Abstract
Context:
Genomic stratification can impact prostate cancer (PC) care through diagnostic, prognostic, and predictive biomarkers that aid in clinical decision-making. The temporal and spatial genomic heterogeneity of PC together with the challenges of acquiring metastatic tissue biopsies hinder implementation of tissue-based molecular profiling in routine clinical practice. Blood-based liquid biopsies are an attractive, minimally invasive alternative.
Objective:
To review the clinical value of blood-based liquid biopsy assays in PC and identify potential applications to accelerate the development of precision medicine.
Evidence acquisition:
A systematic review of PubMed/MEDLINE was performed to identify relevant literature on blood-based circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and extracellular vesicles (EVs) in PC.
Evidence synthesis:
Liquid biopsy has emerged as a practical tool to profile tumor dynamics over time, elucidating features that evolve (genome, epigenome, transcriptome, and proteome) with tumor progression. Liquid biopsy tests encompass analysis of DNA, RNA, and proteins that can be detected in CTCs, ctDNA, or EVs. Blood-based liquid biopsies have demonstrated promise in the context of localized tumors (diagnostic signatures, risk stratification, and disease monitoring) and advanced disease (response/resistance biomarkers and prognostic markers).
Conclusions:
Liquid biopsies have value as a source of prognostic, predictive, and response biomarkers in PC. Most clinical applications have been developed in the advanced metastatic setting, where CTC and ctDNA yields are significantly higher. However, standardization of assays and analytical/clinical validation is necessary prior to clinical implementation
Keywords: Prostate cancer, Genomics, Liquid biopsy, Circulating tumor cell, Circulating tumor DNA, Extracellular vesicles, Precision medicine
Patient summary:
Traces of tumors can be isolated from blood samples from patients with prostate cancer either as whole cells or as DNA fragments. These traces provide information on tumor features. These minimally invasive tests can guide diagnosis and treatment selection.
1. Introduction
Recent studies have provided insight into the molecular landscape of prostate cancer (PC), identifying prognostic biomarkers, actionable targets, and drug resistance biomarkers. The difficulty of obtaining suitable tumor material for molecular testing is one of the reasons hampering clinical implementation of genomic profiling. Primary prostate tumor biopsies, although routine, are often scant in yield, and fixation procedures impact DNA quality. Biopsies of osteoblastic metastatic lesions, on the contrary, are technically challenging and distressing for the patient. Moreover, PC evolves over time as a consequence of therapy-induced selective pressure. Secondary resistance to standard-of-care androgen receptor (AR) targeting agents often involves genomic changes in a polyclonal manner that may be relevant for the selection of subsequent lines of therapy. A single tumor biopsy, from either primary or metastatic lesions, is limited in its ability to capture spatial heterogeneity, especially for repeated longitudinal assessments. Indeed, primary PC is among the most spatially heterogeneous and clonally complex cancer types [1].
The concept of a liquid biopsy encompasses the analysis of tumor material present in a bodily fluid [2]. This material can exist as biomolecules (eg, circulating tumor DNA [ctDNA], RNA, proteins, and mitochondrial DNA), circulating tumor cells (CTCs), or extracellular vesicles (EVs). Liquid biopsies have emerged as an attractive way to study tumor molecular landscapes in a minimally invasive manner, allowing for real-time snapshots of the overall tumor burden. Additionally, liquid biopsy–based biomarkers could serve as early endpoints in clinical trials to expedite drug development [3,4].
We review current knowledge of blood-based liquid biopsy components, their impact on clinical decision-making in PC, opportunities for accelerating precision medicine, and the challenges of implementing such tests in clinical practice.
2. Evidence acquisition
A systematic review of the PubMed/MEDLINE database was performed to identify literature on CTCs, ctDNA, and EVs in PC, published between 2005 and July 2020. Articles involving CTCs, ctDNA, and EVs in blood from PC patients were selected, and references cited within them were also considered.
3. Evidence synthesis
3.1. Circulating tumor cells
CTCs are cancerous cells from primary or metastatic lesions that either have been passively shed or have actively migrated from the tumor into the circulatory system. They can be found as single cells or clusters, the latter having a higher metastatic potential [5].
The half-life of CTCs in circulation is short (<1–2.5 h) [5]. The relatively small number of CTCs in blood remains a challenge for a comprehensive molecular analysis. CTC load increases with disease progression, being very low or near zero for most localized tumors. Typically, CTCs are isolated from a peripheral blood sample of 7.5–10 ml. Nevertheless, some studies have successfully pursued peripheral blood aphaeresis to increase the CTC yield [6]. It remains unclear, however, whether all tumor foci and lesions are represented in the CTC yield.
There are different strategies facilitating CTC isolation from blood samples, based on distinct physical or biological characteristics of CTCs (Table 1). Biological criteria–based methods for CTC isolation rely on selecting cells that express specific antigens (positive selection) and disregarding those that express other antigens (negative selection). This strategy is based on immunoaffinity, using antibodies targeting surface markers of epithelial cells such as EpCAM (CD326), for positive selection, while disregarding normal blood cells based on leukocyte markers such as CD45, CD16, or CD66b. The Food and Drug Administration (FDA)-approved CellSearch system (Menarini-Silicon Biosystems, Castel Maggiore, Italy) relies on a positive immunoaffinity assay based on EpCAM expression, followed by a semiautomated visual identification process based on immunofluorescence. It defines a CTC based on the presence of a DAPI-intact nucleus; lack of expression of CD45; expression of the epithelial cell markers EpCAM and cytokeratins (CKs) 8, 18, and 19; and a diameter of >4 μm [7]. However, several studies have demonstrated that expression of epithelial cell markers varies in CTCs (ie, EpCAM-low cells) [8], indicating that a proportion of CTCs may be missed if selection is based only on EpCAM expression. Therefore, other platforms use EpCAM-independent methods, such as characterizing all nucleated cells and identifying CTCs based on specific tumor-associated protein expression (ie, CK8 and AR) and cell morphology [7,9]. CTCs can also be isolated, leveraging their distinct physical properties as different deformability, density, surface charge, and size compared with nontumoral circulating cells. Several microfluidic devices using this approach have been developed (Table 1) [8,10,11]. DNA and RNA from CTCs can serve as proxies for tumor genomic characterization. Single CTC studies allow for fine dissection of intratumor heterogeneity [12], which might help in understanding therapy resistance; however, single-cell characterization is far from being clinically applicable. Clonal genome-wide copy numbers can be ascertained relatively inexpensively from single nucleotide polymorphism arrays or low-pass whole-genome sequencing (lpWGS). These lower-resolution approaches can also identify features of genomic instability associated with aggressive phenotypes, such as large-scale transitions [13]. Despite the small amount of input material obtained from CTC samples, whole-exome sequencing (WES) approaches to identify mutations have been proved to be feasible [10,14].
