ABSTRACT
REC8 meiotic recombination protein (REC8) is a member of structural maintenance of chromosome (SMC) protein partners, which play an important role in meiosis, antitumor activity, and sperm formation. As the adaptor proteins of RIG-I-like receptor (RLR) signaling and cyclic GMP-AMP synthase (cGAS)-DNA signaling, the activity and stability of MAVS (mitochondrial antiviral signaling protein; also known as VISA, Cardif, and IPS-1) and STING (stimulator of interferon genes; also known as MITA) are critical for innate immunity. Here, we report that REC8 interacts with MAVS and STING and inhibits their ubiquitination and subsequent degradation, thereby promoting innate antiviral signaling. REC8 is upregulated through the JAK-STAT signaling pathway during viral infection. Knockdown of REC8 impairs the innate immune responses against vesicular stomatitis virus (VSV), Newcastle disease virus (NDV), and herpes simplex virus (HSV). Mechanistically, during infection with viruses, the SUMOylated REC8 is transferred from the nucleus to the cytoplasm and then interacts with MAVS and STING to inhibit their K48-linked ubiquitination triggered by RNF5. Moreover, REC8 promotes the recruitment of TBK1 to MAVS and STING. Thus, REC8 functions as a positive modulator of innate immunity. Our work highlights a previously undocumented role of meiosis-associated protein REC8 in regulating innate immunity.
IMPORTANCE The innate immune response is crucial for the host to resist the invasion of viruses and other pathogens. STING and MAVS play a critical role in the innate immune response to DNA and RNA viral infection, respectively. In this study, REC8 promoted the innate immune response by targeting STING and MAVS. Notably, REC8 interacts with MAVS and STING in the cytoplasm and inhibits K48-linked ubiquitination of MAVS and STING triggered by RNF5, stabilizing MAVS and STING protein to promote innate immunity and gradually inhibiting viral infection. Our study provides a new insight for the study of antiviral innate immunity.
KEYWORDS: innate immunity, meiosis, REC8, SUMO, IFN, MAVS, STING, interferons, sumoylation
INTRODUCTION
Innate immune cells can express pattern recognition receptors (PRRs) and then recognize conserved pathogen-associated molecular patterns (PAMPs) (1). Similar to innate immune cells, other host cells also recognize viral PAMPs through PRRs, activate the innate immune response signal pathway, induce the expression of interferons (IFNs), proinflammatory cytokines, and chemokines, and activate the cell’s antiviral response (2). The PRRs mainly include Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), and intracellular DNA receptors (3, 4). TLRs localize in the endosome; TLR3 (also located in the cell membrane) recognizes double-stranded RNA (dsRNA), while TLR7 and TLR8 recognize single-stranded RNA (ssRNA). TLR3 uses the adaptor protein Toll-like receptor adaptor molecule 1 (TRIF) to activate interferon regulatory factor 3 (IRF3) and nuclear factor kappa-light-chain enhancer of activated B cells (NF-κB), while TLR7 and TLR8 use the adaptor protein myeloid differentiation primary response protein (MYD88) to activate the transcription factors IRF3, IRF5, IRF7, and NF-κB and then initiate the transcriptional expression of type I and III IFNs and inflammatory factors (5). RLRs localized in the cytoplasm: RIG-I (retinoic acid-inducible gene 1 protein) senses short-chain dsRNA and ssRNA motifs, while MDA5 (melanoma differentiation-associated protein 5) senses long-chain dsRNA (6). When the ligand binds, RLRs oligomerize and bind to the adaptor protein MAVS (mitochondrial antiviral signaling protein) located on the outer mitochondrial membrane to induce MAVS protein multimerization and then activate the transcription factors IRF3 and NF-κB (7), thereby inducing the expression of type I and III IFNs and inflammatory factors. After cyclic GMP-AMP synthase (cGAS) recognizes endogenous DNA fragments or viral DNA, its synthetase activity is activated to catalyze the synthesis of cyclic GMP-AMP (cGAMP). cGAMP induces the production of type I IFN by stimulating the STING-TBK1-IRF3 signal axis. STING (stimulator of interferon genes) binds to cGAMP and then oligomerizes, which leads to the recruitment and activation of TBK1 kinase (TANK-binding kinase 1), which in turn promotes the phosphorylation of IRF3 (8).
MAVS (also called VISA) and STING (also called MITA) mediate the signal transduction mediated by the PRRs that recognize RNA and DNA in the cytoplasm, respectively. MAVS and STING play an important role in IFN signals. Many studies have shown that viral infection can degrade MAVS and STING. For example, when hepatitis C virus (HCV) infects the cells, its nonstructural protein NS4A can induce MAVS to degrade. V protein of Newcastle disease virus (NDV), NS3 of Zika virus (ZIKV), X protein of hepatitis B virus (HBV), etc., can also promote the degradation of MAVS (9–11). Also, ZIKV’s NS2B3 promotes the degradation of STING (9). The activity and stability of MAVS and STING are regulated by ubiquitination strictly. Many host factors can promote the ubiquitination and degradation of MAVS and STING. MARCH5 promotes K48-dependent ubiquitination degradation at K7 and K500 of MAVS (12); RNF5 promotes K48-dependent ubiquitination degradation at K362 and K461 of MAVS (13); and TRIM25, Smurf1, Smurf2, and AIP4 can induce the K48-dependent ubiquitination degradation of MAVS (14–17). RNF5 promotes K48-dependent ubiquitination degradation at K150 of STING (18). However, there are few reports on host factors stabilizing MAVS and STING.
Structure maintenance chromosome (SMC) protein is a kind of ubiquitous and highly conserved chromosomal ATP synthase (19). At present, three multiprotein complexes are known to be formed with SMC proteins as the core: condensin, cohesin, and the SMC5-SMC6 complex (19). These complexes are involved in the formation and structural maintenance of chromosomes and other dynamic changes, as well as directly in the processes of DNA replication, recombination, and repair (19–21). Studies have found that some SMC proteins play an important role in the host’s defense against viral invasion. Studies have also found that the SMC5-SMC6 complex can inhibit the covalently closed circular DNA (cccDNA) of hepatitis B virus (HBV), and the HBx protein can promote the degradation of the SMC5-SMC6 complex and maintain the replication of HBV (22). SLF2 (SMC5-SMC6 complex localization factor 2) recruits the SMC5/6 complex to compress and silence unintegrated HIV-1 DNA (23). Smc4 promotes NEMO transcription and enhances the activation of NF-κB and IRF3 triggered by TLRs and viruses, leading to the production of proinflammatory cytokines and IFN-β (24). In addition to the SMC proteins, there are few reports on the regulation of pathogen invasion by other non-SMC protein chaperones in the SMC protein complex.
REC8 (REC8 meiotic recombination protein) is an important member of the adhesion protein complex. It plays a key role in chromosome separation and homologous recombination during meiosis of spermatocytes and oocytes (25). In this study, we found that REC8 can be induced by the virus, under viral stimulation, and that the SUMOylation of REC8 transfers it from the nucleus to the cytoplasm; REC8 then binds to MAVS and STING and inhibits the degradation of K48 ubiquitination of MAVS and STING, promoting the innate immune response to viral infection.
RESULTS
REC8 is identified as an interferon-induced gene.
HLCZ01 is a new type of human liver cancer cell line constructed in our laboratory, which presents the complete life cycle of hepatitis B virus (HBV) and hepatitis C virus (HCV) (26). To study how HLCZ01 cells respond to interferon stimulation, we used RNA deep sequencing to analyze the gene profiles in HLCZ01 cells with or without IFN-α treatment. Here, in the RNA sequencing (RNA-Seq) results, we first found that meiotic recombination protein 8 (REC8), which is essential for meiotic process, was upregulated by IFN-α (Fig. 1A and B). Many of our previous studies suggest that interferon-stimulating genes are involved in viral replication (6, 27, 28). Thereby, in this study, we focused on REC8. To confirm the RNA-Seq results, we treated HLCZ01 cells with IFN-α and carried out quantitative reverse transcription-PCR (qRT-PCR), and Western blot results showed that the expression of REC8 was remarkably induced by IFN-α and in a dose- and time-dependent manner (Fig. 1C and D). Poly(I·C) is an interferon inducer which mimics viral double-stranded RNA and is recognized by RIG-I. Upon treatment with poly(I·C), the levels of REC8 mRNA and protein were also significantly elevated (Fig. 1E). These results suggested that REC8 is an interferon-induced gene.
FIG 1.
Viral infection induces the expression of REC8. (A and B) HLCZ01 cells were treated with IFN-α for 12 h before RNA extraction, and RNA-Seq was performed. (C and D) HLCZ01 cells were treated with IFN-α for 4 h at a dose of 100, 200, 500, or 1,000 U/mL (C) or treated with IFN-α (500 U/mL) for the indicated times (D). The mRNA and protein levels of REC8 were analyzed by qRT-PCR and normalized with GAPDH and immunoblotting, respectively. (E) HLCZ01 cells were transfected with 200 ng or 500 ng of poly(I·C) for 12 h. The mRNA and protein levels of REC8 were analyzed by qRT-PCR and normalized with GAPDH and immunoblotting, respectively. (F) HLCZ01 cells were infected with VSV (multiplicity of infection [MOI] = 0.1) for 6 h, 12 h, and 24 h before RNA extraction. The mRNA levels of REC8 and IFTTM1 were analyzed by qRT-PCR and normalized with GAPDH; the protein level of REC8 was analyzed by immunoblotting. (G and H) HLCZ01 cells were infected with NDV (MOI = 0.1) (G) or HSV (MOI = 0.1) (H) for the indicated times before RNA extraction. The mRNA and protein levels of REC8 were analyzed by qRT-PCR and normalized with GAPDH and immunoblotting, respectively. (I) Immunoblot analysis of the indicated proteins in IFNAR1-silenced HLCZ01 or control cells treated with IFN-α (500 U/mL) for 15 or 30 min. (J) IFNAR1-silenced HLCZ01 or control cells were treated with IFN-α (500 U/mL) for 6 h, transfected with poly(I·C) (500 ng) or HCV 3′ UTR RNA (500 ng) for 16 h, or treated with VSV (MOI = 0.1), NDV (MOI = 0.1), or HSV (MOI = 0.1) for 9 h. REC8 mRNA was detected by real-time PCR and normalized with GAPDH. Experiments were independently repeated two or three times with similar results. The results are means and standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 (versus control).
