Abstract
Throughout the world, numerous individuals are infected with Toxoplasma gondii, which may improve immunity against cancer. Furthermore, microRNAs (miRs) may be differentially expressed in the host upon infection with T. gondii. In the present study, RNA-sequencing analysis and reverse transcription-quantitative PCR revealed that miR-429-3p, miR-145a-5p, miR-211-5p, miR-31-3p and miR-135a-5p were determined to be downregulated, while miR-21a-3p, miR-135b-5p, miR-210-5p and miR-146-3p were upregulated in mice post-infection with T. gondii. Antitumor genes [TNF receptor superfamily member 11b, large tumor suppressor kinase (Lats)2 and Lats1] were identified as targets of miR-429-3p, miR-145a-5p, miR-211-5p, miR-31-3p and miR-135a-5p with a luciferase reporter assay. In addition, the protein levels of Lats2 and Lats1 were detected to be higher in T. gondii-infected mice than in the control group. Therefore, these results provide favorable evidence for the suppression of cancer upon T. gondii infection and may give novel ideas for the treatment of tumors.
Keywords: Toxoplasma gondii, miRNA-seq, cancer-associated miRNAs, tumor suppressor genes, suppression
Introduction
Approximately one in three individuals are, or have been previously, infected with Toxoplasma gondii (T. gondii) at varying degrees (1,2). Once the body's immune function is impaired, T. gondii may opportunistically cause diseases (3). Furthermore, T. gondii infection may lead to alterations in the expression of certain microRNAs (miRNAs/miRs) in their hosts. Thus, differentially expressed miRNAs from two genetically distinct strains of T. gondii were able to be developed as diagnostic biomarkers for toxoplasmosis (4). T. gondii infection was reported to specifically increase the levels of key host miRNAs (5). Comparison of splenocyte miRNA expression in pigs during acute and chronic infections indicated that differentially expressed miRNAs have important roles in the host's immune response to T. gondii infection by modulating the expression levels of cellular immunity-related cytokines and immune-related C-type lectins (6). The miRNA data from porcine alveolar macrophages infected with T. gondii first demonstrated the association between miRNAs and macrophages of swine origin (7). Differential brain miRNA expression in mice infected with T. gondii oocysts indicated that T. gondii infection may alter the abundance of miRNAs in the mouse brain, particularly at the chronic stage (8).
Previous studies have indicated that T. gondii infection is able to improve immunity against cancer in the host (9–13). A previous study by our group revealed changes in tumor-related factors after T. gondii infection (14). As infection with T. gondii may change the levels of miRNAs in hosts, it is necessary to analyze the expression of the targets of the cancer-related miRNAs in the host pre- and post-infection with T. gondii.
Materials and methods
Animal experiment
Female BALB/c mice (age, 8 weeks; mean weight, 20.25±0.93 g) were obtained from the Shandong University Laboratory Animal Center. A total of 48 mice were randomly divided into two groups (control group, n=24; infected group, n=24). They were reared in groups of six mice per cage under specific pathogen-free conditions under a 12-h light/dark cycle at 25°C and had ad libitum access to tap water and self-produced mouse feed (8). All animal experiments were approved by the Ethics Committee on Animal Experiments of the Medical School of Shandong University (Jinan, China). Each BALB/c mouse (infected group) was challenged with 10 T. gondii cysts by gavage and the spleens of experimental mice were collected for RNA extraction one month after the challenge. All efforts were made to minimize suffering and humane endpoints were used. Mice with unkempt fur and diarrhea were euthanized. For euthanasia, mice were placed in a chamber and CO2 was administered at a concentration of 60–70% over a 5-min exposure time, after which the cervical dislocation method was at times used to ensure that effective euthanasia had occurred.
Parasite
T. gondii (low virulent Prugniaud strain, obtained from the Department of Pathogen Biology, Anhui Medical University, Hefei, China) was maintained in our laboratory using the passage of cysts in eight-week-old Kunming mice obtained from Shandong University Laboratory Animal Center (mean weight, 42.31±5.31 g).
RNA extraction and high-throughput sequencing
RNA was obtained from different spleen samples (one mouse from the control group and one mouse from the infected group) using TRIzol reagent (Takara Bio, Inc.) according to the manufacturer's instructions. RNA was isolated using the improved cetyltrimethylammonium bromide (CTAB) method, applying isopropanol instead of lithium chloride for RNA precipitation. In brief, one gram of spleen sample was added to liquid nitrogen and ground into a fine powder, which was evenly mixed in 5 ml preheated (65°C) extraction buffer (2% CTAB, 2% polyvinylpyrrolidone, 0.1 M Tris-HCl, 2.0 M NaCl, 25 mM EDTA, 2% beta-mercaptoethanol; pH 8.0). The mixture was incubated for 5 min at 65°C and shaken three times during the incubation period. After a short cooling period, isopropanol (2.5 ml) was added to the mixture, after which the mixture was vortexed for 1 min and centrifuged at 10,900 × g for 15 min at 4°C. DNase was then used to treat the extract and RNA was precipitated at 25°C for 10 min using the same volume of isopropanol. The extracted RNA was first resuspended in an equal volume of phenol/chloroform/isopropanol mixture (25:24:1) and then in an equal volume of chloroform/isopropanol (24:1). Both a 0.1 volume of 3M NaOAC (pH 5.2) and 2.5 volumes of cold ethanol were added to the mixture to precipitate the RNA overnight at −20°C. The quality of the prepared RNA from each sample was analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). The fragmentation buffer was added to cut the mRNA into short fragments (200–700 nucleotides). First-strand cDNA was synthesized using the templates of the short fragments. DNA polymerase I (New England Biolabs, Inc.), dNTPs, RNase H (Invitrogen; Thermo Fisher Scientific, Inc.) and buffer were used to synthesize the second-strand cDNA. The fragments were purified using the QiaQuick PCR kit and washed with EB buffer (consists of sodium chloride, magnesium chloride, HEPES and sucrose) for end repair prior to adding polyA tails and adaptors. Fragments of suitable size were detected by agarose gel electrophoresis and amplified using PCR (8), after which the products were sequenced using Illumina HiSeq™ 2000 (Illumina, Inc.).
