Abstract
Macrolide 2′-phosphotransferase [MPH(2′)] transfers the γ phosphate of ATP to the 2′-OH group of macrolide antibiotics. The role of aspartic acids in the putative ATP-binding site of MPH(2′)II was investigated through the substitution of alanine for aspartate by site-directed mutagenesis. D200A, D209A, D219A, and D231A mutant strains were unable to inactivate the substrate oleandomycin, while a D227A mutant retained 7% of the activity of the original enzyme.
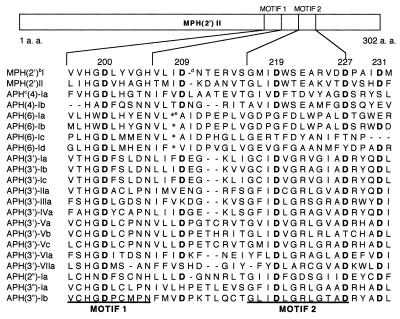
Reported macrolide resistance mechanisms are as follows: (i) methylation (2) of adenine in 23S rRNA (2058 in Escherichia coli), (ii) efflux protein (13), and (iii) inactivation of macrolide by erythromycin esterase (1, 3, 17) or macrolide 2′-phosphotransferase [MPH(2′)]. MPH(2′) is divided into MPH(2′)I (14, 18) and MPH(2′)II (10, 15) on the basis of its substrate specificity and primary amino acid sequence. The phosphotransferases are encoded by mphA and mphB, respectively. The former inactivates 14-membered ring macrolides more effectively than 16-membered ring macrolides, whereas the latter does not show this substrate preference. The primary amino acid sequence similarity between MPH(2′) and aminoglycoside phosphotransferase (APH) is poor, but the C-terminal regions have highly conserved motifs 1 and 2 in common (Fig. 1). They are the putative ATP-binding sites of APH, in which several functional amino acids have already been identified (4–8, 23, 24).
FIG. 1.
Amino acid alignment of conserved motifs I and II (15, 19) in the MPH(2′) and APH family. a. a., amino acid. Superscripts: b, MPH(2′); c, APH; d, deletion of an amino acid; e, insertion of an amino acid. Aspartic acid residues (D) are in boldface.
From these reports, it was expected that there are functional amino acids in the same region as MPH(2′). Five aspartic acids (D200, D209, D219, D227, and D231) were noted, and among these, three were highly conserved not only in bacterial phosphotransferase but also in the eukaryotic protein kinase family (4). To identify the functional aspartic acids, they were each replaced with alanine by site-directed mutagenesis.
(This work was presented in part at the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Diego, Calif., 25 September 1998.)
E. coli TG1 (22), a derivative of K-12, was employed for DNA technology, for measurement of the macrolide susceptibility of cells bearing cloned mphB, and as a host for crude-enzyme preparation.
Bacteria were grown on LB broth and agar (12). Sources for antibiotics were the following: oleandomycin, erythromycin, roxithromycin, troleandomycin, spiramycin, and tylosin, Sigma, St. Louis, Mo.; kitasamycin, rokitamycin, and josamycin, Wako Pure Chemical Industries Ltd., Osaka, Japan; clarithromycin, Taisho Pharmaceutical Co., Ltd., Tokyo, Japan; azithromycin, Pfizer Pharmaceutical Co., Ltd., Tokyo, Japan. ATP disodium salt was purchased from Wako Pure Chemical Industries Ltd., Osaka, Japan, and restriction enzymes and DNA modification enzymes were purchased from Toyobo Co., Osaka, Japan.
The MPH(2′)II-encoding gene, mphB, used in this study was cloned from E. coli CU1, which was isolated in 1997 from clinical material in Japan (20, 21). An approximately 1.0-kb DNA fragment carrying mphB was inserted into the multiple cloning site of pHSG398 (Takara Shuzo Co., Ltd., Tokyo, Japan), and the resultant plasmid was designated pKTA321. Using the specially designed primers ECMPHBF-1 (5′-GCG ATAGAATTCAAGGAGAAATAATATGACCGTAGTCA CGACCGCCGAT-3′) and ECMPHBR-1 (5′-GTTTTCCCAGTCACGACGTTGT-3′), mphB was amplified by the PCR method. The DNAs so produced were digested with EcoRI and PstI. The resulting DNA fragments were inserted into the EcoRI-PstI site of pKF18k (Takara Shuzo Co., Ltd.) to construct a template, designated pKFB280, for site-directed mutagenesis.
