Abstract
Presence of nuclear atypia during histological investigation is often a cause of concern for pathologists while identifying tumor and non-tumor cells in a biopsy sample of oral mucosa. Nuclear atypia is observed in severe inflammation, ulcers and reactive changes. Therefore, additional methods, such as immunohistochemistry, may help precise diagnosis. When the atypia is suggestive of tumorous or reactive origin, the lesion is diagnosed as atypical squamous epithelium (ASE). When there is severe nuclear atypia in the mucosa, such as in disorders of nuclear polarity, large nuclei, and clear nucleolus, the lesion is diagnosed as carcinoma in situ (CIS). However, it is not easy to distinguish ASE and CIS using hematoxylin and eosin staining. The present study aimed to distinguish ASE from CIS using immunohistochemistry. A total of 32 biopsy samples of either ASE or CIS cases were selected and the level of casein kinase 1ε (CK-1ε), differentiated embryonic chondrocyte gene 1 (DEC1), proliferating cell nuclear antigen (PCNA) and CD44, which are four protein markers which have been previously linked to cancer progression, were analyzed. CK-1ε and CD44 expression was higher in CIS samples than in ASE samples. However, DEC1 expression was lower in CIS samples than in ASE samples. PCNA expression was not markedly different between the two groups. Additionally, it was found that DEC1-overexpressing cells had decreased levels of CK-1ε and CD44 compared with control cells, while CK-1ε-overexpressing cells had relatively unchanged levels of CD44, DEC1 and PCNA. These results suggested that DEC1 negatively regulates the expression of CK-1ε and CD44. Thus, DEC1, CK-1ε, and CD44 were identified as mechanistically linked and clinically relevant protein biomarkers, which could help distinguish ASE and CIS.
Keywords: atypical squamous epithelium, carcinoma in situ, DEC1, CK-1ε, PCNA, CD44
Introduction
Advanced oral squamous cell carcinoma (OSCC) is characterized by invasion of surrounding tissues, metastasis to lymph nodes, resistance to chemotherapy, and poor prognosis (1,2). Most OSCC cases develop from oral epithelial dysplasia (OED) and carcinoma in situ (CIS) (2,3). Clinically, the mass or ulcer is obtained from the tongue, gingiva, oral floor and buccal mucosa of the patients (1–4). Histologically, sampled tissue is diagnosed as atypical squamous epithelium (ASE), OED, CIS, or SCC, depending on the grade of nuclear atypia and stromal invasion (2,3,5). However, it is often difficult to determine ASE, OED, or CIS based on hematoxylin and eosin (H&E) findings alone (4–6). Therefore, immunohistochemistry may be helpful in such cases (4–6).
Casein kinase 1ε (CK-1ε) is an enzyme of the casein kinase family and regulates cell proliferation, migration, and circadian rhythm (7–9). The level of CK-1ε in ovarian cancer cells is higher than that in non-cancerous cells (10). Differentiated embryonic chondrocyte gene 1 (DEC1) is a basic helix-loop-helix transcription factor, which regulates inflammation, apoptosis, migration epithelial mesenchymal transition (EMT), cell differentiation, and circadian rhythm (11–16). It has been revealed that DEC1 is highly expressed in tumor cells compared with non-tumor cells of the cervical, pancreatic and oral lineages (12,13,15). Proliferating cell nuclear antigen (PCNA) is a nuclear marker of G1/S phase cell cycle regulation (17). It was recently demonstrated that differences in the localization of positive PCNA staining may distinguish between conventional squamous cell carcinoma and basaloid squamous cell carcinoma (4). CD44 is a known cancer stem cell marker that regulates cell proliferation and migration and is highly expressed in dysplasia and OSCC compared with non-tumor cells (18–20). However, this has not been revealed in ASE and CIS. The present study aimed to immunohistochemically examine the expression levels of CK-1ε, DEC1, PCNA and CD44 in biopsy samples of ASE and CIS.
Materials and methods
Tissue preparation
Histological biopsy specimens between December 2012 and December 2018 were retrieved from the archives of the Department of Diagnostic Pathology of Wakayama Medical University (Wakayama, Japan) according to the guidelines of the Japanese Society of Pathology. The present study was approved (approval no. 1715) by the Research Ethics Committee of Wakayama Medical University and histological specimens were retrieved from Wakayama Medical University hospital archives. Oral informed consents were provided by all patients for the use of their tissues.
