Abstract
Hepatocellular carcinoma (HCC) is one of the most frequently encountered malignant tumor types and to improve its treatment, effective prognostic biomarkers are urgently required. Cell cycle dysregulation is a significant feature of cancer progression. The aim of the present study was to estimate the expression levels of forkhead box protein M1 (FOXM1) and polo-like kinase 1 (PLK1), both of which have essential roles in cell cycle regulation, and determine their prognostic value in HCC. To this end, FOXM1 and PLK1 expression levels were assessed in The Cancer Genome Atlas and International Cancer Genome Consortium Japan HCC cohorts, and the associations between their co-expression were determined via Pearson's correlation analysis. Furthermore, the overall survival and disease-free survival in these cohorts for different FOXM1 and PLK1 expression statuses were analyzed. In vitro knockdown experiments were also performed using Huh7 cells. The results obtained indicated overexpression of FOXM1 and PLK1 in HCC tumor tissues as well as a positive correlation between FOXM1 and PLK1 expression. The results also suggested that both FOXM1 and PLK1 are required for HCC cell proliferation. In addition, upregulation of FOXM1 and PLK1 was indicated to be associated with poor prognosis of patients with HCC. However, only their coordinated overexpression was identified as an independent prognostic factor for HCC.
Keywords: forkhead box protein M1, polo-like kinase 1, hepatocellular carcinoma, cell cycle
Introduction
In 2020, 905,677 patients were diagnosed with liver cancer and this malignancy was accountable for 830,180 cancer-associated mortalities worldwide (1). Hepatocellular carcinoma (HCC), which is a multigene disease with heterogeneous pathological mechanisms and clinical manifestation, accounts for 75–85% of primary liver cancer cases (1) and is a major health problem worldwide. It has been observed that the use of ultrasound monitoring every 6 months (with or without α-fetoprotein for its treatment) is associated with improved early detection as well as improved overall survival (OS); however, in clinical practice, implementation-related limitations frequently result in a high proportion of HCC cases only detected at late stages (2). Furthermore, despite the improvements associated with the use of antiangiogenic drugs and immunotherapy over the past decade, HCC prognosis is limited (3). The major unmet challenges related to HCC treatments include advancements in the treatment at earlier stages of the disease, applying the treatment to patients with liver dysfunction, the discovery and validation of predictive biomarkers and the development of more effective combinatorial or sequential treatment approaches (3,4). Therefore, the mechanisms of HCC require to be explored and the identification of valuable biomarkers is urgently required.
Genome-wide expression profiling has enabled the analysis of patient heterogeneity within a short period. It has been proposed that the expression of numerous genes, including forkhead box protein M1 (FOXM1) (5,6) and polo-like kinase 1 (PLK1) (7,8), may serve as a putative prognostic biomarker for HCC. Although several studies have indicated that FOXM1 and PLK1 overexpression are associated with poor cancer prognosis (9–11), the underlying mechanisms have remained to be fully elucidated. Furthermore, to the best of our knowledge, the effect of the association between FOXM1 and PLK1 on the development and prognosis of HCC has not been reported in any previous study.
FOXM1 belongs to a large family of Fox transcription factors, all of which have a conserved domain attached to DNA (winged helix) (12). Furthermore, it has an important role in regulating cell cycle progression via the stimulation of the genes that are critical for G1-S and G2-M transition, including S-phase kinase-associated protein 2, PLK1, centromeric protein A and survivin (13,14), and is itself regulated during the cell cycle process. Transcriptional activation of FOXM1 depends on cyclin-dependent kinase and PLK1 kinase mediates its phosphorylation (15–17). Furthermore, FOXM1 is frequently expressed at a higher level than normal in a variety of human cancers (18–20). Furthermore, several studies have demonstrated that FOXM1 is a key transcription factor that is associated with HCC (21,22). It has also been reported that elevated FOXM1 expression is associated with a poor prognosis of the disease (23,24).
Cell cycle disorders are essential for tumor development and protein kinases, which have important roles in regulating the cell cycle, are valuable targets for cancer therapy. Specifically, PLK1, a member of the serine/threonine kinase family, promotes cell mitosis in mammalian cells (25,26). It has also been identified as an essential mitotic kinase that controls mitotic entry, spindle assembly, centrosome maturation and cytokinesis (27,28). PLK1 overexpression was reported to cause cell cycle overrides in tumor cells, resulting in the survival, enhanced proliferation and immune evasion of cancer cells (29–31). It has also been observed that its expression is increased in numerous cancer types, including lung, bladder, breast and liver cancers (32–34). In addition, several studies have indicated that PLK1 overexpression may serve as an important prognostic factor for HCC (35,36); however, the underlying associated mechanisms have remained elusive. Selective inhibitors of PLK1 potently cause mitotic arrest and induce tumor cell apoptosis (37–39), indicating that PLK1 is a potential target for antitumor treatment.
PLK1, a target of FOXM1, is required for FOXM1 transcription activation and the formation of a feedback loop (14,16). In addition, PLK1 and FOXM1 are essential for the cell cycle process and are related to HCC prognosis. However, the association between this feedback loop and HCC has remained to be investigated. Therefore, in the present study, the prognostic values of PLK1 and FOXM1 overexpression in HCC were estimated using data from The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium Japan (ICGC JP) HCC cohorts. Furthermore, molecular analyses and cell proliferation assays were performed to examine the role of PLK1 and FOXM1 in Huh7 cells and the results suggested that the feedback loop is required for the proliferation of Huh7 cells.
Materials and methods
Clinical cohorts
Sequencing and clinical data of patients with HCC were obtained from two public cohorts, namely TCGA (http://xena.ucsc.edu) and ICGC (https://dcc.icgc.org). A total of 373 and 243 patients from the TCGA and ICGC JP cohorts, respectively, were included in the analysis. Patients with incomplete OS or disease-free survival (DFS) information were excluded. OS was defined as the time from the date of initial pathological diagnosis to the time of death or last follow-up, while DFS was defined as the time from first treatment to the time of tumor recurrence or death. The TCGA cohort was used as the exploration cohort (clinicopathological information is provided in Table I), while the ICGC JP cohort was used for validation.
Table I.
Baseline of characteristics of the patients (n=373).
| Characteristic | N (%) |
|---|---|
| Age, years | |
| <50 | 70 (18.8) |
| 50-59 | 99 (26.5) |
| 60-69 | 121 (32.4) |
| ≥70 | 83 (22.3) |
| Sex | |
| Male | 252 (67.6) |
| Female | 121 (32.4) |
| Stage | |
| I | 173 (49.6) |
| II | 86 (24.6) |
| III–IV | 90 (25.8) |
| Virus status | |
| None | 199 (56.2) |
| HBV | 99 (28.0) |
| HCV | 48 (13.8) |
| HBV+HCV | 7 (2.0) |
HBV, hepatitis B virus; HCV, hepatitis C virus.
