ABSTRACT
Emerging strains of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the coronavirus disease 2019 (COVID-19) pandemic, that show increased transmission fitness and/or immune evasion are classified as “variants of concern” (VOCs). Recently, a SARS-CoV-2 variant first identified in November 2021 in South Africa has been recognized as a fifth VOC, termed “Omicron.” What makes this VOC so alarming is the high number of changes, especially in the viral Spike protein, and accumulating evidence for increased transmission efficiency and escape from neutralizing antibodies. In an amazingly short time, the Omicron VOC has outcompeted the previously dominating Delta VOC. However, it seems that the Omicron VOC is overall less pathogenic than other SARS-CoV-2 VOCs. Here, we provide an overview of the mutations in the Omicron genome and the resulting changes in viral proteins compared to other SARS-CoV-2 strains and discuss their potential functional consequences.
KEYWORDS: Omicron, SARS-CoV-2, variants of concern, Spike, COVID-19, BA.1, BA.2
INTRODUCTION
On 24 November 2021 the South African minister of health reported the emergence of a new and rapidly spreading variant of SARS-CoV-2 discovered by the Network for Genomic Surveillance in South Africa. Just 2 days later, the WHO declared this novel variant the fifth SARS-CoV-2 variant of concern (VOC), designated Omicron (B.1.1.529) (1). Despite very limited information on transmissibility and pathogenicity of Omicron, it was declared a VOC much faster than in previous cases for Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2) VOCs. The alarming finding that triggered rapid action was the strikingly high number of mutations, especially in the viral Spike protein, which mediates the infection of human cells and is the major target of the humoral immune response (2).
The Omicron VOC was first detected in samples obtained in Botswana and South Africa in the second week of November 2021, with the first Omicron VOC sample collected at the beginning of November 2021 in Africa (3). When we submitted the initial version of the manuscript in December, the knowledge about the features of this VOC was very preliminary and the share of Omicron infections was much lower than that of the Delta VOC. By the time of revision about 4 weeks later, the Omicron VOC had already outcompeted the Delta VOC and was responsible for ∼63% of new SARS-CoV-2 infections worldwide (https://nextstrain.org/ncov/gisaid/global). This agrees with initial epidemiological data indicating a more rapid emergence of the Omicron compared to the Delta VOC in South Africa (4). According to the WHO, Omicron had already been detected in about 77 countries across the world by mid-December, initially with many cases among travelers returning from Africa (5). Meanwhile, Omicron cases have been observed in about 150 countries around the globe, and it is well established that this VOC is highly transmissible and shows an increased ability to infect convalescent and vaccinated individuals (6–9). In accordance with this, numerous recent studies report strongly reduced susceptibility of the Omicron VOC to neutralizing antibodies induced by previous SARS-CoV-2 infection or vaccination (10–15). However, several studies indicate that vaccinated individuals boosted with mRNA-based vaccines elicit reduced but still potent-enough neutralizing humoral immune responses against the Omicron VOC (13, 16–19). In addition, most known T-cell epitopes are conserved, suggesting that the T-cell response in previously infected and/or vaccinated individuals will remain effective against the Omicron VOC (20–23). Modest increases in the transmissibility of the Omicron VOC compared to the Delta VOC have been observed in the golden Syrian hamster model (24). Most people infected with Omicron in Denmark and Norway were reported to be symptomatic (7, 8). More recent evidence suggests, however, that Omicron infections are associated with more frequent asymptomatic carriage, milder symptoms, and decreased hospitalization and fatality rates compared to infections with other SARS-CoV-2 VOCs (25–28). This agrees with recent data showing that Omicron causes reduced inflammatory processes and attenuated replication in mice and hamsters (29–31). Thus, accumulating evidence suggests that the Omicron VOC is less virulent than the Delta VOC.
The knowledge about the transmissibility, pathogenesis, and immune evasion capacity of the Omicron VOC will continue to explode in the coming weeks. Here, we focus on the properties of the Omicron VOC which have triggered global concerns: the type and potential consequences of the mutations that distinguish it from other SARS-CoV-2 VOCs.
INDEPENDENT EVOLUTION OF THE OMICRON VOC
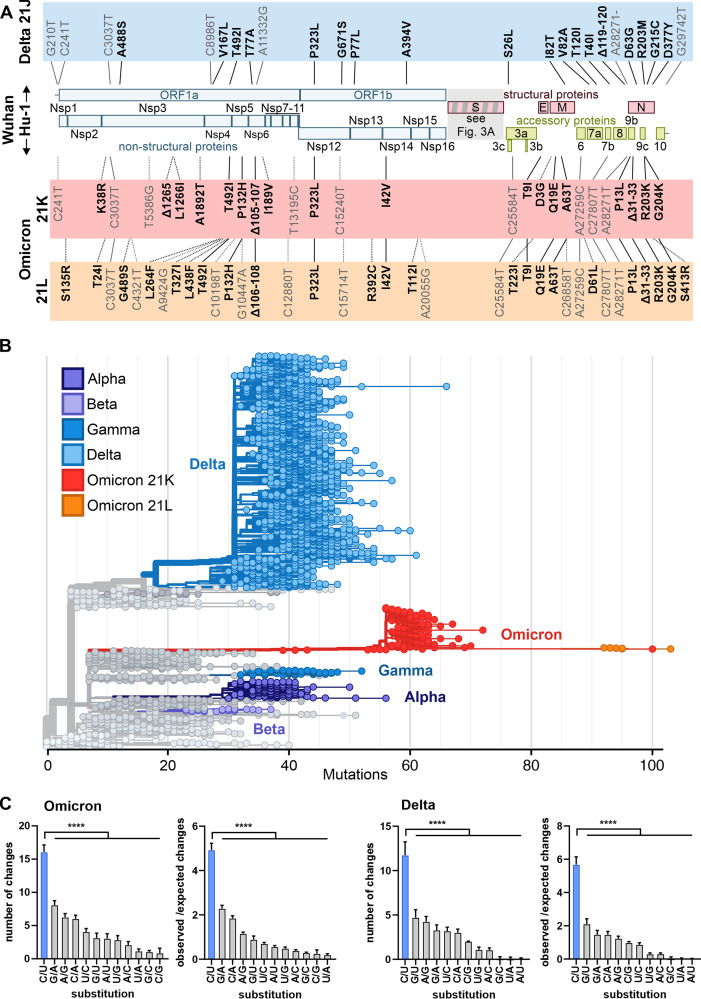
With about 30,000 nucleotides (Fig. 1A), coronaviruses, such as SARS-CoV-2, have the largest genome of all known RNA viruses. Despite the large size, the integrity of the genome is retained by the proofreading function of the viral polymerase (32). Specifically, the nonstructural protein 14 (Nsp14) and its cofactor Nsp10 exert exonuclease activity that substantially reduces the accumulation of mutations in the viral genome (33). The widespread view of SARS-CoV-2 as a highly variable pathogen is thus somewhat misleading (34, 35). For example, the genomes of different subtypes of HIV-1 may differ by up to 15% (36), while the Omicron VOC differs in less than 0.2% of its nucleotides from the early pandemic Wuhan strain of SARS-CoV-2. Nonetheless, the massive spread of SARS-CoV-2, with currently >339 million documented infections since its first occurrence in Wuhan at the end of 2019, is associated with the emergence of several variants showing an increased ability to spread in the human population, with Omicron showing the highest number of alterations from the 2019 isolate.
FIG 1.
Features of the Omicron genome and potential consequences of changes outside the Spike gene. (A) Schematic depiction of the SARS-CoV-2 genome and highlighting of non-Spike defining mutations (according to CoVariants data retrieved on 17 January 2022) in the Delta (blue, prevalent 21J clade; https://covariants.org/variants/21J.Delta) and Omicron (red, 21K, also known as BA.1, https://covariants.org/variants/21K.Omicron; orange, 21J, also known as BA.2, https://covariants.org/variants/21L.Omicron; shared mutations are indicated by solid lines) VOCs compared to the Wuhan Hu-1 isolate. Nonsynonymous (bold) and synonymous (nonbold) mutations are indicated. Positions are given relative to the start of the altered protein (nonsynonymous mutations) or the first nucleotide of the genome (synonymous mutations). Nonstructural proteins, Nsp1 to -16 (blue). Structural proteins, Spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N) (red). Accessory proteins, ORF3 to -10 (green). (B) Phylogenetic analysis of representative SARS-CoV-2 isolates scaled according to their divergence compared to the Wuhan Hu-1 sequence. Retrieved from Nextstrain on 18 January 2022 (https://nextstrain.org/ncov/gisaid/global?m=div) and modified. Color coding according to VOC as indicated. (C) Types of single nucleotide substitutions in SARS-CoV-2 Omicron and Delta VOCs compared to Wuhan Hu-1 reference strain. Shown are absolute numbers and ratios of changes observed to changes expected from random mutations based on SARS-CoV-2 nucleotide composition. Sequences were obtained from GISAID and NCBI virus databases (96, 97) and aligned to Wuhan Hu-1 strain (NCBI: NC_045512.2) using Clustal Omega (98). Mean of n = 475 (Delta) or n = 77 (Omicron) + standard deviation. P < 0.0001 (****), one-way paired analysis of variance with multiple comparisons to C/U set.
