ABSTRACT
In our previous study, we found that a new type of Chikungunya virus particle with a complete capsid deletion (ΔC-CHIKV) is still infectious in BHK-21 cells and demonstrated its potential as a live attenuated vaccine candidate. However, the low yield as well as the disability to propagate in vaccine production cell line Vero of ΔC-CHIKV are not practical for commercial vaccine development. In this study, we not only achieved the successful propagation of the viral particle in Vero cells, but significantly improved its yield through construction of a chimeric VEEV-ΔC-CHIKV and extensive passage in Vero cells. Mechanistically, high production of VEEV-ΔC-CHIKV is due to the improvement of viral RNA packaging efficiency conferred by adaptive mutations, especially those in envelope proteins. Similar to ΔC-CHIKV, the passaged VEEV-ΔC-CHIKV is safe, immunogenic, and efficacious, which protects mice from CHIKV challenge after only one shot of immunization. Our study demonstrates that the utilization of infectious capsidless viral particle of CHIKV as a vaccine candidate is a practical strategy for the development of alphavirus vaccine.
IMPORTANCE Chikungunya virus (CHIKV) is one of important emerging alphaviruses. Currently, there are no licensed vaccines against CHIKV infection. We have previously found a new type of Chikungunya virus particle with a complete capsid deletion (ΔC-CHIKV) as a live attenuated vaccine candidate that is not suitable for commercial vaccine development with the low viral titer production. In this study, we significantly improved its production through construction of a chimeric VEEV-ΔC-CHIKV. Our results proved the utilization of infectious capsidless viral particle of CHIKV as a safe and practical vaccine candidate.
KEYWORDS: Chikungunya virus, alphavirus, capsid, vaccine
INTRODUCTION
Chikungunya virus (CHIKV) is an important re-emerging mosquito-transmitted pathogen within the Alphavirus genus of the Togaviridae family. It generally causes high fever, headache, rashes, myalgia, arthralgia, and occasionally crippling arthritis that may persist for months or even years (1). More severe symptoms, including encephalitis, hemorrhagic disease, and mortality, have also been reported during recent epidemics (2). Recently, perinatal CHIKV infection with severe outcomes has been reported (3–6). Since a large outbreak started in Kenya in 2004, CHIKV has been rapidly spreading throughout Asia, Africa, Europe, and the Americas (7–10), and become a global health threat for public health.
Various CHIKV vaccines have been developed using different strategies, including live-attenuated, inactivated, subunit, DNA, and virus-like particle vaccines (11). Among them, live-attenuated vaccines (LAVs) are considered to be most effective due to their ability to mimic viral natural infection, stimulating a robust and sustained immune response after vaccination (11). Up to now, except the live attenuated CHIKV vaccine derived from attenuated strain CHIK 181/Clone 25 that was halted during clinical evaluation due to insufficient and/or unstable attenuation, only two CHIKV LAVs, including a recombinant measles virus (MV) vector-based CHIKV vaccine (named MV-CHIKV) (12) and a Δ5nsP3 attenuated vaccine with a large deletion of viral nsP3 replicase (named VAL1553) (13), have entered clinical trials in humans. We previously developed a live attenuated CHIKV vaccine with a complete capsid deletion (ΔC-CHIKV) (14); it is infectious in BHK-21 cells and could propagate in culturing cells using glycoproteins (E2-E1) as the only structural proteins (14). Despite satisfactory immunogenic response and safety, two major problems have to be solved before commercial production of ΔC-CHIKV vaccine, including the low yield in cell culture and the replication deficiency in Vero cells, which are the most widely accepted cell line for vaccine production.
In this study, a chimeric VEEV-ΔC-CHIKV was constructed using the Venezuelan equine encephalitis virus (VEEV) replicon as the backbone to express the glycoproteins of CHIKV. Comparing our original ΔC-CHIKV, VEEV-ΔC-CHIKV, yielded viral titers as high as 5 × 106 PFU/mL in Vero cells after extensive serial passages. Further genomic sequencing and reverse genetic analysis revealed that such remarkable enhancement in viral titers may be attributed to increased amount of mature VEEV-ΔC-CHIKV virions conferred by the mutations in glycoproteins during passaging. VEEV-ΔC-CHIKV is also avirulent and immunogenic in mouse models, and a single dose inoculation is enough to protect mice from CHIKV infection.
RESULTS
Construction of chimeric VEEV-ΔC-CHIKV.
As described previously, ΔC-CHIKV could propagat in BHK-21 cells more efficiently. However, when Vero cells were infected with ΔC-CHIKV, no obvious infectious particles were detected by plaque assay (Fig. 1F). To examine whether ΔC-CHIKV released infectious viral particles from Vero cells, we quantified viral RNAs in both the supernatant and cell lysates from Vero cells infected with ΔC-CHIKV (Fig. 1B). Although viral RNAs could be detected within supernatant, most of viral RNAs were maintained within the infected cells. At the same time, we infected Vero cells with increased amounts of ΔC-CHIKV produced from BHK-21 cells, and increased immunofluorescence assay (IFA) positive cells were observed using an anti-E2 polyclonal antibody (Fig. 1C). These results indicated that the viral particles released from Vero cells were not sufficient to initiate a new round of infection. We then attempted to adapt ΔC-CHIKV on Vero cells through multiple independent serial passages, whereas we still failed to recover infectious ΔC-CHIKV virus on Vero cells. Given that VEEV has more efficient virion formation in comparison with other alphaviruses (15, 16), we replaced the backbone of ΔC-CHIKV with VEEV replicon but still retained CHIKV antigen glycoproteins (E3-E2-6K-E1) to generate chimeric VEEV-ΔC-CHIKV (Fig. 1A). Equal amounts of in vitro transcribed VEEV-ΔC-CHIKV and ΔC-CHIKV recombinant RNAs were transfected into BHK-21 cells for IFA using an anti-E2 polyclonal antibody. Similar to ΔC-CHIKV, VEEV-ΔC-CHIKV propagated efficiently in BHK-21 cells, producing increasing numbers of IFA positive cells from 24 h posttransfection (hpt) to 72 hpt (Fig. 1D). VEEV-ΔC-CHIKV and ΔC-CHIKV recovered from transfected BHK-21 cells were designated as P0, and used to passage independently on BHK-21 and Vero cells for three rounds. Each passage was designated as P1, P2. and P3. In contrast to only scattered IFA positive cells observed in P1 ΔC-CHIKV-passaged Vero cells, VEEV-ΔC-CHIKV produced widespread IFA positive cells in each passage of Vero cells (Fig. 1E). The viral growth kinetics of both viruses at P0 were also compared in BHK-21 and Vero cells (Fig. 1F). In BHK-21 cells, a time-dependent increase in viral titers was observed in both VEEV-ΔC-CHIKV and ΔC-CHIKV with peak titers of 105 and 103 PFU/mL at 72 h postinfection (hpi), respectively. In Vero cells, no infectious particles were detected in ΔC-CHIKV infected cells until 72 hpi while VEEV-ΔC-CHIKV generated viral titers of 103 PFU/mL at 72 hpi. The results demonstrated that this chimeric VEEV-ΔC-CHIKV was more efficiently able to propagate in Vero cells.
FIG 1.
