ABSTRACT
Infectious bursal disease virus (IBDV), which targets bursa B lymphocytes, causes severe immunosuppressive disease in chickens, inducing huge economic losses for the poultry industry. To date, the functional receptor for IBDV binding and entry into host cells remains unclear. This study used mass spectrometry to screen host proteins of chicken bursal lymphocytes interacting with VP2. The chicken transmembrane protein cluster of differentiation 44 (chCD44) was identified and evaluated for its interaction with IBDV VP2, the major capsid protein. Overexpression and knockdown experiments showed that chCD44 promotes replication of IBDV. Furthermore, soluble chCD44 and the anti-chCD44 antibody blocked virus binding. The results of receptor reconstitution indicated that chCD44 overexpression conferred viral binding capability in nonpermissive cells. More important, although we found that IBDV could not replicate in the chCD44-overexpressed nonpermissive cells, the virus could enter nonpermissive cells using chCD44. Our finding reveals that chCD44 is a cellular receptor for IBDV, facilitating virus binding and entry in target cells by interacting with the IBDV VP2 protein.
IMPORTANCE Infectious bursal disease virus (IBDV) causes severe immunosuppressive disease in chickens, inducing huge economic losses for the poultry industry. However, the specific mechanism of IBDV invading host cells of IBDV was not very clear. This study shed light on which cellular protein component IBDV is used to bind and/or enter B lymphocytes. The results of our study revealed that chCD44 could promote both the binding and entry ability of IBDV in B lymphocytes, acting as a cellular receptor for IBDV. Besides, this is the first report about chicken CD44 function in viral replication. Our study impacts the understanding of the IBDV binding and entry process and sets the stage for further elucidation of the infection mechanism of IBDV.
KEYWORDS: IBDV, chicken CD44, cellular receptor, virus binding, virus entry
INTRODUCTION
Infectious bursal disease virus (IBDV), a double-stranded RNA virus, belongs to the genus Avibirnavirus of the Birnaviridae family, a causative agent of infectious bursal disease in young chickens-especially 3∼8-week-old chickens (1). The most important pathological manifestation in IBDV infection is bursa of Fabricius (BF) destruction and immunosuppression, leading to the death of young chickens, the failure of vaccination, and the decline of egg production (2). With the emergence of virus mutants, IBDV has become an important pathogen, causing heavy economic losses in the poultry industry (3).
Viruses rely on hosts to complete their life cycle and bind specific receptors to enter the host cells (4, 5). Studying viral receptors is vital to understanding infection mechanisms and developing antiviral agents. For instance, ACE2 has been identified as a major entry receptor for SARS-CoV-2 and has been used as a target for the development of therapeutic antibodies (6), antiviral drugs (7), and vaccines (8) against this virus. The target cell of IBDV is immature B lymphoid cells in the BF (9, 10). However, IBDV can also replicate in macrophages (11), monocytes (12), and natural killer cells (13). Furthermore, the DT40 chicken B lymphoid cell line, which was originated from the bursa, can be persistently infected by IBDV (14). Different virulent IBDV strains have also been shown to complete their entire life cycle in DT40 cell lines, such as very virulent IBDV (vvIBDV), cell-adapted IBDV (caIBDV), and novel virulent IBDV. That said, vvIBDV could not replicate in DF1 cells, the most common cells in caIBDV studies (15).
As a nonenveloped virus, the outermost part of IBDV is the primary capsid protein VP2 (16, 17), responsible for cell–host interaction and cell tropism determination (15, 18). IBDV is a single-layer capsid virus, and VP2 is the only component of the virus capsid. Therefore, VP2 is responsible for cellular receptor binding (19, 20). Previous studies revealed that the expression of the mature form of VP2 could be assembled into subviral particles (SVPs), which could form the 12 pentamers found within the IBDV particle (17, 21). The spatial structure of VP2 comprises the following parts: the base domain (B domain), the shell domain (S domain), and the projection domain (P domain) (17, 22). The P domain (aa 206–350, hypervariable region) plays a pivotal role in interacting with receptor components, viral virulence determination, and neutralizing antibody epitope recognition (23, 24).
In previous studies of the IBDV receptor, some membrane proteins, such as cHsp90α (25), α4β1 integrin (19), surface IgM (26, 27), CD74 isoform Ii-2 (28), and Hsc70 (29) were confirmed to interact with IBDV VP2 and promote virus binding to DF-1 or DT40 cells. Regardless, neither the functional receptor nor the constituents of the complete IBDV receptor complex have been clarified. In this study, IBDV strains with different virulence were used to infect B lymphocytes, after which the primary bursal lymphocytes of chickens were screened for the putative receptor interacting with the capsid protein VP2. This study sheds light on the underlying mechanism that IBDV uses cellular protein components to bind and/or enter B lymphocytes via specific overexpression and knockdown experiments.
RESULTS
Screening and identification for host protein of bursal lymphocytes interacting with IBDV VP2.
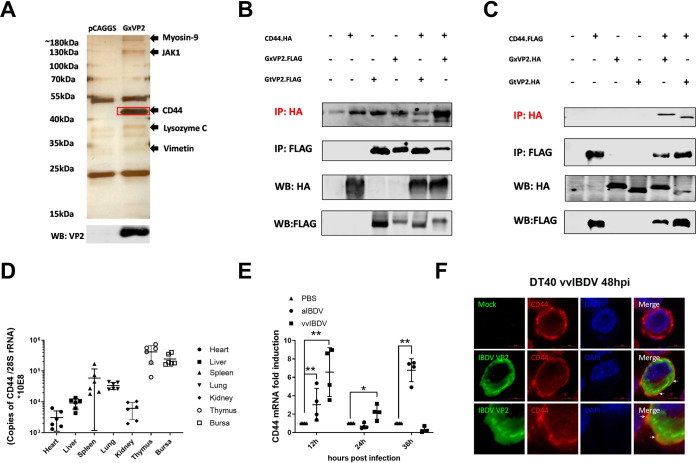
VP2, as the primary capsid protein of IBDV, is recognized as the bridge between IBDV and cellular receptors (29). To identify putative receptor components, we screened the bursal lymphocytes protein which interacted with IBDV VP2 by mass spectrometry (MS) analysis. Figure 1A indicates the different sizes of VP2 interacting proteins shown in the silver stain bands (different bands from pCAGGS as the empty vector transfection group). The results showed that a series of VP2 protein interactions were screened, including myosin-9, JAK1, the cluster of differentiation 44 (CD44), lysozyme C, and vimentin. After excluding the nontransmembrane protein, chicken CD44 (chCD44) was selected for the follow-up experiments. As a transmembrane glycoprotein, chCD44 was located in the most obvious band of ∼43 kDa.
FIG 1.
Screening and identification for host protein of bursal lymphocytes interacting with IBDV VP2. (A) Possible VP2-interacting proteins were identified and listed by LC-MS analysis, including chicken CD44 (chCD44) at ∼43 kDa. (B-C) Co-immunoprecipitation (Co-IP) results indicated the interaction between chCD44 extracellular domain and capsid protein VP2. Western blot analysis using anti-HA-tag antibody shows the bands corresponding to the chCD44 extracellular domain (B). Western blot analysis using the antibody against the HA-tag shows the bands corresponding to the vvIBDV (Gx strain) and aIBDV (Gt strain) VP2 (C). (D) ChCD44 transcription levels in organs (Heart, liver, spleen, lung, kidney, thymus, and the bursa of Fabricius) of 3-week-old SPF chickens. (E) Transcription level of chCD44 in SPF chicken bursa at different time points after vvIBDV or aIBDV infection. ChCD44 mRNA levels were significantly induced by vvIBDV at 12 and 24 hpi and aIBDV at 12 and 36 hpi (P < 0.05), compared with the mock-infected group. Data represent means ± standard deviations (n = 3). (F) Colocalization between VP2 and chCD44 in vvIBDV infected DT40 cells. DT40 cells were infected by vvIBDV (mock infection group as negative control) and conducted the confocal analysis. The results indicated that chCD44 (red fluorescence) was co-localized with IBDV VP2 (green fluorescence, lower panel), while VP2 (green fluorescence) was not detected in the mock-infected groups (upper panel). White arrows were pointing to the co-localtion between chCD44 and VP2.