Our initial alignment of 77 Omicron full-genome sequences available at GISAID (www.gisaid.org) by early December revealed that this VOC differs by about 55 mutations (range 48 to 92; mean 54.6 ± 5.2 sequences) from the initial Wuhan Hu-1 strain of SARS-CoV-2 (Fig. 1A). This number is not that much higher than that found for the dominating Delta VOC, ranging from about 21 to 44 substitutions (34.4 ± 2.6; n = 475). What is alarming is the distribution of mutations, with about 30 of them being located in the viral Spike gene that is critical for virus infection and the key target of protective immune responses. In addition, most nucleotide changes distinguishing the Omicron VOC from the early Wuhan Hu-1 strain are nonsynonymous (37), i.e., associated with amino acid changes, suggesting high positive selection pressure for change. Meanwhile, almost half a million Omicron sequences are available at the GISAID database, revealing significant diversification. For example, the 21L clade of Omicron (https://covariants.org/variants/21L.Omicron) shares 38 mutations across its genome with the much more common 21K clade (https://covariants.org/variants/21K.Omicron), but both also contain an additional 27 and 20 unique changes, respectively (Fig. 1B). The significance of these sequence variations for viral spread and pathogenicity remains to be determined.
Phylogenetic analysis shows that the Omicron VOC clearly did not emerge from other VOCs, including the Delta VOC (Fig. 1B). This is unexpected, as it would seem plausible that zoonotic pathogens, such as SARS-CoV-2, adapt to the new human host in a stepwise process, i.e., they initially acquire key mutations promoting viral replication and spread and subsequently changes associated with more subtle advantages. An alternative scenario and concern are that different SARS-CoV-2 variants may recombine to combine, for example, the increased infectiousness and reduced sensitivity to neutralization of the Delta VOC (13) with the lower susceptibility of the Alpha VOC to interferon-inducible innate immune responses (38, 39). As discussed below, a substantial number of changes found in the Delta VOC have been previously detected in other SARS-CoV-2 strains. However, phylogenetic analysis suggests that these evolved independently by convergent evolution. This raises the question of how the Omicron VOC could evolve unnoticed to accumulate all the present concerning changes. Currently, three possibilities are discussed. One theory is efficient replication in an immunocompromised individual over a long period of time. In support of such a possibility, it has been reported that SARS-CoV-2 replicated for more than 6 months and accumulated many changes detected in VOCs in a young South African woman with HIV infection (40). Another possibility is that Omicron has been circulating in the human population for some time. However, its emergence may have gone unnoticed in one or several countries with less surveillance of COVID-19 and sequencing of emerging variants compared to South Africa. Finally, some researchers raised the possibility that the Omicron may have evolved in a nonhuman species from which it spilled back to humans (41). Coronaviruses are notorious for their ability to cross species barriers, and recent findings suggest that SARS-CoV-2 can be detected, e.g., in household pets and farm and captive animals as well as wildlife (42). It has been suggested that SARS-CoV-2 VOCs show an expanded host range and thus possess an increased capacity to generate new animal reservoirs (43, 44). However, the fact that a significant portion of changes in the Omicron genome is identical to human-specific adaptations found in other VOCs and the preliminary evidence for high replication and transmission fitness suggest emergence in humans. Currently, however, none of these possibilities can be dismissed entirely, and further research is required to elucidate the origin of the Omicron VOC.
POSSIBLE CAUSES OF THE MUTATIONS IN THE OMICRON GENOME
As mentioned above, the proofreading activity of the Nsp14 exoribonuclease of SARS-CoV-2 usually repairs errors during replication. However, several antiviral restriction factors are known to target and introduce mutations in viral RNAs. For example, APOBEC proteins deaminate cytosine to uracil, resulting in C-to-U or G-to-A changes depending on whether the plus or minus strand of the viral RNA is targeted (45). Notably, about 30% of all nucleotide substitutions in the Delta and Omicron VOC compared to an early pandemic reference strain are C to U (Fig. 1C). This rate is about 5-fold higher than expected from random changes. G-to-A changes are also very common and occur at a 2-fold-increased frequency (Fig. 1C). It has been previously suggested that APOBEC-induced mutations contribute to SARS-CoV-2 evolution (46–48). Until recently, experimental evidence that APOBEC proteins induce C-to-U and G-to-A mutations in coronavirus genomes has been reported only for the common cold coronavirus NL63 under overexpression conditions (49), but recent data from overexpression analyses support that APOBEC proteins also affect SARS-CoV-2 replication and fitness (50).
Adenosine deaminases acting on RNAs (ADARs) convert adenosine residues to inosines leading to changes of A to G or U to C if editing happens on the opposite strand. It has been proposed that ADAR A-to-I editing might be one of the factors driving SARS-CoV-2 evolution (51), but direct experimental proof is pending. Compared to the early reference SARS-CoV-2 strain, both types of ADAR signature changes are close to the expected ratio in the Omicron genome (Fig. 1C). Another antiviral factor that may exert selective pressure on viral evolution is the zinc finger antiviral protein (ZAP), which preferentially targets CpG dinucleotides in viral RNA sequences (52). Despite strong CpG suppression, SARS-CoV-2 is still inhibited by endogenous ZAP expression in human lung cells (53). However, there is little change in the level of CpG suppression between the early pandemic SARS CoV-2 and VOCs, with consensus Omicron mutations leading to a loss of 2 CpGs and gain of four new CpG among the 52 nucleotide changes. Thus, APOBEC proteins, but not ADAR or ZAP, were most likely a driving force behind the evolution of nucleotide changes in the Omicron VOC.
NON-SPIKE ALTERATIONS IN THE OMICRON GENOME
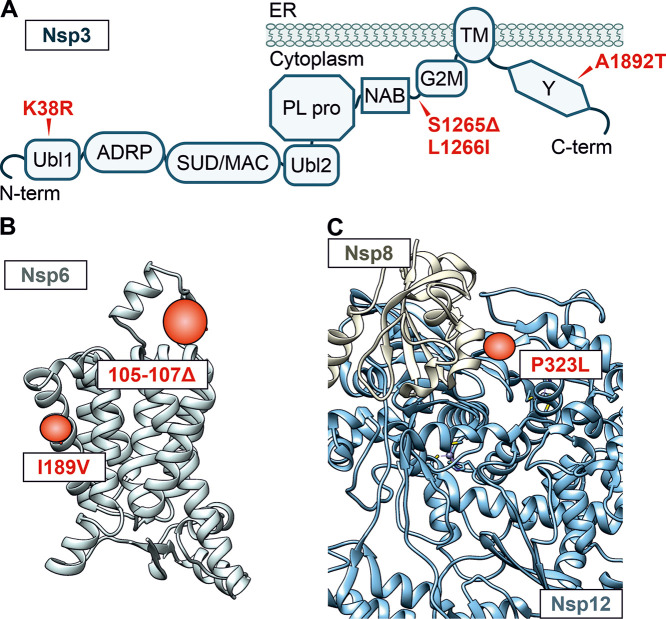
A total of 18 amino acid-changing mutations in the genome of more common 21K clade of the Omicron VOC are located outside the Spike gene (Fig. 1A). Two-thirds of the genome of SARS-CoV-2 encode 16 nonstructural proteins with functions including innate immune evasion, viral RNA replication, proteolytic activity, and proofreading function (54, 55). A variety of changes, such as K38R, Δ1265, and A1892T in Nsp3; P132H in Nsp5; I189V in Nsp6; P323L in Nsp12; and I42V in Nsp14, are present only in the Omicron VOC (Fig. 1A), and their potential functional consequences remain to be determined. Several mutations affect Nsp3, which deISGylates the RNA sensor MDA5 and signal transduction factors to antagonize innate immune responses (19, 23). However, the functional impact of K38R, Δ1265, L1266I, and A1892T changes in Omicron Nsp3 (Fig. 2A) is currently unknown since neither the protease domain nor known interaction interfaces are altered. The other main protease of SARS-CoV-2, Nsp5, targeting the sensor retinoic acid-inducible gene I (RIG-I) and the signal adaptor mitochondrial antiviral-signaling protein (MAVS) known to promote innate immune escape (54, 56), has a P132H mutation. This alteration is located neither in the active site nor in the dimerization interface, and its functional consequences remain to be determined. In support of a selective advantage, substitution of T492I in Nsp4 involved in viral replication is also present in the Lambda, Delta, and Gamma VOCs of SARS-CoV-2. A mutation similar to the 3-amino-acid deletion (Δ105–107) in a loop in the Omicron Nsp6 (Fig. 2B) predicted by alpha-fold (57) is also found in the Alpha, Beta, and Gamma VOCs and may be used for PCR-based differentiation from other SARS-CoV-2 variants. In addition, it has been hypothesized that this mutation may promote innate immune evasion of the virus (58). Nsp7, Nsp8, and Nsp12 comprise the active RNA-dependent RNA polymerase complex of SARS-CoV-2. Notably, the P323L mutation in Nsp12 is at the interface with Nsp8 (Fig. 2C) (59).