A total of 20 cases of ASE and 12 cases of CIS were selected. Diagnoses were performed by at least two pathologists. In the present study, ASE tissues with or without inflammatory and benign lesions were used. CIS with obvious nuclear atypia was used but invasive OSCC was not included. Clinical and histological information are presented in Table I. The percentage of intensity score was calculated using Microsoft excel.
Table I.
Immunohistochemical detection of CK-1ε, DEC1, PCNA and CD44 in ASE and CIS.
| Case | Age/Sex | Lesions | Diagnosis | CK-1ε | DEC1 | PCNA | CD44 |
|---|---|---|---|---|---|---|---|
| 1 | 60/F | Tongue | ASE | 1 | 3 | 2 | 1 |
| 2 | 55/M | Tongue | ASE | 1 | 3 | 2 | 2 |
| 3 | 75/F | Tongue | ASE | 1 | 2 | 1 | 1 |
| 4 | 62/M | Tongue | ASE | 1 | 3 | 1 | 1 |
| 5 | 70/M | Tongue | ASE | 1 | 2 | 2 | 1 |
| 6 | 67/F | Tongue | ASE | 1 | 3 | 3 | 3 |
| 7 | 76/F | Tongue | ASE | 1 | 3 | 3 | 1 |
| 8 | 28/F | Gingiva | ASE | 1 | 2 | 1 | 1 |
| 9 | 69/F | Gingiva | ASE | 1 | 2 | 1 | 1 |
| 10 | 65/F | Gingiva | ASE | 1 | 3 | 1 | 1 |
| 11 | 58/F | Gingiva | ASE | 1 | 3 | 2 | 1 |
| 12 | 59/F | Gingiva | ASE | 1 | 3 | 2 | 1 |
| 13 | 77/F | Gingiva | ASE | 1 | 3 | 2 | 1 |
| 14 | 84/F | Gingiva | ASE | 2 | 2 | 2 | 3 |
| 15 | 74/M | Buccal | ASE | 1 | 3 | 2 | 1 |
| 16 | 68/F | Buccal | ASE | 1 | 2 | 2 | 2 |
| 17 | 74/M | Buccal | ASE | 1 | 3 | 3 | 1 |
| 18 | 76/F | Buccal | ASE | 1 | 3 | 2 | 2 |
| 19 | 77/F | Oral floor | ASE | 1 | 3 | 2 | 1 |
| 20 | 54/F | Palate | ASE | 2 | 3 | 2 | 1 |
| 21 | 67/M | Tongue | CIS | 3 | 1 | 3 | 2 |
| 22 | 78/M | Tongue | CIS | 3 | 3 | 2 | 2 |
| 23 | 68/M | Tongue | CIS | 2 | 1 | 3 | 2 |
| 24 | 60/M | Tongue | CIS | 3 | 1 | 3 | 3 |
| 25 | 55/F | Tongue | CIS | 3 | 2 | 3 | 2 |
| 26 | 74/M | Gingiva | CIS | 3 | 1 | 2 | 1 |
| 27 | 69/F | Gingiva | CIS | 2 | 1 | 3 | 2 |
| 28 | 71/M | Gingiva | CIS | 2 | 1 | 3 | 3 |
| 29 | 80/F | Buccal | CIS | 2 | 1 | 2 | 3 |
| 30 | 69/F | Buccal | CIS | 3 | 3 | 2 | 3 |
| 31 | 84/F | Palate | CIS | 3 | 1 | 2 | 3 |
| 32 | 78/F | Palate | CIS | 2 | 1 | 2 | 1 |
CK-1ε, DEC1, PCNA and CD44 intensity was classified as follows: 1, weak; 2, moderate; and 3, strong. CK-1ε, casein kinase 1ε; DEC1, differentiated embryonic chondrocyte gene 1; PCNA, proliferating cell nuclear antigen; ASE, atypical squamous epithelium; CIS, carcinoma in situ; F, female; M, male.
Immunohistochemistry
CK-1ε, DEC1, PCNA and CD44 expression levels in ASE and CIS tissues were profiled using a Discovery Auto-Stainer with automated protocols (software Nex-ES v10.6) (Ventana Medical Systems, Inc.; Roche Diagnostics) as previously described (4).