Co-immunoprecipitation (Co-IP) assay was used to detect the interaction between VP2 and the extracellular domain of chCD44 (aa 1∼299). The 293T cells were transfected with chCD44-flag (or chCD44-HA) and VP2-HA (or VP2-flag) of the vvIBDV Gx/aIBDV Gt viral strains. Co-IP results showed that the bands corresponded to the chCD44 extracellular domain (Fig. 1B) or VP2 (Fig. 1C) in the flag Co-IP assay. The above results revealed that CD44 could interact with IBDV VP2.
The following organs of specific pathogen-free (SPF) chickens, including heart, liver, spleen, lung, kidney, thymus, and bursa, were detected for transcription levels of chCD44 to explore chCD44's organ distribution in vivo. As shown in Fig. 1D, chCD44 expression was most abundant in the thymus and BF lymphoid organs, compared with the heart, liver, spleen, lung, and kidney. CD44 transcription level in the spleen was lower than the first two organs yet higher than the other four organs (heart, liver, lung, and kidney).
Then vvIBDV Gx strain, attenuated IBDV (aIBDV) Gt strain, or PBS was infected to SPF chickens intranasally to detect the chCD44 transcription level in target organs after infection. The target organ (BF) samples for IBDV were collected at 12, 24, and 36 h postinfection (pi) and detected by RT-qPCR assays for chCD44 transcription. ChCD44 mRNA levels were significantly induced by vvIBDV at 12 and 24 hpi, and aIBDV at 12 and 36 hpi (P <0.05), compared with the mock-infected group (Fig. 1E).
DT40 cells were infected by vvIBDV to confirm the endogenous interaction between CD44 and VP2, and mock infection groups were added as negative control (Fig. 1F). After 48 h of infection, cells were conducted for confocal analysis to detect the co-localization between VP2 and CD44. The results indicated that chCD44 (red fluorescence) was co-localized with VP2 (green fluorescence, Fig. 1F, lower panel), while VP2 (green fluorescence) was not detected in the mock-infected groups (Fig. 1F, upper panel).
Role of chicken CD44 in IBDV replication.
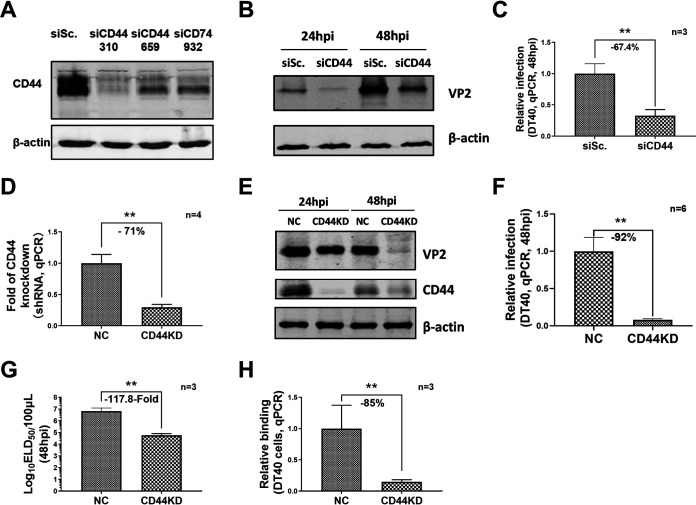
Western blot, RT-qPCR, and ELD50 analysis were performed to detect vvIBDV replication after up or downregulation of chCD44 expression in DT40 cells to identify whether chCD44 participated in IBDV replication. In the chCD44 knockdown experiment, chCD44 was effectively downregulated by CD44 siRNA 310 (Fig. 2A). DT40 cells were transfected with chCD44 siRNA 310 (siRNA control as the negative control) and then infected by 1 multiplicity of infection (MOI) vvIBDV Gx strain after 24 h of transfection. The Western blot results showed that the expression of VP2 in the chCD44 knockdown group was declining between 24∼48 hpi (Fig. 2B). Besides, the RT-qPCR results revealed that chCD44 knockdown could reduce the replication level of IBDV at 48 hpi (–67.4%, P < 0.05, Fig. 2C).
FIG 2.
ChCD44 knockdown suppressed IBDV replication. Knockdown efficiency of chCD44 and its effect on vvIBDV replication in DT40 cells. (A) Knockdown efficiency detection for chCD44 siRNA interference by Western blotting. The siCD44 310 has the best knockdown efficiency compared to other siCD44 groups. (B-C) ChCD44 was knockdown by siRNA interference and infected by 1 MOI vvIBDV infection after 24 h of transfection. The Western blot results indicate that the IBDV VP2 expression was downregulated at 24∼48 hpi in the chCD44 siRNA interference groups, compared with siRNA negative control (B). RT-qPCR analysis shows that IBDV copy number decreases significantly at 48 hpi (67.4% decrease compared with siRNA negative control, P < 0.05; C). (D) ChCD44 shRNA was used to establish a chCD44-knockdown DT40 cell line. RT-qPCR results indicated that chCD44 knockdown was effective. (E-H) 1 MOI vvIBDV infection in CD44KD and shRNA negative control cell lines. Western blot shows the VP2 expression of vvIBDV in CD44KD cell lines declined at 48 hpi (E). RT-qPCR analysis revealed that IBDV copy number decreased at 48 hpi (92% decrease compared with shRNA negative control, P < 0.05; F). ELD50 results indicated that IBDV titer was downregulated at 48 hpi compared with shRNA negative control (117.8-fold decrease, P < 0.05; G). The binding capability of vvIBDV to CD44KD and shRNA control cell lines. The virus binding capability of CD44KD cell lines was significantly lower than shRNA negative control (85% decrease compared with NC, P < 0.05; H). Data represent means ± standard deviations (n = 3).
To further verify the effect of chCD44 on virus replication, the chCD44 knockdown cell line (based on shRNA for chCD44) was constructed by red-fluorescent protein (RFP)-tagged lentivirus infection and fluorescent-labeled cell sorting. The results of RT-qPCR (71% downregulated, Fig. 2D) and Western blot (Fig. 2E) indicated that the levels of chCD44 of the knockdown group were significantly lower than that in the shRNA control group. The chCD44 knockdown cell lines and shRNA control cell lines (negative control, the same below) were infected by 1 MOI vvIBDV. As shown in Fig. 2E, Western blot indicated that VP2 expression declined between 24∼48 hpi compared with the infected shRNA control. Consistent with chCD44 expression, RT-qPCR analysis revealed that the IBDV VP5 copy numbers decreased between 48 hpi (92% decrease, P < 0.05, Fig. 2F). Furthermore, the titer of IBDV was downregulated in chCD44 knockdown cell lines at 48 hpi in the ELD50 assay (117.8-fold decrease, P < 0.05, Fig. 2G) compared with the shRNA control group. These results indicated that the replication level of IBDV decreased significantly in chCD44 knockdown cell lines. To further confirm the effect of chCD44 on virus binding to DT40 cells, chCD44 knockdown and shRNA control cell lines were incubated with vvIBDV, and the relative binding ratio of IBDV was detected by RT-qPCR analysis. The IBDV binding capability was downregulated by 85% in the chCD44 knockdown group, compared with the shRNA control (Fig. 2H and P < 0.05).