Bioinformatics analysis and identification of miRNA targets
A total of two rat miRNA transcriptome libraries were sequenced using the Illumina HiSeq 2000 platform. Details of the raw data are listed in Table I. High-quality 20±21 nucleotide-long reads were processed with the CleaveL pipeline for small RNA target identification, as previously described (15). Sequences of ribosomal RNAs, transfer RNAs, small nucleolar RNAs and small nuclear RNAs were retrieved from the RNA families database (xfam.org). The control (S01) and infected (S02) sample data were analyzed separately. P≤0.01 and fold change ≥2 were used to identify significant differentially expressed genes. The miRDB software (16) was used to predict the target genes of differentially expressed miRNAs.
Table I.
Raw data before filtering of sequencing results.
| Group | Raw data (bp) | Raw reads | Q30 (%) | Clean reads |
|---|---|---|---|---|
| S01 | 1407676653 | 27601503 | 98.17 | 27295019 |
| S02 | 1398651642 | 27424542 | 98.15 | 27008080 |
Gene ontology (GO) functional enrichment and kyoto encyclopedia of genes and genomes (KEGG) pathway analysis
The selected sequence was constructed using BLASTX and the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/guide/sequence-analysis/) to better observe the function of miRNA targets and the metabolic regulatory networks related to mouse miRNAs. BLASTX searches using the InterPro and KEGG databases were used to collect the predicted target proteins with an E-value of 1×1030. Target gene function and metabolic pathways of miRNAs were verified using the best hits. Finally, the terms in the categories molecular function, biological process and cellular component of target genes were obtained using the GO and InterPro databases (https://ngdc.cncb.ac.cn/databasecommons/). The GO terms and KEGG pathways with P≤0.01 and fold change ≥2 were considered significant.
miRNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
miRNAs were extracted from spleen samples using the miRNeasy Serum/Plasma Kit (Qiagen GmbH) according to the manufacturer's protocol. Subsequently, miRNA was reverse transcribed into cDNA using the One Step PrimeScript & Reg miRNA cDNA Synthesis Kit (Takara Bio, Inc.). PCR was performed with the Prime-Script™ RT reagent kit (Takara Bio, Inc.) according to the manufacturer's protocol and in an ABI 7000 real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the following thermocycling conditions: 95°C for 5 sec and 58°C for 20 sec for 35 cycles, and 72°C for 30 sec. The experiments were performed once and set up in triplicate. The mRNA levels of large tumor suppressor kinase (Lats)2, Lats1 and TNF receptor superfamily member 11b (Tnfrsf11b) were detected by qPCR with the 2−∆∆Cq quantification method (17) and the primers are listed in Table II.
Table II.
Primers used for quantitative PCR (5′-3′).
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Lats1 | GTGCAACATTCAATTAACCG | TCCAGACAGAGGTCTTCCTA |
| Lats2 | TGAGCAGATTGTGCGAGTCA | GCGGCGGGGCCCTCGTAGTT |
| Tnfrsf11b | TGTGCTGCGCACTCCTGGTGC | TGCAGTGCTGTTTTAGGTAGG |
| β-actin | TAGGCACCAGGGTGTGATGG | GTGCCAGATCTTCTCCATGTC |
Lats, large tumor suppressor kinase; Tnfrsf11b, TNF receptor superfamily member 11b.
Luciferase activity assay
Selected sequences from wild-type (WT) 3′-UTRs were cloned into the pMir-reporter vector (Ambion; Thermo Fisher Scientific, Inc.) and the mutant 3′-UTRs were generated by altering the predicted miR-31-5p, miR-135a-5p, miR-145a-5p, miR-204-5p, miR-211-5p and miR-429-3p 3′-UTR binding sites via a two-step PCR approach (the template was obtained from mouse miRNAs) (18). The miRNA mimics are listed in Table III. 293-T cells were purchased from FuXiang Biotechnology Co. 293-T cells were cultivated in Dulbecco's modified Eagles medium (DMEM) with antibiotics and Fetal Bovine Serum at 37°C in a humidified 5% CO2 atmosphere. Cells were co-transfected with either a WT or mutant 3′-UTR reporter vector and miR-31-5p, miR-135a-5p, miR-145a-5p, miR-204-5p, miR-211-5p and miR-429-3p mimics or negative control constructs and were cultured for 24 h. The cells were then assessed to ascertain their luciferase activity using a dual-luciferase reporter assay system (Promega Corporation). The experiments were performed once and set up in triplicate.
Table III.
Sequences for miRNA mimics.
| miRNA | Mimic |
|---|---|
| miR-31-5p | AGGCAAGAUGCUGGCAUAGCUG |
| miR-135a-5p | UAUGGCUUUUUAUUCCUAUGUGA |
| miR-145a-5p | GUCCAGUUUUCCCAGGAAUCCCU |
| miR-204-5p | UUCCCUUUGUCAUCCUAUGCCU |
| miR-211-5p | UUCCCUUUGUCAUCCUUUGCCU |
| miR-429-3p | UAAUACUGUCUGGUAAUGCCGU |
miRNA/miR, microRNA.