FIG 2.
Alterations in Nsp3, Nsp6, and Nsp8/Nsp12. (A) Schematic depiction of the domain arrangement of Nsp3. Ubl1, ubiquitin-like domain 1; ADRP, ADP-ribose-1″-phosphate phosphatase; SUD/MAC, SARS-CoV unique/macrodomain; Ubl2, ubiquitin-like domain 2; PL pro, papain-like protease; NAB, nucleic acid-binding domain; G2M, group 2-specific marker; TM, transmembrane domain; Y, Y domain. Mutations in the Omicron consensus sequence compared to Hu-1 are highlighted in red, and position is indicated with an arrow. (B) Alpha-fold prediction of the Nsp6 structure. Regions that are altered in the Omicron VOC are highlighted in red. Respective amino acid deletions (positions 105 to 107) or changes (I189V) are labeled in the box. (C) Structural representation of the complex between Hu-1 Nsp8 (tan) and Nsp12 (blue) derived from cryo-EM data (PDB 6YYT). The alteration in the Omicron VOC at the interface is highlighted in red (P323L).
In addition to the Spike (S) protein, the 3′ end of the SARS-CoV-2 genome encodes the envelope (E), membrane (M), and nucleocapsid (N) proteins as well as a variety of accessory factors thought to be involved in innate immune evasion. Alterations are found in all three non-Spike structural proteins: E (T9I), M (D3G, Q19E, A63T), and N (P13L, Δ31–33, R203K, G204R). Three of the four changes in the N protein of Omicron (P13L, R203K, G204R) have been previously noticed. Specifically, it has been reported that P13 is located in an important T-cell epitope and that the P13L change may help the virus to escape cellular immunity (60). Changes similar to R203K and G204R in N are also found in the Alpha and Gamma VOCs and may be associated with increased viral loads and subgenomic RNA expression (61). However, the functional relevance of the T9I change in the E protein as well as the D3G, Q19E, and A63T substitutions in the M protein is currently unclear. Compared to other VOCs, the Omicron VOC has a surprisingly low number of mutations in its accessory genes. Only ORF9b contains changes of P10S and Δ27–29 but may just be altered as it overlaps the gene encoding the N protein.
The less prevalent 21L clade of the Omicron VOC shares 15 amino acid changes with the 21K clade outside Spike (Fig. 1A). Among them are Nsp4 (T492I), Nsp5 (P132H), Nsp12 (P323L), E (T9I), M (Q19E and A63T), and N (P13L, Δ31–33, R203K, G204R). Additionally, there are 12 amino acid changes specific for the 21L clade of the Omicron VOC: Nsp1 (S135R), Nsp3 (T24I, G489S), Nsp4 (L264F, T327I, L438F), Nsp6 (Δ106–108), Nsp13 (R392C), Nsp15 (T112I), ORF3a (T223I), ORF6 (D61L), and N (S413R). Three of the mutations in 21L, which are not present in 21K, share similarities with mutations known from earlier VOCs: Nsp3 (G489S, similar to the A488S mutation found in the Delta VOC), Nsp4 (L438F, similar to the Nsp4 L438P mutation found in the Lambda VOC), and Nsp6 (the Δ106–108 triple deletion has shifted 1 amino acid to the C terminus from the mutation in the 21K clade and now mimics the mutation found in the Alpha, Beta, and Gamma VOCs). However, the possible functional relevance remains to be determined.
Accumulating evidence suggests that the Delta VOC remains sensitive to interferons (38). In comparison, the Alpha VOC, which has completely disappeared in the meantime, showed reduced sensitivity to the inhibitory effects of interferons (38, 39). It will be important to further investigate how the mutations in the structural and nonstructural proteins of the Omicron VOC affect viral replication fitness and evasion of innate antiviral immune responses.
ALTERATIONS IN THE OMICRON SPIKE PROTEIN
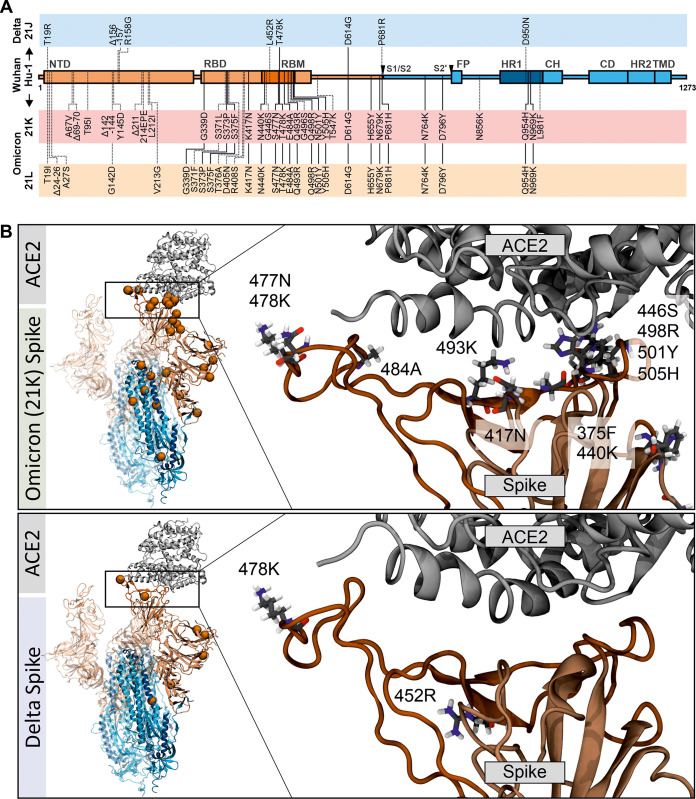
Although only ∼12.8% of the SARS-CoV-2 genome encodes the Spike protein (Fig. 1A), this region contains >60% of all mutations distinguishing the Omicron VOC from the original Wuhan Hu-1 SARS-CoV-2 strain (Fig. 3A). The Spike protein is the key determinant of virus transmission and immune evasion (62, 63). Thus, the strikingly high number of changes makes the Omicron VOC so alarming. The Spike protein mediates infection of human cells via binding to the human cellular angiotensin-converting enzyme (ACE) 2 receptor (64) and several cofactors (65, 66). For activation, Spike needs to be cleaved into the active S1 and S2 subunits by the cellular protease furin and subsequently by transmembrane protease serine subtype 2 (TMPRSS2) or cathepsin L at the S2′ site to liberate the viral fusion peptide that anchors the virion to the cell (64, 67) (Fig. 3A). In addition to determining infectivity, tropism, and transmission fitness, the Spike protein is the primary target of SARS-CoV-2 vaccines and antibodies of convalescent patients.
FIG 3.
Localization and potential interactions of amino acid changes in the Omicron Spike. (A) Schematic depiction of SARS-CoV-2 Spike, its domains, and amino acid alterations in the Delta 21J and Omicron 21K and 21L VOCs compared to the Wuhan Hu-1 sequence. S1 subunit: N-terminal domain (NTD) and receptor-binding domain (RBD) (orange). Receptor-binding motif, RBM (dark orange). S2 subunit: fusion peptide (FP) (blue), heptad repeat 1 (HR1) (dark blue), central helix (CH), connector domain (CD), heptad repeat 2 (HR2), and transmembrane domain (TM) (blue). Shared amino acid exchanges are indicated by solid lines. (B) Position of changes in the three-dimensional structure of the Spike/ACE2 complex based on the CoV-RDB (99). (Left) Overview of the positions of the mutations (orange) in Omicron and Delta Spikes, as indicated. (Right) Focused view of the mutations in the RBD. The mutated amino acid is shown in atomic stick representation. Shown are sample snapshots from reactive molecular dynamics simulations performed to relax the structure as reported previously (100). Initial atomic positions were based on PDB 7A96 (101).
The Omicron 21K and 21L Spike proteins share 20 amino acid changes compared to the original Wuhan Hu-1 strain, with an additional 19 21K- and 14 21L-specific mutations, respectively (Fig. 3A), although even higher numbers of mutations might be found in some viral isolates (68). In comparison, the Delta VOC differs by only nine amino acid changes from the initial viral isolate, two of which (E478K and D614G) are shared by the Omicron VOC (Fig. 3A). Below, we discuss the variation found in the Spike protein of the dominant 21K Omicron VOC.
