CD3+ and CD68+ cells correlate only weakly with each other.4
According to previous observations, EBV status can affect the cellular composition of the tumor. Particularly high proportion of TAMs16,35 and activated CD8+ T cells have been associated with EBV positivity.36 We did not find any correlation between EBV status and immune cell clusters, suggesting that EBV status does not drive the TME diversity between these clusters; nor did we find association between histological subtypes and immune clusters. Instead, the patients older than 45 years had increased TAM content in their tumors; nonetheless, these 4 immune clusters did not translate into outcome differences. Considering that TAM and CTL proportions seem to be high in the same cHL samples, the adverse prognostic impact of high CTL content reported in some previous studies8,9 might also reflect high TAM content.
While checkpoint molecules are generally expressed in nonmalignant immune cells19 in the tumor tissue, they can also be expressed in malignant cells. In cHL, HRS cells express high levels of PD-L120,21,37 and, to some extent, TIM-3,38 but rarely LAG-338 and not IDO-1.39 IDO-1 is expressed particularly in antigen-presenting macrophages and dendritic cells.32 However, we found that in cHL, IDO-1 is expressed in T cells more frequently than in TAMs, similarly to PD-1, LAG-3, and TIM-3. This comparison could not be made for PD-L1 because the PD-L1 antibody was not included in the same panel with T-cell markers, but we found that almost half of the PD-L1 is expressed in TAMs, which is slightly less than previously reported.40
Clustering of the checkpoint molecule–expressing cells revealed that different checkpoint molecules were highly expressed in separate samples. In cHL samples with increased PD-1 expression, other checkpoints proteins (PD-L1, IDO-1, LAG-3, and TIM-3) were expressed at lower levels, whereas IDO-1+, PD-L1+, and TIM-3+ cases clustered together. The clustering demonstrated that expression of each checkpoint molecule was independent of the cell type. Interestingly, high expression of at least 1 of the studied checkpoint molecules translated to unfavorable OS. This finding emphasizes that the impact of checkpoint-mediated immunosuppression in cHL is clinically meaningful. High checkpoint molecule expression was associated with other than nodular sclerosis histology and B2M positivity, and it remained an independent prognostic factor in multivariable model and, after stratification, with unbalanced baseline characteristics, histological subtype, and B2M status. Validation of the adverse prognostic impact of checkpoint molecule cluster on survival based on the corresponding gene expression profiles in independent cohorts of patients with both primary and relapsed cHL underlines the clinical importance of the findings.
We did not find any correlation between T-cell signature and checkpoint molecule expression. However, the genes, which have previously been associated with the cytolytic score in B-cell lymphomas and in cHL,41 were upregulated in the samples with high checkpoint molecule expression. In addition, GZMH and GZMK, which encode proteins expressed in natural killer cells, were among the upregulated genes. The findings suggest that tumors having high checkpoint protein expression are paradoxically also immunologically hot tumors with high cytolytic activity, and by expressing checkpoint molecules, these tumors might protect themselves from host active immune defense. This finding also provides a potential explanation for the association of B2M positivity and high checkpoint expression because CTL response requires HLA I-mediated antigen presentation. However, previous studies have demonstrated that in cHL, PD-1 blockade does not associate with CTL response because instead of the HLA I status, HLA II positivity predicts the response to PD-1 antibody in R/R cHL.21 Additionally, anti–PD-1–based treatment at first line does not activate cytotoxic immune response.42 Furthermore, PD-L1+ HRS cells are in closer proximity to PD-1+CD4+ T cells than PD-1+CD8+ T cells.40 Increased expression of CCL17 and CCL22 in cHLs samples with low checkpoint expression might in turn reflect more pronounced Treg- than checkpoint-mediated immune evasion mechanisms. Similarly, previous studies have found 2 complementary immunosuppression mechanisms in cHL consisting of active PD-1- Th1 Tregs and exhausted PD-1+ Th1 effector cells.43
The accurate role and origin of upregulated IFNG expression in patients with high checkpoint expression remains to be shown. In addition to cytolytic activity, IFNG expression might reflect the variable functional status of the checkpoint protein–expressing cells. On the other hand, IFNG is known to promote tumor progression by inducing expression of PD-L1 and triggering IDO-1 activity.44
In phase I/II trials, 65% to 87% of the patients with R/R cHL have responded to PD-1 inhibition with nivolumab or pembrolizumab.45-48 Earlier preclinical studies have demonstrated that anti–LAG-3 or anti–TIM-3 can act synergistically with anti–PD-1 and enhance each other’s effects.49,50 On the other hand, it has been reported recently that PD-1 is not coexpressed in most LAG3+CD4+ T cells in cHL.24 Furthermore, according to our results, distinct checkpoint proteins are expressed in separate cHLs, proposing that patients might benefit from targeting different checkpoint molecules and that potential synergy of PD-1 and LAG-3/TIM-3 inhibition might not be useful in treatment of cHL. Further studies are needed to answer this question. The finding that high checkpoint molecule expression associates with inferior OS with no impact on FFTF implies that it predicts failure to salvage rather than to primary therapy and that it might be beneficial to combine checkpoint inhibitors with primary chemotherapy in patients with high checkpoint molecule expression.
