Abstract
TYW1 is a radical S-adenosyl-l-methionine (SAM) enzyme that catalyzes the condensation of pyruvate and N-methylguanosine-containing tRNAPhe, forming 4-demethylwyosine-containing tRNAPhe. Homologues of TYW1 are found in both archaea and eukarya; archaeal homologues consist of a single domain, while eukaryal homologues contain a flavin binding domain in addition to the radical SAM domain shared with archaeal homologues. In this study, TYW1 from Saccharomyces cerevisiae (ScTYW1) was heterologously expressed in Escherichia coli and purified to homogeneity. ScTYW1 is purified with 0.54 ± 0.07 and 4.2 ± 1.9 equiv of flavin mononucleotide (FMN) and iron, respectively, per mole of protein, suggesting the protein is ∼50% replete with Fe–S clusters and FMN. While both NADPH and NADH are sufficient for activity, significantly more product is observed when used in combination with flavin nucleotides. ScTYW1 is the first example of a radical SAM flavoenzyme that is active with NAD(P)H alone.
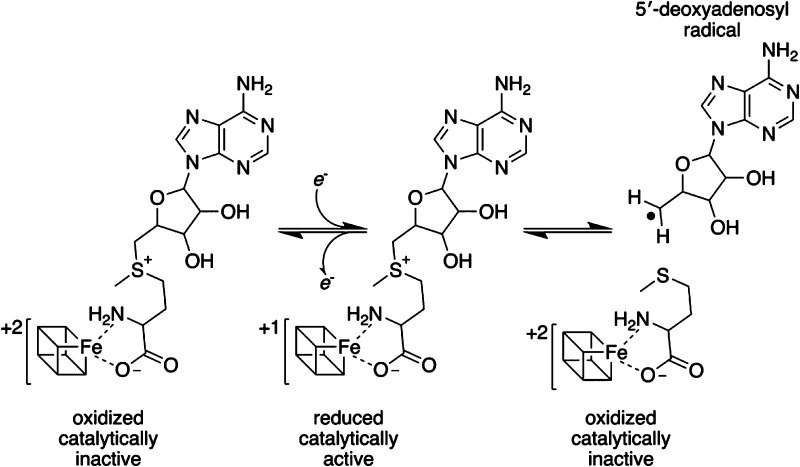
The radical S-adenosyl-l-methionine (SAM) superfamily of enzymes is comprised of more than 100000 members1 that catalyze a wide variety of transformations on substrates that vary from small molecules to macromolecules.2 Radical SAM (RS) enzymes all share a 4Fe–4S cluster coordinated by a cysteine-rich motif, which is typically comprised of CxxxCxxC.3 Three of the Fe ions in the cluster ligate the conserved cysteine residues in the motif, while the remaining Fe ligates to the α-amino and α-carboxy moieties of SAM in the catalytic complex.4−18 Cleavage of SAM by the reduced cluster in the +1 oxidation state results in the formation of methionine and a 5′-deoxyadenosyl radical (dAdo•), which in the majority of cases, initiates catalysis by H atom abstraction (see Scheme 1).5,19 Most characterized RS enzymes use SAM stoichiometrically, whereas in some cases, SAM is utilized as a cofactor and re-formed at the end of the catalytic cycle.2 Additional differences have also been noted, including variations in the sequence of the cluster binding motif,8,20 an alternative site of cleavage to produce the 3-amino-3-carboxylpropyl radical,21,22 or radical addition in place of H atom abstraction.23,24
Scheme 1. Reductive Cleavage of SAM by a 4Fe–4S Cluster.
A key requirement for activation of all RS enzymes is the obligate reduction from the resting +2 state of the Fe–S cluster to the catalytically active +1 oxidation state. Most in vitro studies employ dithionite as a reductant,25−35 though other non-natural reducing systems such as Ti(III) citrate20,36 and various mediators have also been shown to be effective37,38 in some but not all cases.39 Since the demonstration that ribonucleotide reductase40 can be activated by flavodoxin/flavodoxin reductase with NADPH as the electron source, this reducing system (from Escherichia coli) has also been employed as a proxy for the cellular reducing system.25,29,41,42 This has led to the generalization that a flavodoxin-like protein is most likely involved in the activation of RS enzymes in vivo. It is somewhat remarkable that the E. coli flavodoxin homologue has been successfully used to reconstitute activity in a wide variety of RS enzymes, as there is no reason to expect that the surfaces that drive the interactions between the flavodoxin homologue and RS enzymes are identical.37 Indeed, structural studies in one system highlight significant differences between the surfaces, and biochemical studies of the same suggest that optimal reduction may require a cognate flavodoxin.43
Many organisms encode several flavodoxin-like proteins, and to the best of our knowledge, a connection between a particular redox partner and a RS protein has been made in only two systems. In the first, studies with a RS enzyme involved in the formation of the diphthamide post-translational modification identified a protein proposed to be its reductant.44 In Saccharomyces cerevisiae, the Dph1/2 complex installs the diphthamide modification and Dph3, a CSL zinc finger-type protein, is the reductant in this process. Dph3 is an iron-containing protein that, when reduced, stimulates the formation of diphthamide.44 Unlike the Dph system, the reductant in other cases is not clear, and it is possible that many cellular reductants can facilitate reduction of the RS protein. For example, Thermatoga maritima does not encode any flavodoxin homologues, but it harbors five ferredoxins, which with the ferredoxin-NADP+ oxidoreductase from the same organism support the activity of the RS enzyme MiaB.45
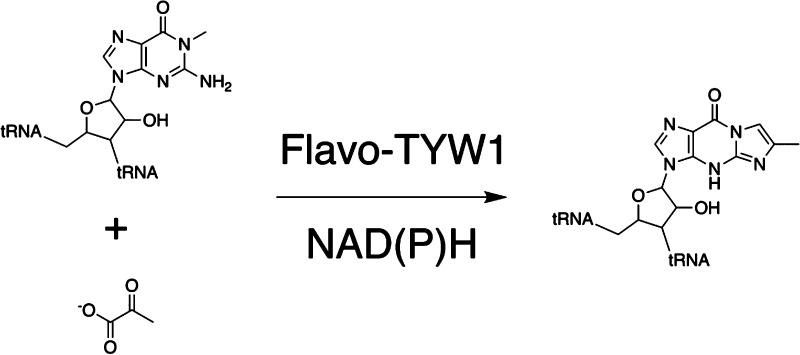
The tricyclic modified base wybutosine and its analogues are found at position 37 in tRNAPhe of many eukaryal and archaeal species.46,47 TYW1 catalyzes the key step in the pathway, which entails the condensation of pyruvate and N-methylguanosine (m1G)-containing tRNAPhe to install 4-demethylwyosine (imG-14) (Scheme 2).48
Scheme 2. Reaction Catalyzed by TYW1.