Cell culture and treatment
CA9-22 cells (cat. no. JCRB0625) were obtained from the Japanese Cancer Research Resources Bank and used until passage five. CA9-22 cells stably overexpressing DEC1 (CA9-22DEC1) and control empty vector-transfected cells (CA9-22vector) were evaluated for stable cells as previously described (21). These cells were cultured in Dulbecco's modified Eagle's medium (Sigma-Aldrich; Merck KGaA) as previously described (4,21). Transient plasmid transfection of a FLAG-tagged CK-1ε was performed using FuGENE HD (Promega Corporation) as previously described (22).
Western blotting
CA9-22 cells were lysed using M-PER lysis buffer (Thermo Fisher Scientific, Inc.) and protein concentration was determined by bicinchoninic acid (BCA) assay. 40 µg of protein was loaded on 12.5% SDS-polyacrylamide gels. The separated proteins were subsequently transferred onto PVDF membranes, which were incubated with primary antibodies overnight at 4°C. The incubation with secondary antibodies was performed for 1 h at room temperature. Western blotting was performed as previously described (4). AE-9300 Ez capture MG (ATTO) was used to capture images. Three independent biological replicates were performed.
Antibodies
The following commercial antibodies were used: CK-1ε (1:200, mouse monoclonal; cat. no. sc-373912; Santa Cruz Biotechnology, Inc.), DEC1 (1:400; rabbit polyclonal; cat. no. NB100-1800; Novus Biologicals, LLC), PCNA (1:1,000; mouse monoclonal; cat. no. sc-56), HCAM (CD44; 1:200; mouse monoclonal; sc-7297; both from Santa Cruz Biotechnology, Inc.), FLAG (1:1,000; mouse monoclonal; cat. no. F3165) and actin (1:10,000; mouse monoclonal, A5441; both from Sigma-Aldrich; Merck KGaA). Anti-rabbit and mouse HRP IgG secondary antibodies (1:5,000; cat. nos. 17502 and 17601, respectively) were purchased from Immuno-Biological Laboratories, Co., Ltd.
Results
Histological features of ASE and CIS
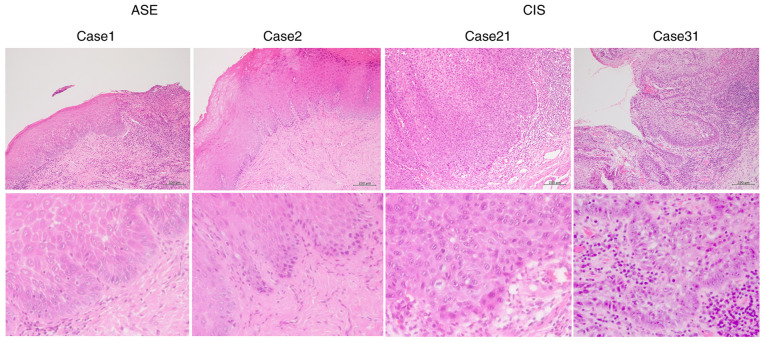
Representative histological H&E-stained images for cases 1 and 2 of ASE, and 21 and 31 of CIS, respectively, are revealed in Fig. 1. Mild nuclear atypia was observed in the ASE samples, whereas severe nuclear atypia was observed in the CIS samples.
Figure 1.
Histological features of ASE and CIS. Representative hematoxylin and eosin stain images of cases 1 and 2 of ASE and 21 and 31 of CIS. Top panel, ×100 magnification. Bottom panel, ×400 magnification. Scale bars, 200 µm. ASE, atypical squamous epithelium; CIS, carcinoma in situ.