Viral infection triggers the production of type I and type III IFNs. To test whether REC8 can be induced by viruses, we infected HLCZ01 cells with vesicular stomatitis virus (VSV), NDV, and herpes simplex virus (HSV). As the results showed, VSV, NDV and HSV enhanced the expression of REC8 (Fig. 1F to H). IFNs activate the JAK/STAT signaling pathway and then induce the expression of interferon-stimulated genes (ISGs), which are mainly involved in host antiviral responses (29). The early stages of VSV infection are often thought to be too early to see the response downstream of IFNAR. To rule out the possibility that VSV directly induces REC8 expression, we measured the expression of IFITM1 as a positive control. As shown in Fig. 1F, IFITM1 was also induced by VSV infection at 6 h, meaning that the interferon response had occurred at 6 h after VSV infection in our system. To further investigate whether the induction of REC8 by viruses depends on the IFN/JAK/STAT pathway, we knocked down the expression of IFNAR1 in HLCZ01 cells. The phosphorylation of STAT1 was significantly impaired in IFNAR1-silenced cells upon IFN-α treatment (Fig. 1I). qRT-PCR results showed that knockdown of IFNAR1 attenuated the expression of REC8 triggered by IFN-α, poly(I·C), NDV, VSV, and HSV (Fig. 1J). These data supported the idea that RCE8 is an ISG and the induction of REC8 depends on the JAK/STAT pathway.
REC8 positively regulates the innate immune response to viral infection and inhibits viral replication.
During viral infection, host cells secrete IFNs to promote the expression of ISGs, thereby inhibiting virus replication. As REC8 can be induced by viral infection, we assumed that REC8 may play an important role in the innate immune response to viral infection. To prove our conjecture, we overexpressed or silenced REC8 in HLCZ01 cells and detected the expression of host antiviral factors. First, we treated HLCZ01 cell with the RNA mimics poly(I·C) and the HCV 3′ untranslated region (UTR); overexpression of REC8 significantly promoted poly(I·C), and the HCV 3′ UTR triggered the expression of IFN-β, interleukin 28A (IL-28A), IL-29, and ISG12a mRNA in HLCZ01 cells (Fig. 2A). To further verify the role of REC8 in the innate immune response to RNA mimics, we designed several small interfering RNAs (siRNAs) against REC8 and found that the third one had a significant silencing effect on REC8, so we used the third one for the following study (Fig. 2B). The results showed that silencing REC8 significantly inhibited the expression of IFN-β, IL-28A, IL-29, and ISG12a triggered by poly(I·C) and the HCV 3′ UTR (Fig. 2C). Moreover, during VSV and NDV infection, the ectopic expression of REC8 in HLCZ01 cells significantly enhanced the expression of IFN-β, IL-28A, IL-29, and ISG12a (Fig. 2D and F). In contrast, knockdown of REC8 suppressed VSV- and NDV-induced cytokine expression (Fig. 2E and G). To exclude cell specificity, we repeated the experiment in A549 cells. As shown in Fig. 3A, overexpression of REC8 promoted the expression of IFN-β, IL-28A, IL-29, and ISG12a induced by VSV and NDV. Knockdown of REC8 attenuated the expression of these IFNs and ISG triggered by VSV and NDV infection (Fig. 3B and C). These data suggested that REC8 promotes the innate immune response to RNA virus infection.
FIG 2.
REC8 positively regulates the innate immune response to RNA viral infection. (A) HLCZ01 cells were transfected with pcDNA3.1a vector or pcDNA3.1a-REC8 for 40 h and then transfected with 500 ng of HMW poly(I·C) or HCV 3′ UTR RNA, respectively, for the indicated times. IFN-β, IL-28A, IL-29, and ISG12a mRNAs were measured by qRT-PCR and normalized with GAPDH. (B) HLCZ01 cells were transfected with three different siRNAs targeting REC8 for 48 h. REC8 protein was analyzed by immunoblotting. (C) HLCZ01 cells were transfected with siRNAs targeting REC8 for 40 h and then transfected with 500 ng of HMW poly(I·C) or HCV 3′ UTR RNA for 8 h. IFN-β, IL-28A, IL-29, and ISG12a mRNAs were measured by qRT-PCR and normalized with GAPDH. (D and F) HLCZ01 cells were transfected with pcDNA3.1a vector or pcDNA3.1a-REC8 for 40 h and then infected with VSV (MOI = 0.01) (D) or NDV (MOI = 0.05) (F) for the indicated times. IFN-β, IL-28A, IL-29, and ISG12a mRNAs were measured by qRT-PCR and normalized with GAPDH. (E and G) HLCZ01 cells were transfected with siRNA targeting REC8 for 40 h and then infected with VSV (MOI = 0.01) (E) or NDV (MOI = 0.01) (G) for the indicated times. IFN-β, IL-28A, IL-29, and ISG12a mRNAs were measured by qRT-PCR and normalized with GAPDH. Experiments were independently repeated two or three times, with similar results. The results are means and standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 (versus control).
FIG 3.
REC8 promotes innate immune response to DNA viral infection and inhibits viral replication. (A) A549 cells were transfected with pcDNA3.1a vector or pcDNA3.1a-REC8 for 40 h and then infected with VSV (MOI = 0.01), NDV (MOI = 0.05), or HSV (MOI = 0.01) for 8 h. The mRNA levels of IFN-β, IL-28A, IL-29, and ISG12a were tested by real-time PCR and normalized with GAPDH. (B) A549 cells were infected with Sh-Vector lenti-viral or Sh-REC8 lenti-viral. and REC8 protein was tested by Western blotting. (C) A549 cells were infected with Sh-Vector lenti-viral or Sh-REC8 lenti-viral. for 48 h and then infected with VSV (MOI = 0.01), NDV (MOI = 0.05), or HSV (MOI = 0.01) for 8 h. The mRNA levels of IFN-β, IL-28A, IL-29, and ISG12a were tested by real-time PCR and normalized with GAPDH. (D) REC8-silenced THP-1 cells were treated with ISD60 (1 μg/mL), cGAMP (5 μM), or HSV (MOI = 0.05) for 9 h. IFN-β and ISG15 mRNAs were measured by qRT-PCR and normalized with GAPDH. (E and F) HLCZ01 cells were transfected with pcDNA3.1a vector or pcDNA3.1a-REC8 for 40 h and then infected with VSV (MOI = 0.01) (E) or NDV (MOI = 0.05) (F) for the indicated times. RNA levels of VSV (E) and NDV (F) were measured by qRT-PCR and normalized with GAPDH. (G) HCV RNA and protein were detected by qRT-PCR and Western blotting in REC8-overexpressing HLCZ01 cells infected with HCV (MOI = 0.5) for 2 or 3 days. (H) Microscopy imaging of REC8-overexpressing HLCZ01 cells infected with VSV carrying green fluorescent protein (VSV-GFP) (MOI = 0.01) for 12 h. (I and J) HLCZ01 cells were transfected with siRNA targeting REC8 for 40h and then infected with VSV (MOI = 0.01) (I) or NDV (MOI = 0.05) (J) for the indicated times. RNA levels of VSV and NDV were measured by qRT-PCR and normalized with GAPDH. Experiments were independently repeated two or three times, with similar results. The results are means and standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 (versus control).
We next examined the effect of REC8 on the innate immune response to DNA virus and found that it was consistent with the results of RNA virus infection. Overexpression or knockdown of REC8 augmented or attenuated the expression of IFN-β, IL-28A, IL-29, and ISG12a induced by HSV infection in A549 cells (Fig. 3A and C). Moreover, upon treatment with cGAMP, ISD60, or HSV infection, knockdown of REC8 inhibited the production of IFN-β and ISG15 in THP1 cells (Fig. 3D). These data suggested that REC8 promotes the innate immune response to DNA virus infection.
The interferon response is important for the host cell to inhibit viral replication. Next, we tested the effect of REC8 on virus replication. As the results showed, the ectopic expression of REC8 significantly reduced the RNA levels of VSV, NDV, and HCV, and the NS5A and core protein levels of HCV were also decreased (Fig. 3E to G). Fluorescence staining also showed that REC8 decreased the abundance of green fluorescent protein (GFP)-tagged HSV (Fig. 3H). In contrast, knockdown of REC8 promoted the replication of VSV and NDV (Fig. 3I and J). All these data suggested that REC8 positively regulates the innate immune response to viral infection and inhibits viral replication.
REC8 promotes the activation of IRF3 and NF-κB signaling.
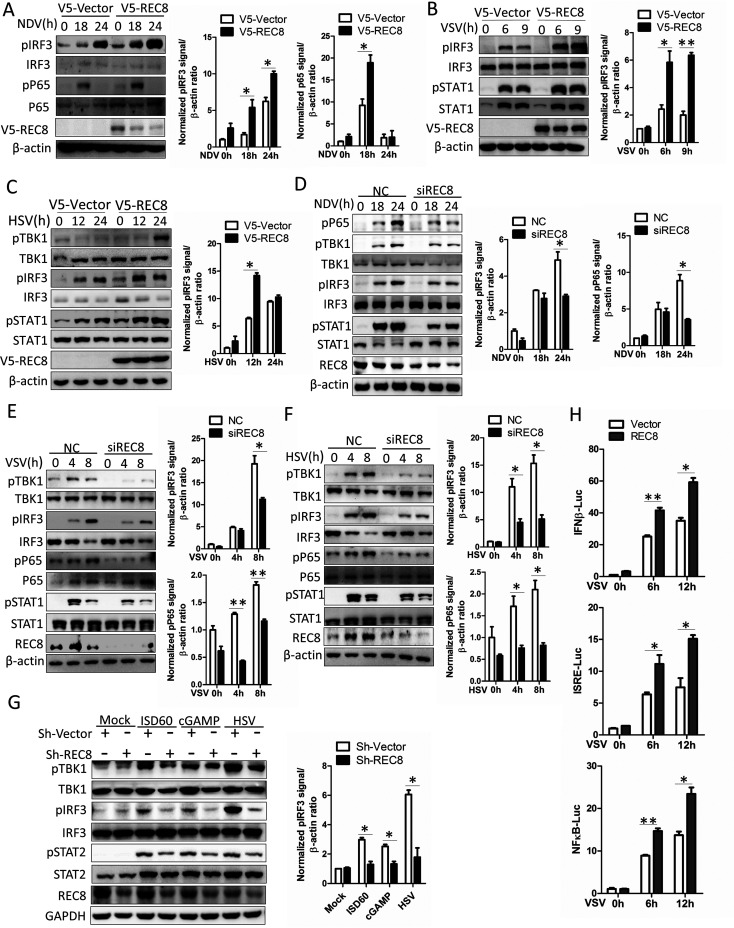
During viral infection, the pattern recognition receptor recognizes viral nucleic acid and then activates the IRF3 signaling pathway to promote the phosphorylation of IRF3 by TBK1 and activates the NF-κB signaling pathway to induce P65 phosphorylation, thereby promoting the production of interferons and other antiviral factors (2). To explore the effect of REC8 on the activation of the IRF3 signal and the NF-κB signal, we infected A549 cells with NDV, VSV, and HSV. As the results showed, overexpression of REC8 augmented the phosphorylation of IRF3 (Fig. 4A to C), and the phosphorylation of P65 was also upregulated in REC8-overexpressing A549 cells infected with NDV (Fig. 4A). Moreover, the phosphorylation of STAT1, which is the downstream molecule of IRF3 signal, also shows an upregulation trend in REC8-overexpressing A549 cells infected with VSV and HSV (Fig. 4B and C). In contrast, knockdown of REC8 suppressed the phosphorylation of IRF3, P65, and STAT1 in A549 cells that were infected with NDV, VSV, and HSV (Fig. 4D to F). To further confirm the results, we knocked down REC8 in THP1 cells and then treated the cells with the DNA mimics ISD60, cGAMP, and HSV. Similarly, knockdown of REC8 suppressed the activation of IRF3 signal (Fig. 4G). Moreover, the ectopic expression of REC8 enhanced the activation of the promoter activity of IFN-β, interferon-sensitive response element (ISRE), and NF-κB in VSV-infected HEK293T cells (Fig. 4H). All these data suggested that REC8 promotes the activation of IRF3 and NF-κB.
