ABSTRACT
SARS-CoV-2 variants of concern (VoC) are impacting responses to the COVID-19 pandemic. Here, we utilized passive immunization using human convalescent plasma (HCP) obtained from a critically ill COVID-19 patient in the early pandemic to study the efficacy of polyclonal antibodies generated to ancestral SARS-CoV-2 against the Alpha, Beta, and Delta VoC in the K18 human angiotensin converting enzyme 2 (hACE2) transgenic mouse model. HCP protected mice from challenge with the original WA-1 SARS-CoV-2 strain; however, only partially protected mice challenged with the Alpha VoC (60% survival) and failed to save Beta challenged mice from succumbing to disease. HCP treatment groups had elevated receptor binding domain (RBD) and nucleocapsid IgG titers in the serum; however, Beta VoC viral RNA burden in the lung and brain was not decreased due to HCP treatment. While mice could be protected from WA-1 or Alpha challenge with a single dose of HCP, six doses of HCP could not decrease mortality of Delta challenged mice. Overall, these data demonstrate that VoC have enhanced immune evasion and this work underscores the need for in vivo models to evaluate future emerging strains.
IMPORTANCE Emerging SARS-CoV-2 VoC are posing new problems regarding vaccine and monoclonal antibody efficacy. To better understand immune evasion tactics of the VoC, we utilized passive immunization to study the effect of early-pandemic SARS-CoV-2 HCP against, Alpha, Beta, and Delta VoC. We observed that HCP from a human infected with the original SARS-CoV-2 was unable to control lethality of Alpha, Beta, or Delta VoC in the K18-hACE2 transgenic mouse model of SARS-CoV-2 infection. Our findings demonstrate that passive immunization can be used as a model to evaluate immune evasion of emerging VoC strains.
KEYWORDS: SARS-CoV-2, COVID-19, variants of concern, Alpha, Beta, Delta, K18-hACE2 transgenic mice, convalescent plasma, modeling COVID-19, passive immunity
INTRODUCTION
The evolution of Severe Acute Respiratory Syndrome CoV-2 (SARS-CoV-2) variants of concern (VoC) has been a source of escalating epidemiological alarm in the currently ongoing coronavirus disease 2019 (COVID-19) pandemic. SARS-CoV-2 VoC have emerged and are thought to be more infectious and more lethal than the early 2020 original Wuhan-Hu-1 or USA-WA1/2020 (WA-1) strains (1–3). The VoC B.1.1.7, also known as Alpha variant (first identified in the United Kingdom) (4), and B.1.351 also known as Beta variant (first identified in South Africa) (5), were two SARS-CoV-2 VoC that rapidly spread around the world and exhibited high levels of infectivity and therapeutic resistance (3, 6–11). Both VoC contain important mutations in the receptor binding domain (RBD) of the spike (S) viral glycoprotein (4, 5) that are predicted to impact binding to the human angiotensin converting enzyme 2 (hACE2) viral receptor and enhance viral entry into host cells (12–16). In particular, Alpha contains the D614G and N501Y mutations in the SARS-CoV-2 S RBD which are theorized to increase the ability of the virus to bind to hACE2 (12, 14). Beta possesses these key mutations in the S RBD, in addition to the K417N and E484K mutations which are not directly implicated in altered viral transmission and hACE2 binding (16, 17). In December 2020, the VoC, B.1.617.2 (Delta) of SARS-CoV-2 first appeared in India, becoming quickly the global predominant circulating variant; however, this distinction could be soon displaced by the novel Omicron variant (18–20). The most common Delta variant has two important mutations on the viral S RBD, L452R and T478K, allowing for increased infectivity, transmissibility, as well as its ability of escaping neutralizing antibodies (21–23). The culmination of high infectivity, therapeutic resistance, and key changes in their viral genome suggests that VoC may have an impact on pathogenicity in animal models of SARS-CoV-2, with a subsequent impact on evaluating vaccines and therapeutics.
The K18-hACE2 transgenic mouse model (24) of SARS-CoV-2 infection was established by several groups in 2020 (25–27). K18-hACE2 transgenic mice challenged with SARS-CoV-2 exhibited significant morbidity and mortality, viral tropism of the respiratory and central nervous systems, elevated systemic chemokine and cytokine levels, significant tissue pathologies, and altered gross clinical measures (26–29). The generation of this mouse model has led to numerous studies of SARS-CoV-2 challenge for a variety of purposes including understanding SARS-CoV-2 related immunity, and therapeutic/vaccine testing (25, 30–35). As the world experiences an increase in the number of SARS-CoV-2 VoC, it is imperative to adapt existing preclinical animal infection models to these newly emerging VoC. Specifically, it is critical to understand if the K18-hACE2 transgenic mouse model first, is useful for studying SARS-CoV-2 VoC infection dynamics and second, if it exhibits any differences after challenge with newly emerged SARS-CoV-2 VoC. An investigation of these key points will provide context for studies important for developing new therapeutics and prophylactics as the COVID-19 pandemic continues and as new VoC emerge.
Neutralizing antibodies against SARS-CoV-2 induced by either natural infection or vaccination serve as an important component of protection against secondary SARS-CoV-2 infection (36); however, according to the WHO and recent data, Omicron variant appears to be able to easily infect fully vaccinated individuals. The S protein is a major target of neutralizing antibodies, with RBD encompassing 90% of the neutralizing antibodies within convalescent-phase sera (37, 38). Emergence of new VoC with mutations in the S protein and in the RBD could decrease the efficacy of neutralizing antibodies not originally generated against the VoC. Studies have shown that N-terminal domain S and RBD monoclonal antibodies generated against the original SARS-CoV-2 strain lose neutralization activity against VoC especially when administered as a monotherapy (7, 9, 39). Human convalescent plasma (HCP) also has demonstrated a decrease in neutralizing antibody efficacy against the VoC that specifically harbor the E484K mutation in the S RBD (7, 9). Here, we evaluated the polyclonal antibodies of HCP obtained from a patient infected with the original strain of SARS-CoV-2 against the Alpha, Beta, and Delta VoC in the K18-hACE2 transgenic mouse model. Our findings indicate that when compared with the original WA-1 strain, Alpha, Beta and Delta VoC are more resistant to HCP polyclonal antibodies in the K18-hACE2 transgenic mouse model. This passive immunity model allows for comparison of in vivo activity of human antibodies, extends upon in vitro studies and will likely assist in understanding immunity among VoC.
RESULTS
Evaluating human antibodies against original SARS-CoV-2 for their ability to protect VoC challenged mice.
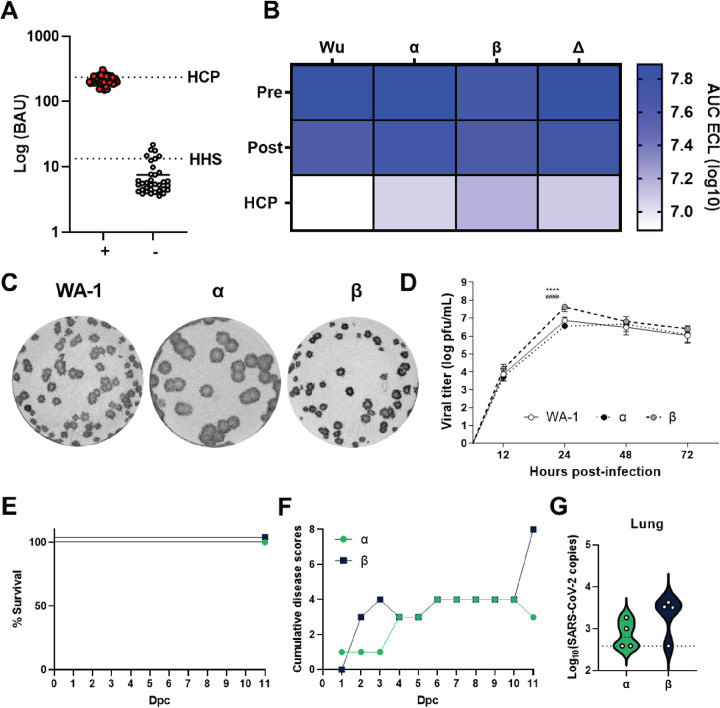
The emergence of SARS-CoV-2 VoC requires re-investigation of their pathogenesis and unique properties. Our goal for this part of the study was to determine if ancestral virus specific antibodies raised in humans would be able to provide protection against Alpha and Beta VoC challenge in K18-hACE2-mouse challenge model. HCP was extensively used early in the COVID-19 pandemic, but currently it is no longer used as a standard of care. The selected HCP for these studies originated from a patient with severe COVID-19 disease in 2020 and contained 236 antibody binding units (WHO COVID-19 International Standard; BAU). This HCP was compared with other 48 HCP samples from COVID-19 patients taken back in spring of 2020 (Fig. 1A). Next, the selected HCP was compared with serum obtained from pre-vaccine and post Pfizer mRNA vaccinated healthy volunteers. The selected HCP sample was able to neutralize Wuhan, Alpha, Beta, and Delta RBD to ACE2 binding using the MSD hACE2-RBD in vitro neutralization assay (Fig. 1B). These data indicate that the selected HCP had high binding and neutralization capacity. In vitro cell culture growth experiments were performed to characterize the Alpha and Beta VoC. The Beta variant appeared to have a modest increase in PFU/ml after 24 h of growth in vitro (Fig. 1CD); however, it had a relatively similar growth curve compared to the original WA-1 strain and Alpha VoC. One caveat about using Alpha or Beta challenge strains in mice, is that it is possible the mutations in RBD will allow for binding and engagement of the mouse ACE2 receptor. Mouse adapted SARS-CoV-2 strains are used to challenge wild type, non-transgenic mice (40), and VoC strains are known to replicate in wild-type mice (41). We performed a challenge study with Alpha and Beta VoC in wild type C57BL6/J mice; however, morbidity or mortality was not observed (Fig. 1E). We observed low disease scores, and very little detectable viral RNA in the lungs of the wild type challenged mice (Fig. 1FG). Based on these data, we do not believe there is much concern about using Alpha or Beta in mice because it appears their ability to infect through mouse ACE2 is limited.