Taken together, our findings provide novel, more accurate information on the composition of different immune cells, checkpoint molecules, and their relationship in the heterogeneous cHL TME. Furthermore, the data recognize the prognostic impact of T cell- and TAM-mediated checkpoint molecules on the survival of patients with cHL.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the DNA Sequencing and Genomics Laboratory unit, Institute of Biotechnology, University of Helsinki, Finland, for the NanoString analyses and Annabrita Schoonenberg (Institute for Molecular Medicine Finland) and the Digital and Molecular Pathology Unit supported by Helsinki University and Biocenter Finland for the mIHC stainings. Anne Aarnio is acknowledged for technical assistance.
Funding support for this article was provided by the grants from the Academy of Finland (S.L.), Finnish Cancer Organizations (S.L.), Sigrid Juselius Foundation (S.L.), University of Helsinki (S.L.), Helsinki University Hospital (S.L.), University of Helsinki, Doctoral Programme in Clinical Research (K.K.), Finnish Society for Oncology (K.K.), and Orion Research Foundation (K.K.). Open acces funded by Helsinki University Library.
Authorship
Contribution: K.K., S.-K.L., T.P., and S.L. conceptualized the study; M.-L.K.-L., C.S., and F.C.C. provided the materials; K.K., S.-K.L., M.-L.K.-L., T.P., and S.L. designed the methodology; K.K., S.-K.L., and T.P. created the formal analysis; T.P. and S.L. supplied the resources; K.K., S.-K.L., and S.L. prepared and wrote the original manuscript; K.K., S.-K.L., C.S., F.C.C., T.P., and S.L. reviewed and edited the manuscript; S.L. supervised the study; and all authors read and agreed to the published version of the manuscript.
Conflict-of-interest disclosure: S.L. received research funding from Bayer, Celgene, Genmab, Novartis, Roche, and Takeda outside the submitted work; received honoraria as a consultant for Incyte, Novartis, Roche, Takeda, and Merck; and is a consultant for Orion. K.K. received travel and conference expenses from Sanofi-Genzyme, Janssen-Cilag, MSD, and Gilead. C.S. is a consultant for Seattle Genetics, Curis Inc., Roche, AbbVie, Juno Therapeutics, and Bayer; received research funding from Bristol Myers Squibb, Epizyme, and Trillium Therapeutics; and is a co-inventor on a patent (“Method for determining lymphoma type”) using NanoString technology. The remaining authors declare no competing financial interests.
Correspondence: Sirpa Leppä, Department of Oncology, Haartmaninkatu 8, FI-0014 University of Helsinki, Helsinki, Finland; e-mail: sirpa.leppa@helsinki.fi.
