Abstract
Coronavirus Disease 2019 (COVID-19) is one of the three lethal coronavirus outbreaks in the recent two decades and a serious threat to global health all over the world. The principal feature of the COVID-19 infection is the so-called "cytokine storm" exaggerated molecular response to virus distribution, which plays massive tissue and organ injury roles. Immunological treatments, including monoclonal antibodies and vaccines, have been suggested as the main approaches in treating and preventing this disease. Therefore, a proper investigation of the roles of antigen-presenting cells (APCs) in the aforementioned immunological responses appears essential. The present review will provide detailed information about APCs' role in the infection and pathogenesis of SARS-CoV-2 and the effect of monoclonal antibodies in diagnosis and treatment.
Keywords: Antigen Presenting Cell, B cells, COVID-19, DC, Macrophages, SARS-CoV-2
1. Introduction
Recently, a virus from the Coronaviridae family with a higher spreading rate than the other family members has caused a pandemic that has affected and endangered global health ever since. This virus classified in the Betacoronavirine subfamily started its course in Wuhan, China, and has infected millions of people, causing a state called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1]. Like the former pathogenic respiratory coronavirus outbreaks, the novel coronavirus disease causes abundant inflammatory responses leading to respiratory system damage and lung failure, probably generated by cytokine storms [2]. Cytokine storm is a condition of the immune system response. Many agents and immune cells are pervasively activated and release a vast amount of chemokines and cytokines, which induce hyper inflammation [3]. It is mainly associated with multiple organ damage and a high fatality rate. Several cytokines or chemokines including type I and II interferons (IFNs), IL-6, interleukin (IL)-1, tumor necrosis factor (TNF)-α, CCL2, or monocyte chemotactic protein-1 (MCP-1), along with several immunosuppressive cytokines like IL-10 and transformation growth factor-β (TGF-β) have been associated with cytokine storms [4]. Cytokine storm has been seen in diverse clinical conditions and diseases like some hematological diseases [5].
Moreover, it is generated by various infectious illnesses and can lead to treatment resistance. Oxygen exchange disorders, increased pulmonary edema, reduces pulmonary diffusion, and causes a loss in lung compliance follow a cytokine storm during acute respiratory distress syndrome (ARDS); and consequently leads to respiratory tissue destruction and then lethal hypoxia [6]. It has been proposed that cytokine storm is triggered in Coronavirus Disease 2019 (COVID-19)-associated pneumonia [7]. Likewise, several immune cells like dendritic cells (DCs), macrophages, and B cells and their stimulation and activation are of great importance in the cytokine storm's pathophysiology. Although more of the COVID-19 cases present mild pulmonary symptoms, almost 20% of the cases demonstrate intensive pulmonary dysfunction [8].
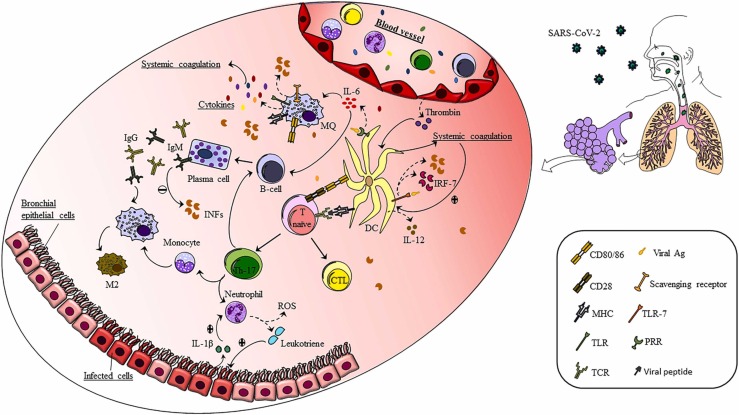
Furthermore, only some cases develop pneumonia which requires oxygenation for their treatment. The reason why only a percentage of SARS-CoV-2 infected patients demonstrate intensive inflammatory status has not yet been discovered [8]. COVID-19 may infect peculiar cells, including macrophages, endothelial vessels, or alveolar wall cells. The transmission of the virus to different kinds of cells may stimulate the initiation of immune responses producing the cytokine storm. In this study, the probable involvement of DCs, B cells, and macrophages in the pathology of COVID-19 is discussed ( Fig. 1).
Fig. 1.
The role of APCs in the progression of COVID-19 disease. After the infection with SARS-CoV-2 binds to the target cell, the innate immune system and innate immune cells such as dendritic cells, macrophages, and granulocytes are activated. These cells, in turn, secrete a complex of pro-inflammatory cytokines that activate the humoral and cellular immune systems. The activation of B cells, and the hypersecretion of antibodies, causes an over-response of the immune system, resulting in tissue damage. T cells also lead to excessive penetration of neutrophils and monocytes into the area of infection, resulting in lung tissue damage and clinical symptoms exacerbation.
2. Overview of SARS-CoV-2infection
The SARS-CoV-2 genome consists of five main open reading frames (ORF) that encode four crucial structural proteins, including nucleocapsid (NP), spike, envelope, and membrane protein (S), and a nonstructural amplification. NPs bind to the viral RNA through direct binding, and their number is very high. The presence of high IgG levels against NP protein in COVID-19 patients indicates this protein's antigenic potential in stimulating the immune system via producing vaccines. On the other hand, identification and binding of SARS-CoV-2 to the target cells are made by trimeric glycoproteins (S). The S protein contains the functional part called S1, the N-terminal region on the outer surface of the virus, which has a receptor-binding domain (RBD) which can identify and bind to its receptor and the C-terminal region (S2, which is associated with the viral envelope and plays a role in entering the cells in the fusion process) [9], [10], [11]. The infection occurs through steps such as target cell identification, maturation, cleavage of protein S, and finally, the entry of the RNA genome into the target cells [12]. The first step in virus entry is binding S1 to the cellular receptor ACE2 [13], [14]. According to a variety of laboratory techniques, the SARS-CoV-2 RBD has a strong tendency to bind to ACE2 receptors; which means that although the SARS virus binds more strongly to this receptor than the SARS-CoV-2, there is a higher affinity for SARS-CoV-2, which is why it has caused a much bigger pandemic due to its low mortality compared to SARS [15]. In this regard, one of the reasons for the higher tendency of SARS-CoV-2 to ACE2 compared to SARS-CoV is the activation of SARS-CoV-2 spike’s RBD by host protease enzyme, change from curved to standing position, and consequently effective spike attachment to the relevant ligand [15]. The virus can detect and enter the target cells by attaching the S protein of SARS-CoV-2 to the ACE2 receptor. This enzyme is expressed in large quantities on the surface of type 2 pneumocytes, airways, endothelial cells, alveolar macrophages, and the surface of nasal epithelial cells [16], [17], [18]. The high transmission of SARS-CoV-2 can be attributed to the increased affinity of S1 to ACE2 in this virus compared to the other SARS-CoVs, which is probably 10–20 times higher [16]. After the SARS-CoV-2 invades the cell and completes its proliferation and replication cycle, the immune system members, by identifying the pathogen, activate and trigger the inflammatory immune responses [19]. The stimulation of the innate immune system's activity occurs under the influence of phagocytic cells and various APCs like granulocytes, macrophages, and dendritic cells (DCs) [19]. Immune components such as pathogen-associated molecular patterns (PAMPs), which are commonly involved in identifying infectious agents, and others like damage-associated molecular patterns (DAMPs), are utilized for pathogens beyond the reach of the immune system. Together, they recognize pathogenic agents by internal and external receptors such as pattern-recognition receptors(PRRs) and Toll-like receptors (TLRs) with the expression of receptors for type I, type III IFNs, and interferon regulating factor (IRF-7), inducing immune responses [19], [20], [21], [22], [23]. Expression of the cellular PRR receptor sets mentioned on dendritic cells causes a rapid detection of foreign pathogenic nucleic acids of CoVs and leads to the generation of type I IFNs, especially IFN-α and type III IFNs [24], [25]. Studies have reported that high innate immune system activity levels and the overproduction of inflammatory cytokines cause acute organ tissue injuries. High levels of IL-1β due to programmed death of the infected cells stimulate neutrophils by the activity of inflammatory caspases and activate T cells [26], [27]. High concentrations of neutrophils are observed in the respiratory tract of COVID-19 patients, indicating the toxicity of neutrophils and their degranulation by the secretion of leukotrienes and reactive oxygen species (ROS) cause acute lung damage. Neutrophils can also be involved in systemic viral spreading by damaging endothelial cells, one of the main targets of SARS-CoV-2 [20], [26], [28], [29]. One of the destructive cytokines in COVID-19 linked with severe clinical conditions is IL-6, which is mainly associated with macrophages and dendritic cells (DCs); not only indicating the widespread involvement of the innate immune system in the occurrence of the infection but also revealing the significant role of APCs [30].
3. DCs and the pathogenesis of COVID-19
DCs are professional APCs and are members of the innate immune response against pathogens, activating the acquired immune system. Through the IL-17 pathway, these cells play a crucial role in inducing a potent immune response mediated by CD4+ Th17 cells by stimulating neutrophils and monocytes [4], [31]. There are generally three subtypes of DCs in the lung; these subtypes include myeloid (mDC), conventional DCs (cDC1 is associated with CD141+ which is expressed in the mucosa and vascular wall and is the responsive factor to Th1. cDC2 is associated with CD1c+ which presents in the human lamina propria) and plasmacytoid DCs (pDCs), which are present in most lung tissues, including parenchyma, alveolar septum, and airways, producing type I interferon in response to viral infections [32], [33], [34]. After detecting and capturing the antigen by immature DCs, these cells become mature DCs, and by processing and delivering the antigen to class I and class II MHC molecules, they activate and differentiate naive T cells, turning them into cytotoxic T lymphocyte (CD8+T-cells) and helper T-cells (CD4+T-cells), and then maturing B cells in the lymph nodes [35]. Mature DCs regulate the expression of CD80, CD86, and CCR7 (C-C Motif Chemokine Receptor 7) for functional properties. Thus, in addition to their high ability to stimulate T lymphocyte responses, DCs can secrete pro-inflammatory cytokines such as IL-12 and TNF-α, decreasing T-cell counts and disrupting immune response. IL-12 induces infiltration of NK cells and production of IFN-γ by NK and T cells [36].