FIG 1.
Characterization of early pandemic human convalescent plasma and in vitro characterization of SARS-CoV-2 variants. (A) RBD human IgG Binding antibody units (BAU) of SARS-CoV-2 + (red dots) compared to SARS-CoV-2 – patients (white dots). HCP dotted line indicate the BAU of the human convalescent plasma from a severe COVID-19 patient utilized in passive immunization studies in K18-hACE2 transgenic mice. HHS dotted line indicate the BAU of the healthy human serum used in passive immunization studies in K18-hACE2 transgenic mice. (B) ACE2-RBD neutralization was assayed, and the human convalescent plasma utilized was more capable of neutralizing receptor binding than mRNA vaccinated human sera. The heat map depicts the log10 AUC of electro chemiluminescent (ECL) values. (C) Plaque morphology of SARS-CoV-2 WA-1, Alpha or Beta infected VeroE6 cells. (D) Viral replication of SARS-CoV-2 variants in VeroE6 cells over time was quantified. Statistical analysis of viral replication was completed by two-way ANOVA followed by Tukey’s multiple comparison test, or RM ANOVA followed by Tukey’s multiple comparison test. **** = P < 0.0001 relative to WA-1, #### = P < 0.0001 relative to Alpha. C57BL6/J Mice were infected with 105 PFU SARS-CoV-2 VoC monitored for survival (E) and disease score (F). (G) Challenge with Alpha or Beta variants resulted in low detectable virus at day 11 post challenge. Dotted line represents limit of detection.
Effects of HCP treatment on disease progression in mice challenged with SARS-CoV-2 VoC.
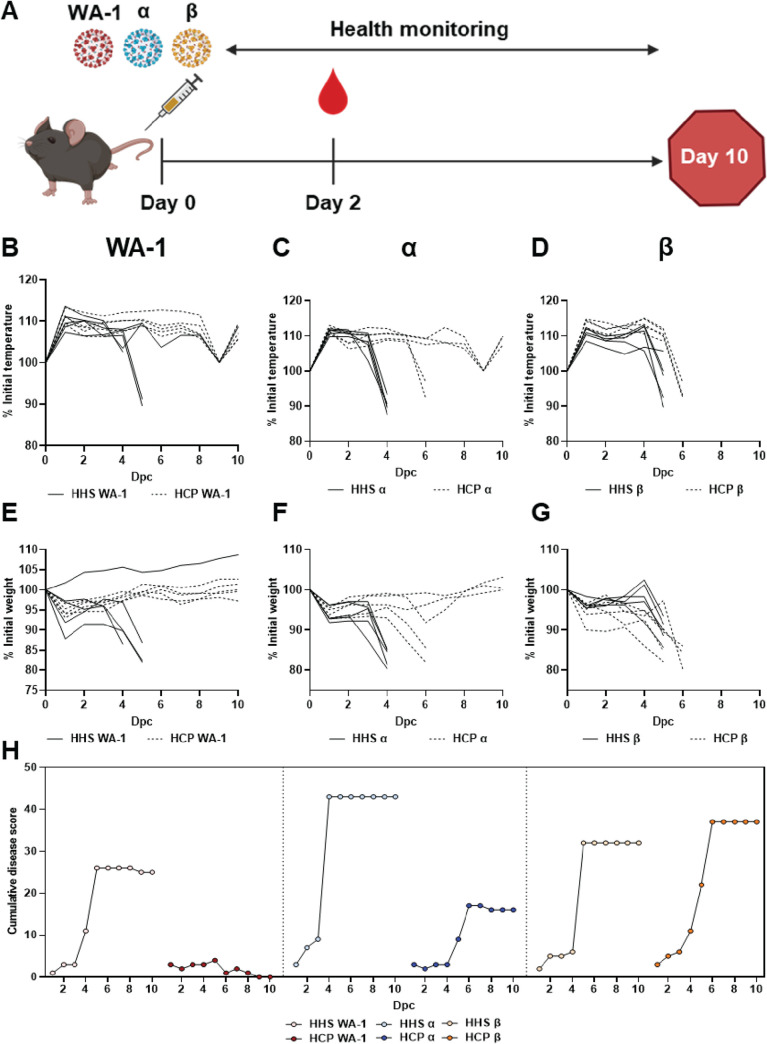
K18-hACE2 transgenic mice were passively immunized with HCP via intraperitoneal administration at day 0 and subsequently challenged with 105 PFU (lethal dose) of WA-1, Alpha, or Beta VoC (Fig. 2A). WA-1 challenged mice that received human serum from healthy individuals (HHS) exhibited a temperature drop, weight loss, and high cumulative disease scores (Fig. 2B, E, H). Mice treated with HCP had normal temperature regulation, maintained weight, and had low disease scores (Fig. 2B, E, H). Protection from WA-1 lethal challenge in HCP treated mice was expected since convalescent humans have immunity against re-challenge. Challenge with Alpha VoC in HHS treated mice resulted in high temperature loss by day 4 post challenge, up to 20% weight loss, and high cumulative disease scores (Fig. 2C, F, H). However, Alpha VoC challenged mice treated with HCP maintained body temperature in three of five animals and similar trends were observed for their body weight loss (Fig. 2CF). These data suggested that HCP was less successful at protecting mice from Alpha VoC challenge compared to WA-1. Disease scores also reflected these observations as HCP treatment was unable to fully suppress disease (Fig. 2H). Unlike WA-1 or Alpha VoC challenged mice, Beta VoC challenged mice treated with HCP compared to HHS had no significant differences by any metric measured (Fig. 2D, G, H). HCP treatment was unsuccessful in preventing disease and morbidity induced by the Beta VoC. Collectively, these data showed that HCP treatment was able to fully protect against WA-1; partially protect against Alpha VoC; but failed to protect against Beta VoC (Fig. 3AB).
FIG 2.
Effect of convalescent plasma treatment on SARS-CoV-2 VoC challenge in K18-hACE2 transgenic mice. (A) Passive immunization and SARS-CoV-2 challenge schematic. Mice were challenged with 105 PFU of SARS-CoV-2 WA-1 and VoC and simultaneously treated intraperitoneally with 500 μl HHS or HCP on day 0. Mice were monitored for temperature (B-D), body weight (E-G) and cumulative clinical score (H) over the 7-day course of infection.
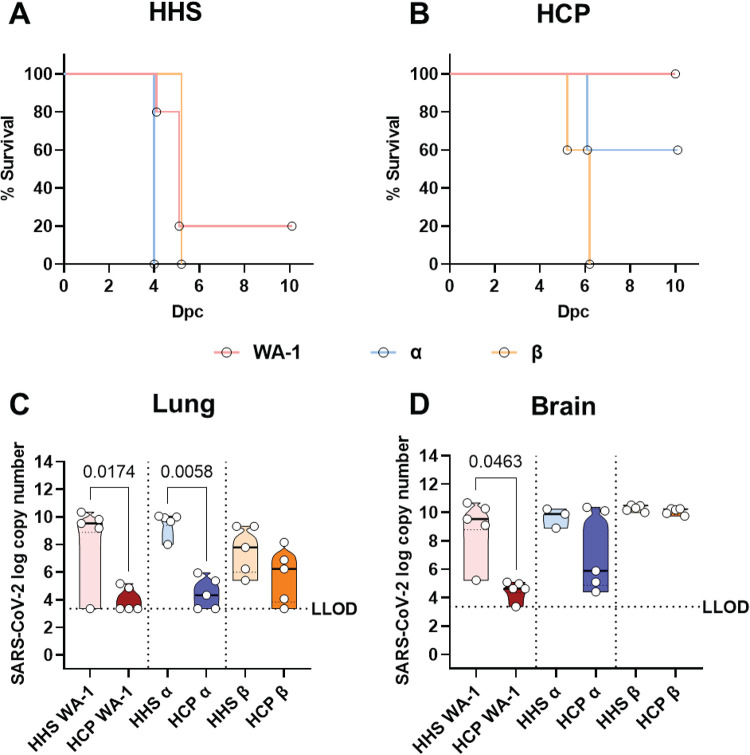
FIG 3.
