ABSTRACT
The spatiotemporal variation of several carbapenemase-encoding genes (CRGs) was investigated in the influent and effluent of municipal WWTPs, with or without hospital sewage input. Correlations among gene abundances, bacterial community composition, and wastewater quality parameters were tested to identify possible predictors of CRGs presence. Also, the possible role of wastewaters in mirroring clinical resistance is discussed. The taxonomic groups and gene abundances showed an even distribution among wastewater types, meaning that hospital sewage does not influence the microbial diversity and the CRG pool. The bacterial community was composed mainly of Proteobacteria, Firmicutes, Actinobacteria, Patescibacteria, and Bacteroidetes. Acinetobacter spp. was the most abundant group and had the majority of operational taxonomic units (OTUs) positively correlated with CRGs. This agrees with recent reports on clinical data. The influent samples were dominated by blaKPC, as opposed to effluent, where blaIMP was dominant. Also, blaIMP was the most frequent CRG family observed to correlate with bacterial taxa, especially with the Mycobacterium genus in effluent samples. Bacterial load, blaNDM, blaKPC, and blaOXA-48 abundances were positively correlated with BOD5, TSS, HEM, Cr, Cu, and Fe concentrations in wastewaters. When influent gene abundance values were converted into population equivalent (PE) data, the highest copies/1 PE were identified for blaKPC and blaOXA-48, agreeing with previous studies regarding clinical isolates. Both hospital and non-hospital-type samples followed a similar temporal trend of CRG incidence, but with differences among gene groups. Colder seasons favored the presence of blaNDM, blaKPC and blaOXA-48, whereas warmer temperatures show increased PE values for blaVIM and blaIMP.
IMPORTANCE Wastewater-based epidemiology has recently been recognized as a valuable, cost-effective tool for antimicrobial resistance surveillance. It can help gain insights into the characteristics and distribution of antibiotic resistance elements at a local, national, and even global scale. In this study, we investigated the possible use of municipal wastewaters in the surveillance of clinically relevant carbapenemase-encoding genes (CRGs), seen as critical antibiotic resistance determinants. In this matter, our results highlight positive correlations among CRGs, microbial diversity, and wastewater physical and chemical parameters. Identified predictors can provide valuable data regarding the level of raw and treated wastewater contamination with these important antibiotic resistance genes. Also, wastewater-based gene abundances were used for the first time to observe possible spatiotemporal trends of CRGs incidence in the general population. Therefore, possible hot spots of carbapenem resistance could be easily identified at the community level, surpassing the limitations of health care-associated settings.
KEYWORDS: carbapenemases, wastewaters, surveillance, wastewater-based epidemiology, carbapenem-resistance genes predictors
INTRODUCTION
A major health threat among antibiotic-resistant bacteria (ARB) is represented by multidrug resistant Gram-negative microorganisms, especially those that have developed resistance to carbapenems. These antimicrobials are seen as last resort drugs recommended as empirical treatment for critically ill patients (1). Carbapenem resistance can be mediated by multiple, different mechanisms. These include carbapenemase production, decreased cell wall permeability, overexpression of efflux pumps, and changes in penicillin-binding proteins. Carbapenemase production is the most concerning mechanism due to its increasing prevalence. It is linked to the easily transmissible nature of carbapenemase genes (CRGs), with the majority being carried on mobile genetic elements (MGE) (2). Here, they can be associated with genes conferring resistance to other classes of antimicrobials, thus leading to multidrug resistance (2). Carbapenemases are grouped into three Ambler classes (i) Class A (quite rare, detected mainly in Enterobacteriaceae) includes Klebsiella pneumoniae carbapenemase (KPC), Serratia marcescens enzyme (SME), imipenemase (IMP), non-metallocarbapenemase-A (NMC), and Guiana extended-spectrum (GES) enzymes. Members of this family are located either on chromosomes or on plasmids. (ii) Class B (observed in Entrobacteriaceae, P. aeruginosa, and A. baumanii) includes New-Delhi metallo-beta-lactamase (NDM), Verona integron-borne metallo-beta-lactamase (VIM), and imipenemase (IMP) types. These are frequently associated with mobile genetic elements. (iii) Class D (typical for A. baumanii, but found also in other Enterobacterales taxa) includes exclusively the OXA enzymes (reference 3 and references therein).
According to the World Health Organization and the EU One Health Action Plan, antimicrobial resistance (AMR) needs to be tackled in a One Health scheme. This includes surveillance efforts outside the clinic (4, 5), using multidisciplinary approaches that complement the more classic epidemiological models. Under the One Health umbrella, a key component for AMR surveillance is municipal wastewater. It contains waste products, including ARBs and antibiotic resistance genes (ARGs), from all members of a given community. Thus, wastewater-based epidemiology (WBE) has recently been recognized as a valuable, cost-effective tool to gain insight into the characteristics and distribution of ARBs and ARGs at a local, national, and even global scale (6–8). WBE could hence be utilized as a complementary surveillance technique. Current efforts are focused on evaluating its potential toward health-related monitoring schemes, for providing early warning of impending risks (9).
Among the CRG families, a considerable clinical concern is caused collectively by KPC, VIM, IMP, NDM, and OXA-48 types (10). They are frequently associated with the Gram-negative members of the ESKAPE group of microorganisms, some being the primary cause of nosocomial infections globally, with a wide range of multidrug resistance patterns (11, 12). These genes have a demonstrated potential for transfer via MGE, are conferring resistance to critical antibiotics, and are frequently encountered in wastewaters. Thus, they fit important criteria to be considered for long-term monitoring within WBE (13, 14). In addition, the presence of carbapenem-resistant bacteria and CRGs in wastewaters represents an important health issue from an ecosystem services perspective. These contaminants are continuously released into the natural environment especially due to wastewater treatment plant effluents (15, 16). Recent studies have highlighted the transmission of carbapenemase-producing bacteria from lotic ecosystems to humans, with a potential impact on public health (17). Despite these matters, to our knowledge, there are no comprehensive studies exploiting the true potential of communal wastewaters in tracking circulating CRGs, in relation to microbial diversity and wastewater characteristics and in comparison to clinical data.
In this context, the overall objectives of this study were (i) to provide insight into the spatiotemporal variation of clinically important CRGs and the associated microbial communities in wastewaters; (ii) to investigate the impact of hospital sewage on shaping the overall microbial communities and CRGs pool, and the role of WWTP types in the spread of these important ARGs into lotic ecosystems; (iii) to evaluate the capacity of bacterial taxa and wastewater quality parameters, routinely monitored during the treatment process, in forecasting CRGs diversity and abundance; and (iv) to discuss the possible use of wastewater-based gene abundance data in mirroring clinical resistance patterns.
RESULTS AND DISCUSSION
Hospital sewage input does not influence the microbial diversity and CRGs abundance in the investigated WWTPs.
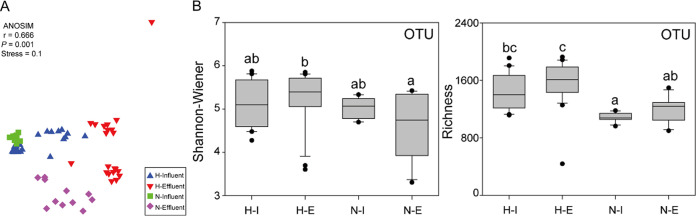
The bacterial diversity was assessed based on operational taxonomic unit (OTU) clustering and revealed a total of 7,138 OTUs, with little differences between bacterial communities from WWTP1 and WWTP2 (ANOSIM test, R = 0.405, P < 0.001). Therefore, as both these WWTPs receive communal and hospital wastewaters, they were grouped in the hospital influent/effluent (H-I/H-E) categories. In contrast, WWTP3 samples were classified as non-hospital influent/effluent (N-I/N-E), as this treatment plant does not receive hospital sewage. When groups and wastewater types (influent and effluent) were compared (ANOSIM test), either significant differences or differences with some similarities were observed in the cases of H-I versus H-E (R = 0.667, P < 0.01), H-E versus N-E (R = 0.691, P < 0.01), and N-I versus N-E (R = 0.549, P < 0.01), respectively. However, in the case of overall H versus N and H-I versus N-I, high levels of similarity were observed (R = 0.231, P < 0.01 and R = 0.193, P < 0.05, respectively), emphasizing that the presence of hospital wastewaters in WWTPs had no significant influence on the overall microbial community composition, an observation previously also made by Sorgen et al. (18). This is sustained by the nonmetric multidimensional scaling (NMDS) ordination of bacterial communities based on Bray-Curtis similarity (Fig. 1A), highlighting a similar pattern of biodiversity for both H-I and N-I and only minor differences between H-E and N-E.
FIG 1.
(A) Nonmetric multidimensional scaling (NMDS) ordination of bacterial communities based on Bray-Curtis similarity. (B) The comparison of bacterialcommunity diversity indices for different groups. H-I, hospital influent; H-E, hospital effluent; N-I, non-hospital influent; N-E, non-hospital effluent. Different letters (a, b, c) above bars indicate significant difference at P < 0.05 based on nonparametric Kruskal-Wallis tests.
The percentages of unique OTUs observed, i.e., 792 OTUs (11%) H-I, 1,217 OTUs (17%) H-E, 274 OTUs (4%) N-I, and 464 OTUs (6.5%) N-E, highlight a richer diversity in the effluent of both H and N wastewaters. However, by comparing the entire bacterial communities from H and N (Shannon–Wiener and species richness indices) (Fig. 1B), the most diverse group is represented by the H WWTP type. The differences between H and N in terms of diversity may be a consequence of the lower number of inhabitants associated to the N-type WWTP, since the composition of a bacterial community is directly proportional to the microbiome of the overall population (19).
