Abstract
Shewanella algae is widely distributed in marine and freshwater habitats, and has been proved to be an emerging marine zoonotic and human pathogen. However, the genomic characteristics and pathogenicity of Shewanella algae are unclear. Here, the whole-genome features of 55 S . algae strains isolated from different sources were described. Pan-genome analysis yielded 2863 (19.4 %) genes shared among all strains. Functional annotation of the core genome showed that the main functions are focused on basic lifestyle such as metabolism and energy production. Meanwhile, the phylogenetic tree of the single nucleotide polymorphisms (SNPs) of core genome divided the 55 strains into three clades, with the majority of strains from China falling into the first two clades. As for the accessory genome, 167 genomic islands (GIs) and 65 phage-related elements were detected. The CRISPR-Cas system with a high degree of confidence was predicted in 23 strains. The GIs carried a suite of virulence genes and mobile genetic elements, while prophages contained several transposases and integrases. Horizontal genes transfer based on homology analysis indicated that these GIs and prophages were parts of major drivers for the evolution and the environmental adaptation of S. algae . In addition, a rich putative virulence-associated gene pool was found. Eight classes of antibiotic-associated resistance genes were detected, and the carriage rate of β-lactam resistance genes was 100 %. In conclusion, S. algae exhibits a high intra-species diversity in the aspects of population structure, virulence-associated genes and potential drug resistance, which is helpful for its evolution in pathogenesis and environmental adaptability.
Keywords: Shewanella algae, diversity, genome characteristics, virulence-associated genes, drug resistance, environmental adaptability
Data Summary
The sources and genomic sequences used throughout this study have been deposited in the National Centre for Biotechnology Information (NCBI), under the assembly accession numbers provided in Tables S1 and S2 (available in the online version of this article).
Impact Statement.
S. algae have become dominant species associated with human and aquatic livestock disease in the genus of Shewanella . Cases of bacteremia, sepsis, and infective endocarditis have also been reported. However, the evolution, biodiversity and pathogenic potential of S. algae are poorly understood. Through a comparative pan-genomic analysis of S. algae strains, we identified new pathogenic genomic islands, prophages, and virulence factors, suggesting that independent acquisition of these mobile genetic elements could play an important role in the evolution and virulence of S. algae . This study provides important insights into intra-species divergence and environmental adaptability underlying genomic sequences. The relationship between genome features and environmental adaptation strategies is an essential part for understanding the ecological functions of S. algae in the marine environments. The investigation of bacterial resistance shows that S. algae plays an important role as a reservoir of antibiotic resistance genes in the nature, which provides a scientific basis for surveillance and control of infectious diseases caused by S. algae .
Introduction
Shewanella spp. are Gram-negative, oxidase-positive, H2S-producing, facultative anaerobe bacteria that are ubiquitously distributed in seafood, marine and freshwater environments, even in deep-sea and polar regions [1]. Shewanella spp. are characterized by prominent biodiversity, and they have evolved to develop complex lifestyles. The most outstanding ecological functions are bioremediation and application in the microbial fuel cells production [2, 3]. Various anaerobic respiration pathways and extracellular electron transfer are the metabolic features of the genus Shewanella [1, 4]. At the time of writing, this genus contains 74 recognized species (http://www.bacterio.net/shewanella.html).
Despite the beneficial roles, some species like Shewanella algae and Shewanella putrefaciens have been identified as opportunistic pathogens [5–7]. The proliferation and the clinical case reports of Shewanella spp. have raised the concern of microbiologists. Reported illnesses included skin and soft tissue infections, otitis media and bacteremia. By 2012, over 50 case reports of human infection caused by the genus Shewanella had been found in PubMed database [1]. Up to now, the cumulative number of Shewanella infections in literature has already exceeded 300. Recent advances in the taxonomy and phylogenetic relatedness of Shewanella spp. supported the concept that most human infections were caused by S. algae [5, 8].