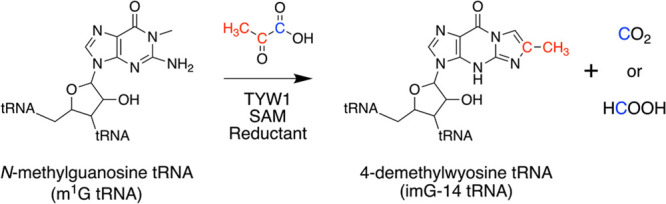
Biochemical studies show that TYW1 catalyzes a complex radical-mediated condensation and ring closure to convert m1G to imG-14.48−52 The incorporation of C2 and C3 into imG-14 was shown by pyruvate isotopologues.48 The methyl moiety of the substrate m1G was identified as the site of H atom abstraction by tracing the isotope from a deuterated analogue to 5′-deoxyadenosine (dAdoH).50 These findings led to a paradigm in which H atom abstraction by dAdo• from the methyl group of m1G initiates the transformation. On the basis of sequence conservation and in vivo complementation experiments, a Lys residue was proposed to play a role in activating the pyruvate substrate, possibly as a Schiff base.48,50,53 This proposal was subsequently confirmed by biochemical studies that identified the modified Lys, and an X-ray crystal structure that revealed electron density consistent with a pyruvate–Lys adduct in the active site.49 The structure also revealed the position of a second 4Fe–4S cluster, a so-called auxiliary cluster, with an open coordination site that is engaged with the nitrogen of the Schiff base and the oxygen of the carboxylate of pyruvate.49 The eventual conversion to imG-14 requires the loss of C1 of pyruvate, the fate of which has not been established. However, the intimate interaction between the auxiliary cluster and the pyruvate suggests that the cluster is central to this process.49
Interestingly, while the archaeal homologues of TYW1 are single-domain proteins, the eukaryotic homologues consist of a flavodoxin_1 domain that is appended to the N-terminus of the RNA-modifying TYW1 domain (see Figure 1).54 The NCBI conserved domains tool55 identifies residues 207–354 of S. cerevisiae TYW1 (ScTYW1) as the flavodoxin_1 domain, pfam 00258 (E value of 1.21 × 10–23). Members of this protein family, such as anaerobic nitric oxide reductase, sulfite reductase, and flavodoxin, are typically flavoproteins that bind either flavin mononucleotide (FMN) or flavin adenine dinucleotide (FAD).56 The eukaryal TYW1 homologues are, to the best of our knowledge, the only members of the RS superfamily that encode a flavodoxin_1 domain attached to the RS domain. The physiological significance of the flavodoxin_1 domain is not known. However, one hypothesis that may be consistent with this observation is that the flavodoxin_1 domain functions as an in situ reducing system to activate the RS enzyme, which may suggest that fused proteins represent functional linkages.57
Figure 1.
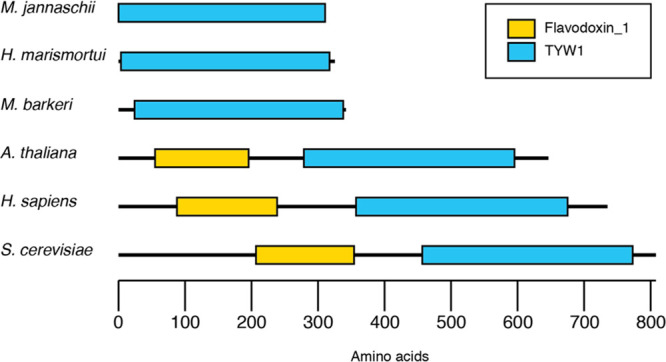
Comparison of TYW1 homologues from archaea and eukarya. The mammalian homologues are unique in that they harbor a flavodoxin_1 domain (yellow) appended to the RNA modification domain (blue).
In this paper, we describe the purification and characterization of the eukaryal TYW1 from S. cerevisiae. In addition to the Fe–S clusters, the protein is shown to be purified with FMN. Reconstitution of the catalytic activity requires the addition of only NAD(P)H, which supports the notion that the flavodoxin_1 domain serves to reduce the RS cluster to the +1 oxidation state and support turnover. Eukaryotic TYW1 is the first example of a RS flavoprotein.
Results and Discussion
Expression and Purification of ScTYW1
Residues 1–45 of ScTYW1 (UniProtKB Q08960) are predicted to be a signal and transmembrane domain. Therefore, the protein expressed and used in these studies consisted of residues 46–810 to enhance solubility. A codon-optimized gene encoding residues 46–810 of ScTYW1 was expressed with an N-terminal His6 tag and TEV protease site, resulting in protein that is at least 90% pure following purification (Figure 2).
Figure 2.
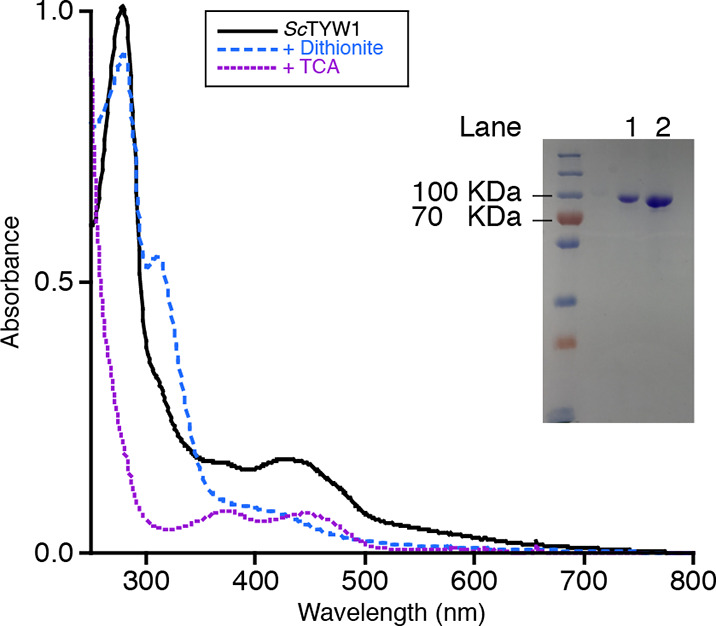
Ultraviolet–visible spectra and a 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel of purified ScTYW1. The black trace is purified and reconstituted ScTYW1; the blue dashed trace is after the addition of dithionite, and the purple dotted trace is for the supernatant following TCA treatment. Lanes 1 and 2 of the SDS–PAGE gel contain 5 and 10 μg of protein, respectively.