Cell lines and cell culture
The human Huh7 cell line (American Type Culture Collection) was cultured in a growth medium consisting of Dulbecco's Modified Eagle's Medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 4.5 g/l glucose, 10% fetal bovine serum (FBS; Shanghai ExCell Biology, Inc.) and 1% penicillin/streptomycin at 37°C in a humidified atmosphere with 5% CO2.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. Complementary DNA (cDNA) was synthesized using the ReverTra Ace qPCR RT Master Mix with a gDNA Remover kit (Toyobo Life Science) according to the manufacturer's instructions. qPCR was then performed in a 25-µl volume reaction mixture containing 12.5 µl AceQ Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd.), 2 µl template cDNA (100 ng/µl) and 1 µM of primers in a LightCycler 96 (Roche Diagnostics Co., Ltd.). The thermocycling conditions were as follows: 95°C for 10 min, followed by 40 cycles of 94°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec. GAPDH was used as a normalization control. Each experiment was performed independently at least three times and the fold change in the expression of each gene was calculated using the 2−ΔΔCq method (40). Primers for qPCR were obtained from BioSune Biotechnology Co., Ltd. All primers were designed to cross an intron-exon junction sequence to minimize genomic DNA contamination. The primer sequences were as follows: qPCR-PLK1-forward (F), 5′-AAGAGATCCCGGAGGTCCTA-3′ and qPCR-PLK1-reverse (R), 5′-GCTGCGGTGAATGGATATTT-3′; qPCR-FOXM1-F, 5′-CGTGGATTGAGGACCACTTT-3′ and qPCR-FOXM1-R, 5′-TCTGCTGTGATTCCAAGTGC-3′; qPCR-GAPDH-F, 5′-ACAACTTTGGTATCGTGGAAGG-3′ and qPCR-GAPDH-R, 5′-GCCATCACGCCACAGTTTC-3′.
Immunoblotting
Cells were lysed in radioimmunoprecipitation assay cell lysis buffer (Beyotime Institute of Biotechnology) containing protease inhibitors (cat. no. HY-K0010; MedChem Express) and phosphatase inhibitor cocktail (cat. no. 78427; Thermo Fisher Scientific, Inc.). Protein concentrations were determined via a bicinchoninic acid protein assay (Pierce; Thermo Fisher Scientific, Inc.). Proteins were separated using 10% SDS-PAGE [gels using 1X running buffer and transferred to polyvinylidene difluoride membranes (MilliporeSigma)]. Afterwards, the membranes were blocked by 5% skimmed milk (Anchor; Fonterra Co-operative Group) at room temperature for 1 h and incubated with primary antibodies against FOXM1 (rabbit; cat. no. A2493; dilution, 1:1,000; Abclonal), PLK1 (rabbit; cat. no. 208G4; dilution, 1:1,000; Cell Signaling Technology, Inc.) and GAPDH (mouse; cat. no. AC002; dilution, 1:10,000; Abclonal) at 4°C overnight. Subsequently, the membranes were cleaned twice with TBS with 0.1% Tween-20 and incubated with secondary goat anti-rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, HRP (dilution, 1:5,000; cat. no. G21234; Thermo Fisher Scientific, Inc.) or goat anti-mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, HRP (dilution, 1:5,000; cat. no. G21040; Thermo Fisher Scientific, Inc.) at room temperature for 1 h. Proteins were detected via enhanced chemiluminescence (Vazyme Biotech Co., Ltd.) with a digital luminescent image analyzer (Tanon-4200; Tanon Science and Technology Co., Ltd.). The intensities of protein bands were semi-quantified using ImageJ software (ImageJ bundled with 64-bit Java 1.8.0_172; National Institutes of Health).
RNA interference
Human FOXM1 small interfering RNA (siRNA), human PLK1 siRNA and control siRNA were obtained from Shanghai GenePharma, Co., Ltd. The siRNA oligonucleotides were transfected into Huh7 cells (at 70% confluence) using Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. siRNAs were mixed with Opti-MEM reduced serum medium (Gibco; Thermo Fisher Scientific, Inc.) and Lipofectamine RNAiMAX and incubated for 20 min at room temperature. Subsequently, siRNA/Lipofectamine in Opti-MEM was diluted 1:5 (corresponding to a final concentration of 50 nM) in differentiation medium, and added to the cells. Thereafter, cells were incubated for 48 h at 37°C with 5% CO2. The following siRNAs were used for the experiments, which had the following sequences: siFOXM1-1, 5′-GCUGGGAUCAAGAUUAUUATT-3′; siFOXM1-2, 5′-GGCUGCACUAUCAACAAUATT-3′; siPLK1-1, 5′-CCCUCACAGUCCUCAAUAATT-3′; siPLK1-2, 5′-GGCAACCAAAGUCGAAUAUTT-3′; si negative control (NC), 5′-ACGUGACACGUUCGGAGAATT-3′.
Cell counting kit 8 (CCK-8) assay
A CCK-8 kit (Vazyme Biotech Co., Ltd.) was used to measure the proliferation of Huh7 cells. A total of 1,000 cells in a volume of 100 µl per well were cultured in six replicate wells in a 96-well plate in medium containing 10% FBS and 1% penicillin-streptomycin at 37°C with 5% CO2 for 6 h. When cells had adhered, CCK-8 reagent (10 µl) was added to 90 µl DMEM to generate the working solution, of which 100 µl was added per well and incubated for 2 h. This assay was performed at 0, 24, 48, 72 and 96 h. The optical densities were measured at a spectral wavelength of 450 nm using a microplate reader (Thermo Fisher Scientific, Inc.). Six replicates were analyzed for each time point.
5-Ethynyl-2′-deoxyuridine (EdU)-DNA synthesis assay
To measure the DNA replication activity of the Huh7 cells, a Cell-Light EdU Apollo488 in Vitro Kit (cat. no. C10310-3; Guangzhou RiboBio Co., Ltd.) was used. Huh7 cells were seeded in 96-well plates at a density of 8×103 cells per well. After 24 h, the cell culture medium was replaced with 50 µM EdU solution diluted with growth culture medium, followed by incubation for 2 h. The cells were then processed using the Cell-Light EdU Apollo488 in Vitro Kit according to the manufacturer's protocol. Images were captured with an Olympus fluorescence microscope (BX53; Olympus Corporation).
Cell-cycle analysis
A total of 1×106 cells were washed twice with PBS and fixed overnight with 1 ml pre-cooled 75% ethanol at 4°C. Thereafter, cells were collected by centrifugation (500 × g; 5 min; 4°C), washed twice in PBS and incubated with propidium iodide (5 ug/ml, Sigma) and RNase A (0.1 mg/ml; Thermo Fisher Scientific, Inc.) for 30 min at 4°C in the dark. A 40-µm screen filter was then used to filter the cell suspension and remove any adhesive cells. This was followed by flow cytometry to analyze the DNA content using the BD LSRFortessa system (BD Biosciences). FlowJo_v10 software (Tree Star, Inc.) was used to estimate the proportion of cells in the G0/G1, S and G2/M phases.
Pharmacological inhibitor of PLK1
The pharmacological PLK1 inhibitor BI 2536 (HY-50698) was purchased from MedChemExpress. The drugs were reconstituted in DMSO and aliquots were stored at −20°C. An equivalent amount of DMSO was used for each experiment as a vehicle control. For cell proliferation and cell cycle assays, Huh7 cells were incubated with 10 nM BI 2536 for 24 h.