Table 1 –
Isolation platforms for CTCs in PC
| Technology | CTC definition | Application in PC |
|---|---|---|
| Antibody-based positive selection | ||
| CellSearch | EpCAM+, CD45−, CK8+, CK18+, CK19+, DAPI+ | [57] |
| Adna Test | EpCAM+, PSA+/PSMA+/EGFR+ | [15] |
| CTC-Chip | EpCAM+, specific antigen+ | [89] |
| CTC-iChip | >3.8 μm size, EpCAM+ | [90] |
| IsoFlux | CD45−, DAPI+, PanCK+, or EpCAM+ | [91] |
| MagSweeper | EpCAM+, CD45−, DAPI + | [10,92] |
| CellCollector | EpCAM+, CD45−, PanCK+, PSA+, Hoechst+ | [74] |
| NanoVelcro | EpCAM+, CD4−, PanCK+, morphological verification | [93] |
| Antibody-based negative selection | ||
| EasySep | CD45− | [94] |
| RosetteSep | CD45−, CD66b−, glycophorin A−, density | [74] |
| EPISPOT | CD45−, CD66b−, glycophorin A−, PSA+, FGF2+ | [74] |
| CTC-iChip | CD45−, CD16−, CD66b− | [90] |
| Selection free | ||
| Epic Sciences | PanCK+, CD45−, DAPI+, AR+ | [9] |
| AccuCyte | DAPI+, PanCK+, CD45−, CD66b−, CD11b−, CD14−, CD34−, EpCAM+ | [95] |
| Physical properties | ||
| ApoStream | Dielectrophoretic field flow | [96] |
| Celsee Diagnostics | >7.5 μm, deformability | [97] |
| ISET | ≥8 μm | [95] |
CTC = circulating tumor cell; EGFR = epidermal growth factor receptor; PC = prostate cancer; PSA = prostate-specific antigen; PSMA = prostate-specific membrane antigen.
Positive (+) and/or negative (−) expression of different capture/detection antigens, or physical properties, is used as a criterion for CTC isolation by different technologies or platforms.
The study of aberrant AR transcripts in CTCs, particularly those derived from AR splice variants, has attracted notable attention due to its potential clinical relevance for AR-targeting agents [15,16]. Interrogation of specific transcripts using in situ padlock probes is an opportunity for targeted transcriptomic approaches [17]. Beyond AR, multiplex assays enable comprehensive profiling of tumor transcriptomics from CTCs. These range from multiplex quantitative polymerase chain reaction approaches [18,19] to single-cell RNA-seq analysis [11]. In addition, methylome analysis in PC CTCs has generated profiles that resemble those derived from metastatic biopsies [20]. Lastly, protein expression in CTCs can also serve as a putative predictive biomarker. For instance, the detection of nuclear versus cytoplasmic AR-V7 has been correlated with a response to AR signaling inhibitors (ARSi) [21]. Another example is prostate-specific membrane antigen (PSMA) expression in CTCs [22], which could be relevant for the development of PSMA-based radiopharmaceuticals [23].
3.2. Circulating tumor DNA
Cell-free DNA (cfDNA) comprises short double-stranded DNA fragments (<200 bp) shed into the circulation from apoptosis or necrosis of normal and tumor cells. In healthy individuals, cfDNA fragments have a dominant peak at 167 bp, supporting a model where cfDNA is associated with the nucleosome core particle and linker histones [24]. In cancer patients, tumor-mutated alleles can be observed in DNA fragments shorter than nucleosomal DNA [25]. In the bloodstream, cfDNA has a short half-life estimated between 16 min and a few hours [26].
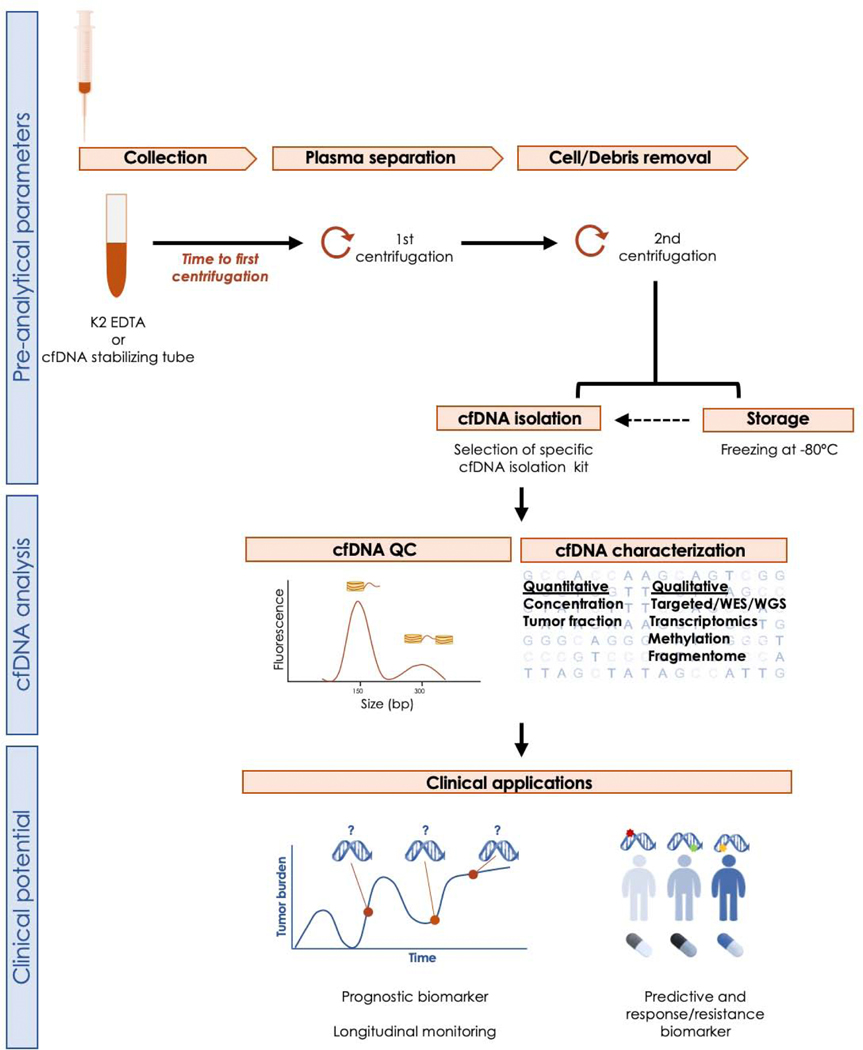
As both normal and tumor cells shed DNA into the blood, tumor DNA is diluted into circulating nontumoral DNA primarily coming from hematopoietic cells [24]. The subset of cfDNA arising from a tumor is known as ctDNA or “cfDNA tumor fraction” [27]. The dilution of ctDNA in nontumoral cfDNA is a significant confounding factor, and tumor fraction is likely to be a more reliable biomarker [26]. Several preanalytical conditions are required to maximize cfDNA yield and quality [28], as depicted in Figure 1. Once cfDNA is isolated and quantified, inference of the tumor fraction relies on computational analysis of the frequency of reads that carry tumor-specific aberrations (eg, point mutations, copy number alterations, and genomic rearrangements). Fragments of cfDNA originated in the tumor tend to have smaller sizes than those of nontumoral cfDNA. Hence, size selection of cfDNA can enrich for tumor content in ctDNA analysis [25]. This specific fragmentation pattern of cfDNA has also been postulated to be cancer type specific and could potentially be used for diagnosis [29].