Currently, multidrug resistance has been increasingly reported in the genus Shewanella [9–12]. Several antibiotic resistance determinants have been detected in this genus. Yousfi et al. discovered multidrug-resistant (MDR) plasmid in S. xiamenensis [10]. Shewanella spp. were resistant to different antibiotic classes, including β-lactams, quinolones and aminoglycosides. However, the genetic characterization of antimicrobial resistance in Shewanella spp. remains limited.
In recent years, the rapid development of high-throughput sequencing technology has brought opportunities for the genetic research of pathogens [13]. The improved sensitivity and accuracy of whole-genome sequencing technology are important for understanding the genome characteristics of pathogens. As increasing species of the genus Shewanella are sequenced, there are more studies focused on the whole genus rather than specific species. Fang et al. have constructed multilocus sequence analysis (MLSA) methods to accurately identify Shewanella strains at the species level [14]. Thorell et al. used the whole-genome sequencing to redefine Shewanella taxonomy [15]. As a predominant human pathogen in the genus Shewanella , S. algae is still largely unknown for its genomic characteristics, virulence factors and antimicrobial resistance. Therefore, in this study, we sequenced the genomes of 12 new S. algae strains and combined the genome sequences of 43 S . algae strains from public databases, to illustrate the intra-species hereditary and present a comprehensive analysis of S. algae .
Methods
Strain information
A total of 55 S . algae strains were used in this study, including 43 strains from public database (genomes with less than 200 contigs were selected) and 12 newly sequenced strains isolated from different sources in China (deposited in the Centre for Human Pathogenic Culture Collection, China CDC). These strains were isolated from China (n=38, 69.1 %), Tanzania (n=1, 1.8%), Korea (n=4, 7.3 %), Peru (n=1, 1.8 %), France (n=3, 5.5 %), Japan (n=3, 5.5 %), USA (n=2, 3.6 %), Argentina (n=1, 1.8 %), and unknown regions (n=2, 3.6 %), respectively. Also, according to the sources of isolation, there were 32 clinical strains and 23 environmental strains. It is worth noting that the S. haliotis strain JCM 14758 from GenBank was included, because S. haliotis has been recently identified as a synonym of S. algae by the previous research [14]. The 12 morphologically well-characterized strains in this study were initially identified by API 20E and 16S rRNA gene analysis. Detailed information of these strains was listed in Table S1.
Genome sequencing
The genomic DNA was extracted by Wizard Genomic DNA Extraction Kit (Madison, WI, Promega, USA) following the manufacturer’s instructions. The HiSeq sequencer (Illumina HiSeq2000, San Diego, CA, USA) was used to perform 250 bp paired-end whole-genome sequencing with 150× coverage. FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to evaluate the quality of the reads [16]. Low-quality reads were discarded if the quality scores of ≥3 consecutive bases were ≤Q30. SOAP de novo (version 2.04) was used to assemble the clean data of each strain [17]. After removing contigs with less than 500 bp, QUAST (version 5.0.1) software was applied to evaluate the quality of assembled genomes [18].
Average nucleotide identity (ANI) analysis
ANI analysis was used to evaluate the evolutionary distance of bacteria at the genomic level based on a Perl script previously described [19]. ANI was calculated between the reference genome and the query genome, and then the ANI value representing the BLASTn matches was computed with the cutoff set as ≥30 % sequence identity and ≥70 % length coverage. The strains with the ANI value >95 % were considered as the same species [20].
Pan-genome analysis
The genomes of 55 S . algae strains were collected together as a local database. Prokka (version 1.12) [21] was used to annotate genomes and produce standards-compliant *.gff output files for each sample. Roary pan-genome pipeline [22] with an identity cutoff i≥95 % was used. R package of pheatmap was used for the heatmap of the presence or absence of all these pan genes.