Cofactor Analysis
Trichloroacetic acid (TCA) precipitation of ScTYW1 purified with no added flavin nucleotide(s) revealed a supernatant with an absorbance spectrum consistent with the presence of a flavin cofactor (Figure 2). High-performance liquid chromatography analysis, along with comparison to authentic FMN and FAD standards, identified the flavin cofactor as FMN. Subsequently, FMN was added to the purification to obtain more complete cofactor incorporation. Quantification of the FMN content of the final protein revealed 0.54 ± 0.07 mol of FMN per mole of protein (average of three independent purifications). The iron content of ScTYW1 is 4.2 ± 1.9 mol of iron per mole of protein (average of three independent purifications). The preparation used in this work contained 0.58 mol of FMN and 2.6 mol of iron per mole of protein. TYW1 harbors two 4Fe–4S clusters, which are required for activity. The stoichiometry of Fe and flavin suggests that the protein is generally no more than 50% replete with each cofactor.
Ultraviolet–Visible (UV–vis) Spectroscopy of ScTYW1
The UV–visible spectrum of 10 μM ScTYW1 is shown in Figure 2 (black solid line). While RS enzymes generally exhibit a broad shoulder at 420 nm due to 4Fe–4S clusters, ScTYW1 has a prominent peak at 450 nm instead. Addition of a 10-fold molar excess of dithionite results in the bleaching of this spectral feature (blue dashed line) and a spectrum that is consistent with a reduced 4Fe–4S cluster. The supernatant obtained following denaturation with TCA and removal of precipitated protein, however, has the characteristic features of oxidized flavin, with peaks at ∼380 and 450 nm. These data unambiguously show that in contrast to all other RS enzymes, ScTYW1 harbors flavin.
Activity of ScTYW1 with Dithionite
The activity of TYW1 has been demonstrated with homologues from the archaeal species Methanocaldococcus jannaschii and Pyrococcus abyssi using dithionite with methyl viologen and dithionite alone, respectively, as the reductant.48,51 Consequently, initial activity assays with ScTYW1 were performed using dithionite as the reductant. The tRNA substrate used in this study was extracted from S. cerevisiae strain ΔYPL207W harboring a deletion in the gene that encodes TYW1.54,58 The assays contained (1,2,3-13C3)-pyruvate to avoid overlap with a contaminating species that elutes with a retention time and m/z values similar to those of imG-14. Figure S.4 shows the extracted ion chromatogram at m/z 324.1–324.2 (the expected m/z value of the product with three 13C atoms is 324.187) of the digested RNA extracted from the complete reaction mixture and control experiments with one of the reaction components removed. The modified base is produced when all of the components are present (blue trace). When ScTYW1, dithionite, pyruvate, SAM, or tRNA is removed, there is no product produced. These observations demonstrate that ScTYW1 can be reduced by dithionite and that it catalyzes the same overall reaction as the two previously characterized homologues of TYW1.
Activity of ScTYW1 with Different Reductants
As eukaryotic homologues of TYW1 contain a “flavodoxin_1” domain, we hypothesized that this enzyme may be able to use the reduced nicotinamide cofactors (NADH and NADPH) as the reductant directly. In addition to NADH and NADPH alone, dithionite, FMN, FAD, and FMN and FAD in combination with NADH and NADPH were tested as reductants. Initially, 100 μM FMN or FAD was used in the assays. When activity was detected, a series of FMN concentrations (from 0 to 60 μM) were tested to determine the optimal concentration to include in the assays. All of the concentrations above 10 μM produced approximately the same amount of product after 4 h (Figure S.5). This observation is intriguing. Recall that the stoichiometry of FMN to the protein is 50%. These experiments were carried out in the presence of 15 μM enzyme, and the flavin concentration profiles show that addition of 10 μM FMN is sufficient to restore maximal activity. The simplest interpretation of this is that the FMN that is supplied during the assays can reconstitute the activity of the protein in situ.
Because control experiments indicated that maximal activity could be observed at ∼10 μM FMN, all subsequent experiments were carried out in the presence of 20 μM FMN or FAD. After incubation with the protein, the RNA was extracted and digested to nucleosides and analyzed by LC-MS. Figure 3 shows the extracted ion chromatogram at m/z 324.1–324.2. As in the control experiments described above, imG-14 was formed in the presence of dithionite. However, the product is also observed in the absence of dithionite, as long as NADPH or NADH is present (Figure 3, inset). Substantially more product is formed when NADH or NADPH is present in addition to FAD or FMN. However, FAD or FMN alone does not support product formation. These data unambiguously show that eukaryotic TYW1 does not require any strong reductants (such as dithionite) for activity and utilizes pyridine nucleotides to support turnover.
Figure 3.
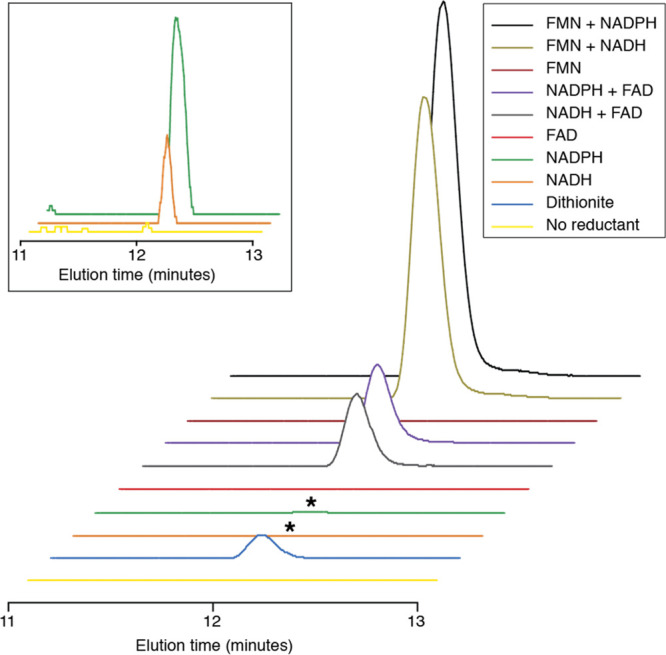
Extracted ion chromatogram at m/z 324.1–324.2 of the digested RNA when ScTYW1 is incubated with the reductants shown for 4 h. The inset shows the traces for NADPH, NADH, and no reductant on a smaller scale.