Immunohistochemical detection of CK-1ε, DEC1, PCNA, and CD44 in ASE and CIS
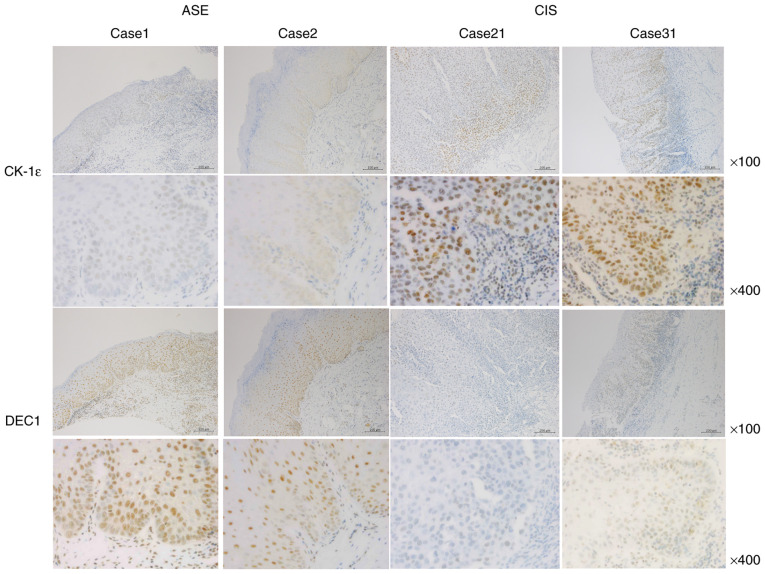
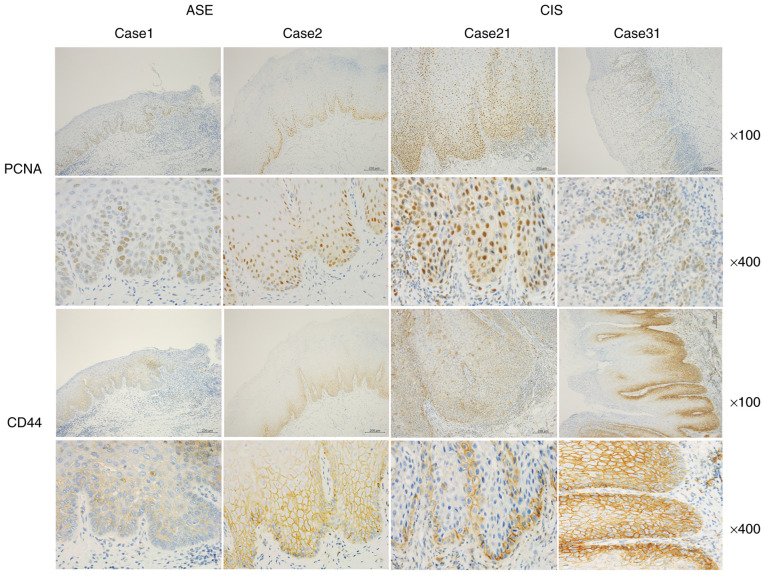
Representative images of CK-1ε and DEC1 immunoreactivities for cases 1 and 2 of ASE and 21 and 31 of CIS are shown in Fig. 2. A total of three grades (1, weak; 2, moderate; and 3, strong) of intensity were defined by immunohistochemistry. CK-1ε was weakly expressed in the nuclei of ASE samples (100% with intensity levels 1–2), whereas it was strongly expressed in the nuclei of CIS samples (100% with intensity levels 2–3) (Tables I and II). By contrast, DEC1 was strongly expressed in ASE and weakly expressed in CIS. All ASE samples stained with DEC1 had an intensity of 2 or 3, while 88% of CIS samples stained with DEC1 had intensities of 1 or 2. Representative images of PCNA and CD44 immunoreactivities for cases 1 and 2 of ASE and 21 and 31 of CIS are presented in Fig. 3. PCNA was weakly and moderately expressed in the nucleus of ASE, and moderately and strongly expressed in the nucleus of CIS. Overall, 85% of the ASE samples stained with PCNA had intensities of 1 or 2, while 60% had an intensity of 2. Collectively, 100% of the CIS samples stained with PCNA had intensities of 2 and 3. Furthermore, 90% of ASE samples stained with CD44 had intensities of 1 and 2, while 84% of the CIS samples had intensities of 2 and 3.
Figure 2.
Immunoreactivities of CK-1ε and DEC1 in ASE and CIS. Representative images for immunoreactivities of CK-1ε and DEC1 in cases 1 and 2 of ASE and 21 and 31 of CIS. Top panel, ×100 magnification. Bottom panel, ×400 magnification. Scale bars, 200 µm. CK-1ε, casein kinase 1ε; DEC1, differentiated embryonic chondrocyte gene 1; ASE, atypical squamous epithelium; CIS, carcinoma in situ.
Table II.
Percentage of intensity score of CK-1ε, DEC1, PCNA and CD44 in ASE and CIS.
| Intensity | CK-1ε (%) | DEC1 (%) | PCNA (%) | CD44 (%) |
|---|---|---|---|---|
| ASE | ||||
| 1 | 90 | 0 | 25 | 75 |
| 2 | 10 | 30 | 60 | 15 |
| 3 | 0 | 70 | 15 | 10 |
| CIS | ||||
| 1 | 0 | 75 | 0 | 16 |
| 2 | 42 | 8 | 50 | 42 |
| 3 | 58 | 17 | 50 | 42 |
CK-1ε, casein kinase 1ε; DEC1, differentiated embryonic chondrocyte gene 1; PCNA, proliferating cell nuclear antigen; ASE, atypical squamous epithelium; CIS, carcinoma in situ.