Core genome comparison
Coding sequences prediction was carried out using Prodigal (version 2.6.3) [23]. Then a non-redundant homologous gene set was computed for 55 strains using CD-HIT [24]. Next, BLAST+ was used to search the homologous genes in the non-redundant homologous gene set of each strain with the cutoff value of ≥90 % sequence identity and ≥60 % length coverage. If a homologous gene found in the non-redundant homologous gene set had just one copy in each strain and existed in all strains, it was considered as a core gene. The core genes were then aligned, and Gubbins (http://github.com/sanger-pathogens/gubbins) was used as a recombination-removal tool to reorganize the core genome. PhyML (version 3.1) [25] was used to construct the phylogenetic trees by maximum-likelihood method based on all these core SNPs (bootstrap replications, 1000). The strain RQs-106 with complete genome was selected as a reference to explore the number of SNPs for other strains. Population structure was defined using FastBaps (https://github.com/gtonkinhill/fastbaps) by a fast hierarchical Bayesian analysis [26].
Genomic islands (GIs), prophages, plasmids, CRISPR and unique genes analysis
Accessory genes were analysed using three online databases with default parameters. IslandViewer (http://www.pathogenomics.sfu.ca/islandviewer/) is a widely-used webserver for the prediction and interactive visualization of GIs (regions with probable horizontal origin). It integrates with IslandPath-DIMOB, SIGI-HMM and IslandPick to make accurate and complementary GI predictions [27]. PHASTER (PHAge Search Tool Enhanced Release) (http://phaster.ca) was used for rapid identification and annotation of prophage sequences in bacterial genomes and plasmids. An online search was performed against a custom prophage/phage database containing protein sequences from NCBI phage database developed by Srividhya et al. [28]. Plasmid replicons were identified online (http://www.genomicepidemiology.org/). CRISPRCasFinder (https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder) was used for the detection of CRISPRs (clustered regularly interspaced short palindromic repeats) and Cas (CRISPR-associated proteins) genes in the submitted sequence data. A blast comparison was performed against NCBI-NR database using the protein sequences of strain-unique genes with default parameters. The R package of plotrix was used to form the flower plot of unique genes.
Virulence-related genes analysis
The Virulence Factor Database (VFDB) [29] was used to predict virulence-related genes by blast+. The parameters included an E-value of 1e-5, a minimum identity and coverage of 50% and 70 %, respectively.
Antimicrobial resistance genes analysis
Potential antimicrobial resistance genes were predicted by Comprehensive Antibiotic Research Database (CARD) (http://arpcard.mcmaster.ca). The parameters of BLAST+ were an E-value of 1e-5, the percentage of sequence identity ≥80 % and the percentage of length coverage ≥80 %.
Antimicrobial susceptibility testing
The Antimicrobial Susceptibility Testing (AST) panel for aerobic Gram-negative bacilli (Shanghai Fosun Long March Medical Science Co., Ltd., China) was used to perform the antimicrobial susceptibility testing by the micro-broth dilution method. Each antibiotic has a concentration gradient on the drug sensitive plate. The bacteria suspension was added to the plate, and then after an incubation period of 18 to 20 h, the results were interpreted. The minimal inhibitory concentration (MIC) values were obtained. There is no CLSI breakpoints for Shewanella, and the results of susceptible (S), intermediate (I), and drug resistance (R) were interpreted according to the Enterobacteriaceae standards of the American Committee for Clinical Laboratory Standardization (CLSI) [30]. Escherichia coli ATCC 25922 was used as a control for chloramphenicol, tetracyclines, sulfonamides, and trimethoprim-sulfamethoxazole.