Twelve of the 31 changes in the S1 region of the Omicron Spike compared to the Wuhan Hu-1 strain are located in the N-terminal domain (NTD), 15 in the ACE2 receptor-binding domain (RBD), and five at the C terminus (Fig. 3A). A total of 10 amino acid alterations are located in the receptor-binding motif (RBM) that makes direct contact with the human ACE2 receptor (Fig. 3B) and may affect Spike affinity for ACE2 (69–72). A significant portion of mutations in the S1 subunit (ΔH69/V70, T95I, ΔY144, K417N, T478K, N501Y, D614G, H655Y, and P681H) has previously been observed in other VOCs (68). For some of them, phenotypic effects such as increased transmission and immune evasion have been reported. For example, deletion of ΔH69/V70 may not only increase viral infectivity and Spike processing (73) but also be associated with negative results of S-assays within, for example, TaqPath tests. The Omicron Spike also contains a deletion of residues 143 to 145 and a 3-amino-acid insertion of “EPE” at position 214, which may provide further means to determine the prevalence of the Omicron 21K VOC in PCR-based assays. Mutations at position E484A have also been observed in other SARS-CoV-2 VOCs and may be associated with immune escape (74). Two other alterations in the RBM of the Omicron Spike, i.e., Q498R and N501Y, have been reported to increase the binding affinity to ACE2 (70). In addition, a mutation that is located in proximity to the S1-S2 furin cleavage site (H655Y) and has been previously observed in the Gamma VOC and during in vivo passage of SARS-CoV-2 in various animal species may be associated with increased Spike cleavage as well as escape from human monoclonal antibodies (75, 76). Two adjacent changes (N679K and P681H) increase S1/S2 cleavage by furin in pseudoparticles and may enhance replication fitness and type I interferon resistance (77–79). It has been reported, however, that the Omicron Spike is processed less efficiently in SARS-CoV-2 virions compared to other VOCs (80). Thus, the efficiency of Omicron Spike processing and the functional roles of the H655Y, N679K, and P681H changes need further study. Six mutations are found in the S2 subunit of the S protein. Three of them are located in the heptad repeat 1 (HR1) region required for 6-helix bundle formation and subsequent fusion. Most changes in the Omicron S protein have not been found in other SARS-CoV-2 VOCs, and the effect of such a complex combination of amino acid changes is difficult to assess. However, the localization of these mutations (Fig. 3B), as well as accumulating epidemiological data, suggests high transmission fitness and efficient immune evasion of the Omicron VOC (4, 6, 15).
The results of in silico analyses of the potential impact of the mutations in the Omicron Spike on its interaction with the ACE2 receptor are controversial. Utilizing an ab initio quantum mechanical model established to assess the role of residue 484 in the RBD of the Delta S with ACE2 (81), changes of K417N, N440K, G446S, S477N, T478K, E484A, Q493K, G496S, Q498R, N501Y, and Y505H in the S1 region of RBD that might affect the interaction of the Omicron Spike with the ACE2 receptor were examined (69). The results suggest that the Omicron Spike binds with higher affinity to ACE2 compared to Delta and that the interacting residues differ significantly from those of other SARS-CoV-2 variants. Improved binding was predicted to be largely due to increased electrostatic interactions. Notably, their modeling suggests some trade-off, i.e., some interactions known to play key roles in the interaction between S and ACE2 are lost but these detrimental changes are overcompensated by novel interactions. Another report provides first structural support from cryo-electron microscopy (cryo-EM) analysis that residues R493, S496, and R498 in the RBD of the Omicron Spike form new salt bridges and hydrogen bonds with ACE2 that (over)compensate for the disruptive effect of a K417N mutation (82). While some in silico analyses predict that the Omicron Spike binds human ACE2 with enhanced affinity compared to the Delta VOC (69, 83), weaker binding has also been reported (84). Altogether, further studies are required to clarify whether an increased affinity for human ACE2 contributes to the rapid spread of the Omicron VOC.
Numerous recent studies show that the Omicron VOC evades many but not all neutralizing antibodies induced by vaccination or previous SARS-CoV-2 infection (12, 14, 85–89). Initial epidemiological findings as well as first experimental studies suggest an up-to-40-fold-decreased efficiency of neutralizing sera against the Omicron VOC (12–14). Sera from individuals boosted with RNA vaccines showed substantial neutralizing activity against the Omicron VOC (16–18), supporting a significant benefit of multiboost vaccination strategies against highly divergent emerging SARS-CoV-2 variants. Notably, some of the changes in the Omicron Spike are identical to or occur at similar positions as the ∼20 mutations in an artificial “polymutant” Spike protein that was essentially fully resistant against antibodies found in vaccinated or convalescent individuals (90). The alterations in the Omicron Spike confer resistance to several Spike-targeting monoclonal antibodies (REGN 10933 and 10897) used in the clinic. Other therapeutic agents, however, such as remdesivir, molnupiravir, nirmatrelvir, hydroxycytidine, Paxlovid, Acriflavine (ACF), PF-07321332, and soluble ACE2 remain active against the Omicron VOC at least in vitro (91–93). Several reports indicate that Omicron entry may have shifted toward the TMPRSS2-independent endosomal route (77, 94, 95). Along these lines, TMPRSS2 inhibitors potently inhibit the Delta VOC but are less effective against the Omicron VOC (94). It has been suggested that the Omicron VOC may have thus an altered tropism and that reduced TMPRSS2 usage of the Omicron Spike favors infection of the upper respiratory tract and reduces the risk of invasion of the lung (94, 95). Consequently, the Omicron VOC replicates less efficiently in lower respiratory tract organoids and Calu-3 cells but shows a similar replication kinetic as the Delta VOC in nasal epithelial cells (77, 95).
PERSPECTIVES
The disturbing properties of the Omicron VOC raised alarm across the globe. Intense investigation of this new VOC is ongoing, and certainly many open questions regarding the spread, transmissibility, pathogenicity, and immune evasion properties have already been answered or will be answered soon. The origin of Omicron may not be easy to elucidate. Some hints come from the virus itself because pathogens will select for changes that are advantageous for them. Accordingly, there was only little pressure to alter the nonstructural and accessory genes but a strong selection pressure to further optimize the Spike protein. The changes in the RBD of the Spike protein suggest that Omicron evolved an improved ability to exploit the human ACE2 receptor for efficient spread. The numerous potential immune evasion mutations suggest emergence of the virus in the presence of selection pressure by antibodies found in people who had been vaccinated or previously infected with SARS-CoV-2. Thus, while currently none of the scenarios discussed above can be excluded, the emergence of the Omicron VOC in an environment characterized by high SARS-CoV-2 infection rates together with little surveillance and sequencing is a plausible scenario. This also illustrates that the COVID-19 pandemic is a global challenge and that it will be critical to improve health care systems and vaccination rates in developing countries.
Accumulating evidence suggests that even double-dose-vaccinated and convalescent people are not well protected against the Omicron VOC (7, 8). However, preexisting immunity most likely still prevents severe COVID-19. Even if protection against the Omicron VOC infection is impaired, it is important to consider that tools to control this pandemic are available: within an amazingly short time frame, scientists have developed convenient and affordable tests and highly effective vaccines against SARS-CoV-2 have been generated and can be adapted to the Omicron VOC. However, they have to be generated and distributed worldwide in sufficient quantity. Hopefully, access combined with accurate and open scientific communication to the public will result in vaccination rates that are high enough to curb the pandemic. The Omicron VOC is highly transmissible but seems to be less pathogenic than other SARS-CoV-2 VOCs. Together with the increasing immunity of the population through vaccination and previous infections, this raises hopes that severe COVD-19 will become rare and that SARS-CoV-2 will become endemic, similarly to seasonal coronaviruses. However, the Omicron VOC will still cause severe COVID-19 and death, especially in risk groups and in unvaccinated individuals. Vaccination seems to confer significant protection from severe disease (15) and is thus highly encouraged.
ACKNOWLEDGMENTS
We thank Dré van der Merwe for comments and suggestions. Knowledge about the Omicron VOC is increasing at an amazing speed, and we apologize to all colleagues whose studies could not be mentioned due to space limitations.
This study was supported by DFG grants to F.K. (CRC 1279, SPP 1923), D.K. (KM 5/1-1), and K.M.J.S. (CRC1279, SPP1923, SP1600/6-1). F.K. and K.M.J.S. were further supported by the BMBF (Restrict SARS-CoV-2 and IMMUNOMOD).
Contributor Information
Frank Kirchhoff, Email: frank.kirchhoff@uni-ulm.de.
Hector C. Aguilar, Cornell University
REFERENCES
- 1.World Health Organization. 2021. Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern.