FIG 4.
REC8 promotes the activation of IRF3 and NF-κB signaling. (A to C) A549 cells were transfected with the indicated plasmids for 40 h and then infected with NDV (MOI = 0.5), VSV (MOI = 0.1), or HSV (MOI = 0.5) for the indicated times. Immunoblot assays were performed with the indicated antibodies. (D to F) A549 cells were transfected with control siRNA or REC8 siRNA for 36 h and then infected with NDV (MOI = 0.5), VSV (MOI = 0.1), or HSV (MOI = 0.5) for the indicated times. Immunoblot assays were performed with the indicated antibodies. (G) THP-1 cells and REC8-silenced THP-1 cells were treated with ISD60 (1 μg/mL), cGAMP (10 μM), or HSV (MOI = 0.1) for 9 h. Immunoblot assays were performed with the indicated antibodies. (H) Luciferase activity of lysates in HEK293T cells cotransfected with IFN-β–Luc, ISRE-Luc, or NF-κB–Luc and p3X-Flag-CMV-vector or p3X-Flag-REC8 for 15 h and then infected with VSV for 9 h. The results are presented relative to the luciferase activity in control cells. Experiments were independently repeated 2 or 3 times with similar results. The results are means and standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 (versus control).
REC8 targets and interacts with MAVS and STING.
To investigate the molecular target of REC8 in enhancing IFN signaling, we cotransfected REC8 with IFN-β–Luc or ISRE-Luc and MDA5, RIG-I-N, cGAS, STING, MAVS, TBK1, and IRF3-5D (S385D, S386D, S396D, S398D, S402D, S404D, and S405D) in HEK293T cells. Overexpression of REC8 enhanced the activation of promoter activity of IFN-β and ISRE by RIG-I-N, MDA5, MAVS, cGAS, and STING, whereas the activation of the IFN-β promoter or ISRE promoter by TBK1 or IRF3-5D was unaffected (Fig. 5A and B). Taken together, these data suggested that REC8 promotes the IFN signaling pathway upstream of TBK1.
FIG 5.
REC8 targets and interacts with MAVS and STING. (A and B) HEK293T cells were cotransfected with REC8 plasmids, pRL-CMV, IFN-β luciferase (A) or ISRE luciferase (B), and MDA5, N-RIG-I, MAVS, TBK1, IRF3-5D, cGAS, or STING plasmid for 24 h. Luciferase assays were then performed. (C and D) HEK293T cells were cotransfected with the plasmids V5-REC8 and Flag-RIG-I, Flag-MDA5, Flag-MAVS, Flag-TBK1, or Flag-IRF3 (C) or Flag-cGAS, Flag-STING, or Flag-P65 (D) for 48 h. Co-IP and immunoblotting were performed with anti-Flag, anti-V5, and anti-β-actin. (E) HEK293T cells were transfected with p3×Flag-REC8 for 48 h. Cellular lysates were immunoprecipitated with anti-Flag or IgG. Immunoprecipitates were analyzed by Western blotting with anti-Flag or anti-MAVS. (F) HEK293T cells were transfected with p3×Flag-REC8 for 48 h. Cellular lysates were immunoprecipitated with anti-MAVS or IgG. Immunoprecipitates were analyzed by Western blotting with anti-Flag or anti-MAVS. (G) HLCZ01 cells were infected with VSV for the indicated times. Immunoprecipitation and Western blot analysis were performed with antibodies against MAVS, REC8, ISG15, and β-actin. (H) THP1 cells were infected with HSV for the indicated times. Immunoprecipitation and Western blotting were performed with antibodies against the indicated proteins. (I) A549 cells were infected with VSV (MOI = 0.05) for 6 h. Fluorescent images were obtained with a fluorescence microscope with a 60× lens objective. Quantitative colocalization were analyzed by Image-Pro Plus 6.0. Experiments were independently repeated two or three times, with similar results. The results are means and standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 (versus control).
To determine the key factors in IFN signaling pathway that could be targeted by REC8, we cotransfected pcDNA3.1a-REC8 with Flag-tagged innate immune signaling components into HEK293T cells. The coimmunoprecipitation (co-IP) experiments showed that REC8 interacts with MAVS and STING but not RIG-I, MDA5, cGAS, TBK1, and IRF3 (Fig. 5C and D). Moreover, the association of exogenous Flag-REC8 with the endogenous MAVS was also confirmed in HEK293T cells (Fig. 5E and F). Furthermore, the interaction between endogenous REC8 and MAVS was enhanced upon VSV infection (Fig. 5G). Also, the interaction between REC8 and STING was enhanced in THP-1 cells during infection with HSV (Fig. 5H). To visually observe whether REC8 and MAVS are colocalized, we performed immunofluorescence staining and analyzed the colocalization signal; then, we obtained the coefficients, including mask of colocalizing object (pixel scatter plot of protein colocalization signals), Pearson’s correlation coefficient (Rr) (Rr = 0.867071), and overlap coefficient (R) (R = 0.891098), as shown in Fig. 5I, which indicated that REC8 colocalized with MAVS in VSV-infected A549 cells (30).
To determine which segment of REC8 interacts with MAVS or STING, we divided REC8 into three segments, including the N-terminal conserved structure, the C-terminal conserved structure, and the middle unknown structure (Fig. 6A). Unexpectedly, the middle unknown domain of REC8 interacted with MAVS, but not the N-terminal conserved structure or the C-terminal conserved structure (Fig. 6B). Moreover, full-length REC8 interacted with STING, while no segment of REC8 interacted with STING (Fig. 6C). Based on these observations, we speculate that the interaction of REC8 with MAVS and STING may be species specific. To better understand the molecular mechanism of REC8-mediated MAVS or STING function, we constructed a series of truncations of MAVS and STING and cotransfected them with full-length REC8 in HEK293T cells. We found that REC8 interacts with DIII of MAVS, and the 111–221 section of STING is the key section for the STING-REC8 interaction (Fig. 6D to G). Interestingly, both DIII of MAVS and the 111–221 segment of STING contain transmembrane-related domains, indicating that REC8 may have similar mechanisms of binding with MAVS and STING. Taken together, these data suggested that REC8 targets and interacts with MAVS and STING.
FIG 6.
REC8 interacts with the transmembrane domain of MAVS and STING. (A) Schematic illustration of REC8 truncations. (B) HEK293T cells were cotransfected with Flag-MAVS and the indicated domains of REC8 for 48 h. Co-IP and immunoblotting were performed with the indicated antibodies. (C) HEK293T cells were cotransfected with Flag-STING and the indicated domains of REC8 for 48 h. Co-IP and immunoblotting were performed with the indicated antibodies. (D) Schematic illustration of MAVS truncations. (E) HEK293T cells were cotransfected with V5-REC8 and the indicated domains of MAVS for 48 h. Co-IP and immunoblotting were performed with the indicated antibodies. (F) Schematic illustration of STING truncations. (G) HEK293T cells were cotransfected with Flag-REC8 and the indicated domains of STING for 48 h. Co-IP and immunoblotting were performed with the indicated antibodies. Experiments were independently repeated two or three times.
SUMOylation of REC8 triggers cytoplasmic translocation upon viral infection.
Previous studies have shown that REC8 mainly plays a role in chromatin remodeling during meiosis, which occurs in the nucleus. Our data showed that REC8 could interact with MAVS and STING in the cytoplasm to regulate the innate immune response to viral infection. We speculate that the subcellular localization of REC8 may change during viral infection. Nuclear and cytoplasmic separation experiments demonstrated that REC8 is mainly located in the nucleus in the resting state (Fig. 7A). During viral infection, the content of REC8 protein in the cytoplasm increases markedly and the REC8 protein content in the nucleus decreases (Fig. 7A). Previous studies have shown that PCBP2 can be SUMOylated, causing its cytoplasmic translocation during viral infection (31). We wanted to know whether the change of REC8 subcellular location relies on a similar mechanism. UBC9 is the only E2 ligase for protein SUMOylation. Therefore, we first explored whether REC8 can interact with UBC9. As shown in Fig. 7B, REC8 interacted with UBC9, and the association of REC8 with UBC9 was enhanced under viral infection. Moreover, SUMO2/3-mediated REC8 SUMOylation was markedly augmented upon VSV or HSV infection (Fig. 7C and D).
FIG 7.
The SUMOylation of REC8 triggers cytoplasmic translocation upon viral infection. (A) A549 cells were incubated with VSV for the indicated times, followed by cytoplasmic and nuclear separation. Lysates were immunoblotted with anti-REC8, anti-LamiB1, and anti-GAPDH. (B) HEK293T cells were cotransfected with p3×Flag-UBC9 and pV5-REC8 for 42 h and then infected with VSV (MOI = 0.1) for 6 h. Co-IP and immunoblotting were performed with the indicated antibodies. (C and D) HEK293T cells were cotransfected with pFlag-UBC9, pV5-REC8, and pHA-SUMO2/3 for 42 h and then infected with VSV (MOI = 0.1) (C) or HSV (MOI = 0.1) (D) for the indicated times. IP and immunoblotting were performed with the indicated antibodies. (E) HEK293T cells and UBC9-silenced HEK293T cells were transfected with Flag-REC8 for 42 h and then infected with VSV (MOI = 0.1) or HSV (MOI = 0.1) for 6 h. IP and immunoblotting were performed with the indicated antibodies. (F) A549 cells and UBC9-silenced A549 cells were incubated with VSV (MOI = 0.01) for 6 h and 9 h, followed by cytoplasmic and nuclear separation. Lysates were immunoblotted with the indicated antibodies. (G) A549 cells and UBC9-silenced A549 cells were infected with sh-REC8 lentivirus for 48 h and then infected with VSV (MOI = 0.01) or HSV (MOI = 0.01) for 6 h. Immunoblotting was performed with the indicated antibodies. (H) A549 cells and UBC9-silenced A549 cells were infected with sh-REC8 lentivirus for 48 h and then infected with VSV (MOI = 0.01) or HSV (MOI = 0.01) for 6 h. UBC9, IFN-β, and ISG15 mRNAs were measured by qRT-PCR and normalized with GAPDH. Experiments were independently repeated 2 or 3 times. The results are means and standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 (versus control).