Growth characterization of VEEV-ΔC-CHIKV particles in BHK-21 and Vero cells. (A) Schematic illustration of CHIKV-WT, ΔC-CHIKV, and VEEV-ΔC-CHIKV genomes. (B) Viral RNA copies detected both in supernatant and cell lysates of Vero cells infected with ΔC-CHIKV at an MOI of 0.01. (C) Immunostaining of ΔC-CHIKV infected Vero cells with different MOI using CHIKV E2 polyclonal antibodies. (D) Immunostaining of BHK-21 cells transfected with equal amounts of VEEV-ΔC-CHIKV or ΔC-CHIKV RNAs (1 μg) at the indicated times posttransfection with CHIKV E2 polyclonal antibodies. (E) IFA analysis of CHIKV-E2 expression using anti-CHIKV E2 rabbit polyclonal antibodies at different passages of VEEV-ΔC-CHIKV and ΔC-CHIKV viruses in both BHK-21 and Vero cells at 72 h postinfection. (F) Growth curves of the recombinant VEEV-ΔC-CHIKV (P0) and ΔC-CHIKV viruses (P0) as well as WT-CHIKV in both BHK-21 and Vero cells. The recombinant VEEV-ΔC-CHIKV and ΔC-CHIKV viruses obtained from BHK-21 cells were used to infect BHK-21 and Vero cells at an MOI of 0.01, and the cell supernatants were collected at the indicated times for plaque assay using BHK-21 cells. Error bars indicate the standard derivation (SD) of three independent experiments. The dashed line indicated the detection limit.
Extensive passage improves the yield of infectious VEEV-ΔC-CHIKV particles in Vero Cells.
Although VEEV-ΔC-CHIKV could propagate in Vero cells, the maximum viral titer obtained in Vero cells was only 103 PFU/mL at 72 hpi at an MOI of 0.01 (Fig. 1F, P0). To improve the production efficiency of VEEV-ΔC-CHIKV, VEEV-ΔC-CHIKV was subjected to serial passage on Vero cells. The viral growth kinetics were measured every 10 rounds of passage. Surprisingly, the viral titers of VEEV-ΔC-CHIKV gradually increased by around 5,000-fold from 103 PFU/mL (P0) to 5 × 106 PFU/mL (P50) after passages (Fig. 2A), but the plaque morphology and size remained nearly unchanged after 50 rounds of passages (Fig. 2C and D). Since there was no further improvement in viral production efficiency from P30 to P50, the viral passage was stopped at P50.
FIG 2.
Generation of higher titer VEEV-ΔC-CHIKV virus in Vero cells by extensive passaging. (A) Growth curves comparison of different passages (P0, P10, P20, P30, P40, P50) of VEEV-ΔC-CHIKV in Vero cells. Vero cells were infected at an MOI of 0.01, and the cell supernatants were collected at the indicated times for determination of virus titers by plaque assay using BHK-21 cells. The data are representative of three independent experiments, and error bars indicate the SD. (B) IFA analysis of CHIKV-E2 and Capsid in WT or the passaged VEEV-ΔC-CHIKV infected Vero cells. (C) Plaque morphology comparison between CHIKV-WT and VEEV-ΔC-CHIKV at P0, P30, and P50. BHK-21 cells were infected with indicated viruses, and plaques were developed after 72 h. (D) Plaque diameter comparison of VEEV-ΔC-CHIKV virus at P0, P30, and P50 in BHK-21 cells at 72 hpi. ns, not significant. (E) Immunogenicity comparison between CHIKV-WT and P50 passaged VEEV-ΔC-CHIKV viruses. Two independent experiments were performed in triplicate. Data represent the mean ± SD of triplicate measurements in a representative experiment. The asterisks denote statistical differences between the indicated groups. ns, not significant. (F) Infectivity of VEEV-ΔC-CHIKV in Huh-7 (hepatoma), A549 (lung adenocarcinoma), and MRC-5 (lung fibroblast cell line) cell lines. The above all cell lines were infected with VEEV-ΔC-CHIKV at an MOI of 1. At 36 h postinfection, CHIKV-E2 expression was detected using anti-CHIKV E2 polyclonal antibodies.
To confirm whether the passaged VEEV-ΔC-CHIKV still retained capsid deletion and antigenicity similar to wild-type (WT) viral particles, IFAs and neutralization assay were carried out with passaged VEEV-ΔC-CHIKV, respectively. In comparison with WT CHIKV, consistent E2-positive and capsid-negative IFA results (Fig. 2B) were observed in both P0 and the passaged VEEV-ΔC-CHIKV (P30 and P50). Neutralization assay showed that the sera against WT CHIKV could efficiently block viral infection of VEEV-ΔC-CHIKV-P50 in a dose-dependent manner as WT virus (Fig. 2E). These results indicated that, accompanied with an increase in viral yield during viral passage, there was no change for either capsid deletion or virion antigenicity.
In addition, we explored the infectivity of passaged VEEV-ΔC-CHIKV in different human cell lines. We selected Huh-7 (hepatoma), A549 (lung adenocarcinoma), SKOV3 (ovarian tumor), A375 (melanoma), and L-02 (hepatocyte) as well as MRC-5 (human embryonic lung fibroblasts) cell lines to test if they are permissive for VEEV-ΔC-CHIKV infection at higher MOI of 1. At 36 hpi, viral E2 proteins were detected by IFA. We found that only Huh-7, A549, and MRC-5 are permissive for VEEV-ΔC-CHIKV infection with different efficiency (Fig. 2F) which indicated that VEEV-ΔC-CHIKV could not propagate efficiently within human cells.
Adapted mutations accumulated during passaging account for the increase in VEEV-ΔC-CHIKV titers by improving the efficiency of viral assembly.
In our previous study, it was demonstrated that ΔC-CHIKV produced large amounts of empty particles with higher ratio of total viral particles to infectious particles compared with WT CHIKV (14). Here, we speculate that the enhancement of viral titers of the passaged VEEV-ΔC-CHIKV may result from increased viral assembly efficiency. Supporting the speculation, it was found that P50 VEEV-ΔC-CHIKV had much lower particle-to-PFU ratio than P0 VEEV-ΔC-CHIKV as 104 PFU contained similar amounts of envelope proteins to those of 10 PFU P0 VEEV-ΔC-CHIKV (Fig. 3A). To further confirm our speculation, VEEV-ΔC-CHIKV from P0 and P50 were collected for sucrose gradient purification as previously described (14). Each fraction from top to bottom was reclaimed (Fraction 1 to 16) and subjected to WB assay for quantification of viral proteins. It has been demonstrated that top fractions contain empty particles, in contrast to middle fractions with infectious particles (14). As shown in Fig. 3B and C, infectious particles and empty particles resided in Fraction 7–11 and Fraction 1–3, respectively. As expected, most P50 VEEV-ΔC-CHIKV were centered in Fraction 7–11 in contrast to the smear-like distribution of P0 particles in Fraction 1–13, confirming that the increase in viral assembly efficiency is responsible for higher viral titers of the passaged VEEV-ΔC-CHIKV.
FIG 3.
The passaged VEEV-ΔC-CHIKV produces more infectious virus particles. (A) Western blotting of CHIKV E1 expression of different viral loads of CHIKV-WT and VEEV-ΔC-CHIKV at P0, P10, and P50 using CHIKV E1 polyclonal antibodies. (B) Western blotting of sucrose density gradient fractionations of VEEV-ΔC-CHIKV at P0 and P50 produced in Vero cells. 105 PFU P0 and 107 PFU P50 viruses were separated on 20%–60% linear sucrose density gradients. Sixteen fractions were harvested from the top (Fraction 1) to the bottom (Fraction 16) of the gradient. Each fraction from P0 and P50 VEEV-CHIKV was subjected to Western blotting assay using E1-specific antibody. (C) Analysis of the intensity of viral E1 protein bands in each fraction using ImageJ software.