Results
General features of S. algae genomes
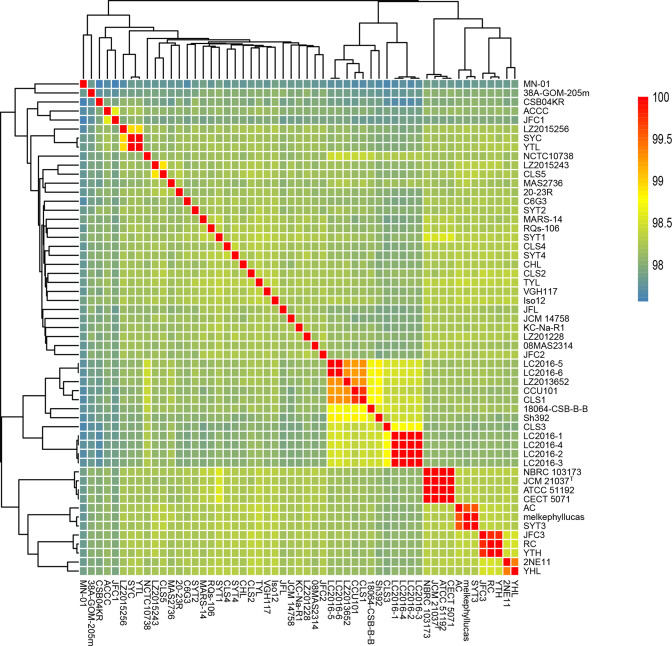
The genome sizes of the 55 strains ranged from 4.60 Mb (strain CLS3) to 5.20 Mb (strain KC-Na-R1), and the G+C content ranged from 52.5 mol% to 53.1 mol%. The general information of the 55 genomes was summarized in Table S2. Based on the matrix of ANI values, the dendrograms were drawn (Fig. 1). The ANI values of the 55 strains ranged from 97.7–99.9 %, providing accurate identification at the species level. The relatively high ANI values were found among three groups (LC2016-1/LC2016-2/LC2016-3/LC2016-4; NBRC 103673/JCM 21037T/ATCC 51192/CECT 5071; JFC3/RC/YTH).
Fig. 1.
Average nucleotide identity (ANI) values for the whole genomic sequences of the 55 S . algae strains.
Pan genome analysis
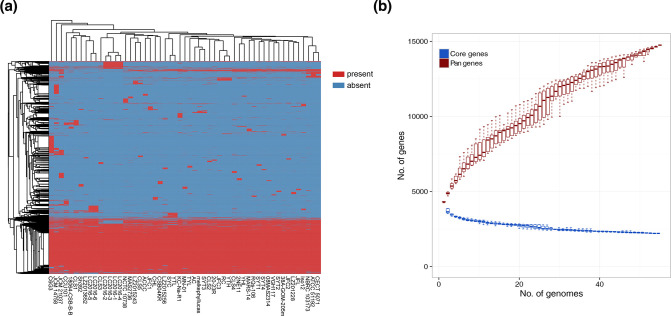
A total of 14780 genes, which represented the pan-genome of 55 S . algae strains, were found, with the CDSs ranging from 4105 (strain CLS3) to 5694 (strain C6G3) (Fig. 2a). Among them, 2863 (19.4 %) genes were shared by all strains and comprised the core genome. The COG functional annotation of the core genome focused on genes related to the basic lifestyle such as metabolism and energy production. The length of the core genome was 1 506 597 bp, and a total of 120 559 SNPs were detected. Taking the complete genome of strain 2NE11 as a reference, the number of SNPs ranged from 7496 (strain YHL) to 25 395 (strain MN-01), with the average number of 21 162. The dilution curve showed that the number of novel gene families in the pan-genome constantly increased when more genomes were considered, while the number of conserved genes constituting the core genome gradually stabilized (Fig. 2b).
Fig. 2.
Pan genome analysis of 55 S . algae strains and dilution curves of core/pan genes. (a): The heatmap based on pan genome sequences. On the top was the cluster of strains, and on the left was the cluster of pan genes. The red colour represented the presence of genes, while the blue colour represented the absence. (b): The dilution curves of core/pan genes.