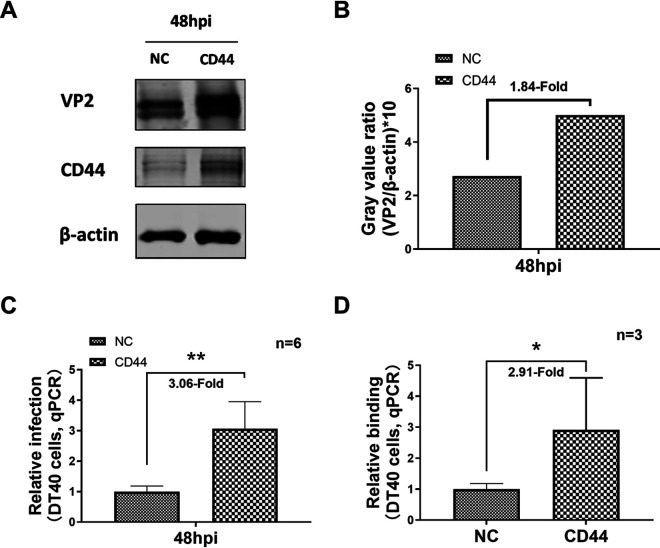
To further evaluate the function of chCD44 in IBDV replication, chCD44-overexpressed and nonoverexpressed (as negative control) DT40 cell lines were constructed by green-fluorescent protein (GFP)-tagged lentivirus infection and fluorescent-labeled cell sorting. Western blot showed that chCD44 expression was significantly increased in the chCD44-overexpressed cell line (Fig. 3A). ChCD44-overexpressed and control cell lines were infected by vvIBDV (MOI = 1), and the cell samples were collected at 48 hpi. Western blot results showed that IBDV VP2 protein levels were upregulated at 48 hpi, compared to the nonoverexpressed cell line (Fig. 3A–B). As expected, RT-qPCR analysis indicated that the IBDV replication level was increased in the chCD44-overexpressed group compared with the nonoverexpressed cell line at 48 hpi (3.06-fold increase, P < 0.05, Fig. 3C). ChCD44-overexpressed DT40 cell lines were co-treated with vvIBDV to confirm whether chCD44 overexpression promotes virus binding to DT40 cells. The binding capability of IBDV was upregulated in the chCD44-overexpressed cells by 2.91-fold (P < 0.05), compared to the nonoverexpressed cell line (Fig. 3D).
FIG 3.
CD44 overexpression promotes IBDV replication. (A–C) The IBDV VP2 expression was upregulated at 48 hpi in the overexpressed group compared with the nonoverexpressed group (A). Gray value ratio of VP2/β-actin for Fig. 2A (B). The viral load of IBDV was increased in the chCD44-overexpressed cell lines at 48 hpi than the nonoverexpressed group by RT-qPCR analysis (P < 0.05) (C). (D) 50 MOI vvIBDV was added to the CD44 overexpression cell lines and incubated for 1 h at 4°C, and the binding capability of vvIBDV to CD44-overexpressed cell lines were detected by RT-qPCR assay. IBDV copy number was increased in the overexpressed cell lines than the nonoverexpressed group by RT-qPCR analysis (P < 0.05).
Altogether, these results demonstrated that the knockdown of chCD44 could significantly inhibit vvIBDV replication, while chCD44 overexpression enhanced the replication of vvIBDV in DT40 cells.
A soluble form of chCD44 extracellular domain and chCD44 antibody reduced IBDV binding capability to DT40 cells.
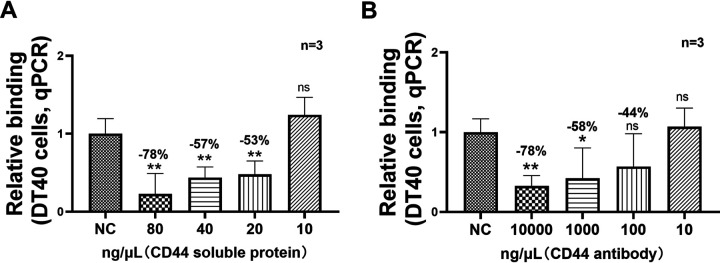
To further confirm whether chCD44 plays a pivotal role in the early replication process for IBDV, a blocking assay was conducted on DT40 cells in the following study. The viability of DT40 cells was not affected in the detected concentrations of the blockade assay (data not shown). In the protein blocking assay, eukaryotic expressed his-tagged chCD44 protein (extracellular domain) was respectively incubated with vvIBDV. RT-qPCR results indicated that viral binding was inhibited by 20, 40, and 80 ng/μL protein, and the blocking percentage ranged between 53 and 78% in a concentration-dependent manner (Fig. 4A and P < 0.05).
FIG 4.
A soluble form of CD44 extracellular domain and CD44 antibody reduced IBDV binding capability to DT40 cells. (A) In the CD44 soluble protein blocking assay, soluble CD44 protein (20, 40, or 80 ng/μL) was able to inhibit the binding capability of IBDV (blocking percentage range: 53–78%, P < 0.05). (B) In the antibody blocking assay, RT-qPCR analysis indicates a decrease in the binding ability of IBDV at the dilution of 102, 103, and 104 ng/μL in a concentration-dependent manner (blocking percentage range: 44–78%). Data represent means ± standard deviations (n = 3).
In the antibody blocking assay, chCD44 rabbit polyclonal antibody or rabbit IgG (negative control) were used, and RT-qPCR results confirmed that viral binding was decreased at the dilutions of 102, 103, and 104 ng/μL in a concentration-dependent manner. The antibody blocking percentage was ranged between 44 and 78% (Fig. 4B and P < 0.05). The above results showed that the CD44 soluble protein and CD44 polyclonal antibody could effectively inhibit IBDV binding to the target B lymphocytes.
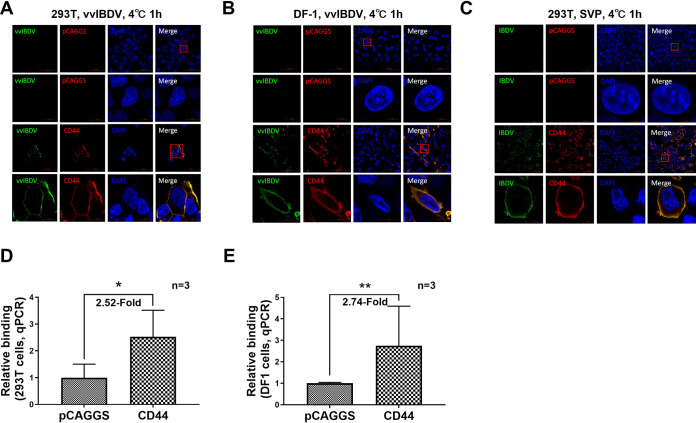
ChCD44 confers the binding ability to vvIBDV nonpermissive cell lines.