Chemically synthesized mutant primers for site-directed mutagenesis (11) in the mphB gene were designed from published sequence data and purchased from Life Technologies, Inc. The sequences of the primers were 5′-GATTCATGGCGCCGTACATGCCGG-3′, 5′-ACTATGATCGCGAAGGATGCCAATG-3′, 5′-AATGTGACAGGCCTAATCGCTTGGAC-3′ 5′-AAGGTTACAGCTGTTTCGCATGAC-3′, and 5′-GTTTCGCACGCGTTTATTTTCAAC-3′, and they were used to create D200A, D209A, D219A, D227A, and D231A mutants, respectively. The underlined letters indicate base mismatches compared to the wild-type sequence. A Mutan-Super Express kit (Takara Shuzo Co., Ltd.) with E. coli MV1184 for the Oligonucleotide-directed Dual Amber-Long and Accurate (ODA-LA) method was used for construction of mutant mphB by using pKFB280 as a template. The cycling program consisted of an initial incubation at 94°C for 5 min; 30 cycles of 93°C for 1 min, 52°C for 2 min, and 72°C for 2 min; and a final step of 72°C for 6 min. The mutant genes were completely sequenced by the chain termination method using a DSQ-1000 DNA sequencer (Shimadzu Co., Kyoto, Japan) and specific fluorescein-labelled primers designated RV22-FITC (5′-CACACAGGAAACAGCTATGACC-3′) and M422-FITC (5′-CCAGGGTTTTCCCAGTCACGCC-3′) to confirm the desired change in the nucleotide sequence. These mutant plasmids were digested with Eco47III and PstI. The resulting approximately 520-bp DNA fragment was used to displace the corresponding region of mphB on pKTA321.
Macrolide-inactivating activity was measured (10) in samples consisting of 50 μl of 40 mM ATP, 50 μl of macrolide antibiotics, and 400 μl of crude extract diluted with TMK buffer (0.06 M KCl, 0.01 M MgCl2, 0.006 M 2-mercaptoethanol in 0.1 M Tris-HCl buffer, pH 7.8) that were mixed and allowed to react at 37°C for 0, 0.5, 1, 2, and 4 h. A 30-μl sample of the reaction mixture was spotted on a paper disk (8-mm diameter; thin; TOYO Filter Co., Ltd., Tokyo, Japan), and the disk was heated in a microwave oven to stop the reaction. The residual potency of the antibiotics was determined by microbioassay using Bacillus subtilis ATCC 6633 (9) as an indicator organism on a nutrient soft agar upper layer and a nutrient agar lower layer.
Change in D200.
In a previous study of aminoglycoside 3′-phosphotransferase IIa [APH(3′)IIa], D190, which corresponds to D200 of MPH(2′), was replaced with glutamine (Q). The affinity of D190Q for ATP was the same as that of the wild type, and there was a slight retention of enzymatic activity (6).
It has also been reported that a D190A mutant of APH(3′)IIIa had no enzymatic activity (4), and it was proposed that D190 of APH(3′)IIIa was a general base activating the 3′-OH group to attack the γ phosphate of ATP.
In the case of MPH(2′)II, the specific enzyme activity of D200A was less than 0.1% of the original activity, thereby demonstrating that D200 is essential for the catalytic activity of MPH(2′)II. These results suggested that D200 might similarly be a general base activating the 2′-OH group of macrolide antibiotics.
Changes in D209, D219, and D231.
In earlier work, D208 and D220 of APH(2′)IIa [corresponding to D219 and D231 of MPH(2′)II, respectively] were each replaced with glycine (6). The results suggested that D208 and D220 are involved in the binding of ATP. Our testing determined that the specific activities of D219A and D231A for oleandomycin were less than 0.1 U (nanomoles of oleandomycin inactivated per hour), which suggested that D219 and D231 were an essential for enzymatic activity. These aspartic acids were highly conserved in motif II in the same way as D208 and D220 of APH(3′), so that the role of D219 and D231 might be correspondingly similar.
The specific activity of the D209A mutant was also less than 0.1 U, which suggested that D209 is crucial for catalysis. The amino acid corresponding to D209 in MPH(2′)II has not been studied in APH and other phosphotransferases, so further experimentation is needed to clarify the precise role of this residue.
Change in D227.