Figure 3.
Immunoreactivities of PCNA and CD44 in ASE and CIS. Representative images for immunoreactivities of PCNA and CD44 in cases 1 and 2 of ASE and 21 and 31 of CIS. Top panel, ×100 magnification. Bottom panel, ×400 magnification. Scale bars, 200 µm. PCNA, proliferating cell nuclear antigen; ASE, atypical squamous epithelium; CIS, carcinoma in situ.
DEC1 overexpression decreases the expression of CK-1ε and CD44 in oral cancer cells
It was further examined whether DEC1 overexpression in CA9-22 cells (CA9-22DEC1) affected the levels of CK-1ε and CD44. CA9-22-DEC1-overexpressing cells exhibited decreased expression of CK-1ε and CD44 (Fig. 4). In addition, it was previously reported that DEC1 overexpression decreases the expression of PCNA (21). The transient overexpression of CK-1ε was effective, but it had little effect on the expression of CD44, DEC1 and PCNA. The present study suggested immunohistochemical strategies for distinguishing between ASE and CIS in Fig. 5.
Figure 4.
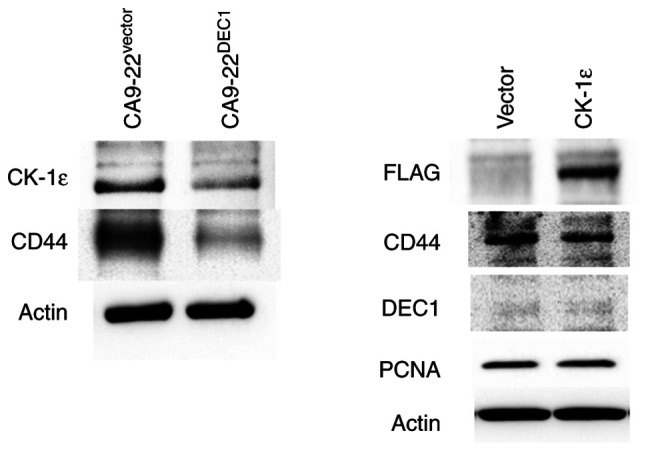
DEC1 overexpression decreases the expression of CK-1ε and CD44 in oral cancer CA9-22 cells. Representative western blots of CK-1ε, CD44, and actin in cells with a control vector or cells stably overexpressing DEC1 (left panel). Representative western blots of FLAG, CD44, DEC1, PCNA and actin in cells expressing control vector or cells transiently overexpressing CK-1ε. DEC1, differentiated embryonic chondrocyte gene 1; CK-1ε, casein kinase 1ε; PCNA, proliferating cell nuclear antigen.
Figure 5.
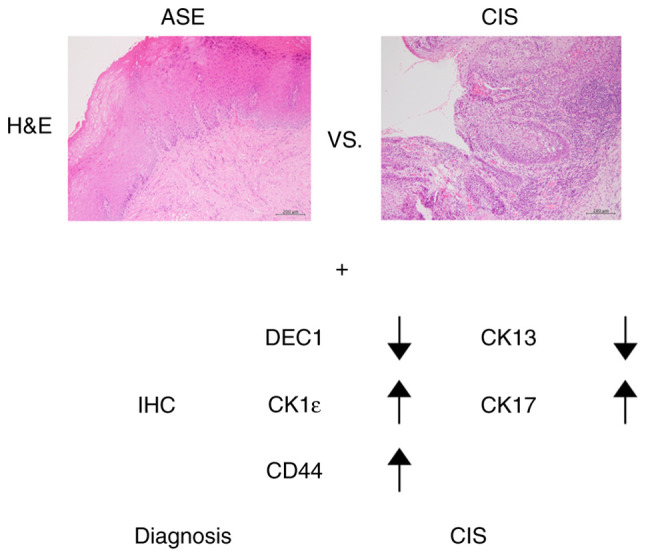
Pathological differentiation of ASE and CIS. Immunohistochemical findings of CK13, CK17, DEC1, CK-1ε and CD44 may help distinguish between ASE and CIS. A diagnosis of CIS using IHC is indicated by decreased expression of CK13 and DEC1 and increased expression of CK17, CK-1ε and CD44. ASE, atypical squamous epithelium; CIS, carcinoma in situ; DEC1, differentiated embryonic chondrocyte gene 1; CK-1ε, casein kinase 1ε; IHC, immunohistochemistry; H&E, hematoxylin and eosin staining.