Overall, the absolute and relative abundance of different groups showed an even distribution of CRGs among all the tested wastewater samples (NMDS analysis based on ANOSIM test), except for H-I versus H-E, where statistically significant differences were noted (Table 1; Fig. S1 in the supplemental material). As in the case of microbial diversity, we can conclude that the presence of hospital sewage does not influence the abundance of CRGs in communal wastewaters. Similar results were obtained by Pallares-Vega et al. (20) and Blaak et al. (8), the latter focused specifically on carbapenemase-producing Enterobacteriaceae.
TABLE 1.
Analysis of similarity (ANOSIM) for absolute and relative CRGs abundances among sample types
| Sample type | Absolute CRG copy no. |
Relative CRG copy no. (CRG/16S rRNA gene) |
||
|---|---|---|---|---|
| R | P | R | P | |
| Influent vs Effluent | 0.324a | 0.001 | 0.478a | 0.001 |
| Hospital vs Non-Hospital | 0.01 | 0.38 | 0.117a | 0.006 |
| H-I vs H-E | 0.623a | 0.001 | 0.64a | 0.001 |
| H-I vs N-I | 0.383a | 0.002 | 0.194b | 0.012 |
| H-I vs N-E | 0.334a | 0.003 | 0.224a | 0.005 |
| H-E vs N-I | 0.355a | 0.002 | 0.863a | 0.001 |
| H-E vs N-E | 0.208b | 0.017 | 0.244a | 0.009 |
| N-I vs N-E | 0.174a | 0.005 | 0.335a | 0.001 |
P < 0.01.
P < 0.05.
Spatio-temporal variation of bacterial diversity and abundance of carbapenemases in wastewaters.
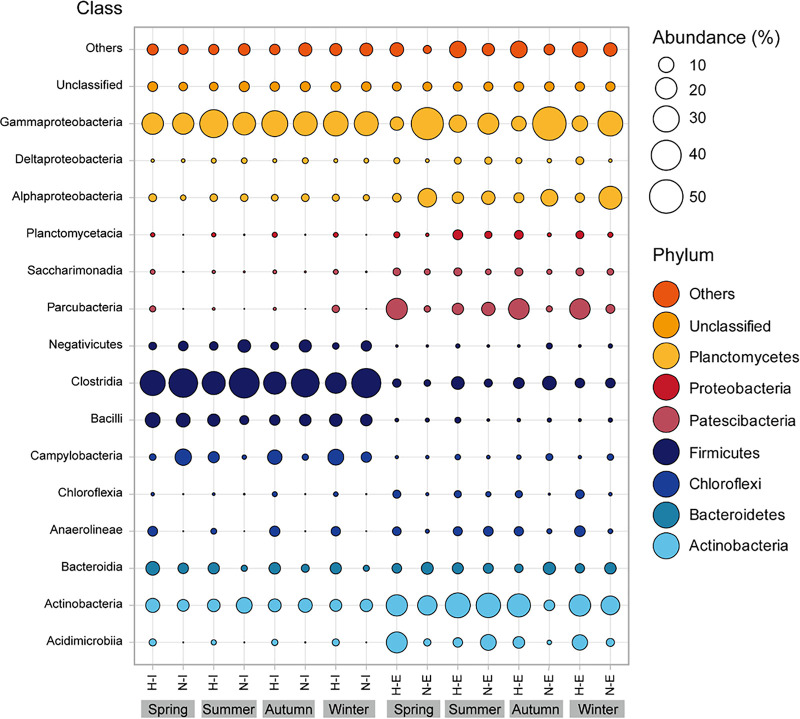
An in-depth taxonomic analysis showed an average of 37 phyla, 78 classes, 201 orders, and 317 bacterial families commonly found in all wastewater samples. The main bacteria phyla (>1% of total reads) were Proteobacteria (29%), Firmicutes (21%), Actinobacteria (19%), Patescibacteria (9%), and Bacteroidetes (5%). Also, the unclassified bacteria represented 4% , and 4% were minor phyla that each contributed less than 1% to the total community (Fig. 2).
FIG 2.
Class level seasonal breakdown of the relative abundances of bacterial taxonomic groups. Classes belonging to the same phyla are represented by the same color. Bubble size corresponds to the relative abundance, and only major taxonomic groups (>1% total abundance) were included in the graphic. H-I, hospital influent; H-E, hospital effluent; N-I, non-hospital influent; N-E, non-hospital effluent. Phyla and classes with abundances <1% are designated as “Others.”
A similar distribution of dominant bacterial phyla, with some differences in abundance, was observed recently by several authors in different sites worldwide. For example, Proteobacteria (62%), Firmicutes (20%), Bacteroidetes (12%), and Actinobacteria (1.7%) dominated the influent sewage from multiple WWTPs across the U.S. (21). Also, the investigations of influent wastewater from a Chinese WWTP have shown that Proteobacteria (90%) and Firmicutes (33%) were key phyla in these samples (22). A comparison conducted in Poland between raw and treated sewage highlighted that Proteobacteria was the most abundant phylum (50%), especially Campylobacteraceae and Moraxella families (23). In addition, a Spanish study investigated the bacterial community from WWTP biofilm, Firmicutes and Gammaproteobacteria being the most abundant groups, having Aeromonas (18%) and Acinetobacter (8%) as their key species (24).
Proteobacteria stand out as the most abundant group in the investigated wastewaters. They are indicators of human fecal contamination and are frequently associated with wastewater habitats (25). The Firmicutes phylum is usually present in wastewaters with high levels of antibiotic pollution, as it is known for its ability to survive in extreme environmental conditions (22, 26). Actinobacteria, the third most abundant group in the samples explored in this study, was shown to be involved in the decomposition of organic matter during the wastewater treatment process (27). Other less predominant bacterial phyla such as Bacteroidetes, Chloroflexi, Epsilonbacteraeota, and Planctomycetes had a lower contribution (<10%) to the overall bacterial community. They have previously been detected in various wastewaters (28–30) and activated sludge (31).
Although these bacterial taxa were present in all the wastewater samples, some differences in terms of frequency among the investigated groups was observed. Proteobacteria and Firmicutes were more abundant in the N type sequencing libraries (40% and 27%, respectively) compared to H wastewater (24% and 17%, respectively). In the latter, Actinobacteria, Patescibacteria, and Chloroflexi were more prevalent (22%, 11%, and 5%, respectively) (Fig. 2). The phyla Bacteroidetes and Epsilonbacteraeota were almost evenly distributed among groups, regardless of the hospital wastewater input, with a relative abundance of 5% in H and 4% in N for the former and 4% in H and 3% in N types for the latter. Besides these similarities found in the H and N groups, a moderate variation between influent and effluent was observed. Proteobacteria were significantly plentiful in N-E with a relative abundance of 52%, compared to 16% in H-E. Notably, the abundance of Firmicutes decreased considerably from 31% in H-I and 48% in N-I to 6% in both effluent types. Epsilonbacteraeota followed the same trend, presenting a reduced abundance after wastewater treatment (Fig. 3), from 9% H-I and 4% N-I to 0.4% H-E and 2% N-E. The relative abundances of the remaining taxa increased during the treatment process in all tested wastewaters. These results agree with other studies performed that show an increased presence of Actinobacteria in N-E wastewaters, as opposed to Chloroflexi and Planctomyces in H-E (19, 32), a possible consequence of the wastewater treatment process, during the activated sludge step (33). Overall, seasonal variation had a minimal impact on microbial diversity, except for a slight increase for Proteobacteria and Patescibacteria during winter and Actinobacteria in the summer (Fig. 2).
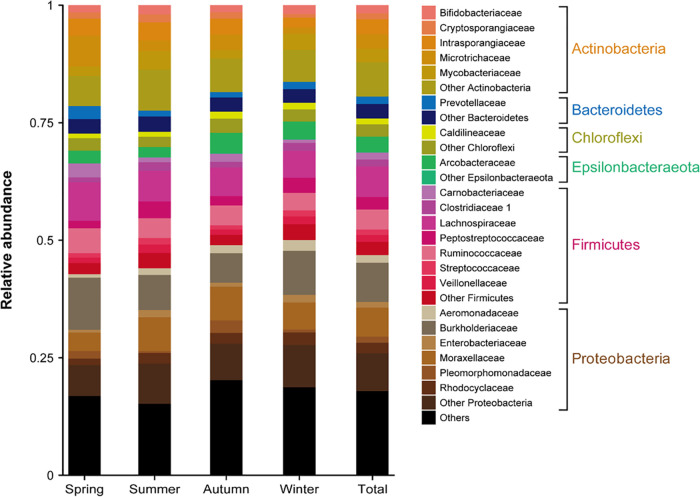
FIG 3.
Total and seasonal family level taxonomic breakdown of bacterial communities. Families belonging to the same dominant phylum are presented. Only taxonomic groups with more than 1% total abundance were included in the bar chart.
Within these phyla, 21 out of the observed 317 families were predominant (Fig. 3), such as Burkholderiaceae (8%), Moraxellaceae (6%), Lachnospiraceae (6%), Ruminococcaceae (4%), and Arcobacteraceae (3%), while 18% of families remained unclassified and 24% were designated as “other” (Fig. 3). However, each of these nonclassified families comprised less than 1% of total sequence abundance. Some bacterial families showed a small variation across seasons: Peptostreptococcaceae, Rhodocyclaceae, Clostridiaceae, Cryptosporangiaceae, Enterobacteriaceae, and Streptococcaceae increased in summer; Burkholderiaceae, Lachnospiraceae, Ruminococcaceae, Prevotellaceae, Carnobacteriaceae were better observed in spring; and winter temperatures favored the growth of Moraxellaceae and Arcobacteraceae. While colder temperatures can drastically reduce bacterial diversity (34), recent investigations have shown that the frequency of Rhodocyclaceae, Enterobacteriaceae, and Prevotellaceae families increased in the spring and summer (23), these findings agreeing with the results observed here.