Population structure analysis
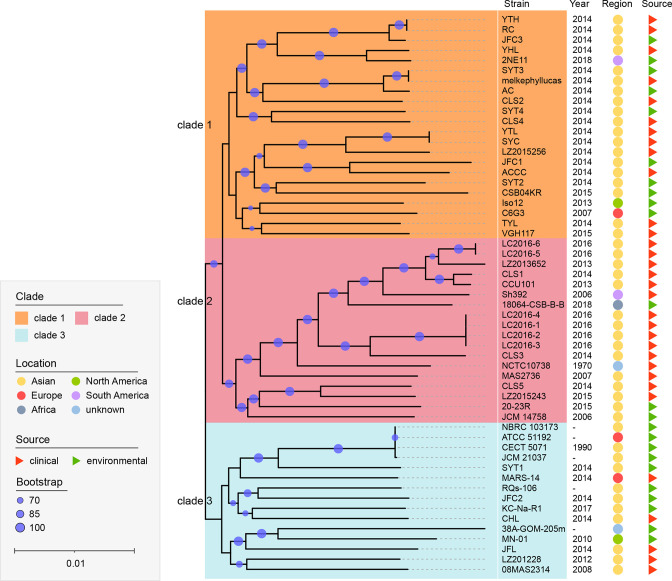
A robust core genome sequences based ML-tree was inferred to investigate the phylogenetic relationship of the 55 S . algae strains (Fig. 3). Three clades were identified according to the hierarchical Bayesian analysis by FastBaps. Clade 1 contained the largest number of strains (n=22), followed by clade 2 (n=18) and clade 3 (n=15). Among the 38 strains isolated from China, 31 strains were clustered in clade 1 and clade 2. Most strains derived from clinical patients appeared to be clustered in clade 2. The distribution of pairwise SNP distances (Fig. S1) showed that most isolates in clade 2 had a closer pairwise evolutionary relationship than those in clade 1 and clade 3.
Fig. 3.
Phylogenetic tree of core genome sequences by maximum-likelihood method. The strains were divided into six clades. The isolation year, regions and sources were shown on the right. Regions were circled in different colours and sources are triangulated (green for environmental sources and red for clinical sources). The robustness of tree topologies was evaluated with 1000 bootstrap replications. The scale represented a nucleotide substitution rate of 0.01 for each site.
Inter-species relationship between S. algae and other phylogenetically closed Shewanella spp
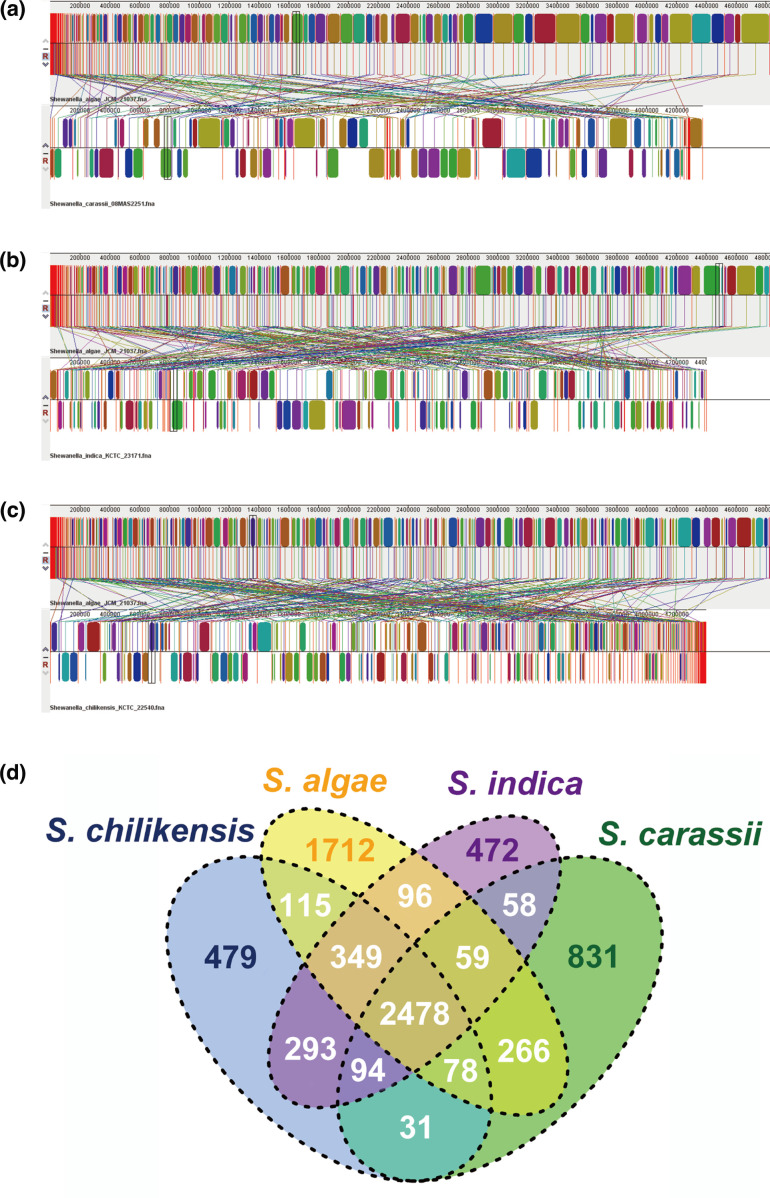
Comparative genomic analysis of S. algae JCM 21037T and three strains of other Shewanella spp. with the closed genetic relationships ( S. carassii 08MAS2251T, S. chilikensis KCTC 22540T and S. indica KCTC 23171T) was performed, according to the report of Thorell et al. [15]. We preliminarily evaluated the rearrangement and duplication events within the genomes through genome collinearity comparison (Fig. 4a–c). The three-genome alignments yielded 61, 65, and 68 Mauve blocks, respectively. The overall