Kinetic profiles of the reaction support the observation that pyridine nucleotides support turnover by TYW1. A time course with aliquots removed at 1, 2, and 4 h was performed on samples containing either dithionite, NADH, NADPH, or FMN and FAD in combination with NADH or NADPH (Figure 4). RNA was digested to the nucleoside level and analyzed by LC-MS. imG-14 forms with either NADPH or NADH alone. imG-14 forms in a time-dependent manner with reducing systems containing FMN and NAD(P)H or FAD and NAD(P)H. Activity is also observed with dithionite alone, as shown in Figure 3. By contrast, control reactions show that while NAD(P)H alone does not support the activity of the archaeal homologue (UniProtKB Q57705), this enzyme is similarly active with FMN and NAD(P)H or FAD and NAD(P)H (Figure S.6). These data demonstrate that the addition of nicotinamide cofactors alone is sufficient to support turnover of eukaryotic TYW1, via the appended flavodoxin_1 domain.
Figure 4.
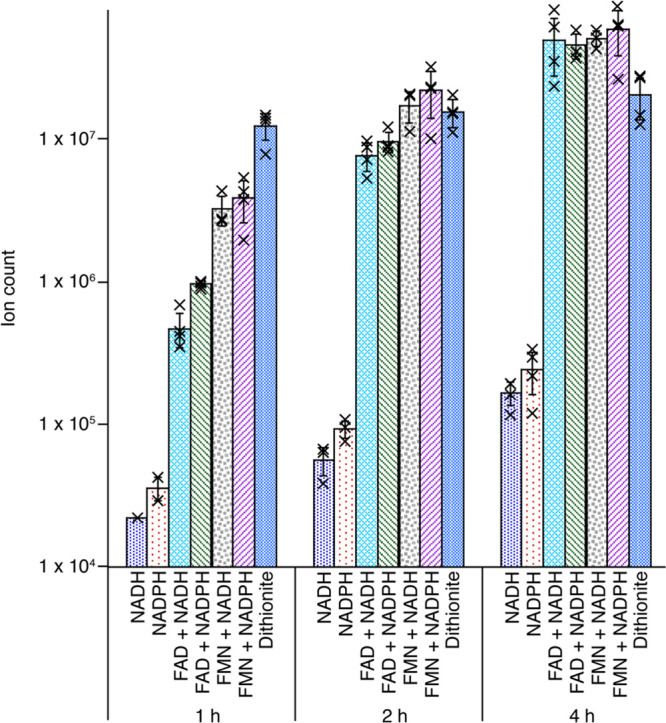
Time and reductant dependence of ScTYW1 activity. The reaction mixtures contained the indicated reducing systems. Samples were analyzed 1, 2, or 4 h after initiation of the reaction. The symbol X represents an individual data point, and the error bars represent one standard deviation from the mean. Note that the y-axis has a log scale.
We note that the data show that at each time point, NADH or NADPH alone produces at least 70-fold less product than dithionite or the flavin/pyridine nucleotide reducing system. All of the assays were carried out in the presence of the same concentration of enzyme (15 μM). NAD(P)H is a two-electron reductant necessitating the initial transfer of reducing equivalents to the bound flavin, prior to one-electron reduction of the cluster. Direct comparisons of product formed do not account for the concentrations of reductants and the differences in their midpoint potentials. In this context, the observation that NAD(P)H alone can support the activity of ScTYW1 is notable.
We were initially surprised by the observation that FMN or FAD could support protein activity in the presence of NADH or NADPH with both MjTYW1 and ScTYW1, suggesting that pyridine nucleotides could directly reduce the flavin nucleotide in solution. To probe this directly, equal volumes of 4 mM NADPH and 40 μM FMN were placed in the chambers of a split-cell quartz cuvette. The absorbance spectrum prior to mixing shows the expected features of the flavin in the range of 400–500 nm (Figure S.7A). When the contents of the two chambers were mixed, there is a time-dependent reduction of the absorbance at ∼450 nm, which results from reduction of the FMN. Control experiments in which NADPH was omitted show no change in the absorbance spectrum over time (Figure S.7B). This shows that flavin can be reduced by NAD(P)H non-enzymatically, providing an explanation for why when both are present, we observe activity with MjTYW1. There is precedent in the literature for NAD(P)H and flavin nucleotides reducing heme in hemoglobin and myoglobin.59 Flavin and nicotinamide cofactors in combination have also been used to reduce azo dyes non-enzymatically.60
Overall, these results support the role of the flavodoxin_1 domain in mediating the reductive activation of the protein. The eukaryal TYW1 characterized here is the first example of a RS flavoprotein. The data show that ScTYW1 is purified with FMN and that unlike the archaeal TYW1, NAD(P)H is sufficient for observing activity with the eukaryal protein.
Conclusions
The RS superfamily consists of more than 100000 members that are distributed throughout all kingdoms of life.1 A common feature of the RS superfamily is the need for a one-electron reductant to reduce a 4Fe–4S cluster from its resting state of +2 to the active state of +1. The reduced cluster then reductively cleaves SAM to form the highly reactive intermediate dAdo• (with the exception of a few characterized enzymes21−24), which abstracts a hydrogen atom from the substrate, leading to the formation of a multitude of diverse products formed by this superfamily.