To verify the importance of SUMOylation modification in the regulation of innate immunity by REC8, we knocked down UBC9 in 293T cells. Silencing of UBC9 dramatically inhibited the SUMOylation of REC8 upon viral infection (Fig. 7E). Moreover, the cytoplasmic translocation of REC8 was also inhibited during viral infection (Fig. 7F). Importantly, knockdown of REC8 impaired the activation of IRF3 and STAT1 triggered by VSV or HSV and subsequently inhibited the expression of IFN-β and ISG15 induced by VSV and HSV (Fig. 7G and H). However, knockdown of UBC9 abolished the inhibition of the phosphorylation of IRF3 and STAT1 or the expression of IFN-β and ISG15 in REC8-silenced cells under VSV and HSV infection (Fig. 7G and H). These results indicated that SUMOylation of REC8 is essential for the innate immune response to viral infection. Interestingly, knockdown of UBC9 inhibited the phosphorylation of IRF3 and STAT1 triggered by VSV or HSV, while the induction of IFN-β, ISG15 by VSV and HSV was increased (Fig. 7G and H). We speculate that UBC9 may regulate the innate immune response in multiple ways.
Viruses promote the SUMOylation of REC8 at Lys30 and Lys530.
To examine which lysines of REC8 can undergo SUMOylation during viral infection, we used GPS-SUMO 2.0 to predict the SUMOylation sites of REC8 (32). As determined by the software, the K30 and K530 sites of REC8 may undergo SUMOylation. To confirm this assumption, we constructed REC8 mutants (K30R, K530R, and K30R K530R). Upon VSV infection, the SUMOylation of wild-type (WT) REC8 was increased but not the mutant REC8 (K30R, K530R, and K30R K530R) (Fig. 8A), suggesting that the K30 and K530 sites might be the main SUMOylation sites of REC8. Moreover, the specific mutants (K30R, K530R, and K30R K530R) of REC8 had weaker promotion of the activation of IRF3 and STAT1 than WT REC8 in VSV- and HSV-infected A549 cells (Fig. 8B and C). Consistently, the induction of IFN-β by viral infection was also inhibited (Fig. 8D), and its antiviral function was abolished (Fig. 8E). All these data supported the idea that the SUMOylation of REC8 at K30 and K530 has a regulatory function in the regulation of the innate immune response to viral infection.
FIG 8.
Viral infection promotes the SUMOylation of REC8 at Lys30 and Lys530. (A) HEK293T cells were cotransfected with Flag-REC8 or REC8 mutants (K30R, K530R, and K30R K530R) and HA-SUMO2/3 for 42 h and infected with VSV (MOI = 0.1) for 6 h. IP and immunoblotting assays were performed with the indicated antibodies. (B and C) A549 cells were transfected with Flag-REC8 or REC8 mutants (K30R, K530R, and K30R K530R) for 42 h and infected with VSV (MOI = 0.01) (B) or HSV (C) (MOI = 0.01) for 6 h. Immunoblotting assays were performed with the indicated antibodies. (D) A549 cells were transfected with Flag-REC8 or REC8 mutants (K30R, K530R, and K30R K530R) for 42 h and infected with VSV (MOI = 0.01) or HSV (MOI = 0.01) for 6 h. IFN-β mRNA was measured by qRT-PCR and normalized with GAPDH. (E) A549 cells were transfected with Flag-REC8 or REC8 mutants for 42 h and infected with GFP-VSV (MOI = 0.01) and GFP-HSV (MOI = 0.01) for 12 h. Images were taken under a fluorescence microscope. Experiments were independently repeated two or three times. The results are means and standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 (versus control).
REC8 stabilizes MAVS and STING by weakening the K48-linked ubiquitination of MAVS and STING.
The stability of MAVS and STING controls the immune signaling transduction. We found that ectopic expression of REC8 stabilized MAVS and STING proteins. Consistently, the proteasome inhibitor MG132 further augmented REC8-mediated upregulation of endogenous MAVS and STING protein (Fig. 9A). Moreover, after we used cycloheximide (CHX) to inhibit protein translation, REC8 also significantly stabilized MAVS and STING and inhibited their degradation (Fig. 9B). Although knockdown of REC8 did not change the levels of MAVS and STING proteins in A549 cells in the resting state, during infection with VSV or HSV, knockdown of REC8 reduced the protein levels of MAVS and STING (Fig. 9C and D). This is because the endogenous REC8 is located in the nucleus in the resting state and cannot affect MAVS and STING in the cytoplasm. Furthermore, silencing of UBC9 reversed downregulation of MAVS and STING mediated by knocking down of REC8 in VSV- and HSV-infected cells (Fig. 9C and D). These results supported the idea that REC8 stabilizes MAVS and STING proteins in VSV or HSV-infected cells. Interestingly, we found that knockdown of UBC9 also decreased the MAVS and STING protein levels in infected cells (Fig. 9C and D). We think that MAVS and STING might be SUMOylated and stabilized during viral infection.
FIG 9.
REC8 stabilizes MAVS and STING by weakening the K48-linked ubiquitination of MAVS and STING. (A) A549 cells were transfected with pcDNA3.1a vector or pcDNA3.1a-REC8 plasmid for 42 h and then treated with or without MG132 (25 μM) for 6 h. Immunoblot assays were performed with the indicated antibodies. (B) A549 cells were transfected with pcDNA3.1a vector or pcDNA3.1a-REC8 plasmid for 36 h and then treated with CHX (25 μM) for the indicated times. Immunoblot assays were performed with the indicated antibodies. (C and D) A549 cells and UBC9-silenced A549 cells were infected with VSV (MOI = 0.1) (C) or HSV (MOI = 0.1) (D) for 6 h. Immunoblotting was performed with the indicated antibodies. (E and F) HEK293T cells transfected with WT or K6O-, K11O-, K27O-, K33O-, K48O-, or K63O-linked HA-Ub plasmid with Flag-MAVS (E) or Flag-STING (F) and V5-REC8 for 42 h and then treated with MG132 (25 μM) for 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (G) HEK293T cells were cotransfected with HA-Ub plasmids and V5-REC8 for 42 h and then infected with VSV (MOI = 0.1) for 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (H) THP1 cells were transfected with V5-REC8 for 42 h and then infected with HSV (MOI = 0.1) for 6 h and 9 h. Ubiquitination and immunoblotting assays were performed with anti-STING, anti-V5, anti-Ub, and anti-GAPDH. (I) HEK293T cells were transfected with Flag-REC8 or REC8 mutant (K30R, K530R, and K30R K530R) for 42 h and treated with MG132 (25 μM) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (J) 293T cells were cotransfected with Flag-REC8 or REC8 mutants (K30R, K530R, and K30R K530R) and Flag-STING for 42 h and treated with MG132 (25 μM) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. Experiments were independently repeated two or three times.
K48-linked ubiquitination is related to protein degradation. Therefore, we investigated whether REC8 may regulate the ubiquitination of MAVS and STING. As shown in Fig. 9E and F, REC8 inhibited K48-linked ubiquitination of MAVS and STING but not the K6-, K11-, K27-, K29-, K33- and K63-linked ubiquitination of MAVS and STING. Moreover, the ectopic expression of REC8 significantly inhibited the ubiquitination of endogenous MAVS during infection with VSV (Fig. 9G). Similarly, overexpression of REC8 weakened the ubiquitination of STING in HSV-infected THP1 cells at 9 h (Fig. 9H).
The above data showed that the SUMO modification of REC8 at K30 and K530 promotes the regulation of REC8 in the innate immune response to viral infection. To test whether the SUMO modification of these two SUMO sites of REC8 affects the ubiquitination of MAVS and STING, we cotransfected pcDNA3.1a-REC8 (WT), pcDNA3.1a-REC8 (K30R), pcDNA3.1a-REC8 (K530R), or pcDNA3.1a-REC8 (K30R K530R) with ubiquitin plasmid carrying the HA tag (HA-ub)- and Flag-tagged MAVS or STING. As the results showed, the K30 K530 REC8 mutant lost the inhibitory effect of WT REC8 on the K48-linked ubiquitination of MAVS and STING (Fig. 9I to J). These results indicated that the SUMOylation of REC8 regulates the ubiquitination of MAVS and STING.
MAVS has 14 lysine sites. To determine at which lysine site ubiquitination of MAVS is inhibited by REC8, we transfected HA-Ub- and Myc-tagged WT MAVS or Myc-tagged MAVS mutants [Myc-MAVS (K7R), (K10R), (K136R), (K270R),(K279R), (K311R), (K325R), (K331R), (K348R), (K362R), (K371R), (K420R), (K461R) or (K500R)] into 293T cells. REC8 did not inhibit the ubiquitination of Myc-MAVS (K362R) (Fig. 10A). Moreover, REC8 impaired the K48-dependent ubiquitination of WT MAVS, while it did not affect that of MAVS (K362R) (Fig. 10B). These data suggested that REC8 weakened the K48-linked polyubiquitination of MAVS on Lys362.
FIG 10.
REC8 weakens the K48-linked ubiquitination of MAVS at K362 and STING at K150/K370 and promotes the recruitment of TBK1 to MAVS and STING. (A) HEK293T cells were cotransfected with Myc-MAVS or MAVS mutants (K7R, K10R, K136R, K270R, K297R, K311R, K325R, K331R, K348R, K362R, K371R, K420R, K461R, and K500R) with pcDNA3.1a-vector or pcDNA3.1a-REC8 for 42 h and treated with MG132 (25 μM) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (B) HEK293T cells were cotransfected with Myc-MAVS (WT) or Myc-MAVS (K362R) with the indicated plasmid for 42 h and treated with MG132 (25 μM) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (C) HEK293T cells were cotransfected with Flag-STING or STING mutants (K20R, K137R, K150R, K224R, K236R, K289R, K338R, K347R, and K370R) with pcDNA3.1a-vector or pcDNA3.1a-REC8 for 42 h and treated with MG132 (25 μM) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (D) HEK293T cells were cotransfected with Flag-STING or Flag-STING (K150R) or Flag-STING (K370R) and the indicated plasmids for 42 h and treated with MG132 (25 μM) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (E) HEK293T cells were cotransfected with Flag-MAVS and V5-REC8 for 48 h. Immunoprecipitation and Western blotting were performed with antibodies against the indicated proteins. (F) HEK293T cells were cotransfected with Flag-STING and V5-REC8 for 48 h. Immunoprecipitation and Western blotting were performed with antibodies against the indicated proteins. (G) A549 cells and REC8-silenced A549 cells were infected with VSV for the indicated times. Immunoprecipitation and Western blotting were performed with antibodies against the indicated proteins. (H) 293T cells were cotransfected with V5-REC8 or REC8 mutants and Flag-MAVS for 48 h. Immunoprecipitation and Western blotting were performed with antibodies against the indicated proteins. (I) 293T cells were cotransfected with V5-REC8 or REC8 mutants and Flag-STING for 48 h. Immunoprecipitation and Western blotting were performed with antibodies against the indicated proteins. Experiments were independently repeated two or three times.