Increased virus production capabilities indicate some genetic changes occurring during VEEV-ΔC-CHIKV passage. The whole genome of P50 VEEV-ΔC-CHIKV was sequenced, and 14 nucleotide mutations were identified besides the original engineered capsid deletion (Fig. 4A). Specifically, these 14 nucleotide mutations resulted in three silent mutations in nonstructural protein 1 (nsP1), six amino acid changes in the other three nonstructural proteins (nsP2, nsP3, and nsP4), and five amino acid substitutions in envelope proteins (Fig. 4A). All mutations within nsPs and envelope proteins were designated as nsPsmut and Emut, respectively, and their effects on the yield of infectious VEEV-ΔC-CHIKV particles were examined by reverse genetic analysis. DNA fragments containing nsPsmut or Emut were generated through reverse transcription PCR (RT-PCR) using the extracted genomic RNA of P50 VEEV-ΔC-CHIKV, and were used to replace the corresponding fragments in the cDNAs clone of VEEV-ΔC-CHIKV. Recombinant viruses were rescued from BHK-21 cells transfected with in vitro transcribed RNAs containing nsPsmut, Emut, or (nsPs+E)mut (Fig. 4B) and used to infect Vero cells at an MOI of 0.01. At different time points, the supernatants were collected for plaque assay. As shown in Fig. 4D, Emut mutations dramatically improved viral productions of VEEV-ΔC-CHIKV compared with nsPsmut mutations, and the combination of Emut and nsPsmut ((nsPs+E)mut) further increased viral titer to a level almost equivalent to that of P50 VEEV-ΔC-CHIKV. Besides, plaque morphologies of nsPsmut and Emut as well as (nsPs+E)mut viruses were all similar to P50 virus (Fig. 4C), which was in line with the results that increase of viral titers after passaging mainly results from increased viral assembly efficiency, as explored in Fig. 3. Overall, the above results demonstrated that the mutations within both structural and nonstructural proteins, especially the structural protein mutations, are responsible for the improvement of viral production of P50 VEEV-ΔC-CHIKV.
FIG 4.
Reverse genetic analysis of the adaptive mutations in P50 VEEV-ΔC-CHIKV. (A) Nucleotide and amino acid changes in the P50 VEEV-ΔC-CHIKV genome. (B) Diagram illustration of reconstruction of VEEV-ΔC-CHIKV mutants with the mutations in either structural proteins (nsPsmut), structural proteins (Emut) or both ((nsPs+E)mut). The titers at 72 hpt in Vero cells for each construct are given on the corresponding line. (C) Plaque morphology comparison among different recombinant viruses. (D) Growth curves of the recovered constructs. Vero cells were infected with the indicated viruses at an MOI of 0.01 and the cell supernatants were collected at the indicated times for plaque assay in BHK-21 cells. The data are representative of two independent experiments, and error bars indicate the SD.
To further explore the effective effect of structural protein mutations on viral production, we examined the function of Emut mutations in the background of ΔC-CHIKV instead of VEEV-ΔC-CHIKV. The Emut mutations were introduced into ΔC-CHIKV-eGFP constructs described in a previous study (17). Equal amounts (1 μg) of in vitro transcribed RNA of ΔC-CHIKV-eGFP and ΔC-CHIKV-Emut-eGFP were transfected into Vero cells, respectively. The results showed that ΔC-CHIKV-Emut-eGFP RNA produced increasing the number of eGFP positive cells from 48 to 120 h posttransfection (hpt), with much higher positive rates than ΔC-CHIKV-eGFP RNA at each time point, indicating that Emut mutations could also enhance the viral production of ΔC-CHIKV in Vero cells (Fig. 5A).
FIG 5.
The whole glycoprotein mutations instead of E2 G82R mutation alone account for the enhanced viral titer. (A) Function of Emut mutations in the background of ΔC-CHIKV. Schematic illustration of ΔC-CHIKV-eGFP and ΔC-CHIKV-Emut-eGFP constructs were presented in the top panel. Equal amounts (1 μg) of in vitro transcribed RNA of ΔC-CHIKV-eGFP and ΔC-CHIKV-Emut-eGFP were transfected into Vero cells, respectively. At indicated time points after transfection, eGFP positive cells were detected using a NIKON upright fluorescence microscope (Tokyo, Japan), as shown in the bottom panel. (B) Effect of E2 G82R mutation on the viral titer. The E2 G82R mutation was engineered into the original VEEV-ΔC-CHIKV constructs, as shown in the top panel. Equal amounts (1 μg) of in vitro transcribed RNAs of VEEV-ΔC-CHIKV and VEEV-ΔC-CHIKV-E2 G82R were transfected into Vero cells, respectively. The expressions of viral E2 protein were detected by IFA at indicated time points posttransfection, as shown in the bottom panel. (C) Viral production of VEEV-ΔC-CHIKV and the E2-G82R mutant. The genomes/mL data and vital titers were determined by triplicate qPCRs and standard plaque assay, respectively. Three independent experiments were performed and the average values are shown.
It is notable that adapted mutation at position 82 of the E2 protein (E2-G82R) is positively charged, which was found to enhance cell binding efficiency through heparin binding and contribute to the efficiency of viral production with a decreased viral genome/PFU ratio (18). Therefore, we specifically tested the role of the adapted mutation E2-G82R on viral yield. E2-G82R mutation was introduced into the original VEEV-ΔC-CHIKV constructs. Equal amounts (1 μg) of VEEV-ΔC-CHIKV and VEEV-ΔC-CHIKV-G82R RNA transcribed in vitro were transfected into Vero cells, respectively, and expression of viral E2 protein at indicated time points posttransfection were detected by IFA. Although similar amounts of IFA positive cells were observed at each time point posttransfection (Fig. 5B), we found that VEEV-ΔC-CHIKV-G82R (3 × 103 PFU/mL) produced higher viral titer than VEEV-ΔC-CHIKV (103 PFU/mL). Furthermore, we directly examined the viral genomes and infectious virus particles of the two viruses to calculate the genome/PFU ratio. VEEV-ΔC-CHIKV- G82R had a genome/PFU ratio of 625, which was 2-fold lower than VEEV-ΔC-CHIKV with a genome/PFU ratio of 1328. In agreement with the previous study, our data demonstrated that the G82R mutation in E2 could improve viral production with a decreased genome/PFU ratio (18). However, E2-G82R alone could not fully recover VEEV-ΔC-CHIKV production efficiency, and additional underlying mechanisms needs to be explored in our future studies.
The passaged VEEV-ΔC-CHIKV is highly attenuated in C57BL/6 mice.