Survival and viral RNA burden of serum-treated K18-hACE2 transgenic mice challenged with SARS-CoV-2 VoC. Kaplan-Meyer survival curves of mice challenged with Alpha, Beta, or WA-1 treated with HHS (A) or early pandemic SARS-CoV-2 HCP (B). Viral copy numbers in the lung (C) and brain (D) of challenged mice. LLOD = lower limit of detection based on a standard curve. Statistical significance of survival curves was assessed with the Mantel-Cox test. For HHS, WA-1 vs Alpha P = 0.0143; WA-1 vs Beta P = 0.9372 and Alpha vs Beta P = 0.0027. For HCP, WA-1 vs Alpha P = 0.1336; WA-1 vs Beta P = 0.0031 and Alpha vs Beta P = 0.0290. Statistical significance between viral copy number was assessed by a Kruskal-Wallis test followed by Dunn’s multiple-comparison test. n > 3 subjects per group. P values for significant differences are reported.
Effects of HCP treatment on viral RNA burden in lungs and brain of challenged mice.
To determine the viral distribution between the lungs and brain of challenged mice, qRT-PCR was used to quantify nucleocapsid copy number. HCP treatment significantly decreased viral RNA down to the lower limit of detection in the lung of the WA-1 and Alpha challenged treated with HHS mice compared to HHS (Fig. 3C). Beta variant challenged mice had two logs lower RNA compared to WA-1 and Alpha HHS treated mice and HCP treatment was able to decrease two of the mice down to the lower limit of detection (Fig. 3C). A lethal dose of SARS-CoV-2 WA-1 is known to infiltrate the brain of K18-hACE2 transgenic mice (26, 27). As expected, brain WA-1 viral copy numbers were decreased due to HCP treatment (Fig. 3D). Similarly, three of five Alpha VoC challenged mice treated with HCP had low viral RNA detected in their brains (Fig. 3D), which corresponded to their survival data (Fig. 3AB). Surprisingly, HCP treatment did not decrease brain Beta VoC virus RNA copies, further demonstrating the ability of Beta VoC to break through antibody protection that was derived against original Wuhan or WA-1-like viruses (Fig. 3D).
Human IgG levels in convalescent plasma treated K18-hACE2 transgenic mice challenged with SARS-CoV-2 VoC.
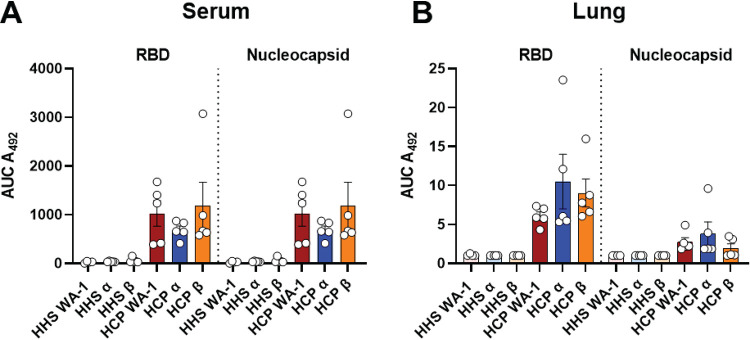
To determine the level of IgGs delivered to HHS and HCP treated mice, we analyzed whether human anti-SARS-CoV-2 IgGs were present within the lung and sera of animals treated with HCP or HHS through the course of infection (Fig. 4AB). Data demonstrated that significant quantities of human anti-SARS-CoV-2 IgGs targeting both the RBD and nucleocapsid proteins were present at 2 days post challenge in HCP-treated relative to HHS-treated mice (data not shown) as well as at euthanasia in the sera and lung (Fig. 4AB). Overall, these data indicated that passive immunization with HCP resulted in persistence of SARS-CoV-2 specific human antibodies in mice through the experimental time frame studied.
FIG 4.
Human anti-SARS-CoV-2 IgGs in serum-treated K18-hACE2 transgenic mice challenged with SARS-CoV-2 VoC at euthanasia. Area under the curve (AUC) analyses of anti-RBD IgG levels in the serum (A) or lung (B) of HHS or HCP VoC challenged mice. Statistical significance between AUCs was assessed by a Kruskal-Wallis test followed by Dunn’s multiple-comparison test. n > 3 subjects per group.
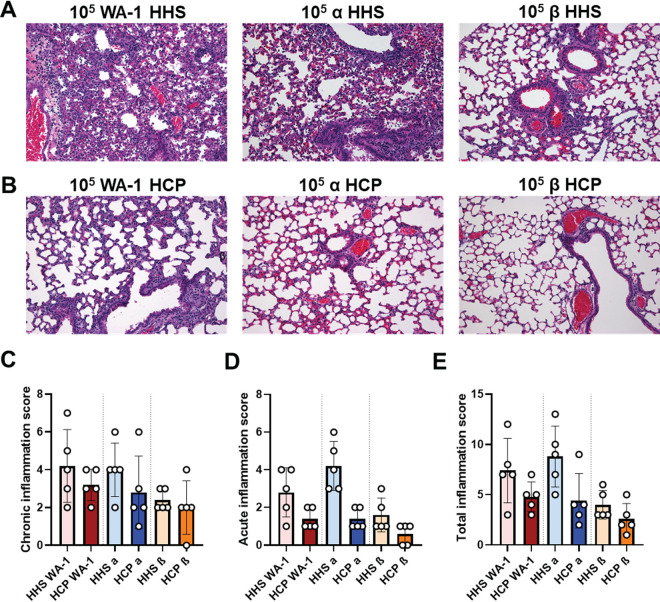
HCP treatment lowered chronic and acute inflammation in the lung caused by SARS-CoV-2 challenge.
Histopathology analysis was performed to characterize disease manifestation in the lung due to inflammation caused by WA-1, Alpha, or Beta challenge during HHS and HCP treatments (Fig. 5). Chronic inflammation was denoted as presence of lymphocytes, plasma cells and alveolar macrophages, whereas acute inflammation was characterized by neutrophils and edema in the lung parenchyma, vasculature, and bronchi. Total inflammation was determined by the addition of chronic and acute inflammation scores. HHS treatment groups challenged with WA-1, Alpha or Beta VoC had the highest chronic and acute inflammation scores in the lung parenchyma and surrounding blood vessels compared to the HCP treated mice (Fig. 5A, C–E). HHS treated mice challenged with WA-1 or Alpha VoC had the highest average total inflammation scores of 7.4 and 8.8, respectively; whereas Beta VoC challenged mice had an average total inflammation score of 4.0 (Fig. 5C, D, E). HCP treatment groups challenged with WA-1, Alpha or Beta VoC also had mixed chronic and acute inflammation albeit lower total inflammation compared to HHS treated mice (Fig. 5B–E). HCP treated mice challenged with WA-1 had the highest average total inflammation score (4.0), characterized by more chronic inflammation than acute (Fig. 5C, D). Mice treated with HCP and challenged with Alpha VoC had an average inflammation score of 4.4 and decreased acute inflammation compared to HHS treatment (Fig. 5D-E). Interestingly, HHS and HCP treated mice challenged with Beta VoC had low lung inflammation (Fig. 5C-E), which correlated with the low viral RNA burden of Beta VoC (Fig. 3C). Overall, HHS treated, and SARS-CoV-2 challenged mice had elevated levels of both chronic and acute inflammation compared to the HCP treated and challenged mice.
FIG 5.
Histopathological analysis of VoC challenged lungs. Left lobes of lungs from HHS and HCP treated and SARS-CoV-2 challenged mice were subjected to hematoxylin and eosin staining (A) ×200 magnification of the lung in HHS treated and SARS-CoV-2 challenged mice (B) ×200 magnification of the lung in HCP treated and SARS-CoV-2 challenged mice (C) Total chronic inflammation scores of each mouse. (D) Total acute inflammation score of each mouse. (E) Total inflammation score (chronic + acute) for each mouse. All statistical analysis was performed using Kruskal-Wallis test with Dunn’s multiple-comparison test.
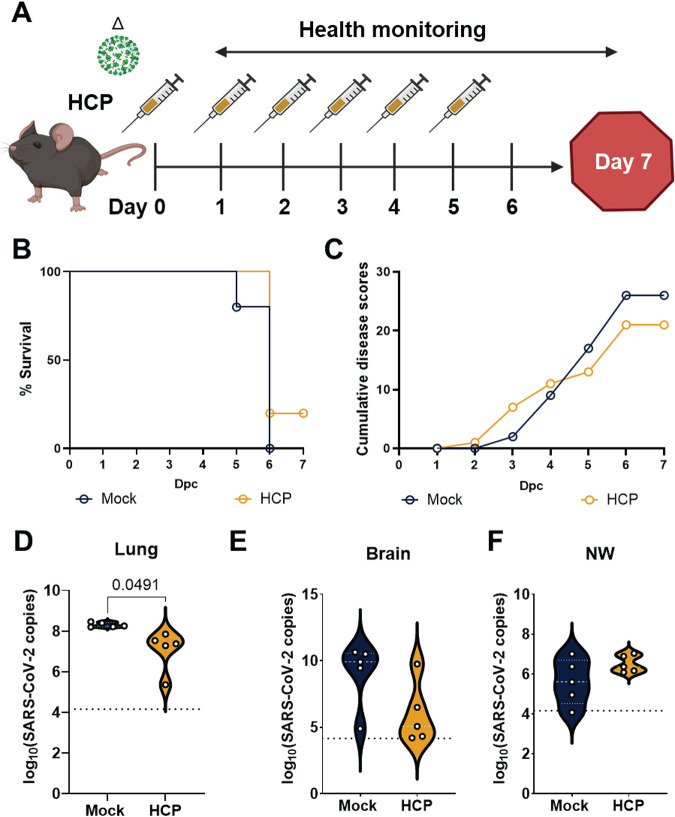
HCP passive immunization was insufficient to protect against Delta VoC challenge.