chromosome structures differed considerably among these strains. The lack of conservation appeared to be the result of a large inversion and multiple expansions of repetitive regions between the genomes of strains. Also, the Venn diagram of the shared and unique genes demonstrated that S. algae JCM 21037T, S. carassii 08MAS2251T, S. chilikensis KCTC 22540T and S. indica KCTC 23171T strains shared 2478 core genes (Fig. 4d), and these four strains harboured 1712, 831, 479, and 472 unique genes, respectively. An additional gene set, ranging from 31 to 293 genes, was shared by any two strains, while the number of genes that were shared by any three strains varied from 59 to 349.
Fig. 4.
The collinearity and the phylogeny among Shewanella spp. (a): General comparisons between S. algae JCM 21037T and S. carassii 08MAS2251T presented by Mauve software. (b): General comparisons between S. algae JCM 21037T and S. indica KCTC 23171T presented by Mauve software. (c): General comparisons between S. algae JCM 21037T and S. chilikensis KCTC 22540T presented by Mauve software. The Mauve parameter settings were default. (d): Venn diagram of the shared and unique genes found in S. algae JCM 21037T and other three Shewanella genomes.
GIs, prophages, plasmids, CRISPR and unique genes analysis
One hundred and sixty-seven GIs from six complete genomes of S. algae strains (2NE11: n=37, RQs-106: n=37, VGH117: n=25, 18064-CSB-B-B: n=24, CCU101: n=14, KC-Na-R1: n=30) were detected. The detected genes were related to virulence, metabolism, resistance, structure, fitness factors, modification-restriction systems, transport and toxins. GIs related to mobile element protein, integrase, transposase, acetyltransferase and transcriptional regulator were found in all six strains. Multidrug resistance transporter and heat shock protein were found in both RQs-106 and VGH117.