Discussion
In the present study, the expression of CK-1ε, DEC1, PCNA, and CD44 was examined in ASE and CIS biopsy samples. It was previously reported that DEC1 expression was increased in invasive cancer cells compared with that in non-cancerous cells (13). Notably, DEC1 expression was lower in CIS than in ASE samples. In addition, DEC1 was reported to be more highly expressed in spindle lesions of EMT than in conventional invasive lesions (12,15). Although DEC1 expression was lower in CIS compared with ASE samples, invasive processes such as EMT from CIS may induce DEC1 expression. It is well known that transforming growth factor beta (TGFβ) induces EMT and DEC1 expression in various cancer cells (15,23–27). Consistent with this finding, DEC1 expression was higher in ASE and invasive OSCC samples than that in CIS samples (12,13,15).
It has been reported that DEC1 promotes tumor progression, EMT, metastasis, and anti-apoptotic activity via hypoxia-inducible factor-1α, TGFβ and SMAD3 (14,21). CD44 promotes tumor proliferation, invasion and anti-apoptotic functions (28). DEC1 negatively regulates the expression of stem cell markers SOX2 and c-MYC in cervical cancer cells (12). Consistent with this, it was observed that the cancer stem cell marker CD44 was decreased in CA9-22-DEC1-overexpressing cells. In addition, opposite expression patterns of DEC1 and CD44 were observed in ASE and CIS samples. These results suggested that DEC1 negatively regulated CD44 expression, although both the expression levels of DEC1 and CD44 were higher in invasive cancer cells. DEC1 regulates target genes by binding E-boxes and sp1 sites (14,16,22). Notably, overexpression of circadian clock gene Period2 (PER2) decreased CD44 expression in lung cancer cells (29). Since DEC1 negatively regulates PER2 through E-boxes (14,16), it is possible that DEC1 regulates CD44 via E-boxes. It was hypothesized that DEC1 suppresses CD44 expression to slow the invasive progression of OSCC. Future studies are required to clarify how DEC1 regulates CD44. It was revealed that CK-1ε expression is similar to that of CD44. Low expression levels of CK-1ε have been associated with poor prognosis in advanced OSCC (30). These previous results suggested that CK-1ε expression is inversely correlated with DEC1 in CIS and in invasive OSCC. Therefore, DEC1 negatively regulates CK-1ε. In the present study, no marked changes in PCNA were identified in ASE and CIS. However, it was observed that DEC1 overexpression decreased the PCNA expression in CA9-22 cells, as previously described (21). It was hypothesized that DEC1 negatively regulates PCNA in invasive OSCC but not in CIS. Clarification on how these molecules functionally regulate ASE and CIS, using a mouse model, is warranted in future studies.
CK13 and CK17 have been previously used to distinguish between ASE and CIS (1,6). In CIS, the expression of CK13 is decreased whereas that of CK17 is increased. Τhe use of additional immunohistochemical markers such as CK-1ε, DEC1, CD44, CK13 and CK17 is suggested to distinguish ASE and CIS. However, changes in these immunohistochemical markers are often observed upon severe inflammation, such as in active ulcers and in ASE. Furthermore, the usefulness of biopsies to distinguish between ASE and CIS is limited since ASE occasionally exhibits severe nuclear atypia. Therefore, the difference between ASE and CIS should be clarified using resected samples. Another limitation was the small sample size and the absence of statistical analysis to quantify results in the present study. As a result, a larger number of samples needs to be examined in future studies.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- ASE
atypical squamous epithelium
- CIS
carcinoma in situ
- EMT
epithelial to mesenchymal transition
- H&E
hematoxylin and eosin
- OED
oral epithelial dysplasia
- OSCC
oral squamous cell carcinoma
- DEC1
differentiated embryonic chondrocyte gene 1
- CK-1ε
casein kinase 1ε
- PCNA
proliferating cell nuclear antigen
Funding Statement
The present study was supported by JSPS KAKENHI (grant no. 16K09624).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