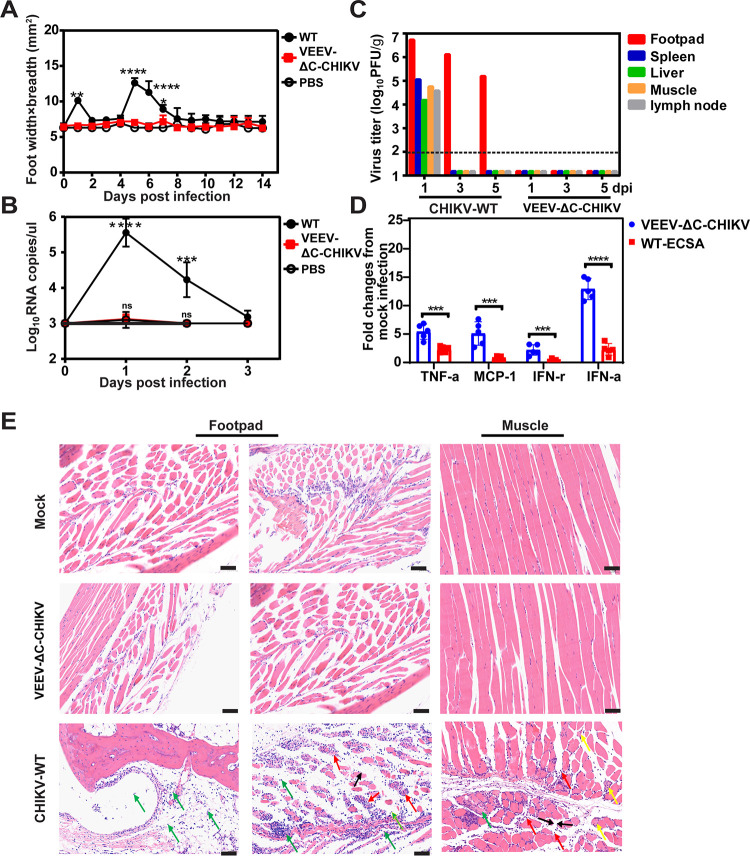
An adult wild-type mouse model of CHIKV arthritis has been established with C57BL/6 mice (19), which recapitulates symptoms of CHIKV infection of humans with clear signs of self-limiting foot swelling and histological evidence of acute and persistent arthritis, tenosynovitis, and myositis (19). Thus, 6-week-old C57BL/6 mice were infected subcutaneously (s.c.) in the hind foot with 105 PFU of WT CHIKV and P50 VEEV-ΔC-CHIKV, respectively. The signs of footpad swelling and the induced viremia were monitored. Similar to mock-infected mice, no footpad swelling and viremia were observed in mice inoculated with P50 VEEV-ΔC-CHIKV (Fig. 6A and B). At the same time, no viruses were detected in different tissues of P50 VEEV-ΔC-CHIKV infected mice (Fig. 6C). In contrast, the mice infected with WT CHIKV developed foot swelling with two peaks at approximately 1 and 5 days postinfection (Fig. 6A), and viremia (Fig. 6B). High virus loads were also detected in the feet, muscle, spleen, liver, and lymph nodes of WT CHIKV infected mice on day 1 postinfection (Fig. 6C).
FIG 6.
Pathogenicity of VEEV-ΔC-CHIKV in the C57BL/6 mouse model. C57BL/6 mice (6-week-olds, n = 5 per group) were injected s.c. in the ventral/lateral side of the hind foot with 105 PFU of WT CHIKV (ECSA strain), VEEV-ΔC-CHIKV, and the same volume of PBS (negative control), respectively. (A) Footpad swelling. (B) Viremia. (C) Viral distribution in organs of infected mice. Organs from infected mice were collected and homogenized on days 1, 3, and 5 postinfection. The amounts of viruses were quantified using plaque assay. (D) Expression of cytokine or chemokine in ankle tissue of mice infected with both viruses were analyzed using qRT-PCR. Results were normalized to the level of the housekeeping geneβ-actin and expressed as fold changes compared to the levels in the mock control samples. Student's t test was used for statistical analysis: ***, P < 0.001; ****, P < 0.0001. (E) Histology of feet and muscle from WT CHIKV (ECSA strain), VEEV-ΔC-CHIKV, or PBS infected C57BL/6 mice. Scale bar represents 500 μm for footpad left panel, and 50 μm for footpad right panel and muscle panel. The row labeled “footpad” shows no inflammation in the observed ankle joint of VEEV-ΔC-CHIKV and mock-infected mice, while WT CHIKV (ECSA strain) infected mice exhibited muscle edema (black arrow) and necrosis of numerous muscle cells (red arrow) as well as large amounts of inflammatory cell infiltration in the muscle, synovial membrane, and articular cavity (green arrow). In infected muscle analysis, there was no obvious inflammation or necrosis with VEEV-ΔC-CHIKV and mock-infected mice, whereas a large amount of inflammation (yellow arrow) and necrosis (red arrow) was detected in WT CHIKV infected mice. Also, some fibrous connective tissue hyperplasia was seen (green arrow).
To further investigate the attenuation effect of VEEV-ΔC-CHIKV in mice, the footpads were collected at the peak of swelling (5 dpi) for histological analysis (Fig. 6E). Consistently, there was no inflammation observed in the quadriceps muscle or ankle joint of VEEV-ΔC-CHIKV and mock-infected mice, while WT CHIKV infected mice demonstrated extensive inflammation and myositis in muscle and around the ankle joint. Previous studies have demonstrated that capsid is one of the important virulence determinants of alphavirus (20–23) and necessary for inhibition of type-I interferon (IFN) gene expression (20). Additionally, the type-I IFN response also plays a key role in controlling CHIKV infection, and capsid is sufficient to antagonize type-I IFN responses by degrading cGAS (24). To determine whether different IFN responses could be observed between VEEV-ΔC-CHIKV and WT-CHIKV at early time points post viral infection, we quantified IFN-α mRNA levels at 3 hpi. Consistent with capsid deletion, high levels IFN-α were detected in the footpads of mice infected with VEEV-ΔC-CHIKV in contrast to WT-CHIKV (Fig. 6D). Additionally, higher monocyte chemoattractant protein-1 (MCP-1), interferon gamma (IFN-γ), and tumor necrosis factor alpha (TNF-α) were also observed at 3 hpi in the footpad of mice infected with VEEV-ΔC-CHIKV relative to WT-CHIKV. Abortion of type-I IFN inhibition at early viral infection provides an alternative explanation for attenuation of VEEV-ΔC-CHIKV with capsid deletion. Collectively, these data demonstrate that VEEV-ΔC-CHIKV is highly attenuated in C57BL/6 mice.
A single dose of VEEV-ΔC-CHIKV can provide strong protection against CHIKV infection in C57BL/6 mice.
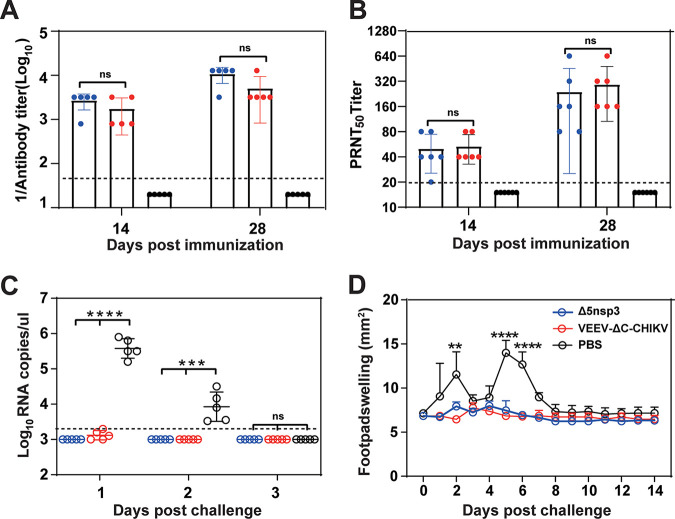
To evaluate the immunogenicity and protection efficacy of P50 VEEV-ΔC-CHIKV, 4-week-old C57BL/6 mice were inoculated with 104 PFU of VEEV-ΔC-CHIKV or Δ5nsp3 vaccine candidate through the s.c. route. Δ5nsp3 was constructed with a large deletion of 62 amino acids in the nsP3 gene. The sequence of the missing nucleotides ranges from amino acid residues 1656 to 1717 of the P1234 polyprotein (25). The negative controls were inoculated with phosphate-buffered saline (PBS) alone. Apparent seroconversion was observed at 14 and 28 days postinfection (Fig. 7A). Comparable neutralizing antibody titers of 1:160–1:320 were observed among mice infected with Δ5nsp3 and VEEV-ΔC-CHIKV (Fig. 7B), although higher IgG antibodies were induced in mice inoculated with Δ5nsp3 (1:3,200–1:12,800) than those of VEEV-ΔC-CHIKV (1:3,200) (Fig. 7A), which indicated that VEEV-ΔC-CHIKV induces neutralizing antibody as efficiently as Δ5nsp3 in the C57BL/6 mouse model. At 30 days postimmunization, all the mice were challenged with 2.5 × 105 PFU of WT CHIKV (ECSA strain) (26). The mice from the Δ5nsp3 and VEEV-ΔC-CHIKV immunized groups were completely protected from challenge without any signs of illness such as footpad swelling or viremia, in contrast with PBS-immunized mice (Fig. 7C and D). Collectively, these data demonstrated that VEEV-ΔC-CHIKV is immunogenic and a single dose immunization could protect mice from CHIKV infection.