Among the dominant bacterial families, some important pathogenic and water pollution indicator taxa (35) could be identified. Acinetobacter spp., sometimes a major constituent of bacterial communities in wastewaters (36), had the highest number of OTUs (115, 1.61% of total) in all tested wastewaters. It was followed by Bacteroides, Mycobacterium, Streptococcus, Clostridium sensu stricto, Arcobacter, Aeromonas, and Eubacterium, each with more than 30 OTUs (0.74%–0.5% of total OTUs). Other bacterial taxa such as Clostridium perfringens, Escherichia-Shigella, Enterococcus spp., and Streptococcus spp. were also observed. These are commonly found in human-associated or human-impacted water habitats and considered fecal pollution indicators (35, 37). Even though Aeromonas spp, and Pseudomonas spp. were less common in the wastewater samples investigated here (32 and 22 OTUs, respectively), they are considered environmental bacteria susceptible to developing antibiotic resistance (38), some included in the WHO AMR priority pathogens list (39). The presence of Legionella (28 OTUs), Leptospira (3 OTUs), and Mycobacterium (38 OTUs) genera in the wastewaters could represent a potential health risk once they enter the receiving rivers, as they are considered important waterborne pathogens (38). Although less abundant, Serratia marcescens (3 OTUs) and Bacillus spp. (4 OTUs) may be used as indicator taxa for cadmium (Cd), lead (Pb), pesticides, and detergent contamination (35).
The Procrustes test (Fig. 4) based on Bray-Curtis similarity metrics (r = 0.44–0.73) supports the idea of a significant correlation between CRGs and the bacterial community, especially for N-I/N-E groups. Also, these correlations were analyzed based on seasonal distribution, and the results have shown a slightly uniform pattern for H-I, N-I, and N-E groups in all seasons. In the case of H-E, a different distribution was observed in the winter samples, probably a consequence of the sharp increase of blaIMP relative abundance during that season.
FIG 4.
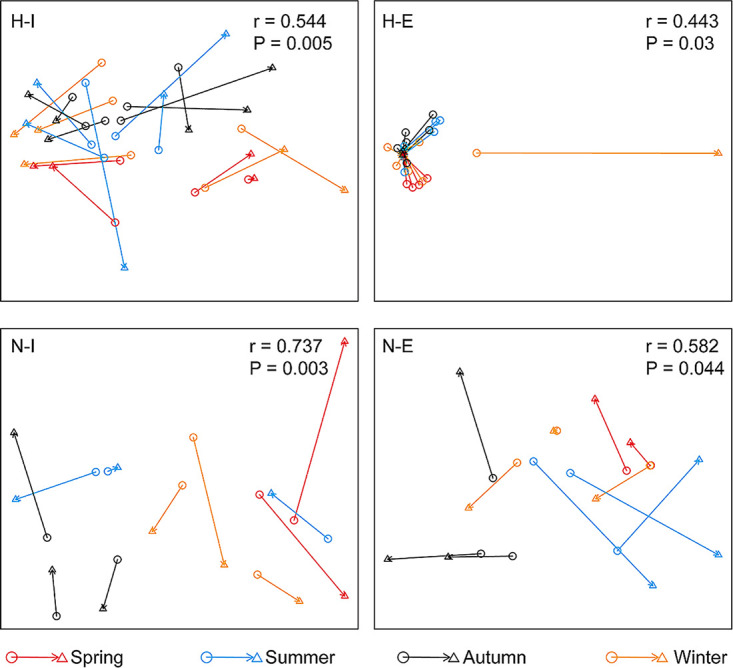
Procrustes test showing the correlation between CRG profiles (circle) and bacterial communities (triangle) based on Bray-Curtis similarity metrics. H-I, hospital influent; H-E, hospital effluent; N-I, non-hospital influent; N-E, non-hospital effluent.
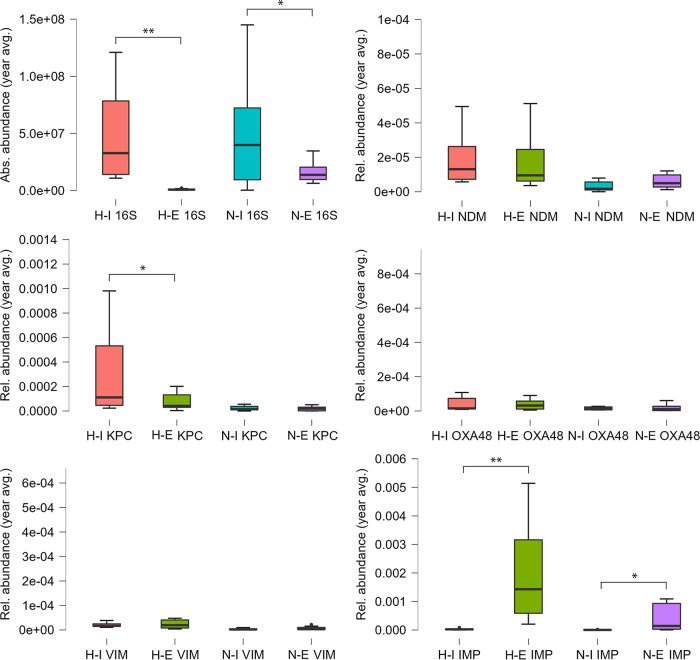
A variate gene distribution pattern emerged between influent and effluent of H and N WWTPs (Fig. 5) (paired t test and Wilcoxon test). There was a significant difference between the means of 16S rRNA gene absolute abundances for both H-type (97% reduction; t(10) = 3.95, P < 0.01, Wilcoxon P < 0.01) and N-type (64% reduction; t(10) = 2.79, P < 0.05, Wilcoxon P < 0.05) WWTPs. Regarding CRGs, significant differences between influent and effluent relative abundances were observed for blaKPC in the H-type samples, i.e., a 75% decrease (t(9) = 2.501, P < 0.05, Wilcoxon P < 0.01), and for blaIMP for both H-type (3,704% increase; t(10) = 3.77, P < 0.01, Wilcoxon P < 0.01) and N-type (8,000% increase; t(10) = 3.03, P < 0.05, Wilcoxon P < 0.01). There was no significant difference between the means of relative abundances of the influent and effluent measures for the remainder of genes tested (blaNDM, blaOXA-48, and blaVIM).
FIG 5.
Comparative analysis (yearly average) of absolute (16S rRNA gene) and relative (target CRG copies/16S rRNA gene copies) gene abundances in the influent and effluent of hospital and non-hospital type WWTPs. H-I, hospital influent; H-E, hospital effluent; N-I, non-hospital influent; N-E, non-hospital effluent. NDM, blaNDM; KPC, blaKPC; OXA48, blaOXA-48; VIM, blaVIM; IMP, blaIMP. *, P < 0.05; **, P < 0.01.
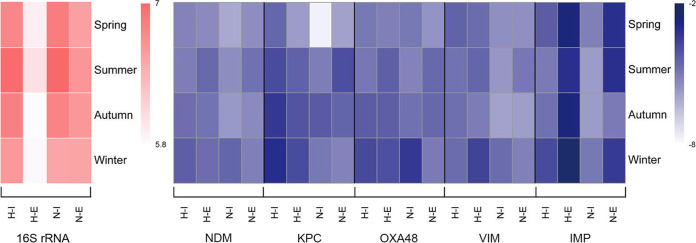
On a seasonal level, differences could be observed for both absolute and relative abundances of genes (Fig. 6; Tables S1 and S2). However, they were not significant, most likely due to the low number of samples taken during each season. The highest bacterial load (16S rRNA gene copy numbers) was observed in summer, as opposed to winter, when the lowest values were recorded. A similar trend was observed by Caucci et al. (40). On the contrary, Caltagirone et al. (41) noticed increases in bacterial counts from the beginning of the winter season in Italy, reaching the highest value in early spring. Overall, there are few records on seasonal variations of microbial diversity in wastewaters; thus, future investigations are required to investigate the drivers of bacterial cell abundances in these water habitats.
FIG 6.
Heatmap of seasonal absolute (16S rRNA, in red) and relative (target CRG copies/16S rRNA gene copies, in blue) abundances of genes; data were log transformed. H-I, hospital influent; H-E, hospital effluent; N-I; non-hospital influent; N-E, non-hospital effluent. NDM, blaNDM; KPC, blaKPC; OXA48, blaOXA-48; VIM, blaVIM; IMP, blaIMP.
In the H-type, blaKPC relative abundances were reduced after treatment on average by 78% during all seasons. In the N samples, a 44% reduction was observed during winter and autumn, and an increase in spring and summer, with 97% and 89%, respectively. A seasonal variation was highlighted in the case of blaNDM as well in the influent versus effluent of WWTPs, with a 27–57% decrease during spring, autumn, and winter in the H types and 76% in winter, for the N samples. In summer, the relative abundance increased with 61% for H and 54–77% in spring, summer, and autumn for N samples. For blaOXA-48, their values also decreased during spring, autumn, and winter in the H wastewaters (11–32%), opposed to an increase of 70% in summer. In N samples, blaOXA-48 relative abundances were reduced in spring and winter by 77–97% and increased in summer and autumn by 40–75%. Reduction of blaVIM relative abundances was 36–41% in spring and autumn in H samples and 0.6–69% in the N samples collected in spring and winter. In summer and winter for the H and summer and autumn for the N wastewaters, an increase in the relative abundances of 69% and 65%, respectively, was observed. When compared to the other four CRG families, blaIMP gene abundances showed a significant increase after wastewater treatment in all sample types and seasons (on average with 98% in H and 95% in N samples) (Fig. 6; Table S2). Future investigations are required, built around a more frequent sampling scheme, to test the hypothesis of significant seasonal variation of CRGs in communal wastewaters.