Western blot analysis
In brief, the samples were treated with RIPA lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing 1 mM protease inhibitor phenylmethanesulfonyl fluoride and centrifuged at 12,100 × g at 4°C for 10 min. The supernatant was then separated and mixed with 50 µl of SDS-PAGE sample buffer and boiled for 5 min, after which 5 µg (the protein concentration was determined by bicinchoninic acid protein assay kit) per lane was loaded onto the polyacrylamide gel and SDS-PAGE was performed with a 10% gel. Proteins were transferred onto polyvinylidene fluoride membranes (Beyotime Institute of Biotechnology) via electrophoresis, performed at 80 V for 3 h, using a Bio-Rad transfer system (Bio-Rad Laboratories, Inc.). The membranes were blocked for 2 h with skimmed milk (Beyotime Institute of Biotechnology) at room temperature and probed with the corresponding antibody (rabbit) diluted in blocking buffer at 1:10,000 (anti-LATS1, cat. no. ab70561; anti-LATS2, cat. no. ab111054; Anti-Tnfrsf11b, cat. no. ab183910; all from Abcam) at 4°C for 12 h. The membrane was then incubated for 2 h at room temperature with horseradish peroxidase-labeled goat anti-rabbit IgG antibody (cat. no. 18772; MilliporeSigma), diluted in blocking buffer at 1:20,000, and signals were detected with a super-sensitive signal enhanced chemiluminescence system (Beyotime Institute of Biotechnology). The levels of Lats2, Lats1 and Tnfrsf11b proteins were measured in infected and control mice in this experiment. The experiments were performed once and set up in triplicate.
Statistical analysis
Values are expressed as the mean ± standard deviation. Statistically significant differences between two groups were determined using Student's t-test. ANOVA and least-significant difference test were used for comparison among multiple groups. The results of the western blot were analyzed using ImageJ software (v1.51; National Institutes of Health). P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of miRNAs in the presence or absence of T. gondii
A total of 1,899 miRNAs were identified after a series of filtering. Of these, 327 miRNAs were identified as known miRNAs, whereas 477 were identified as homologs in miRbase. Specifically, the expression of 1,000 miRNAs was detected in the control group, among which 320 miRNAs were known miRNAs and 362 were novel miRNAs. Furthermore, the expression of 996 miRNAs was detected in the infected group, of which 304 were known miRNAs and 202 were novel miRNAs. Of the 1,899 miRNAs identified, 114 were specifically expressed in the control group, while 110 were specifically expressed in the infected group. Furthermore, it was observed that most of the known miRNAs, such as miR-99a-5p, miR-143-5p and miR-27b-5p, were highly expressed in the control and infected groups, which was not unexpected. However, various novel miRNAs exhibited low expression or were expressed in only one group (Table SI).
Differentially expressed miRNAs following exposure to T. gondii
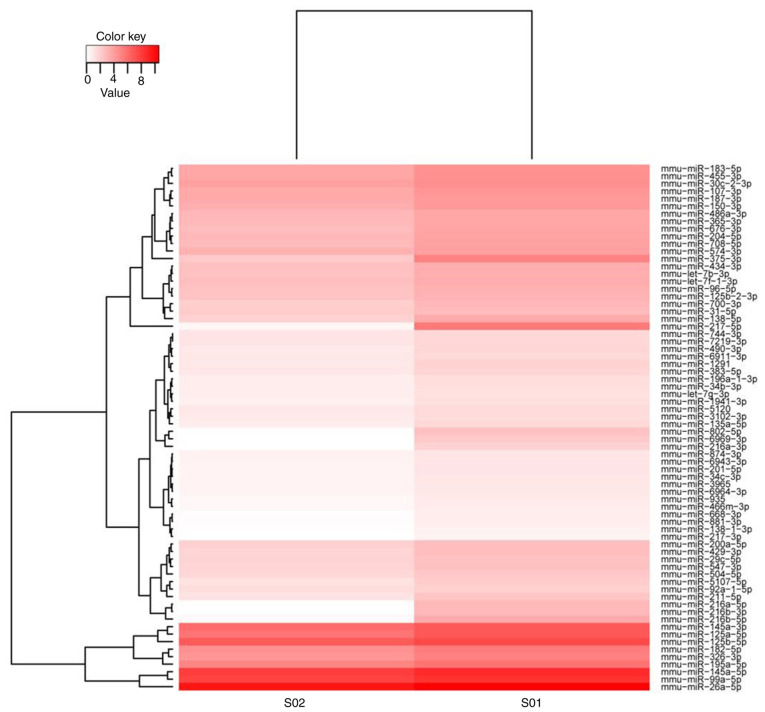
The differentially expressed miRNAs identified are listed in Table SII. The expression levels of a total of 398 miRNAs were significantly changed (P<0.05; cut-off criterion: Fold change ≥2) after T. gondii challenge. Compared with the control group, 111 miRNAs were upregulated and 287 miRNAs were downregulated in the infected group. The heatmap indicated that 72 miRNAs were markedly differentially expressed (P<0.01; cut-off criterion: Fold change ≥3) between the infected and control groups. Among these differentially expressed miRNAs, 18 were upregulated and 54 were downregulated (Fig. 1).
Figure 1.
Heat map of the differentially expressed miRNAs. Groups: S01, Control group; S02, T. gondii-infected group. miRNA/miR, microRNA.