To further confirm if chCD44 could confer binding ability in nonpermissive cell lines, 293T and DF1 cells (both nonpermissive cells for vvIBDV) were used in the following experiments. 293T and DF1 cells were transfected with either HA-tagged chCD44 or pCAGGS (as negative control) before binding with vvIBDV. The confocal results indicated no virus was detected in 293T (Fig. 5A, upper panel) or DF1 cells (Fig. 5B, upper panel). In the chCD44 overexpression group, chCD44 (red fluorescence) could co-localize with IBDV (green fluorescence) both on the cell membrane of 293T cells (Fig. 5A, lower panel) and DF1 cells (Fig. 5B, lower panel). Consistent with confocal results, RT-qPCR analysis showed that chCD44 could also promote vvIBDV binding to nonpermissive cells, with a 2.52-fold increase in 293T cells (Fig. 5D) and a 2.74-fold increase in DF1 cells (Fig. 5E; both P < 0.05), compared with negative control groups.
FIG 5.
ChCD44 confers the binding ability to vvIBDV nonpermissive cell lines. (A–B) 293T (A) and DF-1 cells (B) (both nonpermissive for vvIBDV) were overexpressed by full-length CD44 protein or pCAGGS empty vector (as negative control) before binding with vvIBDV. To ensure the integrity of the cell membrane and facilitate the observation of virus localization on the cell membrane, cells were processed without membrane permeabilization for confocal analysis. No virus (green fluorescence) was detected in the empty vector control (upper panel). In the CD44 overexpression group, CD44 (red fluorescence) on the cell membrane was co-localized with the IBDV (green fluorescence, lower panel). (C) ChCD44 or the empty vector-transfected cells were incubated with SVPs from vvIBDV VP2. No binding was observed in the empty vector group (upper panel). CD44 (red fluorescence) was co-localized with SVPs (green fluorescence) on the cell membrane (lower panel). (D-E) RT-qPCR analysis revealed that CD44 overexpression promoted the binding capability of IBDV to 293T (D) or DF-1 (E) cells compared with the negative control group (2.52-fold increase in 293T cells and 2.74-fold increase in DF-1 cells, both P < 0.05). Data represent means ± standard deviations (n = 3).
To further confirm whether IBDV binds to chCD44 on the cell membrane via the viral VP2 protein, SVPs from the vvIBDV Gx strain VP2 expressed in Pichia pastoris (21) were used in the following experiments: 293T cells were transfected by full-length HA-chCD44 or pCAGGS empty vector (as negative control), then attached with SVPs. The results demonstrated that SVPs (green fluorescence) were co-localized with chCD44-overexpressing (red fluorescence) on the nonpermissive cell membranes but were not co-localized to negative control cells (Fig. 5C), indicating that IBDV binds to the chCD44-overexpressing nonpermissive cells via the primary capsid protein VP2.
The results revealed that chCD44 could confer binding ability to vvIBDV nonpermissive cell lines via interaction with IBDV VP2.
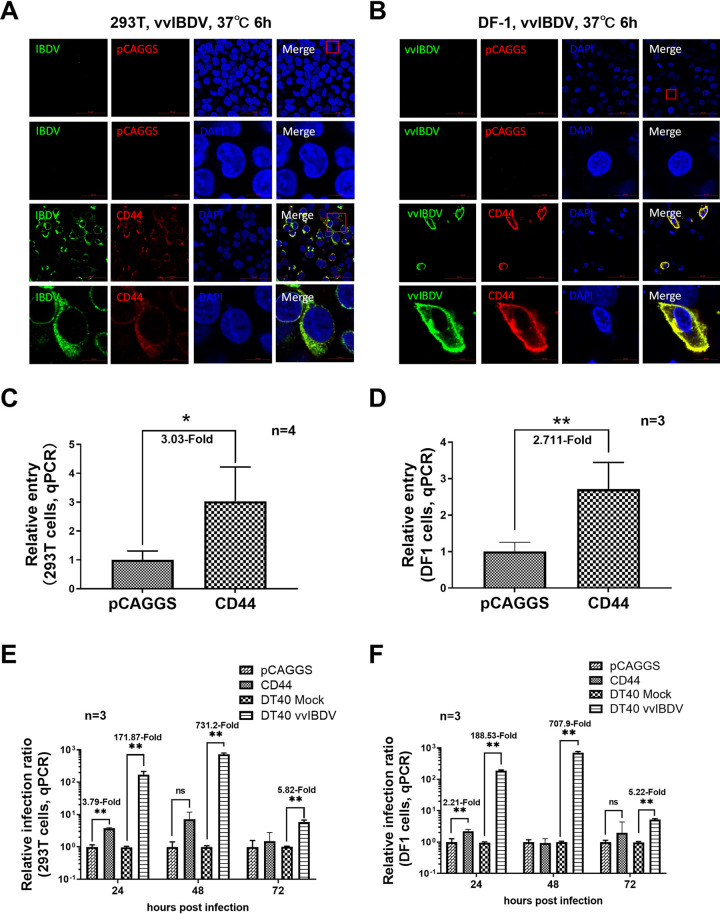
ChCD44 confers entry ability to vvIBDV nonpermissive cell lines.
To further detect whether chCD44 could confer entry ability of vvIBDV in the nonpermissive cell lines, 293T and DF-1 cells were transfected with HA-tagged full-length chCD44 plasmid (or the empty vector as negative control). Cells were incubated with vvIBDV after 24 h of transfection and processed using confocal and RT-qPCR analysis. The results indicated that chCD44 (red fluorescence) was located both on the cell membrane and cytoplasm after membrane permeation treatment and co-localized with IBDV (green fluorescence; Fig. 6A and B, lower panel), while no virus (green fluorescence) was detected in the negative control groups (Fig. 6A and B, upper panel). RT-qPCR results indicated that the entry of vvIBDV into CD44-overexpressed nonpermissive cells was upregulated by 3.03-fold in 293T cells (Fig. 6C) and 3.03-fold in DF-1 cells (Fig. 6D), compared with the negative control group (normalized to 1), which is consistent with the confocal results.
FIG 6.
ChCD44 confers entry ability to vvIBDV nonpermissive cell lines. (A-B) ChCD44 confers entry ability to vvIBDV nonpermissive cell lines (confocal assay). 293T (A) and DF-1 cells (B) were overexpressed by full-length CD44 protein before incubating with 50 MOI IBDV at 37°C for 6 h. Cells were processed with membrane permeabilization for confocal analysis. In the CD44 overexpression group, CD44 (red fluorescence) was able to co-localize with IBDV (green fluorescence, lower panel) in the cell plasmid. No virus (green fluorescence) was detected in the empty vector control (upper panel). (C–D) ChCD44 confers entry ability to vvIBDV nonpermissive cell lines (RT-qPCR assay). RT-qPCR results indicate that the entry of vvIBDV into CD44-overexpressed nonpermissive cells was upregulated by 3.03-fold in 293T cells (C) and 2.71-fold in DF-1 cells (D), compared with the empty vector-transfected control (normalized to 1), which is consistent with confocal results. (E–F) ChCD44 did not confer replication ability to vvIBDV nonpermissive cell lines (RT-qPCR). 293T and DF-1 cells were transfected by HA-tagged full-length chicken CD44 plasmid (or the empty vector as negative control). After 24 h transfection, cells were incubated with 50 MOI vvIBDV at 37°C for 24, 48, or 72 h (vvIBDV infected DT40 cells as positive control), then processed for RT-qPCR analysis. RT-qPCR analysis shows the viral load of vvIBDV was only upregulated significantly at 24 hpi, but not 48 and 72 hpi (P < 0.05, pCAGGS group were normalized to 1) in 293T (3.74-fold; E) and DF-1 cells (2.21-fold; F). The viral copies were significantly promoted at 24∼72 hpi in vvIBDV infected DT40 groups in both Fig. 6E and F. Data represent means ± standard deviations (n = 3).