Delta VoC contains mutations on the RBD that compromise antibody neutralization (23). We further evaluated whether polyclonal antibodies in the HCP generated from an original virus immune plasma could protect mice from a lethal Delta VoC challenge. Here, we used a challenge dose of 104 PFU/dose of Delta VoC instead of a 105 PFU/dose as we previously used for WA-1, Alpha, and Beta VoC. In pilot studies, we demonstrated that 104 PFU/dose of Delta VoC resulted in 100% morbidity in K18-hACE2 transgenic mice (data not shown). Thus, mice were administered HCP (n = 5) or PBS (n = 5) intraperitoneally and concurrently intranasally challenged with a lethal Delta VoC dose on day 0 (Fig. 6A). HCP treated mice received treatment for 5 consecutive days after the first dose on day 0. All mice were monitored for disease for 7 days (Fig. 6A). Mice that did not receive HCP treatment succumbed to Delta VoC challenge by day 6 and had elevated cumulative disease scores (Fig. 6B, C). However, only 20% of mice that received 6 treatments of HCP survived the Delta VoC challenge and had disease scores similar to untreated mice (Fig. 6B, C). Viral RNA burden mirrored survival and disease scores for both HCP treated and untreated mice. Lung, brain, and nasal wash (NW) of the HCP treated mice had similar levels of viral RNA compared to untreated mice indicating that HCP treatment did not block viral replication (Fig. 6D–F). Overall, polyclonal antibodies generated against the ancestral SARS-CoV-2 strain did not protect mice from Delta VoC challenge suggesting that the Delta VoC is resistant to polyclonal antibodies generated against Wuhan-lineage virus strains.
FIG 6.
HCP passive immunization was insufficient to protect against Delta variant challenge. (A) Experimental workflow of passive immunization study with HCP and challenge with lethal dose of Delta variant (104 PFU/dose). (B) Kaplan Meier survival curve comparing Delta challenged mice that received either 1XDPBS vehicle or HCP. (C) Cumulative disease scores comparing Delta challenged mice that received either 1XDPBS vehicle or HCP. SARS-CoV-2 nucleocapsid RNA copies in A) lung, B) brain, and C) nasal wash of untreated and HCP treated and challenged mice. All statistical analysis was performed using Welch’s t test. P = 0.0491 (lumg, D), P = 0.0692 (brain, E), and P = 0.1603 (NW, F).
DISCUSSION
SARS-CoV-2 VoC are constantly evolving and dramatically impacting the ongoing COVID-19 pandemic. Since the beginning of the pandemic, three major infection waves have occurred: (i) original virus, (ii) Alpha variant, and (iii) Delta variant, with a recent wave starting directed by the novel Omicron (B.1.1.529) VoC. Approved vaccines are implemented all around the world with 8 billion total doses administered meaning 1 dose per person in the world. However, there are massive inequities in vaccine coverage with the United States/Canada, Latin America, Asia-Pacific, and Europe with ∼60–70% vaccination with one dose, whereas Africa is only at 10% coverage with one dose. Overall, the world is at 56% vaccine coverage with one dose. All current vaccines are designed against the original virus spike antigen sequence, but two major waves have been fueled by the Alpha and Delta VoC. Vaccine redevelopment will always be a challenge and new VoC have been constantly arising.
Alpha and Beta VoC spike antigens were extensively studied by binding and neutralization assays that suggested antibodies generated by infection or vaccination would be able to provide protection. Ultimately relatively low numbers of vaccine breakthrough occurred. To confirm the in vitro predictions regarding Alpha and Beta VoC, we designed this study to use a passive immunization model in K18-hACE2 transgenic mice to compare antibody dependent immunity between original virus versus VoC. Our observations suggested that Alpha VoC is partially neutralized in K18-hACE2 transgenic mice treated with HCP (Fig. 2 and 3), where the Beta VoC was not sufficiently neutralized to prevent lethality in this model system (Fig. 2 and 3). HCP treatment dramatically decreased viral RNA burden in the lungs and brain in WA-1 and Alpha VoC challenged mice, but minimal to no decrease was observed in mice challenged by the Beta VoC (Fig. 3C and D), which likely contributed to the morbidity and mortality caused by the Beta VoC. Low viral burden in the lung correlated with low chronic or acute inflammations scores (Fig. 5). Antibody breakthrough and aggressive pathogenesis suggested that the Beta VoC was going to likely be a variant of high concern. When Beta variant appeared, it was able to impact vaccine trial efficacy studies and seemed poised to infect vaccinated people (42–44). However, the Beta VoC peaked at a total of 12% genome worldwide frequency by April 2021. Thus, it seems likely that the Beta VoC was not highly transmissible, and our passive immunization model does not take this variable into consideration.
HCP as a treatment was used widely since the onset of the COVID-19 pandemic (45–47), but its efficacy was questionable (48–51) and convalescent plasma therapy for COVID-19 has been largely replaced as a treatment by monoclonal antibodies. In this study, we used HCP from an early pandemic COVID-19+ severe disease patient to understand how antibodies generated against the original SARS-CoV-2 strain would function against Alpha and Beta VoC. In December 2020, the Delta VoC appeared in India and by mid-2021, this VoC was the dominant variant found in genomic surveillance. To build upon our observations regarding Alpha and Beta VoC in the HCP passive immunity model with K18-hACE2 transgenic mice, we next aimed to evaluate the Delta VoC. Our pilot studies indicated a massive histopathological and inflammatory gene expression in Delta VoC versus Alpha VoC challenged mice (data not shown). We reasoned that the Delta VoC was more aggressive, would likely need a lower dose to be fully virulent compared to WA-1 strain and would also require more HCP to neutralize the virus in vivo. Thus, we challenged mice with a lower dose of 104 PFU and HCP treatment was provided daily out to 6-days post challenge. Unexpectedly, even though we provided 6X more HCP, mice were morbid with high disease scores and high viral burden (Fig. 6). It is now well appreciated that Delta VoC can cause breakthrough cases in previously infected as well as vaccinated humans (52, 53). Currently, with the highly mutated Omicron VoC, passive immunity and active immunization studies in pre-clinical models will be important to determine the breakthrough capacity of this new VoC. Furthermore, HCP or MAb passive studies can inform the scientific community about enhanced virulence or immune subversion of VoC and we anticipate this passive model can be applied going forward for rapid responses to characterize new variants.
In summary, this study provides insights into differences in SARS-CoV-2 VoC pathogenicity in K18-hACE2 transgenic mice in relation to antibody immunity. Passive immunization of mice with human antibodies can allow for robust characterization of breakthrough capacity (9, 20). This study demonstrates increased disease pathology for mice challenged with Alpha or Beta VoC, and the lack of protection from HCP in mice challenged with Beta or Delta VoC. These data corroborate observations about Beta and Delta VoC in human populations. The human convalescent plasma passive immunity model presented here can be useful in supporting in vitro studies and facilitate decision making and planning of research priorities around the overall immune evasion characteristics of SARS-CoV-2 variants.
MATERIALS AND METHODS
Ethics and biosafety.
The HCP used in this study was obtained under West Virginia University (WVU) IRB no. 2004976401 (54). HCP was obtained from a single individual with PCR-confirmed SARS-CoV-2 infection in March 2020. Experiments with live SARS-CoV-2 were conducted in Biosafety Level 3 (BSL-3) at Texas Biomedical Research Institute (TBRI IBC BSC20-004) or at WVU (IBC 20-09-03). All BSL-3 animal experiments were conducted under WVU IACUC protocol no. 2009036460.
Assessment of human IgGs against WA-1 SARS-CoV-2 S RBD and N.
Human IgGs against WA-1 SARS-CoV-2 S RBD and N were quantified using ELISA as described (54). WA-1 S RBD (2 μg/ml) or N (1 μg/ml) proteins were coated on plates and blocked with 3% milk in 0.1% Tween 20 +PBS (PBS-T). Plates were washed three times with PBS-T (200 μl) and virus inactivated samples (25 μl) from human plasma or infected mice were added to 100 μl of sample buffer (1% milk + 0.1% Tween 20 diluted in PBS) and serially diluted (5-fold) down the plates. The final row was left with 100 μl of sample buffer as a negative control. Plates were incubated for 10 min at room temperature shaking at 60 rpm and subsequently washed four times with PBS-T (200 μl). Secondary antibody (100 μl 1:500 anti-human IgG HRP, Invitrogen 31410) was added and plates were incubated for 10 min at room temperature shaking at 60 rpm. After incubation, plates were washed five times with PBS-T (200 μl) and SigmaFAST OPD (Sigma-Aldrich P9187, 100 μl) was added to each well of the plate. OPD development was stopped with 25 μl of 3 M hydrochloric acid and plates were read at an absorbance of 492 nm on a Synergy H1 plate-reader. Binding antibody units (BAU) were calculated based on the NIBSC 1st WHO International Standard (NIBSC code 20/136). Area under the curve analysis was completed in GraphPad Prism v.9.