Furthermore, mDCs are the leading producers of pro-inflammatory cytokines, one of the essential cytokines being IL-6; stimulates alternative macrophage pathway (M2) activity and leads to acute pulmonary fibrosis [37]. Studies of pDCs in SARS-CoV patients indicated that these cells have the ability to release IFN I, which helps to suppress the viral infection [38]. Increased mature DCs in bronchoalveolar lavage may suggest that these efficient cells are present in the lungs and may predict their response to SARS-CoV-2 infection [39]. The effect of DCs, which have moderate expressions of ACE2, is more than lymphocytes and lower than endothelial/epithelial cells. Their interaction with their specific receptor, dipeptidyl peptidase 4 (DPP4) by micropinocytosis in MERS-CoV disease, extensively produces pro-inflammatory cytokines TNF -α and IL-6 [40]. Analysis of DC levels in the blood and lung tissues of patients with COVID-19 showed an overall decrease in these DC subsets (CD1c+, CD141+, and pDCs+) and the accumulation of CD1c+ in the lung [41]. Observation of relatively significant reductions in blood DC cells and their dysfunction in the patients improved from acute conditions, and high increases in cDC/pDC ratio in acute patients can be due to decreased interferon production and initial weakness in innate immunity against SARS-CoV-2 infection resulting in the reduced numbers of DCs [42]. With a deleterious influence on the population of DCs, SARS-COV-2 can cause innate and acquired immune system deficiency [43]. Plasmacytoid DCs, on the other hand, increase IFN-α production by secreting a chemokine-like protein, but they also inhibit CD8+ Tcell activation by delaying their response, reducing virus clearance [44].
COVID-19 autopsy reports of deaths from sepsis or multiple organ failure showed high macro-thrombosis levels [45]. Also, increased blood coagulation factor, D-dimer, is considered a prognosis agent for higher coagulation and increased mortality in COVID-19 [46]. The leading cause of blood clotting is assumed to be the cytokine storm [47]. Confirmation of SARS-COV-2 particles in endothelial cells by electron microscopy showed that the virus could also infect blood vessel organoids [48] and cause thrombin activation, proinflammation, and sepsis. SARS-COV-2 can change the expression of cytokines, chemokines, and chemokine receptors by entering DCs, which are the most critical immune monitoring agents in delivering antigens to reacting components of the acquired immune system [49].
Furthermore, the coagulation pathway may stimulate cytokine storms in COVID-19 by activating the proteinase-activated receptors (PARs), which are, in turn, activated by thrombin [50]. Virus-infected endothelial cells in which the presence of thrombin-PAR-1 has occurred cause DCs to migrate and be activated, resulting in a DC response that increases the severity of inflammation and systemic coagulation [51]. In addition to being associated with inflammation and coagulation, DC's function in COVID-19 patients can reduce the number of circulating pDC and CD141+ cells [42].
3.1. Macrophages and the pathogenesis of COVID-19
Macrophages, mononuclear phagocytes, are the primary producers of COVID-19-associated inflammation and hyper inflammation [52], [53], [54], [55], [56], [57]. These cells develop a wide array of intracellular and plasma membrane receptors. These receptors act as sensors of micro-organisms, soluble agents, and dying cells and have a specific role in recognition, signaling, activation, and migration pathways [58]. Moreover, macrophages are known as professional phagocytes. They use complements, Fcs (fragment crystallizable region), non-opsonic lectin-like receptors, Toll-like receptors, and scavenger receptors for ingestion and effectively clearing the body from adverse agents and tissue debris. Moreover, macrophages are competent biosynthetic and secretory cells, participating in humoral and cellular; innate and adaptive immunity, antimicrobial defenses, and inflammation [53]. They act as two-edged swords; for instance, they participate in inducing repair mechanisms and the homeostasis of tissues and on the other hand, can increase tissue damage. They readily react with different cells via interacting with soluble substances, including enzymes, chemokines, and cytokines, activating complement cascades and plasma coagulation [56]. Furthermore, they produce reactive nitrogen, oxygen, and arachidonate metabolites, which engage in the host's inflammation and contribute to the resolution of the disease [59]. They are classified as M1-type (pro-inflammatory) and M2-type (anti-inflammatory, reparative) macrophages [60].
3.1.1. The role of macrophages in mild and asymptomatic COVID-19infections
Current studies have shown that the outcome of COVID-19 infection is determined by severe inflammatory and anti-viral macrophage responses and other innate and adaptive mechanisms [52]. COVID-19 infections occur via upper respiratory airway infection and inhalation of air droplets which may be asymptomatic to mild array; this depends on IgA protection and mucosal immunity in the upper respiratory tract. Studies on other respiratory viruses, for instance, influenza virus, indicate that, in addition to IgA, IgG could also play an essential role in virus neutralization events in the respiratory tract. Their relative contribution is associated with the physiologic portion in which these components most often are found there, and IgG takes parting protects the lower respiratory tract while IgA protecting of chiefly the upper respiratory tract [61]. So, a remarkable decrease in IgA2 in symptomatic COVID-19 patients is related to their worse condition [62].
Some studies suggest that macrophages are not infected with the virus as they do not present SARS-CoV-2 receptors and co-receptors, even if macrophages do not express transmembrane protease serine 2 (TMPRSS2) and ACE2 receptors in local tissues and are not straightly infected with SARS-CoV-2; they take part in inflammatory cytokine responses against SARS-CoV-2-infected cells via endosomal, cytosolic, and surface receptors and can release anti-viral interferons (IFNs) [63]. In SARS-CoV-2 infections, lectin-like receptors (primarily CDs) of DCs, tissue macrophages, and monocytes can attach to the virus and transport it to local lymph nodes [64].
Transportation of viral particles by droplets and lectins (attachment receptors) to the lungs causes the capillary endothelium and ACE2 + alveolar epithelium infection [65]. In this regard, the uptake of endothelial and epithelial cell debris could cause viral genome detection in cytoplasmic alveolar macrophages. Besides, antibodies against SARS-CoV-2 opsonize the viruses, inducing macrophages to uptake viral particles via Fc-receptor-mediated endocytosis [57]. Many primary infection signs can be an indirect consequence of viral-induced epithelial cell lysis, mediated by ACE2 and different co-receptors, like TMPRSS2 [66]. The uptake of cell debris and viral particles via inhibited lung macrophages can initiate inflammasome activation by purinergic receptor P2×7 (P2RX7), producing Type1 IFN, IL-1b, IL-6, and TNF [67], which participate in lethargy, pain, headache, and fever.
Moreover, raised levels of C-reactive proteins (CRP), induced by IL-6 in hepatocytes, can act as plasma biomarkers of COVID-19. Other products are also generated by macrophages, including chemokines such as MCP-1 (CCL2) and IL-8 (CXCL8), recruiting plenty of macrophages and monocytes into infection sites. Through viremia, SARS-CoV-2 spreads to other organs; such as, brain, gut, kidney, and heart, where recruited and resident macrophage populations are located. These mechanisms lead to organ-specific dysfunctions, like cardiac arrhythmia and myocarditis. These dysfunctions are produced due to interactions between the infected cardiomyocytes and macrophages [68]. SARS-CoV-2 viremia creates intravascular coagulation and promotes micro Thromb emboli distribution to the brain, heart, and lungs; through these steps, degraded fibrin particles, D-dimers, are revealed in blood. This event is increased by anti-viral IgG and IgM [56], [67]. The SARS-CoV-2 infection causes immune system failure and caused IFNs and other pro-inflammatory responses become dysregulated in circulating monocytes and macrophages. Like other viruses, SARS-CoV-2 and SARS-CoV have developed mechanisms to diminish their exposure to IFN-I. In these viruses infections, agents were recognized to block the generation of IFN-β. It seems SARS-CoV-2 is extremely sensitive to IFN-I while the anti-viral of IFN-Is on SARS-CoV is moderate. Infected cells with SARS-CoV-2, IFN-I leading to increasing STAT-1 amount and ISG generation [69].
Cause of innate immune system failure in battle of SARS-CoV-2 limiting of IFN-β generation by-products and proteins of the SARS-CoV-2 virus. Within cells, three principal pattern recognition receptors (PRRs) sense RNA viruses which Include: RIG-I-like receptors (RLRs), Toll-like receptors (i.e., TLR-3, −7, −8), and NOD-like receptors (NLRs) [70]. To recognize the mechanisms that obstacle IFN-β generation via activation of IRF3/7, various research groups transfected cells separately with whole the SARS-CoV-2 viral genes and with either MDA5, MAVS or RIG I [71], [72]. Among all SARS-CoV-2 proteins entered into cells, ORF6 and nsp14 as important suppressors of IFN-β. Yuen et al. as well as recognized nsp15and 13. In this way, Lei et al., was distinguished the M protein, nsp12, and nsp1 proteins as strong suppressors of the MAVS pathway, resulting in the prevention of IFN-β generation. In both studies, ORF6 was identified as one of the most potent inhibitors of IFN-β generation. It was noticed as the only SARS-CoV-2 gene was limited to the activity of an interferon-stimulated response element (ISRE) promoter. Lei et al. also distinguished nsp14 and nsp1 as potent suppressors of the induction of an ISRE promotor. Another investigation by Li et al. exhibited that the viral nucleocapsid ORF6 and ORF8 proteins are rugged suppressors of IFN-β generation and caused to suppress the IFN-I innate immune response [72]. Also, Konno et al. discovered ORF3b as a potent antagonist against IFN-I generation [73]. Therefore, it seems that the immune system can not limit the effects of local infection [74], [75], [76], [77]. These events and compromised lungs oxygenation may cause systemic vascular and immune dysfunction. In younger patients, a more moderate disease leads to chilblain-like inflammations in fingers and toes. The rare multisystem Kawasaki-like inflammatory syndrome in children has resulted from uncommon immune responses to SARS-CoV-2 [78]. Regardless of platelet activation, monocytes and macrophages (mononuclear phagocytes) play an essential role in generating tissue factors and other procoagulants [79], factors 5 and 13. Fibrinolysis via inflammatory vascular macrophages and endothelium is mediated by urokinase release, transforming plasminogen into plasmin. This phenomenon is regulated by the inhibitors present in plasma, such as alpha2 macroglobulin. Intra nasal or inhaled type-1 IFN and anticoagulants may be beneficial inhibitors of local viral infection and thrombosis, restricting the infection's progression into severe forms [80]. Also, some of the studies revealed T cell reduction and an overall increase in innate cell lineages that cuased increasing inflammatory cytocines, an early promoting in cytokine levels was related to severe disease [81]. In addition to the immune system, genetic characteristics and nutrition of patient may play a role in limiting local infection[82], [83].
3.1.2. The role of macrophages in severe COVID-19infections
Mild COVID-19 infections can progress to severe type after approximately 1–2 weeks from primary infection. Epithelial cells of COVID-19 patients displayed an average triple increase in expression of receptor ACE2 of SARS-CoV-2, which associated to elevation interferon signals by immune cells [84].