The specific activity of the D227A mutant was greatly reduced, but measurable activity (7%) was retained (Table 1). This suggested that D227 is not essential for, but clearly affects, enzymatic activity. The substrate specificity of D227A for various macrolides was examined (Table 1). The data showed that substitution at D227 resulted in a much less significant alteration of the substrate specificity of MPH(2′)II for 14-membered ring macrolides, such as erythromycin, troleandomycin (16), roxithromycin, and clarithromycin and 15-membered ring macrolides such as azithromycin, in contrast to that for 16-membered ring macrolides such as spiramycin and rokitamycin. In the latter two cases, activity was decreased by at least 25-fold compared with that of the wild type. In the other 16-membered ring macrolides, the relative activities of D227A demonstrated a 12-fold reduction for kitasamycin, similar activity for josamycin, and a fourfold reduction for tylosin compared with that of the wild type. These results suggested that D227 participated in the recognition of 16-membered ring macrolides, especially kitasamycin, spiramycin, and rokitamycin.
TABLE 1.
Substrate specificity of crude enzyme extract containing wild-type or D227A mutant MPH(2′)II
| Macrolide | Relative activity (%)a
|
|
|---|---|---|
| Wild type | D227A mutant | |
| 14 membered | ||
| Oleandomycin | 100 | 100 |
| Troleandomycin | 104 | 58 |
| Erythromycin | 83 | 83 |
| Clarithromycin | 46 | 49 |
| Roxithromycin | 50 | 9 |
| 15 membered azithromycin | 88 | 27 |
| 16 membered | ||
| Kitasamycin | 202 | 16 |
| Spiramycin | 75 | 85 |
| Josamycin | 75 | 2 |
| Rokitamycin | 50 | 2 |
| Tylosin | 55 | 13 |
The specific activity of the wild type enzyme for oleandomycin was 121 nmol/h/mg of protein and that of the D227A mutant was 8.5 nmol/h/mg of protein.
In spite of structural differences among macrolides, specific activities for 14- and 16-membered ring macrolides with the original enzyme were not so very different from each other (about 50∼200% of that of oleandomycin). Additionally, there is poor homology between MPH(2′) and erythromycin esterase in their primary amino acid sequences, so that it is difficult to identify the 14-membered ring recognition site in these macrolide-modifying enzymes. Kitasamycin, josamycin, and rokitamycin have a bulky side chain at the 4" position of l-mycarose, whereas spiramycin and tylosin have a small OH group at the same position. On the other hand, 14- and 15-membered ring macrolides do not have l-mycarose at the 4′ position of d-desosamine.
On the basis of this information, we predict that MPH(2′)II might more strongly interact with the sugar moiety than the lactone ring and we speculate that D227 makes a pocket where the desosamine and mycarose moieties fit. To identify the exact part of the macrolide which interacts with D227, a more detailed examination will follow in our laboratory.
Acknowledgments
This study was supported by a grant from the Ministry of Health and Welfare, Japan, 1998, for molecular characterization of antibiotic resistance and development of methods for rapid detection of drug-resistant bacteria.
REFERENCES
- 1.Andremont A, Sancho-Garnier H, Tancrede C. Epidemiology of intestinal colonization by members of the family Enterobacteriaceae highly resistant to erythromycin in a hematology-oncology unit. Antimicrob Agents Chemother. 1986;29:1104–1107. doi: 10.1128/aac.29.6.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Andremont A, Courvalin P. Distribution of erythromycin esterase and rRNA methylase genes in members of the family Enterobacteriaceae highly resistant to erythromycin. Antimicrob Agents Chemother. 1987;31:404–409. doi: 10.1128/aac.31.3.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur M, Courvalin P. Contribution of two different mechanisms to erythromycin resistance in Escherichia coli. Antimicrob Agents Chemother. 1986;30:694–700. doi: 10.1128/aac.30.5.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hon W C, Mckay G A, Thompson P R, Sweet R M, Young D S C, Wright G D, Berghuis A M. Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell. 1997;89:887–895. doi: 10.1016/s0092-8674(00)80274-3. [DOI] [PubMed] [Google Scholar]
- 5.Kocabiyik S, Mullins C, Breeding C, Perlin M H. Structure-function analyses for aminoglycoside 3′-phosphotransferase II (APH(3′)II) SAAS Bull Biochem Biotech. 1992;5:58–63. [PubMed] [Google Scholar]