- 2.Dai L, Gao GF. 2021. Viral targets for vaccines against COVID-19. Nat Rev Immunol 21:73–82. 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callaway E. 2021. Heavily mutated Omicron variant puts scientists on alert. Nature 600:21. 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- 4.Grabowski F, Kochańczyk M, Lipniacki T. 2022. The spread of SARS-CoV-2 variant Omicron with the doubling time of 2.0–3.3 days can be explained by immune evasion. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.08.21267494v2. [DOI] [PMC free article] [PubMed]
- 5.Bai Y, Du Z, Xu M, Wang L, Wu P, Lau E, Cowling BJ, Meyers LA. 2021. International risk of SARS-CoV-2 Omicron variant importations originating in South Africa. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.07.21267410v1. [DOI] [PMC free article] [PubMed]
- 6.Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, Dushoff J, Mlisana K, Moultrie H. 2021. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv. https://www.medrxiv.org/content/10.1101/2021.11.11.21266068v2. [DOI] [PMC free article] [PubMed]
- 7.Espenhain L, Funk T, Overvad M, Edslev SM, Fonager J, Ingham AC, Rasmussen M, Madsen SL, Espersen CH, Sieber RN, Stegger M, Gunalan V, Wilkowski B, Larsen NB, Legarth R, Cohen AS, Nielsen F, Lam JUH, Lavik KE, Karakis M, Spiess K, Marving E, Nielsen C, Wiid Svarrer C, Bybjerg-Grauholm J, Olsen SS, Jensen A, Krause TG, Müller L. 2021. Epidemiological characterisation of the first 785 SARS-CoV-2 Omicron variant cases in Denmark, December 2021. Euro Surveill 26(50). 10.2807/1560-7917.ES.2021.26.50.2101146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandal LT, MacDonald E, Veneti L, Ravlo T, Lange H, Naseer U, Feruglio S, Bragstad K, Hungnes O, Ødeskaug LE, Hagen F, Hanch-Hansen KE, Lind A, Watle SV, Taxt AM, Johansen M, Vold L, Aavitsland P, Nygård K, Madslien EH. 2021. Outbreak caused by the SARS-CoV-2 Omicron variant in Norway, November to December 2021. Euro Surveill 26(50). 10.2807/1560-7917.ES.2021.26.50.2101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrews N, Stowe J, Kirsebom F, Toffa S, Rickeard T, Gallagher E, Gower C, Kall M, Groves N, O’Connell A-M, Simons D, Blomquist PB, Zaidi A, Nash S, Aziz NIBA, Thelwall S, Dabrera G, Myers R, Amirthalingam G, Gharbia S, Barrett JC, Elson R, Ladhani SN, Ferguson N, Zambon M, Campbell CN, Brown K, Hopkins S, Chand M, Ramsay M, Bernal JL. 2021. Effectiveness of COVID-19 vaccines against the Omicron (B.1.1.529) variant of concern. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.14.21267615v1. [DOI] [PMC free article] [PubMed]
- 10.Lu L, Mok BW-Y, Chen L-L, Chan JM-C, Tsang OT-Y, Lam BH-S, Chuang VW-M, Chu AW-H, Chan W-M, Ip JD, Chan BP-C, Zhang R, Yip CC-Y, Cheng VC-C, Chan K-H, Jin D-Y, Hung IF-N, Yuen K-Y, Chen H, To KK-W. 2021. Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin Infect Dis. 10.1093/cid/ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Li Q, Liang Z, Li T, Liu S, Cui Q, Nie J, Wu Q, Qu X, Huang W, Wang Y. 2022. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg Microbes Infect 11:1–5. 10.1080/22221751.2021.2017757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilhelm A, Widera M, Grikscheit K, Toptan T, Schenk B, Pallas C, Metzler M, Kohmer N, Hoehl S, Helfritz FA, Wolf T, Goetsch U, Ciesek S. 2021. Reduced neutralization of SARS-CoV-2 Omicron variant by vaccine sera and monoclonal antibodies. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.07.21267432v4. [DOI] [PMC free article] [PubMed]
- 13.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Le Guen J, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Péré H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Lorière E, Rey FA, Schwartz O. 2021. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596:276–280. 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann M, Krüger N, Schulz S, Cossmann A, Rocha C, Kempf A, Nehlmeier I, Graichen L, Moldenhauer A-S, Winkler MS, Lier M, Dopfer-Jablonka A, Jäck H-M, Behrens GMN, Pöhlmann S. 2021. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cele S, Jackson L, Khoury DS, Khan K, Moyo-Gwete T, Tegally H, San JE, Cromer D, Scheepers C, Amoako DG, Karim F, Bernstein M, Lustig G, Archary D, Smith M, Ganga Y, Jule Z, Reedoy K, Hwa S-H, Giandhari J, Blackburn JM, Gosnell BI, Abdool Karim SS, Hanekom W, NGS-SA, COMMIT-KZN Team, von Gottberg A, Bhiman JN, Lessells RJ, Moosa M-YS, Davenport MP, de Oliveira T, Moore PL, Sigal A. 2021. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basile K, Rockett RJ, McPhie K, Fennell M, Johnson-Mackinnon J, Agius JE, Fong W, Rahman H, Ko D, Donavan L, Hueston L, Lam C, Arnott A, Chen SC-A, Maddocks S, O’Sullivan MV, Dwyer DE, Sintchenko V, Kok J. 2021. Improved neutralization of the SARS-CoV-2 Omicron variant after Pfizer-BioNTech BNT162b2 COVID-19 vaccine boosting. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.12.472252v1. [DOI] [PMC free article] [PubMed]
- 17.Garcia-Beltran WF, Denis KJS, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML, Berrios C, Ofoman O, Chang CC, Hauser BM, Feldman J, Gregory DJ, Poznansky MC, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB. 2021. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.14.21267755v1. [DOI] [PMC free article] [PubMed]
- 18.Gruell H, Vanshylla K, Tober-Lau P, Hillus D, Schommers P, Lehmann C, Kurth F, Sander LE, Klein F. 2021. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.14.21267769v1. [DOI] [PMC free article] [PubMed]
- 19.Cong Z, Evans JP, Qu P, Faraone J, Zheng Y-M, Carlin C, Bednash JS, Zhou T, Lozanski G, Mallampalli R, Saif LJ, Oltz EM, Mohler P, Xu K, Gumina RJ, Liu S-L. 2021. Neutralization and stability of SARS-CoV-2 Omicron variant. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.16.472934v1.
- 20.Redd AD, Nardin A, Kared H, Bloch EM, Abel B, Pekosz A, Laeyendecker O, Fehlings M, Quinn TC, Tobian AA. 2021. Minimal cross-over between mutations associated with Omicron variant of SARS-CoV-2 and CD8+ T cell epitopes identified in COVID-19 convalescent individuals. bioRxiv. 10.1101/2021.12.06.471446. [DOI] [PMC free article] [PubMed]
- 21.Ahmed SF, Quadeer AA, McKay MR. 2021. SARS-CoV-2 T cell responses are expected to remain robust against Omicron. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.12.472315v1. [DOI] [PMC free article] [PubMed]
- 22.Gao Y, Cai C, Grifoni A, Müller TR, Niessl J, Olofsson A, Humbert M, Hansson L, Österborg A, Bergman P, Chen P, Olsson A, Sandberg JK, Weiskopf D, Price DA, Ljunggren H-G, Karlsson AC, Sette A, Aleman S, Buggert M. 2022. Ancestral SARS-CoV-2-specific T cells cross-recognize Omicron. Nat Med. 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeton R, Tincho MB, Ngomti A, Baguma R, Benede N, Suzuki A, Khan K, Cele S, Bernstein M, Karim F, Madzorera SV, Moyo-Gwete T, Mennen M, Skelem S, Adriaanse M, Mutithu D, Aremu O, Stek C, du Bruyn E, Mescht MAVD, de Beer Z, de Villiers TR, Bodenstein A, van den Berg G, Mendes A, Strydom A, Venter M, Grifoni A, Weiskopf D, Sette A, Wilkinson RJ, Bekker L-G, Gray G, Ueckermann V, Rossouw T, Boswell MT, Bihman J, Moore PL, Sigal A, Ntusi NAB, Burgers WA, Riou C. 2021. SARS-CoV-2 spike T cell responses induced upon vaccination or infection remain robust against Omicron. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.26.21268380v1.
- 24.Yuan S, Ye Z-W, Liang R, Tang K, Zhang AJ, Lu G, Ong CP, Poon VK-M, Chan CC-S, Mok BW-Y, Qin Z, Xie Y, Sun H, Tsang JO-L, Yuen TT-T, Chik KK-H, Chan CC-Y, Cai J-P, Luo C, Lu L, Yip CC-Y, Chu H, To KK-W, Chen H, Jin D-Y, Yuen K-Y, Chan JF-W. 2022. The SARS-CoV-2 Omicron (B.1.1.529) variant exhibits altered pathogenicity, transmissibility, and fitness in the golden Syrian hamster model. bioRxiv. https://www.biorxiv.org/content/10.1101/2022.01.12.476031v1. [DOI] [PubMed]
- 25.Vihta K-D, Pouwels KB, Peto TE, Pritchard E, House T, Studley R, Rourke E, Cook D, Diamond I, Crook D, Matthews PC, Stoesser N, Eyre DW, Walker AS, Team C-19 IS. 2022. Omicron-associated changes in SARS-CoV-2 symptoms in the United Kingdom. medRxiv. https://www.medrxiv.org/content/10.1101/2022.01.18.22269082v1. [DOI] [PMC free article] [PubMed]
- 26.Garrett N, Tapley A, Andriesen J, Seocharan I, Fisher LH, Bunts L, Espy N, Wallis CL, Randhawa AK, Ketter N, Yacovone M, Goga A, Bekker L-G, Gray GE, Corey L. 2022. High rate of asymptomatic carriage associated with variant strain Omicron. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.20.21268130v2.