Hyper-inflammation is the hallmark of this event. Overall, COVID-19-associated hyper inflammation is closely linked to clinical manifestations like macrophage activation syndrome (MAS) symptoms specified by edema, increased vascular permeability, pneumonia, and hypoxia, expanding the requirement of oxygen or mechanical ventilation. The virus produces a systemic viral infection in ACE2+ epithelia and endothelia, leading to hypotension, ischemia, widespread cell death, and organ failure. Blood analysis revealed increased monocytes, lymphopenia, and polymorphonuclear leukocytosis [85], [86]. Increased numbers of pro-inflammatory monocyte-derived macrophages in the bronchoalveolar fluid of patients with severe COVID-9 infection were associated to a worse infection status, whereas the presence of CD8+ T cells in moderate disease was correlated to a mild grade [87]. The recruitment of myeloid cells contributes to coagulopathy and hyperinflammatory processes of ARDS and cytokines [88]. Cytokines are generated during the immune response then attract macrophages and neutrophils as pro-inflammatory cells to the infection location to induce an inflammatory response. However, these responses are critical to 34wvirus clearance, and they can also destruction normal host tissues [89]. It is unclear which factors assist to these events: enhanced viral production probably induced by the quick and weak neutralization of IgM, complements, or IgG; and the reduction or absence of macrophage's anti-viral mechanisms - straightly or secondarily leads to lymphopenia, immense local endothelial and epithelial necrosis, hyperactivation of recently recruited immature mononuclear phagocytes such as monocytes, or a "perfect storm," comprised of multiple of the above phenomena. This event could be initiated by the induction of ACE2 receptors on particular populations of macrophages and monocytes, resulting in their straight infection. At this stage, draining lymph nodes and spleen are intensely engaged at the infection sites [90]. CD169 and other monocytes' receptors transport virus particles on their surface, then disperse them throughout the body with a "Trojan horse" mechanism. It is not yet clear that virus-transmitting monocytes are infected or not [91]. It is unknown whether or not the infected lymphocytes die by necrosis or apoptosis induced by the activated macrophages and lymphocytes products such as TNF; or by phagocytosis. In this section, we summarize the records indicating macrophages' participation in severe COVID-19; we then investigate pathogenic macrophage mechanisms, which may be targeted for treatment.
Prior studies of SARS1/MERS showed many aspects of macrophage involvement in ARDS occurrence [92], [93]. Also, tissue and blood analysis of COVID-19 patients showed vigorous phagocytic activity in macrophages and monocyte abnormality [90]. Myelopoiesis was detected in bronchoalveolar macrophages and monocytes using single-cell RNA and fluorescence-activated cell sorting (FACS) [86], [94].
They corroborated the presence of activated and recruited macrophages among interstitial and alveolar lung cells [57]. Plasma membrane opsonins and other receptors participate in monocytic hyperactivation, dysregulated inflammation, and defective phagocytic removal of necrotic and apoptotic cell remnants and debris. These receptors act via NF-kB, DNA, RNA, and other signaling and sensing pathways; they generate many secretory products. Some pro-inflammatory and anti-viral products are targeted for anti-inflammatory therapy, such as IL-1, IFNs, TNF, and IL-6 [54], [63], [75], [95].
Furthermore, macrophages and monocytes react with plasma cascades; they interact via unregulated adhesion molecules, platelets, other lymphoid and myeloid cells, and system-wide endothelia and epithelia. Macrophages participate in the thrombotic and hyperinflammatory activities in severe COVID-19 infections, and the dysregulated activation of macrophages can then worsen the condition of the patients [56], [67], [96]. Virus-mediated cell damage, necroptosis, necrosis, and apoptosis are recognized by opposing macrophage receptors associated with effector responses. These comprise activator and suppressor pathways of IL-1 generation, secretion, and processing [76], [95] produced by tissue damage, caspase activation, and the inflammasomes. An imperfect type 1 IFNs response is determined by low IFN-α and no IFN-β production and activity. Overall, SARS-CoV-2 infection results in attenuation of IFN-I production, especially inhibition of IFN-β and increasing IL-6 and TNF-α. Also, activation of NF-kB consequence low production of IFN-Is and high generation of pro-inflammatory cytokines in SARS-CoV-2 infection. Imperfect IFN-I generation in severe COVID-19 infection can result in the dysfunction of pro-repair action airway macrophages and exhaustion of CD8+ and CD4+ T cells. IFN- I has boosted T cell survival, and the early generation of IFN-Is elevates effective T cell responses. In contrast, the postponed response may prevent T cell proliferation or cause their exit disruption, thus leading to their functional exhaustion [97], [98]. Imperfect type 1 IFN response is correlated with persistent viremia and intense inflammatory responses, somewhat driven by IL-6, NF-kB, and TNF [74]. Endogenous oxidized phospholipids regulate the mechanisms of hyper inflammation and macrophage metabolism. By the uptake of inflammatory lipids produced from damaged and dying cells, the CD14 instigates phagocyte hyperactivation [99]. The utilization and generation of macrophages are extremely sensitive to hypoxia-inducible factor 1 (HIF-1) responses, hypoxia, and iron accessibility; these conditions effect on M1 polarization and effector functions.
The activation of macrophages effectively triggers coagulation and complement cascades and angiotensin and bradykinin pathways, destroying vascular permeability and tone and intensifying edema and inflammation. Moreover, infection with the SARS-CoV-2 virus causes pyroptosis, a kind of cell death triggered by the activation of inflammasome cascades in the NLR Family Pyrin Domain Containing 3 (NLRP3) [100], [101]. One of the downstream indications of pyroptosis is IL-1; a rise in IL-1 in the serum can indicate pyroptosis in COVID-19-associated lung inflammation. Pyroptosis leads to the release of DAMPs, which prompt other macrophages to generate pro-inflammatory chemokines and cytokines [101]. Inflammation signals received by the macrophages, in turn, attract immune cells like T cells into the inflammation site.
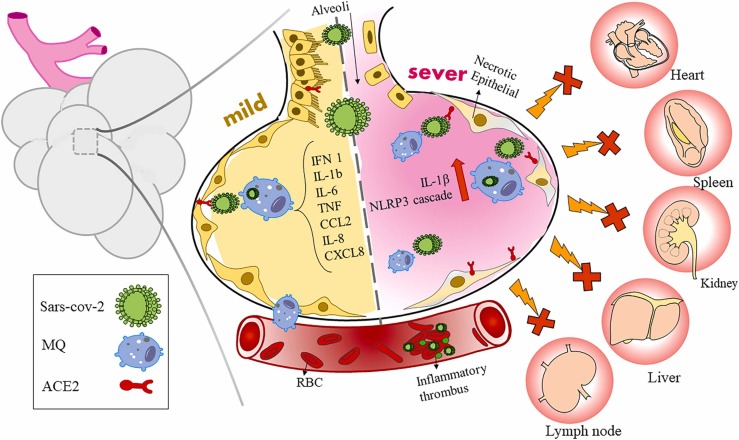
Consequently, local inflammation is provoked by SARS-CoV-2 infection, activating the macrophages at the site of inflammation and disseminating the condition quickly to the whole lung, probably due to the excellent presentation of the virus entry receptors in lung tissues such as ACE2 and TMPRSS2 [102]. The accumulation of immune cells speeds up lung inflammation progression to ARDS; these patients can not limit local inflammation in their lungs, thus spreading the disease to multiple organs, causing tissue damage and death [14], [102] ( Fig. 2). So, host immune response against severe SARS-CoV-2 disease has an essential anti-viral function and causes concurrent pathogenic damage of tissues and organs, which ascertains the disease progression, severity, and consequence [103].
Fig. 2.
The role of pulmonary macrophages in mild and severe cases. After the migration of monocyte-derived macrophages from the capillaries into the lung alveoli, macrophages are produced by the uptake of virus-infected cytokine-infected epithelial cell debris and inflammatory-induced chemokines. Gradually, with the intensification of inflammation, over-activation of macrophages due to the production of inflammatory lipids from damaged cells, the accumulation of immune cells at the site of infection and disruption of the ratio of monocytes to macrophages, the expression of ACE2 receptors on the surface of these cells increased, so the viruses spread to other organs and disrupts them.
3.1.3. Lung macrophages and the pathogenesis of COVID-19
Another reason for severe SARS-CoV-2 infection, mentioned above, is the type of macrophages in the induction of inflammation. Macrophages are one of the principal cells of the body's immune system in response to SARS-CoV-2 infection, and they also participate in the development of inflammation and the progression of COVID-19 pathogenesis [104]. Inhabitant macrophages in the upper respiratory tract and the lungs, known as sentinel cells, are the primary cells of the immune system to encounter the invasion of viruses. Macrophages can limit primary viral replications by producing IFN-I responses. They also recruit extra immune cells by driving severe inflammatory reactions [105], [106]. The inflammatory response is necessary to begin further immune responses against SARS-CoV-2 infections; therefore, the additional inflammation created by SARS-CoV-2 produces a cytokine storm participating in the mortalities of COVID-19 [67]. Respiratory systems, especially lungs, have two macrophage populations; alveolar macrophages (AMs) and interstitial macrophages [107]. The interstitial macrophages have at least two subsets which among them, the nerve-associated and airway-associated macrophages (NAMs) subset may limit inflammatory responses against the SARS-CoV-2 infection. NAMs found in the lung's interstitial tissue are strongly related to innervating nervous tissue; they also have various growth factor dependency and ontology compared to AMs (118). No specific studies are comparing NAMs and AMs done with COVID19. NAMs are detected as essential macrophages restricting viral-induced inflammation in some viral infections such as influenza, while AMs are pro-inflammatory and necessary for viral elimination [108]. The evidence suggests that AMs are pro-inflammatory and anti-viral, while NAMs limit excessive inflammation and harmful effects. NAMs, inhibit IL-6 generation during influenza infections, showing that NAMs can be a significant controlling point in IL-6 amounts and control hyper-inflammation and cytokine storm in COVID-19 cases [108]. So, IL-6 inhibition by administration of JAK inhibitors in patients with COVID-19 can be an appropriate therapeutic method. The main target of JAK inhibitors is IL-6, which is a crucial pro-inflammatory cytokine in severe cases of COVID-19 [109].