References
- 1.Küppers R. The biology of Hodgkin’s lymphoma. Nat Rev Cancer. 2009;9(1):15-27. [DOI] [PubMed] [Google Scholar]
- 2.Aldinucci D, Borghese C, Casagrande N. Formation of the immunosuppressive microenvironment of classic Hodgkin lymphoma and therapeutic approaches to counter it. Int J Mol Sci. 2019;20(10):2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wein F, Küppers R. The role of T cells in the microenvironment of Hodgkin lymphoma. J Leukoc Biol. 2016;99(1):45-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jachimowicz RD, Pieper L, Reinke S, et al. Whole-slide image analysis of the tumor microenvironment identifies low B-cell content as a predictor of adverse outcome in patients with advanced-stage classical Hodgkin lymphoma treated with BEACOPP. Haematologica. 2021;106(6):1684-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wein F, Weniger MA, Höing B, et al. Complex immune evasion strategies in classical Hodgkin lymphoma. Cancer Immunol Res. 2017;5(12):1122-1132. [DOI] [PubMed] [Google Scholar]
- 6.Marshall NA, Christie LE, Munro LR, et al. Immunosuppressive regulatory T cells are abundant in the reactive lymphocytes of Hodgkin lymphoma. Blood. 2004;103(5):1755-1762. [DOI] [PubMed] [Google Scholar]
- 7.Re D, Küppers R, Diehl V. Molecular pathogenesis of Hodgkin’s lymphoma. J Clin Oncol. 2005;23(26):6379-6386. [DOI] [PubMed] [Google Scholar]
- 8.Alvaro T, Lejeune M, Salvadó MT, et al. Outcome in Hodgkin’s lymphoma can be predicted from the presence of accompanying cytotoxic and regulatory T cells. Clin Cancer Res. 2005;11(4):1467-1473. [DOI] [PubMed] [Google Scholar]
- 9.Koreishi AF, Saenz AJ, Persky DO, et al. The role of cytotoxic and regulatory T cells in relapsed/refractory Hodgkin lymphoma. Appl Immunohistochem Mol Morphol. 2010;18(3):206-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollander P, Rostgaard K, Smedby KE, et al. An anergic immune signature in the tumor microenvironment of classical Hodgkin lymphoma is associated with inferior outcome. Eur J Haematol. 2018;100(1):88-97. [DOI] [PubMed] [Google Scholar]
- 11.Greaves P, Clear A, Coutinho R, et al. Expression of FOXP3, CD68, and CD20 at diagnosis in the microenvironment of classical Hodgkin lymphoma is predictive of outcome. J Clin Oncol. 2013;31(2):256-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schreck S, Friebel D, Buettner M, et al. Prognostic impact of tumour-infiltrating Th2 and regulatory T cells in classical Hodgkin lymphoma. Hematol Oncol. 2009;27(1):31-39. [DOI] [PubMed] [Google Scholar]
- 13.Vardhana S, Younes A. The immune microenvironment in Hodgkin lymphoma: T cells, B cells, and immune checkpoints. Haematologica. 2016; 101(7):794-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steidl C, Lee T, Shah SP, et al. Tumor-associated macrophages and survival in classic Hodgkin’s lymphoma. N Engl J Med. 2010;362(10): 875-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamper P, Bendix K, Hamilton-Dutoit S, Honoré B, Nyengaard JR, d’Amore F. Tumor-infiltrating macrophages correlate with adverse prognosis and Epstein-Barr virus status in classical Hodgkin’s lymphoma. Haematologica. 2011;96(2):269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azambuja D, Natkunam Y, Biasoli I, et al. Lack of association of tumor-associated macrophages with clinical outcome in patients with classical Hodgkin’s lymphoma. Ann Oncol. 2012;23(3):736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kayal S, Mathur S, Karak AK, et al. CD68 tumor-associated macrophage marker is not prognostic of clinical outcome in classical Hodgkin lymphoma. Leuk Lymphoma. 2014;55(5):1031-1037. [DOI] [PubMed] [Google Scholar]
- 19.Ok CY, Young KH. Checkpoint inhibitors in hematological malignancies. J Hematol Oncol. 2017;10(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roemer MG, Advani RH, Ligon AH, et al. PD-L1 and PD-L2 genetic alterations define classical Hodgkin lymphoma and predict outcome. J Clin Oncol. 2016;34(23):2690-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roemer MGM, Redd RA, Cader FZ, et al. Major histocompatibility complex class II and programmed death ligand 1 expression predict outcome after programmed death 1 blockade in classic Hodgkin lymphoma. J Clin Oncol. 2018;36(10):942-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollander P, Kamper P, Smedby KE, et al. High proportions of PD-1+ and PD-L1+ leukocytes in classical Hodgkin lymphoma microenvironment are associated with inferior outcome. Blood Adv. 2017;1(18):1427-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karihtala K, Leivonen SK, Brück O, et al. Prognostic impact of tumor-associated macrophages on survival is checkpoint dependent in classical Hodgkin lymphoma. Cancers (Basel). 2020;12(4):877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoki T, Chong LC, Takata K, et al. Single-cell transcriptome analysis reveals disease-defining T-cell subsets in the tumor microenvironment of classic Hodgkin lymphoma. Cancer Discov. 2020;10(3):406-421. [DOI] [PubMed] [Google Scholar]