Statistical analysis
Statistical analysis was performed using R software (version 4.0.3) for survival analysis and Cox analysis, and GraphPad Prism (version 8.0; GraphPad Software, Inc.) for others. Kaplan-Meier curves were used to estimate OS and DFS. The log-rank test was used to compare patient survival times between high and low gene expression groups and hazard ratios (HRs) were calculated using the Cox proportional hazards model. The combined expression of FOXM1 and PLK1 was dependent on FOXM1 expression and PLK1 expression, thus they were analyzed respectively in Multivariable Cox analysis. Due to the crossing of survival curves, the ‘TSHRC’ package (v0.1-6; http://CRAN.R-project.org/package=TSHRC) of R software, which is a two-stage procedure for comparing HR functions, particularly suited for situations where HR functions cross, was used to perform a two-stage test rather than the log-rank test (41,42). Pearson's correlation coefficient was calculated to analyze the correlation between FOXM1 and PLK1 expression. Differences between 2 groups were analyzed using the unpaired Student's t-test with or without Welch's correction. One-way ANOVA followed by Tukey's post-hoc test was used for comparisons between multiple groups. Values are expressed as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Baseline characteristics
The TCGA cohort was used as the exploration cohort and the ICGC JP cohort was used for validation. The baseline characteristics of the TCGA data set analyzed in the present study are summarized in Table I. The mean age of TCGA cohort was 59.5 years and the percentage of men was 67.6%. The most prevalent hepatitis virus was HBV. The ICGC JP cohort contained sequencing and clinical data of 243 patients with HCC from Japan. The mean age of the ICGC cohort was 67.5 years and the percentage of men was 74.9%. Most patients presented with primary tumors (98.8%) and approximately half of the patients had stage II tumors (45.3%).
Overexpression of FOXM1 and PLK1 is associated with poor prognosis for HCC
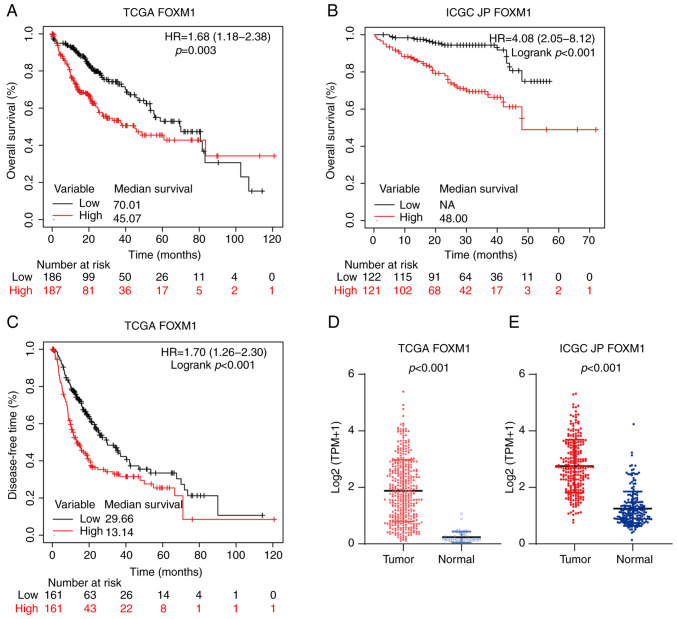
The expression of FOXM1 and PLK1 in HCC and their effect on survival were first analyzed (Fig. 1). Compared with non-tumor liver tissues, the tumor tissues exhibited significantly higher FOXM1 expression in the TCGA (P<0.001; Fig. 1D) and ICGC JP (P<0.001; Fig. 1E) cohorts. To investigate the prognostic value of FOXM1 and PLK1 expression in HCC, the patients with HCC were divided into low and high expression groups at the 50th percentile. In addition, in the TCGA cohort, patients with high FOXM1 expression levels (FH) had shorter OS [HR, 1.68; 95% confidence interval (CI), 1.18-2.38; P=0.003; Fig. 1A] and DFS (HR, 1.70; 95% CI, 1.26-2.30; P<0.001; Fig. 1C). Consistently, FH was associated with poor prognosis in the ICGC JP cohort (HR, 4.08; 95% CI, 2.05-8.12; P<0.001; Fig. 1B).
Figure 1.
Overexpression of FOXM1 is associated with a poor prognosis in HCC. Kaplan-Meier survival curve estimates of overall survival in the (A) TCGA (n=373) and (B) ICGC JP (n=243) cohorts. The patients were divided into two subgroups of high and low FOXM1 expression. (C) Kaplan-Meier survival curve estimates of disease-free survival in the TCGA cohort (n=322). Expression of FOXM1 in HCC and non-tumor liver tissues determined by transcriptome sequencing from the (D) TCGA and (E) ICGC JP datasets. FOXM1, forkhead box protein M1; HR, hazard ratio; TCGA, The Cancer Genome Atlas; ICGC JP, International Cancer Genome Consortium Japan; HCC, hepatocellular carcinoma; NA, not available.
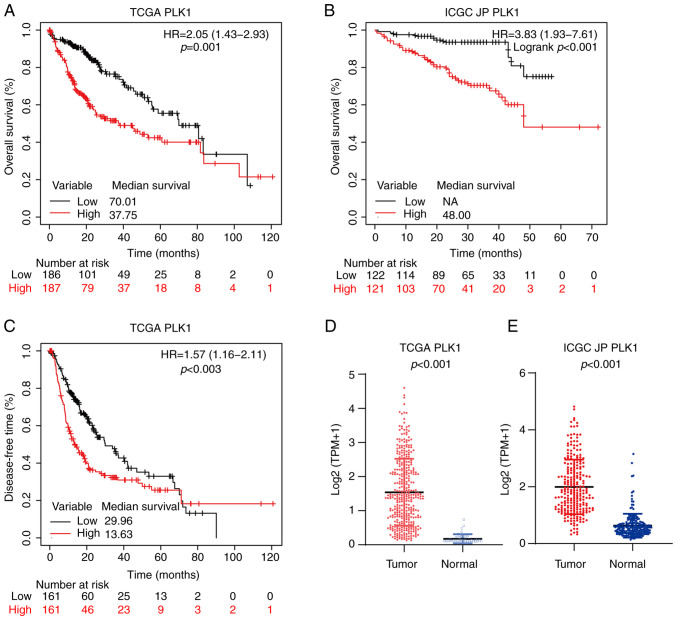
As presented in Fig. 2, similar to FOXM1, PLK1 was highly expressed in HCC tumor tissues in both the TCGA (P<0.001; Fig. 2D) and ICGC JP (P<0.001; Fig. 2E) cohorts. In addition, in the TCGA cohort, patients with high PLK1 expression level (PH) had shorter OS (HR, 2.05; 95% CI, 1.43-2.93; P<0.001; Fig. 2A) and DFS (HR, 1.57; 95% CI, 1.16-2.11; P=0.003; Fig. 2C). In the ICGC JP cohort, patients with PH also had significantly poorer OS (HR, 3.83; 95% CI, 1.93-7.61; P<0.001; Fig. 2B).