GO and KEGG enrichment analyses of cancer-related target genes
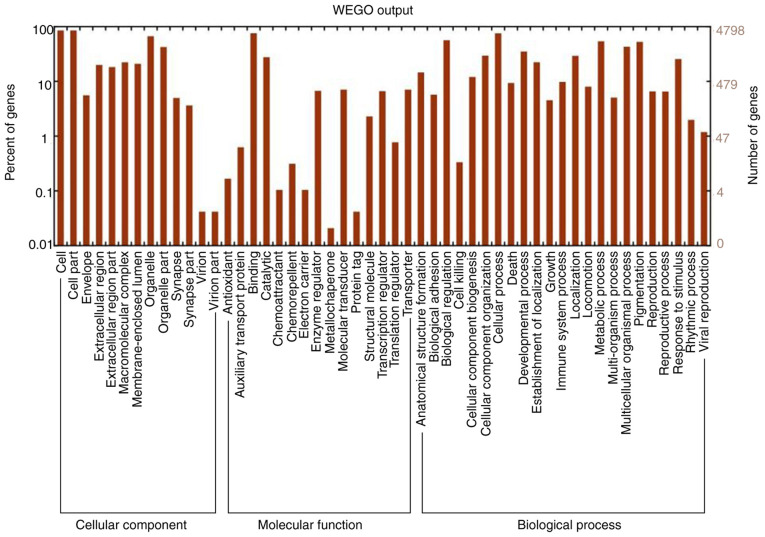
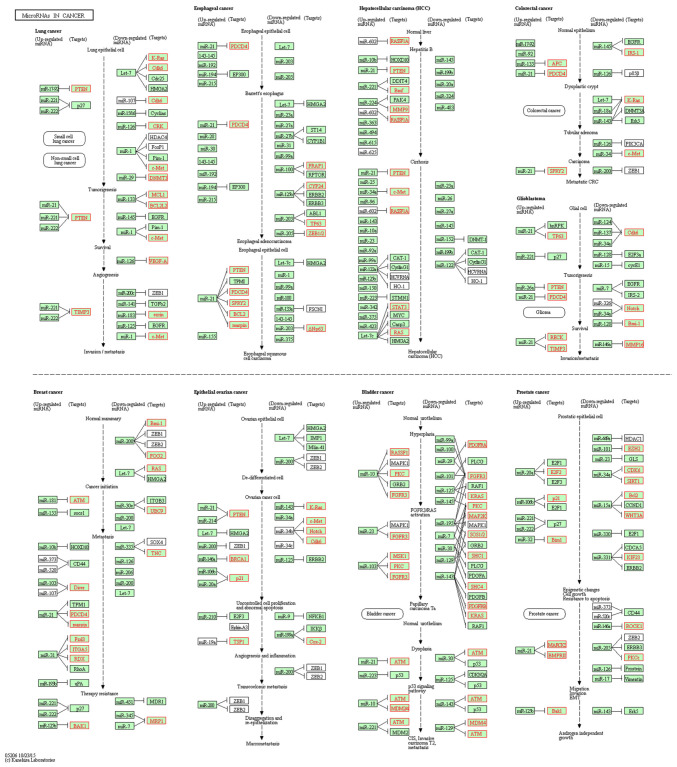
As presented in Fig. 2, the top enriched GO terms for cancer-related genes were determined. Furthermore, KEGG enrichment analysis was performed using the target mRNAs of the differentially expressed miRNAs. The functional terms and numbers of genes in the categories cellular component, molecular function and biological process are listed in Fig. 2. In the category cellular component, genes were associated with cell part, envelope and organelle. In the category molecular function, terms associated with binding, catalytic activity, enzyme regulation and molecular transduction were significant. In the category biological process, significantly enriched terms were associated with biological regulation, cellular process, developmental process, death, metabolic process and pigmentation. In Fig. 3, KEGG pathways associated with cancer signaling are presented. Various miRNAs, including miR-145, miR-31, miR-21, miR-107, miR-125, miR-135 and miR-96, were analyzed. Compared with the control group, the genes PDCD4, PKC, ITGA5 and ATM were upregulated, while genes such as CYP24, Fzd3, p21 and RDX were downregulated in the infected group (Table SIII). Certain genes did not change (e.g. ERBB2, RAF1 and ERBB3) in the infected group.
Figure 2.
Gene Ontology classification of assembled unigenes. The functional enrichment of differentially expressed genes was classified into 3 categories: Molecular function, cellular component and biological process. The x-axis indicates the specific category of genes in those main categories. The y-axis indicates the number of genes in a category.
Figure 3.
Kyoto Encyclopedia of Genes and Genomes enriched miRNAs predicted to be involved in cancer signaling pathways, including pathways in lung cancer, esophageal cancer, hepatocellular carcinoma, colorectal cancer, glioblastoma, epithelial ovarian cancer, bladder cancer, prostate cancer and breast cancer. Red indicates differential expression in the infected group compared with the control group.
Prediction of cancer-related genes targeted by differentially expressed miRNAs
Through the degradation of target gene mRNAs or the inhibition of the translation of target transcripts, miRNAs have important roles in cell proliferation and differentiation, apoptosis and a variety of diseases. A total of 8,116 target genes were analyzed for 45 differentially expressed cancer-related genes using miRDB (Table SIV). On the one hand, miRNAs are able to target a variety of cancer-related genes. For instance, miR-211-5p targets Tnfaip8, C1qtnf3, Lats2, Lats1 and St7; miR-145a-5p targets Tnfrsf11b, Etaa1 and C1qtnf9; and miR-429-3p targets Lats2, Lrp1b, Mtus1 and Tnfrsf11b. On the other hand, a single cancer-related gene may be targeted by multiple miRNAs. For instance, the genes Tnfaip8, C1qtnf3, Lats2, Lats1 and St7 may all be targeted by both miR-211-5p and miR-204-5p. Furthermore, miR-211-5p, miR-31-5p, miR-135a-5p, miR-204-5p and miR-429-3p are all able to target the Lats2 gene. The Lats1 gene is able to be targeted by four miRNAs (miR-201-5p, miR-211-5p, miR-204-5p and miR-107-3p), whereas Lzts3 is only targeted by miR-138-5p.