3.1.4. COVID-19 and the downregulation of macrophage antigen presentation
Some studies display COVID-19 patients with ARDS have a condition of immune disbalance, which cuased to dysregulation of both innate and adaptive immune responses that may be a worse disease course [109]. Human macrophages and monocytes may be straightly infected by SARS-CoV-2 via ACE-2 or indirectly infected through ACE-2-independent pathways using DC-SIGN, L-SIGN, CD147, ADE, and phagocytosis of apoptotic bodies which contain viruses [110], [111]. These events indicate that the virus straightly manipulates and controls the macrophages to escape the immune system. It is not clear whether SARS-CoV-2 affects the activity and function of macrophages or not, while the infection of macrophages by other coronaviruses (CoVs) imposes modified functional status. MERS-CoV infected macrophages express multiple CD80, CD86, and major histocompatibility complex I (MHC I), while significant histocompatibility complex (MHC) II is absent. This event suggests that MERS-CoV defects the presentation of MHC II [112], [113]. Moreover, the downregulation of MHC II was newly proved in B cells and monocytes of COVID-19 cases [114]. The MHC II downregulation mechanisms are induced via epigenetic alterations of the infected cells [115]. Different CoVs share the epigenetic downregulation of MHCs in a process that CoV-EMC downregulates the MHC IIs through epigenetic reprogramming [116]. This event is not joint, as SARS-CoV does not routinely limit MHC II's antigen presentation. Down-regulation of MHC I in HIV infection by Nef protein is an instance that confirms that viruses can effectively regulate MHC I and inhibit MHC pathways and escape clearance [117].
The results of SARS-CoV-2 infection studies presented some possible insights about different mechanisms through which the virus can interfere with macrophages' activity and function. Current studies showed that the interferon signaling blockage would restrict the interferon-stimulated gene expression (ISG). Therefore, cytokine-mediated MHC II's face is reduced in the macrophages [118]. The nonstructural protein 5 (Nsp5) of SARS-CoV-2 interacts with histone deacetylase 2 (HDAC2) as the epigenetic regulator and thus regulates cytokines' production and MHC II's expression [119], [120], [121], whether SARS-CoV-2 enhances or inhibits the array [122]. Other viruses use a different mechanism to inhibit antigen presentation to other cells; for instance, Nef protein of HIV limits antigen presentation by returning MHC I's trafficking towards the Golgi apparatus [122], [123]; furthermore, Nsp10 of SARS-CoV-2 reacts to adaptor protein 2 (AP2), an endocytosis regulator, and transfers MHC II to the compartment of antigen loading. While many immune evasion mechanisms remain chiefly theoretic, the potency of SARS-CoV-2 to infect macrophages and react with main macrophage activity proteins proposes a powerful capability to modulate and regulate macrophage function via systemic immune responses [123], [124].
3.1.5. Macrophages and recovery
Host immune response against acute SARS-CoV-2 infection plays an anti-viral role. It leads to simultaneous pathogenic injury of organs and tissues, especially in the lungs of patients with COVID-19, which determines the disease severity and progression, and outcome.
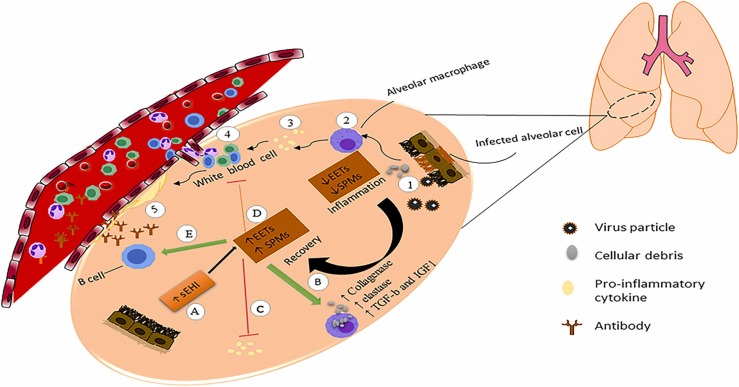
Macrophages are the most effective cells in the cellular activities which participate in the recovery of COVID-19 infection; they clear necrotic tissues and debris via phagocytosis and secretion of collagenase and elastase and generate TGF-β and IGF1 to enhance the recovery of lung tissue. Macrophages produce potent lipid metabolites which limit inflammation. These metabolites are specialized proresolvin mediators (SPMs) and comprise protectins, maresins, and resolvins. They are bioactive lipid autacoids that determine the duration and extent of the inflammation and induce endogenous resolving via phagocytosis of cellular debris and antagonize the release of pro-inflammatory cytokines/chemokines [59], [125]. They also confront inflammation without causing immunosuppression and via increasing efferocytosis of dying neutrophils. It seems SPMs may play a vital role in handling this lung disease because SPM effectively induse the resolution of infectious inflammation. They are also organ protective that which has been proven in animal disease models. Many cells and tissues have been suitable and susceptible targets for treating inflammation with SPMs to stimulate dynamic elimination of inflammation [126].
Similarly, anti-viral B lymphocytic activity induced by SPMs has been predicted to be a helpful therapy for COVID-19 infection [127]. Besides, 17-hydroxydocosahexaenoic acid (17-HDHA), as an SPM precursor, has been presented as an effective adjuvant for some vaccines, especially the influenza vaccine, as they elevate the activity of adaptive immunity against infections [128], [129]. Another mediator for inflammation clearance is arachidonic acid-derived epoxyeicosatrienoic acids (EETs). These effective mediators increase cellular remnants' clearance and activate anti-inflammatory processes, limiting the production of several critical inflammatory cytokines [130], [131]. Also, the generation of SPMs is increased by EETs and various other epoxy fatty acids produced by altering arachidonic acid metabolism (136). As soluble epoxide hydrolase (sEH) quickly metabolizes EETs, the use of sEH inhibitors (sEHIs) stabilizes EETs during severe inflammation and suppresses the pro-inflammatory responses of macrophages in lung inflammation; decreasing cytokine storm and infiltration of immune cells. Also, EETs can limit the formation of hyaline membranes, which play essential roles in ARDS generation [132]. As a result, sEHIs restore lung function, making them an appealing potential treatment strategy for COVID-19. Also, SPMs and sEHIs regulate NF-κB as a transcription regulator and decrease its levels [132], [133] ( Fig. 3). Therefore, using SPMs or SPM-associated precursors combined with COVID-19 treatments and vaccines can be a new and beneficial treatment approach. Recovery can take a long time after severe infection, and it involves lethargy, fever, tissue damage, and lack of organ function [134]. In addition to tissue recovery and restoring lung function after the primary infection, SARS-CoV-2 infection can also help remission some diseases, including Hodgkin's lymphoma. This event can be justified via a phenomenon related to activation of natural killer cells mediated by cytokines generated in response to the infection cross-reactivity of pathogen-specific T cells with tumor antigens [135].
Fig. 3.
Role of macrophage in recovery COVID-19 infection Inflammation: Alveolar cells infected by viral particle and apoptotic and necrotic debris accumulated (1) macrophages uptake debris (2) debris motivate the generation of pro-inflammatory cytokines of macrophages (3) pro-inflammatory cytokines increase infiltration of immune cell (4) infiltration of the immune cell increased hyaline membrane formation and cause acute respiratory distress syndrome (5). Recovery: EETs and SPMs are reduced in a severe inflammatory condition. During recovery, EETs increase, then EETs increase the production of SPMs (A). SPMs stimulate macrophage phagocytosis (B), reduce pro-inflammatory cytokine(C), inhibit leukocytosis, and thereby decrease the infiltration of leukocytes (D) and may facilitate the adaptive immune response and enhance the production of anti-SARS-CoV-2 antibodies (E).
3.2. B cell response
TCR and BCR analyses manifested a high level of clonal expansion in the case with severe COVID-19, particularly B lymphocytes, and the clonally expanded B cells significantly expressed genes associated with lymphocyte activation and inflammatory responses [136].
B cells express antigen peptides in the MHC II molecules. Several characteristics allow the B cells to present antigens efficiently: BCR-mediated endocytosis enables them to concentrate small amounts of specific antigens. Then, B cells can find T cells in secondary lymphoid organs shortly after antigen entrance [137]. SARS-CoV-2 activates both innate and adaptive immune responses. Antigen presentation eventually stimulates the body's cellular and humoral immune responses, i.e., virus-specific T and B cells. Most COVID-19 patients develop a humoral reaction during the first two weeks of infection [138]. In some patients, inappropriate humoral responses are associated with ineffective SARS-CoV-2 clearance, highlighting B cells' critical role for viral clearance [139]. Human B cells are typically classified into different types: immature or transitional cells that have recently emigrated from the bone marrow; naïve B cells or mature lymphocytes which their specific antigen has never stimulated; and differentiated cells (memory B cells and antibody-secreting plasma cells) [140].
3.3. B cells and the pathogenesis of COVID-19
3.3.1. B cell subsets
The differences between B cells allow us to identify the severity of patients' conditions with their specific subpopulations. Inflammation biomarkers, respiratory parameters, and clinical scores have revealed associations with some of these subsets. The seriousness of COVID-19 is associated with the changes occurring in the B cells' subpopulations, either immature or terminally differentiated. Furthermore, the existing relationship between B cell subset frequencies and clinical and laboratory parameters proposes that these lymphocytes could serve as potential biomarkers and even active participants in the adaptive anti-viral response against SARS-CoV-2. B cell numbers remain unchanged or may decrease in the COVID-19 patients [141]. The numbers of transitional B cell subsets decrease with the disease's severity but are usually increased in mild cases. Other non- typical subsets such as double-negative B cells have also revealed essential changes according to the severity of the disease [142]. Pathological mature B cells are called double-negative (DN) B cells lacking the expression of CD27 and immunoglobulin D biomarkers (IgD-/CD27-). These cells are associated with systemic lupus erythematosus (SLE), HIV, rheumatoid arthritis, and other diseases presenting significant immune system pathology [143]. Through autoantibody production and cytokine release, DN B cells boost the progression of these diseases. DN2 B cells (IgD-/CD27-/CXRC5-/CD21+), through an extra-follicular pathway, enhance COVID-19 pathogenesis at early stages of the disease; this pathway was previously identified in SLE [144]. In these cells, the activated toll-like receptor 7 (TLR7) drives the expression of T-bet and CD11c and, through uncharacterized mechanisms, promotes the severity of COVID-19 [145]. Circulating leukocytes do not express high levels of pro-inflammatory chemokines and cytokines, suggesting that the COVID-19 cytokine storm is driven by the cells within the lung [146]. Antibody-secreting cells' (ASC) percentage is increased significantly in COVID-19 patients. When the ASCs' subpopulation was analyzed, a discrete increment of these cells was detected in moderate COVID-19 patients, significantly magnifying the severity of the illness.