STING contains 9 lysine residues. We transfected HA-Ub with Flag-tagged wild-type STING [Flag-STING (WT)] or 9 Flag-tagged STING mutants [Flag-STING (K20R), (K137R), (K150R), (K224R), (K236R), (K289R), (K338R), (K347R), or (K370R)] into HEK293T cells. REC8 did not attenuate the ubiquitination of STING with the K150R mutation and K370R mutation (Fig. 10C). REC8 inhibited the K48-linked polyubiquitination of STING (WT) but did not affect the K48-linked polyubiquitination of STING (K150R) and STING (K370R) (Fig. 10D). These data suggested that REC8 weakened the K48-linked polyubiquitination of STING on Lys150 and Lys370. Taken together, these data suggested that REC8 stabilizes MAVS and STING and weakens the K48-linked ubiquitination of MAVS and STING.
REC8 promotes the recruitment of TBK1 to MAVS and STING.
We speculated that the inhibition of REC8 on K48-linked polyubiquitination of MAVS on K362 and STING on K150 and K370 may promote their interaction with TBK1. As expected, overexpression of REC8 enhanced the recruitment of TBK1 by MAVS or STING (Fig. 10E and F). Knockdown of REC8 impaired the recruitment of TBK1 by MAVS upon VSV stimulation (Fig. 10G). Moreover, the K30 K530 double mutation of REC8 weakened the ability of REC8 to promote the recruitment of TBK1 by MAVS or STING (Fig. 10H and I). These data suggested that REC8 promotes the recruitment of TBK1 to MAVS and STING.
REC8 inhibits RNF5-triggered ubiquitination of MAVS and STING.
REC8 is not a deubiquitinating enzyme. We speculated that REC8 may inhibit the effect of E3 ligases on the ubiquitination of MAVS and STING. Here, we chose RNF5 (the E3 that affects the ubiquitination of K48 in MAVS and STING) and other MAVS-related E3 ligases, such as TRIM21, AIP4 and TRAF3, to test whether REC8 affects the ubiquitination of MAVS or STING by these E3 ligases. Co-IP experiments showed that REC8 interacted with RNF5 and that the conserved domain at the N terminus of REC8 interacted with RNF5 (Fig. 11A and B). To investigate whether the effect of REC8 on the expression of MAVS and STING depends on RNF5, we silenced RNF5 in A549 cells. As the result showed, overexpression of REC8 increased the protein levels of MAVS and STING in A549 cells but not in RNF5-knockdown A549 cells (Fig. 11C). Consistently, overexpression of REC8 increased the mRNA levels of IFN-β in VSV-infected A549 cells but not in RNF5 knockdown A549 cells infected with VSV (Fig. 11D). Moreover, overexpression of REC8 inhibited the ubiquitination of MAVS and STING triggered by RNF5 (Fig. 11E and G), while in RNF5 knockdown HEK293T cells, REC8 does not show any influence on the ubiquitination of MAVS and STING (Fig. 11F and H). We wondered whether REC8 inhibits the ubiquitination and degradation of MAVS and STING by RNF5 through competitive binding to MAVS or STING. As shown in Fig. 11I and J, REC8 did not inhibit the interaction of MAVS or STING with RNF5. We speculate that REC8 may regulate the effects of RNF5 on MAVS and STING through other mechanisms. Taken together, these data support the idea that REC8 inhibits RNF5-triggered ubiquitination of MAVS and STING.
FIG 11.
REC8 inhibits RNF5-triggered ubiquitination of MAVS and STING. (A) HEK 293T cells were cotransfected with Flag-TRIM21, Flag-RNF5, Flag-AIP4, or Flag-TRAF3 and V5-REC8 for 48 h. Co-IP and immunoblotting were performed with the indicated antibodies. (B) HEK293T cells were cotransfected with the plasmids encoding RNF5 and the indicated domains of REC8 for 48 h. Co-IP and immunoblotting were performed with the indicated antibodies. (C) A549 cells and RNF5-silenced A549 cells were transfected with V5-REC8 for 48 h. Immunoblotting was performed with the indicated antibodies. (D) A549 cells and RNF5-silenced A549 cells were transfected with V5-REC8 for 42 h and infected with VSV for an additional 6 h. IFN-β mRNA was measured by qRT-PCR and normalized with GAPDH. (E) HEK293T cells were cotransfected with V5-REC8, HA-Ub, and RNF5 plasmids for 42 h and treated with MG132 (25 μM) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (F) HEK293T cells and RNF5 knockdown HEK293T cells were cotransfected with V5-REC8 and HA-Ub for 42 h and treated with MG132 (25 μM) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (G) HEK293T cells were cotransfected with V5-REC8, Flag-STING, HA-Ub, and RNF5 plasmids for 42 h and treated with MG132 (25 μM) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (H) HEK293T cells and RNF5-knockdown HEK293T cells were cotransfected with V5-REC8, Flag-STING, and HA-Ub for 42 h and treated with MG132 (25 μM) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (I) 293T cells were cotransfected with V5-REC8 and Flag-MAVS for 48 h. Immunoprecipitation and Western blot analysis were performed with antibodies against the indicated proteins. (J) 293T cells were cotransfected with V5-REC8 and Flag-STING for 48 h. Immunoprecipitation and Western blot analysis were performed with antibodies against the indicated proteins. Experiments were independently repeated two or three times. The results are means and standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 (versus control).
DISCUSSION
During viral infection, host cells produce a large number of IFNs, and the presence of IFNs induces ISGs, inhibiting virus replication (33). MX1, CH25H, and IFITMs can inhibit viruses from entering host cells (34–37). Some ISGs inhibit virus replication by blocking the synthesis of viral proteins, such as ZAP, IFITs, and PKR (38–40). Viperin and tetherin inhibit the release of the virus (41, 42). In addition to affecting the life cycle of viruses, many ISGs affect the activation of host innate immunity. For example, RLRs (RIG-I and MDA5) that recognize pathogenic RNA are encoded by interferon-stimulating genes (4). Some interferon regulatory factors (IRF1, IRF9, etc.) can also be induced by interferon (43). TRIM21, OTDU4, and TRIM56 can affect the protein activity or stability of MAVS or STING, regulating the activation of the innate immune response (27, 44, 45).
Our RNA sequencing data showed that REC8, an important member of the adhesion protein complex, which plays a key role in chromosome separation and homologous recombination during meiosis of spermatocytes and oocytes (46, 47), can be induced by IFN-α. Our results showed that REC8 can be induced by VSV, NDV, HSV and other viruses, which depends on the JAK-STAT signaling pathway. So far, no studies demonstrate that other genes related to meiosis can be induced by viral infection and IFNs. Therefore, we speculate that REC8 plays an important role in regulating the host’s innate immune response to viral infection.
REC8 mainly plays an important role in meiosis, and its function in different species is very conservative (46, 48, 49). REC8 affects meiosis in plants, yeast extract, mammals, and other eukaryotes (46, 50–53). Also, a few reports show that REC8 plays a role in regulating the occurrence and development of tumors. REC8 is a member of the tumor/testis antigens (54). In gastric cancer, melanoma, and other malignant tumors, due to promoter methylation modification, REC8 expression is low, which affects cell cycle, angiogenesis, invasion and metastasis, and clinical prognosis (55–57). Here, we found that REC8 promotes the activation of IRF3 and NF-κB signals induced by RNA and DNA viruses, leading to the production of proinflammatory cytokines and IFNs, eventually inhibiting the replication of RNA and DNA viruses.
As adaptor proteins, the activity and protein stability of MAVS and STING directly affect immune signal transduction. Protein ubiquitination and deubiquitination are important ways to affect the biological functions of MAVS and STING. RNF5, PCBP2, AIP4, Smurf1, and TRIM25 promote the K48-linked ubiquitination of MAVS, thereby promoting MAVS degradation (13, 15–17, 58). TRIM31 enhances the K63-linked ubiquitination of MAVS and the activation of downstream signals (59). TRIM21 triggers the K27-linked ubiquitination of MAVS and promotes the recruitment of TBK1 to MAVS (27). RNF5 can promote the K48-linked ubiquitination of MAVS (13). Unlike MAVS, there are few reports on the K48-linked ubiquitination of STING. E3 ligase, including MUL1, AMFR, TRIM32, TRIM56, and deubiquitinating enzymes such as USP13 and USP49 affect the K27- or K63-linked ubiquitination of STING, which in turn affects the recruitment of TBK1 (45, 60–64). In the existing studies, there are few reports of stabilizing MAVS and STING proteins. We find that REC8 interacts with MAVS or STING upon viral stimulation, inhibiting the K48-linked polyubiquitination of MAVS on K461 and STING on K150 and K370. Our results show that REC8 interacts with RNF5, a common E3 of MAVS and STING, and inhibits the ubiquitination and degradation of MAVS and STING by RNF5. REC8 does not compete with RNF5 to bind to MAVS and STING. REC8 may inhibit the degradation of MAVS and STING by RNF5 in other ways. In addition to their role in innate immunity, MAVS and STING are also critical in antitumor immunity (65–68). Although previous studies have shown that REC8 acts as a tumor suppressor gene in different tumors (55–57), the inhibition mechanism is unclear. Here, we assume that REC8 may target MAVS and STING to promote antitumor immunity and then inhibit tumor growth. This is what we will research next.
In eukaryotic cells, there are at least three SUMO proteins, including SUMO1, SUMO2, and SUMO3 (69). SUMO proteins function in a manner similar to that of ubiquitin (70). However, unlike ubiquitin, which targets proteins for degradation, these proteins are involved in various cellular processes, such as nuclear transport, transcriptional regulation, apoptosis, and protein stability (69–73). One study showed that upon stimulation with RNA viruses, IRTKS can recruit UBC9 to SUMOylate PCBP2, which in turn promotes the transport of PCBP2 from the nucleus to the cytoplasm (31). We found that viral infection enhances the interaction between REC8 and UBC9 and increases the level of SUMOylation of REC8, promoting the translocation of REC8 from the nucleus to the cytoplasm. Knockdown of UBC9 decreases the level of SUMOylation of REC8 and inhibits its nuclear transport. Our study supports the idea that REC8 is a new substrate for SUMOylation. Additionally, we found a seemingly contradictory result. Silencing UBC9 inhibited the phosphorylation of IRF3 by viral infection, but the mRNA level of type I interferon was upregulated (Fig. 7G and H). Previous studies have shown that the SUMOylation of RIG-I and MDA5 promoted the phosphorylation activation of IRF3 induced by viral infection (74, 75). Here, we assume that silencing UBC9 inhibits the SUMOylation of interferon signal activators such as RIG-I and MDA5, so it inhibits the phosphorylation of IRF3. However, another study showed that knockdown of UBC9 or SUMO2/3 can directly induce the expression of type I interferon and ISG and does not depend on IRF3 and IFNAR1 in THP1 cells (76). Therefore, we speculate that the upregulation of interferon after we silenced UBC9 in A549 cells may be based on a similar mechanism, and that is why the p-IRF3 level was decreased while the mRNA level of IFN was increased. However, how the specific host cell balances this relationship remains to be studied in depth.