Figure 2.
Overexpression of PLK1 is associated with poor prognosis in HCC. Kaplan-Meier survival curve estimates of overall survival in the (A) TCGA (n=373) and (B) ICGC JP (n=243) cohorts. The patients were divided into two subgroups of high and low PLK1 expression. (C) Kaplan-Meier survival curve estimates of DFS in TCGA (n=322) cohort. The expression of PLK1 in HCC and non-tumor liver tissues determined by transcriptome sequencing from the (D) TCGA and (E) ICGC JP datasets. PLK1, polo-like kinase 1; HR, hazard ratio; TCGA, The Cancer Genome Atlas; ICGC JP, International Cancer Genome Consortium Japan; HCC, hepatocellular carcinoma; NA, not available.
To assess the independent predictive value of FH and PH, logistic regression with a multivariate Cox proportional hazards model was utilized. After adjusting for age, sex, stage and virus status, stage and virus status were identified as independent prognostic factors for OS. However, neither FH (HR, 1.14; 95% CI, 0.67-1.95; P=0.634; Table II), nor PH (HR, 1.73; 95% CI, 1.00-2.99; P=0.052; Table II) were significantly and independently associated with a shorter OS.
Table II.
Cox regression proportional hazards model for the analysis of the prognostic values of FOXM1 expression and PLK1 expression for overall survival in patients with hepatocellular carcinoma.
| Univariate | Multivariate | |||
|---|---|---|---|---|
|
|
|
|||
| Characteristic | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (continuous) | 1.01 (1.00-1.03) | 0.089 | 1.01 (0.99-1.03) | 0.292 |
| Sex (female vs. male) | 1.26 (0.88-1.80) | 0.200 | 1.08 (0.72-1.63) | 0.712 |
| Stage | ||||
| II vs. I | 1.42 (0.87-2.31) | 0.164 | 1.11 (0.66-1.88) | 0.691 |
| III–IV vs. I | 2.82 (1.86-4.28) | <0.001 | 2.00 (1.26-3.18) | 0.003 |
| Virus status (vs. none) | ||||
| HBV | 0.34 (0.20-0.56) | <0.001 | 0.43 (0.24-0.78) | 0.005 |
| HCV | 0.95 (0.56-1.61) | 0.847 | 1.15 (0.64-2.05) | 0.646 |
| HBV+HCV | 0.42 (0.10-1.74) | 0.233 | 0.37 (0.08-1.61) | 0.184 |
| FOXM1 (high vs. low) | 1.68 (1.18-2.38) | 0.004 | 1.14 (0.67-1.95) | 0.634 |
| PLK1 (high vs. low) | 2.05 (1.43-2.93) | <0.001 | 1.73 (1.00-2.99) | 0.052 |
Patients with HCC were divided into the low expression group and the high expression group at the 50th percentile. HBV, hepatitis B virus; HCV, hepatitis C virus; FOXM1, forkhead box protein M1; PLK1, polo-like kinase 1; HR, hazard ratio.
Combined overexpression of FOXM1 and PLK1 is associated with poor HCC prognosis
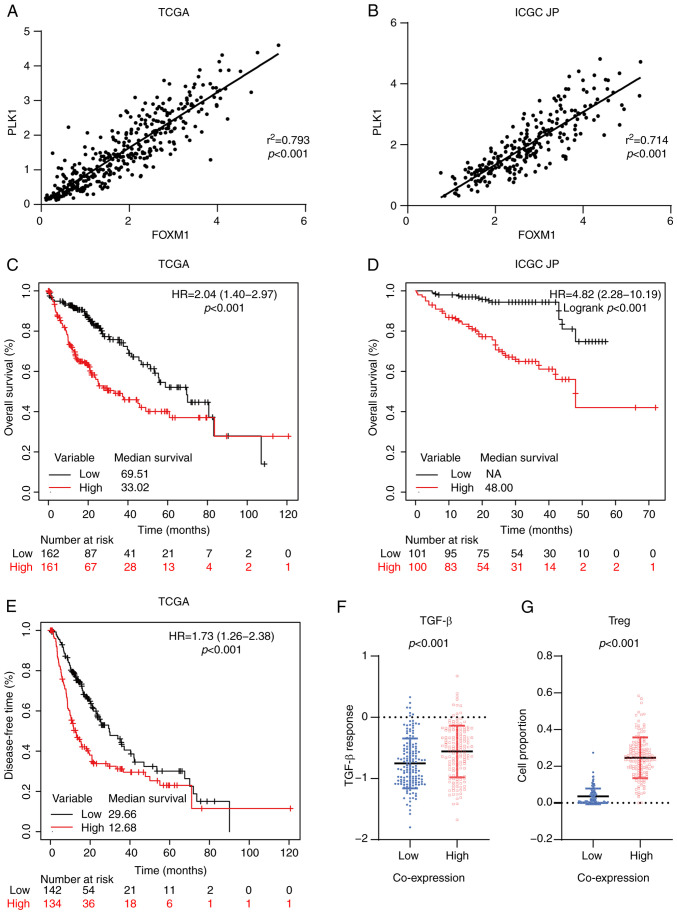
FOXM1 and PLK1 are essential for cell cycle progression. PLK1 itself is a target of FOXM1 and phosphorylation of FOXM1 mediated by PLK1 is required for FOXM1 transcription activation (14,16). In both the TCGA (r2=0.793, P<0.001; Fig. 3A) and ICGC JP (r2=0.714, P<0.001; Fig. 3B) cohorts, a positive linear correlation between FOXM1 and PLK1 expression was observed, consistent with the presence of a feedback loop between them in HCC tissues. In the TCGA cohort, patients with combined high expression of FOXM1 and PLK1 (FH-PH) exhibited significantly shorter OS (HR, 2.04; 95% CI, 1.40-2.97; P<0.001; Fig. 3C) and DFS (HR, 1.73; 95% CI, 1.26-2.38, P<0.001; Fig. 3E). The median OS corresponding to the FH-PH (33.02; Fig. 3C) group was shorter than that corresponding to the FH (45.07; Fig. 1A) and PH (37.75; Fig. 2A) groups. These observations were validated in the ICGC JP cohort (Fig. 3B). Furthermore, the prognostic value of FH-PH expression in patients with HCC was confirmed using logistic regression with a multivariate Cox proportional hazards model. The results indicated that FH-PH expression was the most significant independent predictor of OS (HR, 1.94; 95% CI, 1.31-2.89; P=0.001; Table III).
Figure 3.