Studies dealing specifically with the spatiotemporal variation of CRGs in WWTPs are scarce worldwide. It is a known fact that the treatment process may promote the increase of gene abundance, as observed in some effluents of hospital and municipal wastewater treatment plants from Singapore, with high relative abundances of beta-lactam ARG types, especially blaKPC and blaOXA-48 (42). Compared to our results, other studies described a similar pattern of CRG abundances in wastewaters. For instance, a Chinese study observed high copy numbers of blaKPC and blaIMP in the effluent samples of urban WWTPs, while blaVIM and blaOXA-48 were not detected (43). Moreover, these resistance genes appeared more frequently in H-type wastewaters, especially blaKPC, usually associated with clinical isolates (44, 45). Also, blaKPC alongside blaNDM and blaOXA-48 were previously reported in hospital effluents from Spain, having relative abundance values higher or similar (4.8·10−2 blaKPC, 6.86·10−4 blaNDM, 1.59·10−6 blaOXA-48) to those found in our study, in both the H and N sample types (46). Furthermore, a seasonal effect was also observed in wastewaters from Germany and India, with significantly increased relative abundances in winter for blaOXA-48 and blaVIM (47), or blaNDM (48). When compared to the other CRG groups, we observed that blaIMP had a different seasonal pattern in our samples, being abundant all year round. This CRG family is frequently encountered in wastewaters regardless of seasonal change or wastewater type (49). Increased CRG abundances during colder seasons could be linked to higher rates of overall antibiotic prescriptions (40), the Romanian population being an important consumer of antibiotics, including beta-lactams (50). However, the correlation between antimicrobial consumption and increased CRGs presence in wastewaters was not considered in this study, thus needing further confirmation.
Bacterial taxa and water quality parameters as possible predictors of carbapenemases in wastewaters.
The investigated bacterial communities included 136 different OTUs, positively correlated with one, two, or three CRGs per OTU (Table S3). The most frequent CRG family observed to correlate with bacterial taxa was blaIMP (60 OTUs), followed by blaNDM (37 OTUs), blaVIM (23 OTUs), blaKPC (20 OTUs), and blaOXA-48 (8 OTUs) (Fig. S2). In the H-I samples, a very strong correlation (Spearman’s r > 0.8; P < 0.01) could be observed among several OTUs and blaNDM (29 OTUs), followed by blaVIM (23 OTUs), blaKPC (8 OTUs), blaOXA-48 (5 OTUs), and blaIMP (2 OTUs). The N influent is clearly dominated by OTUs strongly correlated (Spearman’s r > 0.8; P < 0.01) to blaIMP (58 OTUs), distantly followed by blaKPC (11 OTUs), blaNDM (8 OTUs), and blaOXA-48 (3 OTUs). The taxa belonging to the Proteobacteria phylum, especially the Acinetobacter genus, represented the majority of OTUs (14.1%) associated with CRGs, mostly in combinations of two CRGs for the same OTU (blaNDM + blaVIM or blaNDM + blaKPC). Our findings mirror those of documented clinical resistance, Romania being one of the leading places regarding the number of carbapenem-resistant Acinetobacter spp. invasive isolates within the EU/EEA countries (51). A significant association between CRGs and some high-risk pathogens like Acinetobacter has been previously reported for wastewaters (19).
The positive correlations between different OTUs and CRGs underlined that blaIMP is the most frequent gene associated with several bacterial groups within Proteobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Patescibacteria, Planctomycetes, and Verrucomicrobia in the investigated wastewaters. The strong association with representatives of the Actinobacteria phylum, especially the Mycobacterium genus (r = 0.82), which had a significant presence in the effluent, might be responsible for the increased number of blaIMP gene copies in the treated wastewaters (Fig. 5). Thus, this taxonomic group could be used as a possible predictor of increased levels of blaIMP in wastewaters, but this needs to be confirmed in future studies targeting Actinobacteria isolates. Even though blaIMP was observed before in association with Pseudomonas aeruginosa, Klebsiella pneumoniae, or Acinetobacter baumanii (52, 53), no such positive correlations could be observed in our study. The second most frequent CRG family was blaNDM, which was positively correlated with taxa belonging to Acinetobacter (r = 0.9), Flavobacterium (r = 0.88), Moraxella (r = 0.82), and Streptococcus genera (r = 0.86). Notably, a strong correlation was observed between the blaNDM gene and the presence of Candidatus Accumulibacter (r = 0.82), an uncharacterized group from Betaproteobacteria. This group is responsible for the accumulation of significant amounts of intracellular polyphosphate, used in the wastewater treatment process for the removal of biological phosphorus (54). Representatives of this group are known to contain antibiotic resistance genes such as tetA and sul1 (55), but, to our knowledge, the presence of carbapenem resistance in this novel clade has not been proposed before. The last three CRGs studied, blaVIM, blaKPC, and blaOXA-48, were less frequent, but still positively correlated with the presence of some high-risk pathogens such as Acinetobacter (associated with all three genes, r = 0.89), Aeromonas (with blaKPC, r = 0.83), and Arcobacter (with blaOXA-48, r = 0.8). Some recent studies described a positive association between blaKPC and Acinetobacter or Aeromonas (56, 57), and between blaVIM and Acinetobacter isolates (58), but the presence of blaOXA-48 gene in Arcobacter needs to be confirmed. Overall, future studies should be performed, using methods that provide a more comprehensive analysis of correlations among bacterial biodiversity and the CRG pool (e.g., metagenomic sequencing) in wastewaters.
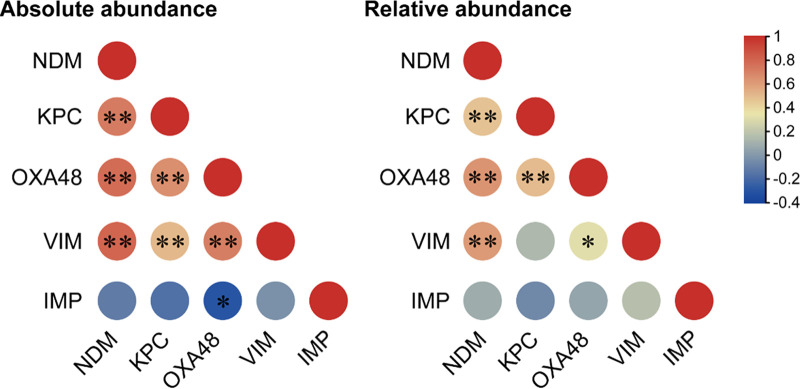
Regarding the co-occurrence of CRGs, a very strong correlation between blaNDM and blaKPC, blaOXA-48, blaVIM (Spearman’s r = 0.6–0.8; P < 0.01), for both absolute and relative abundances, and a weak to moderate correlation between blaIMP and blaOXA-48 (r = 0.2; P < 0.05), was observed (Fig. 7). Similar patterns of CRG co-occurrence were previously observed, such as blaOXA-48 with blaVIM in some wastewater effluents (49), blaNDM and blaOXA-58 in Acinetobacter pitti (59), or the combination of two, three, or four CRGs in Klebsiella pneumoniae isolated from wastewaters (60).
FIG 7.
Co-occurrence of CRGs in wastewater samples as tested by the Spearman's rank correlation coefficient. NDM, blaNDM; KPC, blaKPC; OXA48, blaOXA-48; VIM, blaVIM; IMP, blaIMP. *, P < 0.05; **, P < 0.01.
To gain a general perspective on the relationships among wastewater physical and chemical data and the relative abundance of CRGs, statistical analyses were carried out using the average overall values of wastewater parameters expressed as seasonal variation and sample types (H, hospital receiving, and N, non-hospital receiving WWTPs) (Table S4). The pH values were constant throughout the seasons in all tested wastewaters, ranging between 7.4 and 7.7. A similar trend was observed for NH4+, NO3–, NO2–, TP, SO42–, Cl–, detergent, Cd, Cr, Cu, Fe, Ni, Pb, and Zn. Water parameters like COD, BOD5, TSS, TN, dissolved solids, and HEM had higher levels in winter, colder temperatures being known to promote the rise of various wastewater constituent concentrations (61). Nonetheless, these increased values may indicate high levels of organic residues and may stimulate the growth of pathogenic bacteria and, implicitly, the associated CRG abundance (62).