To further detect whether chCD44 could confer replication ability to IBDV, 293T and DF-1 cells were transfected by HA-tagged full-length chCD44 plasmid (or the empty vector as negative control) and then incubated with vvIBDV. IBDV infected DT40 cells were conducted as the positive control. The RT-qPCR analysis revealed that the viral copies were only significantly increased in the overexpressed group at 24 hpi, both in the 293T (3.74-fold; Fig. 6E) and DF-1 cells (2.21-fold; Fig. 6F), but not significantly promoted at 48 and 72 hpi comparing with the negative control group. The viral copies were significantly promoted at 24∼72 hpi in vvIBDV infected DT40 groups in both Fig. 6E and F. The results revealed that overexpressed chCD44 in nonpermissive cells could not confer the replication ability of vvIBDV.
Altogether, the results above indicated that although chCD44 could not support IBDV replication in nonpermissive cells, it could confer entry ability to IBDV.
DISCUSSION
To find the cellular receptor of IBDV and further clarify the early replication mechanism of the virus, the interaction between the host proteins of bursal lymphocytes with the viral major capsid protein was investigated. Here, IBDV VP2 was used to screen for the host membrane proteins of bursal lymphocytes involved in IBDV-host interaction. The results confirmed that the transmembrane protein chCD44 interacted with VP2 and promoted IBDV replication in DT40 cells. Furthermore, chCD44 could confer virus binding and entry ability into nonpermissive cells, indicating that chCD44 is a cellular receptor of IBDV. As the cellular receptor of IBDV plays an important role in the occurrence of infectious diseases, this finding is significant for studying the IBDV infection mechanism.
Virus binding and entry into host cells are the first major steps of infection, mainly relying on the combination of the virus and cellular receptors (5, 30). Previous studies reported that chicken Hsp90 was identified on the cell membrane of DF-1 cells while anti-Hsp90 antibody could block the IBDV infection in DF-1 cells (25). Furthermore, α4β1 integrin was confirmed to interact with the VP2 IDA (Ile-Asp-Ala) motif in the virus binding process, acting as a binding receptor for IBDV in DF-1 cells (19). The sIgM λ light chain was also confirmed to promote IBDV binding and infection (26, 27). Our previous study demonstrated that the chicken CD74 isoform Ii-2 was a novel binding receptor for IBDV, which promotes IBDV binding to target cells via VP2 protein in DT40 cells (28). However, no successful receptor reconstruction experiments confer effective IBDV replication in nonpermissive cells. Due to the different cellular tropisms in different virulent IBDV strains, the identification of IBDV receptors has become more complex. Primary bursal lymphocytes were chosen to screen the putative IBDV receptor in our study as different virulent strains of IBDV could infect them. A series of interacting proteins were screened, and the transmembrane protein chCD44 was found to interact with primary capsid protein VP2 and significantly promote IBDV replication in target cells.
CD44 is a complex transmembrane glycoprotein, which is widely distributed and expressed by lymphocytes and nonlymphoid cells (31). This protein interacts with ligands and signaling receptors in the cell membrane and cell plasma, such as hyaluronan (32), c-Src kinase (33), collagen (34), fibronectin (35), and laminin (31), to act as a signaling hub. Human CD44 (huCD44) participated in physiological processes and pathological processes, such as tumorigenesis and viral infection (36). It therefore also plays a critical role in the pathogenic process of multiple viral diseases, such as HPV, HBV, HCV, and HIV-1, exacerbating the injury, promoting virus replication, or capturing viruses (32, 37–39). ChCD44 is a 396 aa protein (Table 1) with low homology with other species, with only 61.2% identity to human and chimpanzee CD44, 61.6% to monkey, 51.5% to wolf, and 54.3% to mouse. However, there is no relevant report on the function of chCD44. As the previous study reported, IBDV infection could upregulate the phosphorylation of c-Src to promote viral entry (33). HuCD44 has been reported to be coupled to c-Src kinase to phosphorylate c-Src (40, 41). With only 61.2% identity to huCD44, whether chCD44 has a similar effect as huCD44 to phosphorylate c-Src and promote IBDV entry is worth exploring. Furthermore, huCD74 could interact with huCD44 as a receptor complex for MIF, a pro-inflammatory factor (42). In our previous study, chCD74 was confirmed as a binding receptor for IBDV (28). However, whether chCD44 has a similar function to chCD74 or constitutes part of the receptor complex with chCD74 to promote IBDV infection needs further study.
TABLE 1.
Amino acid sequence for full-length chicken CD44a
aBlack font represents the extracellular domain; red font represents the transmembrane domain; blue font represents the intracellular domain.
To determine the role of chCD44 in early infection of IBDV, we detected both the binding and entry-stage process through receptor reconstitution experiments. The results showed that chCD44 could confer both binding and entry ability to vvIBDV in CD44-overexpressed 293T cells, indicating that vvIBDV might not hijack the host factors of human 293T cells for the post-entry process. To further explore whether the inability of IBDV to replicate in the 293T cells via chCD44 might be due to the homology difference between the human and chicken host proteins, we also chose the chicken DF-1 cell line (nonpermissive cells for vvIBDV) for receptor reconstitution assays. Unfortunately, vvIBDV could not complete the whole life cycle in the chCD44-overexpressed DF-1 cells. ChCD44 might only confer replicative ability to vvIBDV in primary B lymphocytes or B cell lines in vitro due to molecular chaperones specific to these cell lines. In this regard, the nonpermissive cells might lack these necessary molecular chaperones related to the post-entry process of vvIBDV, thus causing the viral particles to degrade without assembly or release. Besides, we detected an abundance of chCD44 in the lymphoid organs, which was consistent with the histotropism of IBDV (43).
At present, vvIBDV was confirmed to replicate only in the primary bursal lymphocytes and DT40 cells in vitro, but cell-adapted IBDV could replicate in more kinds of cells, such as chicken embryo fibroblasts (CEF), Vero and DF-1 cells. For these reasons, the VP2 protein of both very virulent IBDV (vvIBDV) and attenuated IBDV (aIBDV) were used in the co-ip experiment, and the results confirmed the interaction between different virulent of IBDV VP2 and chCD44 protein. There was no significant difference in the interactions between various IBDV strains and chCD44. In terms of the notable difference in cell tropism between vvIBDV and caIBDV, we speculate that caIBDV might use more kinds of receptors than vvIBDV for a wider cell tropism due to the amino acid difference of VP2. Chicken CD44 might not be the only receptor for caIBDV entry. The details still need to be verified by further study.
In summary, our study identified chCD44 from bursal lymphocytes as a cellular receptor for IBDV, revealing the novel function of chCD44 in the early stage of its viral replication.
MATERIALS AND METHODS
Cell cultures, viruses, proteins, and antibodies.
DT40 cells (a chicken lymphoma cell line) were generously gifted by Prof. Venugopal Nair of the Pirbright Institute and routinely maintained at 37°C in a 5% CO2 atmosphere in RPMI 1640 medium (Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS), 2% chicken serum (Gibco, Billings, MT, USA), 1% GlutaMAX (Gibco), and 50 μM β-mercaptoethanol (Gibco). 293T and DF1 cells were maintained in DMEM (Sigma) supplemented with 10% FBS (Gibco) in an atmosphere of 5% CO2 at 37°C. Primary bursal lymphocytes were isolated from bursas of 3-week-old chickens, using previously reported methods by Dulwich et al. (10), and maintained at 42°C in a 5% CO2 atmosphere.