Coordinated overexpression of FOXM1 and PLK1 is associated with poor prognosis in HCC. Correlation between FOXM1 and PLK1 expression in HCC in the (A) TCGA and (B) ICGC JP cohorts based on Pearson's correlation analysis. Kaplan-Meier survival curve estimates of overall survival in the (C) TCGA (n=323) and (D) ICGC JP (n=201) cohorts. The patients were divided into two subgroups: High FOXM1+PLK1 and low FOXM1+PLK1. (E) Kaplan-Meier survival curve estimates of disease-free survival in the TCGA cohort (n=276). (F) Transforming growth factor-β response score of HCC in the TCGA cohort, stratified by the co-expression status of FOXM1 and PLK1. (G) Tumor-infiltrating Tregs in HCC in the TCGA cohort, stratified based on the co-expression status of FOXM1 and PLK1. FOXM1, forkhead box protein M1; PLK1, polo-like kinase 1; HR, hazard ratio; TCGA, The Cancer Genome Atlas; ICGC JP, International Cancer Genome Consortium Japan; HCC, hepatocellular carcinoma; NA, not available; Tregs, regulatory T cells.
Table III.
Cox regression proportional hazards model for the analysis of the prognostic value of combined expression of FOXM1 and PLK1 for overall survival in patients with hepatocellular carcinoma.
| Univariate | Multivariate | |||
|---|---|---|---|---|
|
|
|
|||
| Characteristic | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (continuous) | 1.01 (1.00-1.03) | 0.089 | 1.01 (1.01-1.03) | 0.150 |
| Sex (female vs. male) | 1.26 (0.88-1.80) | 0.200 | 1.22 (0.79-1.89) | 0.366 |
| Stage | ||||
| II vs. I | 1.42 (0.87-2.31) | 0.164 | 1.03 (0.59-1.81) | 0.905 |
| III–IV vs. I | 2.82 (1.86-4.28) | <0.001 | 1.97 (1.20-3.26) | 0.008 |
| Virus status (vs. none) | ||||
| HBV | 0.34 (0.20-0.56) | <0.001 | 0.49 (0.25-0.96) | 0.038 |
| HCV | 0.95 (0.56-1.61) | 0.847 | 1.07 (0.58-1.98) | 0.821 |
| HBV+HCV | 0.42 (0.10-1.74) | 0.233 | 0.44 (0.10-2.01) | 0.289 |
| Combined FOXM1+PLK1 (high vs. low) | 2.02 (1.39-2.95) | <0.001 | 1.94 (1.31-2.89) | 0.001 |
Patients with HCC were divided into the low expression group and the high expression group at the 50th percentile. The combined expression of FOXM1 and PLK1 was defined as patients with high expression of FOXM1 and PLK1 or low expression of FOXM1 and PLK1. HR, hazard ratio; HBV, hepatitis B virus; HCV, hepatitis C virus; FOXM1, forkhead box protein M1; PLK1, polo-like kinase 1.
Cancer immunotherapy has revolutionized cancer treatment. Antibodies against programmed death 1/ligand 1 and cytotoxic T-lymphocyte-associated protein 4 are effective for the treatment of HCC (43–45). Given that the immune microenvironment has an important role in the response to immunotherapy (46,47), the impact of FH-PH expression on the immune microenvironment in patients with HCC was evaluated. Low activation of the transforming growth factor (TGF)-β pathway is associated with better clinical outcomes for patients with cancer (32). Thus, the TGF-β response score (48) were applied in patients with HCC and it was observed that the FH-PH group had a relatively higher score (P<0.001; Fig. 3F). Furthermore, CIBERSORT (49) has been used to analyze immune cell infiltration. Its application in the present study suggested that regulatory T (Treg) cells were significantly enriched in the FH-PH group (P<0.001; Fig. 3G). Of note, Treg cells are able to inhibit T-cell proliferation and cytokine production and have a critical role in preventing tumor immune response (50,51). These results suggested that FH-PH expression is associated with a suppressive immune microenvironment and may hamper immunotherapy treatment in HCC.
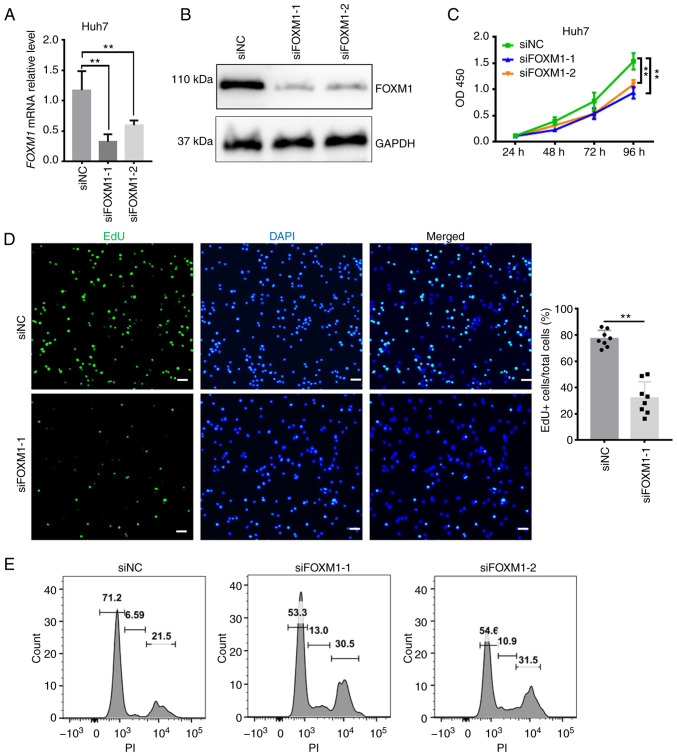
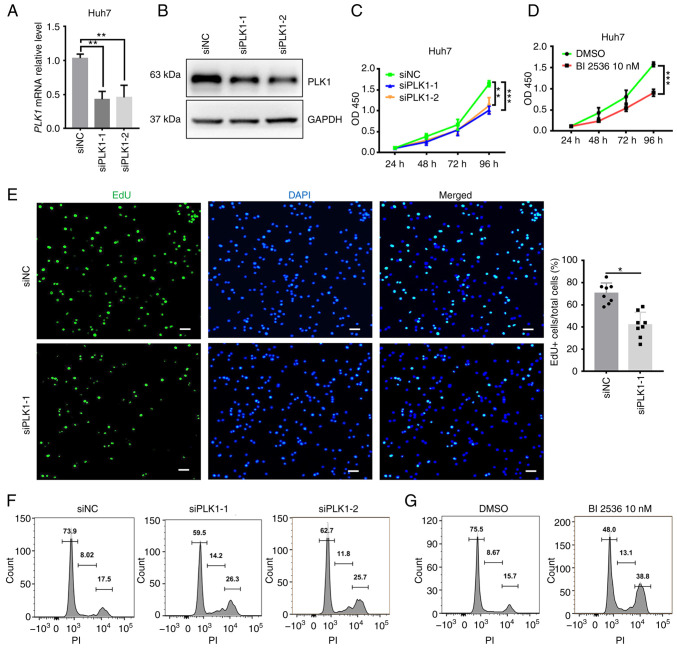
FOXM1 and PLK1 are required for HCC cell proliferation
Given that the overexpression of FOXM1 and PLK1 was associated with poor outcomes of HCC and based on their key regulatory roles in cell cycle progression, it was investigated whether FOXM1 and PLK1 influence HCC progression (Figs. 4 and 5). siRNA targeting FOXM1 and PLK1 was synthesized and the knockdown efficiency was measured in Huh7 cells (Figs. 4A and B and 5A and B). The cell proliferation rate was estimated using CCK-8 and EdU assays. FOXM1 knockdown decreased Huh7 cell viability (Fig. 4C), as well as the percentage of EdU-positive Huh7 cells (Fig. 4D), indicating a lower proportion of cells entering the DNA replication phase of the cell cycle. PLK1 knockdown exerted a similar effect on Huh7 cells (Fig. 5C and E). To confirm the requirement of PLK1 for Huh7 cell proliferation, a pharmacological inhibitor of PLK1, BI 2536 (38), was applied during the CCK-8 assay. In the presence of the inhibitor, a significant decrease in the proliferation of Huh7 cells was observed (Fig. 5D). Furthermore, flow cytometry suggested that knockdown of FOXM1 (Fig. 4E) or PLK1 (Fig. 5F) resulted in a marked increase in the proportion of cells in S and G2/M phase, as well as a decrease in the proportion of cells in G1 phase at 24 h after transfection. It appears that, as more cells progress away from G0/G1 phase, the cells are trying to proliferate but they exhibit cell cycle arrest in S and G2/M phase. Consistent with this observation, Huh7 cells treated with BI 2536 also exhibited a marked increase in the proportion of cells in S and G2/M phase (Fig. 5G). These results suggested that FOXM1 and PLK1 are required for HCC cell proliferation. Insufficient FOXM1 or PLK1 were thus indicated to hamper cell cycle progression as well as the proliferation of HCC cells, providing additional evidence that the coordinated overexpression of FOXM1 and PLK1 is an independent prognostic factor for HCC.