Based on the Spearman`s correlation between water parameters and the absolute abundances of target genes (Table S5), we observed that COD had a moderate influence on the gene abundances, whereas BOD (biological oxygen demand) and TSS (total suspended solids) show either moderate or strong correlation with the 16S rRNA (BOD: r = 0.62; P < 0.01; TSS: r = 0.61; P < 0.01), blaOXA-48 (BOD: r = 0.68; P < 0.01), and blaNDM (TSS: r = 0.6; P < 0.01) genes. As BOD, representing the overall organic material, together with temperature and water flow, are the most relevant factors affecting the bacterial community abundance (63), they might indirectly influence the abundance of CRGs in the investigated wastewater samples from our study. A water parameter that to our knowledge has not been previously investigated as a possible indicator of cell and ARGs abundances in wastewaters is HEM (n-Hexane Extractable Material). Here, strong correlations were observed for both 16S rRNA and blaOXA-48 (r = 0.68; P < 0.01 and r = 0.71; P < 0.01, respectively). A similar pattern emerged for heavy metals as well, strong correlations being observed between 16S rRNA and Fe (r = 0.63; P < 0.01), blaNDM and Cr (r = 0.6; P < 0.01), and blaOXA-48 with Cu (r = 0.595; P < 0.01). Even though the association between metal and antibiotic resistance is documented in wastewaters, for example, that of Cu and blaNDM-1 carrying Enterobacteriaceae (64) or Acinetobacter baumanii (65), Cr was not previously reported as a possible indicator of increased blaNDM abundance in these water habitats. Even though blaIMP showed strong positive correlations with several bacterial taxonomic groups, an opposite trend was observed between this CRG family and almost all water parameters. The permutational multivariate analysis of variance (PERMANOVA) also highlighted the possible intricate relationships among the investigated CRGs, the bacterial community, BOD5, TSS, HEM, water flow, and other physical and chemical parameter average values (Table S6).
Monitoring circulating CRGs in the human population using wastewater-based gene abundance data.
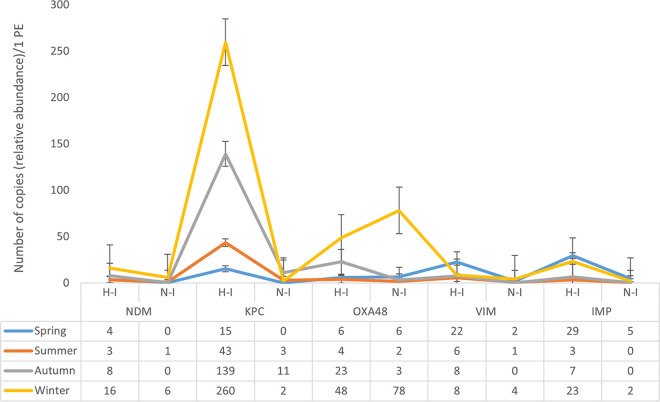
Recent studies acknowledge that untreated wastewater is a good indicator of the prevalence of circulating ARBs and ARGs, including carbapenem resistance genes, in a given community (6, 8). We converted BOD5 values into population equivalents (PE) (1 PE equates to 60 g of BOD5 per person per day) and calculated the number of relative CRG copies/1 PE/day, for each of the sampling months (Fig. 8). Both H- and N-type samples follow a similar temporal trend, with blaNDM, blaKPC, and blaVIM being observed throughout the year. blaKPC and blaNDM were most frequent in the colder months (early autumn until late winter), as opposite to blaVIM, which was mostly present during spring and early summer. For blaOXA-48, warmer months showed very low copies/1 PE, their numbers starting to increase mid-autumn toward the winter months, especially for non-hospital input wastewaters. The blaIMP group appeared in elevated values during spring, late autumn, and winter months. The highest copies/1 PE were identified for blaKPC and blaOXA-48, with the difference that H-I was dominated by blaKPC (with 260 ± 217 copies/1 PE in winter), while blaOXA-48 was predominant in N-I (78 ± 58 copies/1 PE, also during winter). These results agree with previous studies showing that blaKPC and blaOXA-48 are frequently encountered in clinical isolates (66–68). Concerning the other CRG families investigated, blaVIM and blaIMP seemed to have similar abundances/PE, followed by blaNDM. The top values were noticed either in spring (blaVIM), spring and winter (blaIMP), or winter (blaNDM) (Fig. 8), being associated mostly with the H-type population. With regard to overall clinical resistance, blaKPC, blaOXA-48, followed by blaNDM are frequently encountered in patients from Cluj County (Dr. Mirela Flonta, personal communication). Even though blaIMP and blaVIM had similar or slightly higher abundances/1 PE than blaNDM, they seem to be rarely identified in clinical isolates. This might suggest that more classic epidemiological models can lead to an underestimation of circulating ARGs, as they are limited by the reliance on patient-level sampling. In comparison, wastewater-based epidemiology could provide more substantial data at the community level, as it can surpass the restriction of health care-associated settings. As recent information on antimicrobial resistance in clinical bacterial isolates is biased toward COVID-19 patients, future studies are required. They need to combine clinical and environmental data to prove this hypothesis for carbapenem-resistance determinants.
FIG 8.
Seasonal distribution of CRGs relative abundances (target CRG copies/16S rRNA gene copies) calculated per population equivalent (PE). H-I, hospital influent; N-I, non-hospital influent. NDM, blaNDM; KPC, blaKPC; OXA48, blaOXA-48; VIM, blaVIM; IMP, blaIMP.
Even though WBE is gaining ground in tracking ARGs, there are certain limitations that should be taken into consideration and tackled in future investigations that use communal wastewaters. For instance, monitoring the antibiotic resistance level trends in the community using WBE methods cannot provide information about the nonhuman sources of bacteria and genes. Some authors have shown that livestock, slaughterhouses, domestic pets, food waste (69), wildlife, and animal farms sewage can contain carbapenem-resistant bacteria and CRGs (70), but not in significant amounts. For example, Europe registered a prevalence of <1% carbapenem-resistant Enterobacteriaceae among livestock and pets (71). Also, seasonal variation may depend on sampling design (69), an important factor to be considered when designing the monitoring scheme. Overall, wastewater surveillance can be a sensitive and high throughput method to detect carbapenem resistance in the general population (8), especially when combining culture-dependent and molecular microbiology.
Overall, our study has shown that the presence of hospital sewage input does not influence the overall bacterial diversity and CRGs pool in municipal wastewaters. The bacterial community was composed mainly of Proteobacteria, Firmicutes, Actinobacteria, Patescibacteria, and Bacteroidetes. Acinetobacter spp. was the most abundant group and had the majority of OTUs positively correlated with the presence of CRGs. This agrees with recent reports on clinical data. The influent samples were dominated by blaKPC, as opposed to effluent, in which blaIMP was dominant. Also, blaIMP was the most frequent CRG family observed to correlate with bacterial taxa, especially with the Mycobacterium genus in effluent samples. Bacterial load, blaNDM, blaKPC, and blaOXA-48 were positively correlated with BOD5, TSS, HEM, Cr, Cu, and Fe concentrations in wastewaters. When influent gene abundance values were converted into population equivalent (PE) data, the highest copies/1 PE were identified for blaKPC and blaOXA-48, agreeing with previous studies showing that these CRGs are frequently encountered in clinical isolates. Both hospital- and non-hospital-type samples followed a similar temporal trend of CRG incidence, but with differences among gene groups. Colder seasons favored the presence of blaNDM, blaKPC, and blaOXA-48, whereas warmer temperatures show increased PE values for blaVIM and blaIMP.
MATERIALS AND METHODS
Sample collection and processing.
Raw (influent) and treated (effluent) 24-h-composite wastewater samples were collected monthly for a year (2019–2020) from three different wastewater treatment plants (WWTPs), noted as WWTP1, WWTP2, and WWTP3, located in the Cluj County, Romania. WWTP1 processes around 115,000 m3 of wastewater/24 h from an average of 400,000 inhabitants, WWTP2 receives water from around 20,000 people and can process 3,456 m3/24 h, and WWTP3 is treating 864 m3/24 h, from an average of 10,000 inhabitants. Furthermore, besides water from the city, WWTP1 receives wastewater from several hospitals, WWTP2 collects water from a single hospital, and WWTP3 has no hospital input. No animal farm nor meat processing facilities release wastewater into the three sewage systems. To test the possible seasonal variation in the CRGs load and microbial diversity, the samples were grouped and analyzed according to each season, as follows: spring (March and May; April was not sampled due to COVID-19 lockdown), summer (June, July, August), autumn (September, October, November) and winter (December, January, February).
Influent and effluent wastewater samples were collected in 1,000 mL sterile bottles and transported to the Environmental Microbiology Laboratory at the Institute of Biological Research Cluj-Napoca (ICB Cluj). A volume of 40 mL from the influents and 300 mL from the effluents was filtered on 0.22 μm sterile filters (Sartorius, Germany) in triplicate, and the filters were stored at −20°C for subsequent analysis. DNA extraction from each filter was performed using the Quick-DNA Fecal/Soil Microbe Miniprep Kit (ZymoResearch, Irvine, CA, USA), according to the manufacturer’s instructions. The concentration and quality of the extracted DNA were determined with a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA). After quantification, the triplicates were pooled to form a representative sample, and resulting DNA samples were stored at –20°C until further molecular investigations.
Physical and chemical parameters routinely monitored by the WWTPs were provided by the water company, following the standard methods of analysis: pH (SR ISO 10523: 2012); COD-Mn (Chemical Oxygen Demand-Mn, STAS 3002: 1985); COD-Cr (Chemical Oxygen Demand-Cr, SR ISO 6060: 1996); BOD5 (biological oxygen demand), determination of biochemical oxygen consumption after n days (BODn) using the dilution and seeding method with allylthiourea (SR EN 1899-1: 2003) and using undiluted samples (SR EN 1899-2: 2002); TSS (total suspended solids, STAS 6953/1981); dissolved solids (STAS 6953/1981); NH4+ (SR ISO 7150-1: 2001); NO3- (PSLE – 12 Ed 5 rev. 3 SR ISO 7890-1); NO2- (SR EN 26777:2002/C91: 2006); TN (total nitrogen, PSLE – 19 Ed 04 R2) using Hach Lange LCK 311 method; TP (total phosphorus, SR EN ISO 6878: 2005); SO42- (PSLE – 14 Ed 05 R1 EPA 375.4:2003); Cl- (PSLE – 19 Ed 04 R2) using Hach Lange LCK 311 method; HEM (n-Hexane Extractable Material, Oil and Grease, EPA Method - 821: 2010); detergent (SR EN 903: 2003), determination of anionic surfactants by measuring the methylene blue index MBAS; Cd, Cr, Cu, Ni, Pb (EPA 3015A:2007, EPA 7010:2007, EPA 7000A:1992) by performing microwave assisted acid digestion of aqueous samples and extracts, graphite furnace atomic absorption spectrophotometry, and atomic absorption methods; and Fe, Zn (SR ISO 8288: 2001, SR 13315: 1996/C91: 2008) using flame atomic absorption spectrometry (FAAS).