The vvIBDV Gx strain was obtained and preserved in our laboratory, and the viral loads were detected by performing a 50% egg lethal dosage (ELD50) (44). The aIBDV Gt strain, also known as caIBDV, was preserved in our laboratory, and the viral loads were detected by median cell culture infective dose (TCID50) in DF1 cells (40, 44).
The SVPs originated from the vvIBDV Gx strain VP2 were expressed and preserved by our laboratory and detected by Western blotting, SDS-PAGE, as well as electronic microscopy experiments (21).
The IBDV VP2 monoclonal antibodies were stocked in our laboratory. The anti-β-actin, HA tag, Flag tag antibodies were purchased from Sigma. The custom C-terminal 6× His tag chCD44 protein and rabbit anti-chCD44 polyclonal antibody (targeting full-length chCD44, Table 1) was obtained from the GenScript Company (Nanjing, China).
Animals.
36 SPF chickens (3-week-old; purchased from National Poultry Laboratory Animal Resource Center) were infected with either 200 μL vvIBDV (1 × 102 ELD50), 200 μL aIBDV (1.05 × 103 TCID50, same virus copies with vvIBDV based on RT-qPCR detection), or PBS (pH 7.0) per chicken to establish a chicken infection model. The samples of organs (including heart, liver, spleen, lung, kidney, thymus, and bursa) were collected for RNA extraction and quantitative PCR experiments. This experiment was approved by the Committee on the Ethics of Animal Experiments at the Harbin Veterinary Research Institute (Harbin, China), Chinese Academy of Agricultural Sciences (animal-ethics committee approval number: Heilongjiang-SYNK-2017-009) and performed following the Guidelines for Experimental Animals of the Ministry of Science and Technology (Beijing, China). The chickens were cared for following humane procedures.
Protein precipitation and mass spectrometry.
Primary bursal cells were lysed, incubated with eukaryotic expressed GxVP2 protein, and the following procedure was as previously described (28). Differential bands between VP2 and the negative control (empty vector pCAGGS transfected groups) were detected by SDS-PAGE and silver staining (Thermo Fisher Scientific, Waltham, MA, USA) and subjected to MS. The subsequent MS analysis was completed by the Hoogen-bio Company (Shanghai, China).
Co-IP analysis.
293T cells were co-transfected with bait and prey plasmids (with Flag- or HA-tag) in 6-well plates using lipofectamine LTX (Invitrogen, Waltham, MA, USA). Then total protein was lysed from the transfected 293T cells by NP40 buffer (with protease inhibitor cocktail; Thermo Fisher Scientific) on ice after 48 h. According to the manufacturer's instructions, the co-IP analysis was performed by ANTI-FLAG M2 Affinity Gel (Sigma). Western blot subsequently detected proteins using monoclonal antibodies against Flag and HA (Sigma).
RNA extraction and quantitative PCR.
Specific primers and TaqMan probes for chicken 28S rRNA (41), IBDV viral load (45), and chCD44 (#4351372, Assay number: Gg03310495_m1; Thermo Fisher Scientific) were synthesized or purchased from Thermo Fisher Scientific. Total RNA was extracted using the RNeasy minikit (Qiagen, Hilden, Germany). As previously described, the RNA products were reverse transcribed to cDNA and detected by quantitative PCR (28). The absolute quantification method was used to analyze the quantification of chCD44 or the IBDV viral load. The measured IBDV or chCD44 were normalized to copies of 28S cDNA measured from the same samples, and then the negative control group was normalized to 1. All samples (including the standards, negative controls, and infected) were examined in triplicate on the same plate.
ELD50.
Supernatants of vvIBDV-infected DT40 cells were collected for ELD50 detection. Samples were diluted by PBS and then inoculated on the chorioallantoic membrane of 9-day-old SPF eggs (100-μL sample per egg, purchased from the Harbin Veterinary Research Institute). The mortality was observed daily for 7 dpi. The ELD50 value was determined by Reed and Muench method (46). All the chicken embryos were cared for following humane procedures.
RNA interference assays.
The following siRNAs we used to downregulate chCD44: Negative control, sense 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense 5′-ACGUGACACGUUCGGAGAATT-3′; CD44-gga-310, sense 5′-AUUUACAAACUUUCAGCAATT-3′, antisense 5′-UUGCUGAAAGUUUGTAAAUTT-3′, CD44-gga-659: sense 5′-AAGACUUUCCUGUGAAACAT-3′, antisense 5′-UGUUUCACAGGAAAGUCUUTT-3′; and CD44-gga-932, sense 5′-CAUUGAUCCUUGCCGUGUGTT-3′, antisense 5′-CACACGGCAAGGAUCA.
AUGTT-3′ (all synthesized by Gene Pharma, Shanghai, China). According to the manufacturer's instructions, cells (5 × 106) were electro-transfected with 100 pmol siRNA by the Amaxa Cell Line Nucleofector Kit T (Lonza, Basel, Switzerland). The RNA interference efficiency was verified 48 h after transfection by Western blotting assay.
At 24 h after transfection, DT40 cells were infected by 1 MOI vvIBDV at 42°C for 5 h, washed three times by PBS (pH 7.0), then DT40 complete medium was added. Cells were collected at 24 or 48 hpi for Western blot, 48 hpi for RT-qPCR, and ELD50 analysis to detect vvIBDV replication.
For the virus-binding assay, DT40 cells were pre-chilled at 4°C for 30 min and incubated with 50 MOI vvIBDV at 4°C for 1 h after 24 h transfection. Cells were washed five times with PBS (pH 7.0) and collected for RT-qPCR analysis to detect vvIBDV-binding capability.
Construction of chCD44 knockdown or overexpression cell line.
The lentivirus packaging for chCD44 knockdown and overexpression was performed by Gene Pharma. For the knockdown cell line, the shRNA sequence consisted of sense 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense 5′-UUGCUGAAAGUUUGTAAAUTT-3′. For the overexpression cell line, the full-length of chCD44 was overexpressed. Both cell lines were constructed according to the following protocol: DT40 cells were infected by lentivirus, and the infected cells were screened by RFP (knockdown) or GFP (overexpression) fluorescence-activated cell sorting (SONY-SH800S; Sony, Tokyo, Japan). The knockdown or overexpression efficiency of chCD44 was assessed by RT-qPCR and/or Western blot analysis.
Antibody and protein blocking assay of viral binding.
The protein blocking assay was used to investigate the inhibition of IBDV attachment by chCD44-his soluble protein. Different dilutions of chCD44-his protein (10, 20, 40, and 80 ng/μL) were incubated with 50 MOI vvIBDV for 3 h at 4°C and then added to chilled DT40 cells for 1 h at 4°C. Cells were then washed 5 times on ice and collected for RT-qPCR analysis.
Different dilutions of the anti-chCD44 antibodies (101, 102, 103, and 104 ng/μL) for the antibody blocking assay or 104 ng/μL rabbit IgG (negative control) were added prechilled DT40 cells for 3 h at 4°C. Cells were incubated with 50 MOI IBDV for 1 h at 4°C, washed 5 times on ice afterward, and collected for RT-qPCR to detect the IBDV-binding capability.
Confocal imaging.