- 6.Kocabiyik S, Perlin M H. Site-specific mutation of conserved C-terminal residues in aminoglycoside 3′-phosphotransferase II: phenotypic and structural analysis of mutant enzymes. Biochem Biophys Res Commun. 1992;185:925–931. doi: 10.1016/0006-291x(92)91715-3. [DOI] [PubMed] [Google Scholar]
- 7.Kocabiyik S, Perlin M H. Altered substrate specificity by substitution at Tyr 218 in bacterial aminoglycoside 3′-phosphotransferase-II. FEMS Microbiol Lett. 1992;93:199–202. doi: 10.1016/0378-1097(92)90529-w. [DOI] [PubMed] [Google Scholar]
- 8.Kocabiyik S, Perlin M H. Amino acid substitutions within the analogous nucleotide binding loop (P-loop) of aminoglycoside 3′-phosphotransferase II. Int J Biochem. 1994;26:61–66. doi: 10.1016/0020-711x(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 9.Kono M, Ohmiya K, Kanda T, Noguchi N, O’Hara K. Purification and characterization of chromosomal streptomycin-adenylyltransferase from derivative of Bacillus subtilis Marburg 168. FEMS Microbiol Lett. 1987;40:233–238. [Google Scholar]
- 10.Kono M, O’Hara K, Ebisu T. Purification and characterization of macrolide 2′-phosphotransferase type II from a strain of Escherichia coli highly resistant to macrolide antibiotics. FEMS Microbiol Lett. 1992;97:89–94. doi: 10.1016/0378-1097(92)90369-y. [DOI] [PubMed] [Google Scholar]
- 11.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 13.Matsuoka M, Endou K, Saitoh S, Katoh M, Nakajima Y. A mechanism of resistance to partial macrolide and streptogramin B antibiotics in Staphylococcus aureus clinically isolated in Hungary. Biol Pharm Bull. 1995;18:1482–1486. doi: 10.1248/bpb.18.1482. [DOI] [PubMed] [Google Scholar]
- 14.Noguchi N, Emura A, Matsuyama H, O’Hara K, Sasatsu M, Kono M. Nucleotide sequence and characterization of erythromycin resistance determinant that encodes macrolide 2′-phosphotransferase I in Escherichia coli. Antimicrob Agents Chemother. 1995;39:2359–2363. doi: 10.1128/aac.39.10.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noguchi N, Katayama J, O’Hara K. Cloning and nucleotide sequence of the mphB gene for macrolide 2′-phosphotransferase II in Escherichia coli. FEMS Microbiol Lett. 1996;144:197–202. doi: 10.1111/j.1574-6968.1996.tb08530.x. [DOI] [PubMed] [Google Scholar]
- 16.O’Hara K. Reaction mechanism of macrolide 2′-phosphotransferase from Escherichia coli to the 2′-modified macrolide antibiotics. Jpn J Antibiot. 1993;46:818–826. [PubMed] [Google Scholar]
- 17.O’Hara K, Yamamoto K. Reaction of roxithromycin and clarithromycin with macrolide-inactivating enzymes from highly erythromycin-resistant Escherichia coli. Antimicrob Agents Chemother. 1996;40:1036–1038. doi: 10.1128/aac.40.4.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Hara K, Kanda T, Ohmiya K, Ebisu T, Kono M. Purification and characterization of macrolide 2′-phosphotransferase from a strain of Escherichia coli that is highly resistant to erythromycin. Antimicrob Agents Chemother. 1989;33:1354–1357. doi: 10.1128/aac.33.8.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaw K J, Rather P N, Hare R S, Miller G H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57:138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniguchi K, Nakamura A, Tsurubuchi K, Ishii A, O’Hara K, Sawai T. Appearance in Japan of highly macrolide-resistant E. coli producing macrolide 2′-phosphotransferase. Microbios. 1999;97:137–144. [PubMed] [Google Scholar]
- 21.Taniguchi K, Nakamura A, Tsurubuchi K, O’Hara K, Sawai T. Identification of Escherichia coli clinical isolates producing macrolide 2′-phosphotransferase by a highly sensitive detection method. FEMS Microbiol Lett. 1998;167:191–195. doi: 10.1111/j.1574-6968.1998.tb13227.x. [DOI] [PubMed] [Google Scholar]
- 22.Taylor J W, Ott J, Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphothioate-modified DNA. Nucleic Acids Res. 1985;13:8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson P R, Hughes D W, Wright G D. Mechanism of aminoglycoside 3′-phosphotransferase type IIIa: His 188 is not a phosphate-accepting residue. Chem Biol. 1996;3:747–755. doi: 10.1016/s1074-5521(96)90251-3. [DOI] [PubMed] [Google Scholar]
- 24.Thompson P R, Hughes D W, Nicholas N P, Wright G D. Spectinomycin kinase from Legionella pneumophila. J Biol Chem. 1998;273:14788–14795. doi: 10.1074/jbc.273.24.14788. [DOI] [PubMed] [Google Scholar]