Figure 4.
FOXM1 is required for Huh7 cell proliferation. Knockdown efficiency of FOXM1 confirmed using (A) reverse transcription-quantitative PCR and (B) western blot analysis. Decrease in the proliferation rate of Huh7 cells following FOXM1 knockdown as indicated by (C) a Cell Counting Kit-8 assay and (D) EdU staining (scale bars, 50 µm). (E) DNA content analyzed using flow cytometry 24 h after transfection with siFOXM1 in Huh7 cells. Results are provided in histograms. Quantified values are expressed as the mean ± standard deviation of three independent experiments. **P<0.01 vs. siNC. FOXM1, forkhead box protein M1; siRNA, small interfering RNA; siNC, negative control siRNA; siFOXM1, siRNA targeting FOXM1; PI, propidium iodide; EdU, 5-ethynyl-2′-deoxyuridine; OD450, optical density at 450 nm.
Figure 5.
PLK1 is required for Huh7 cell proliferation. Knockdown efficiency of PLK1 confirmed using (A) reverse transcription-quantitative PCR and (B) western blot analysis. Cell proliferation rate analyzed using a Cell Counting Kit-8 assay after (C) transfection with siPLK1 or (D) treatment with 10 nM BI 2536 in Huh7 cells. (E) Decrease in the proliferation rate of Huh7 cells following PLK1 knockdown as indicated by EdU staining (scale bars, 50 µm). DNA content of Huh7 cell analyzed via flow cytometry 24 h after (F) transfection with siPLK1 or (G) treatment with 10 nM BI 2536. Results are provided in histograms. Quantified values are expressed as the mean ± standard deviation of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 vs. siNC. PLK1, polo-like kinase 1; siRNA, small interfering RNA; siNC, negative control siRNA; siPLK1, siRNA targeting PLK1; PI, propidium iodide; EdU, 5-ethynyl-2′-deoxyuridine; OD450, optical density at 450 nm.
Discussion
HCC is the primary malignancy of the liver and complete surgical resection is the only curative approach. However, most patients with HCC are only diagnosed when the disease is already in the advanced stage, which is unsuitable for surgery. In addition, the prognosis associated with the systemic treatment of HCC is poor (3) and valuable prognostic biomarkers are urgently required.
Cell cycle dysfunction is a marked feature of tumor cells (52) and several regulators of the cell cycle have been proposed as putative prognostic biomarkers for cancer (53). In this regard, FOXM1 and PLK1, which are essential cell cycle regulators with prognostic value in HCC, have been extensively studied. Consistent with previous reports (23,35), the present results suggested that both FOXM1 and PLK1 were overexpressed in tumor tissues and associated with poor prognosis in patients with HCC. However, after adjusting for age, sex, stage and virus status, a multivariate Cox proportional hazards model indicated that neither FOXM1 nor PLK1 is able to independently serve as a prognostic factor for HCC.
PLK1 is a target of FOXM1 and is required for FOXM1 transcriptional activation. A positive linear correlation between FOXM1 and PLK1 expression was observed in HCC tissues. Insufficient FOXM1 or PLK1 may hamper the cell cycle and it may be speculated that the coordinated overexpression of FOXM1 and PLK1 may serve as an independent prognostic factor for HCC. The present results indicated that FH-PH expression was associated with significantly shorter OS and DFS in patients with HCC. In addition, after adjusting for age, sex, stage and virus status, an FH-PH expression status was indicated to be the most significant prognostic factor for patients with HCC. Furthermore, an FH-PH expression status was associated with lower TGF-β response scores and a higher number of infiltrating Treg cells, suggesting that patients with HCC with FH-PH expression status harbor a suppressed immune microenvironment that leads to treatment failure.
In the present study, the requirement for FOXM1 and PLK1 expression in HCC cells was investigated via in vitro knockdown of FOXM1 and PLK1 in Huh7 cells. It was observed that either FOXM1 or PLK1 knockdown was able to hamper cell cycle progression as well as the proliferation of Huh7 cells. The antitumor activity of pharmacological inhibitors of PLK1 has been evaluated in clinical trials, but the efficacy was limited (54–56). The limited antitumor activity may be attributed to the low PLK1 or FOXM1 expression levels in these patients. For these patients, PLK1 was not the essential tumor-growth driving factor. Thus, the present results suggested that, to improve therapeutic efficacy in the future, it may be necessary to perform clinical trials involving patients with coordinated overexpression of FOXM1 and PLK1.
In conclusion, the present results indicated that FOXM1 and PLK1 were overexpressed in HCC tumor tissues and exhibited a positive linear correlation. FOXM1 and PLK1 are required for HCC cell proliferation. The present results also indicated that FH and PH expression were associated with poor prognosis for HCC; however, only the coordinated overexpression of FOXM1 and PLK1 was able to serve as an independent prognostic factor for HCC. Therefore, targeting FOXM1 or PLK1 is a potential treatment for improving HCC prognosis.
Acknowledgements
Not applicable.