FIG 7.
VEEV-ΔC-CHIKV protects the C57BL/6 mouse from CHIKV infection. Four groups of 4-weeks-old C57BL/6 mice (n = 5 per group) were immunized s.c. in the ventral/lateral side of the hind foot once with either 104 PFU of Δ5nsP3 or VEEV-ΔC-CHIKV; PBS was used as mock immunization. On day 30 postimmunization, mice were challenged s.c. in feet with 2.5 × 105 PFU of WT CHIKV (ECSA strain). (A) Total anti-CHIKV IgG antibodies and (B) NAb titers in mouse sera at the indicated days postimmunization. (C) Viremia and (D) footpad swelling postchallenge. All the data represent mean ± SD of group mice, and the horizontal dotted line represents the limit of detection. Student's t test or two-way ANOVA was used to determine statistical differences between groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, respectively.
VEEV-ΔC-CHIKV is safe as a CHIKV vaccine candidate.
The IFNAR−/− mouse lacks functional type I interferon receptors and is a lethal murine model with apparent diseases for CHIKV pathogenesis study (27, 28). Here, we used it to assess the safety of VEEV-ΔC-CHIKV vaccine candidate. Six-week-old IFNAR−/− mice were infected subcutaneously (s.c.) in the hind footpad with different doses of VEEV-ΔC-CHIKV (104, 105, and 106 PFU) and 102 PFU of WT CHIKV, respectively, and were monitored for illness. The mice infected with VEEV-ΔC-CHIKV all survived (Fig. 8A) without any signs of illness with an LD50 more than 106 PFU. In contrast, the mice infected with WT CHIKV (102 PFU) all died within 8 days postinfection (Fig. 8A) with footpad swelling (Fig. 8B), weight loss (Fig. 8C), and apparent viremia (Fig. 8D). Overall, these data suggested that VEEV-ΔC-CHIKV is a safe live vaccine with high attenuation even in IFNAR−/− mice.
FIG 8.
Safety evaluation of VEEV-ΔC-CHIKV in the IFNAR−/− mouse model. Four groups of 6-weeks-old IFNAR−/− mice (n = 5 per group) were infected s.c. in the ventral/lateral side of the hind foot with different doses of WT CHIKV or VEEV-ΔC-CHIKV. The PBS infection was used as mock immunization. Animal survival (A), footpad swelling (B), and weight loss (C) as well as viremia (D) were monitored daily until 14 days postinoculation. Student's t test or two-way ANOVA was used to determine statistical differences between groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, respectively.
DISCUSSION
In our previous study, we provided a proof of concept that the infectious capsidless CHIKV viral particles (ΔC-CHIKV) can be used as a new type of live attenuated vaccine (14). ΔC-CHIKV itself can propagate in BHK-21 cells using envelopes as the single structural proteins (14), which is different from traditional viral-like replicon particle (VRP) vaccines produced through a complementation/packaging system with structural proteins expressed in trans for packaging (29). In this study, we achieved high titer ΔC-CHIKV in vaccine production cell line Vero through construction of chimeric VEEV-ΔC-CHIKV in which the VEEV replicon was used as the backbone to express CHIKV envelope proteins. The robust viral production in Vero cells after extensive passage, high attenuation in the IFNAR−/− mice model, and complete protection from CHIKV challenge with a single dose immunization highlights the feasibility of such vaccine strategy in the development of safe and effective vaccines against alphavirus infection.
Different from ΔC-CHIKV, VEEV-ΔC-CHIKV was not only able to propagate in Vero cells upon entry (Fig. 1C), but yielded a much higher titer (>106 PFU/mL) after extensive passage (Fig. 2A). Although what causes such discrepancies, or in other words, what the underlying mechanism of the formation of these infectious capsidless VEEV-ΔC-CHIKV or ΔC-CHIKV particles is, remains unclear, two lines of evidence from our studies indicate that both host and viral factors contribute to this: (i) the different abilities of either ΔC-CHIKV or the parental VEEV-ΔC-CHIKV to propagate in BHK-21 and Vero cells observed in our studies (Fig. 1C) imply that some BHK-21 cell-specific host factors may assist their efficient propagation in host cells; (ii) the mutations in VEEV nsPs and CHIKV envelope proteins accumulated during passaging greatly improved viral production (Fig. 4), providing the straightforward evidence from viral factors. Additionally, the event that P0 VEEV-ΔC-CHIKV was able to propagate in Vero cells, a condition lacking BHK-21 host factors, also indicates that some specific viral factors in VEEV-ΔC-CHIKV could compensate for the functions of BHK-21-specific cellular factors. The underlying mechanism is worth exploring in future studies.
Despite utilizing a different replicon backbone, VEEV-ΔC-CHIKV displayed similar antigenicity to WT CHIKV (Fig. 2E), and still retained capsid deletion even after 50 rounds of passage in Vero cells (Fig. 2B). Likewise, it elicited robust immune responses to confer complete protection of C57BL/6 mice against WT CHIKV challenge (Fig. 7), and was highly attenuated in both immunocompetent and immunocompromised mice (Fig. 6 and 8). It should be noted that the VEEV replacement in nsPs and 5′- and 3′-untranslated regions did not lead to additional visible virulence in mice.
In summary, developing infectious capsidless alphavirus vaccine candidate is a practical strategy to combat against alphavirus infections, and it is worthwhile to further test the VEEV-ΔC-CHIKV vaccine candidate in nonhuman primates to evaluate its suitability for human usage.
MATERIALS AND METHODS
Ethics statement.
C57BL/6 mice and immunocompromised IFNAR−/− mice were provided by the Animal Centre of Wuhan Institute of Virology. All the mice were housed in compliance with the recommendations of National Institutes of Health Guidelines for the Care and Use of Experimental Animals. All animal experiments were conducted in an animal biosafety level 3 (ABSL-3) facility at Wuhan Institute of Virology under a protocol approved by the Laboratory Animal Ethics Committee of the Wuhan Institute of Virology, Chinese Academy of Sciences (permit number: WIVA26201701).
Cell lines, viruses, and antibodies.
Vero cells and BHK-21 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Germany) containing 10% fetal bovine serum (FBS), 100 U/mL of penicillin, and 100 μg/mL of streptomycin at 37°C with 5% CO2.