Meso Scale Discovery COVID-19 ACE2 Neutralization assay.
SARS-CoV-2 challenged serum was analyzed using the SARS-CoV-2 Plate 11 Multi-Spot 96-well, 10 spot plate following the manufacturer protocol (catalog #: K15458U-2) on the MSD QuickPlex SQ120. The 10 spots contained RBD from different SARS-CoV-2 VoC: (i) B.1427, B.1.429, B.1.526.1; (ii) B.1.351, B.1.351.1; (iii) B.1.525, B.1.526, B.1.618, P.2, R.1; (iv) P.1; (v) B.1.526.2; (vi) B.1.17; (vii) B.1.17+E484K, P.3; (viii) B.1.617, B.1.617.1, B.1.617.3; (ix) AY.3, AY.4, AY.5, AY.6, AY.7, AY.12, AY.14, B.1.617.2, B.1.617.2+Δ144; and (x) A (WT). Three dilutions of serum, 1:10, 1:50 and 1:100 were analyzed for each mouse to perform Area Under the Curve analysis on the electrochemiluminescence using GraphPad Prism v.9.
Viral growth and in vitro analysis of SARS-CoV-2 replication.
SARS-CoV-2 USA-WA-1/2020 (NR-52281) (WA-1), B.1.1.7/Alpha (NR-54000), and B.1.351/Beta (NR-54008) strains were obtained from BEI Resources, and SARS-CoV-2 Delta variant B.1.617.2 hCoV-19/USA/WV-WVU-WV118685/2021 (GISAID Accession ID: EPI_ISL_1742834) was obtained from a patient sample at WVU. These strains were propagated in Vero E6 cells (ATCC-CRL-1586) as described (26, 55). Vero E6 cells for viral titrations (6-well plate, 106 cells/well) were infected with 10-fold serial dilutions of SARS-CoV-2. At 72 h post infection, cells were fixed overnight with 10% formalin (Sigma HT501128-4L), permeabilized and immunostained with 1 μg/ml of a SARS-CoV cross-reactive N protein antibody 1C7C7, kindly provided by Dr. Thomas Moran at the Icahn School of Medicine at Mount Sinai. For viral growth kinetics, Vero E6 cells (6-well plate, 106 cells/well, triplicates) were infected (multiplicity of infection, MOI 0.01) with SARS-CoV-2 WA-1, Alpha or Beta. At the indicated times after viral infection (12, 24, 48 and 72 h), tissue culture samples were collected and titrated by plaque assay as described (26).
Genome sequencing of SARS-CoV-2 VoC.
SARS-CoV-2 viral RNA from all stocks used for in vitro analyses was deep sequenced according to the method described (56). Briefly, we generated libraries using KAPA RNA HyperPrep Kit (Roche KK8541) with a 45 min adapter ligation incubation including 6-cycle of PCR with 100 ng RNA and 7 mM adapter concentration. Samples were sequenced on an Illumina Hiseq X machine. Raw reads were quality filtered using Trimmomatic v0.39 (57) and mapped to a SARS-CoV-2 reference genome (GenBank Accession No. MN985325) with Bowtie2 v2.4.1 (58). Genome coverage was quantified with MosDepth v0.2.6 (59). We genotyped each sample for low frequency VoC with LoFreq* v2.1.3.1 (60) and filtered sites with allele frequencies less than 20%. SARS-CoV-2 viral RNA from stocks used for K18-hACE2 transgenic mouse infection was deep sequenced and reads were aligned to the MN908947.3 reference genome using BWA v.0.7.17 (61) and trimmed for base-calling quality using iVar v.1.3.1 (62) with default parameters. Consensus sequence and individual mutations relative to the reference genome were determined using iVar, with a minimum allele frequency of 30% used as a threshold for calling a mutation. Coverage was computed using samtools mpileup v.1.11 (63). Lineage was confirmed using pangolin v.2.3.5 and pangoLEARN v.2021-03-16 (64). Authentication of the Beta stock was performed using metagenomic sequencing as described (65, 66). Viral RNA was treated with Turbo DNase I (Thermo Fisher). cDNA was generated from random hexamers using SuperScript III reverse transcriptase, second strand was generated using Sequenase 2.0, and cleaned using 0.8× Ampure XP beads purification on a SciClone IQ (Perkin Elmer). Sequencing libraries were generated using two-fifths volumes of Nextera XT on ds-cDNA with 18 cycles of PCR amplification. Libraries were cleaned using 0.8×Ampure XP beads and pooled equimolarly before sequencing on an Illumina NovaSeq (1 × 100bp run). Raw fastq reads were trimmed using cutadapt (-q 20) (67). To interrogate potential resistance alleles, reference-based mapping to NC_045512.2 was carried out using our modified Longitudinal Analysis of Viral Alleles (LAVA; https://github.com/michellejlin/lava) (68) pipeline. LAVA constructs a candidate reference genome from early passage virus using bwa (61), removes PCR duplicates with Picard, calls variants with VarScan (69, 70), and converts these changes into amino acid changes with Annovar (71). The genome sequence for strain Beta is accession number QWE88973. The genome sequence of the Beta contained the expected mutations spike and has a wild type furin cleavage site. A 52aa deletion was observed in orf7a; however, it is not expected that this deletion has any impact on the in vivo infection capacity of this strain as orf7a mutants are observed in surveillance. Beta VoC was able to effectively colonize and cause morbidity in experiments presented in this study.
Challenge of K18-hACE2 transgenic mice with SARS-CoV-2 VoC and treatment with HCP.
SARS-CoV-2 WA-1 and Alpha and Beta VoC were thawed from −80°C and diluted in infection medium (Dulbecco’s Modified Eagle Medium 4/.5g/L glucose + 2% fetal bovine serum + 1% HEPES + 1% penicillin/streptomycin at 100 units/μg/ml) to a concentration of 106 PFU/ml in the WVU BSL-3 facility. Delta VoC was diluted to a 104 PFU/dose from a 2.4 × 105 PFU/ml stock in 1X Dulbecco’s phosphate-buffered saline. Male 8 to 10 weeks old B6.Cg-Tg(K18-hACE2)2Prlmn/J mice (Jackson Laboratory 034860) were anesthetized with a single intraperitoneal dose of ketamine (Patterson Veterinary 07–803-6637, 80 mg/kg) + xylazine (Patterson Veterinary 07–808-1947, 8.3 mg/kg) and 50 μl infectious dose was administered with a pipette intranasally, 25 μl per nare. HCP, 500 μl, or healthy human sera (HHS) with known anti-SARS-CoV-2 IgGs and neutralizing Abs (nAbs) were administered intraperitoneally at this time. For the Delta VoC challenge study, 500 μl HCP was administered for 6 consecutive days (Fig. 6A). Mice were monitored until awake and alert.
Cumulative disease scoring of SARS-CoV-2 challenged mice.
Mice were scored daily on a scale encompassing appearance (score of 0–2), eye health (score of 0–2), respiration (score of 0–2), activity (score of 0–3) and weight loss (score of 0–5). Appearance included visual identification of a combination of mild to severe piloerection (0–2) or lack of grooming (0–2). Eye health scores were defined by observation of squinting (1), prolonged eye closure not related to sleep (2), or eye discharge (0–2) depending on severity. The maximal combined score for eye health was 2. Respiration (assessed visually) outside the range of 80–240 breaths per minute required mandatory euthanasia and scored as 2. Respiration that was abnormal in regularity was scored as 1. Activity was scored as slow (1), immobile (2), or collapsed and immobile (3). Weight loss was scored as 0–5% (0), 5–10% (1), 10–15% (2), 15–20% (3), >20% (4, 5). All mice with weight loss greater than 20% were humanely euthanized. Rectal temperature was also monitored daily throughout the experiments.
Euthanasia and necropsy of SARS-CoV-2 challenged mice.
Euthanasia was conducted by administering 200 μl of pentobarbital (Patterson Veterinary 07–805-9296, 390 mg/kg diluted in 0.9% sterile NaCl) and cardiac puncture. Blood was aliquoted into gold serum separator tubes (BD 365967) and centrifuged at 15,000 × g for 5 min. Serum was removed and stored in 1.5 ml tubes at −80°C until needed. Lungs were removed from animals and the right lobes of the lung were homogenized in 1 ml of PBS in Miltentyi C tubes (Miltenyi Biotec 130-096-334) using the m_lung_02 program on a Miltenyi gentleMACS tissue dissociator. An aliquot of each lung homogenate (300 μl) was added to 100 μl of TRIReagent (Zymo Research R2050-1-200) and stored at −80°C. Remaining homogenates (300 μl) were spun down at 15,000 × g and the supernatants collected. Pellets were frozen at −80°C until use. Brain tissue was removed from animals and split down the mid-line. The right brain was added to 1 ml of PBS in Miltenyi C tubes and homogenized using the m_lung_02 program. An aliquot of each homogenate (500 μl) was added 167 μl aliquots of TRIReagent and stored at −80°C until use. Remaining homogenates were frozen at −80°C until use. To inactivate virus from tissue samples, 1% vol/vol Triton X-100 (Sigma-Aldrich T8787) (27) was added to each sample and incubated for 1 h at room temperature. Inactivated samples were then removed from the BSL-3 High Containment facility.