Fig. 1 –
Workflow depicting preanalytical, analytical, and postanalytical steps for blood-based cfDNA studies. After venipuncture, blood is collected in a tube containing anticoagulants (EDTA and citrate are preferred to heparin). The time from sample acquisition to processing is critical, as cfDNA degrades within few hours. To overcome this problem, tubes containing different DNA stabilizers are available; the use of these tubes is particularly relevant in large multicenter studies with centralized analysis, or in general when the sample is not processed at the point of collection. A two-step centrifugation process is recommended to separate the plasma component, from which the cfDNA will be extracted. If the cfDNA is not extracted immediately, plasma can be stored at −80°C for prolonged periods, although repeated freeze-thawing cycles compromise cfDNA quality by increasing the amount of nontumor DNA contamination. After cfDNA isolation, quality control (QC) testing to assess cfDNA concentration and fragment size is performed prior to characterization. cfDNA = cell-free DNA; WES = whole-exome sequencing; WGS = whole-genome sequencing.
Tumor fraction in cfDNA usually increases in later disease stages and with a higher tumor burden. For example, in two studies of metastatic castration-resistant PC (mCRPC), the median ctDNA fraction was in the range of 15–20%, although interpatient variability was high [30,31].
The lpWGS uses copy number ratios to calculate tumor fraction, with a lower bound of detection of about 3%. However, this method might result in false-negative results for copy number–quiet tumors [32,33]. Targeted sequencing approaches, on the contrary, are a more affordable strategy to deliver high-read depths at specific regions in the genome but assume a priori that at least one somatic mutation would be present within a targeted region to infer ctDNA proportion; the probability of detecting mutations increases with the size of the targeted region.
Highly sensitive assays, such as droplet digital polymerase chain reaction (ddPCR), can detect point mutations with sensitivity ranging from 0.001% to 1%. These assays can detect AR mutations [34] and copy number gains [35]. However, its application is limited to the analysis of individual or a small set of known mutations (multiplexed ddPCR). These can be particularly useful for longitudinally monitoring tumor adaptation to targeted therapies, especially for hotspot mutations or for mutations previously determined by larger-scale sequencing.
Using targeted sequencing, Wyatt et al [36] showed good concordance between ctDNA and metastatic tissue biopsy for alterations in selected PC driver genes. Others have also used WES on ctDNA showing high agreement with tissue biopsies, although WES requires a minimum tumor fraction, probably above 10% [30,37]. Identification of low-frequency events, such as subclonal mutations, is more likely to be masked in samples with a low tumor fraction. Indeed, inference of the clonal versus subclonal origin of a mutation requires capturing enough ctDNA alleles in order to define clonality thresholds. By allocating sequencing depth to fewer genomic regions, targeted sequencing allows the study of low allele frequency events.
Methylation profiling of cfDNA is likely to be more sensitive than somatic alteration profiling for the detection of ctDNA, since there are millions of methylation marks available to profile but only a few thousand somatic alterations [38]. Recent studies in tumor [39] and cfDNA [40,41] samples have identified methylation-based PC subtypes and changes during disease progression.
3.3. Extracellular vesicles
EVs are secreted vesicles with a lipid bilayer and a typical size between 50 nm and 1 μm. Regarding their origin, EVs usually derive from the plasma membrane (eg, microvesicles) or, alternatively, have an endosomal origin (eg, exosomes). Some recent works have also described smaller (exomeres, ~35 nm [42]) and larger (oncosomes, 1–10 μm [43]) EVs with important roles in cancer. The International Society for EVs (ISEV) recommends that EVs be classified according to: (1) size (small [<100 nm], medium [100–200 nm], or large [>200 nm] EVs), (2) biochemical composition (eg, CD63+/CD81+), or (3) cell of origin [44]. EVs can contain proteins, lipids, metabolites, RNA (mRNA and miRNAs), and DNA as cargo. EVs play a key role in cell-to-cell communication during cancer progression and metastasis, as well as in triggering immune responses [45–47]. In addition, EVs have been associated with metastasis or relapse in cancer patients and can serve as diagnostic and prognostic markers; these can also be used for detecting therapeutic targets [47]. To date, few studies have investigated the relevance of blood EVs in PC [48,49], with most evidence coming from urine-derived EV studies [50,51]. EV size poses a significant challenge to the accuracy and reliability of their isolation and quantification [52]. Ultracentrifugation is currently considered the gold standard method, but to increase specificity, additional techniques such as ultrafiltration, density gradients, and chromatography can be implemented [44]. Different approaches have been used for the molecular characterization of EVs and their cargo, including transcriptomic analysis (ddPCR, real-time PCR, and RNA-seq) for the analysis of prostate-specific antigen (PSA), PCA3, ERG, AR, or AR-V7 [50,53,54] as well as WGS of larger EVs [43].