Additionally, these cells are positively correlated with the clinical phenotypes seen in COVID-19-infected cells, differentiating into plasma cells that produce a vast antibody repertoire, protecting against almost every pathogen. The proportions and the numbers of classical memory B cells (cMBC, CD21+/CD27+/CD10−) were significantly reduced in patients with severe COVID-19 compared to the control group and recovered individuals. Additionally, the analysis of B-cell subsets revealed a significant expansion of atypical memory cells (atMBC, CD21lo/CD27−/CD10−), both in numbers and frequencies in the patients with SARS-CoV-2 compared to the control group and recovered individuals. Significantly, the portion atMBC was higher in the deceased patients than in those who had survived [147]. Atypical B-cell subsets (atMBCs) appear during chronic infections, including HIV, HCV, HBV, malaria, and in autoimmune settings such as lupus. Because of their association with ongoing infections, it has been proposed that they may be associated with impaired B-cell function. However, this issue and the significance of atMBC arising during viral infections are still questionable.
3.3.2. Extra follicular B cell response
Evidence shows that extrafollicular (EF) B cell responses in critically ill COVID-19 patients are similar to previously reported autoimmune settings. It is now established that EF B cell responses can undergo affinity maturation and generate long-living plasma and memory cells even independently to the T cells. Extrafollicular activation is strongly correlated with high SARS-CoV-2-neutralizing antibodies, antibody class-switching, and poor clinical outcomes. Disease severity and poor clinical outcomes in COVID-19 are closely correlated with the intense activation of the EF B cell pathways despite the presence of high COVID-19 neutralizing antibody titers. This subject is validated by recent works identifying a lack of germinal center formation in the patients' spleen and lymph nodes showing severe illness outcomes. The immunological outlook associated with these effector B cells' mobilization in COVID-19 is highly related to active autoimmune processes [148].
3.3.3. Lack of germinal center (GC) responses
Germinal centers (GCs) are transitory microanatomical environments that form after antigen-activated B cells receive help from a follicular T helper (TFH) cell, a specialized CD4+ T cell subset. Within Germinal centers, B cells undergo clonal extension and affinity maturation, differentiating into long-living plasma cells or memory cells with the help of TFH cells [149].
A new study reported that within ten days of the onset of respiratory symptoms in severe COVID-19 patients, lymph and splenic node's germinal centers and lymphoma 6 expressing B cells (Bcl-6+) are remarkably absent. The lack of GCs and Bcl6 expressing B cells or TFH cells, which are indispensable for generating GCs, suggests a mechanism for short-lasting and low titer antibody responses observed in COVID-19 patients. Severe SARS-CoV-2 infections blunt the germinal center responses, which is likely to dampen the generation of long-lasting antibody responses [149]. SARS infections were thought to be caused by the lack of GC responses [150]. The short -lasting antibody responses are probably the result of thymus are probably the result of thymus independent (TI) B cell activation. TI is a event during that some of the bacterial lipopolysaccharides and polysaccharides and polymeric proteins can stimulate B lymphocytes without contribution of helper T cells [151].
3.4. Antibody
Humoral immunity is an antibody-mediated immune response. B cells differentiate into plasma cells after being stimulated by T helper cells, producing virus-specific antigen antibodies, including immunoglobulin (Ig) G and (Ig) M. Antibodies have a multifactorial role in viral clearance. Antibodies can be adjusted to viral proteins expressed on the infected cell's surface and help the clearance of the infected cells, directing NK cells to kill the infected cells via antibody-dependent cytotoxicity. Antibodies can also bind freely-circulating virions, facilitating their Fc receptor-mediated clearance. Antibodies against viral entry mediators such as the SARS-CoV-2 S protein can also prevent the interactions between the viral protein and the host's target cells, subsequently controlling the infection [152]. SARS-CoV immunogenic epitopes are S and N proteins recognized by CD8+ and CD4+ T cells. The viral antigen is recognized by APCs, which stimulate the body's humoral immunity via virus-specific B and plasma cells and produce neutralizing antibodies. The RBD domain of SARS-CoV-2 Spike (S) is the primary target of SARS-CoV-2 neutralizing antibodies, as was the case for SARS-CoV [153].
During B cells' development, antigen-antibody interactions occur at the appropriate B cell surfaces, driving antibody isotype switching (through class switching recombination), virus-specific cell proliferation, cell maturation germinal center formations in mucosal and systemic sites. For respiratory viruses, each antibody and cell present in the upper respiratory tract tissues is the receptor for a different antigenic target [154].
3.5. Antibody enhancement
While antibodies are crucial for the clearance of viral pathogens, inappropriate activation of B cells may contribute to the severity of SARS-CoV-2's pathophysiology; some antibodies may even enhance the infection state. There is a connection between the severity of the disease and antibody titers. Patients who presented higher antibody titers experienced the more severe disease. Severe disease may boost memory B cells' development, leading to higher antibody titers in the following exposures. It is not clear whether higher titer levels increase the protection to re-infection. Also, DN-B cells produced during chronic and autoimmune infections are not GC-induced; however, they show a change in the T cell-dependent isotype class [148]. Moreover, according to a research by Woodruff, M.C et al., in cases with severe COVID-19, DN B cells lacked CD21 and CD11c, and also regulate the CXCR3 gene, which leads to higher DN B cell usage in IFN-induced inflammatory tissue despite lacke CXCR5 gene expression (Th1) [148]. Given that immunocompromised individuals do not have a prolonged SARS-CoV-2 infection, the higher accumulation of DN cells in COVID-19 patients could be attributed to an abnormal inflammatory axis [155]. On the other hand, an antibody response is needed to clear the SARS-CoV-2 infection, patients who generated an early antibody response to the S protein had a higher risk of death than those who experienced late antibody responses. This unexpected outcome is thought to be due to the expanded inflammation and impaired healing within the lungs, as an increased polarization of alveolar macrophages into pro-inflammatory M1 phenotypes occurred [156]. This might contribute to antibody-dependent enhancement (ADE), which has been seen in MERS-CoV and SARS-CoV. Early antibody synthesis is enhanced by viral entrance into macrophages and other immune cells, allowing virion uptake through phagocytic Fc receptors [157]. The necessity of antibody response duration has a numerus impact on recombinant antibodies and plasma treatment as therapeutic options. Antibody-based therapies may need to be used later in the course of the disease as it is believed that early high titers of neutralizing antibodies are associated with more severe clinical outcomes in SARS-CoV-2 patients [158].
3.6. Autoantibodies
Immune-mediated inflammation and tissue damage could result from rapid autoimmune responses and the production of self-reactive autoantibodies [159]. In COVID-19 patients, some rare autoantibody production cases against the coagulation protein S have been reported, as was seen in the macro- or micro-circulation thrombosis during the varicella disease [160]. Recent studies on severe acute COVID-19 have reported a high prevalence of antinuclear antibodies [159]. Specific autoantibodies, such as anti-2GPI, anti-cardiolipin (a-CL), and lupus anticoagulant, have previously been associated to the thromboembolic consequences seen in many COVID-19 patients [161]. It's not surprising that cytokines like interleukin-6 might trigger autoimmunity and autoinflammatory reactions in the cytokine storm, most likely through molecular mimicry or pre-existing natural B cell clones [162]. IgG auto-antibodies against type I IFNs were found in nearly 15% of severe COVID-19 patients in a recent study, suggesting that the generation of auto-antibodies in these patients may be attributed to life-threatening complications and mortality, and may be a reason for the low serum IFN levels seen in some severe cases [163].
3.7. Monoclonal antibodies
The coronavirus disease 2019 universal crisis has attracted researchers' attention to prevent and treat SARS-CoV-2 infection. Most of the studies have focused on convalescent plasma infusions, developing vaccines, and new anti-viral agents for prevention and treatment. Neutralizing antibodies are a clef ingredient of protective immunity for most viral diseases. In the disease caused by SARS-CoV-2, neutralizing monoclonal antibodies can lead to the design and development of appropriate drugs and vaccines with therapeutic and prophylactic activities [164]. In addition, the use of monoclonal antibodies in other viral diseases such as Ebola virus infection respiratory syncytial viral pneumonia has been considered [165]. Given the emergence of the SARS-CoV-2 virus, monoclonal antibodies have been reported to be recovered from patients' B cells or individuals with a history of SARS-CoV-1, as well as by immunizing mice [164]. Also, due to the similarity of SARS-CoV-2 and SARS-CoV-1 virus cell receptors, such as angiotensin 2 converting enzyme (ACE2), and the role of viral S protein in entering the host cell, this protein has been targeted to produce mAb [166].
In this way, from the summer of 2020, several SARS-CoV-2 monoclonal antibodies were reported to have reached clinical trials. Clinical trials are used to prevent the progression of the disease, such as treating people with SARS-CoV-2 infection with different stages of the disease [164]. It has also been observed that, given the long half-life of most monoclonal antibodies, for example, one injection dose for IgG1, approximately three weeks, is sufficient. It has also been reported that one of the limitations of using monoclonal antibodies during the COVID-19 pandemic is the unknown function of IgG on body tissues, including lung tissue, which is the main target organ of infection [164]. In this regard, monoclonal antibodies were designed to target only viral protein sequences; in addition, the use of a combination of two monoclonal antibodies targets different locations of viral proteins such as S protein [164].
On the other hand, monoclonal antibodies have some benefits. They can be an alternative to vaccination in people who cannot be vaccinated or need immediate prevention before or after exposure to the virus. However, modifications have been made to increase the half-life of these drugs and the ability to infusion intramuscularly or subcutaneously [167].
Furthermore, these drugs, in turn, lead to several side effects, such as nausea, diarrhea, dizziness, headache, and vomiting. Also, in the second phase of clinical trials, 1.9% of people treated with bamlanivimab 1.8% in combination therapy with bamlanivimab and etesevimab showed injection-related reactions; these are very mild and are not dose-dependent [168], [169].
In this regard, in the United States, three anti-SARS-CoV-2 drugs have been approved to treat non-hospitalized patients with mild to moderate symptoms, such as bamlanivimab as a single treatment bamlanivimab with etesevimab, or casirivimab with imdevimab as a combination therapy [169]. The following are some of their features;.
3.7.1. REGN-COV2therapy
The monoclonal antibodies of casirivimab and imdevimab, called REGN-COV2, act as a potent neutralizing IgG1 mAb with unmodified Fc regions. Antibodies bind to two distinct and non-overlapping sites in RBD. Combining these two antibodies is more effective because a mutation in the S protein of the SARS-CoV-2 virus cannot limit or inhibit the effects of both antibodies [169], [170]. In addition, experiments have shown that this drug compound retains its effects against mutations in protein S; they also induced cytotoxicity and phagocytosis in virus-infected cells. Also, in humans, this combination reduces viral load, incidence, and severity of damage to lung tissue [169], [170].