Evaluating viral copy number in SARS-CoV-2 challenged tissues.
RNA from homogenized virus-inactivated lung and brain tissues of SARS-CoV-2 infected mice was extracted using the Direct-zol RNA MiniPrep Kit (Zymo Research R2051) following the manufacturer’s instructions. RT-PCR and qPCR were performed by generating a master mix of: 10 μl of TaqMan RT-PCR Mix from the Applied Biosystems TaqMan RNA to CT One Step Kit (Thermo-Fisher Scientific 4392938), 900 nM (1.8 μl) of (ATGCTGCAATCGTGCTACAA) forward nucleocapsid primer (27), 900 nM (1.8 μl) of (GACTGCCGCCTCTGCTC) reverse nucleocapsid primer (27), 250 nM (0.5 μl) of TaqMan probe (56-FAM/TCAAGGAAC/ZEN/AACATTGCCAA/3IABkFQ), 0.5 μl of TaqMan RT enzyme from the Applied Biosystems TaqMan RNA to CT One Step Kit (Thermo-Fisher Scientific 4392938), 100 ng of RNA, and RNase/DNase free water to make a 20 μl total reaction volume. Samples were run in triplicate in Microamp Optical 96-well Fast Reaction Plates (Thermo-Fisher Scientific 4306737) through the following protocol: reverse transcription at 48°C for 15 min, activation of AmpliTaq Gold DNA polymerase at 95°C for 10 min, and 50 cycles of 95°C denaturing for 10 s followed by 60°C annealing for 60 s. Samples were run on an Applied Biosystems StepOnePlus real-time PCR system. Samples with undetectable virus were assigned a value of 1. CT values and copy numbers were calculated and analyzed in Microsoft Excel and GraphPad Prism v.9.0.0.
Lung histopathology.
Left lobes of lungs were fixed in 10 ml of 10% neutral buffered formalin. Fixed lungs were paraffin embedded into 5-μm sections. Sections were stained with hematoxylin and eosin and sent to iHisto for pathological analysis. Lungs were scored by a pathologist for chronic and acute inflammation in the lung parenchyma, blood vessels, and airways. Pathologist was blinded to the experimental groups but was aware of groups that were challenged with SARS-CoV-2. Each mouse was scored individually using a standard qualitative toxicologic scoring criteria: 0 = none; 1 = minimal; 2 = mild; 3 = moderate; 4 = marked; 5 = severe. Chronic inflammation was marked by lymphocytes, plasma cells, and alveolar macrophages in the parenchyma, blood vessels and airway. Acute inflammation was scored by the presence of neutrophils and edema in the parenchyma, blood vessels and airway.
Statistical analyses.
All statistical tests were performed on groups with n ≥ 5 in GraphPad Prism v.9.0.0. To compare two-groups, student’s t-tests were used. To compare three or more groups, one-way ANOVA (parametric data) or Kruskal-Wallis (non-parametric data) were used followed by Tukey’s (parametric data) or Dunn’s (non-parametric data) multiple comparisons tests. To compare grouped data, two-way ANOVA with no correction was performed followed by Tukey’s multiple comparison test. To assess statistical differences between Kaplan-Meyer curves, Mantel-Cox log-rank tests were performed.
ACKNOWLEDGMENTS
We would like to express our gratitude to Laura Gibson and Clay Marsh for enabling this research through resources and support. This project was supported by the Vaccine Development Center at the West Virginia University Health Sciences Center. F.H.D. and the VDC are supported by the Research Challenge Grant no. HEPC.dsr.18.6 from the Division of Science and Research, WV Higher Education Policy Commission. MSD analyses were supported financially by the West Virginia University Flow Cytometry & Single Cell Core Facility, which is supported by the National Institutes of Health equipment grant number S10OD016165 and the Institutional Development Awards (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant numbers P30GM121322 (TME CoBRE) and P20GM103434 (INBRE).
A.M.H., F.H.D., I.M., J.B.T., C.Y., and L.M.S. designed the experiments. C.Y performed in vitro analyses of SARS-CoV-2 VoC, and J.S., A.G., and R.K.P. performed sequencing. M.T.W. and I.M. propagated virus for animal experiments. A.M.H., T.Y.W., B.P.R., K.S.L., O.A.M., M.C., H.A.C., and F.H.D. challenged and euthanized animals, dissected organs, and prepared them for analyses. A.M.H., B.P.R., and N.A.R. performed ELISA against SARS-CoV-2 antigens and A.M.H., T.Y.W., H.A.C., and O.A.M. performed qPCR to determine viral load. J.D. analyzed sequence data from viral passages used for infection of K18-hACE2 transgenic mice. A.M.H., F.H.D., and T.Y.W. analyzed, formatted, and represented data for publication. All authors contributed to the writing and revision of the manuscript.
We declare no competing interests.
Contributor Information
F. Heath Damron, Email: fdamron@hsc.wvu.edu.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Angyal A, Brown RL, Carrilero L, Green LR, Groves DC, Johnson KJ, Keeley AJ, Lindsey BB, Parsons PJ, Raza M, Rowland-Jones S, Smith N, Tucker RM, Wang D, Wyles MD, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Sheffield COVID-19 Genomics Group . 2020. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toyoshima Y, Nemoto K, Matsumoto S, Nakamura Y, Kiyotani K. 2020. SARS-CoV-2 genomic variations associated with mortality rate of. J Hum Genet 65:1075–1082. 10.1038/s10038-020-0808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Challen R, Brooks-Pollock E, Read JM, Dyson L, Tsaneva-Atanasova K, Danon L. 2021. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: Matched cohort study. BMJ 372:n579–10. 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, Connor T, Peacock T, Robertson DL, Volz E. 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations COVID-19 Genomics Consortium UK (CoG-UK). https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 [Google Scholar]
- 5.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K, von Gottberg A, Walaza S, Allam M, Ismail A, Mohale T, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Petruccione F, Sigal A, Hardie D, Marais G, Hsiao M, Korsman S, Davies M-A, Tyers L, Mudau I, York D, Maslo C, Goedhals D, Abrahams S, Laguda-Akingba O, Alisoltani-Dehkordi A, Godzik A, Wibmer CK, Sewell BT, Lourenço J, Alcantara LCJ, Pond SLK, Weaver S, Martin D, Lessells RJ, Bhiman JN, Williamson C, de Oliveira T. 2020. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. [Google Scholar]
- 6.Davies NG, Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, Russell TW, Tully DC, Washburne AD, Wenseleers T, Gimma A, Waites W, Wong KLM, Van Zandvoort K, Silverman JDWorking CC-, Consortium C-GUKC, Diaz-ordaz K, Keogh R ,. 2021. Estimated transmissibility and impact of SARS-CoV-2 lineage B. 1. 1. 7 in England. Science 372: eabg3055. 10.1126/science.abg3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD, Graham BS, Mascola JR, Chang JY, Yin MT, Sobieszczyk M, Kyratsous CA, Shapiro L, Sheng Z, Huang Y, Ho DD. 2021. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593:130–135. 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 8.Galloway SE, Prabasaj P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A, Slayton RB, Tong S, Silk BJ, Armstrong GL, Biggerstaff M, Dugan VG. 2021. Emergence of SARS-CoV-2 B.1.1.7 lineage—United States, December 29, 2020–January 12, 2021. MMWR Morb Mortal Wkly Rep 70:95–99. 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen RE, Zhang X, Case JB, Winkler ES, Liu Y, VanBlargan LA, Liu J, Errico JM, Xie X, Suryadevara N, Gilchuk P, Zost SJ, Tahan S, Droit L, Turner JS, Kim W, Schmitz AJ, Thapa M, Wang D, Boon ACM, Presti RM, O’Halloran JA, Kim AHJ, Deepak P, Pinto D, Fremont DH, Crowe JE, Corti D, Virgin HW, Ellebedy AH, Shi P-Y, Diamond MS. 2021. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med 27:717–726. 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Regeneron. 2021. Fact sheet for health care providers emergency use authorization (EUA) of REGEN-COVTM (casirivimab with imdevimab). FDA Tarrytown, NY: Regeneron. [Google Scholar]
- 11.Eli Lilly. 2021. Fact sheet for health care providers emergency use authorization (EUA) of bamlanivimab and etesevimab. Indianapolis, IN: Eli Lilly. [Google Scholar]
- 12.Tian F, Tong B, Sun L, Shi S, Zheng B, Wang Z, Dong X, Zheng P. 2021. Mutation N501Y in RBD of Spike Protein Strengthens the Interaction between COVID-19 and its Receptor ACE2. bioRxiv 192021.02.14.431117. [Google Scholar]
- 13.Shah M, Ahmad B, Choi S, Woo HG. 2020. Mutations in the SARS-CoV-2 spike RBD are responsible for stronger ACE2 binding and poor anti-SARS-CoV mAbs cross-neutralization. Comput Struct Biotechnol J 18:3402–3414. 10.1016/j.csbj.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozono S, Zhang Y, Ode H, Sano K, Tan TS, Imai K, Miyoshi K, Kishigami S, Ueno T, Iwatani Y, Suzuki T, Tokunaga K. 2021. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat Commun 12. 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bozdaganyan ME, Sokolova OS, Shaitan KV, Kirpichnikov MP, Orekhov PS. 2021. Effects of mutations in the receptor-binding domain of SARS-CoV-2 spike on its binding affinity to ACE2 and neutralizing antibodies revealed by computational analysis. bioRxiv. [DOI] [PMC free article] [PubMed]