RT-qPCR confirmation of differentially expressed miRNAs
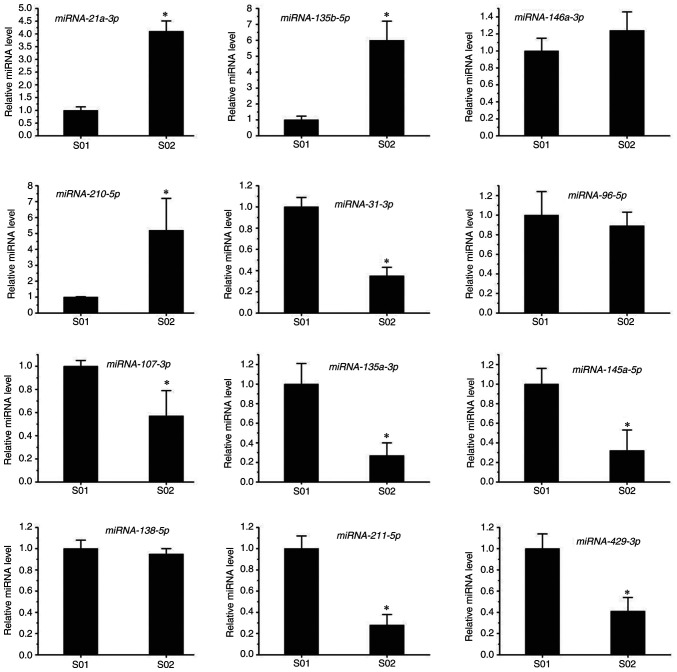
To further confirm the miRNA-seq results, RT-qPCR was performed in the present study. Candidate miRNAs were selected from cancer metabolic signaling pathways. Compared with the control group, miR-210-5p, miR-135b-5p, miR-21a-3p and miR-146a-3p were upregulated, while miR-135a-5p, miR-125a-5p, miR-145a-5p, miR-107-3p, miR-211-5p, miR-429-3p, miR-96-5p and miR-31-3p were downregulated in the infected group (Fig. 4). The RT-qPCR results suggested that the levels of miR-210-5p, miR-135b-5p and miR-21a-3p were significantly upregulated after T. gondii infection, whereas no difference in miR-146a-3p levels was observed in the infected group. The levels of miR-210-5p and miR-135b-5p in the infected group were five times higher than those in the control group. In addition, the levels of miR-135a-5p, miR-125a-5p, miR-145a-5p and miR-31-3p were markedly downregulated after infection with T. gondii, while no difference in the levels of miR-96-5p or miR-138-5p was observed between the infected and control groups. The levels of miR-135a-5p and miR-145a-5p in the infected group were only one-quarter of those in the control group. These results were in line with those of the RNA-seq.
Figure 4.
Validation of the difference in expression of the miRNAs using reverse transcription-quantitative PCR. *P<0.05 vs. S01. Groups: S01, Control group; S02, Infected group.
Luciferase activity assay
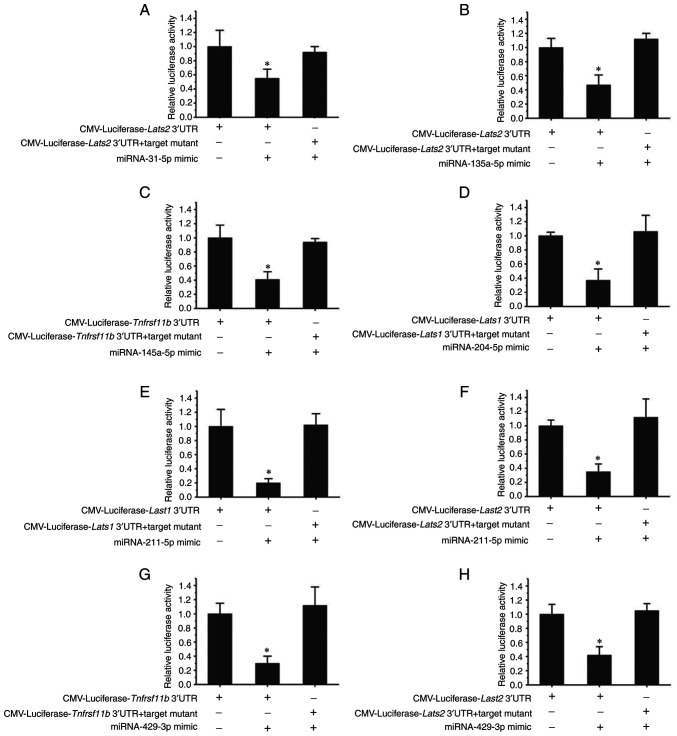
Targets of miR-31-5p, miR-135a-5p, miR-145a-5p, miR-204-5p, miR-211-5p and miR-429-3p were verified by luciferase activity assays in 293T cells (Fig. 5). While miR-31-5p, miR-135a-5p, miR-211-5p and miR-429-3p were confirmed to be able to target the Lats2 gene, miR-204-5p and miR-211-5p target the Lats1 gene. In addition, miR-145a-5p and miR-429-3p were indicated to be targets of Tnfrsf11b. Furthermore, miR-211-5p was able to target the Lats1 and Lats2 genes, and miR-429-3p was confirmed to target the Tnfrsf11b and Lats2 genes.
Figure 5.
Verification of the targets of miR-31-5p, miR-135a-5p, miR-145a-5p, miR-204-5p, miR-211-5p and miR-429-3p by luciferase activity assay. (A and B) The same target (Lats2) was verified for (A) miR-31-5p and (B) miR-135a-5p. (C) Tnfrsf11b was verified as a target of miR-145a-5p. (D) Lats1 was verified as a target of miR-204-5p. (E) Lats2 and (F) Lats1 were verified as targets of miR-211-5p. (G) Lats2 and (H) Tnfrsf11b were verified as targets of miR-429-3p. *P<0.05 vs. other groups. CMV, cytomegalovirus; miRNA/miR, microRNA; Lats, large tumor suppressor kinase; Tnfrsf11b, TNF receptor superfamily member 11b.