The WT CHIKV and ΔC-CHIKV were recovered from corresponding infectious cDNA clones using Vero cells or BHK-21 cells as described previously (14). CHIKV ECSA strains were isolated from human serum during the Pakistan outbreak of 2016–2017 and propagated on Vero cells (26).
Murine polyclonal antiserum against CHIKV capsid proteins and anti-CHIKV E2 rabbit polyclonal antibodies was generated in-house. Fluorescein isothiocyanate (FITC)/horseradish peroxidase (HRP)-conjugated goat anti-mouse/rabbit IgG was purchased from Proteintech (China).
Construction of VEEV-ΔC-CHIKV and its variants.
The infectious clone of the VEEV strain TC83, designated as pACYC-VEEV-TC83, which carries an AscI restriction site downstream of the subgenomic promoter and a PacI restriction site upstream of the 3’UTR, was used to construct the replicon capable of expressing CHIKV envelop proteins. The sequence of CHIKV envelop proteins was amplified using WT CHIKV infectious clone as the template and cloned into pACYC-VEEV-TC83 at AscI and PacI restriction sites to replace the structural genes of VEEV, generating the pACYC-VEEV-ΔC-CHIKV plasmid. The resulting plasmid pACYC-VEEV-ΔC-CHIKV was verified by DNA sequencing. For the construction of VEEV-ΔC-CHIKV variants containing different mutations in nonstructural or/and structural proteins (namely, nsPsmut, Emut, or (nsPs+E)mut mutants), the extracted P50 VEEV-ΔC-CHIKV RNA was subjected to RT-PCR to generate the following fragments spanning the restriction sites: RsrII-BglII, Bsu36I-AscI, and AscI-PacI. These fragments containing the P50 VEEV-ΔC-CHIKV mutations were then used to replace the corresponding fragments in pACYC-VEEV-ΔC-CHIKV to generate the nsPsmut, Emut, and (nsPs+E)mut variants, respectively.
RNA transcription and transfection.
The VEEV-ΔC-CHIKV/variants cDNA plasmids were subjected to sequential NotI linearization and in vitro transcription using mMESSENGER mMACHINE T7 Kit (Ambion, USA) following the manufacturer’s protocols. The recombinant viral RNAs were then transfected into Vero or BHK-21 cells with DMRIE-C (Invitrogen, USA) according to the manufacturer’s instructions. At indicated time points posttransfection, the supernatants containing viruses were collected, aliquoted, and stored at −80°C for the following experiments.
Plaque assay.
BHK-21 cells were seeded into 24-well plates at a density of 1 × 105 cells per well 1 day before plaque assay. A series of 1:10 dilutions were made by mixing 15 μL of virus sample with 135 μL of DMEM. Then, 100 μL of each dilution was added to individual well of 24-well plates containing confluent BHK-21 cells. The plates were incubated at 37°C with 5% CO2 for 1 h before the layer of 2% methyl cellulose was added. After 3 days of incubation at 37°C with 5% CO2, the cells were fixed with 3.7% formaldehyde and then stained with 1% crystal violet. Plaque morphology and numbers were recorded after washing the plates with tap water.
Indirect immunofluorescence assay (IFA).
Vero or BHK-21 cells were seeded into six-well plates containing coverslips and transfected with viral genomic RNA or infected with the indicated viruses. At the indicated time points, the coverslips containing transfected or infected cells were collected, washed with PBS, and fixed with cold (−20°C) 5% acetic acid in acetone for 15 min at room temperature. After washing with PBS three times, the fixed cells were incubated with the primary antibodies diluted in PBS (mice anti-CHIKV E2 protein polyclonal antibody, 1:250, made in-house) for 1 h. The cells were washed three times with PBS and then incubated with goat anti-mouse IgG antibodies conjugated with FITC (fluorescein isothiocyanate; 1:125 dilution in PBS, Proteintech, China) at room temperature for another hour. The cells on the coverslips were mounted with 90% glycerol and examined under a fluorescence microscope. The fluorescent images were taken at 200× magnification with a NIKON upright fluorescence microscope (Tokyo, Japan).
Purification of VEEV-ΔC-CHIKV virions.
Vero cells were infected with VEEV-ΔC-CHIKV-P0 or VEEV-ΔC-CHIKV-P50 at an MOI of 0.01. At 120 hpi (VEEV-ΔC-CHIKV-P0) or 48 hpi (VEEV-ΔC-CHIKV-P50), the supernatants were reclaimed through sequential centrifugation at 400 g for 10 min and 5,000 g for 20 min at 4°C and the clarified supernatants were concentrated through polyethylene glycol-8000 (PEG 8000) precipitation at a final concentration of 8% at 4°C overnight. Following centrifugation at 14,000 g for 1 h at 4°C, the pellet was gently resuspended in PBS. Then, the suspension was overlaid on a 20% to 60% linear sucrose gradient in PBS, and subjected to ultracentrifugation at 34,000 rpm for 3 h at 4°C using an MLS-50 rotor in an Optima MAX-XP ultracentrifuge (Beckman, USA). The virus fraction was recovered from the gradient. Sixteen fractions of 500 μL volume were collected from the top to the bottom and analyzed by Western blotting assay.
Western blotting.
Purified VEEV-ΔC-CHIKV were boiled at 95CHIKVsis min, followed by separation by 10% SDS-PAGE and then electro-transferred onto a polyvinylidene fluoride (PVDF) membrane (0.2 μm; Millipore). The membranes were blocked with 5% skim milk in TBST (50 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 20, pH 7.4) for 1 h at room temperature. The blocked membranes were then incubated with the primary antibodies (Mice anti-CHIKV E1 protein polyclonal antibody, 1:2000, made in-house) at room temperature for another 1 h. After being washed three times with TBST, the membranes were incubated with horseradish peroxidase (HRP) conjugated secondary goat antimouse or goat antirabbit antibodies (Proteintech, China) at room temperature for 1 h. Following three washes with TBST, the protein bands were visualized with a chemiluminescent HRP-conjugated antibody detection reagent (Millipore, USA).
Viral RNA extraction and viral genome sequencing.
Viral RNA was extracted from P50 VEEV-ΔC-CHIKV infected cells using TRIzol reagent (TaKaRa, China) following the manufacturer's protocol. The extracted RNA was used to perform four one-step RT-PCRs by using a series of overlapping primers covering the complete viral genome of VEEV-ΔC-CHIKV. The RT-PCR products were purified with gel extraction kit and subjected to DNA sequencing for viral genome alignment. Primers for RT-PCRs were as follows: VEEV-5′UTR-F: 5′-ATG GGC GGC GCA TGA GAG AAG C-3′, VEEV-2600-R: 5′-AGT CAC AGA TTT AGT GCA ACG G-3′; VEEV-2401-F: 5′-TTG ACG AAG CTT TTG CTT GTC-3′, VEEV-5400-R: 5′-GCG CCA GAA ACT CCA TAC TC-3′; VEEV-5201-F: 5′-CCA CCA GGT GCT GCA AGT CG-3′, CHIKV-E2-T12I-R: 5′-GCT AAG TAT GGT CTT ATG GCT TTA TAG AC-3′.
Enzyme linked immunosorbent assay (ELISA).
Indirect ELISA was used to detect the CHIKV-specific IgG antibody titer of serum samples collected from immunized mice. The 96-well microtiter plates were coated with inactivated WT CHIKV at 2–8°C overnight, and blocked with 5% skim milk for 1 h at room temperature. Diluted sera were added to each well and incubated for 2 h at 37°C. After 3 times in a PBS wash, the plates were incubated with goat antimouse antibodies conjugated with HRP for another 1 h at 37°C. The plate was developed using TMB, following the addition of 1 M H2SO4 to stop the reaction, and read at 450 nm by ELISA plate reader for final data.