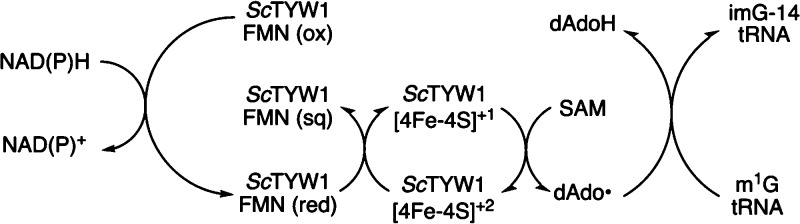
Eukaryotic homologues of TYW1 are the first RS enzymes identified to contain a fused flavin binding domain. Initially, we hypothesized that the flavodoxin_1 domain would contain a flavin cofactor that could potentially be reduced directly by NAD(P)H. The reduced flavin would in turn reduce the 4Fe–4S cluster, which would then go on to produce dAdo• and the product (see Scheme 3). ScTYW1 as purified contains FMN, confirming the domain annotated as flavodoxin_1 binds flavin. We discovered, while preparing this work, a structure of the flavodoxin-like domain of Schizosaccharomyces japonicus TYW1 had been deposited in the Protein Data Bank (entries 6PUP and 6PUQ). These structures show the flavodoxin-like domain in complex with FMN. Figure 4 shows that ScTYW1 is active with both NADPH and NADH. While it is unusual for an enzyme to be active with both nicotinamide cofactors as they are usually specific to one cofactor due to binding constraints, there are examples among nicotinamide-utilizing systems.61−63
Scheme 3. Proposed Pathway for Delivery of Equivalents from NAD(P)H to Support Turnover by TYW1.
When FMN or FAD was added to reaction mixtures containing NAD(P)H, substantially more product was formed. One explanation for this could lie in the stoichiometry of the cofactors. The previously studied homologues of TYW1 contain two 4Fe–4S clusters,49,51 one of which binds the cofactor and the other the pyruvate substrate. Both are required for activity, and the cluster binding Cys residues are conserved in the eukaryotic homologue. However, ScTYW1 contains approximately 4 mol of iron per mole of protein. This is consistent with the protein containing one 4Fe–4S cluster, on average, instead of the expected two. In addition, flavin analysis revealed there is approximately 0.5 mol of FMN per mole of protein, so both cofactors are present in only 50% of the protein. On the basis of cofactor content, we would estimate that ∼13% of the protein is fully replete (two 4Fe–S clusters and one FMN). The increased activity in the presence of added flavin could simply result from exogenous cofactor binding and reconstitution of the protein. Another potential explanation for the increased activity is that pyridine nucleotides reduce the flavin in solution, which in turn reduces the Fe–S cluster in ScTYW1 directly or via the bound FMN.
We cannot exclude the possibility that in vivo, additional proteins are engaged in the activation of ScTYW1. This would be reminiscent of the flavodoxin/flavodoxin reductase system found in bacterial species such as E. coli. In these systems, NADPH reduces a flavin cofactor in flavodoxin reductase, which in turn reduces the flavin cofactor in flavodoxin. Assuming that the ScTYW1-bound FMN represents flavodoxin, if the flavin reductase is missing, the flavin cofactor in solution could be substituting.
In most RS enzymes, in addition to the cluster that activates the SAM, there are additional iron–sulfur-containing auxiliary clusters that are essential for activity.64 In TYW1, the auxiliary cluster binds pyruvate and is required for the catalytic cycle.49,51,52 A small subset of RS enzymes also contains additional cofactors. For example, the class B RS methylases employ cobalamin.20,36,65 Lysine 2,3-aminomutase is a rare example of a pyridoxal phosphate (PLP)-dependent RS enzyme, where the PLP serves to stabilize and catalyze the interchange of groups.66 The FMN domain in ScTYW1 does not serve a catalytic role but is used for the reductive activation of the enzyme. Nevertheless, this discovery expands the cofactor repertoire of RS enzymes to include flavin.
Acknowledgments
The authors thank Professor Squire J. Booker (The Pennsylvania State University, University Park, PA) for generously providing both pDB1282 (originally constructed by Dennis Dean at Virginia Tech) and pPH151 (originally constructed by Hermann Schindelin at the State University of New York, Stony Brook). E. coli strain DM22-(pK8) was a gift from George Markham. Sequencing was performed at the DNA Sequencing Core Facility of the University of Utah.
Glossary
Abbreviations
- SAM
S-adenosyl-l-methionine
- ScTYW1
S. cerevisiae TYW1
- FMN
flavin mononucleotide
- NADPH
nicotinamide adenine dinucleotide phosphate
- NADH
nicotinamide adenine dinucleotide
- RS
radical S-adenosyl-l-methionine
- dAdo•
5′-deoxyadenosyl radical
- dAdoH
5′-deoxyadenosine
- m1G
N-methylguanosine
- imG-14
4-demethylwyosine
- FAD
flavin adenine dinucleotide
- TCA
trichloroacetic acid.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.1c00349.
Detailed experimental procedures and Figures S.1–S.7 showing the DNA and protein sequence of ScTYW1, the dependence of ScTYW1 activity on the presence of SAM, reductant, etc., the dependence of ScTYW1 activity on the concentration of FMN, comparison of archaeal and eukaryotic TYW1, and reduction of NADPH with FMN (PDF)
Accession Codes
The research reported here was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Grant R35 GM126956.
The authors declare no competing financial interest.