Protein changes
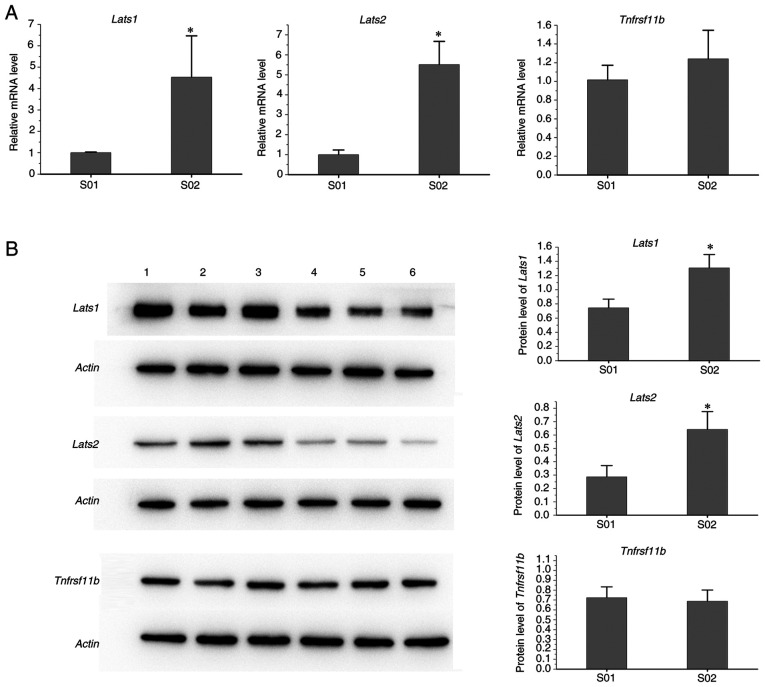
As presented in Fig. 6, the protein levels of Lats1 and Lats2 in spleen samples of infected mice were higher than those in the control group, while the levels of Tnfrsf11b protein from the infected group were similar to those of the control mice.
Figure 6.
Validation of the differentially expressed genes by reverse transcription-quantitative PCR and western blot. (A) mRNA levels of Lats2, Lats1 and Tnfrsf11b. (B) Protein levels of Lats2, Lats1 and Tnfrsf11b. Lanes 1–3 are for individual mice from the S02 group and lanes 4–6 from the S01 group. *P<0.05 vs. S01. Groups: S01, Control group; S02, Infected group. Lats, large tumor suppressor kinase; Tnfrsf11b, TNF receptor superfamily member 11b.
Discussion
T. gondii is most likely to manipulate pathways involved in host innate immunity, biosynthesis and transferase activity (7). Furthermore, the levels of miRNAs in host cells were previously reported to be acutely changed after T. gondii infection; compared with uninfected DC2.4 cells, 3,434 differentially expressed miRNAs were obtained from T. gondii-infected DC2.4 cells after high-throughput sequencing (19). Host miRNA expression is altered by T. gondii, which is reflected in the differences in regulation of important biological processes that are related to host responses to both chronic and acute T. gondii infection (6). miRNAs of porcine macrophages were reported to be differentially expressed after T. gondii infection (7); this knowledge may aid in the investigation of T. gondii infections. Hu et al (8) indicated that T. gondii infection, particularly at the chronic stage, may change the abundance of miRNAs in the mouse brain. Another previous study suggested that miRNAs may be related to the communication between hosts and T. gondii and several specific miRNAs have been proposed and confirmed (20). miRNA expression in the brain of mice may change with the invasion of cyst-forming T. gondii (21). Chronic and acute infections may cause differential expression of miRNAs in the host. In the present study, compared with the control group, 207 miRNAs were upregulated and 414 miRNAs were downregulated in the T. gondii-infected group. The fact that certain miRNAs were differentially expressed upon T. gondii infection was consistent with previous studies.
Infection with T. gondii may stimulate immunity against tumors (11,13,22). Furthermore, the levels of tumor-related mRNAs were altered in T. gondii-infected mice from a previous study by our group (14). In the present study, the levels of numerous miRNAs were changed in infected mice compared to the control group. Specifically, the levels of cancer-related miRNAs, such as miR-135a-5p, miR-135b-5p, miR-145a-5p, miR-146a-3p, miR-21a-3p, miR-107-3p, miR-31-3p, miR-96-5p, miR-210-5p, miR-211-5p and miR-429-3p, were altered in the infected mice. miRNAs have important roles in cancer. miR-135a-5p, a significant tumor regulator, may affect the development of diverse cancers by impacting multiple genes in oncogenic pathways (23–25). miR-135b-5p was able to enhance doxorubicin sensitivity in breast cancer cells by targeting anterior gradient 2 protein (26). miR-96-5p was reported to promote the migration and proliferation of ovarian cancer cells by suppressing caveolae 1 (27). In the present study, the RT-qPCR results indicated that the levels of miR-210-5p, miR-135b-5p and miR-21a-3p were significantly upregulated after infection with T. gondii, while the levels of miR-135a-5p, miR-125a-5p, miR-145a-5p, miR-211-5p, miR-429-3p and miR-31-3p were markedly downregulated after T. gondii challenge. miR-135a-5p was previously reported to inhibit head and neck squamous cell carcinoma (HNSCC) cell proliferation and promote apoptosis by directly targeting HOXA10, suggesting the importance of miR-135a-5p in HNSCC treatment (28). miR-125a-5p was downregulated in colorectal cancer tissues and cell lines and inhibited colorectal cancer cell proliferation, migration and invasion; it also reduced the ability of human umbilical vein endothelial cells to form tubes (29). Overexpression of miR-31-3p inhibited malignant behaviors and epithelial-to-mesenchymal transition of cervical cancer cells in vitro (30).
In the present study, cancer-related genes (Lats2, Lats1 and Tnfrsf11b) were targeted by miR-429-3p, miR-211-5p, miR-145a-5p, miR-31-3p, miR-204-5p and miR-135a-5p. In a previous study, the expression of both Lats2 and Lats1 was significantly downregulated in human breast cancer and loss of either one accelerated mammary tumorigenesis in mice (31). Lats2 was able to inhibit oncogenic Wnt/β-catenin-mediated transcription by disrupting β-catenin/BCL9 interactions, suggesting that it may be an important target for anti-cancer therapies (32). Nucleocytoplasmic translocation of Lats1 protein and upregulated expression of Lats1 mRNA were observed during tumorigenesis of HNSCC (33). In addition, cancer-related miRNAs were altered in mice after T. gondii infection. The altered miRNAs affected cancer-related genes, suggesting that the challenge with T. gondii may alter the host's resistance to tumors. In the present study, the levels of miR-429-3p, miR-211-5p, miR-145a-5p, miR-31-3p and miR-135a-5p were downregulated after infection, indicating that a smaller number of antitumor genes (Lats2, Lats1 and Tnfrsf11b) may be targeted in infected mice compared with others. Furthermore, the protein levels of Lats2 and Lats1 were markedly increased in infected mice compared with the control group, suggesting that the altered miRNAs may enhance the expression of antitumor genes. Consequently, T. gondii infection may increase host resistance to tumors, which is consistent with the results of the previous study by our group (14). Furthermore, previous studies suggested that infection with T. gondii may improve immunity against cancer in the host (9–12). High expression of antitumor genes may have an important role in immunity.