Based on our data, we propose a working model of REC8-mediated regulation of MAVS and STING during innate antiviral signaling (Fig. 12). REC8 is induced by viral infection. Upon virus infection, REC8 is SUMOylated at K30 and K530 and then is translocated from the nucleus to the cytoplasm. REC8 interacts with MAVS and STING in the cytoplasm and inhibits K48-linked ubiquitination of MAVS and STING triggered by RNF5, stabilizing MAVS and STING protein to promote innate immunity and gradually inhibit viral infection. Our work reveals a new mechanism for the host to stabilize MAVS and STING proteins during viral infection. Moreover, as a key protein of meiosis, REC8 participates in the innate immune response, revealing a certain connection between meiosis and antiviral innate immunity. Is there such a possibility? Viral infection and the abuse of IFNs will have a huge impact on meiosis, which in turn affects the formation of sperm and eggs, leading to infertility.
FIG 12.
Working model of REC8 in the regulation of the innate immune response to viral infection. REC8 is induced by viral infection. Upon virus infection, REC8 is SUMOylated at K30 and K530 and then translocated from the nucleus to the cytoplasm. REC8 interacts with MAVS and STING in the cytoplasm and inhibits K48-linked ubiquitination of MAVS and STING triggered by RNF5, stabilizing MAVS and STING proteins to induce the expression of IFNs, thereby inhibiting viral infection.
MATERIALS AND METHODS
Cells.
HLCZ01 cells were established in our lab at Hunan University (26). HEK293T cells were purchased from Boster. A549 and THP-1 cells were obtained from the American Type Culture Collection. HLCZ01 cells were cultured in collagen-coated tissue culture plates containing Dulbecco’s modified Eagle medium (DMEM)–F-12 medium supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Gibco), 40 ng/mL of dexamethasone (Sigma), insulin-transferrin-selenium (ITS) (Lonza), penicillin, and streptomycin. HEK293T and A549 cells were propagated in DMEM supplemented with 10% FBS, l-glutamine, nonessential amino acids, penicillin, and streptomycin. THP-1 cells were propagated in RPMI 1640 medium supplemented with 10% FBS and 0.05 mM β-mercaptoethanol.
Antibodies and reagents.
Antibodies used include anti-REC8 (Abcam, ab246985), anti-MAVS (Santa Cruz, sc-166583), anti-UBC9 (Santa Cruz, sc-271057), anti-RNF5 (Santa Cruz, sc-81716), anti-STING (CST, 13647S), anti-TBK1 (CST, 38066S), anti-p-TBK1 (CST, 5483S), anti-IRF3 (CST, 4302S), anti-p-IRF3 (CST, 4947S), anti-P65 (CST, 8242S), anti-p-P65 (CST, 3033S), anti-STAT1 (CST, 14995S), anti-p-STAT1 (CST, 9167S), anti-STAT2 (CST, 72604S), anti-p-STAT2 (CST, 88410S), anti-LMNB1 (CST, 17416S), anti-SUMO2/3 (CST, 4971T), anti-Myc (CST, 2276S), anti-HA (Abcam, ab236632), anti-GAPDH (Millipore, MAB374), anti-V5 (Invitrogen, R960-25). Anti-Flag and anti-β-actin were obtained from Sigma. MG132 (APExBIO Technology, C3348), CHX (CST, 2112S), puromycin (Thermo, A1113803), and 2′-3′-cGAMP (APExBIO Technology, B8362) were also used.
Nuclear and cytoplasmic extraction.
Nuclear and cytoplasmic separation of cells was done using a standard protocol as previously described (28).
Real-time PCR assay.
Total RNA was isolated with the TRIzol reagent (Invitrogen, Carlsbad, CA), and qPCR analysis was used to measure mRNA levels of the indicated genes as previously described (6). The qPCR primers are listed in Table 1.
TABLE 1.
Primers for real-time PCR
| Primer | Sequence |
|---|---|
| IFNβ-(F) | 5′-CAGCATTTTCAGTGTCAGAAGC-3′ |
| IFNβ-(R) | 5′-TCATCCTGTCCTTGAGGCAGT-3′ |
| IL-28A (F) | 5′-GCCTCAGAGTTTCTTCTGC-3′ |
| IL-28A (R) | 5′-AAGGCATCTTTGGCCCTCTT-3′ |
| IL-29 (F) | 5′-CGCCTTGGAAGAGTCACTCA-3′ |
| IL-29 (R) | 5′-GAAGCCTCAGGTCCCAATTC-3′ |
| IL6-(F) | 5′-CTCAATATTAGAGTCTCAACCCCCA-3′ |
| IL6-(R) | 5′-GAGAAGGCAACTGGACCGAA-3′ |
| TNFα-(F) | 5′-AGAACTCACTGGGGCCTACA-3′ |
| TNFα-(R) | 5′-GCTCCGTGTCTCAAGGAAGT-3′ |
| GAPDH (F) | 5′-AATGGGCAGCCGTTAGGAAA-3′ |
| GAPDH (R) | 5′-GCGCCCAATACGACCAAATC-3′ |
| ISG12a (F) | 5′-TGCCATGGGCTTCACTGCGG-3′ |
| ISG12a (R) | 5′-CTGCCCGAGGCAACTCCACC-3′ |
| NDV (F) | 5′-TCACAGACTCAACTCTTGGG-3′ |
| NDV (R) | 5′-CAGTATGAGGTGTCAAGTTCTTC-3′ |
| ISG15 (F) | 5′-CACCGTGTTCATGAATCTGC-3′ |
| ISG15 (R) | 5′-CTTTATTTCCGGCCCTTGAT-3′ |
| VSV (F) | 5′-CAAGTCAAAATGCCCAAGAGTCACA-3′ |
| VSV (R) | 5′-TTTCCTTGCATTGTTCTACAGATGG-3′ |
| REC8 (F) | 5′-CATCCCACCAGAAGAACGG-3′ |
| REC8 (R) | 5′-GCACCAAAGGCATCTCCAT-3′ |
| CXCL9 (F) | 5′-GGTGTTCTTTTCCTCTTGGGC-3′ |
| CXCL9 (R) | 5′-TTCTCACTACTGGGGTTCCTTG-3′ |
| RNF5-RT-F | 5′-TTAAAAACTCCACCCCGCCC-3′ |
| RNF5-RT-R | 5′-CAAATGGCTGGAATCCCCCTC-3′ |
| UBC9-RT-F | 5′-GAGGGAAGTCCCGAGACAAA-3′ |
| UBC9-RT-R | 5′-ATGTTCAAAGTCCCTCGGGC-3′ |
| IFITM1-RT-F | 5′-CGGCTCTGTGACAGTCTACC-3′ |
| IFITM1-RT-R | 5′-CTGCTGTATCTAGGGGCAGG-3′ |
siRNA.
The siRNAs targeting REC8 were transfected into cells by using Lipofectamine 2000 (Lipo2000; Life Technologies), followed by immunoblot analysis or qPCR. The siRNA sequences used in this study are as follows: siREC8(1#), 5′-CCGGCUGCUUUGCCACCAUTT-3′; siREC8(2#), 5′-CCGCGUCUAUUCUCAACAATT-3′; siREC8(3#), 5′-GCUCUCAGCGCAACAGAUUTT-3′.
shRNA.
Double-stranded oligonucleotides corresponding to the target sequences were cloned into the pGreenPuro short hairpin RNA (shRNA) expression lentivector plasmids. The sequence 5′-GCGAGGAAATCCTCAATTACG-3′ was targeted for human REC8. The sequence 5′-GCAGAGGCCTACACGATTTAC-3′ was targeted for human UBC9. The sequence 5′-GAGTGTCCAGTATGTAAAGCT-3′ was targeted for human RNF5.
shRNA-transduced stable A549 cells, HEK293T cells, and THP-1 cells.
The 293T cells were transfected with two packaging plasmids (PAPX2 and PMD-2G) together with REC8-shRNA, UBC9-shRNA, or RNF5-shRNA lentiviral plasmid—by using Lipo2000. Twenty-four hours after transfection, the cells were incubated with new medium. The recombinant-virus-containing medium was collected every 8 h. The recombinant-virus-containing medium was filtered with a 0.45-μm filter (Millex) and then added to cultured A549 cells, HEK293T cells, and THP-1 cells. The infected cells were selected with puromycin (1 mg/mL for A549 cells and HEK293T cells or 0.5 mg/mL for THP-1 cells) for at least 7 days before additional experiments were performed.
Viruses.
VSV, Sendai virus (SeV), and HSV were incubated with 293T cells, followed by supernatant collection 24 h later. HCV was collected as previously described (6).
Luciferase assay.
Luciferase reporter vectors were cotransfected with pRL-CMV and other plasmids into 293T cells by using Lipo2000. Luciferase assays were performed with guidelines provided by the manufacturer (Promega).
Plasmids.
REC8 and its mutants, RNF5, UBC9, AIP4, and TRAF3 were subsequently cloned into the pcDNA3.1a vector or the p3×FLAG-CMV vector by standard molecular biology techniques. IFN-β, ISRE, and NF-κB luciferase reporter plasmids, mammalian expression plasmids for Myc- and Flag-tagged MAVS and its mutants, ubiquitin and its mutants, MDA5, RIG-I, RIG-I-N, TBK1, IRF3, IRF3-5D, and TRIM21 were described in our previous study (27). HA-SUMO1, HA-SUMO2, and HA-SUMO3 plasmids and HA- and Flag-tagged STING (MITA) and its mutants were kindly provided by Shu Hongbing (Wuhan University, Wuhan, China).
Coimmunoprecipitation and immunoblotting.
Cells were washed with ice-cold PBS and lysed in IP lysis buffer containing protease inhibitor cocktail. The cell lysates (400 μg) were incubated with the indicated antibodies at 4°C for 8 h and protein G agarose for 4 h. The immunoprecipitates were washed 3 times with PBS and subjected to immunoblot analysis.
Immunofluorescence staining.