3.7.2. Bamlanivimab therapy
Bamlanivimab is derived from the plasma of people with COVID-19 during recovery; as a type of IgA1 mAb, it is a potent neutralizer with an unmodified Fc region [169], [171]. By binding to the RBD region of S protein and participating in its epitopes in high and low conformations, this mAb leads to an appropriate effect in monotherapy [169]. Furthermore, bamlanivimab was studied in primary human macrophage cells and SARS-CoV-2 virus-infected immune cell lines to assess the theoretical risk of ADE and was found not to lead to productive viral infection [169]. Also, to evaluate the prophylactic effect of the drug, it was tested with bamlanivimab for 24 h before exposure of macaque rosacea to the virus, and led to a significant reduction in viral load and exerted its anti-viral effect [171].
3.7.3. Bamlanivimab and etesevimab therapy
In another study, bamlanivimab was combined with etesevimab, in which IgG1 binds to the S protein with a modified Fc region, resulting in effective neutralization [168], [172]. Besides, bamlanivimab and etesevimab have been reported to be combined and leading to a 70% reduction in hospitalization rates, mortality, and viral load [168], [169].
3.7.4. Meplazumab therapy
Meplazumab is a monoclonal IgG2 antibody, also known as HP6H8 and GTPL11026. It is an anti-CD147 and was developed by Jiangsu Pacific Meinuoke Bio-Pharmaceutical. This cellular marker is one of the leading causes of inflammation, as a receptor for the SARS-CoV-2 virus and making the mechanism of action of targeting drugs pleiotropic [165].
3.7.5. Drug-resistant monoclonal antibodies in SARS- CoV-2strains
According to clinical studies on bamlanivimab, mutations in the viral S protein have been shown to cause resistant mutations in the drug. In addition, casirivimab, in combination with imdevimab, can lead to drug-resistant viral variants [169].
Also, bamlanivimab and imdevimab lead to the neutralization activity of the SARS-CoV-2 strain of UK origin; as a result, these mAbs maintain their performance against the new UK strain [173], [174]. However, in general, monoclonal antibodies such as bamlanivimab, bamlanivimab, etesevimab, and finally casirivimab and imdevimab, in turn, reduce the viral load in the early stages of infection and people with mild to moderate clinical symptoms [169]. As a result, mAbs, combined with other drugs, serve as a prevention and treatment strategy.
4. Conclusion
COVID-19 is a concerned universal threat with the potential to cause severe illness in the lungs of infected patients [175]. Up to now, SARS-CoV-2 has infected millions of people worldwide and caused more than one million confirmed deaths. The COVID-19 pandemic caused by SARS-CoV-2 is spreading worldwide at an alarming rate. Some cases of COVID-19 are life-threatening, and the disease poses a serious threat to global health and safety. Controlling the spread of the pandemic as soon as possible is an emergency issue. SARS-CoV-2 has caused more infections and deaths than SARS-CoV or MERS-CoV, and its outcome is likely to be determined by the extent of the host immune system's imbalance. In other words, it is the exaggerated immune response, or the immunopathogenic response, which is responsible for severe pneumonia and consequently respiratory failure and death.
The relationship between a virus and its host is complicated; various factors involving the virus and the host are involved in viral infection and pathogenesis. In this review, we focused on the interactions between antigen-presenting cells and the immunopathogenic mechanisms of the SARS-CoV2. The treatment strategies for the patients with COVID-19 should be carefully designed based on viral biochemical features and immunopathogenesis. In this regard, the COVID-19 pandemic has led to research programs to develop appropriate strategies to limit the spread of infection and reduce mortality. Monoclonal antibodies and vaccines are appropriate strategies for effective prevention and treatment [164]. Also, the cross-reactive mAbs can be a valuable tool for research and the development of diagnostic methods for SARS-CoV-2 infection [166]. As the knowledge of the virus grows, more effective approaches emerge to deal with this infection. We hope that the virus can be defeated soon by prevention, cutting off the chain of transmission, and effective treatments with professional medical researchers' efforts. Finally, preventive vaccination is highly recommended to prevent future emerging coronavirus-related epidemics or pandemics.
Funding
We did not support a specific source of funding for this article.
CRediT authorship contribution statement
Hossein Bannazadeh Baghi: Design the study, review the manuscript, corresponding author. Rana Farzi: Literature search, design figure, manuscript preparation, writing- original draft, review of the manuscript. Parisa Shiri Aghbash: Literature search, design the figure, manuscript preparation, writing- review and editing, review of the manuscript. Negar Eslami: Design the figure, manuscript preparation. Arezou Azadi: Manuscript preparation. Ali Shamekh: Edit and review the manuscript. Nima Hemmat: Edit and review the manuscript. Taher Entezari-Maleki: Edit and review the manuscript.
Declaration of competing interest
The authors declared that there is no conflict of interest related to this study.
Acknowledgment
This project was supported by the Infectious and Tropical Diseases Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
References
- 1.Pormohammad A., et al. Comparison of confirmed COVID‐19 with SARS and MERS cases‐Clinical characteristics, laboratory findings, radiographic signs and outcomes: a systematic review and meta‐analysis. Rev. Med. Virol. 2020 doi: 10.1002/rmv.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gómez-Rial J., et al. Role of monocytes/macrophages in Covid-19 pathogenesis: implications for therapy. Infect. Drug Resist. 2020;13:2485–2493. doi: 10.2147/IDR.S258639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behrens E.M., Koretzky G.A. Cytokine storm syndrome: looking toward the precision medicine era. Arthritis Rheumatol. 2017;69(6):1135–1143. doi: 10.1002/art.40071. [DOI] [PubMed] [Google Scholar]
- 4.Aghbash P.S., et al. The role of Th17 cells in viral infections. Int. Immunopharmacol. 2021;91 doi: 10.1016/j.intimp.2020.107331. [DOI] [PubMed] [Google Scholar]
- 5.Crayne C.B., et al. The immunology of macrophage activation syndrome. Front. Immunol. 2019;10:119. doi: 10.3389/fimmu.2019.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang X., et al. The role of macrophages in the pathogenesis of ALI/ARDS. Mediat. Inflamm. 2018 doi: 10.1155/2018/1264913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGonagle D., et al. Interleukin-6 use in COVID-19 pneumonia related macrophage activation syndrome. Autoimmun. Rev. 2020 doi: 10.1016/j.autrev.2020.102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otsuka R., Seino K.-i. Macrophage activation syndrome and COVID-19. Inflamm. Regen. 2020;40(1):1–6. doi: 10.1186/s41232-020-00131-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemmat N., et al. The roles of signaling pathways in SARS-CoV-2 infection; lessons learned from SARS-CoV and MERS-CoV. Arch. Virol. 2021:1–22. doi: 10.1007/s00705-021-04958-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pillay T.S. Gene of the month: the 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. J. Clin. Pathol. 2020;73(7):366. doi: 10.1136/jclinpath-2020-206658. [DOI] [PubMed] [Google Scholar]
- 13.Jia H.P., et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79(23):14614. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarkesh A., et al. Extrapulmonary clinical manifestations in COVID-19 patients. Am. J. Trop. Med. Hyg. 2020;103(5):1783–1796. doi: 10.4269/ajtmh.20-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang J., et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukassen S., et al. SARS‐CoV‐2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39(10) doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varga Z., et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395(10234):1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao C., Zhao W. NLRP3 Inflammasome—a key player in antiviral responses. Front. Immunol. 2020;11:211. doi: 10.3389/fimmu.2020.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vardhana S.A., Wolchok J.D. The many faces of the anti-COVID immune response. J. Exp. Med. 2020;217(6) doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu Y.-F., et al. A review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. Sci. 2020;21(7):2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly A., et al. Immune cell profiling of IFN-λ response shows pDCs express highest level of IFN-λR1 and are directly responsive via the JAK-STAT pathway. J. Interferon Cytokine Res. 2016;36(12):671–680. doi: 10.1089/jir.2015.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Megjugorac N.J., Gallagher G.E., Gallagher G. Modulation of human plasmacytoid DC function by IFN‐λ1 (IL‐29) J. Leukoc. Biol. 2009;86(6):1359–1363. doi: 10.1189/jlb.0509347. [DOI] [PubMed] [Google Scholar]
- 24.Colonna M., Trinchieri G., Liu Y.-J. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 2004;5(12):1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 25.Kumagai Y., et al. Alveolar macrophages are the primary interferon-α producer in pulmonary infection with RNA viruses. Immunity. 2007;27(2):240–252. doi: 10.1016/j.immuni.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 26.Barnes B.J., et al. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J. Exp. Med. 2020;217(6) doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parisi V., Leosco D. Precision medicine in COVID-19: IL-1β a potential target. JACC Basic Transl. Sci. 2020;5(5):543–544. doi: 10.1016/j.jacbts.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soy M., et al. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin. Rheumatol. 2020:1. doi: 10.1007/s10067-020-05190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemmat N., et al. Neutrophils, crucial, or harmful immune cells involved in coronavirus infection: a bioinformatics study. Front. Genet. 2020;11:641. doi: 10.3389/fgene.2020.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKechnie J.L., Blish C.A. The innate immune system: fighting on the front lines or fanning the flames of COVID-19? Cell Host Microbe. 2020;27(6):863–869. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunter C.A., Jones S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015;16(5):448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 32.Li L., et al. Classical dendritic cells regulate acute lung inflammation and injury in mice with lipopolysaccharide‑induced acute respiratory distress syndrome. Int. J. Mol. Med. 2019;44(2):617–629. doi: 10.3892/ijmm.2019.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collin M., Bigley V. Human dendritic cell subsets: an update. Immunology. 2018;154(1):3–20. doi: 10.1111/imm.12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsson M., Beignon A.-S., Bhardwaj N. DC-virus interplay: a double edged sword. Semin. Immunol. 2004;16(3):147–161. doi: 10.1016/j.smim.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Yadava K., Marsland B.J. Lymphoid follicles in chronic lung diseases. Thorax. 2013;68(6):597–598. doi: 10.1136/thoraxjnl-2012-203008. [DOI] [PubMed] [Google Scholar]