Numerous miRNAs have been indicated to be altered in mice infected with T. gondii. Cancer-related miRNAs (miR-429-3p, miR-211-5p, miR-145a-5p, miR-31-3p and miR-135a-5p) were downregulated, suggesting that a smaller amount of antitumor genes were targeted and more antitumor proteins were expressed. In the present study, the findings were favorable for the suppression of cancer and may provide novel ideas for the treatment of tumors. The current study aimed to explore the tumor factors affected by T. gondii. Although certain cancer-related proteins were altered in mice after T. gondii infection, the exact implication and type of cancer this may affect remain elusive and require further exploration. In further studies, a variety of mouse tumor models may be used as experimental models. Attenuated T. gondii may be inoculated prior to and after tumorigenesis. The tumor growth of mice may be observed and the changes of tumor factors detected. The tumor model with the greatest impact may be obtained by comparison. The upstream and downstream molecules of the tumor factor may be detected to explore the possible pathways through which T. gondii affects the growth of tumors in the host.
In conclusion, cancer-associated miRNAs were altered in mice after T. gondii infection. Antitumor genes (Tnfrsf11b, Lats2 and Lats1) were changed accordingly. In addition, a larger amount of Lats2 and Lats1 protein expression was detected in T. gondii-infected mice than in the control group, indicating that T. gondii may enhance host immunity against cancer by enhancing the abundance of antitumor proteins. These findings are favorable for the suppression of cancer and may provide novel ideas for the treatment of tumors.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
This work was supported by the National Foundation of Natural Science of China (grant no. 81902079) and the Science Foundation of Shandong Province (grant no. ZR2013HM033).
Availability of data and materials
The sequencing data generated in the present study may be obtained from the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/sra; accession no. PRJNA803770). The other datasets used and/or analyzed in the present study are available from the corresponding author on reasonable request.
Authors' contributions
GL conceived and designed the study and critically revised the manuscript. LW performed the experiments and drafted the manuscript. NW and YHZ contributed to the analysis and interpretation of data. GL and YHZ check and approve the authenticity of the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Institutional Animal Care and Use Committee of Shandong University (no. 2011–0015). The animals were kept and the experiments were performed in accordance with the committee's criteria for the care and use of laboratory animals. This was also in accordance with the NIH's Guide for the Care and Use of Laboratory Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kochanowsky JA, Koshy AA. Toxoplasma gondii. Curr Biol. 2018;28:R770–R771. doi: 10.1016/j.cub.2018.05.035. [DOI] [PubMed] [Google Scholar]
- 2.Coutermarsh-Ott S. Toxoplasma gondii as a model of in vivo host-parasite interactions. Methods Mol Biol. 2019;1960:237–247. doi: 10.1007/978-1-4939-9167-9_21. [DOI] [PubMed] [Google Scholar]
- 3.Lima TS, Lodoen MB. Mechanisms of human innate immune evasion by Toxoplasma gondii. Front Cell Infect Microbiol. 2019;9:103. doi: 10.3389/fcimb.2019.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menard KL, Haskins BE, Denkers EY. Impact of Toxoplasma gondii infection on host non-coding RNA responses. Front Cell Infect Microbiol. 2019;9:132. doi: 10.3389/fcimb.2019.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medina L, Castillo C, Liempi A, Guerrero-Muñoz J, Rojas-Pirela M, Maya JD, Prieto H, Kemmerling U. Trypanosoma cruzi and Toxoplasma gondii induce a differential MicroRNA profile in human placental explants. Front Immunol. 2020;11:595250. doi: 10.3389/fimmu.2020.595250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou Z, Liu D, Su S, Wang L, Zhao Z, Ma Y, Li Q, Jia C, Xu J, Zhou Y, Tao J. Comparison of splenocyte microRNA expression profiles of pigs during acute and chronic toxoplasmosis. BMC Genomics. 2019;20:97. doi: 10.1186/s12864-019-5458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li SY, Yang J, Wang LY, Du F, Zhao JL, Fang R. Expression profile of microRNAs in porcine alveolar macrophages after Toxoplasma gondii infection. Parasit Vectors. 2019;12:65. doi: 10.1186/s13071-019-3297-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu RS, He JJ, Elsheikha HM, Zhang FK, Zou Y, Zhao GH, Cong W, Zhu XQ. Differential brain MicroRNA expression profiles after acute and chronic infection of mice with Toxoplasma gondii oocysts. Front Microbiol. 2018;9:2316. doi: 10.3389/fmicb.2018.02316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders KL, Fox BA, Bzik DJ. Attenuated Toxoplasma gondii therapy of disseminated pancreatic cancer generates long-lasting immunity to pancreatic cancer. Oncoimmunology. 2015;5:e1104447. doi: 10.1080/2162402X.2015.1104447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders KL, Fox BA, Bzik DJ. Attenuated Toxoplasma gondii stimulates immunity to pancreatic cancer by manipulation of myeloid cell populations. Cancer Immunol Res. 2015;3:891–901. doi: 10.1158/2326-6066.CIR-14-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Poppoe F, Chen J, Yu L, Deng F, Luo Q, Xu Y, Cai Y, Shen J. Macrophages polarized by expression of ToxoGRA15II inhibit growth of hepatic carcinoma. Front Immunol. 2017;8:137. doi: 10.3389/fimmu.2017.