- 16.Laffeber C, de Koning K, Kanaar R, Lebbink JH. 2021. Experimental evidence for enhanced receptor binding by rapidly spreading SARS-CoV-2 variants. bioRxiv. [DOI] [PMC free article] [PubMed]
- 17.Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Tuekprakhon A, Nutalai R, Wang B, Paesen GC, Lopez-Camacho C, Slon-Campos J, Hallis B, Coombes N, Bewley K, Charlton S, Walter TS, Skelly D, Lumley SF, Dold C, Levin R, Dong T, Pollard AJ, Knight JC, Crook D, Lambe T, Clutterbuck E, Bibi S, Flaxman A, Bittaye M, Belij-Rammerstorfer S, Gilbert S, James W, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR. 2021. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. 2021 Cell 184:2348–2361.e6. 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar R, Lo M, Saha R, Dutta S, Chawla-Sarkar M. 2021. S glycoprotein diversity of the Omicron variant. bioRxiv. 10.1101/2021.12.04.21267284. [DOI] [Google Scholar]
- 19.Cele S, Jackson L, Khan K, Khoury D, Moyo-Gwete T, Tegally H, Scheepers C, Amoako D, Karim F, Bernstein M, Lustig G, Archary D, Smith M, Ganga Y, Jule Z, Reedoy K, San JE, Hwa S-H, Giandhari J, Blackburn JM, Gosnell BI, Karim SA, Hanekom W, Team C-K, von Gottberg A, Bhiman J, Lessells RJ, Moosa M-YS, Davenport M, de Oliveira T, Moore PL, Sigal A, NGS-SA . 2021. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv. 10.1101/2021.12.08.21267417. [DOI] [Google Scholar]
- 20.Vanblargan LA, Errico JM, Halfmann PJ, Zost SJ, Crowe JE, Purcell LA, Kawaoka Y, Corti D, Fremont DH, Diamond MS, Biotechnology V, Louis S, Mo U. 2021. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by 1 several therapeutic monoclonal antibodies 2 3. bioRxiv. 10.1101/2021.12.15.472828. [DOI] [PMC free article] [PubMed]
- 21.Mlcochova P, Kemp SA, Dhar MS, Papa G, Meng B, Ferreira IATM, Datir R, Collier DA, Albecka A, Singh S, Pandey R, Brown J, Zhou J, Goonawardane N, Mishra S, Whittaker C, Mellan T, Marwal R, Datta M, Sengupta S, Ponnusamy K, Srinivasan Radhakrishnan V, Abdullahi A, Charles O, Chattopadhyay P, Devi P, Caputo D, Peacock T, Wattal C, Goel N, Satwik A, Vaishya R, Agarwal M. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Chiara Silacci-Fegni 13:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.T M Ferreira IA, Kemp SA, Datir R, Saito A, Meng B, Rakshit P, Takaori-Kondo A, Kosugi Y, Uriu K, Kimura I, Shirakawa K, Abdullahi A, Agarwal A, Ozono S, Tokunaga K, Sato K, Gupta RK. 2021. SARS-CoV-2 B.1.617 mutations L452R and E484Q are not synergistic for antibody evasion. J Infect Dis 224:989–994. 10.1093/infdis/jiab368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Le Guen J, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Péré H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Lorière E, Rey FA, Schwartz O. 2021. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nat 2021 5967871 596:276–280. 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 24.McCray PB, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, Netland J, Jia HP, Halabi C, Sigmund CD, Meyerholz DK, Kirby P, Look DC, Perlman S. 2007. Lethal Infection of K18-hACE2 Mice Infected with Severe Acute Respiratory Syndrome Coronavirus. J Virol 81:813–821. 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreau GB, Burgess SL, Sturek JM, Donlan AN, Petri WA, Mann BJ. 2020. Evaluation of K18-hACE2 mice as a model of SARS-CoV-2 infection. Am J Trop Med Hyg 103:1215–1219. 10.4269/ajtmh.20-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oladunni FS, Park JG, Pino PA, Gonzalez O, Akhter A, Allué-Guardia A, Olmo-Fontánez A, Gautam S, Garcia-Vilanova A, Ye C, Chiem K, Headley C, Dwivedi V, Parodi LM, Alfson KJ, Staples HM, Schami A, Garcia JI, Whigham A, Platt RN, Gazi M, Martinez J, Chuba C, Earley S, Rodriguez OH, Mdaki SD, Kavelish KN, Escalona R, Hallam CRA, Christie C, Patterson JL, Anderson TJC, Carrion R, Dick EJ, Hall-Ursone S, Schlesinger LS, Alvarez X, Kaushal D, Giavedoni LD, Turner J, Martinez-Sobrido L, Torrelles JB. 2020. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat Commun 11. 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, Fox JM, Chen RE, Earnest JT, Keeler SP, Ritter JH, Kang LI, Dort S, Robichaud A, Head R, Holtzman MJ, Diamond MS. 2020. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol 21:1327–1335. 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yinda CK, Port JR, Bushmaker T, Owusu IO, Purushotham JN, Avanzato VA, Fischer RJ, Schulz JE, Holbrook MG, Hebner MJ, Rosenke R, Thomas T, Marzi A, Best SM, de Wit E, Shaia C, van Doremalen N, Munster VJ. 2021. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog 17:e1009195–21. 10.1371/journal.ppat.1009195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng J, Wong LYR, Li K, Verma AK, Ortiz ME, Wohlford-Lenane C, Leidinger MR, Knudson CM, Meyerholz DK, McCray PB, Perlman S. 2021. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589:603–607. 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassan AO, Kafai NM, Dmitriev IP, Fox JM, Smith BK, Harvey IB, Chen RE, Winkler ES, Wessel AW, Case JB, Kashentseva E, McCune BT, Bailey AL, Zhao H, VanBlargan LA, Dai YN, Ma M, Adams LJ, Shrihari S, Danis JE, Gralinski LE, Hou YJ, Schäfer A, Kim AS, Keeler SP, Weiskopf D, Baric RS, Holtzman MJ, Fremont DH, Curiel DT, Diamond MS. 2020. A Single-Dose Intranasal ChAd Vaccine Protects Upper and Lower Respiratory Tracts against SARS-CoV-2. Cell 183:169–184.e13. 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandey K, Acharya A, Mohan M, Ng CL, Reid SP, Byrareddy SN. 2021. Animal models for SARS-CoV-2 research: A comprehensive literature review. Transbound Emerg Dis 68:1868–1885. 10.1111/tbed.13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu R, Americo JL, Cotter CA, Earl PL, Erez N, Peng C, Moss B. 2021. One or two injections of MVA-vectored vaccine shields hACE2 transgenic mice from SARS-CoV-2 upper and lower respiratory tract infection. Proc Natl Acad Sci USA 118:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silvas J, Morales-Vasquez D, Park J-G, Chiem K, Torrelles JB, Platt RN, Anderson T, Ye C, Martinez-Sobrido L. 2021. Contribution of SARS-CoV-2 accessory proteins to viral pathogenicity in K18 hACE2 transgenic mice. bioRxiv 6. [Google Scholar]
- 34.Sarkar J, Guha R. 2020. Infectivity, virulence, pathogenicity, host-pathogen interactions of SARS and SARS-CoV-2 in experimental animals : a systematic review. Vet Res Commun 44:101–110. 10.1007/s11259-020-09778-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumari P, Rothan HA, Natekar JP, Stone S, Pathak H, Strate PG, Arora K, Brinton MA, Kumar M. 2020. Neuroinvasion and encephalitis following intranasal inoculation of SARS-CoV-2 in K18-hACE2 mice. bioRxiv. 10.1101/2020.12.14.422714. [DOI] [PMC free article] [PubMed]
- 36.Sette A, Crotty S. 2021. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 184:861–880. 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piccoli L, Park YJ, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, Silacci-Fregni C, Pinto D, Rosen LE, Bowen JE, Acton OJ, Jaconi S, Guarino B, Minola A, Zatta F, Sprugasci N, Bassi J, Peter A, De Marco A, Nix JC, Mele F, Jovic S, Rodriguez BF, Gupta SV, Jin F, Piumatti G, Lo Presti G, Pellanda AF, Biggiogero M, Tarkowski M, Pizzuto MS, Cameroni E, Havenar-Daughton C, Smithey M, Hong D, Lepori V, Albanese E, Ceschi A, Bernasconi E, Elzi L, Ferrari P, Garzoni C, Riva A, Snell G, Sallusto F, Fink K, Virgin HW, Lanzavecchia A, Corti D, Veesler D. 2020. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell 183:1024–1042.e21. 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brouwer PJ, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, A Okba NM, Claireaux M, Kerster G, H Bentlage AE, van Haaren MM, Guerra D, Burger JA, Schermer EE, Verheul KD, van der Velde N, van der Kooi A, van Schooten J, van Breemen MJ, L Bijl TP, Sliepen K, Aartse A, Derking R, Bontjer I, Kootstra NA, Joost Wiersinga W, Vidarsson G, Haagmans BL, Ward AB, de Bree GJ, Sanders RW, van Gils MJ. 2020. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369:364–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S, Chandele A, Sharma A. 2021. Current status of therapeutic monoclonal antibodies against SARS-CoV-2. PLoS Pathog 17:e1009885. 10.1371/journal.ppat.1009885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dinnon KH, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Hou YJ, Adams LE, Gully KL, Brown AJ, Huang E, Bryant MD, Choong IC, Glenn JS, Gralinski LE, Sheahan TP, Baric RS. 2020. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586:560–566. 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montagutelli X, Prot M, Levillayer L, Salazar EB, Jouvion G, Conquet L, Donati F, Albert M, Gambaro F, van der Behillil S, Enouf V, Rousset D, Jaubert J, Rey F, van der Werf S, Simon-Loriere E. 2021. The B.1.351 and P.1 variants extend SARS-CoV-2 host range to mice. bioRxiv:1–16.