FS performed experiments, pathological diagnosis and wrote the draft of the manuscript. SO and NS performed immunohistochemistry and western blotting. FS, SO and NS confirmed the authenticity of all the raw data. YM checked pathological diagnosis. UKB and KO helped acquisition of data and interpretation of data. UKB, KO and YM revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved (approval no. 1715) by the Research Ethics Committee of Wakayama Medical University (Wakayama, Japan) and histological specimens were retrieved from Wakayama Medical University hospital archives. Oral informed consents were provided by all patients for the use of their tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ikeda M, Shima K, Kondo T, Semba I. Atypical immunohistochemical patterns can complement the histopathological diagnosis of oral premalignant lesions. J Oral Biosci. 2020;62:93–98. doi: 10.1016/j.job.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Speight PM, Farthing PM. The pathology of oral cancer. Br Dent J. 2018;225:841–847. doi: 10.1038/sj.bdj.2018.926. [DOI] [PubMed] [Google Scholar]
- 3.Brouns E, Baart J, Karagozoglu Kh, Aartman I, Bloemena E, van der Waal I. Malignant transformation of oral leukoplakia in a well-defined cohort of 144 patients. Oral Dis. 2014;20:e19–e24. doi: 10.1111/odi.12095. [DOI] [PubMed] [Google Scholar]
- 4.Sato F, Bhawal UK, Tojyo I, Fujita S, Murata SI, Muragaki Y. Differential expression of claudin-4, occludin, SOX2 and proliferating cell nuclear antigen between basaloid squamous cell carcinoma and squamous cell carcinoma. Mol Med Rep. 2019;20:1977–1985. doi: 10.3892/mmr.2019.10417. [DOI] [PubMed] [Google Scholar]
- 5.Odell E, Kujan O, Warnakulasuriya S, Sloan P. Oral epithelial dysplasia: Recognition, grading and clinical significance. Oral Dis. 2021;27:947–976. doi: 10.1111/odi.13993. [DOI] [PubMed] [Google Scholar]
- 6.Nobusawa A, Sano T, Negishi A, Yokoo S, Oyama T. Immunohistochemical staining patterns of cytokeratins 13, 14, and 17 in oral epithelial dysplasia including orthokeratotic dysplasia. Pathol Int. 2014;64:20–27. doi: 10.1111/pin.12125. [DOI] [PubMed] [Google Scholar]
- 7.Fish KJ, Cegielska A, Getman ME, Landes GM, Virshup DM. Isolation and characterization of human casein kinase I epsilon (CKI), a novel member of the CKI gene family. J Biol Chem. 1995;270:14875–14883. doi: 10.1074/jbc.270.25.14875. [DOI] [PubMed] [Google Scholar]
- 8.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/S0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Zhou L, Wang Y, Peng Q, Li H, Zhang X, Su Z, Song J, Sun Q, Sayed S, et al. The CK1δ/ε-AES axis regulates tumorigenesis and metastasis in colorectal cancer. Theranostics. 2021;11:4421–4435. doi: 10.7150/thno.53901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez N, Yang J, Hasselblatt K, Liu S, Zhou Y, Rauh-Hain JA, Ng SK, Choi PW, Fong WP, Agar NY, et al. Casein kinase I epsilon interacts with mitochondrial proteins for the growth and survival of human ovarian cancer cells. EMBO Mol Med. 2012;4:952–963. doi: 10.1002/emmm.201101094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Sato F, Tanimoto K, Rajeshwaran N, Thangavelu L, Makishima M, Bhawal UK. The potential roles of Dec1 and Dec2 in Periodontal inflammation. Int J Mol Sci. 2021;22:10349. doi: 10.3390/ijms221910349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato F, Bhawal UK, Sugiyama N, Osaki S, Oikawa K, Muragaki Y. Potential role of DEC1 in cervical cancer cells involving overexpression and apoptosis. Clocks Sleep. 2020;2:26–38. doi: 10.3390/clockssleep2010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhawal UK, Sato F, Arakawa Y, Fujimoto K, Kawamoto T, Tanimoto K, Ito Y, Sasahira T, Sakurai T, Kobayashi M, et al. Basic helix-loop-helix transcription factor DEC1 negatively regulates cyclin D1. J Pathol. 2011;224:420–429. doi: 10.1002/path.2878. [DOI] [PubMed] [Google Scholar]