3.4. Moving liquid biopsies toward clinical management of PC
3.4.1. Quantitative prognostic and response biomarkers to accelerate drug development
Drug approval in the metastatic PC (mPC) setting is based on improvements in overall survival (OS) and/or radiographic progression-free survival (rPFS) [55]. CTC and cfDNA/ctDNA kinetics have value as prognostic and response biomarkers in mPC, offering faster readouts for clinical trials and allowing acceleration of the development of the most promising drugs.
Pioneering work by Cristofanilli et al [56] demonstrated the prognostic value of CTC enumeration in breast cancer, with an optimal threshold of five or more CTCs in 7.5 ml of blood, using the CellSearch system. Two studies in patients with PC prior to initiating chemotherapy demonstrated that (1) patients with five or more CTCs per 7.5 ml of blood, referred to as an “unfavorable profile” or a “high CTC count”, had shorter OS (prognostic biomarker), and (2) achievement of a decrease in CTC counts after therapy initiation (fewer than five CTCs per 7.5 ml; CTC conversion) correlated with OS (response biomarker) and was a stronger predictor than PSA changes [57,58]. Using the COU-301 registration trial of abiraterone as a model, Scher et al [59] confirmed the value of CTC counts and their changes over time, as prognostic and response biomarkers. CTC counts combined with lactate dehydrogenase (LDH) levels were shown to be a surrogate of OS, endorsing their use as intermediate biomarkers in mPC clinical trials. In a retrospective meta-analysis including data from five independent randomized clinical trials, Heller et al [60] demonstrated that CTC0 (change from detectable to undetectable CTCs) and CTC conversion consistently achieved higher C-index than percentage PSA decreases to discriminate OS, supporting that CTC kinetics could outperform PSA changes as a response biomarker.
Since then, CTC counts have been incorporated as a biomarker of response in several phase II trials, including the proof-of-concept studies for olaparib in PC [61], where CTC kinetics strongly correlated with rPFS. Further studies have also shown that relative changes in CTC counts (>30% decrease from baseline) could be potential surrogate endpoints [62]. Moreover, CTC kinetics could assist in therapy switch decisions as indicators of disease progression; increases in CTC counts after 10–12 wk of therapy significantly correlate with reduced rPFS and OS [63,64]. Of interest, large tumor–derived EVs expressing the same cell surface capture markers as CTCs (ie, EpCAM) can be coisolated with them and studied in platforms such as CellSearch. Enumeration of these EpCAM + EV may have a prognostic value in mCRPC, to further stratify patients with favorable CTC counts [49]. Additional studies are needed to confirm how the combination of different liquid biopsy approaches can improve patient stratification.
The cfDNA yield, and in particular ctDNA, increases as cancer progresses, in association with markers of overall tumor burden (PSA, LDH, and alkaline phosphatase). In an analysis of 571 patients from the FIRSTANA and PROSELICA phase III trials of taxane-based therapies [65], cfDNA baseline levels were an independent prognostic factor of rPFS and OS. In addition, absolute and relative changes in cfDNA levels on therapy correlated with PSA responses. Similarly, in the TOPARP-A trial, a 50% drop in cfDNA levels on olaparib therapy strongly correlated with rPFS and OS [66].
In a randomized trial of abiraterone acetate versus enzalutamide, ctDNA was quantifiable by WES and deep targeted 72-gene sequencing. Higher ctDNA fractions (>30%) were associated with clinical markers of tumor burden, including PSA, LDH, and alkaline phosphatase. Tumor fraction was prognostic, with ctDNA >30% presenting the worst rPFS, followed by a fraction between 2% and 30%, and patients with no detectable ctDNA experiencing longer times to progression [30]. In the hormone-naïve mPC setting, ctDNA levels appeared to diminish rapidly during the initial weeks of androgen deprivation therapy [67].
3.4.2. Clinical applications in localized prostate cancer
Current models for estimating the risk of relapse after definitive local therapy rely on pretreatment serum PSA abundance, International Society of Urological Pathology grade on biopsy, and clinical T category. Different biomarkers (ie, PTEN status, TP53 mutations, and cribriform histology) and tissue-based molecular signatures (ie, Decipher and Oncotype DX) have been proposed to improve patient stratification in localized PC, and in some cases these have been included in clinical guidelines, but their impact on therapeutic decision-making is still limited [68]. Tools supporting precise stratification of localized PC are needed, particularly in the case of longitudinal monitoring of patients on active surveillance protocols. Several studies have demonstrated the presence of disseminated cancer cells, particularly in the bone marrow [69,70], among patients with localized PC, providing the rationale for studying disease dissemination via liquid biopsies. Davis et al [71] studied the presence of CTCs in patients with localized PC (n = 97) with the CellSearch platform. CTCs were detected in a similar proportion of biopsy-positive patients (21%) to a control cohort of negative biopsy patients (20%); moreover, when present, counts were low (fewer than one to three CTCs per sample). In a more recent study, Salami et al [72] identified CTCs using the Epic Sciences platform in 33/45 (73%) patients with high-risk localized PC prior to receiving treatment. Biochemical recurrence was associated with higher baseline AR-positive CTC counts. Xu et al [73] showed that identification of CTCs and CTCRNA–based signatures could improve detection of clinically significant PC. Kuske et al [74] combined three independent CTC assays (CellSearch, CellColector, and EPISPOT) and found a cumulative positivity rate of 81% in patients with nonmetastatic high-risk PC; however, only 21% harbored five or more CTCs per 7.5 ml of blood. This work suggests that composite biomarker assays might increase our capacity to interrogate liquid biopsies in localized PC. In sum, the small number of CTCs in the blood of patients with clinically localized PC makes potential clinical applications challenging.