Supplementary Material
References
- Holliday G. L.; Akiva E.; Meng E. C.; Brown S. D.; Calhoun S.; Pieper U.; Sali A.; Booker S. J.; Babbitt P. C. (2018) Atlas of the Radical SAM Superfamily: Divergent Evolution of Function Using a “Plug and Play” Domain. Methods Enzymol. 606, 1–71. 10.1016/bs.mie.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick J. B.; Duffus B. R.; Duschene K. S.; Shepard E. M. (2014) Radical S-Adenosylmethionine Enzymes. Chem. Rev. 114, 4229–4317. 10.1021/cr4004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofia H. J.; Chen G.; Hetzler B. G.; Reyes-Spindola J. F.; Miller N. E. (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29, 1097–1106. 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsby C. J.; Hong W.; Broderick W. E.; Cheek J.; Ortillo D.; Broderick J. B.; Hoffman B. M. (2002) Electron-Nuclear Double Resonance Spectroscopic Evidence That S-Adenosylmethioine Binds in Contact with the Catalytically Active [4Fe-4S]+ Cluster of Pyruvate Formate-Lyase Activating Enzyme. J. Am. Chem. Soc. 124, 3143–3151. 10.1021/ja012034s. [DOI] [PubMed] [Google Scholar]
- Chen D.; Walsby C.; Hoffman B. M.; Frey P. A. (2003) Coordination and Mechanism of Reversible Cleavage of S-Adenosylmethionine by the [4Fe-4S] Center in Lysine 2,3-Aminomutase. J. Am. Chem. Soc. 125, 11788–11789. 10.1021/ja036120z. [DOI] [PubMed] [Google Scholar]
- Bridwell-Rabb J.; Zhong A.; Sun H. G.; Drennan C. L.; Liu H.-w. (2017) A B12-dependent radical SAM enzyme involved in oxetanocin A biosynthesis. Nature 544, 322–326. 10.1038/nature21689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruender N. A.; Grell T. A.; Dowling D. P.; McCarty R. M.; Drennan C. L.; Bandarian V. (2017) 7-Carboxy-7-deazaguanine Synthase: A Radical S-Adenosyl-l-methionine Enzyme with Polar Tendencies. J. Am. Chem. Soc. 139, 1912–1920. 10.1021/jacs.6b11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling D. P.; Bruender N. A.; Young A. P.; McCarty R. M.; Bandarian V.; Drennan C. L. (2014) Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism. Nat. Chem. Biol. 10, 106–112. 10.1038/nchembio.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman P. J.; Grove T. L.; Booker S. J.; Drennan C. L. (2013) X-ray analysis of butirosin biosynthetic enzyme BtrN redefines structural motifs for AdoMet radical chemistry. Proc. Natl. Acad. Sci. U. S. A. 110, 15949–15954. 10.1073/pnas.1312228110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman P. J.; Grove T. L.; Sites L. A.; McLaughlin M. I.; Booker S. J.; Drennan C. L. (2013) X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification. Proc. Natl. Acad. Sci. U. S. A. 110, 8519–8524. 10.1073/pnas.1302417110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vey J. L.; Yang J.; Li M.; Broderick W. E.; Broderick J. B.; Drennan C. L. (2008) Structural basis for glycyl radical formation by pyruvate formate-lyase activating enzyme. Proc. Natl. Acad. Sci. U. S. A. 105, 16137–16141. 10.1073/pnas.0806640105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovitch F.; Nicolet Y.; Wan J. T.; Jarrett J. T.; Drennan C. L. (2004) Crystal Structure of Biotin Synthase, an S-Adenosylmethionine-Dependent Radical Enzyme. Science 303, 76–79. 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y.; Zallot R.; Grove T. L.; Payan D. J.; Martin-Verstraete I.; Šepic S.; Balamkundu S.; Neelakandan R.; Gadi V. K.; Liu C.-F.; Swairjo M. A.; Dedon P. C.; Almo S. C.; Gerlt J. A.; de Crécy-Lagard V. (2019) Discovery of novel bacterial queuine salvage enzymes and pathways in human pathways. Proc. Natl. Acad. Sci. U. S. A. 116, 19126–19135. 10.1073/pnas.1909604116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove T. L.; Himes P. M.; Hwang S.; Yumerefendi H.; Bonanno J. B.; Kuhlman B.; Almo S. C.; Bowers A. A. (2017) Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides. J. Am. Chem. Soc. 139, 11734–11744. 10.1021/jacs.7b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell T. A. J.; Kincannon W. M.; Bruender N. A.; Blaesi E. J.; Krebs C.; Bandarian V.; Drennan C. L. (2018) Structural and spectroscopic analyses of the sporulation killing factor biosynthetic enzyme SkfB, a bacterial AdoMet radical sactisynthase. J. Biol. Chem. 293, 17349–17361. 10.1074/jbc.RA118.005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quitterer F.; List A.; Eisenreich W.; Bacher A.; Groll M. (2012) Crystal Structure of Methylornithine Synthase (PylB): Insights into the Pyrrolysine Biosynthesis. Angew. Chem., Int. Ed. 51, 1339–1342. 10.1002/anie.201106765. [DOI] [PubMed] [Google Scholar]
- Boal A. K.; Grove T. L.; McLaughlin M. I.; Yennawar N. H.; Booker S. J.; Rosenzweig A. C. (2011) Structural Basis for Methyl Transfer by a Radical SAM Enzyme. Science 332, 1089–1092. 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolet Y.; Pagnier A.; Zeppieri L.; Martin L.; Amara P.; Fontecilla-Camps J. C. (2015) Crystal Structure of HydG from Carboxydothermus hydrogenoformans: A Trifunctional [FeFe]-Hydrogenase Maturase. ChemBioChem 16, 397–402. 10.1002/cbic.201402661. [DOI] [PubMed] [Google Scholar]
- Frey P. A.; Hegeman A. D.; Ruzicka F. J. (2008) The Radical SAM Superfamily. Crit. Rev. Biochem. Mol. Biol. 43, 63–88. 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- Kim H. J.; McCarty R. M.; Ogasawara Y.; Liu Y.-n.; Mansoorabadi S. O.; LeVieux J.; Liu H.-w. (2013) GenK-Catalyzed C-6’ Methylation in the Biosynthesis of Gentamicin: Isolation and Characterization of a Cobalamin-Dependent Radical SAM Enzyme. J. Am. Chem. Soc. 135, 8093–8096. 10.1021/ja312641f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demick J. M.; Lanzilotta W. N. (2011) Radical SAM Activation of the B12-Independent Glycerol Dehydratase Results in Formation of 5′-Deoxy-5′-(methylthio)adenosine and Not 5′-Deoxyadenosine. Biochemistry 50, 440–442. 10.1021/bi101255e. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Zhu X.; Torelli A. T.; Lee M.; Dzikovski B.; Koralewski R. M.; Wang E.; Freed J.; Krebs C.; Ealick S. E.; Lin H. (2010) Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature 465, 891–896. 10.1038/nature09138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S.; Kudo F.; Rohmer M.; Eguchi T. (2020) Characterization of Radical SAM Adenosylhopane Synthase, HpnH, which Catalyzes the 5′-Deoxyadenosyl Radical Addition to Diploptene in the Biosynthesis of C35 Bacteriohopanepolyols. Angew. Chem., Int. Ed. 59, 237–241. 10.1002/anie.201911584. [DOI] [PubMed] [Google Scholar]