- 27.Christensen PA, Olsen RJ, Long SW, Snehal R, Davis JJ, Saavedra MO, Reppond K, Shyer MN, Cambric J, Gadd R, Thakur RM, Batajoo A, Mangham R, Pena S, Trinh T, Kinskey JC, Williams G, Olson R, Gollihar J, Musser JM. 2022. Signals of significantly increased vaccine breakthrough, decreased hospitalization rates, and less severe disease in patients with COVID-19 caused by the Omicron variant of SARS-CoV-2 in Houston, Texas. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.30.21268560v3. [DOI] [PMC free article] [PubMed]
- 28.Wolter N, Jassat W, Walaza S, Welch R, Moultrie H, Groome M, Amoako DG, Everatt J, Bhiman JN, Scheepers C, Tebeila N, Chiwandire N, du Plessis M, Govender N, Ismail A, Glass A, Mlisana K, Stevens W, Treurnicht FK, Makatini Z, Hsiao N, Parboosing R, Wadula J, Hussey H, Davies M-A, Boulle A, von Gottberg A, Cohen C. 2021. Early assessment of the clinical severity of the SARS-CoV-2 Omicron variant in South Africa. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.21.21268116v1. [DOI] [PMC free article] [PubMed]
- 29.Bentley EG, Kirby A, Sharma P, Kipar A, Mega DF, Bramwell C, Penrice-Randal R, Prince T, Brown JC, Zhou J, Screaton GR, Barclay WS, Owen A, Hiscox JA, Stewart JP. 2021. SARS-CoV-2 Omicron-B.1.1.529 variant leads to less severe disease than Pango B and Delta variants strains in a mouse model of severe COVID-19. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.26.474085v2.
- 30.Diamond M, Halfmann P, Maemura T, Iwatsuki-Horimoto K, Iida S, Kiso M, Scheaffer S, Darling T, Joshi A, Loeber S, Foster S, Ying B, Whitener B, Floyd K, Ujie M, Nakajima N, Ito M, Wright R, Uraki R, Li R, Sakai Y, Liu Y, Larson D, Osorio J, Hernandez-Ortiz J, Čiuoderis K, Florek K, Patel M, Bateman A, Odle A, Wong L-Y, Wang Z, Edara VV, Chong Z, Thackray L, Ueki H, Yamayoshi S, Imai M, Perlman S, Webby R, Seder R, Suthar M, Garcia-Sastre A, Schotsaert M, Suzuki T, Boon A, Kawaoka Y, Douek D, Moliva J, Sullivan N, Gagne M, Ransier A, Case J, Jeevan T, Franks J, Fabrizio T, DeBeauchamp J, Kercher L, Seiler P, Singh G, Warang P, Gonzalez-Reiche AS, Sordillo E, van Bakel H, Simon V. 2022. The SARS-CoV-2 B.1.1.529 Omicron virus causes attenuated infection and disease in mice and hamsters. Res Sq. https://www.researchsquare.com/article/rs-1211792/v1.
- 31.McMahan K, Giffin V, Tostanoski LH, Chung B, Siamatu M, Suthar MS, Halfmann P, Kawaoka Y, Piedra-Mora C, Martinot AJ, Kar S, Andersen H, Lewis MG, Barouch DH. 2022. Reduced pathogenicity of the SARS-CoV-2 Omicron variant in hamsters. bioRxiv. https://www.biorxiv.org/content/10.1101/2022.01.02.474743v1. [DOI] [PMC free article] [PubMed]
- 32.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res 117:17–37. 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saramago M, Bárria C, Costa VG, Souza CS, Viegas SC, Domingues S, Lousa D, Soares CM, Arraiano CM, Matos RG. 2021. New targets for drug design: importance of nsp14/nsp10 complex formation for the 3’-5’ exoribonucleolytic activity on SARS-CoV-2. FEBS J 288:5130–5147. 10.1111/febs.15815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckerle LD, Becker MM, Halpin RA, Li K, Venter E, Lu X, Scherbakova S, Graham RL, Baric RS, Stockwell TB, Spiro DJ, Denison MR. 2010. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog 6:e1000896. 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robson F, Khan KS, Le TK, Paris C, Demirbag S, Barfuss P, Rocchi P, Ng W-L. 2020. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol Cell 79:710–727. 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G, Piampongsant S, Faria NR, Voet A, Pineda-Peña A-C, Khouri R, Lemey P, Vandamme A-M, Theys K. 2015. An integrated map of HIV genome-wide variation from a population perspective. Retrovirology 12:18. 10.1186/s12977-015-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bansal K, Kumar S. 2021. Mutational cascade of SARS-CoV-2 leading to evolution and emergence of omicron variant. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.06.471389v1. [DOI] [PMC free article] [PubMed]
- 38.Nchioua R, Schundner A, Klute S, Noettger S, Zech F, Koepke L, Graf A, Krebs S, Blum H, Kmiec D, Frick M, Kirchhoff F, Sparrer KMJ. 2021. The Delta variant of SARS-CoV-2 maintains high sensitivity to interferons in human lung cells. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.11.16.468777v1.
- 39.Thorne LG, Bouhaddou M, Reuschl A-K, Zuliani-Alvarez L, Polacco B, Pelin A, Batra J, Whelan MVX, Ummadi M, Rojc A, Turner J, Obernier K, Braberg H, Soucheray M, Richards A, Chen K-H, Harjai B, Memon D, Hosmillo M, Hiatt J, Jahun A, Goodfellow IG, Fabius JM, Shokat K, Jura N, Verba K, Noursadeghi M, Beltrao P, Swaney DL, Garcia-Sastre A, Jolly C, Towers GJ, Krogan NJ. 2021. Evolution of enhanced innate immune evasion by the SARS-CoV-2 B.1.1.7 UK variant. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.06.06.446826v1.
- 40.Karim F, Moosa MYS, Gosnell BI, Cele S, Giandhari J, Pillay S, Tegally H, Wilkinson E, San JE, Msomi N, Mlisana K, Khan K, Bernstein M, Manickchund N, Singh L, Ramphal U, COMMIT-KZN Team, Hanekom W, Lessells RJ, Sigal A, de Oliveira T. 2021. Persistent SARS-CoV-2 infection and intra-host evolution in association with advanced HIV infection. medRxiv. https://www.medrxiv.org/content/10.1101/2021.06.03.21258228v1.
- 41.Wei C, Shan K-J, Wang W, Zhang S, Huan Q, Qian W. 2021. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.14.472632v1. [DOI] [PMC free article] [PubMed]
- 42.Sharun K, Dhama K, Pawde AM, Gortázar C, Tiwari R, Bonilla-Aldana DK, Rodriguez-Morales AJ, de la Fuente J, Michalak I, Attia YA. 2021. SARS-CoV-2 in animals: potential for unknown reservoir hosts and public health implications. Vet Q 41:181–201. 10.1080/01652176.2021.1921311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stone S, Rothan HA, Natekar JP, Kumari P, Sharma S, Pathak H, Arora K, Aurani TT, Kumar M. 2021. SARS-CoV-2 variants of concern infect the respiratory tract and induce inflammatory response in wild-type laboratory mice. bioRxiv. https://biorxiv.org/cgi/content/short/2021.09.29.462373. [DOI] [PMC free article] [PubMed]
- 44.Shuai H, Chan JF-W, Yuen TT-T, Yoon C, Hu J-C, Wen L, Hu B, Yang D, Wang Y, Hou Y, Huang X, Chai Y, Chan CC-S, Poon VK-M, Lu L, Zhang R-Q, Chan W-M, Ip JD, Chu AW-H, Hu Y-F, Cai J-P, Chan K-H, Zhou J, Sridhar S, Zhang B-Z, Yuan S, Zhang AJ, Huang J-D, To KK-W, Yuen K-Y, Chu H. 2021. Emerging SARS-CoV-2 variants expand species tropism to murines. EBioMedicine 73:103643. 10.1016/j.ebiom.2021.103643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salter JD, Bennett RP, Smith HC. 2016. The APOBEC protein family: united by structure, divergent in function. Trends Biochem Sci 41:578–594. 10.1016/j.tibs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azgari C, Kilinc Z, Turhan B, Circi D, Adebali O. 2021. The mutation profile of SARS-CoV-2 is primarily shaped by the host antiviral defense. Viruses 13:394. 10.3390/v13030394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simmonds P. 2020. Rampant C→U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: causes and consequences for their short- and long-term evolutionary trajectories. mSphere 5:e00408-20. 10.1128/mSphere.00408-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mourier T, Sadykov M, Carr MJ, Gonzalez G, Hall WW, Pain A. 2021. Host-directed editing of the SARS-CoV-2 genome. Biochem Biophys Res Commun 538:35–39. 10.1016/j.bbrc.2020.10.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milewska A, Kindler E, Vkovski P, Zeglen S, Ochman M, Thiel V, Rajfur Z, Pyrc K. 2018. APOBEC3-mediated restriction of RNA virus replication. Sci Rep 8:5960. 10.1038/s41598-018-24448-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X, Kim K, Calabrese P, Wang S, Qin C, Rao Y, Feng P. 2021. APOBEC-mediated editing of SARS-CoV-2 genomic RNA impacts viral replication and fitness. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.18.473309v1. [DOI] [PMC free article] [PubMed]
- 51.Song Y, He X, Yang W, Tang T, Zhang R. 2021. ADAR mediated A-to-I RNA editing affects SARS-CoV-2 characteristics and fuels its evolution. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.07.22.453345v1.