- 36.Costela-Ruiz V.J., et al. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Page C., et al. Induction of alternatively activated macrophages enhances pathogenesis during severe acute respiratory syndrome coronavirus infection. J. Virol. 2012;86(24):13334–13349. doi: 10.1128/JVI.01689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cervantes-Barragan L., et al. Control of coronavirus infection through plasmacytoid dendritic-cell–derived type I interferon. Blood. 2007;109(3):1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong Y., et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu H., et al. Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology. 2014;454–455:197–205. doi: 10.1016/j.virol.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez-Cerrillo I., et al. Differential redistribution of activated monocyte and dendritic cell subsets to the lung associates with severity of COVID-19. medRxiv. 2020 [Google Scholar]
- 42.Zhou R., et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53(4):864–877. doi: 10.1016/j.immuni.2020.07.026. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campana P., et al. Dendritic cells and SARS-CoV-2 infection: still an unclarified connection. Cells. 2020;9(9):2046. doi: 10.3390/cells9092046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wikstrom M.E., et al. A chemokine-like viral protein enhances alpha interferon production by plasmacytoid dendritic cells but delays CD8+ T cell activation and impairs viral clearance. J. Virol. 2013;87(14):7911–7920. doi: 10.1128/JVI.00187-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carsana L., et al. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infect. Dis. 2020;20(10):1135–1140. doi: 10.1016/S1473-3099(20)30434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L., et al. D‐dimer levels on admission to predict in‐hospital mortality in patients with Covid‐19. J. Thromb. Haemost. 2020;18(6):1324–1329. doi: 10.1111/jth.14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monteil V., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905–913. doi: 10.1016/j.cell.2020.04.004. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.José R., et al. Regulation of neutrophilic inflammation in lung injury induced by community-acquired pneumonia. Lancet. 2015;385:S52. doi: 10.1016/S0140-6736(15)60367-1. [DOI] [PubMed] [Google Scholar]
- 49.Lau Y.-L., Peiris J., Law H. Role of dendritic cells in SARS coronavirus infection. Hong. Kong Med. J. 2012;18(Suppl 3):28–30. [PubMed] [Google Scholar]
- 50.Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. Med. 2020;8(6):e46–e47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niessen F., et al. Dendritic cell PAR1–S1P3 signalling couples coagulation and inflammation. Nature. 2008;452(7187):654–658. doi: 10.1038/nature06663. [DOI] [PubMed] [Google Scholar]
- 52.Wang, J., et al., Cytokine storm and leukocyte changes in mild versus severe SARS‐CoV‐2 infection: Review of 3939 COVID‐19 patients in China and emerging pathogenesis and therapy concepts. J. Leukocyte Biol. [DOI] [PMC free article] [PubMed]
- 53.Gordon S., Plüddemann A. Tissue macrophages: heterogeneity and functions. BMC Biol. 2017;15(1):53. doi: 10.1186/s12915-017-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feldmann M., et al. Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. Lancet. 2020;395(10234):1407–1409. doi: 10.1016/S0140-6736(20)30858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen W., et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6(1):1–18. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teuwen L.A., et al. Author Correction: COVID-19: the vasculature unleashed (Nature Reviews Immunology,(2020), 10.1038/s41577-020-0343-0) Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bost P., et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020 doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guilliams M., Mildner A., Yona S. Developmental and functional heterogeneity of monocytes. Immunity. 2018;49(4):595–613. doi: 10.1016/j.immuni.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 59.D. Panigrahy. et al. Inflammation resolution: a dual-pronged approach to averting cytokine storms in COVID-19? 2020. [DOI] [PMC free article] [PubMed]
- 60.Martinez F.O., Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000prime Rep. 2014;6 doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Z., et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021;13(577):eabf1555. doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stephenson E., et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021;27(5):904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.E. Toniato, R. Ross, S.K. Kritas, How to reduce the likelihood of coronavirus-19 (CoV-19 or SARS-CoV-2) infection and lung inflammation mediated by IL-1. 2020. [DOI] [PubMed]
- 64.Bedin A.-S., et al. Monocyte CD169 expression as a biomarker in the early diagnosis of COVID-19. medRxiv. 2020 [Google Scholar]
- 65.Lempp F.A., et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature. 2021;598(7880):342–347. doi: 10.1038/s41586-021-03925-1. [DOI] [PubMed] [Google Scholar]
- 66.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020:1–8. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hulsmans M., et al. Macrophages facilitate electrical conduction in the heart. Cell. 2017;169(3):510–522. doi: 10.1016/j.cell.2017.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schreiber G. The role of type I interferons in the pathogenesis and treatment of COVID-19. Front. Immunol. 2020:2599. doi: 10.3389/fimmu.2020.595739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jensen S., Thomsen A.R. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 2012;86(6):2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Altay O., et al. Combined metabolic cofactor supplementation accelerates recovery in mild-to-moderate COVID-19. medRxiv. 2020 doi: 10.1002/advs.202101222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lei X., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11(1):1–12. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lecca S., et al. Heterogeneous habenular neuronal ensembles during selection of defensive behaviors. Cell Rep. 2020;31(10) doi: 10.1016/j.celrep.2020.107752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hadjadj J., et al. Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients. MedRxiv. 2020 doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee J.S., et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5(49) doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Italiani P., et al. Profiling the course of resolving vs. persistent inflammation in human monocytes: the role of IL-1 family molecules. Front. Immunol. 2020;11:1426. doi: 10.3389/fimmu.2020.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lombardi A., et al. Early phases of COVID-19 are characterized by a reduction in lymphocyte populations and the presence of atypical monocytes. Front. Immunol. 2020:11. doi: 10.3389/fimmu.2020.560330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Verdoni L., et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020 doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang N., et al. Expression of factor V by resident macrophages boosts host defense in the peritoneal cavity. J. Exp. Med. 2019;216(6):1291–1300. doi: 10.1084/jem.20182024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gordon S., Unkeless A.C., Cohn Z.A. Induction of macrophage plasminogen activator by endotoxin stimulation and phagocytosis: evidence for a two-stage process. J. Exp. Med. 1974;140(4):995–1010. doi: 10.1084/jem.140.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lucas C., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.James P.T., et al. The role of nutrition in COVID-19 susceptibility and severity of disease: a systematic review. J. Nutr. 2021;151(7):1854–1878. doi: 10.1093/jn/nxab059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pairo-Castineira E., et al. Genetic mechanisms of critical illness in Covid-19. Nature. 2021;591(7848):92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 84.Chua R.L., et al. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38(8):970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 85.Lombardi A., et al. Early phases of COVID-19 are characterized by a reduction of lymphocyte populations and the presence of atypical monocytes. medRxiv. 2020 doi: 10.3389/fimmu.2020.560330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mann E.R., et al. Longitudinal immune profiling reveals distinct features of COVID-19 pathogenesis. medRxiv. 2020 [Google Scholar]
- 87.Liao M., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 88.Chen P.-M., et al. Up-regulation of FOXM1 by E6 oncoprotein through the MZF1/NKX2-1 axis is required for human papillomavirus–associated tumorigenesis. Neoplasia. 2014;16(11):961–971. doi: 10.1016/j.neo.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang L., et al. COVID-19: immunopathogenesis and Immunotherapeutics. Signal Transduct. Target. Ther. 2020;5(1):1–8. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feng Z., et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. MedRxiv. 2020 [Google Scholar]
- 91.Park M.D. Macrophages: a Trojan horse in COVID-19? Nat. Rev. Immunol. 2020;20(6) doi: 10.1038/s41577-020-0317-2. 351-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Memish Z.A., et al. Middle East respiratory syndrome. Lancet. 2020;395(10229):1063–1077. doi: 10.1016/S0140-6736(19)33221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Channappanavar R., Perlman S. MERS Coronavirus. Springer; 2020. Evaluation of activation and inflammatory activity of myeloid cells during pathogenic human coronavirus infection; pp. 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Daamen A.R., et al. Comprehensive transcriptomic analysis of COVID-19 blood, lung, and airway. Sci. Rep. 2021;11(1):1–19. doi: 10.1038/s41598-021-86002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Conti P., et al. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34(2):327–331. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 96.Giamarellos-Bourboulis E.J., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000. doi: 10.1016/j.chom.2020.04.009. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Blanco-Melo D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Acharya D., Liu G.Q., Gack M.U. Dysregulation of type I interferon responses in COVID‐19. Nat. Rev. Immunol. 2020;20:397–398. doi: 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zanoni I., et al. By capturing inflammatory lipids released from dying cells, the receptor CD14 induces inflammasome-dependent phagocyte hyperactivation. Immunity. 2017;47(4):697–709. doi: 10.1016/j.immuni.2017.09.010. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang, M., Cell pyroptosis, a potential pathogenic mechanism of 2019-nCoV infection. Available at SSRN 3527420, 2020.