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pyo KH, Lee YW, Lim SM, Shin EH. Immune adjuvant effect of a Toxoplasma gondii profilin-like protein in autologous whole-tumor-cell vaccination in mice. Oncotarget. 2016;7:74107–74119. doi: 10.18632/oncotarget.12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox BA, Sanders KL, Rommereim LM, Guevara RB, Bzik DJ. Secretion of rhoptry and dense granule effector proteins by nonreplicating Toxoplasma gondii uracil auxotrophs controls the development of antitumor immunity. PLoS Genet. 2016;12:e1006189. doi: 10.1371/journal.pgen.1006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu G, Zhou J, Zhao YH, Li QL, Gao YY, Wang L. Transcriptome sequencing investigated the tumor-related factors changes after T. gondii infection. Front Microbiol. 2019;10:181. doi: 10.3389/fmicb.2019.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang T, Pan H, Wang J, Yang W, Cheng T, Zhang Q. Identification and profiling of novel and conserved microRNAs during the flower opening process in Prunus mume via deep sequencing. Mol Genet Genomics. 2014;289:169–183. doi: 10.1007/s00438-013-0800-6. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Wang X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 18.Pjevac P, Hausmann B, Schwarz J, Kohl G, Herbold CW, Loy A, Berry D. An economical and flexible dual barcoding, two-step PCR approach for highly multiplexed amplicon sequencing. Front Microbiol. 2021;12:669776. doi: 10.3389/fmicb.2021.669776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li DL, Zou WH, Deng SQ, Peng HJ. Analysis of the differential exosomal miRNAs of DC2.4 dendritic cells induced by Toxoplasma gondii infection. Int J Mol Sci. 2019;20:5506. doi: 10.3390/ijms20215506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acar İE, Saçar Demirci MD, Groß U, Allmer J. The expressed MicroRNA-mRNA interactions of Toxoplasma gondii. Front Microbiol. 2018;8:2630. doi: 10.3389/fmicb.2017.02630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou CX, Ai K, Huang CQ, Guo JJ, Cong H, He SY, Zhu XQ. miRNA and circRNA expression patterns in mouse brain during toxoplasmosis development. BMC Genomics. 2020;21:46. doi: 10.1186/s12864-020-6464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatai H, Lepelley A, Zeng W, Hayden MS, Ghosh S. Toll-like receptor 11 (TLR11) interacts with flagellin and profilin through disparate mechanisms. PLoS One. 2016;11:e0148987. doi: 10.1371/journal.pone.0148987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Jiang WL, Yang JY, Huang J, Kang G, Hu HB, Xie S. Downregulation of lysyl oxidase-like 4 LOXL4 by miR-135a-5p promotes lung cancer progression in vitro and in vivo. J Cell Physiol. 2019;234:18679–18687. doi: 10.1002/jcp.28508. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y, Zheng B, Meng X, Yan Y, He J, Liu Y. LncRNA DANCR promotes the proliferation, migration, and invasion of tongue squamous cell carcinoma cells through miR-135a-5p/KLF8 axis. Cancer Cell Int. 2019;19:302. doi: 10.1186/s12935-019-1016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei X, Yang X, Wang B, Yang Y, Fang Z, Yi C, Shi L, Song D. LncRNA MBNL1-AS1 represses cell proliferation and enhances cell apoptosis via targeting miR-135a-5p/PHLPP2/FOXO1 axis in bladder cancer. Cancer Med. 2020;9:724–736. doi: 10.1002/cam4.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Xia F, Zhang F, Cui Y, Wang Q, Liu H, Wu Y. miR-135b-5p enhances doxorubicin-sensitivity of breast cancer cells through targeting anterior gradient 2. J Exp Clin Cancer Res. 2019;38:26. doi: 10.1186/s13046-019-1024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B, Zhang J, Yang D. miR-96-5p promotes the proliferation and migration of ovarian cancer cells by suppressing caveolae1. J Ovarian Res. 2019;12:57. doi: 10.1186/s13048-019-0533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo LM, Ding GF, Xu WC, Ge H, Jiang Y, Chen XJ, Lu YF. MiR-135a-5p represses proliferation of HNSCC by targeting HOXA10. Cancer Biol Ther. 2018;19:973–983. doi: 10.1080/15384047.2018.1450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Qiu J, Kang H, Wang Y, Qian J. miR-125a-5p suppresses colorectal cancer progression by targeting VEGFA. Cancer Manag Res. 2018;10:5839–5853. doi: 10.2147/CMAR.S161990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jing L, Bo W, Yourong F, Tian W, Shixuan W, Mingfu W. Sema4C mediates EMT inducing chemotherapeutic resistance of miR-31-3p in cervical cancer cells. Sci Rep. 2019;9:17727. doi: 10.1038/s41598-019-54177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furth N, Pateras IS, Rotkopf R, Vlachou V, Rivkin I, Schmitt I, Bakaev D, Gershoni A, Ainbinder E, Leshkowitz D, et al. LATS1 and LATS2 suppress breast cancer progression by maintaining cell identity and metabolic state. Life Sci Alliance. 2018;1:e201800171. doi: 10.26508/lsa.201800171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Chen X, Ding X, Cheng Y, Zhao B, Lai ZC, Al Hezaimi K, Hakem R, Guan KL, Wang CY. LATS2 suppresses oncogenic Wnt signaling by disrupting β-catenin/BCL9 interaction. Cell Rep. 2020;31:107792. doi: 10.1016/j.celrep.2020.107792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Zhao Z, Zhang H, Kong F, Jiang H, Huang K, Zheng H. LATS1 inhibits metastasis and epithelial-mesenchymal transition in head and neck squamous cell carcinoma. Int J Clin Exp Pathol. 2018;11:2053–2063. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data generated in the present study may be obtained from the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/sra; accession no. PRJNA803770). The other datasets used and/or analyzed in the present study are available from the corresponding author on reasonable request.