Plaque reduction neutralization test (PRNT).
The neutralizing activities of serum samples from immunized mice were analyzed by plaque reduction neutralization test (PRNT) as described previously (14). Briefly, approximately 100 PFU of WT CHIKV was preincubated with 2-fold serial dilutions of heat-inactivated mouse sera (starting at 1:10 dilution) for 1 h at 37°C and then the mixture was added to BHK-21 cell monolayers in 12-well plates and removed after 1 h of incubation followed by plaque assay to quantify viral titers. Neutralizing antibody titers (PRNT50) were determined to be the highest serial dilutions for which the virus plaque count was reduced by 50% compared with control.
Mouse experiments.
The pathogenicity of VEEV-ΔC-CHIKV was evaluated in C57BL/6 mice. Cohorts of 6-week-old female C57BL/6 mice were infected subcutaneously (s.c.) in the ventral/lateral side of the hind foot with 105 PFU of WT CHIKV (ECSA strain) or VEEV-ΔC-CHIKV. The animals were monitored for footpad welling and viremia. Footpad swelling induced by WT CHIKV (ECSA strain) or VEEV-ΔC-CHIKV was assessed by measuring the height and width of the peri-metatarsal area of the hind foot using Kincrome digital vernier calipers. At 1, 3, and 5 days postinfection (dpi), tissues of mice infected with WT CHIKV (ECSA strain) or VEEV-ΔC-CHIKV virus including footpad, liver, spleen, muscle, and lymph nodes were harvest for viral examination by plaque assay, and the feet taken at 5 dpi were analyzed for cytokine or chemokine mRNA level by qRT-PCR assay.
For immunization and challenge experiments, 4-week-old female C57BL/6 mice (n = 5) were immunized with 104 PFU of VEEV-ΔC-CHIKV or Δ5nsp3 s.c. in the ventral/lateral side of the hind foot. The same volume of PBS was inoculated as negative control. Sera from mice at 14 and 28 days postimmunization were collected for determination of CHIKV-specific IgG antibody titers and neutralizing antibody titers. On day 30 postimmunization, all the mice were challenged with 2.5 × 105 PFU of WT CHIKV (ECSA strain) s.c. in the ventral/lateral side of the hind foot. Footpad swelling and viremia of mice were monitored.
For safety evaluation of VEEV-ΔC-CHIKV, 6-week-old IFNAR−/− mice (n = 5) were infected s.c in the ventral/lateral side of the hind foot with 102 PFU of WT CHIKV and 104 PFU and 106 PFU of VEEV-ΔC-CHIKV, respectively. The animals were monitored for survival, body weight changes and viremia. Footpad swelling induced by WT CHIKV or VEEV-ΔC-CHIKV was assessed by measuring the height and width of the peri-metatarsal area of the hind foot using Kincrome digital vernier calipers.
RNA extraction and quantification.
To measure viremia, about 50 μL volume of blood was mixed with 1 mL TRIzol reagent followed by RNA extraction according to the manufacturer’s instructions. Viral loads in blood were quantified by qRT–PCR as previously described (26). For the analysis of cytokine or chemokine mRNA levels in infected mouse feet, the feet were cut lengthways and homogenized with DMEM. RNA was extracted using TRIzol reagent followed by SYBR green based one-step real-time RT-PCR assay. Primers: 5′-TTA AAA ACC TGG ATC GGA ACC AA-3′ and 5′-GCA TTA GCT TCA GAT TTA CGG GT-3′ for monocyte chemoattractant protein 1 (MCP-1); 5′-AAT TCG AGT GAC AAG CCT GTA GC-3′ and 5′-AGT AGA CAA GGT ACA ACC CAT CG-3′ for tumor necrosis factor alpha (TNF-α); 5′-ATG AAC GCT ACA CAC TGC ATC-3′ and 5′-CCA TCC TTT TGC CAG TTC CTC-3′ for IFN-γ; 5′-GCT CAA GCC ATC CTT GTG CTA A-3′ and 5′-CAT TGA GCT GAT GGA GGT C-3′ for IFN-α. Data were normalized to the β-actin housekeeping gene, and the fold change in mRNA expression relative to mock-infected PBS-treated samples for each gene was calculated using the threshold cycle (ΔΔCT) method: ΔΔCT = ΔCT of virus infected − ΔCT of mock infected, where ΔCT = CT of gene of interest − CT of housekeeping gene. The fold change for each gene is calculated as 2−ΔΔCT.
Histological analysis.
At 5 dpi, the feet from mice infected with 105 PFU of WT CHIKV or VEEV-ΔC-CHIKV were fixed with 4% paraformaldehyde, embedded in paraffin, sagittal sectioned at 4-μm thickness on a microtome, mounted on APS-coated slides followed by hematoxylin and eosin (H&E) stain for pathological analysis.
Statistical analyses.
All data were analyzed using GraphPadPrism 8.0.2 software and expressed as mean ± standard deviation (SD) unless specified otherwise. Kaplan-Meier tests were used for survival analysis, Student's t test was used to determine differences between two groups, and one-way ANOVA or two-way ANOVA tests were utilized for statistical analyses among multiple groups.
ACKNOWLEDGMENTS
We are grateful to the Center for Animal Experiment staff (Xue-fang An, Fan Zhang, He Zhao, and Li Li) and the BSL-3 laboratory (Hao Tang and Jun Liu) at Wuhan Institute of Virology and Wuhan Key Laboratory of Special Pathogens and Biosafety for their helpful supports during the course of the work.
This work was supported by the National Key Research and Development Program of China (2018YFA0507201). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Han-Qing Ye, Email: yehq@wh.iov.cn.
Bo Zhang, Email: zhangbo@wh.iov.cn.