- 101.Wang S., et al. The mechanisms of NLRP3 inflammasome/pyroptosis activation and their role in Parkinson’s disease. Int. Immunopharmacol. 2019;67:458–464. doi: 10.1016/j.intimp.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 102.Bar-On Y.M., et al. Science forum: SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang J.-Y., et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21(9):1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- 104.Feng Z., et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. medRxiv. 2020 [Google Scholar]
- 105.Lavin Y., et al. Regulation of macrophage development and function in peripheral tissues. Nat. Rev. Immunol. 2015;15(12):731–744. doi: 10.1038/nri3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aghbash P.S., et al. SARS-CoV-2 infection: the role of PD-1/PD-L1 and CTLA-4 axis. Life Sci. 2021;270 doi: 10.1016/j.lfs.2021.119124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.de Groen R.A., et al. IFN‐λ‐mediated IL‐12 production in macrophages induces IFN‐γ production in human NK cells. Eur. J. Immunol. 2015;45(1):250–259. doi: 10.1002/eji.201444903. [DOI] [PubMed] [Google Scholar]
- 108.Ural B.B., et al. Identification of a nerve-associated, lung-resident interstitial macrophage subset with distinct localization and immunoregulatory properties. Sci. Immunol. 2020;5(45) doi: 10.1126/sciimmunol.aax8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee J.S., Shin E.-C. The type I interferon response in COVID-19: implications for treatment. Nat. Rev. Immunol. 2020;20(10):585–586. doi: 10.1038/s41577-020-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Boumaza A., et al. Monocytes and macrophages, targets of severe acute respiratory syndrome coronavirus 2: the clue for coronavirus disease 2019 immunoparalysis. J. Infect. Dis. 2021;224(3):395–406. doi: 10.1093/infdis/jiab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jafarzadeh A., et al. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: lessons from SARS and MERS, and potential therapeutic interventions. Life Sci. 2020;257 doi: 10.1016/j.lfs.2020.118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Channappanavar R., et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Investig. 2019;129(9) doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou J., et al. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J. Infect. Dis. 2014;209(9):1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wilk, A.J., et al., A single-cell atlas of the peripheral immune response to severe COVID-19. medRxiv 2020. CrossRef][PubMed]. [DOI] [PMC free article] [PubMed]
- 115.Menachery V.D., et al. MERS-CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proc. Natl. Acad. Sci. USA. 2018;115(5):E1012–E1021. doi: 10.1073/pnas.1706928115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Josset L., et al. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. MBio. 2013;4(3) doi: 10.1128/mBio.00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Painter M.M., et al. Concanamycin A counteracts HIV-1 Nef to enhance immune clearance of infected primary cells by cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. USA. 2020;117(38):23835–23846. doi: 10.1073/pnas.2008615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Keskinen P., et al. Regulation of HLA class I and II expression by interferons and influenza A virus in human peripheral blood mononuclear cells. Immunology. 1997;91(3):421–429. doi: 10.1046/j.1365-2567.1997.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gordon D.E., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020:1–13. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fang W.-F., et al. Histone deacetylase 2 (HDAC2) attenuates lipopolysaccharide (LPS)-induced inflammation by regulating PAI-1 expression. J. Inflamm. 2018;15(1):1–11. doi: 10.1186/s12950-018-0179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kong X., et al. HDAC2 deacetylates class II transactivator and suppresses its activity in macrophages and smooth muscle cells. J. Mol. Cell. Cardiol. 2009;46(3):292–299. doi: 10.1016/j.yjmcc.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 122.Zhang Y., et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. bioRxiv. 2020 [Google Scholar]
- 123.Dirk B.S., Heit B., Dikeakos J.D. Visualizing interactions between HIV-1 Nef and host cellular proteins using ground-state depletion microscopy. AIDS Res. Hum. Retrovir. 2015;31(7):671–672. doi: 10.1089/aid.2014.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McCormick P.J., Martina J.A., Bonifacino J.S. Involvement of clathrin and AP-2 in the trafficking of MHC class II molecules to antigen-processing compartments. Proc. Natl. Acad. Sci. USA. 2005;102(22):7910–7915. doi: 10.1073/pnas.0502206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Werz O., et al. Human macrophages differentially produce specific resolvin or leukotriene signals that depend on bacterial pathogenicity. Nat. Commun. 2018;9(1):59. doi: 10.1038/s41467-017-02538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Regidor P.-A., et al. Pro resolving inflammatory effects of the lipid mediators of omega 3 fatty acids and its implication in SARS COVID-19. Med. Hypotheses. 2020;145 doi: 10.1016/j.mehy.2020.110340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Regidor P.-A. Covid-19 management with inflammation resolving mediators? Perspectives and potential. Med. Hypotheses. 2020;142 doi: 10.1016/j.mehy.2020.109813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ramon S., et al. The specialized proresolving mediator 17-HDHA enhances the antibody-mediated immune response against influenza virus: a new class of adjuvant? J. Immunol. 2014;193(12):6031–6040. doi: 10.4049/jimmunol.1302795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Morita M., et al. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell. 2013;153(1):112–125. doi: 10.1016/j.cell.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 130.Gilroy D.W., et al. CYP450-derived oxylipins mediate inflammatory resolution. Proc. Natl. Acad. Sci. USA. 2016;113(23):E3240–E3249. doi: 10.1073/pnas.1521453113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Node K., et al. Anti-inflammatory properties of cytochrome P450 epoxygenase-derived eicosanoids. Science. 1999;285(5431):1276–1279. doi: 10.1126/science.285.5431.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schmelzer K.R., et al. Soluble epoxide hydrolase is a therapeutic target for acute inflammation. Proc. Natl. Acad. Sci. USA. 2005;102(28):9772–9777. doi: 10.1073/pnas.0503279102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Serhan C.N. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510(7503):92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Martinez F.O., et al. Monocyte activation in systemic Covid-19 infection: assay and rationale. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Challenor, S. and D. Tucker, SARS-CoV-2-induced remission of Hodgkin lymphoma. Br. J. Haematol. [DOI] [PubMed]
- 136.Fan X., et al. Single-cell RNA-seq and V (D) J profiling of immune cells in COVID-19 patients. MedRxiv. 2020 [Google Scholar]
- 137.Rivera A., et al. Role of B cells as antigen-presenting cells in vivo revisited: antigen-specific B cells are essential for T cell expansion in lymph nodes and for systemic T cell responses to low antigen concentrations. Int. Immunol. 2001;13(12):1583–1593. doi: 10.1093/intimm/13.12.1583. [DOI] [PubMed] [Google Scholar]
- 138.Chen Y., et al. A comprehensive, longitudinal analysis of humoral responses specific to four recombinant antigens of SARS-CoV-2 in severe and non-severe COVID-19 patients. PLoS Pathog. 2020;16(9) doi: 10.1371/journal.ppat.1008796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ye X., et al. Low humoral immune response and ineffective clearance of SARS-Cov-2 in a COVID-19 patient with CLL during a 69-day follow-up. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.01272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Romero‐Ramírez S., et al. Innate‐like B cell subsets during immune responses: beyond antibody production. J. Leukoc. Biol. 2019;105(5):843–856. doi: 10.1002/JLB.MR0618-227R. [DOI] [PubMed] [Google Scholar]
- 141.Shimizu Y. Understanding the immunopathogenesis of COVID-19: Its implication for therapeutic strategy. World J. Clin. Cases. 2020;8(23):5835. doi: 10.12998/wjcc.v8.i23.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sosa-Hernández V.A., et al. B cell subsets as severity-associated signatures in COVID-19 patients. Front. Immunol. 2020;11:3244. doi: 10.3389/fimmu.2020.611004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Matthias P., Rolink A.G. Transcriptional networks in developing and mature B cells. Nat. Rev. Immunol. 2005;5(6):497–508. doi: 10.1038/nri1633. [DOI] [PubMed] [Google Scholar]
- 144.Taefehshokr N., Taefehshokr S., Heit B. Mechanisms of dysregulated humoral and cellular immunity by SARS-CoV-2. Pathogens. 2020;9(12):1027. doi: 10.3390/pathogens9121027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Woodruff M., et al. Critically ill SARS-CoV-2 patients display lupus-like hallmarks of extrafollicular B cell activation. medRxiv. 2020 [Google Scholar]
- 146.Mortaz E., et al. The immune response and immunopathology of COVID-19. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.02037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Oliviero B., et al. Expansion of atypical memory B cells is a prominent feature of COVID-19. Cell. Mol. Immunol. 2020;17(10):1101–1103. doi: 10.1038/s41423-020-00542-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Woodruff M.C., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21(12):1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cañete P.F., Vinuesa C.G. COVID-19 makes B cells forget, but T cells remember. Cell. 2020;183(1):13–15. doi: 10.1016/j.cell.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gu J., et al. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202(3):415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stein K.E. Thymus-independent and thymus-dependent responses to polysaccharide antigens. J. Infect. Dis. 1992;165(Supplement_1):S49–S52. doi: 10.1093/infdis/165-supplement_1-s49. [DOI] [PubMed] [Google Scholar]
- 152.Klasse P. Neutralization of virus infectivity by antibodies: old problems in new perspectives. Adv. Biol. 2014;2014 doi: 10.1155/2014/157895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Moderbacher C.R., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183(4):996–1012. doi: 10.1016/j.cell.2020.09.038. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hurwitz J.L. B cells, viruses, and the SARS-CoV-2/COVID-19 pandemic of 2020. Viral Immunol. 2020;33(4):251–252. doi: 10.1089/vim.2020.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Khanolkar A. Elucidating T cell and B cell responses to SARS-CoV-2 in humans: gaining insights into protective immunity and immunopathology. Cells. 2022;11(1):67. doi: 10.3390/cells11010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu L., et al. Anti–spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4(4) doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang S.-F., et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014;451(2):208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhao J., et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lerma L.A., et al. Prevalence of autoantibody responses in acute coronavirus disease 2019 (COVID-19) J. Transl. Autoimmun. 2020;3 doi: 10.1016/j.jtauto.2020.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Amiral J. Can COVID-19 induce an autoimmune disease associated with long-lasting symptoms and delayed complications. Ann. Clin. Immunol. Microbiol. 2020;2(1) 2020. 1014. [Google Scholar]
- 161.Chen G., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vlachoyiannopoulos P.G., et al. Autoantibodies related to systemic autoimmune rheumatic diseases in severely ill patients with COVID-19. Ann. Rheum. Dis. 2020;79(12):1661–1663. doi: 10.1136/annrheumdis-2020-218009. [DOI] [PubMed] [Google Scholar]
- 163.Bastard P., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370(6515) doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Marovich M., Mascola J.R., Cohen M.S. Monoclonal antibodies for prevention and treatment of COVID-19. J. Am. Med. Assoc. 2020;324(2):131–132. doi: 10.1001/jama.2020.10245. [DOI] [PubMed] [Google Scholar]
- 165.Tuccori M., et al. Anti-SARS-CoV-2 neutralizing monoclonal antibodies: clinical pipeline. mAbs. 2020;12(1) doi: 10.1080/19420862.2020.1854149. Epub 2020/12/16. https://doi. org/10.1080/19420862.2020. 1854149 PMID: 33319649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zheng Z., et al. Monoclonal antibodies for the S2 subunit of spike of SARS-CoV-1 cross-react with the newly-emerged SARS-CoV-2. Eurosurveillance. 2020;25(28) doi: 10.2807/1560-7917.ES.2020.25.28.2000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cohen M.S. Monoclonal antibodies to disrupt progression of early covid-19 infection. Mass Med. Soc. 2021 doi: 10.1056/NEJMe2034495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gottlieb R.L., et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. J. Am. Med. Assoc. 2021;325(7):632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Taylor P.C., et al. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021:1–12. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Baum A., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369(6506):1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jones B.E., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13(593) doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shi R., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 173.P. Wang, et al., Increased Resistance of SARS-CoV-2 Variants B. 1.351 and B. 1.1. 7 to Antibody Neutralization. bioRxiv. 2021. Google Scholar, 2021.
- 174.G. Zhang, et al., The basis of a more contagious 501Y. V1 variant of SARS-COV-2. 2021. [DOI] [PMC free article] [PubMed]
- 175.Choreño-Parra J.A., et al. Clinical and immunological factors that distinguish COVID-19 from pandemic influenza A (H1N1) Front. Immunol. 2021;12:1222. doi: 10.3389/fimmu.2021.593595. [DOI] [PMC free article] [PubMed] [Google Scholar]