A549 cells were seeded on glass coverslips and fixed with 4% methanol for 15 min at 20°C. Cells were blocked with goat serum (diluted in PBS to 1:50) for 60 min and then incubated with mouse monoclonal anti-MAVS antibody and rabbit monoclonal anti-REC8 antibody for 8 h at 4°C. Cells were then washed three times with PBS and stained with fluorescence-labeled secondary antibodies (diluted in PBS to 1:300; Invitrogen) for 60 min. Finally, the coverslips were washed with PBS three times and counterstained with DAPI (4′,6-diamidino-2-phenylindole). Fluorescent images were obtained with a fluorescence microscope (Olympus). Quantitative colocalization were analyzed by Image-Pro Plus 6.0.
Statistical analysis.
Experiments were independently repeated two or three times, with similar results. Statistical analysis was performed by Student’s two-sided t test, and the data are presented as means and standard deviations (SD) for three biological replicates.
ACKNOWLEDGMENTS
We thank Xuetao Cao for SeV, VSV, and HSV. We also thank Chen Liu, Hongbing Shu, Jianguo Wu, Zhengfan Jiang, Deyin Guo, and Chengjiang Gao for kindly sharing research materials.
This work was supported by the National Natural Science Foundation of China (81730064, 82072269, 81571985, and 81902069), the National Science and Technology Major Project (2017ZX10202201), the Hunan Natural Science Foundation (2018JJ3090), and the Postgraduate Scientific Research Innovation Project of Hunan Province (CX20190275).
Contributor Information
Haizhen Zhu, Email: zhuhaizhen69@yahoo.com.
Bryan R. G. Williams, Hudson Institute of Medical Research
REFERENCES
- 1.Zindel J, Kubes P. 2020. DAMPs, PAMPs, and LAMPs in immunity and sterile inflammation. Annu Rev Pathol 15:493–518. 10.1146/annurev-pathmechdis-012419-032847. [DOI] [PubMed] [Google Scholar]
- 2.Luecke S, Sheu KM, Hoffmann A. 2021. Stimulus-specific responses in innate immunity: multilayered regulatory circuits. Immunity 54:1915–1932. 10.1016/j.immuni.2021.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartok E, Hartmann G. 2020. Immune sensing mechanisms that discriminate self from altered self and foreign nucleic acids. Immunity 53:54–77. 10.1016/j.immuni.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cadena C, Hur S. 2019. Filament-like assemblies of intracellular nucleic acid sensors: commonalities and differences. Mol Cell 76:243–254. 10.1016/j.molcel.2019.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzgerald KA, Kagan JC. 2020. Toll-like receptors and the control of immunity. Cell 180:1044–1066. 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Q, Chen S, Tian R, Huang X, Deng R, Xue B, Qin Y, Xu Y, Wang J, Guo M, Chen J, Tang S, Li G, Zhu H. 2018. Long noncoding RNA ITPRIP-1 positively regulates the innate immune response through promotion of oligomerization and activation of MDA5. J Virol 92:e00507-18. 10.1128/JVI.00507-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mills EL, Kelly B, O’Neill LAJ. 2017. Mitochondria are the powerhouses of immunity. Nat Immunol 18:488–498. 10.1038/ni.3704. [DOI] [PubMed] [Google Scholar]
- 8.Chen Q, Sun L, Chen ZJ. 2016. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 17:1142–1149. 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 9.Li W, Li N, Dai S, Hou G, Guo K, Chen X, Yi C, Liu W, Deng F, Wu Y, Cao X. 2019. Zika virus circumvents host innate immunity by targeting the adaptor proteins MAVS and MITA. FASEB J 33:9929–9944. 10.1096/fj.201900260R. [DOI] [PubMed] [Google Scholar]
- 10.Sun Y, Zheng H, Yu S, Ding Y, Wu W, Mao X, Liao Y, Meng C, Ur Rehman Z, Tan L, Song C, Qiu X, Wu F, Ding C. 2019. Newcastle disease virus V protein degrades mitochondrial antiviral signaling protein to inhibit host type I interferon production via E3 ubiquitin ligase RNF5. J Virol 93:e00322-19. 10.1128/JVI.00322-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei C, Ni C, Song T, Liu Y, Yang X, Zheng Z, Jia Y, Yuan Y, Guan K, Xu Y, Cheng X, Zhang Y, Yang X, Wang Y, Wen C, Wu Q, Shi W, Zhong H. 2010. The hepatitis B virus X protein disrupts innate immunity by downregulating mitochondrial antiviral signaling protein. J Immunol 185:1158–1168. 10.4049/jimmunol.0903874. [DOI] [PubMed] [Google Scholar]
- 12.Yoo YS, Park YY, Kim JH, Cho H, Kim SH, Lee HS, Kim TH, Sun Kim Y, Lee Y, Kim CJ, Jung JU, Lee JS, Cho H. 2015. The mitochondrial ubiquitin ligase MARCH5 resolves MAVS aggregates during antiviral signalling. Nat Commun 6:7910. 10.1038/ncomms8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong B, Zhang Y, Tan B, Liu TT, Wang YY, Shu HB. 2010. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J Immunol 184:6249–6255. 10.4049/jimmunol.0903748. [DOI] [PubMed] [Google Scholar]
- 14.Pan Y, Li R, Meng J-L, Mao H-T, Zhang Y, Zhang J. 2014. Smurf2 negatively modulates RIG-I–dependent antiviral response by targeting VISA/MAVS for ubiquitination and degradation. J Immunol 192:4758–4764. 10.4049/jimmunol.1302632. [DOI] [PubMed] [Google Scholar]
- 15.You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z. 2009. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol 10:1300–1308. 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Tong X, Ye X. 2012. Ndfip1 negatively regulates RIG-I-dependent immune signaling by enhancing E3 ligase Smurf1-mediated MAVS degradation. J Immunol 189:5304–5313. 10.4049/jimmunol.1201445. [DOI] [PubMed] [Google Scholar]
- 17.Castanier C, Zemirli N, Portier A, Garcin D, Bidere N, Vazquez A, Arnoult D. 2012. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol 10:44. 10.1186/1741-7007-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. 2009. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30:397–407. 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Hassler M, Shaltiel IA, Haering CH. 2018. Towards a unified model of SMC complex function. Curr Biol 28:R1266–R1281. 10.1016/j.cub.2018.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nichols MH, Corces VG. 2018. A tethered-inchworm model of SMC DNA translocation. Nat Struct Mol Biol 25:906–910. 10.1038/s41594-018-0135-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhlmann F. 2016. SMC complexes: from DNA to chromosomes. Nat Rev Mol Cell Biol 17:399–412. 10.1038/nrm.2016.30. [DOI] [PubMed] [Google Scholar]
- 22.Murphy CM, Xu Y, Li F, Nio K, Reszka-Blanco N, Li X, Wu Y, Yu Y, Xiong Y, Su L. 2016. Hepatitis B virus X protein promotes degradation of SMC5/6 to enhance HBV replication. Cell Rep 16:2846–2854. 10.1016/j.celrep.2016.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupont L, Bloor S, Williamson JC, Cuesta SM, Shah R, Teixeira-Silva A, Naamati A, Greenwood EJD, Sarafianos SG, Matheson NJ, Lehner PJ. 2021. The SMC5/6 complex compacts and silences unintegrated HIV-1 DNA and is antagonized by Vpr. Cell Host Microbe 29:792–805.E6. 10.1016/j.chom.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Wang C, Li N, Liu X, Ren W, Wang Q, Cao X. 2018. Condensin Smc4 promotes inflammatory innate immune response by epigenetically enhancing NEMO transcription. J Autoimmun 92:67–76. 10.1016/j.jaut.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Ma W, Zhou J, Chen J, Carr AM, Watanabe Y. 2021. Meikin synergizes with shugoshin to protect cohesin Rec8 during meiosis I. Genes Dev 35:692–697. 10.1101/gad.348052.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang D, Zuo C, Wang X, Meng X, Xue B, Liu N, Yu R, Qin Y, Gao Y, Wang Q, Hu J, Wang L, Zhou Z, Liu B, Tan D, Guan Y, Zhu H. 2014. Complete replication of hepatitis B virus and hepatitis C virus in a newly developed hepatoma cell line. Proc Natl Acad Sci USA 111:E1264–E1273. 10.1073/pnas.1320071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue B, Li H, Guo M, Wang J, Xu Y, Zou X, Deng R, Li G, Zhu H. 2018. TRIM21 promotes innate immune response to RNA viral infection through Lys27-linked polyubiquitination of MAVS. J Virol 92:e00321-18. 10.1128/JVI.00321-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Li H, Xue B, Deng R, Huang X, Xu Y, Chen S, Tian R, Wang X, Xun Z, Sang M, Zhu H. 2020. IRF1 promotes the innate immune response to viral infection by enhancing the activation of IRF3. J Virol 94:e01231-20. 10.1128/JVI.01231-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zinchuk V, Wu Y, Grossenbacher-Zinchuk O. 2013. Bridging the gap between qualitative and quantitative colocalization results in fluorescence microscopy studies. Sci Rep 3:1365. 10.1038/srep01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia P, Wang S, Xiong Z, Ye B, Huang LY, Han ZG, Fan Z. 2015. IRTKS negatively regulates antiviral immunity through PCBP2 sumoylation-mediated MAVS degradation. Nat Commun 6:8132. 10.1038/ncomms9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J. 2014. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res 42:W325–W330. 10.1093/nar/gku383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrat FJ, Crow MK, Ivashkiv LB. 2019. Interferon target-gene expression and epigenomic signatures in health and disease. Nat Immunol 20:1574–1583. 10.1038/s41590-019-0466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Graf L, Chen T, Liao Q, Bai T, Petric PP, Zhu W, Yang L, Dong J, Lu J, Chen Y, Shen J, Haller O, Staeheli P, Kochs G, Wang D, Schwemmle M, Shu Y. 2021. Rare variant MX1 alleles increase human susceptibility to zoonotic H7N9 influenza virus. Science 373:918–922. 10.1126/science.abg5953. [DOI] [PubMed] [Google Scholar]