Quantitative real-time PCR assay.
Quantitative real-time PCR (qPCR) was used to quantify blaKPC, blaNDM, blaOXA-48, blaVIM, and blaIMP gene families. The copy numbers of the 16S rRNA (rRNA) gene were also analyzed to determine the total bacterial abundance in the collected WWTP samples and for the normalization of CRG abundance data. Gradient PCR with different primer concentrations was initially performed for all qPCR assays to check the annealing temperatures described in the references (Table 2). Quantifications were carried out on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) using Eva Green detection chemistry. All reactions were performed in triplicate, in a total volume of 14 μL, containing 7 μL 1× Sso Fast EvaGreen SuperMix (Bio-Rad), 0.4 mM each forward and reverse primer, 20 ng of DNA, and RNase/DNase-free water to create a final volume of 14 μL. The cycling protocol consisted of an initial denaturation at 98°C for 2 min, followed by 45 cycles at 95°C for 10 s and 60°C for 45 s (blaKPC and blaOXA-48) or 60 s (blaNDM, blaVIM, blaIMP). The optimal annealing condition for the 16S rRNA gene was at 55°C for 60 s. After amplification, a melting curve was constructed in the range of 65 to 95°C to verify the specificity of the amplification products.
TABLE 2.
Primer sequences of carbapenemase-encoding and 16S rRNA genes
| Target gene | Primer sequence 5′–3′ | Amplicons length (bp) | References |
|---|---|---|---|
| bla KPC | F: CAGCTCATTCAAGGGCTTTC R: GGCGGCGTTATCACTGTATT |
196 | 72 |
| bla NDM | F: GATTGCGACTTATGCCAATG R: CGATCCCAACGGTGATATT |
189 | 72 |
| bla OXA-48 | F: AGGCACGTATGAGCAAGATG R: GGCTTGTTTGACAATACGC |
189 | 72 |
| bla VIM | F: GTTTGGTCGCATATCGCAAC R: CCAATTTGCTTYTCAATCTCCG |
155 | 46 |
| bla IMP | F: TCTCRATCTATCCCCACGTATGC R: GCGGACTTTGGCCAAGCTTCTA |
269 | 46 |
| 16S rRNA | 1369F: CGGTGAATACGTTCYCGG 1492R: GGWTACCTTGTTACGACT |
142 | 73, 74 |
Standard curves were generated using 10-fold dilutions of known quantities of cloned target genes. Plasmids containing the target genes were constructed by cloning the purified PCR products (GeneJET Gel Extraction kit, Thermo Scientific) into the pJET1.2/blunt vector using the CloneJET PCR cloning kit (Thermo Scientific). Plasmids were purified using the GeneJET plasmid miniprep kit (Thermo Scientific), and their concentration was determined using Qubit 2.0 fluorometer (LifeTechnologies). Genes were validated by Sanger sequencing at Macrogen Europe (Netherlands) and BLAST search in the GenBank database (https://www.ncbi.nlm.nih.gov/).
The gene copy numbers per reaction were calculated using the slope of the standard curve. Reaction efficiencies for the 16S rRNA and carbapenemase genes, which were assessed using the background-subtracted data and the LinRegPCR software (75), varied in a gene-dependent manner between 90 and 98.5%. The limit of quantification was 12, 19, 10, 16, 13, and 32 gene copy numbers per reaction for the 16S rRNA, blaKPC, blaNDM, blaOXA-48, blaVIM, and blaIMP genes, respectively. Final results in the gene copy numbers per mL of sample (copies/mL) were obtained by taking into account the template volume, DNA elution volume, and volume of filtered water. Additionally, the copy number of each target gene in a sample was normalized to the abundance of the 16S rRNA gene.
Microbial community analysis.
The biodiversity of microbial communities was assessed as a third-party service (Génome Québec, Montréal, Canada) through a Next-Generation Sequencing approach on any Illumina platform, targeting the V3-V4 variable regions of the 16S rRNA gene. Sequence analysis was carried out using mothur v.1.34.1 (76) according to the MiSeq SOP (77) on http://www.mothur.org/wiki/MiSeq_SOP. Sequences were aligned to the SILVA bacterial database (http://academic.oup.com/nar/article/41/D1/D590/1069277/The-SILVA-ribosomal-RNA-gene-database-project) and classified based on the RDP classifier (78). Diversity was assessed based on observed OTUs at 97% sequence similarity after rarefying samples to identical read numbers.
Statistical analysis.
All data except pH were log-transformed before analyses to improve normality. The diversity indices and Bray-Curtis similarity were calculated using the vegan package in R v3.6.1 (79). Analysis of similarities (ANOSIM) and nonmetric multidimensional scaling (NMDS) were performed in Primer v7.0 to explore the significant differences in the composition of different groups (80). Procrustes test based on Bray-Curtis similarity and permutational multivariate analysis of variance (PERMANOVA) were performed using the vegan package in R v3.6.1. Heatmaps were created in R v3.6.1 using the heatmap package (https://mran.microsoft.com/snapshot/2018-08-31/web/packages/pheatmap/pheatmap.pdf). Spearman’s correlations and nonparametric Kruskal-Wallis tests were performed in SPSS v22.0 (IBM Corp., Armonk, NY, USA).
Data availability.
Sequence data have been uploaded to the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/) under the BioProject accession number PRJNA790840.
ACKNOWLEDGMENTS
This work was funded through grant 117/2020 of the Romanian National Authority for Scientific Research and Innovation, CCCDI-UEFISCDI, project number ERANET-BIODIVHEALT-ANTIVERSA, within PNCDI III. A.B. was partially supported also by the core program PN2019–2022-BIODIVERS 3, grant 25N/2019-BIOSERV. A.H. was partially supported also by Norway Grants Call 2019 (RO-NO 2019)—Collaborative Research Projects, Contract No. 28/2020. P.F. acknowledges a China Scholarship Council (CSC) scholarship (202004910327). U.K. and T.U.B. are supported through the ANTIVERSA project funded by the Bundesministerium für Bildung, und Forschung under grant number 01LC1904A and the JPI AMR - EMBARK project funded by the Bundesministerium für Bildung, und Forschung under grant number F01KI1909A. Responsibility for the information and views expressed in the manuscript lies entirely with the authors.
We thank Compania de Apa Somes S.A. for their help and support in the sampling and wastewater characterization and Charlotte Briddon for proofreading the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Cristian Coman, Email: cristian.coman@icbcluj.ro.
Monica Adriana Garcia-Solache, Brown University.