To visualize the location of vvIBDV and chCD44, 293T and DF1 cells were transfected by full-length HA-chCD44, then transfected cells were incubated with 50 MOI vvIBDV or 200 μg SVPs at 4°C for 1 h (binding assay) or 37°C for 6 h (entry assay). Then cells were washed by PBS (PH 7.0) 5 times and fixed with 4% formaldehyde for 30 min. Membrane permeabilization treatment was conducted by 0.1% Triton X-100 for 5 min for the entry assay, but not the binding assay. Then, the cells were blocked by 5% (wt/vol) skim milk at 4°C overnight and incubated with HA-tag antibody and IBDV VP2 monoclonal antibody (1:200) in PBS at 37°C for 1 h and washed by PBS for three times. The cells were then incubated with the mixture of goat anti-rabbit IgG (H+L) cross-adsorbed secondary antibody and Alexa Fluor 546 (Invitrogen) (1:200) to visualize HA-tagged chCD44; goat anti-mouse IgG (H+L) cross-adsorbed secondary antibody and Alexa Fluor 488 (Invitrogen) (1:200) to visualize IBDV and SVP, and DAPI to visualize the cell nuclei at 37°C for 1 h, and then washed 5 times with PBS. Images were captured on an LSM980 confocal laser scanning microscope (Zeiss, Oberkochen, Germany) with Fast Airyscan using the ZEN2 software.
Statistical analysis.
All statistical analyses were performed with unpaired t-tests using Prism v9.0 (GraphPad, San Diego, CA, USA). P < 0.05 was considered statistically significant. The data were reported as means ± standard deviation.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 32102649), Heilongjiang Provincial Natural Science Foundation of China (grant no. TD2019C003), China's Agricultural Research System (grant no. CARS-41).
We declare no competing financial interests.
Contributor Information
Yulong Gao, Email: gaoyulong@caas.cn.
Susana López, Instituto de Biotecnologia/UNAM.
REFERENCES
- 1.Lasher HN, Davis VS. 1997. History of infectious bursal disease in the U.S.A.—the first two decades. Avian Dis 41:11–19. 10.2307/1592439. [DOI] [PubMed] [Google Scholar]
- 2.van den Berg TP, Eterradossi N, Toquin D, Meulemans G. 2000. Infectious bursal disease (Gumboro disease). Rev Sci Tech Oie 19:509–543. 10.20506/rst.19.2.1227. [DOI] [PubMed] [Google Scholar]
- 3.Muller H, Islam MR, Raue R. 2003. Research on infectious bursal disease–the past, the present and the future. Vet Microbiol 97:153–165. 10.1016/j.vetmic.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Arias CF, Silva-Ayala D, López S. 2015. Rotavirus entry: a deep journey into the cell with several exits. J Virol 89:890–893. 10.1128/JVI.01787-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arias CF, López S. 2021. Rotavirus cell entry: not so simple after all. Curr Opin Virol 48:42–48. 10.1016/j.coviro.2021.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Gattinger P, Niespodziana K, Stiasny K, Sahanic S, Tulaeva I, Borochova K, Dorofeeva Y, Schlederer T, Sonnweber T, Hofer G, Kiss R, Kratzer B, Trapin D, Tauber PA, Rottal A, Körmöczi U, Feichter M, Weber M, Focke-Tejkl M, Löffler-Ragg J, Mühl B, Kropfmüller A, Keller W, Stolz F, Henning R, Tancevski I, Puchhammer-Stöckl E, Pickl WF, Valenta R. 2022. Neutralization of SARS-CoV-2 requires antibodies against conformational receptor-binding domain epitopes. Allergy 77:230–242. 10.1111/all.15066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schütz D, Ruiz-Blanco YB, Münch J, Kirchhoff F, Sanchez-Garcia E, Müller JA. 2020. Peptide and peptide-based inhibitors of SARS-CoV-2 entry. Adv Drug Deliv Rev 167:47–65. 10.1016/j.addr.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, Nkolola JP, Liu J, Li Z, Chandrashekar A, Martinez DR, Loos C, Atyeo C, Fischinger S, Burke JS, Slein MD, Chen Y, Zuiani A, Lelis FJN, Travers M, Habibi S, Pessaint L, Van Ry A, Blade K, Brown R, Cook A, Finneyfrock B, Dodson A, Teow E, Velasco J, Zahn R, Wegmann F, Bondzie EA, Dagotto G, Gebre MS, He X, Jacob-Dolan C, Kirilova M, Kordana N, Lin Z, Maxfield LF, Nampanya F, Nityanandam R, Ventura JD, Wan H, Cai Y, Chen B, Schmidt AG, Wesemann DR, Baric RS, et al. 2020. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369:806–811. 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becht H, Muller H. 1991. Infectious bursal disease–B cell dependent immunodeficiency syndrome in chickens. Behring Inst Mitt 89:217–225. [PubMed] [Google Scholar]
- 10.Dulwich KL, Asfor AS, Gray AG, Nair V, Broadbent AJ. 2018. An ex vivo chicken primary bursal-cell culture model to study infectious bursal disease virus pathogenesis. J Vis Exp. 10.3791/58489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khatri M, Palmquist JM, Cha RM, Sharma JM. 2005. Infection and activation of bursal macrophages by virulent infectious bursal disease virus. Virus Res 113:44–50. 10.1016/j.virusres.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Inoue M, Yamamoto H, Matuo K, Hihara H. 1992. Susceptibility of chicken monocytic cell lines to infectious bursal disease virus. J Vet Med Sci 54:575–577. 10.1292/jvms.54.575. [DOI] [PubMed] [Google Scholar]
- 13.Jahromi MZ, Bello MB, Abdolmaleki M, Yeap SK, Hair-Bejo M, Omar AR. 2018. Differential activation of intraepithelial lymphocyte-natural killer cells in chickens infected with very virulent and vaccine strains of infectious bursal disease virus. Dev Comp Immunol 87:116–123. 10.1016/j.dci.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Delgui L, González D, Rodríguez JF. 2009. Infectious bursal disease virus persistently infects bursal B-lymphoid DT40 cells. J Gen Virol 90:1148–1152. 10.1099/vir.0.008870-0. [DOI] [PubMed] [Google Scholar]
- 15.Qi X, Gao H, Gao Y, Qin L, Wang Y, Gao L, Wang X. 2009. Naturally occurring mutations at residues 253 and 284 in VP2 contribute to the cell tropism and virulence of very virulent infectious bursal disease virus. Antiviral Res 84:225–233. 10.1016/j.antiviral.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Coulibaly F, Chevalier C, Gutsche I, Pous J, Navaza J, Bressanelli S, Delmas B, Rey FA. 2005. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120:761–772. 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Lee CC, Ko TP, Chou CC, Yoshimura M, Doong SR, Wang MY, Wang AH. 2006. Crystal structure of infectious bursal disease virus VP2 subviral particle at 2.6A resolution: implications in virion assembly and immunogenicity. J Struct Biol 155:74–86. 10.1016/j.jsb.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 18.van Loon AA, de Haas N, Zeyda I, Mundt E. 2002. Alteration of amino acids in VP2 of very virulent infectious bursal disease virus results in tissue culture adaptation and attenuation in chickens. J Gen Virol 83:121–129. 10.1099/0022-1317-83-1-121. [DOI] [PubMed] [Google Scholar]
- 19.Delgui L, Ona A, Gutierrez S, Luque D, Navarro A, Caston JR, Rodriguez JF. 2009. The capsid protein of infectious bursal disease virus contains a functional alpha 4 beta 1 integrin ligand motif. Virology 386:360–372. 10.1016/j.virol.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 20.Kibenge FS, Dhillon AS, Russell RG. 1988. Biochemistry and immunology of infectious bursal disease virus. J Gen Virol 69:1757–1775. 10.1099/0022-1317-69-8-1757. [DOI] [PubMed] [Google Scholar]