- 52.Takata MA, Gonçalves-Carneiro D, Zang TM, Soll SJ, York A, Blanco-Melo D, Bieniasz PD. 2017. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 550:124–127. 10.1038/nature24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nchioua R, Kmiec D, Müller JA, Conzelmann C, Groß R, Swanson CM, Neil SJD, Stenger S, Sauter D, Münch J, Sparrer KMJ, Kirchhoff F. 2020. SARS-CoV-2 is restricted by zinc finger antiviral protein despite preadaptation to the low-CpG environment in humans. mBio 11:e01930-20. 10.1128/mBio.01930-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayn M, Hirschenberger M, Koepke L, Nchioua R, Straub JH, Klute S, Hunszinger V, Zech F, Prelli Bozzo C, Aftab W, Christensen MH, Conzelmann C, Müller JA, Srinivasachar Badarinarayan S, Stürzel CM, Forne I, Stenger S, Conzelmann K-K, Münch J, Schmidt FI, Sauter D, Imhof A, Kirchhoff F, Sparrer KMJ. 2021. Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. Cell Rep 35:109126. 10.1016/j.celrep.2021.109126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia H, Cao Z, Xie X, Zhang X, Chen JY-C, Wang H, Menachery VD, Rajsbaum R, Shi P-Y. 2020. Evasion of type I interferon by SARS-CoV-2. Cell Rep 33:108234. 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, Qin C, Rao Y, Ngo C, Feng JJ, Zhao J, Zhang S, Wang T-Y, Carriere J, Savas AC, Zarinfar M, Rice S, Yang H, Yuan W, Camarero JA, Yu J, Chen XS, Zhang C, Feng P. 2021. SARS-CoV-2 Nsp5 demonstrates two distinct mechanisms targeting RIG-I and MAVS to evade the innate immune response. mBio 12:e02335-21. 10.1128/mBio.02335-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benvenuto D, Angeletti S, Giovanetti M, Bianchi M, Pascarella S, Cauda R, Ciccozzi M, Cassone A. 2020. Evolutionary analysis of SARS-CoV-2: how mutation of non-structural protein 6 (NSP6) could affect viral autophagy. J Infect 81:e24–e27. 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hillen HS, Kokic G, Farnung L, Dienemann C, Tegunov D, Cramer P. 2020. Structure of replicating SARS-CoV-2 polymerase. Nature 584:154–156. 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 60.de Silva TI, Liu G, Lindsey BB, Dong D, Moore SC, Hsu NS, Shah D, Wellington D, Mentzer AJ, Angyal A, Brown R, Parker MD, Ying Z, Yao X, Turtle L, Dunachie S, COVID-19 Genomics UK (COG-UK) Consortium, Maini MK, Ogg G, Knight JC, ISARIC4C Investigators, Peng Y, Rowland-Jones SL, Dong T. 2021. The impact of viral mutations on recognition by SARS-CoV-2 specific T cells. iScience 24:103353. 10.1016/j.isci.2021.103353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leary S, Gaudieri S, Parker MD, Chopra A, James I, Pakala S, Alves E, John M, Lindsey BB, Keeley AJ, Rowland-Jones SL, Swanson MS, Ostrov DA, Bubenik JL, Das S, Sidney J, Sette A, COVID-19 Genomics UK (COG-UK) Consortium, de Silva TI, Phillips E, Mallal S. 2021. Generation of a novel SARS-CoV-2 sub-genomic RNA due to the R203K/G204R variant in nucleocapsid: homologous recombination has potential to change SARS-CoV-2 at both protein and RNA level. bioRxiv. https://www.biorxiv.org/content/10.1101/2020.04.10.029454v4. [DOI] [PMC free article] [PubMed]
- 62.Letko M, Marzi A, Munster V. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5:562–569. 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein. Cell 181:281–292.e6. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lempp FA, Soriaga LB, Montiel-Ruiz M, Benigni F, Noack J, Park Y-J, Bianchi S, Walls AC, Bowen JE, Zhou J, Kaiser H, Joshi A, Agostini M, Meury M, Dellota E, Jaconi S, Cameroni E, Martinez-Picado J, Vergara-Alert J, Izquierdo-Useros N, Virgin HW, Lanzavecchia A, Veesler D, Purcell LA, Telenti A, Corti D. 2021. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature 598:342–347. 10.1038/s41586-021-03925-1. [DOI] [PubMed] [Google Scholar]
- 66.Prelli Bozzo C, Nchioua R, Volcic M, Koepke L, Krüger J, Schütz D, Heller S, Stürzel CM, Kmiec D, Conzelmann C, Müller J, Zech F, Braun E, Groß R, Wettstein L, Weil T, Weiß J, Diofano F, Rodríguez Alfonso AA, Wiese S, Sauter D, Münch J, Goffinet C, Catanese A, Schön M, Boeckers TM, Stenger S, Sato K, Just S, Kleger A, Sparrer KMJ, Kirchhoff F. 2021. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat Commun 12:4584. 10.1038/s41467-021-24817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao M-M, Yang W-L, Yang F-Y, Zhang L, Huang W-J, Hou W, Fan C-F, Jin R-H, Feng Y-M, Wang Y-C, Yang J-K. 2021. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct Target Ther 6:134. 10.1038/s41392-021-00558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sarkar R, Lo M, Saha R, Dutta S, Chawla-Sarkar M. 2021. S glycoprotein diversity of the Omicron variant. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.04.21267284v2.
- 69.Genovese L, Zaccaria M, Farzan M, Johnson WE, Momeni B. 2021. Investigating the mutational landscape of the SARS-CoV-2 Omicron variant via ab initio quantum mechanical modeling. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.01.470748v1. [DOI] [PMC free article] [PubMed]
- 70.Zahradnik J, Tuekprakhon A, Ginn HM, Duyvesteyn HME, Bahar M, Khan S, Avinoam O, Zhou D, Nutalai R, Supasa P, Wang B, Dejnirattisai W, Liu C, Dijokaite A, Temperton N, Mongkolsapaya J, Fry EE, Ren J, Screaton GR, Schreiber G, Stuart DI. 2021. Receptor binding and escape from Beta antibody responses drive Omicron-B.1.1.529 evolution. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.03.471045v1.
- 71.Golcuk M, Yildiz A, Gur M. 2021. The Omicron variant increases the interactions of SARS-CoV-2 Spike glycoprotein with ACE2. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.06.471377v1. [DOI] [PMC free article] [PubMed]
- 72.Shah M, Woo HG. 2021. Omicron: a heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escape approved COVID-19 therapeutic antibodies. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.04.471200v1. [DOI] [PMC free article] [PubMed]
- 73.Meng B, Kemp SA, Papa G, Datir R, Ferreira IATM, Marelli S, Harvey WT, Lytras S, Mohamed A, Gallo G, Thakur N, Collier DA, Mlcochova P, COVID-19 Genomics UK (COG-UK) Consortium, Duncan LM, Carabelli AM, Kenyon JC, Lever AM, De Marco A, Saliba C, Culap K, Cameroni E, Matheson NJ, Piccoli L, Corti D, James LC, Robertson DL, Bailey D, Gupta RK. 2021. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep 35:109292. 10.1016/j.celrep.2021.109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ku Z, Xie X, Davidson E, Ye X, Su H, Menachery VD, Li Y, Yuan Z, Zhang X, Muruato AE, Grinyo I Escuer A, Tyrell B, Doolan K, Doranz BJ, Wrapp D, Bates PF, McLellan JS, Weiss SR, Zhang N, Shi P-Y, An Z. 2021. Molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape. Nat Commun 12:469. 10.1038/s41467-020-20789-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dieterle ME, Haslwanter D, Bortz RH, Wirchnianski AS, Lasso G, Vergnolle O, Abbasi SA, Fels JM, Laudermilch E, Florez C, Mengotto A, Kimmel D, Malonis RJ, Georgiev G, Quiroz J, Barnhill J, Pirofski L, Daily JP, Dye JM, Lai JR, Herbert AS, Chandran K, Jangra RK. 2020. A replication-competent vesicular stomatitis virus for studies of SARS-CoV-2 Spike-mediated cell entry and its inhibition. Cell Host Microbe 28:486–496.e6. 10.1016/j.chom.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Escalera A, Gonzalez-Reiche AS, Aslam S, Mena I, Pearl RL, Laporte M, Fossati A, Rathnasinghe R, Alshammary H, van de Guchte A, Bouhaddou M, Kehrer T, Zuliani-Alvarez L, Meekins DA, Balaraman V, McDowell C, Richt JA, Bajic G, Sordillo EM, Krogan N, Simon V, Albrecht RA, van Bakel H, Garcia-Sastre A, Aydillo T. 2021. SARS-CoV-2 variants of concern have acquired mutations associated with an increased spike cleavage. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.11.09.467693v1. [DOI] [PMC free article] [PubMed]
- 77.Peacock TP, Brown JC, Zhou J, Thakur N, Newman J, Kugathasan R, Sukhova K, Kaforou M, Bailey D, Barclay WS. 2022. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.31.474653v1.