REFERENCES
- 1.Codjoe FS, Donkor ES. 2017. Carbapenem resistance: a review. Med Sci 6:1. doi: 10.3390/medsci6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gniadek TJ, Carroll KC, Simner PJ. 2016. Carbapenem-resistant non-glucose-fermenting Gram-negative bacilli: the missing piece to the puzzle. J Clin Microbiol 54:1700–1710. doi: 10.1128/JCM.03264-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Workneh M, Yee R, Simner PJ. 2019. Phenotypic methods for detection of carbapenemase production in carbapenem-resistant organisms: what method should your laboratory choose? Clin Microbiol Newsl 41:11–22. doi: 10.1016/j.clinmicnews.2019.01.001. [DOI] [Google Scholar]
- 4.Robinson TP, Bu DP, Carrique-Mas J, Fèvre EM, Gilbert M, Grace D, Hay SI, Jiwakanon J, Kakkar M, Kariuki S, Laxminarayan R, Lubroth J, Magnusson U, Thi Ngoc P, Van Boeckel TP, Woolhouse ME. 2016. Antibiotic resistance is the quintessential One Health issue. Trans R Soc Trop Med Hyg 110:377–380. doi: 10.1093/trstmh/trw048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel JB. 2017. One Health and the politics of antimicrobial resistance. Emerg Infect Dis 23:724–724. doi: 10.3201/eid2304.161871. [DOI] [Google Scholar]
- 6.Aarestrup FM, Woolhouse MEJ. 2020. Using sewage for surveillance of antimicrobial resistance. Science 367:630–632. doi: 10.1126/science.aba3432. [DOI] [PubMed] [Google Scholar]
- 7.Diemert S, Yan T. 2020. Municipal wastewater surveillance revealed a high community disease burden of a rarely reported and possibly subclinical Salmonella enterica serovar Derby strain. Appl Environ Microbiol 86:e00814-20. doi: 10.1128/AEM.00814-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaak H, Kemper MA, Man H, van Leuken JPG, Schijven JF, van Passel MWJ, Schmitt H, Roda Husman AM. 2021. Nationwide surveillance reveals frequent detection of carbapenemase-producing Enterobacterales in Dutch municipal wastewater. Sci Total Environ 776:145925. doi: 10.1016/j.scitotenv.2021.145925. [DOI] [Google Scholar]
- 9.Choi PM, Tscharke BJ, Donner E, O'Brien JW, Grant SC, Kaserzon SL, Mackie R, O'Malley E, Crosbie ND, Thomas KV, Mueller JF. 2018. Wastewater-based epidemiology biomarkers: past, present and future. Trends Anal Chem 105:453–469. doi: 10.1016/j.trac.2018.06.004. [DOI] [Google Scholar]
- 10.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Santajit S, Indrawattana N. 2016. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int 2016:2475067. doi: 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. 2019. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol 10:539. doi: 10.3389/fmicb.2019.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laht M, Karkman A, Voolaid V, Ritz C, Tenson T, Virta M, Kisand V. 2014. Abundances of tetracycline, sulphonamide and beta-lactam antibiotic resistance genes in conventional wastewater treatment plants (WWTPs) with different waste load. PLoS One 9:e103705. doi: 10.1371/journal.pone.0103705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, Bürgmann H, Sørum H, Norström M, Pons MN, Kreuzinger N, Huovinen P, Stefani S, Schwartz T, Kisand V, Baquero F, Martinez JL. 2015. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol 13:310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 15.Mills MC, Lee J. 2019. The threat of carbapenem-resistant bacteria in the environment: evidence of widespread contamination of reservoirs at a global scale. Environ Pollut 255:113143. doi: 10.1016/j.envpol.2019.113143. [DOI] [PubMed] [Google Scholar]
- 16.Alexander J, Hembach N, Schwartz T. 2020. Evaluation of antibiotic resistance dissemination by wastewater treatment plant effluents with different catchment areas in Germany. Sci Rep 10:8952. doi: 10.1038/s41598-020-65635-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laurens C, Jean-Pierre H, Licznar-Fajardo P, Hantova S, Godreuil S, Martinez O, Jumas-Bilak E. 2018. Transmission of IMI-2 carbapenemase-producing Enterobacteriaceae from river water to human. J Glob Antimicrob Resist 15:88–92. doi: 10.1016/j.jgar.2018.06.022. [DOI] [PubMed] [Google Scholar]
- 18.Sorgen A, Johnson J, Lambirth K, Clinton SM, Redmond M, Fodor A, Gibas C. 2021. Characterization of environmental and cultivable antibiotic-resistant microbial communities associated with wastewater treatment. Antibiotics 10:352. doi: 10.3390/antibiotics10040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lira F, Vaz-Moreira I, Tamames J, Manaia CM, Martínez JL. 2020. Metagenomic analysis of an urban resistome before and after wastewater treatment. Sci Rep 10:8174. doi: 10.1038/s41598-020-65031-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pallares-Vega R, Blaak H, van der Plaats R, de Roda Husman AM, Hernandez Leal L, van Loosdrecht MCM, Weissbrodt DG, Schmitt H. 2019. Determinants of presence and removal of antibiotic resistance genes during WWTP treatment: a cross-sectional study. Water Res 161:319–328. doi: 10.1016/j.watres.2019.05.100. [DOI] [PubMed] [Google Scholar]
- 21.Shanks OC, Newton RJ, Kelty CA, Huse SM, Sogin ML, McLellan SL. 2013. Comparison of the microbial community structures of untreated wastewaters from different geographic locales. Appl Environ Microbiol 79:2906–2913. doi: 10.1128/AEM.03448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang ZH, Yang JQ, Zhang DJ, Zhou J, Zhang CD, Su XR, Li TW. 2014. Composition and structure of microbial communities associated with different domestic sewage outfalls. Genet Mol Res 13:7542–7552. doi: 10.4238/2014.September.12.21. [DOI] [PubMed] [Google Scholar]
- 23.Makowska N, Philips A, Dabert M, Nowis K, Trzebny A, Koczura R, Mokracka J. 2020. Metagenomic analysis of β-lactamase and carbapenemase genes in the wastewater resistome. Water Res 170:115–277. [DOI] [PubMed] [Google Scholar]
- 24.Marti E, Jofre J, Balcazar JL. 2013. Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PLoS One 8:e78906. doi: 10.1371/journal.pone.0078906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szekeres E, Baricz A, Chiriac CM, Farkas A, Opris O, Soran ML, Andrei AS, Rudi K, Balcázar JL, Dragos N, Coman C. 2017. Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environ Pollut 225:304–315. doi: 10.1016/j.envpol.2017.01.054. [DOI] [PubMed] [Google Scholar]
- 26.Bäumlisberger M, Youssar L, Schilhabel MB, Jonas D. 2015. Influence of a non-hospital medical care facility on antimicrobial resistance in wastewater. PLoS One 10:e0122635. doi: 10.1371/journal.pone.0122635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anandan R, Dharumadurai D, Manogaran GP. 2016. An introduction to Actinobacteria, p 3–37. In Dhanasekaran D, Jiang Y (ed), Actinobacteria—basics and biotechnological applications. IntechOpen Limited, London, United Kingdom. [Google Scholar]
- 28.Li D, Qi R, Yang M, Zhang Y, Yu T. 2011. Bacterial community characteristics under long-term antibiotic selection pressures. Water Res 45:6063–6073. doi: 10.1016/j.watres.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Cai L, Ju F, Zhang T. 2014. Tracking human sewage microbiome in a municipal wastewater treatment plant. Appl Microbiol Biotechnol 98:3317–3326. doi: 10.1007/s00253-013-5402-z. [DOI] [PubMed] [Google Scholar]
- 30.Cydzik-Kwiatkowska A, Zielińska M. 2016. Bacterial communities in full-scale wastewater treatment systems. World J Microbiol Biotechnol 32:66. doi: 10.1007/s11274-016-2012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao D, Huang R, Zeng J, Yu Z, Liu P, Cheng S, Wu QL. 2014. Pyrosequencing analysis of bacterial community and assembly in activated sludge samples from different geographic regions in China. Appl Microbiol Biotechnol 98:9119–9128. doi: 10.1007/s00253-014-5920-3. [DOI] [PubMed] [Google Scholar]
- 32.Pandit PR, Kumar R, Kumar D, Patel Z, Pandya L, Kumar M, Joshi C. 2021. Deciphering the black box of microbial community of common effluent treatment plant through integrated metagenomics: tackling industrial effluent. J Environ Manage 289:112448. doi: 10.1016/j.jenvman.2021.112448. [DOI] [PubMed] [Google Scholar]
- 33.Ye L, Zhang T. 2013. Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl Microbiol Biotechnol 97:2681–2690. doi: 10.1007/s00253-012-4082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu S, Yao J, Ainiwaer M, Hong Y, Zhang Y. 2018. Analysis of bacterial community structure of activated sludge from wastewater treatment plants in winter. Biomed Res Int 2018:8278970. doi: 10.1155/2018/8278970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hussain QA. 2019. Bacteria: the natural indicator of environmental pollution. Freshw Microbiol 2019:393–420. [Google Scholar]
- 36.Gupta SK, Shin H, Han D, Hur HG, Unno T. 2018. Metagenomic analysis reveals the prevalence and persistence of antibiotic- and heavy metal-resistance genes in wastewater treatment plant. J Microbiol 56:408–415. doi: 10.1007/s12275-018-8195-z. [DOI] [PubMed] [Google Scholar]
- 37.Grøndahl-Rosado RC, Tryland I, Myrmel M, Aanes KJ, Lucy J. 2014. Robertson detection of microbial pathogens and indicators in sewage effluent and river water during the temporary interruption of a wastewater treatment plant. Water Qual Expo Health 6:155–159. doi: 10.1007/s12403-014-0121-y. [DOI] [Google Scholar]
- 38.Numberger D, Ganzert L, Zoccarato L, Mühldorfer K, Sauer S, Grossart HP, Greenwood AD. 2019. Characterization of bacterial communities in wastewater with enhanced taxonomic resolution by full-length 16S rRNA sequencing. Sci Rep 9:9673. doi: 10.1038/s41598-019-46015-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fouz N, Pangesti KNA, Yasir M, Al-Malki AL, Azhar EI, Hill-Cawthorne GA, Abd El Ghany M. 2020. The contribution of wastewater to the transmission of antimicrobial resistance in the environment: implications of mass gathering settings. TropicalMed 5:33. doi: 10.3390/tropicalmed5010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caucci S, Karkman A, Cacace D, Rybicki M, Timpel P, Voolaid V, Gurke R, Virta M, Berendonk TU. 2016. Seasonality of antibiotic prescriptions for outpatients and resistance genes in sewers and wastewater treatment plant outflow. FEMS Microbiol Ecol 92:fiw060. doi: 10.1093/femsec/fiw060. [DOI] [PubMed] [Google Scholar]