Similarly, the representation of ctDNA in early disease settings seems extremely low, challenging any downstream applicability for clinical testing, although the presence of ctDNA in patients with localized PC has been demonstrated in studies of methylation [75], allelic imbalance [76], and LOH [77]. The most comprehensive series to date came from Hennigan et al [78], in which no significant tumor fraction was detected by lpWGS or targeted sequencing, even in patients with high preprostatectomy serum PSA levels who subsequently recurred. Lastly, the field of tumor EVs in blood as PC biomarkers remains relatively unexplored. Park et al [48] used PSMA expression to enrich for tumor-derived EVs from patients with either benign prostatic hyperplasia or localized PC tumors. Interestingly, concentration of PSMA-positive EVs increased from low- to high-risk PC.
3.4.3. Liquid biopsies for precision use of AR targeting agents
Resistance to AR targeting agents typically emerges through multiple alterations affecting AR activity. Liquid biopsy can be repeated over time and represents an attractive opportunity for biomarker stratification for more precise ARSi use. AR amplification and mutations can be detected in ctDNA, and are associated with worse OS, PFS, and PSA response rate [31,34,79]. Carreira et al [80] showed that longitudinally acquired plasma samples allow monitoring of tumor dynamics and emerging drug resistance mechanisms.
Antonarakis et al [81] provided proof-of-concept evidence for the clinical value of AR-V7 detection in CTCs. Up to 39% of mCRPC patients treated with enzalutamide and 19% with abiraterone had AR-V7–positive CTCs; the presence of AR-V7 in CTCs was associated with lower PSA response and shorter biochemical progression-free survival (bPFS). This was further validated in a larger prospective study where mCRPC patients were classified as CTC–, CTC+/AR-V7–, and CTC+/AR-V7+. PSA response rates, bPFS, and OS were shorter in patients positive for CTCs, and even shorter in those with AR-V7+ CTCs [15]. AR-V7 status likely offers both prognostic and predictive information. In particular, nuclear-specific localization of AR-V7 in CTCs was found to impact OS significantly in preARSi blood CTCs but was not associated with differential response to taxanes [21]. The presence of AR-V7 in CTCs was further validated as an independent predictor of poor outcome in the PROPHECY multicenter prospective trial using two isolation approaches (AdnaTest and Epic Sciences assay) [82]. Del Re et al [83] used plasma-derived exosomal RNA to detect AR-V7, with AR-V7 exosome-positive patients having a worse prognosis and shorter response to treatment. AR profiling in CTCs and ctDNA from the same patient offers complementary information that could aid in ARSi treatment decision. However, the polyclonal nature and coexistence of multiple AR aberrations (copy number changes, splice variants, and mutations) limit the negative predictive value of each assay separately [16].
3.4.4. Predictive biomarkers for targeted therapies
The use of ctDNA as a predictive biomarker offers a potential advantage over tissue-based biopsies because ctDNA may comprise material shed by different metastatic lesions [41]. In a randomized phase II trial of ARSi in mCRPC [30], detection of TP53 mutations, DDR gene alterations, or AR amplification in ctDNA was associated with worse outcome. Other studies have confirmed a poor prognosis in patients with TP53 alterations in ctDNA [84].
Several clinical trials in mPC are now using panel-based cfDNA next-generation sequencing (NGS) to enrich their populations for testing targeted agents. Phase II trials of PARP inhibitors, such as TRITON2 (rucaparib) or GALAHAD (niraparib), allowed recruitment based on DDR gene alterations in ctDNA. In the TOPARP-A trial of olaparib, a good correlation in DDR mutation status between plasma and tumor was observed, with ctDNA detecting reversion mutations in BRCA2 and PALB2 upon secondary resistance [66]. Relevant to registration trials of AKT inhibitors in PC, recent work by Herberts et al [85] identified AKT1 and PIK3CA mutations in ctDNA. In addition, the detection of microsatellite instability and tumor somatic hypermutation in ctDNA is associated with MMR gene defects, and could be relevant to patient selection for immune checkpoint inhibitors [27,86].
Overall, although promising, challenges remain when using ctDNA to identify tumor mutations in the clinical setting. For example, a study comparing two different commercially available panels revealed discordant results, probably due to different coverage of the panels, but also due to different sensitivities and specificities for certain alterations [87]. These results highlight the need for pursuing clinical qualification of ctDNA assays in prospective trials. Umbrella studies such as PC-BETS (NCT03385655) and ProBio (NCT03903835) are now testing the clinical value of ctDNA in multiarm clinical trials.
3.5. Perspectives and future directions
Liquid biopsies can accelerate biomarker development for precision care in PC. As novel biomarker-driven therapies are validated, liquid biopsies also represent an inexpensive opportunity to facilitate the implementation of genomic testing into community practice, where metastasis biopsies are less common than in academic centers.
The value of CTC counts as prognostic and response biomarkers has clearly been demonstrated, offering a surrogate biomarker for accelerating drug development and potentially guiding therapeutic decisions. However, cost and access to technology, as well as heterogeneity among studies in terms of CTC definitions and isolation platforms, have complicated the translation of CTC analysis to routine clinical testing. Preliminary studies suggest that ctDNA kinetics may also be a useful prognostic and response biomarker in clinical practice, although further qualification in clinical trials is needed. As both CTCs and ctDNA yield parallel outcomes in tumor burden, applicability in localized disease may be challenging, although as ultrasensitive assays are developed, liquid biopsies might have the potential to assist in monitoring patients after radical therapy or to complement tissue-based biomarkers to improve patient stratification. The use of EVs in a clinical setting holds promise, and could complement CTC and ctDNA analyses, but faces challenges in standardization of isolation methods and downstream applications.
Identification of targetable alterations and emerging resistance biomarkers represents an attractive feature of liquid biopsies, particularly in the advanced disease setting, and could assist in the implementation of precision medicine therapeutics in PC practice. The FDA clearance of the CellSearch system for CTC enumeration was a first-in-class achievement. The recent FDA approval of the Guardant360 CDx and FoundationOne Liquid CDx as cfDNA NGS-based companion diagnostic assays represents a milestone in the field, but also a reminder that liquid biopsy assays need to be analytically validated and clinically qualified to be endorsed for routine clinical use. Other platforms of CTC characterization or cfDNA analysis, as well as assays for cfDNA-based cancer diagnosis are now at different stages of clinical validation. Some protocols allow coisolation of CTCs and ctDNA within the same blood sample [88]; however, these are not used in clinical-grade tests.