- 14.Sato F, Bhawal UK, Yoshimura T, Muragaki Y. DEC1 and DEC2 crosstalk between circadian rhythm and tumor progression. J Cancer. 2016;7:153–159. doi: 10.7150/jca.13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Sato F, Yamada T, Bhawal UK, Kawamoto T, Fujimoto K, Noshiro M, Seino H, Morohashi S, Hakamada K, et al. The BHLH transcription factor DEC1 plays an important role in the epithelial-mesenchymal transition of pancreatic cancer. Int J Oncol. 2012;41:1337–1346. doi: 10.3892/ijo.2012.1559. [DOI] [PubMed] [Google Scholar]
- 16.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 17.Tsai ST, Jin YT. Proliferating cell nuclear antigen (PCNA) expression in oral squamous cell carcinomas. J Oral Pathol Med. 1995;24:313–315. doi: 10.1111/j.1600-0714.1995.tb01191.x. [DOI] [PubMed] [Google Scholar]
- 18.Yu SS, Cirillo N. The molecular markers of cancer stem cells in head and neck tumors. J Cell Physiol. 2020;235:65–73. doi: 10.1002/jcp.28963. [DOI] [PubMed] [Google Scholar]
- 19.Bourguignon LYW, Earle C, Shiina M. Activation of matrix hyaluronan-mediated CD44 signaling, epigenetic regulation and chemoresistance in head and neck cancer stem cells. Int J Mol Sci. 2017;18:1849. doi: 10.3390/ijms18091849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghazi N, Saghravanian N, Taghi Shakeri M, Jamali M. Evaluation of CD44 and TGF-B expression in oral carcinogenesis. J Dent (Shiraz) 2021;22:33–40. doi: 10.30476/DENTJODS.2020.84393.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato F, Otsuka T, Kohsaka A, Le HT, Bhawal UK, Muragaki Y. Smad3 suppresses epithelial cell migration and proliferation via the clock gene Dec1, which negatively regulates the expression of clock genes Dec2 and Per1. Am J Pathol. 2019;189:773–783. doi: 10.1016/j.ajpath.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Sato F, Muragaki Y, Zhang Y. DEC1 negatively regulates AMPK activity via LKB1. Biochem Biophys Res Commun. 2015;467:711–716. doi: 10.1016/j.bbrc.2015.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ooshima A, Park J, Kim SJ. Phosphorylation status at Smad3 linker region modulates transforming growth factor-β-induced epithelial-mesenchymal transition and cancer progression. Cancer Sci. 2019;110:481–488. doi: 10.1111/cas.13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts AB, Tian F, Byfield SD, Stuelten C, Ooshima A, Saika S, Flanders KC. Smad3 is key to TGF-beta-mediated epithelial-to-mesenchymal transition, fibrosis, tumor suppression and metastasis. Cytokine Growth Factor Rev. 2006;17:19–27. doi: 10.1016/j.cytogfr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Kon N, Hirota T, Kawamoto T, Kato Y, Tsubota T, Fukada Y. Activation of TGF-beta/activin signalling resets the circadian clock through rapid induction of Dec1 transcripts. Nat Cell Biol. 2008;10:1463–1469. doi: 10.1038/ncb1806. [DOI] [PubMed] [Google Scholar]
- 26.Ehata S, Hanyu A, Hayashi M, Aburatani H, Kato Y, Fujime M, Saitoh M, Miyazawa K, Imamura T, Miyazono K. Transforming growth factor-beta promotes survival of mammary carcinoma cells through induction of antiapoptotic transcription factor DEC1. Cancer Res. 2007;67:9694–9703. doi: 10.1158/0008-5472.CAN-07-1522. [DOI] [PubMed] [Google Scholar]
- 27.Zawel L, Yu J, Torrance CJ, Markowitz S, Kinzler KW, Vogelstein B, Zhou S. DEC1 is a downstream target of TGF-beta with sequence-specific transcriptional repressor activities. Proc Natl Acad Sci USA. 2002;99:2848–2853. doi: 10.1073/pnas.261714999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Q, Yang C, Gao F. The state of CD44 activation in cancer progression and therapeutic targeting. FEBS J. 2021 Sep 3; doi: 10.1111/febs.16179. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 29.Xiang R, Cui Y, Wang Y, Xie T, Yang X, Wang Z, Li J, Li Q. Circadian clock gene Per2 downregulation in non-small cell lung cancer is associated with tumour progression and metastasis. Oncol Rep. 2018;40:3040–3048. doi: 10.3892/or.2018.6704. [DOI] [PubMed] [Google Scholar]
- 30.Lin SH, Lin YM, Yeh CM, Chen CJ, Chen MW, Hung HF, Yeh KT, Yang SF. Casein kinase 1 epsilon expression predicts poorer prognosis in low T-stage oral cancer patients. Int J Mol Sci. 2014;15:2876–2891. doi: 10.3390/ijms15022876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.