Mark T. Heise, University of North Carolina at Chapel Hill
REFERENCES
- 1.Schwartz O, Albert ML. 2010. Biology and pathogenesis of Chikungunya virus. Nat Rev Microbiol 8:491–500. 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- 2.Das T, Jaffar-Bandjee MC, Hoarau JJ, Krejbich Trotot P, Denizot M, Lee-Pat-Yuen G, Sahoo R, Guiraud P, Ramful D, Robin S, Alessandri JL, Gauzere BA, Gasque P. 2010. Chikungunya fever: CNS infection and pathologies of a re-emerging arbovirus. Prog Neurobiol 91:121–129. 10.1016/j.pneurobio.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Gerardin P, Samperiz S, Ramful D, Boumahni B, Bintner M, Alessandri JL, Carbonnier M, Tiran-Rajaoefera I, Beullier G, Boya I, Noormahomed T, Okoi J, Rollot O, Cotte L, Jaffar-Bandjee MC, Michault A, Favier F, Kaminski M, Fourmaintraux A, Fritel X. 2014. Neurocognitive outcome of children exposed to perinatal mother-to-child Chikungunya virus infection: the CHIMERE cohort study on Reunion Island. PLoS Negl Trop Dis 8:e2996. 10.1371/journal.pntd.0002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Enter BJD, Huibers MHW, van Rooij L, Steingrover R, van Hensbroek MB, Voigt RR, Hol J. 2018. Perinatal outcomes in vertically infected neonates during a Chikungunya outbreak on the Island of Curacao. Am J Trop Med Hyg 99:1415–1418. 10.4269/ajtmh.17-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bandeira AC, Campos GS, Sardi SI, Rocha VF, Rocha GC. 2016. Neonatal encephalitis due to Chikungunya vertical transmission: first report in Brazil. IDCases 5:57–59. 10.1016/j.idcr.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Contopoulos-Ioannidis D, Newman-Lindsay S, Chow C, LaBeaud AD. 2018. Mother-to-child transmission of Chikungunya virus: a systematic review and meta-analysis. PLoS Negl Trop Dis 12:e0006510. 10.1371/journal.pntd.0006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiboutot MM, Kannan S, Kawalekar OU, Shedlock DJ, Khan AS, Sarangan G, Srikanth P, Weiner DB, Muthumani K. 2010. Chikungunya: a potentially emerging epidemic? PLoS Negl Trop Dis 4:e623. 10.1371/journal.pntd.0000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burt FJ, Rolph MS, Rulli NE, Mahalingam S, Heise MT. 2012. Chikungunya: a re-emerging virus. Lancet 379:662–671. 10.1016/S0140-6736(11)60281-X. [DOI] [PubMed] [Google Scholar]
- 9.Dash AP, Bhatia R, Sunyoto T, Mourya DT. 2013. Emerging and re-emerging arboviral diseases in Southeast Asia. J Vector Borne Dis 50:77–84. [PubMed] [Google Scholar]
- 10.Morrison TE. 2014. Re-emergence of chikungunya virus. J Virol 88:11644–11647. 10.1128/JVI.01432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S, Fink D, Hulse A, Pratt RD. 2017. Regulatory considerations in development of vaccines to prevent disease caused by Chikungunya virus. Vaccine 35:4851–4858. 10.1016/j.vaccine.2017.07.065. [DOI] [PubMed] [Google Scholar]
- 12.Ramsauer K, Schwameis M, Firbas C, Mullner M, Putnak RJ, Thomas SJ, Despres P, Tauber E, Jilma B, Tangy F. 2015. Immunogenicity, safety, and tolerability of a recombinant measles-virus-based chikungunya vaccine: a randomised, double-blind, placebo-controlled, active-comparator, first-in-man trial. Lancet Infect Dis 15:519–527. 10.1016/S1473-3099(15)70043-5. [DOI] [PubMed] [Google Scholar]
- 13.Wressnigg N, Hochreiter R, Zoihsl O, Fritzer A, Bézay N, Klingler A, Lingnau K, Schneider M, Lundberg U, Meinke A, Larcher-Senn J, Čorbic-Ramljak I, Eder-Lingelbach S, Dubischar K, Bender W. 2020. Single-shot live-attenuated Chikungunya vaccine in healthy adults: a phase 1, randomised controlled trial. Lancet Infect Dis 20:1193–1203. 10.1016/s1473-3099(20)30238-3. [DOI] [PubMed] [Google Scholar]
- 14.Zhang YN, Deng CL, Li JQ, Li N, Zhang QY, Ye HQ, Yuan ZM, Zhang B. 2019. Infectious Chikungunya virus with a complete capsid deletion: a new approach for CHIKV vaccine. J Virol 93:e00504-19. 10.1128/JVI.00504-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma A, Knollmann-Ritschel B. 2019. Current understanding of the molecular basis of Venezuelan equine encephalitis virus pathogenesis and vaccine development. Viruses 11:164. 10.3390/v11020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volkova E, Gorchakov R, Frolov I. 2006. The efficient packaging of Venezuelan equine encephalitis virus-specific RNAs into viral particles is determined by nsP1–3 synthesis. Virology 344:315–327. 10.1016/j.virol.2005.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng CL, Liu SQ, Zhou DG, Xu LL, Li XD, Zhang PT, Li PH, Ye HQ, Wei HP, Yuan ZM, Qin CF, Zhang B. 2016. Development of neutralization assay using an eGFP Chikungunya virus. Viruses 8:181. 10.3390/v8070181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva LA, Khomandiak S, Ashbrook AW, Weller R, Heise MT, Morrison TE, Dermody TS. 2014. A single-amino-acid polymorphism in Chikungunya virus E2 glycoprotein influences glycosaminoglycan utilization. J Virol 88:2385–2397. 10.1128/JVI.03116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardner J, Anraku I, Le TT, Larcher T, Major L, Roques P, Schroder WA, Higgs S, Suhrbier A. 2010. Chikungunya virus arthritis in adult wild-type mice. J Virol 84:8021–8032. 10.1128/JVI.02603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aguilar PV, Leung LW, Wang E, Weaver SC, Basler CF. 2008. A five-amino-acid deletion of the eastern equine encephalitis virus capsid protein attenuates replication in mammalian systems but not in mosquito cells. J Virol 82:6972–6983. 10.1128/JVI.01283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, Frolov I. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J Virol 81:2472–2484. 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguilar PV, Weaver SC, Basler CF. 2007. Capsid protein of eastern equine encephalitis virus inhibits host cell gene expression. J Virol 81:3866–3876. 10.1128/JVI.02075-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni P, Cheng Kao C. 2013. Non-encapsidation activities of the capsid proteins of positive-strand RNA viruses. Virology 446:123–132. 10.1016/j.virol.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webb LG, Veloz J, Pintado-Silva J, Zhu T, Rangel MV, Mutetwa T, Zhang L, Bernal-Rubio D, Figueroa D, Carrau L, Fenutria R, Potla U, Reid SP, Yount JS, Stapleford KA, Aguirre S, Fernandez-Sesma A. 2020. Chikungunya virus antagonizes cGAS-STING mediated type-I interferon responses by degrading cGAS. PLoS Pathog 16:e1008999. 10.1371/journal.ppat.1008999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hallengard D, Kakoulidou M, Lulla A, Kummerer BM, Johansson DX, Mutso M, Lulla V, Fazakerley JK, Roques P, Le Grand R, Merits A, Liljestrom P. 2014. Novel attenuated Chikungunya vaccine candidates elicit protective immunity in C57BL/6 mice. J Virol 88:2858–2866. 10.1128/JVI.03453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu SQ, Li X, Zhang YN, Gao AL, Deng CL, Li JH, Jehan S, Jamil N, Deng F, Wei H, Zhang B. 2017. Detection, isolation, and characterization of Chikungunya viruses associated with the Pakistan outbreak of 2016–2017. Virol Sin 32:511–519. 10.1007/s12250-017-4059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erasmus JH, Auguste AJ, Kaelber JT, Luo H, Rossi SL, Fenton K, Leal G, Kim DY, Chiu W, Wang T, Frolov I, Nasar F, Weaver SC. 2017. A Chikungunya fever vaccine utilizing an insect-specific virus platform. Nat Med 23:192–199. 10.1038/nm.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plante K, Wang E, Partidos CD, Weger J, Gorchakov R, Tsetsarkin K, Borland EM, Powers AM, Seymour R, Stinchcomb DT, Osorio JE, Frolov I, Weaver SC. 2011. Novel Chikungunya vaccine candidate with an IRES-based attenuation and host range alteration mechanism. PLoS Pathog 7:e1002142. 10.1371/journal.ppat.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed DS, Glass PJ, Bakken RR, Barth JF, Lind CM, da Silva L, Hart MK, Rayner J, Alterson K, Custer M, Dudek J, Owens G, Kamrud KI, Parker MD, Smith J. 2014. Combined alphavirus replicon particle vaccine induces durable and cross-protective immune responses against equine encephalitis viruses. J Virol 88:12077–12086. 10.1128/JVI.01406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]