In addition, research on novel features from liquid biopsy analytes, such as “fragmentomics” (based on ctDNA fragment sizes), tissue-of-origin analysis, and methylation profiling, could potentially be informative in earlier tumor stages. Importantly, since liquid biopsies can reveal a broader landscape of mutations in multiple analytes, integration of these complex multidimensional data into composite biomarkers is a current need and an active area of research in the field.
4. Conclusions
The field of liquid biopsies in PC has advanced exponentially over the last decade, developing prognostic and predictive biomarkers, and holding promise for a minimally invasive means of monitoring the genetics of tumor evolution. Liquid biopsies could guide therapeutic decisions and accelerate the development of precision medicine in PC. However, issues relating to standardization of assay sensitivity and specificity, prospective clinical qualification of different assays, as well as cost and accessibility need to be addressed to endorse their implementation in routine clinical practice.
Acknowledgments
Funding/Support and role of the sponsor: Irene Casanova-Salas was supported by a PERIS fellowship from the Departament de Salut Generalitat de Catalunya with ref. SLT008/18/00185, and by a fellowship from ”la Caixa” Foundation (ID 100010434) and from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement number 847648. The fellowship code is LCF/BQ/PI20/11760033. This work was supported in part by the Intramural Research Program of the National Cancer Institute, NIH; by the NIH/NCI under award number P30CA016042; and through an operating grant from the Early Detection Research Network (1U01CA214194-01) to Paul C. Boutros. The authors affiliated to VHIO (Irene Casanova-Salas, Alejandro Athie, and Joaquin Mateo) acknowledge funding from the Cellex Foundation, and “la Caixa” Foundation (ID 100010434), AECC (Spanish Association Against Cancer) Scientific Foundation, and Fundacion FERO. Joaquin Mateo acknowledges support from the Prostate Cancer Foundation Young Investigator program.
Financial disclosures: Joaquin Mateo certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: P.C. Boutros is a member of the Scientific Advisory Boards of BioSymetrics Inc. and Intersect Diagnostics Inc. M. Del Re is a consultant for Ipsen, Janssen, Sanofi-Aventis, and Novartis. K.J. Pienta is a consultant for Cue Biopharma, Inc.; is a founder of Keystone Biopharma, Inc.; and receives research support from Progenics, Inc. J. Mateo has served on scientific advisory boards from Amgen, AstraZeneca, Clovis Oncology, Janssen, Merck/MSD, and Roche; has participated in speaker bureaus from AstraZeneca, Pfizer, Janssen, Sanofi, and Astellas Pharma; and has received research funding from AstraZeneca and Pfizer Oncology through grants to the institution. The remaining authors have nothing to disclose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Boutros PC, Fraser M, Harding NJ, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet 2015;47:736–45. [DOI] [PubMed] [Google Scholar]
- [2].Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med 2010;16:398–406. [DOI] [PubMed] [Google Scholar]
- [3].Ku SY, Gleave ME, Beltran H. Towards precision oncology in advanced prostate cancer. Nat Rev Urol 2019;16:645–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sumanasuriya S, Omlin A, Armstrong A, et al. Consensus statement on circulating biomarkers for advanced prostate cancer. Eur Urol Oncol 2018;1:151–9. [DOI] [PubMed] [Google Scholar]
- [5].Aceto N, Bardia A, Miyamoto DT, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014;158:1110–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lambros MB, Seed G, Sumanasuriya S, et al. Single-cell analyses of prostate cancer liquid biopsies acquired by apheresis. Clin Cancer Res 2018;24:5635–44. [DOI] [PubMed] [Google Scholar]
- [7].van der Toom EE, Verdone JE, Gorin MA, Pienta KJ. Technical challenges in the isolation and analysis of circulating tumor cells. Oncotarget 2016;7:62754–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol 2014;11:129–44. [DOI] [PubMed] [Google Scholar]
- [9].Beltran H, Jendrisak A, Landers M, et al. The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin Cancer Res 2016;22:1510–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lohr JG, Adalsteinsson VA, Cibulskis K, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol 2014;32:479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Miyamoto DT, Zheng Y, Wittner BS, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 2015;349:1351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Keller L, Pantel K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat Rev Cancer 2019;19:553–67. [DOI] [PubMed] [Google Scholar]
- [13].Malihi PD, Graf RP, Rodriguez A, et al. Single-cell circulating tumor cell analysis reveals genomic instability as a distinctive feature of aggressive prostate cancer. Clin Cancer Res 2020;26:4143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhao L, Lu YT, Li F, et al. High-purity prostate circulating tumor cell isolation by a polymer nanofiber-embedded microchip for whole exome sequencing. Adv Mater 2013;25:2897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Antonarakis ES, Lu C, Luber B, et al. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first- and second-line abiraterone and enzalutamide. J Clin Oncol 2017;35:2149–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].De Laere B, van Dam PJ, Whitington T, et al. Comprehensive profiling of the androgen receptor in liquid biopsies from castration-resistant prostate cancer reveals novel intra-AR structural variation and splice variant expression patterns. Eur Urol 2017;72:192–200. [DOI] [PubMed] [Google Scholar]
- [17].El-Heliebi A, Hille C, Laxman N, et al. In situ detection and quantification of AR-V7, AR-FL, PSA, and KRAS point mutations in circulating tumor cells. Clin Chem 2018;64:536–46. [DOI] [PubMed] [Google Scholar]
- [18].Gorges TM, Kuske A, Röck K, et al. Accession of tumor heterogeneity by multiplex transcriptome profiling of single circulating tumor cells. Clin Chem 2016;62:1504–15. [DOI] [PubMed] [Google Scholar]
- [19].Miyamoto DT, Lee RJ, Kalinich M, et al. An RNA-based digital circulating tumor cell signature is predictive of drug response and early dissemination in prostate cancer. Cancer Discov 2018;8:288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Friedlander TW, Ngo VT, Dong H, et al. Detection and characterization of invasive circulating tumor cells derived from men with metastatic castration-resistant prostate cancer. Int J Cancer 2014;134:2284–93. [DOI] [PubMed] [Google Scholar]