- Mahanta N.; Fedoseyenko D.; Dairi T.; Begley T. P. (2013) Menaquinone Biosynthesis: Formation of Aminofutalosine Requires a Unique Radical SAM Enzyme. J. Am. Chem. Soc. 135, 15318–15321. 10.1021/ja408594p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandarian V. (2018) Radical SAM Enzymes, Elsevier. [Google Scholar]
- Bodea S.; Balskus E. P. (2018) Purification and Characterization of the Choline Trimethylamine-Lyase (CutC)-Activating Protein CutD. Methods Enzymol. 606, 73–94. 10.1016/bs.mie.2018.04.012. [DOI] [PubMed] [Google Scholar]
- Young A. P.; Bandarian V. (2018) TYW1: A Radical SAM Enzyme Involved in the Biosynthesis of Wybutosine Bases. Methods Enzymol. 606, 119–153. 10.1016/bs.mie.2018.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari D. M.; Fedoseyenko D.; Begley T. P. (2018) Mechanistic Studies on the Radical SAM Enzyme Tryptophan Lyase (NosL). Methods Enzymol. 606, 155–178. 10.1016/bs.mie.2018.06.008. [DOI] [PubMed] [Google Scholar]
- Joshi S.; Fedoseyenko D.; Mahanta N.; Begley T. P. (2018) Aminofutalosine Synthase (MqnE): A New Catalytic Motif in Radical SAM Enzymology. Methods Enzymol. 606, 179–198. 10.1016/bs.mie.2018.05.002. [DOI] [PubMed] [Google Scholar]
- McCarthy E. L.; Booker S. J. (2018) Biochemical Approaches for Understanding Iron-Sulfur CLuster Regeneration in Escherichia coli Lipoyl Synthase During Catalysis. Methods Enzymol. 606, 217–239. 10.1016/bs.mie.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.; LaMattina J. W.; Badding E. D.; Gadsby L. K.; Grove T. L.; Booker S. J. (2018) Using Peptide Mimics to Study the Biosynthesis of the Side-Ring System of Nosiheptide. Methods Enzymol. 606, 241–268. 10.1016/bs.mie.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byer A. S.; McDaniel E. C.; Impano S.; Broderick W. E.; Broderick J. B. (2018) Mechanistic Studies of Radical SAM Enzymes: Pyruvate Formate-Lyase Activating Enzyme and Lysine 2,3-Aminomutase Case Studies. Methods Enzymol. 606, 269–318. 10.1016/bs.mie.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M.; Zhang Y.; Lin H. (2018) Methods for Studying the Radical SAM Enzymes in Diphthamide Biosynthesis. Methods Enzymol. 606, 421–438. 10.1016/bs.mie.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushin L. B.; Seyedsayamdost M. R. (2018) Guidelines for Determining the Structures of Radical SAM Enzyme-Catalyzed Modifications in the Biosynthesis of RiPP Natural Products. Methods Enzymol. 606, 439–460. 10.1016/bs.mie.2018.04.016. [DOI] [PubMed] [Google Scholar]
- Pang H.; Yokoyama K. (2018) Lessons From the Studies of a C-C Bond Forming Radical SAM Enzyme in Molydebum Cofactor Biosynthesis. Methods Enzymol. 606, 485–522. 10.1016/bs.mie.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen K. D.; Wang S. C. (2014) Spectroscopic characterization and mechanistic investigation of P-methyl transfer by a radical SAM enzyme from the marine bacterium Shewanella denitrificans OS217. Biochim. Biophys. Acta, Proteins Proteomics 1844, 2135–2144. 10.1016/j.bbapap.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruender N. A.; Young A. P.; Bandarian V. (2015) Chemical and Biological Reduction of the Radical SAM Enzyme 7-Carboxy-7-deazaguanine Synthase. Biochemistry 54, 2903–2910. 10.1021/acs.biochem.5b00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent A.; Guillot A.; Benjdia A.; Chartier G.; Leprince J.; Berteau O. (2016) The B12-Radical SAM Enzyme PoyC Catalyzes Valine Cβ-Methylation during Polytheonamide Biosynthesis. J. Am. Chem. Soc. 138, 15515–15518. 10.1021/jacs.6b06697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr I.; Latham J. A.; Iavarone A. T.; Chantarojsiri T.; Hwang J. D.; Klinman J. P. (2016) Demonstration That the Radical S-Adenosylmethionine (SAM) Enzyme PqqE Catalyzes de Novo Carbon-Carbon Cross-linking within a Peptide Substrate PqqA in the Presence of the Peptide Chaperone PqqD. J. Biol. Chem. 291, 8877–8884. 10.1074/jbc.C115.699918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi V.; Eliasson R.; Fontecave M.; Mulliez E.; Hoover D. M.; Matthews R. G.; Reichard P. (1993) Flavodoxin is required for the activation of the anaerobic ribonucleotide reductase. Biochem. Biophys. Res. Commun. 197, 792–797. 10.1006/bbrc.1993.2548. [DOI] [PubMed] [Google Scholar]
- Lewis J. K.; Bruender N. A.; Bandarian V. (2018) QueE: A Radical SAM Enzyme Involved in the Biosynthesis of 7-Deazapurine Containning Natural Products. Methods Enzymol. 606, 95–118. 10.1016/bs.mie.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer J. D.; Jarrett J. T. (2018) Purification, Characterization and Biochemical Assays of Biotin Synthase From Escherichia coli. Methods Enzymol. 606, 363–388. 10.1016/bs.mie.2018.06.003. [DOI] [PubMed] [Google Scholar]
- Grell T. A. J.; Bell B. N.; Nguyen C.; Dowling D. P.; Bruender N. A.; Bandarian V.; Drennan C. L. (2019) Crystal structure of AdoMet radical enzyme 7-carboxy-7-deazaguanine synthase from Escherichia coli suggests how modifications near [4Fe-4S] cluster engender flavodoxin specificity. Protein Sci. 28, 202–215. 10.1002/pro.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M.; Su X.; Dzikovski B.; Dando E. E.; Zhu X.; Du J.; Freed J. H.; Lin H. (2014) Dph3 Is an Electron Donor for Dph1-Dph2 in the First Step of Eukaryotic Diphthamide Biosynthesis. J. Am. Chem. Soc. 136, 1754–1757. 10.1021/ja4118957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcinas A. J.; Maiocco S. J.; Elliott S. J.; Silakov A.; Booker S. J. (2019) Ferredoxins as interchangeable redox components in support of MiaB, a radical S-adenosylmethionine methylthiotransferase. Protein Sci. 28, 267–282. 10.1002/pro.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. N.; Tener G. M. (1973) Properties of tRNAPhe from Drosophila. Biochim. Biophys. Acta, Nucleic Acids Protein Synth. 312, 267–275. 10.1016/0005-2787(73)90372-9. [DOI] [PubMed] [Google Scholar]