- 35.Majdoul S, Compton AA. 2021. Lessons in self-defence: inhibition of virus entry by intrinsic immunity. Nat Rev Immunol. 10.1038/s41577-021-00626-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zang R, Case JB, Yutuc E, Ma X, Shen S, Gomez Castro MF, Liu Z, Zeng Q, Zhao H, Son J, Rothlauf PW, Kreutzberger AJB, Hou G, Zhang H, Bose S, Wang X, Vahey MD, Mani K, Griffiths WJ, Kirchhausen T, Fremont DH, Guo H, Diwan A, Wang Y, Diamond MS, Whelan SPJ, Ding S. 2020. Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion. Proc Natl Acad Sci USA 117:32105–32113. 10.1073/pnas.2012197117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao X, Sehgal M, Hou Z, Cheng J, Shu S, Wu S, Guo F, Le Marchand SJ, Lin H, Chang J, Guo JT. 2018. Identification of residues controlling restriction versus enhancing activities of IFITM proteins on entry of human coronaviruses. J Virol 92:e01535-17. 10.1128/JVI.01535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi YJ, Bowman JW, Jung JU. 2018. A talented duo: IFIT1 and IFIT3 patrol viral RNA caps. Immunity 48:474–476. 10.1016/j.immuni.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixit U, Pandey AK, Mishra P, Sengupta A, Pandey VN. 2016. Staufen1 promotes HCV replication by inhibiting protein kinase R and transporting viral RNA to the site of translation and replication in the cells. Nucleic Acids Res 44:5271–5287. 10.1093/nar/gkw312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takata MA, Gonçalves-Carneiro D, Zang TM, Soll SJ, York A, Blanco-Melo D, Bieniasz PD. 2017. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 550:124–127. 10.1038/nature24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang YD, Na L, Zhu CH, Shen N, Yang F, Fu XQ, Wang YH, Fu LH, Wang JY, Lin YZ, Wang XF, Wang X, Zhou JH, Li CY. 2014. Equine viperin restricts equine infectious anemia virus replication by inhibiting the production and/or release of viral Gag, Env, and receptor via distortion of the endoplasmic reticulum. J Virol 88:12296–12310. 10.1128/JVI.01379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liberatore RA, Mastrocola EJ, Powell C, Bieniasz PD. 2017. Tetherin inhibits cell-free virus dissemination and retards murine leukemia virus pathogenesis. J Virol 91:e02286-16. 10.1128/JVI.02286-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbalat R, Ewald SE, Mouchess ML, Barton GM. 2011. Nucleic acid recognition by the innate immune system. Annu Rev Immunol 29:185–214. 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 44.Liuyu T, Yu K, Ye L, Zhang Z, Zhang M, Ren Y, Cai Z, Zhu Q, Lin D, Zhong B. 2019. Induction of OTUD4 by viral infection promotes antiviral responses through deubiquitinating and stabilizing MAVS. Cell Res 29:67–79. 10.1038/s41422-018-0107-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. 2010. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33:765–776. 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 46.Hong S, Joo JH, Yun H, Kleckner N, Kim KP. 2019. Recruitment of Rec8, Pds5 and Rad61/Wapl to meiotic homolog pairing, recombination, axis formation and S-phase. Nucleic Acids Res 47:11691–11708. 10.1093/nar/gkz903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva MCC, Powell S, Ladstatter S, Gassler J, Stocsits R, Tedeschi A, Peters JM, Tachibana K. 2020. Wapl releases Scc1-cohesin and regulates chromosome structure and segregation in mouse oocytes. J Cell Biol 219:e201906100. 10.1083/jcb.201906100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dreissig S, Maurer A, Sharma R, Milne L, Flavell AJ, Schmutzer T, Pillen K. 2020. Natural variation in meiotic recombination rate shapes introgression patterns in intraspecific hybrids between wild and domesticated barley. New Phytol 228:1852–1863. 10.1111/nph.16810. [DOI] [PubMed] [Google Scholar]
- 49.Acquaviva L, Boekhout M, Karasu ME, Brick K, Pratto F, Li T, van Overbeek M, Kauppi L, Camerini-Otero RD, Jasin M, Keeney S. 2020. Ensuring meiotic DNA break formation in the mouse pseudoautosomal region. Nature 582:426–431. 10.1038/s41586-020-2327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shahid S. 2020. The rules of attachment: REC8 cohesin connects chromatin architecture and recombination machinery in meiosis. Plant Cell 32:808–809. 10.1105/tpc.20.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lambing C, Tock AJ, Topp SD, Choi K, Kuo PC, Zhao X, Osman K, Higgins JD, Franklin FCH, Henderson IR. 2020. Interacting genomic landscapes of REC8-cohesin, chromatin, and meiotic recombination in Arabidopsis. Plant Cell 32:1218–1239. 10.1105/tpc.19.00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mengoli V, Jonak K, Lyzak O, Lamb M, Lister LM, Lodge C, Rojas J, Zagoriy I, Herbert M, Zachariae W. 2021. Deprotection of centromeric cohesin at meiosis II requires APC/C activity but not kinetochore tension. EMBO J 40:e106812. 10.15252/embj.2020106812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y, Wang Y, Upadhyay S, Xue C, Lin X. 2020. Activation of meiotic genes mediates ploidy reduction during cryptococcal infection. Curr Biol 30:1387–1396.E5. 10.1016/j.cub.2020.01.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Litvinov IV, Cordeiro B, Huang Y, Zargham H, Pehr K, Dore MA, Gilbert M, Zhou Y, Kupper TS, Sasseville D. 2014. Ectopic expression of cancer-testis antigens in cutaneous T-cell lymphoma patients. Clin Cancer Res 20:3799–3808. 10.1158/1078-0432.CCR-14-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu M, Xu W, Su M, Fan P. 2020. REC8 suppresses tumor angiogenesis by inhibition of NF-kappaB-mediated vascular endothelial growth factor expression in gastric cancer cells. Biol Res 53:41. 10.1186/s40659-020-00307-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao J, Geng L, Duan G, Xu W, Cheng Y, Huang Z, Zhou Z, Gong S. 2018. REC8 inhibits EMT by downregulating EGR1 in gastric cancer cells. Oncol Rep 39:1583–1590. 10.3892/or.2018.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu J, Liang Q, Wang J, Wang K, Gao J, Zhang J, Zeng Y, Chiu PW, Ng EK, Sung JJ. 2017. REC8 functions as a tumor suppressor and is epigenetically downregulated in gastric cancer, especially in EBV-positive subtype. Oncogene 36:182–193. 10.1038/onc.2016.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qin Y, Xue B, Liu C, Wang X, Tian R, Xie Q, Guo M, Li G, Yang D, Zhu H. 2017. NLRX1 Mediates MAVS degradation to attenuate the hepatitis C virus-induced innate immune response through PCBP2. J Virol 91:e01264-17. 10.1128/JVI.01264-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu B, Zhang M, Chu H, Zhang H, Wu H, Song G, Wang P, Zhao K, Hou J, Wang X, Zhang L, Gao C. 2017. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat Immunol 18:214–224. 10.1038/ni.3641. [DOI] [PubMed] [Google Scholar]
- 60.Ni G, Konno H, Barber GN. 2017. Ubiquitination of STING at lysine 224 controls IRF3 activation. Sci Immunol 2:eaah7119. 10.1126/sciimmunol.aah7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Q, Liu X, Cui Y, Tang Y, Chen W, Li S, Yu H, Pan Y, Wang C. 2014. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 41:919–933. 10.1016/j.immuni.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J, Hu MM, Wang YY, Shu HB. 2012. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem 287:28646–28655. 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye L, Zhang Q, Liuyu T, Xu Z, Zhang MX, Luo MH, Zeng WB, Zhu Q, Lin D, Zhong B. 2019. USP49 negatively regulates cellular antiviral responses via deconjugating K63-linked ubiquitination of MITA. PLoS Pathog 15:e1007680. 10.1371/journal.ppat.1007680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun H, Zhang Q, Jing YY, Zhang M, Wang HY, Cai Z, Liuyu T, Zhang ZD, Xiong TC, Wu Y, Zhu QY, Yao J, Shu HB, Lin D, Zhong B. 2017. USP13 negatively regulates antiviral responses by deubiquitinating STING. Nat Commun 8:15534. 10.1038/ncomms15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lam KC, Araya RE, Huang A, Chen Q, Di Modica M, Rodrigues RR, Lopes A, Johnson SB, Schwarz B, Bohrnsen E, Cogdill AP, Bosio CM, Wargo JA, Lee MP, Goldszmid RS. 2021. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell 184:5338–5356.E21. 10.1016/j.cell.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liang K, Abt ER, Le TM, Cho A, Dann AM, Cui J, Li L, Rashid K, Creech AL, Wei L, Ghukasyan R, Rosser EW, Wu N, Carlucci G, Czernin J, Donahue TR, Radu CG. 2021. STING-driven interferon signaling triggers metabolic alterations in pancreas cancer cells visualized by [(18)F]FLT PET imaging. Proc Natl Acad Sci USA 118:e2105390118. 10.1073/pnas.2105390118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu X, Wang H, Li X, Guo C, Yuan F, Fisher PB, Wang XY. 2016. Activation of the MDA-5-IPS-1 viral sensing pathway induces cancer cell death and type I IFN-dependent antitumor immunity. Cancer Res 76:2166–2176. 10.1158/0008-5472.CAN-15-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang W, Gong J, Yang H, Wan L, Peng Y, Wang X, Sun J, Li F, Geng Y, Li D, Liu N, Mei G, Cao Y, Yan Q, Li H, Zhang Y, He X, Zhang Q, Zhang R, Wu F, Zhong H, Wei C. 2020. The mitochondrial protein MAVS stabilizes p53 to suppress tumorigenesis. Cell Rep 30:725–738.E4. 10.1016/j.celrep.2019.12.051. [DOI] [PubMed] [Google Scholar]
- 69.Chang HM, Yeh ETH. 2020. SUMO: from bench to bedside. Physiol Rev 100:1599–1619. 10.1152/physrev.00025.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chelbi-Alix MK, Thibault P. 2021. Crosstalk between SUMO and ubiquitin-like proteins: implication for antiviral defense. Front Cell Dev Biol 9:671067. 10.3389/fcell.2021.671067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao X. 2018. SUMO-mediated regulation of nuclear functions and signaling processes. Mol Cell 71:409–418. 10.1016/j.molcel.2018.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodriguez JA. 2014. Interplay between nuclear transport and ubiquitin/SUMO modifications in the regulation of cancer-related proteins. Semin Cancer Biol 27:11–19. 10.1016/j.semcancer.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 73.Nie M, Boddy MN. 2016. Cooperativity of the SUMO and ubiquitin pathways in genome stability. Biomolecules 6:14. 10.3390/biom6010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu MM, Liao CY, Yang Q, Xie XQ, Shu HB. 2017. Innate immunity to RNA virus is regulated by temporal and reversible sumoylation of RIG-I and MDA5. J Exp Med 214:973–989. 10.1084/jem.20161015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mi Z, Fu J, Xiong Y, Tang H. 2010. SUMOylation of RIG-I positively regulates the type I interferon signaling. Protein Cell 1:275–283. 10.1007/s13238-010-0030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crowl JT, Stetson DB. 2018. SUMO2 and SUMO3 redundantly prevent a noncanonical type I interferon response. Proc Natl Acad Sci USA 115:6798–6803. 10.1073/pnas.1802114115. [DOI] [PMC free article] [PubMed] [Google Scholar]