- 21.Wang M, Pan Q, Lu Z, Li K, Gao H, Qi X, Gao Y, Wang X. 2016. An optimized, highly efficient, self-assembled, subvirus-like particle of infectious bursal disease virus (IBDV). Vaccine 34:3508–3514. 10.1016/j.vaccine.2016.02.072. [DOI] [PubMed] [Google Scholar]
- 22.Garriga D, Querol-Audi J, Abaitua F, Saugar I, Pous J, Verdaguer N, Caston JR, Rodriguez JF. 2006. The 2.6-Angstrom structure of infectious bursal disease virus-derived T=1 particles reveals new stabilizing elements of the virus capsid. J Virol 80:6895–6905. 10.1128/JVI.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haywood AM. 1994. Virus receptors: binding, adhesion strengthening, and changes in viral structure. J Virol 68:1–5. 10.1128/jvi.68.1.1-5.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zárate S, Cuadras MA, Espinosa R, Romero P, Juárez KO, Camacho-Nuez M, Arias CF, López S. 2003. Interaction of rotaviruses with Hsc70 during cell entry is mediated by VP5. J Virol 77:7254–7260. 10.1128/JVI.77.13.7254-7260.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin TW, Lo CW, Lai SY, Fan RJ, Lo CJ, Chou YM, Thiruvengadam R, Wang AH, Wang MY. 2007. Chicken heat shock protein 90 is a component of the putative cellular receptor complex of infectious bursal disease virus. J Virol 81:8730–8741. 10.1128/JVI.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chi J, You L, Li P, Teng M, Zhang G, Luo J, Wang A. 2018. Surface IgM λ light chain is involved in the binding and infection of infectious bursal disease virus (IBDV) to DT40 cells. Virus Genes 54:236–245. 10.1007/s11262-018-1535-6. [DOI] [PubMed] [Google Scholar]
- 27.Luo J, Zhang H, Teng M, Fan JM, You LM, Xiao ZJ, Yi ML, Zhi YB, Li XW, Zhang GP. 2010. Surface IgM on DT40 cells may be a component of the putative receptor complex responsible for the binding of infectious bursal disease virus. Avian Pathol 39:359–365. 10.1080/03079457.2010.506211. [DOI] [PubMed] [Google Scholar]
- 28.Liu A, Pan Q, Li Y, Yan N, Wang J, Yang B, Chen Z, Qi X, Gao Y, Gao L, Liu C, Zhang Y, Cui H, Li K, Wang Y, Wang X. 2020. Identification of Chicken CD74 as a Novel Cellular Attachment Receptor for Infectious Bursal Disease Virus in Bursa B Lymphocytes. J Virol 94. 10.1128/JVI.01712-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C, Qin Y, Qian K, Shao H, Ye J, Qin A. 2020. HSC70 is required for infectious bursal disease virus (IBDV) infection in DF-1 cells. Virol J 17:65. 10.1186/s12985-020-01333-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez S, Arias CF. 2006. Early steps in rotavirus cell entry. Curr Top Microbiol Immunol 309:39–66. 10.1007/3-540-30773-7_2. [DOI] [PubMed] [Google Scholar]
- 31.Hibino S, Shibuya M, Engbring JA, Mochizuki M, Nomizu M, Kleinman HK. 2004. Identification of an active site on the laminin alpha5 chain globular domain that binds to CD44 and inhibits malignancy. Cancer Res 64:4810–4816. 10.1158/0008-5472.CAN-04-0129. [DOI] [PubMed] [Google Scholar]
- 32.Bourguignon LYW, Earle C, Shiina M. 2019. Hyaluronan-CD44 interaction promotes HPV 16 E6 oncogene-mediated oropharyngeal cell carcinoma survival and chemoresistance. Matrix Biol 78–79:180–200. 10.1016/j.matbio.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Bourguignon LY, Zhu H, Shao L, Chen YW. 2001. CD44 interaction with c-Src kinase promotes cortactin-mediated cytoskeleton function and hyaluronic acid-dependent ovarian tumor cell migration. J Biol Chem 276:7327–7336. 10.1074/jbc.M006498200. [DOI] [PubMed] [Google Scholar]
- 34.Govindaraju P, Todd L, Shetye S, Monslow J, Puré E. 2019. CD44-dependent inflammation, fibrogenesis, and collagenolysis regulates extracellular matrix remodeling and tensile strength during cutaneous wound healing. Matrix Biol 75–76:314–330. 10.1016/j.matbio.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jalkanen S, Jalkanen M. 1992. Lymphocyte CD44 binds the COOH-terminal heparin-binding domain of fibronectin. J Cell Biol 116:817–825. 10.1083/jcb.116.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heldin P, Kolliopoulos C, Lin CY, Heldin CH. 2020. Involvement of hyaluronan and CD44 in cancer and viral infections. Cell Signal 65:109427. 10.1016/j.cellsig.2019.109427. [DOI] [PubMed] [Google Scholar]
- 37.Iqbal J, Sarkar-Dutta M, McRae S, Ramachandran A, Kumar B, Waris G. 2018. Osteopontin Regulates Hepatitis C Virus (HCV) Replication and Assembly by Interacting with HCV Proteins and Lipid Droplets and by Binding to Receptors alphaVbeta3 and CD44. J Virol 92. 10.1128/JVI.02116-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murakami T, Kim J, Li Y, Green GE, Shikanov A, Ono A. 2018. Secondary lymphoid organ fibroblastic reticular cells mediate trans-infection of HIV-1 via CD44-hyaluronan interactions. Nat Commun 9:2436. 10.1038/s41467-018-04846-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng Y, Chen ZJ, Lan F, He QT, Chen SY, Du YF, Li S, Qin X. 2019. Association of CD44 polymorphisms and susceptibility to HBV-related hepatocellular carcinoma in the Chinese population. J Clin Lab Anal 33:e22977. 10.1002/jcla.22977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi X, Gao Y, Gao H, Deng X, Bu Z, Wang X, Fu C, Wang X. 2007. An improved method for infectious bursal disease virus rescue using RNA polymerase II system. J Virol Methods 142:81–88. 10.1016/j.jviromet.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 41.Ragland WL, Novak R, El-Attrache J, Savic V, Ester K. 2002. Chicken anemia virus and infectious bursal disease virus interfere with transcription of chicken IFN-alpha and IFN-gamma mRNA. J Interferon Cytokine Res 22:437–441. 10.1089/10799900252952226. [DOI] [PubMed] [Google Scholar]
- 42.Shi X, Leng L, Wang T, Wang W, Du X, Li J, McDonald C, Chen Z, Murphy JW, Lolis E, Noble P, Knudson W, Bucala R. 2006. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity 25:595–606. 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haynes BF, Telen MJ, Hale LP, Denning SM. 1989. CD44–a molecule involved in leukocyte adherence and T-cell activation. Immunol Today 10:423–428. 10.1016/0167-5699(89)90040-6. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, Zhang H, Gao H, Fu C, Gao Y, Ju Y. 2007. Changes in VP3 and VP5 genes during the attenuation of the very virulent infectious bursal disease virus strain Gx isolated in China. Virus Genes 34:67–73. 10.1007/s11262-006-0002-y. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Qi X, Gao H, Gao Y, Lin H, Song X, Pei L, Wang X. 2009. Comparative study of the replication of infectious bursal disease virus in DF-1 cell line and chicken embryo fibroblasts evaluated by a new real-time RT-PCR. J Virol Methods 157:205–210. 10.1016/j.jviromet.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]