Funding Statement
This work was supported by the Shandong Province Key Research Program (grant no. 2019GSF108012).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BJ conceived and designed the study and also provided administrative support. WF and HM provided the study materials, performed the experiments and collected the public data. All the authors performed data analysis and interpretation, participated in writing the manuscript and all authors read and approved the final version of the manuscript. WF and HM confirm the authenticity of all the raw data. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol. 2020;72:250–261. doi: 10.1016/j.jhep.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faivre S, Rimassa L, Finn RS. Molecular therapies for HCC: Looking outside the box. J Hepatol. 2020;72:342–352. doi: 10.1016/j.jhep.2019.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Sangro B, Sarobe P, Hervás-Stubbs S, Melero I. Advances in immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2021;18:525–543. doi: 10.1038/s41575-021-00438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song BN, Chu IS. A gene expression signature of FOXM1 predicts the prognosis of hepatocellular carcinoma. Exp Mol Med. 2018;50:e418. doi: 10.1038/emm.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teng L, Wang K, Liu Y, Ma Y, Chen W, Bi L. Based on integrated bioinformatics analysis identification of biomarkers in hepatocellular carcinoma patients from different regions. Biomed Res Int. 2019;2019:1742341. doi: 10.1155/2019/1742341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agarwal R, Narayan J, Bhattacharyya A, Saraswat M, Tomar AK. Gene expression profiling, pathway analysis and subtype classification reveal molecular heterogeneity in hepatocellular carcinoma and suggest subtype specific therapeutic targets. Cancer Genet. 2017:216–217. 37–51. doi: 10.1016/j.cancergen.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Sun W, Su Q, Cao X, Shang B, Chen A, Yin H, Liu B. High expression of polo-like kinase 1 is associated with early development of hepatocellular carcinoma. Int J Genomics. 2014;2014:312130. doi: 10.1155/2014/312130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seyedabadi S, Saidijam M, Najafi R, Mousavi-Bahar SH, Jafari M, MohammadGanji S, Mahdavinezhad A. Assessment of CEP55, PLK1 and FOXM1 expression in patients with bladder cancer in comparison with healthy individuals. Cancer Invest. 2018;36:407–414. doi: 10.1080/07357907.2018.1514504. [DOI] [PubMed] [Google Scholar]
- 10.Dibb M, Han N, Choudhury J, Hayes S, Valentine H, West C, Sharrocks AD, Ang YS. FOXM1 and polo-like kinase 1 are co-ordinately overexpressed in patients with gastric adenocarcinomas. BMC Res Notes. 2015;8:676. doi: 10.1186/s13104-015-1658-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang R, Song Y, Xu X, Wu Q, Liu C. The expression of Nek7, FoxM1, and Plk1 in gallbladder cancer and their relationships to clinicopathologic features and survival. Clin Transl Oncol. 2013;15:626–632. doi: 10.1007/s12094-012-0978-9. [DOI] [PubMed] [Google Scholar]
- 12.Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: Tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13:482–495. doi: 10.1038/nrc3539. [DOI] [PubMed] [Google Scholar]
- 13.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laoukili J, Kooistra MR, Brás A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 15.Chen YJ, Dominguez-Brauer C, Wang Z, Asara JM, Costa RH, Tyner AL, Lau LF, Raychaudhuri P. A conserved phosphorylation site within the forkhead domain of FoxM1B is required for its activation by cyclin-CDK1. J Biol Chem. 2009;284:30695–30707. doi: 10.1074/jbc.M109.007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM, Tindall DJ, Chen J. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol. 2008;10:1076–1082. doi: 10.1038/ncb1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Yuan C, Wu J, Elsayed Z, Fu Z. Polo-like kinase 1-mediated phosphorylation of forkhead box protein M1b antagonizes its SUMOylation and facilitates its mitotic function. J Biol Chem. 2015;290:3708–3719. doi: 10.1074/jbc.M114.634386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pilarsky C, Wenzig M, Specht T, Saeger HD, Grützmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–750. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, Major ML, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Park HJ, Carr JR, Chen YJ, Zheng Y, Li J, Tyner AL, Costa RH, Bagchi S, Raychaudhuri P. FoxM1 in tumorigenicity of the neuroblastoma cells and renewal of the neural progenitors. Cancer Res. 2011;71:4292–4302. doi: 10.1158/0008-5472.CAN-10-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu G, Yan Z, Zhang C, Cheng M, Yan Y, Wang Y, Deng L, Lu Q, Luo S. FOXM1 promotes hepatocellular carcinoma progression by regulating KIF4A expression. J Exp Clin Cancer Res. 2019;38:188. doi: 10.1186/s13046-019-1202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chai N, Xie HH, Yin JP, Sa KD, Guo Y, Wang M, Liu J, Zhang XF, Zhang X, Yin H, et al. FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochem Biophys Res Commun. 2018;500:924–929. doi: 10.1016/j.bbrc.2018.04.201. [DOI] [PubMed] [Google Scholar]
- 23.Egawa M, Yoshida Y, Ogura S, Kurahashi T, Kizu T, Furuta K, Kamada Y, Chatani N, Hamano M, Kiso S, et al. Increased expression of Forkhead box M1 transcription factor is associated with clinicopathological features and confers a poor prognosis in human hepatocellular carcinoma. Hepatol Res. 2017;47:1196–1205. doi: 10.1111/hepr.12854. [DOI] [PubMed] [Google Scholar]
- 24.Sun HC, Li M, Lu JL, Yan DW, Zhou CZ, Fan JW, Qin XB, Tang HM, Peng ZH. Overexpression of Forkhead box M1 protein associates with aggressive tumor features and poor prognosis of hepatocellular carcinoma. Oncol Rep. 2011;25:1533–1539. doi: 10.3892/or.2011.1230. [DOI] [PubMed] [Google Scholar]
- 25.Combes G, Alharbi I, Braga LG, Elowe S. Playing polo during mitosis: PLK1 takes the lead. Oncogene. 2017;36:4819–4827. doi: 10.1038/onc.2017.113. [DOI] [PubMed] [Google Scholar]
- 26.Colicino EG, Hehnly H. Regulating a key mitotic regulator, polo-like kinase 1 (PLK1) Cytoskeleton (Hoboken) 2018;75:481–494. doi: 10.1002/cm.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barr FA, Silljé HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 28.Xie S, Xie B, Lee MY, Dai W. Regulation of cell cycle checkpoints by polo-like kinases. Oncogene. 2005;24:277–286. doi: 10.1038/sj.onc.1208218. [DOI] [PubMed] [Google Scholar]