- [21].Scher HI, Graf RP, Schreiber NA, et al. Nuclear-specific AR-V7 protein localization is necessary to guide treatment selection in metastatic castration-resistant prostate cancer. Eur Urol 2017;71:874–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Miyamoto DT, Lee RJ, Stott SL, et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov 2012;2:995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Autio KA, Dreicer R, Anderson J, et al. Safety and efficacy of BIND-014, a docetaxel nanoparticle targeting prostate-specific membrane antigen for patients with metastatic castration-resistant prostate cancer: a phase 2 clinical trial. JAMA Oncol 2018;4:1344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 2016;164:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mouliere F, Chandrananda D, Piskorz AM, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med 2018;10:eaat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Choudhury AD, Werner L, Francini E, et al. Tumor fraction in cell-free DNA as a biomarker in prostate cancer. JCI Insight 2018;3:e122109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mayrhofer M, De Laere B, Whitington T, et al. Cell-free DNA profiling of metastatic prostate cancer reveals microsatellite instability, structural rearrangements and clonal hematopoiesis. Genome Med 2018;10:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Greytak SR, Engel KB, Parpart-Li S, et al. Harmonizing cell-free DNA collection and processing practices through evidence-based guidance. Clin Cancer Res 2020;26:3104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cristiano S, Leal A, Phallen J, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019;570:385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Annala M, Vandekerkhove G, Khalaf D, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov 2018;8:444–57. [DOI] [PubMed] [Google Scholar]
- [31].Romanel A, Gasi Tandefelt D, Conteduca V, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med 2015;7:312re10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Heitzer E, Ulz P, Belic J, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med 2013;5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ulz P, Belic J, Graf R, et al. Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer. Nat Commun 2016;7:12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Conteduca V, Wetterskog D, Sharabiani MTA, et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: a multi-institution correlative biomarker study. Ann Oncol 2017;28:1508–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jayaram A, Wingate A, Wetterskog D, et al. Plasma androgen receptor copy number status at emergence of metastatic castration-resistant prostate cancer: a pooled multicohort analysis. JCO Precis Oncol 2019;3:PO.19.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wyatt AW, Annala M, Aggarwal R, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Inst 2017;109:djx118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 2017;8:1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shen SY, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018;563:579–83. [DOI] [PubMed] [Google Scholar]
- [39].Zhao SG, Chen WS, Li H, et al. The DNA methylation landscape of advanced prostate cancer. Nat Genet 2020;52:778–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wu A, Cremaschi P, Wetterskog D, et al. Genome-wide plasma DNA methylation features of metastatic prostate cancer. J Clin Invest 2020;130:1991–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Beltran H, Romanel A, Conteduca V, et al. Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J Clin Invest 2020;130:1653–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang H, Freitas D, Kim HS, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol 2018;20:332–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Vagner T, Spinelli C, Minciacchi VR, et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J Extracell Vesicles 2018;7:1505403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 2014;30:255–89. [DOI] [PubMed] [Google Scholar]
- [46].Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 2020;367:eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell 2016;30:836–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Park YH, Shin HW, Jung AR, et al. Prostate-specific extracellular vesicles as a novel biomarker in human prostate cancer. Sci Rep 2016;6:30386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nanou A, Miller MC, Zeune LL, et al. Tumour-derived extracellular vesicles in blood of metastatic cancer patients associate with overall survival. Br J Cancer 2020;122:801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Donovan MJ, Noerholm M, Bentink S, et al. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis 2015;18:370–5. [DOI] [PubMed] [Google Scholar]
- [51].Pellegrini KL, Patil D, Douglas KJS, et al. Detection of prostate cancer-specific transcripts in extracellular vesicles isolated from post-DRE urine. Prostate 2017;77:990–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Junker K, Heinzelmann J, Beckham C, Ochiya T, Jenster G. Extracellular vesicles and their role in urologic malignancies. Eur Urol 2016;70:323–31. [DOI] [PubMed] [Google Scholar]
- [53].Woo HK, Park J, Ku JY, et al. Urine-based liquid biopsy: non-invasive and sensitive AR-V7 detection in urinary EVs from patients with prostate cancer. Lab Chip 2018;19:87–97. [DOI] [PubMed] [Google Scholar]
- [54].Connell SP, Hanna M, McCarthy F, et al. A four-group urine risk classifier for predicting outcome in prostate cancer patients. BJU Int 2019;124:609–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Morris MJ, Molina A, Small EJ, et al. Radiographic progression-free survival as a response biomarker in metastatic castration-resistant prostate cancer: COU-AA-302 results. J Clin Oncol 2015;33:1356–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781–91. [DOI] [PubMed] [Google Scholar]
- [57].de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302–9. [DOI] [PubMed] [Google Scholar]
- [58].Scher HI, Jia X, de Bono JS, et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol 2009;10:233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Scher HI, Heller G, Molina A, et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol 2015;33:1348–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Heller G, McCormack R, Kheoh T, et al. Circulating tumor cell number as a response measure of prolonged survival for metastatic castration-resistant prostate cancer: a comparison with prostate-specific antigen across five randomized phase III clinical trials. J Clin Oncol 2018;36:572–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mateo J, Porta N, Bianchini D, et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARPB): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol 2020;21:162–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lorente D, Olmos D, Mateo J, et al. Decline in circulating tumor cell count and treatment outcome in advanced prostate cancer. Eur Urol 2016;70:985–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lorente D, Olmos D, Mateo J, et al. Circulating tumour cell increase as a biomarker of disease progression in metastatic castration-resistant prostate cancer patients with low baseline CTC counts. Ann Oncol 2018;29:1554–60. [DOI] [PubMed] [Google Scholar]