- 78.Liu Y, Liu J, Johnson BA, Xia H, Ku Z, Schindewolf C, Widen SG, An Z, Weaver SC, Menachery VD, Xie X, Shi P-Y. 2021. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv. 10.1101/2021.08.12.456173. [DOI] [PMC free article] [PubMed]
- 79.Lista MJ, Winstone H, Wilson HD, Dyer A, Pickering S, Galao RP, Lorenzo GD, Cowton VM, Furnon W, Suarez N, Orton R, Palmarini M, Patel AH, Snell L, Nebbia G, Swanson C, Neil SJD. 2021. The P681H mutation in the Spike glycoprotein confers type I interferon resistance in the SARS-CoV-2 alpha (B.1.1.7) variant. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.11.09.467693v1. [DOI] [PMC free article] [PubMed]
- 80.Wang Q, Anang S, Iketani S, Guo Y, Liu L, Ho DD, Sodroski JG. 2021. Functional properties of the spike glycoprotein of the emerging SARS-CoV-2 variant B.1.1.529. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.27.474288v1. [DOI] [PMC free article] [PubMed]
- 81.Zaccaria M, Genovese L, Farzan M, Dawson W, Nakajima T, Johnson W, Momeni B. 2021. Anticipating future SARS-CoV-2 variants of concern through ab initio quantum mechanical modeling. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.11.25.470044v1.
- 82.Mannar D, Saville JW, Zhu X, Srivastava SS, Berezuk AM, Tuttle K, Marquez C, Sekirov I, Subramaniam S. 2021. SARS-CoV-2 Omicron variant: ACE2 binding, cryo-EM structure of Spike protein-ACE2 complex and antibody evasion. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.19.473380v1. [DOI] [PMC free article] [PubMed]
- 83.Rath SL, Padhi AK, Mandal N. 2022. Scanning the RBD-ACE2 molecular interactions in Omicron variant. Biochem Biophys Res Commun 592:18–23. 10.1016/j.bbrc.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu L, Zhou L, Mo M, Liu T, Wu C, Gong C, Lu K, Gong L, Zhu W, Xu Z. 2022. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct Target Ther 7:8. 10.1038/s41392-021-00863-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu L, Iketani S, Guo Y, Chan JF-W, Wang M, Liu L, Luo Y, Chu H, Huang Y, Nair MS, Yu J, Chik KK-H, Yuen TT-T, Yoon C, To KK-W, Chen H, Yin MT, Sobieszczyk ME, Huang Y, Wang HH, Sheng Z, Yuen K-Y, Ho DD. 2021. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.14.472719v2. [DOI] [PubMed]
- 86.VanBlargan LA, Errico JM, Halfmann P, Zost SJ, Crowe JE, Purcell LA, Kawaoka Y, Corti D, Fremont DH, Diamond M. 2021. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.15.472828v1. [DOI] [PMC free article] [PubMed]
- 87.Cameroni E, Saliba C, Bowen JE, Rosen LE, Culap K, Pinto D, Marco AD, Zepeda SK, di Iulio J, Zatta F, Kaiser H, Noack J, Farhat N, Czudnochowski N, Havenar-Daughton C, Sprouse KR, Dillen JR, Powell AE, Chen A, Maher C, Yin L, Sun D, Soriaga L, Gustafsson C, Franko NM, Logue J, Iqbal NT, Mazzitelli I, Geffner J, Grifantini R, Chu H, Gori A, Riva A, Giannini O, Ceschi A, Ferrari P, Franzetti-Pellanda A, Garzoni C, Hebner C, Purcell LA, Piccoli L, Pizzuto MS, Walls AC, Telenti A, Virgin HW, Lanzavecchia A, Veesler D, Snell G, Corti D. 2021. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.12.472269v1. [DOI] [PMC free article] [PubMed]
- 88.Aggarwal A, Stella AO, Walker G, Akerman A, Milogiannakis V, Brilot F, Amatayakul-Chantler S, Roth N, Coppola G, Schofield P, Jackson J, Henry JY, Mazigi O, Langley D, Lu Y, Forster C, McAllery S, Mathivanan V, Fichter C, Hoppe AC, Munier ML, Jack H-M, Cromer D, Darley D, Matthews G, Christ D, Khoury D, Davenport M, Rawlinson W, Kelleher AD, Turville S. 2021. SARS-CoV-2 Omicron: evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.14.21267772v1. [DOI] [PMC free article] [PubMed]
- 89.Xie X, Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, Li T, Yu Y, Wang Y, Wang J, Niu X, Wang P, An R, Liang H, Sun H, Yang S, Cui Q, Liu S, Du S, Zhang Z, Shao F, Huang W, Xiao J, Wang Y, Wang X, Yang X, Li Q, Hao X, Ronghua J. 2021. B.1.1.529 escapes the majority of SARS-CoV-2 neutralizing antibodies of diverse epitopes. Res Sq. https://www.researchsquare.com/article/rs-1148985/v1.
- 90.Schmidt F, Weisblum Y, Rutkowska M, Poston D, DaSilva J, Zhang F, Bednarski E, Cho A, Schaefer-Babajew DJ, Gaebler C, Caskey M, Nussenzweig MC, Hatziioannou T, Bieniasz PD. 2021. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature 600:512–516. 10.1038/s41586-021-04005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vangeel L, Chiu W, Jonghe SD, Maes P, Slechten B, Raymenants J, André E, Leyssen P, Neyts J, Jochmans D. 2022. Remdesivir, molnupiravir and nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.27.474275v2. [DOI] [PMC free article] [PubMed]
- 92.Monteil V, Stephanie D, Klingström J, Thålin C, Kellner MJ, Christ W, Havervall S, Mereiter S, Knapp S, Montserrat N, Braunsfeld B, Kozieradzki I, Ali OH, Hagelkruys A, Stadlmann J, Oostenbrink C, Wirnsberger G, Penninger JM, Mirazimi A. 2021. Clinical grade ACE2 effectively inhibits SARS-CoV-2 Omicron infections. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.12.25.474113v1.
- 93.Bojkova D, Widera M, Ciesek S, Wass MN, Michaelis M, Cinatl J. 2022. Reduced interferon antagonism but similar drug sensitivity in Omicron variant compared to Delta variant SARS-CoV-2 isolates. bioRxiv. https://www.biorxiv.org/content/10.1101/2022.01.03.474773v1. [DOI] [PMC free article] [PubMed]
- 94.Zhao H, Lu L, Peng Z, Chen L-L, Meng X, Zhang C, Ip JD, Chan W-M, Chu AW-H, Chan K-H, Jin D-Y, Chen H, Yuen K-Y, To KK-W. 2022. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect 11:277–218. 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Willett BJ, Grove J, MacLean OA, Wilkie C, Logan N, Lorenzo GD, Furnon W, Scott S, Manali M, Szemiel A, Ashraf S, Vink E, Harvey W, Davis C, Orton R, Hughes J, Holland P, Silva V, Pascall D, Puxty K, da Filipe AS, Yebra G, Shaaban S, Holden MTG, Pinto RM, Gunson R, Templeton K, Murcia P, Patel AH, The COVID-19 DeplOyed VaccinE (DOVE) Cohort Study investigators, The COVID-19 Genomics UK (COG-UK) Consortium, The G2P-UK National Virology Consortium, The Evaluation of Variants Affecting Deployed COVID-19 Vaccines (EVADE) investigators, Haughney J, Robertson DL, Palmarini M, Ray S, Thomson EC. 2022. The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. medRxiv. https://www.medrxiv.org/content/10.1101/2022.01.03.21268111v1.
- 96.Elbe S, Buckland-Merrett G. 2017. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall 1:33–46. 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hatcher EL, Zhdanov SA, Bao Y, Blinkova O, Nawrocki EP, Ostapchuck Y, Schäffer AA, Brister JR. 2017. Virus Variation Resource - improved response to emergent viral outbreaks. Nucleic Acids Res 45:D482–D490. 10.1093/nar/gkw1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tzou PL, Tao K, Nouhin J, Rhee S-Y, Hu BD, Pai S, Parkin N, Shafer RW. 2020. Coronavirus Antiviral Research Database (CoV-RDB): an online database designed to facilitate comparisons between candidate anti-coronavirus compounds. Viruses 12:1006. 10.3390/v12091006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zech F, Schniertshauer D, Jung C, Herrmann A, Cordsmeier A, Xie Q, Nchioua R, Prelli Bozzo C, Volcic M, Koepke L, Müller JA, Krüger J, Heller S, Stenger S, Hoffmann M, Pöhlmann S, Kleger A, Jacob T, Conzelmann K-K, Ensser A, Sparrer KMJ, Kirchhoff F. 2021. Spike residue 403 affects binding of coronavirus spikes to human ACE2. Nat Commun 12:6855. 10.1038/s41467-021-27180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. 2020. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 588:327–330. 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]