- Garel J.-P.; Hentzen D.; Schlegel M.; Dirheimer G. (1976) Structural studies on RNA from Bombyx mori L. Biochimie 58, 1089–1100. 10.1016/S0300-9084(76)80087-9. [DOI] [PubMed] [Google Scholar]
- Young A. P.; Bandarian V. (2011) Pyruvate Is the Source of the Two Carbons That Are Required for Formation of the Imidazoline Ring of 4-Demethylwyosine. Biochemistry 50, 10573–10575. 10.1021/bi2015053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell T. A.; Young A. P.; Drennan C. L.; Bandarian V. (2018) Biochemical and Structural Characterization of a Schiff Base in the Radical-Mediated Biosynthesis of 4-Demethylwyosine by TYW1. J. Am. Chem. Soc. 140, 6842–6852. 10.1021/jacs.8b01493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A. P.; Bandarian V. (2015) Mechanistic Studies of the Radical S-Adenosyl-l-methionine Enzyme 4-Demethylwyosine Synthase Reveal the Site of Hydrogen Atom Abstraction. Biochemistry 54, 3569–3572. 10.1021/acs.biochem.5b00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perche-Letuvée P.; Kathirvelu V.; Berggren G.; Clemancey M.; Latour J.-M.; Maurel V.; Douki T.; Armengaud J.; Mulliez E.; Fontecave M.; García-Serres R.; Gambarelli S.; Atta M. (2012) 4-Demethylwyosine Synthase from Pyrococcus abyssi Is a Radical-S-adenosyl-l-methionine Enzyme with an Additional [4Fe-4S]+2 Cluster That Interacts with the Pyruvate Co-substrate. J. Biol. Chem. 287, 41174–41185. 10.1074/jbc.M112.405019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathirvelu V.; Perche-Letuvée P.; Latour J.-M.; Atta M.; Forouhar F.; Gambarelli S.; García-Serres R. (2017) Spectroscopic evidence for cofactor-substrate interaction in the radical-SAM enzyme TYW1. Dalton Transactions 46, 13211–13219. 10.1039/C7DT00736A. [DOI] [PubMed] [Google Scholar]
- Suzuki Y.; Noma A.; Suzuki T.; Senda M.; Senda T.; Ishitani R.; Nureki O. (2007) Crystal Structure of the Radical SAM Enzyme Catalyzing Tricyclic Modified Base Formation in tRNA. J. Mol. Biol. 372, 1204–1214. 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]
- Noma A.; Kirino Y.; Ikeuchi Y.; Suzuki T. (2006) Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 25, 2142–2154. 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.; Wang J.; Chitsaz F.; Derbyshire M. K.; Geer R. C.; Gonzales N. R.; Gwadz M.; Hurwitz D. I.; Marchler G. H.; Song J. S.; Thanki N.; Yamashita R. A.; Yang M.; Zhang D.; Zheng C.; Lanczycki C. J.; Marchler-Bauer A. (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S.; Mistry J.; Bateman A.; Eddy S. R.; Luciani A.; Potter S. C.; Qureshi M.; Richardson L. J.; Salazar G. A.; Smart A.; Sonnhammer E. L. L.; Hirsh L.; Paladin L.; Piovesan D.; Tosatto S. C. E.; Finn R. D. (2019) The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte E. M.; Pellegrini M.; Ng H.-L.; Rice D. W.; Yeates T. O.; Eisenberg D. (1999) Detecting Protein Function and Protein-Protein Interactions from Genome Sequences. Science 285, 751–753. 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- Waas W. F.; de Crécy-Lagard V.; Schimmel P. (2005) Discovery of a Gene Family Critical to Wyosine Base Formation in a Subset of Phenylalanine-specific Transfer RNAs. J. Biol. Chem. 280, 37616–37622. 10.1074/jbc.M506939200. [DOI] [PubMed] [Google Scholar]
- Brown W. D.; Snyder H. E. (1969) Nonenzymatic reduction and Oxidation of Myoglobin and Hemoglobin by Nicotinamide Adenine Dinucleotides and Flavins. J. Biol. Chem. 244, 6702–6706. 10.1016/S0021-9258(18)63463-5. [DOI] [PubMed] [Google Scholar]
- Morrison J. M.; John G. H. (2013) The non-enzymatic reduction of azo dyes by flavin and nicotinamide cofactors under varying conditions. Anaerobe 23, 87–96. 10.1016/j.anaerobe.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Sellés Vidal L.; Kelly C. L.; Mordaka P. M.; Heap J. T. (2018) Review of NAD(P)H-dependent oxidorecutases: Properties, engineering and application. Biochim. Biophys. Acta, Proteins Proteomics 1866, 327–347. 10.1016/j.bbapap.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Woodyer R.; Simurdiak M.; van der Donk W. A.; Zhao H. (2005) Heterologous Expression, Purification, and Characterization of a Highly Active Xylose Reductase from Neurospora crassa. Appl. Environ. Microbiol. 71, 1642–1647. 10.1128/AEM.71.3.1642-1647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen C. N.; Cartwright J.; Ward J.; Hart S.; Turkenburg J. P.; Ali S. T.; Allen M. J.; Grogan G. (2012) A Flavoprotein Monooxygenase that Catalyses a Baeyer-Villiger Reaction and Thioether Oxidation Using NADH as the Nicotinamide Cofactor. ChemBioChem 13, 872–878. 10.1002/cbic.201200006. [DOI] [PubMed] [Google Scholar]
- Lanz N. D.; Booker S. J. (2015) Auxiliary iron-sulfur cofactors in radical SAM enzymes. Biochim. Biophys. Acta, Mol. Cell Res. 1853, 1316–1334. 10.1016/j.bbamcr.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Bauerle M. R.; Schwalm E. L.; Booker S. J. (2015) Mechanistic Diversity of Radical S-Adenosylmethionine (SAM)-dependent Methylation. J. Biol. Chem. 290, 3995–4002. 10.1074/jbc.R114.607044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovich R. M.; Ruzicka F. J.; Reed G. H.; Frey P. A. (1992) Characterization of Iron-Sulfur Clusters in Lysine 2,3-Aminomutase by Electron Paramagnetic Resonance Spectroscopy. Biochemistry 31, 10774–10781. 10.1021/bi00159a019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.