- 29.He Z, Wu J, Dang H, Lin H, Zheng H, Zhong D. Polo-like kinase 1 contributes to the tumorigenicity of BEL-7402 hepatoma cells via regulation of Survivin expression. Cancer Lett. 2011;303:92–98. doi: 10.1016/j.canlet.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Park JS, Sohn HJ, Park GS, Chung YJ, Kim TG. Induction of antitumor immunity using dendritic cells electroporated with Polo-like kinase 1 (Plk1) mRNA in murine tumor models. Cancer Sci. 2011;102:1448–1454. doi: 10.1111/j.1349-7006.2011.01974.x. [DOI] [PubMed] [Google Scholar]
- 31.Jang HR, Shin SB, Kim CH, Won JY, Xu R, Kim DE, Yim H. PLK1/vimentin signaling facilitates immune escape by recruiting Smad2/3 to PD-L1 promoter in metastatic lung adenocarcinoma. Cell Death Differ. 2021;28:2745–2764. doi: 10.1038/s41418-021-00781-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fristrup N, Ulhøi BP, Birkenkamp-Demtröder K, Mansilla F, Sanchez-Carbayo M, Segersten U, Malmström PU, Hartmann A, Palou J, Alvarez-Múgica M, et al. Cathepsin E, maspin, Plk1, and survivin are promising prognostic protein markers for progression in non-muscle invasive bladder cancer. Am J Pathol. 2012;180:1824–1834. doi: 10.1016/j.ajpath.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 33.Takai N, Hamanaka R, Yoshimatsu J, Miyakawa I. Polo-like kinases (Plks) and cancer. Oncogene. 2005;24:287–291. doi: 10.1038/sj.onc.1208272. [DOI] [PubMed] [Google Scholar]
- 34.Cheng L, Wang C, Jing J. Polo-like kinase 1 as a potential therapeutic target for osteosarcoma. Curr Pharm Des. 2015;21:1347–1350. doi: 10.2174/1381612820999141029162811. [DOI] [PubMed] [Google Scholar]
- 35.He ZL, Zheng H, Lin H, Miao XY, Zhong DW. Overexpression of polo-like kinase1 predicts a poor prognosis in hepatocellular carcinoma patients. World J Gastroenterol. 2009;15:4177–4182. doi: 10.3748/wjg.15.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian L, Yao K, Liu K, Han B, Dong H, Zhao W, Jiang W, Qiu F, Qu L, Wu Z, et al. PLK1/NF-κB feedforward circuit antagonizes the mono-ADP-ribosyltransferase activity of PARP10 and facilitates HCC progression. Oncogene. 2020;39:3145–3162. doi: 10.1038/s41388-020-1205-8. [DOI] [PubMed] [Google Scholar]
- 37.Gumireddy K, Reddy MV, Cosenza SC, Boominathan R, Baker SJ, Papathi N, Jiang J, Holland J, Reddy EP. ON01910, a non-ATP-competitive small molecule inhibitor of Plk1, is a potent anticancer agent. Cancer Cell. 2005;7:275–286. doi: 10.1016/j.ccr.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 38.Steegmaier M, Hoffmann M, Baum A, Lénárt P, Petronczki M, Krssák M, Gürtler U, Garin-Chesa P, Lieb S, Quant J, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 39.Rudolph D, Steegmaier M, Hoffmann M, Grauert M, Baum A, Quant J, Haslinger C, Garin-Chesa P, Adolf GR. BI 6727, a Polo-like kinase inhibitor with improved pharmacokinetic profile and broad antitumor activity. Clin Cancer Res. 2009;15:3094–3102. doi: 10.1158/1078-0432.CCR-08-2445. [DOI] [PubMed] [Google Scholar]
- 40.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 41.Qiu P, Sheng J. A two-stage procedure for comparing hazard rate functions. J R Stat Soc Series B (Stat Methodol) 2008;70:191–208. [Google Scholar]
- 42.Li H, Han D, Hou Y, Chen H, Chen Z. Statistical inference methods for two crossing survival curves: A comparison of methods. PLoS One. 2015;10:e0116774. doi: 10.1371/journal.pone.0116774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duffy AG, Ulahannan SV, Makorova-Rusher O, Rahma O, Wedemeyer H, Pratt D, Davis JL, Hughes MS, Heller T, ElGindi M, et al. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma. J Hepatol. 2017;66:545–551. doi: 10.1016/j.jhep.2016.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, Kudo M, Breder V, Merle P, Kaseb AO, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382:1894–1905. doi: 10.1056/NEJMoa1915745. [DOI] [PubMed] [Google Scholar]
- 45.Ren Z, Xu J, Bai Y, Xu A, Cang S, Du C, Li Q, Lu Y, Chen Y, Guo Y, et al. Sintilimab plus a bevacizumab biosimilar (IBI305) versus sorafenib in unresectable hepatocellular carcinoma (ORIENT-32): A randomised, open-label, phase 2–3 study. Lancet Oncol. 2021;22:977–990. doi: 10.1016/S1470-2045(21)00252-7. [DOI] [PubMed] [Google Scholar]
- 46.Cariani E, Missale G. Immune landscape of hepatocellular carcinoma microenvironment: Implications for prognosis and therapeutic applications. Liver Int. 2019;39:1608–1621. doi: 10.1111/liv.14192. [DOI] [PubMed] [Google Scholar]
- 47.Oura K, Morishita A, Tani J, Masaki T. Tumor Immune microenvironment and immunosuppressive therapy in hepatocellular carcinoma: A review. Int J Mol Sci. 2021;22:5801. doi: 10.3390/ijms22115801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kondĕlková K, Vokurková D, Krejsek J, Borská L, Fiala Z, Ctirad A. Regulatory T cells (TREG) and their roles in immune system with respect to immunopathological disorders. Acta Medica (Hradec Kralove) 2010;53:73–77. doi: 10.14712/18059694.2016.63. [DOI] [PubMed] [Google Scholar]
- 51.Hatzioannou A, Boumpas A, Papadopoulou M, Papafragkos I, Varveri A, Alissafi T, Verginis P. Regulatory T cells in autoimmunity and cancer: A duplicitous lifestyle. Front Immunol. 2021;12:731947. doi: 10.3389/fimmu.2021.731947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matthews HK, Bertoli C, de Bruin RAM. Cell cycle control in cancer. Nat Rev Mol Cell Biol. 2022;23:74–88. doi: 10.1038/s41580-021-00404-3. [DOI] [PubMed] [Google Scholar]
- 53.Kashyap D, Garg VK, Sandberg EN, Goel N, Bishayee A. Oncogenic and tumor suppressive components of the cell cycle in breast cancer progression and prognosis. Pharmaceutics. 2021;13:569. doi: 10.3390/pharmaceutics13040569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mross K, Dittrich C, Aulitzky WE, Strumberg D, Schutte J, Schmid RM, Hollerbach S, Merger M, Munzert G, Fleischer F, Scheulen ME. A randomised phase II trial of the Polo-like kinase inhibitor BI 2536 in chemo-naïve patients with unresectable exocrine adenocarcinoma of the pancreas-a study within the Central European society anticancer drug research (CESAR) collaborative network. Br J Cancer. 2012;107:280–286. doi: 10.1038/bjc.2012.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia-Manero G, Fenaux P, Al-Kali A, Baer MR, Sekeres MA, Roboz GJ, Gaidano G, Scott BL, Greenberg P, Platzbecker U, et al. Rigosertib versus best supportive care for patients with high-risk myelodysplastic syndromes after failure of hypomethylating drugs (ONTIME): A randomised, controlled, phase 3 trial. Lancet Oncol. 2016;17:496–508. doi: 10.1016/S1470-2045(16)00009-7. [DOI] [PubMed] [Google Scholar]
- 56.Pujade-Lauraine E, Selle F, Weber B, Ray-Coquard IL, Vergote I, Sufliarsky J, Del Campo JM, Lortholary A, Lesoin A, Follana P, et al. Volasertib versus chemotherapy in platinum-resistant or -refractory ovarian cancer: A randomized phase II groupe des investigateurs nationaux pour l'Etude des cancers de l'Ovaire study. J Clin Oncol. 2016;34:706–713. doi: 10.1200/JCO.2015.62.1474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.