- [64].De Laere B, Oeyen S, Van Oyen P, et al. Circulating tumor cells and survival in abiraterone- and enzalutamide-treated patients with castration-resistant prostate cancer. Prostate 2018;78:435–45. [DOI] [PubMed] [Google Scholar]
- [65].Mehra N, Dolling D, Sumanasuriya S, et al. Plasma cell-free DNA concentration and outcomes from taxane therapy in metastatic castration-resistant prostate cancer from two phase III trials (FIRSTANA and PROSELICA). Eur Urol 2018;74:283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Goodall J, Mateo J, Yuan W, et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov 2017;7:1006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Vandekerkhove G, Struss WJ, Annala M, et al. Circulating tumor DNA abundance and potential utility in de novo metastatic prostate cancer. Eur Urol 2019;75:667–75. [DOI] [PubMed] [Google Scholar]
- [68].Eggener SE, Rumble RB, Armstrong AJ, et al. Molecular biomarkers in localized prostate cancer: ASCO guideline. J Clin Oncol 2020;38:1474–94. [DOI] [PubMed] [Google Scholar]
- [69].Morgan TM, Lange PH, Porter MP, et al. Disseminated tumor cells in prostate cancer patients after radical prostatectomy and without evidence of disease predicts biochemical recurrence. Clin Cancer Res 2009;15:677–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Köllermann J, Weikert S, Schostak M, et al. Prognostic significance of disseminated tumor cells in the bone marrow of prostate cancer patients treated with neoadjuvant hormone treatment. J Clin Oncol 2008;26:4928–33. [DOI] [PubMed] [Google Scholar]
- [71].Davis JW, Nakanishi H, Kumar VS, et al. Circulating tumor cells in peripheral blood samples from patients with increased serum prostate specific antigen: initial results in early prostate cancer. J Urol 2008;179:2187–91; discussion 2191. [DOI] [PubMed] [Google Scholar]
- [72].Salami SS, Singhal U, Spratt DE, et al. Circulating tumor cells as a predictor of treatment response in clinically localized prostate cancer. JCO Precis Oncol 2019;3:PO.18.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Xu L, Mao X, Grey A, et al. Noninvasive detection of clinically significant prostate cancer using circulating tumor cells. J Urol 2020;203:73–82. [DOI] [PubMed] [Google Scholar]
- [74].Kuske A, Gorges TM, Tennstedt P, et al. Improved detection of circulating tumor cells in non-metastatic high-risk prostate cancer patients. Sci Rep 2016;6:39736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Cortese R, Kwan A, Lalonde E, et al. Epigenetic markers of prostate cancer in plasma circulating DNA. Hum Mol Genet 2012;21:3619–31. [DOI] [PubMed] [Google Scholar]
- [76].Schwarzenbach H, Alix-Panabières C, Müller I, et al. Cell-free tumor DNA in blood plasma as a marker for circulating tumor cells in prostate cancer. Clin Cancer Res 2009;15:1032–8. [DOI] [PubMed] [Google Scholar]
- [77].Sunami E, Shinozaki M, Higano CS, et al. Multimarker circulating DNA assay for assessing blood of prostate cancer patients. Clin Chem 2009;55:559–67. [DOI] [PubMed] [Google Scholar]
- [78].Hennigan ST, Trostel SY, Terrigino NT, et al. Low abundance of circulating tumor DNA in localized prostate cancer. JCO Precis Oncol 2019;3:PO.19.00176. [DOI] [PMC free article] [PubMed]
- [79].Wyatt AW, Azad AA, Volik SV, et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol 2016;2:1598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Carreira S, Romanel A, Goodall J, et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med 2014;6:254ra125. [DOI] [PMC free article] [PubMed]
- [81].Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Armstrong AJ, Halabi S, Luo J, et al. Prospective multicenter validation of androgen receptor splice variant 7 and hormone therapy resistance in high-risk castration-resistant prostate cancer: the PROPHECY study. J Clin Oncol 2019;37:1120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Del Re M, Biasco E, Crucitta S, et al. The detection of androgen receptor splice variant 7 in plasma-derived exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur Urol 2017;71:680–7. [DOI] [PubMed] [Google Scholar]
- [84].De Laere B, Oeyen S, Mayrhofer M, et al. Outperforms other androgen receptor biomarkers to predict abiraterone or enzalutamide outcome in metastatic castration-resistant prostate cancer. Clin Cancer Res 2019;25:1766–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Herberts C, Murtha AJ, Fu S, et al. Activating AKT1 and PIK3CA mutations in metastatic castration-resistant prostate cancer. Eur Urol 2020;78:834–44. [DOI] [PubMed] [Google Scholar]
- [86].Ritch E, Fu SYF, Herberts C, et al. Identification of hypermutation and defective mismatch repair in ctDNA from metastatic prostate cancer. Clin Cancer Res 2020;26:1114–25. [DOI] [PubMed] [Google Scholar]
- [87].Torga G, Pienta KJ. Patient-paired sample congruence between 2 commercial liquid biopsy tests. JAMA Oncol 2018;4:868–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Rothwell DG, Smith N, Morris D, et al. Genetic profiling of tumours using both circulating free DNA and circulating tumour cells isolated from the same preserved whole blood sample. Mol Oncol 2016;10:566–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007;450:1235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Fachin F, Spuhler P, Martel-Foley JM, et al. Monolithic chip for highthroughput blood cell depletion to sort rare circulating tumor cells. Sci Rep 2017;7:10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Nimir M, Ma Y, Jeffreys SA, et al. Detection of AR-V7 in liquid biopsies of castrate resistant prostate cancer patients: a comparison of AR-V7 analysis in circulating tumor cells, circulating tumor RNA and exosomes. Cells 2019;8:688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Cann GM, Gulzar ZG, Cooper S, et al. mRNA-Seq of single prostate cancer circulating tumor cells reveals recapitulation of gene expression and pathways found in prostate cancer. PLoS One 2012;7:e49144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Chen JF, Zhu Y, Lu YT, et al. Clinical applications of NanoVelcro rare-cell assays for detection and characterization of circulating tumor cells. Theranostics 2016;6:1425–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Liu X, Ledet E, Li D, et al. A whole blood assay for AR-V7 and AR. J Urol 2016;196:1758–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].van der Toom EE, Groot VP, Glavaris SA, et al. Analogous detection of circulating tumor cells using the AccuCyte. Prostate 2018;78:300–7. [DOI] [PubMed] [Google Scholar]
- [96].Gupta V, Jafferji I, Garza M, et al. ApoStream(™), a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics 2012;6:24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Gogoi P, Sepehri S, Zhou Y, et al. Development of an automated and sensitive microfluidic device for capturing and characterizing circulating tumor cells (CTCs) from clinical blood samples. PLoS One 2016;11:e0147400. [DOI] [PMC free article] [PubMed] [Google Scholar]