- 42.Shinde V, Bhikha S, Hoosain Z, Archary M, Bhorat Q, Fairlie L, Lalloo U, Masilela MSL, Moodley D, Hanley S, Fouche L, Louw C, Tameris M, Singh N, Goga A, Dheda K, Grobbelaar C, Kruger G, Carrim-Ganey N, Baillie V, de Oliveira T, Lombard Koen A, Lombaard JJ, Mngqibisa R, Bhorat AE, Benadé G, Lalloo N, Pitsi A, Vollgraaff P-L, Luabeya A, Esmail A, Petrick FG, Oommen-Jose A, Foulkes S, Ahmed K, Thombrayil A, Fries L, Cloney-Clark S, Zhu M, Bennett C, Albert G, Faust E, Plested JS, Robertson A, Neal S, Cho I, Glenn GM, Dubovsky F, Madhi SA, 2019nCoV-501 Study Group . 2021. Efficacy of NVX-CoV2373 Covid-19 vaccine against the B.1.351 variant. N Engl J Med 384:1899–1909. 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heath PT, Galiza EP, Baxter DN, Boffito M, Browne D, Burns F, Chadwick DR, Clark R, Cosgrove C, Galloway J, Goodman AL, Heer A, Higham A, Iyengar S, Jamal A, Jeanes C, Kalra PA, Kyriakidou C, McAuley DF, Meyrick A, Minassian AM, Minton J, Moore P, Munsoor I, Nicholls H, Osanlou O, Packham J, Pretswell CH, Ramos ASF, Saralaya D, Sheridan RP, Smith R, Soiza RL, Swift PA, Thomson EC, Turner J, Viljoen ME, Albert G, Cho I, Dubovsky F, Glenn G, Rivers J, Robertson A, Smith K, Toback S, 2019nCoV-302 Study Group . 2021. Safety and efficacy of NVX-CoV2373 Covid-19 vaccine. N Engl J Med 385:1172–1183. 10.1056/NEJMoa2107659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Falsey AR, Sobieszczyk ME, Hirsch I, Sproule S, Robb ML, Corey L, Neuzil KM, Hahn W, Hunt J, Mulligan MJ, McEvoy C, DeJesus E, Hassman M, Little SJ, Pahud BA, Durbin A, Pickrell P, Daar ES, Bush L, Solis J, Carr QO, Oyedele T, Buchbinder S, Cowden J, Vargas SL, Guerreros Benavides A, Call R, Keefer MC, Kirkpatrick BD, Pullman J, Tong T, Brewinski Isaacs M, Benkeser D, Janes HE, Nason MC, Green JA, Kelly EJ, Maaske J, Mueller N, Shoemaker K, Takas T, Marshall RP, Pangalos MN, Villafana T, Gonzalez-Lopez A. 2021. Phase 3 safety and efficacy of AZD1222 (ChAdOx1 nCoV-19) Covid-19 vaccine. N Engl J Med 385:2348–2360. 10.1056/NEJMoa2105290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bloch EM, Shoham S, Casadevall A, Sachais BS, Shaz B, Winters JL, Van Buskirk C, Grossman BJ, Joyner M, Henderson JP, Pekosz A, Lau B, Wesolowski A, Katz L, Shan H, Auwaerter PG, Thomas D, Sullivan DJ, Paneth N, Gehrie E, Spitalnik S, Hod EA, Pollack L, Nicholson WT, Pirofski LA, Bailey JA, Tobian AAR. 2020. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J Clin Invest 130:2757–2765. 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bloch EM. 2020. Convalescent plasma to treat COVID-19. Blood 136:654–655. 10.1182/blood.2020007714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L, Xiong J, Bao L, Shi Y. 2020. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis 20:398–400. 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.NIH. 2021. NIH halts trial of COVID-19 convalescent plasma in emergency department patients with mild symptoms. Bethesda, MD: NIH. [Google Scholar]
- 49.Zhao Q, He Y. 2020. Challenges of convalescent plasma therapy on COVID-19. J Clin Virol 127. 10.1016/j.jcv.2020.104358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casadevall A, Henderson J, Joyner M, Pirofski L. 2021. SARS-Cov2 variants and convalescent plasma: reality, fallacies, and opportunities. J Clin Invest 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cele S, Gazy I, Jackson L, Hwa S-H, Tegally H, Lustig G, Giandhari J, Pillay S, Wilkinson E, Naidoo Y, Karim F, Ganga Y, Khan K, Bernstein M, Balazs AB, Gosnell BI, Hanekom W, Moosa M-YS, Team C, Lessells RJ, de Oliveira T, Sigal A. 2021. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shastri J, Parikh S, Aggarwal V, Agrawal S, Chatterjee N, Shah R, Devi P, Mehta P, Pandey R. 2021. Severe SARS-CoV-2 breakthrough reinfection with delta variant after recovery from breakthrough infection by Alpha variant in a fully vaccinated health worker. Front Med 8:1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lipsitch M, Krammer F, Regev-Yochay G, Lustig Y, Balicer RD. 2021. SARS-CoV-2 breakthrough infections in vaccinated individuals: measurement, causes and impact. Nat Rev Immunol 2021:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horspool AM, Kieffer T, Russ BP, DeJong MA, Wolf MA, Karakiozis JM, Hickey BJ, Fagone P, Tacker DH, Bevere JR, Martinez I, Barbier M, Perrotta PL, Damron FH. 2021. Interplay of antibody and cytokine production reveals CXCL13 as a potential novel biomarker of lethal SARS-CoV-2 infection. mSphere 6. 10.1128/mSphere.01324-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Case JB, Bailey AL, Kim AS, Chen RE, Diamond MS. 2020. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology 548:39–48. 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye C, Chiem K, Park J, Oladunni F, Anderson T, Almazan F, Martinez-Sobrido L. 2020. Rescue of SARS-CoV-2 from a Single Bacterial Artificial Chromosome. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–360. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pedersen BS, Quinlan AR. 2018. Mosdepth : quick coverage calculation for genomes and exomes. Bioinformatics 34:867–868. 10.1093/bioinformatics/btx699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilm A, Poh P, Aw K, Bertrand D, Hui G, Yeo T, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. 2012. LoFreq : a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40:11189–11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows – Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grubaugh ND, Gangavarapu K, Quick J, Matteson NL, De Jesus JG, Main BJ, Tan AL, Paul LM, Brackney DE, Grewal S, Gurfield N, Van Rompay KKA, Isern S, Michael SF, Coffey LL, Loman NJ, Andersen KG. 2019. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Data GP, Sam T, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Toole Á, S E, Underwood A, Jackson B, Hill V, McCrone J, Ruis C, Abu-Dahab K, Taylor B, Yeats C, Du Plessis L, Aanensen D, Holmes E, Pybus O, Rambaut A. 2021. Pangolin: lineage assignment in an emerging pandemic as an epidemiological tool. Virus Evolution 7:veab064. 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Addetia A, Crawford KHD, Dingens A, Zhu H, Roychoudhury P, Huang ML, Jerome KR, Bloom JD, Greninger AL. 2020. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with high attack rate. medRxiv 58:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greninger AL, Waghmare A, Adler A, Qin X, Crowley JL, Englund JA, Kuypers JM, Jerome KR, Zerr DM. 2017. Rule-out outbreak: 24-hour metagenomic next-generation sequencing for characterizing respiratory virus source for infection prevention. J Pediatric Infect Dis Soc 6:168–172. 10.1093/jpids/pix019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17:10–12. 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 68.Jin M, Shean RC, Makhsous N, Greninger AL. 2019. LAVA: a streamlined visualization tool for longitudinal analysis of viral alleles. bioRxiv.
- 69.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. 2009. VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25:2283–2285. 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568–576. 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang K, Li M, Hakonarson H. 2010. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]