- 25.Leivonen SK, Pollari M, Brück O, et al. T-cell inflamed tumor microenvironment predicts favorable prognosis in primary testicular lymphoma. Haematologica. 2019;104(2):338-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott DW, Chan FC, Hong F, et al. Gene expression-based model using formalin-fixed paraffin-embedded biopsies predicts overall survival in advanced-stage classical Hodgkin lymphoma. J Clin Oncol. 2013;31(6):692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan FC, Mottok A, Gerrie AS, et al. Prognostic model to predict post-autologous stem-cell transplantation outcomes in classical Hodgkin lymphoma. J Clin Oncol. 2017;35(32):3722-3733. [DOI] [PubMed] [Google Scholar]
- 28.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carpenter AE, Jones TR, Lamprecht MR, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44-57. [DOI] [PubMed] [Google Scholar]
- 31.Roemer MG, Advani RH, Redd RA, et al. Classical Hodgkin lymphoma with reduced β2M/MHC class I expression is associated with inferior outcome independent of 9p24.1 status. Cancer Immunol Res. 2016;4(11):910-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F, Zhang R, Li S, Liu J. IDO1: an important immunotherapy target in cancer treatment. Int Immunopharmacol. 2017;47:70-77. [DOI] [PubMed] [Google Scholar]
- 33.Goncharova O, Flinner N, Bein J, et al. Migration properties distinguish tumor cells of classical Hodgkin lymphoma from anaplastic large cell lymphoma cells. Cancers (Basel). 2019;11(10):1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhat MY, Solanki HS, Advani J, et al. Comprehensive network map of interferon gamma signaling. J Cell Commun Signal. 2018;12(4):745-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hohaus S, Santangelo R, Giachelia M, et al. The viral load of Epstein-Barr virus (EBV) DNA in peripheral blood predicts for biological and clinical characteristics in Hodgkin lymphoma. Clin Cancer Res. 2011;17(9):2885-2892. [DOI] [PubMed] [Google Scholar]
- 36.Wu R, Sattarzadeh A, Rutgers B, Diepstra A, van den Berg A, Visser L. Erratum: The microenvironment of classical Hodgkin lymphoma: heterogeneity by Epstein-Barr virus presence and location within the tumor. Blood Cancer J. 2018;8:9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green MR, Monti S, Rodig SJ, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116(17):3268-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Halabi L, Adam J, Gravelle P, et al. Expression of the immune checkpoint regulators LAG-3 and Tim-3 in classical Hodgkin lymphoma. Clin Lymphoma Myeloma Leuk. 2020;4:257-266.e253. [DOI] [PubMed] [Google Scholar]
- 39.Choe JY, Yun JY, Jeon YK, et al. Indoleamine 2,3-dioxygenase (IDO) is frequently expressed in stromal cells of Hodgkin lymphoma and is associated with adverse clinical features: a retrospective cohort study. BMC Cancer. 2014;14:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carey CD, Gusenleitner D, Lipschitz M, et al. Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood. 2017;130(22):2420-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dufva O, Pölönen P, Brück O, et al. Immunogenomic landscape of hematological malignancies. Cancer Cell. 2020;38(3):424-428. [DOI] [PubMed] [Google Scholar]
- 42.Reinke S, Bröckelmann PJ, Iaccarino I, et al. Tumor and microenvironment response but no cytotoxic T-cell activation in classic Hodgkin lymphoma treated with anti-PD1. Blood. 2020;136(25):2851-2863. [DOI] [PubMed] [Google Scholar]
- 43.Cader FZ, Schackmann RCJ, Hu X, et al. Mass cytometry of Hodgkin lymphoma reveals a CD4+ regulatory T-cell-rich and exhausted T-effector microenvironment. Blood. 2018;132(8):825-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castro F, Cardoso AP, Goncalves RM, Serre K, Oliveira MJ. Interferon-Gamma at the crossroads of tumor immune surveillance or evasion. Front Immunol. 2018;9:847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015; 372(4):311-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Younes A, Santoro A, Shipp M, et al. Nivolumab for classical Hodgkin’s lymphoma after failure of both autologous stem-cell transplantation and brentuximab vedotin: a multicentre, multicohort, single-arm phase 2 trial. Lancet Oncol. 2016;17(9):1283-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Armand P, Shipp MA, Ribrag V, et al. Programmed death-1 blockade with pembrolizumab in patients with classical Hodgkin lymphoma after brentuximab vedotin failure. J Clin Oncol. 2016;34(31):3733-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen R, Zinzani PL, Fanale MA, et al. ; KEYNOTE-087 . Phase II study of the efficacy and safety of pembrolizumab for relapsed/refractory classic Hodgkin lymphoma. J Clin Oncol. 2017;35(19):2125-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woo SR, Turnis ME, Goldberg MV, et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 2012;72(4):917-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010;207(10):2187-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.