- 41.Caltagirone M, Nucleo E, Spalla M, Zara F, Novazzi F, Marchetti VM, Piazza A, Bitar I, De Cicco M, Paolucci S, Pilla G, Migliavacca R, Pagani L. 2017. Occurrence of extended spectrum beta-lactamases, KPC-Type, and MCR-1.2-producing Enterobacteriaceae from wells, river water, and wastewater treatment plants in Oltrepò Pavese Area. Front Microbiol 8:2232. doi: 10.3389/fmicb.2017.02232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng C, Tay M, Tan B, Le TH, Haller L, Chen H, Koh TH, Barkham TMS, Gin KY. 2017. Characterization of metagenomes in urban aquatic compartments reveals high prevalence of clinically relevant antibiotic resistance genes in wastewaters. Front Microbiol 8:2200. doi: 10.3389/fmicb.2017.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang F, Mao D, Zhou H, Luo Y. 2016. Prevalence and fate of carbapenemase genes in a wastewater treatment plant in Northern China. PLoS One 11:e0156383. doi: 10.1371/journal.pone.0156383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nasri E, Subirats J, Sànchez-Melsió A, Mansour HB, Borrego CM, Balcázar JL. 2017. Abundance of carbapenemase genes (blaKPC, blaNDM and blaOXA-48) in wastewater effluents from Tunisian hospitals. Environ Pollut 229:371–374. doi: 10.1016/j.envpol.2017.05.095. [DOI] [PubMed] [Google Scholar]
- 45.Paulus GK, Hornstra LM, Alygizakis N, Slobodnik J, Thomaidis N, Medema G. 2019. The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. Int J Hyg Environ Health 222:635–644. doi: 10.1016/j.ijheh.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Subirats J, Royo E, Balcázar JL, Borrego CM. 2017. Real-time PCR assays for the detection and quantification of carbapenemase genes (blaKPC, blaNDM, and blaOXA-48) in environmental samples. Environ Sci Pollut Res Int 24:6710–6714. doi: 10.1007/s11356-017-8426-6. [DOI] [PubMed] [Google Scholar]
- 47.Schages L, Wichern F, Kalscheuer R, Bockmühl D. 2020. Winter is coming—impact of temperature on the variation of beta-lactamase and mcr genes in a wastewater treatment plant. Sci Total Environ 712:136499. doi: 10.1016/j.scitotenv.2020.136499. [DOI] [PubMed] [Google Scholar]
- 48.Lamba M, Graham DW, Ahammad SZ. 2017. Hospital wastewater releases of carbapenem-resistance pathogens and genes in urban India. Environ Sci Technol 51:13906–13912. doi: 10.1021/acs.est.7b03380. [DOI] [PubMed] [Google Scholar]
- 49.Khan FA, Söderquist B, Jass J. 2019. Prevalence and diversity of antibiotic resistance genes in Swedish aquatic environments impacted by household and hospital wastewater. Front Microbiol 10:688. doi: 10.3389/fmicb.2019.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robertson J, Vlahović-Palčevski V, Iwamoto K, Högberg LD, Godman B, Monnet DL, Garner S, Weist K, ESAC-Net Study Group, WHO Europe AMC Network Study Group. 2021. Variations in the consumption of antimicrobial medicines in the European region, 2014–2018: findings and implications from ESAC-Net and WHO Europe. Front Pharmacol 12:639207. doi: 10.3389/fphar.2021.639207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.ECDC. 2020. Antimicrobial resistance surveillance in Europe 2019. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). Stockholm, Sweden. [Google Scholar]
- 52.Ghamgosha M, Shahrekizahedani S, Kafilzadeh F, Bameri Z, Taheri RA, Farnoosh G. 2015. Metallo-beta-Lactamase VIM-1, SPM-1, and IMP-1 genes among clinical Pseudomonas aeruginosa species isolated in Zahedan, Iran. Jundishapur J Microbiol 8:e17489. doi: 10.5812/jjm.8(4)2015.17489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abbas S, Ejaz H, Jahan S, Younas S, Alzahrani B, Farraj DAA, Alkufeidy RM. 2019. Molecular detection of blaIMP genes in Metallo-Beta-Lactamase producing clinical Gram-negative isolates. Clin Lab 65. doi: 10.7754/Clin.Lab.2019.190202. [DOI] [PubMed] [Google Scholar]
- 54.He S, McMahon KD. 2011. Microbiology of “Candidatus Accumulibacter” in activated sludge. Microb Biotechnol 4:603–619. doi: 10.1111/j.1751-7915.2011.00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du B, Yang Q, Wang R, Wang R, Wang Q, Xin Y. 2019. Evolution of antibiotic resistance and the relationship between the antibiotic resistance genes and microbial compositions under long-term exposure to tetracycline and sulfamethoxazole. IJERPH 16:4681. doi: 10.3390/ijerph16234681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martínez-Porchas M, Villalpando-Canchola E, Vargas-Albores F. 2016. Significant loss of sensitivity and specificity in the taxonomic classification occurs when short 16S rRNA gene sequences are used. Heliyon 2:e00170. doi: 10.1016/j.heliyon.2016.e00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sekizuka T, Inamine Y, Segawa T, Hashino M, Yatsu K, Kuroda M. 2019. Potential KPC-2 carbapenemase reservoir of environmental Aeromonas hydrophila and Aeromonas caviae isolates from the effluent of an urban wastewater treatment plant in Japan. Environ Microbiol Rep 11:589–597. doi: 10.1111/1758-2229.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davoodi S, Boroumand MA, Sepehriseresht S, Pourgholi L. 2015. Detection of VIM- and IMP-type Metallo-Beta-Lactamase genes in Acinetobacter baumannii isolates from patients in two hospitals in Tehran. Iran J Biotechnol 13:63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou S, Chen X, Meng X, Zhang G, Wang J, Zhou D, Guo X. 2015. “Roar” of blaNDM-1 and “silence” of blaOXA-58 co-exist in Acinetobacter pittii. Sci Rep 5:8976. doi: 10.1038/srep08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teban-Man A, Farkas A, Baricz A, Hegedus A, Szekeres E, Pârvu M, Coman C. 2021. Wastewaters, with or without hospital contribution, harbour MDR, carbapenemase-producing, but not Hypervirulent Klebsiella pneumoniae. Antibiotics (Basel) 10:361. doi: 10.3390/antibiotics10040361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alsulaili A, Al-Buloushi BY, Hamoda MF. 2020. Seasonal variation pattern of physico-chemical and microbial parameters in a wastewater treatment plant. DWT 208:244–260. doi: 10.5004/dwt.2020.26461. [DOI] [Google Scholar]
- 62.Aniyikaiye TE, Oluseyi T, Odiyo JO, Edokpayi JN. 2019. Physico-chemical analysis of wastewater discharge from selected paint industries in Lagos, Nigeria. IJERPH 16:1235. doi: 10.3390/ijerph16071235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Novo A, André S, Viana P, Nunes OC, Manaia CM. 2013. Antibiotic resistance, antimicrobial residues and bacterial community composition in urban wastewater. Water Res 47:1875–1887. doi: 10.1016/j.watres.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 64.Yang QE, Agouri SR, Tyrrell JM, Walsh TM. 2018. Heavy metal resistance genes are associated with blaNDM-1- and blaCTX-M-15-carrying Enterobacteriaceae. Antimicrob Agents Chemother 62:e02642-17. doi: 10.1128/AAC.02642-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thummeepak R, Pooalai R, Harrison C, Gannon L, Thanwisai A, Chantratita N, Millard AD, Sitthisak S. 2020. Essential gene clusters involved in copper tolerance identified in Acinetobacter baumannii clinical and environmental isolates. Pathogens 9:60. doi: 10.3390/pathogens9010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baicus A, Lixandru B, Stroia M, Cirstoiu M, Constantin A, Usein CR, Cirstoiu CF. 2018. Antimicrobial susceptibility and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae strains isolated in an emergency university hospital. Rom Biotechnol Lett 23:13525–13529. [Google Scholar]
- 67.Főldes A, Bilca DV, Székely E. 2018. Phenotypic and molecular identification of carbapenemase-producing Enterobacteriaceae—challenges in diagnosis and treatment. Rev Rom Med Lab 26:221–230. doi: 10.2478/rrlm-2018-0018. [DOI] [Google Scholar]
- 68.Dumitru IM, Dumitrascu M, Vlad ND, Cernat RC, Ilie-Serban C, Hangan A, Slujitoru RE, Gherghina A, Mitroi-Maxim C, Curtali L, Carp DS, Dumitrescu A, Mitan R, Lesanu R, Rugina S. 2021. Carbapenem-resistant Klebsiella pneumoniae associated with COVID-19. Antibiotics (Basel) 10:561. doi: 10.3390/antibiotics10050561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fahrenfeld N, Bisceglia KJ. 2016. Emerging investigators series: sewer surveillance for monitoring antibiotic use and prevalence of antibiotic resistance: urban sewer epidemiology. Environ Sci: Water Res Technol 2:788–799. doi: 10.1039/C6EW00158K. [DOI] [Google Scholar]
- 70.Ebomah KE, Okoh AI. 2020. Detection of carbapenem-resistance genes in Klebsiella species recovered from selected environmental niches in the Eastern Cape Province, South Africa Antibiotics (Basel) 9:425. doi: 10.3390/antibiotics9070425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Köck R, Daniels-Haardt I, Becker K, Mellmann A, Friedrich AW, Mevius D, Schwarz S, Jurke A. 2018. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin Microbiol Infect 24:1241–1250. doi: 10.1016/j.cmi.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 72.Watahiki M, Kawahara R, Suzuki M, Aoki M, Uchida K, Matsumoto Y, Kumagai Y, Noda M, Masuda K, Fukuda C, Harada S, Senba K, Suzuki M, Matsui M, Suzuki S, Shibayama K, Shinomiya H. 2020. Single-tube multiplex polymerase chain reaction for the detection of genes encoding Enterobacteriaceae carbapenemase. Jpn J Infect Dis 73:166–172. doi: 10.7883/yoken.JJID.2019.041. [DOI] [PubMed] [Google Scholar]
- 73.Suzuki MT, Beja O, Taylor LT, DeLong EF. 2001. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ Microbiol 3:323–331. doi: 10.1046/j.1462-2920.2001.00198.x. [DOI] [PubMed] [Google Scholar]
- 74.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom. [Google Scholar]
- 75.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-indexsequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 80.Clarke KR, Gorley RN. 2015. PRIMER v7: user manual/tutorial. PRIMER-E Ltd, Plymouth, UK. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM02711-21_Supp_1_seq11.pdf, PDF file, 0.5 MB (473.9KB, pdf)
Supplemental material. Download SPECTRUM02711-21_Supp_2_seq12.xlsx, XLSX file, 0.02 MB (24.9KB, xlsx)
Data Availability Statement
Sequence data have been uploaded to the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/) under the BioProject accession number PRJNA790840.