Sixty-five different prophage-related elements were detected, in which four genomes (strains KC-Na-R1, RQs-106, JFL and melkephyllucas) had intact prophages. Specifically, 40 (61.5 %) prophage-related sequences with the length between 4.4 and 42 kb were unique in 24 strains (Table S3), while five prophage-related sequences were shared by S. algae strains (Table S4). The detection of the presence of fitness factors encoded inside these sequences showed that 19 sequences carried transposase-like elements and 15 sequences contained integrase-like elements linked to horizontal gene transfer.
Plasmids were found in six strains, in which four strains were isolated from clinical specimens and the remaining two were sourced from environment. Three types of plasmid replicons were predicted. The information about the antimicrobial resistance genes and the functions of products encoded by the virulence-associated genes were described. The strains 18064-CSB-B-B and CCU101 appeared to carry resistance-transfer factors (RTFs). For complete genomes (18064-CSB-B-B and CCU101), the plasmid lengths were defined, otherwise the plasmid replicons were just localized. The detailed information was shown in Table S5.
Credible CRISPR-Cas system (evidence level=4) was predicted in the entire S. algae genomes. Two Cas types were detected, namely Cas-TypeIE for strains NCTC 10738, JFC2, JFL, SYT2 and CHL, and Cas-TypeIF for the others. The number of spacer sequences contained in CRISPR varied greatly among different strains. Specifically, strain C6G3 only harboured CRISPR sequences but not Cas clusters. The detailed information was shown in Table S6.
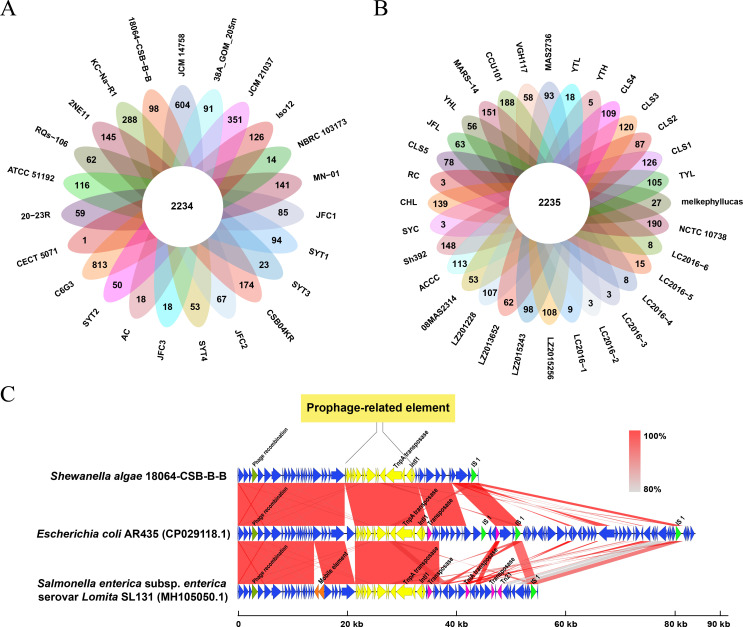
A large number of CDSs were annotated as hypothetical proteins. The number of unique genes in the 55 S . algae strains ranged from one to 813 (Fig. 5a, b). The strain C6G3, isolated from French seabed sediments in 2007, was found to contain the highest number of unique genes (813), while the strain CECT 5071 contained the fewest (one). The average number of unique genes was 75 in the clinical strains (Fig. 5a), and 106 in the environmental strains (Fig. 5b) (Wilcoxon rank sum test, no significant statistical difference, P>0.05).
Fig. 5.
The distribution of unique genes in environmental vs. clinical isolates and the gene clusters composition of prophage-related element. (a): The unique genes distribution of environmental strains. (b): The unique genes distribution of clinical strains. (c): The comparison of nucleotide similarity of the prophage-related element in S. algae 18064-CSB-BB and the homologs in E. coli and Salmonella enterica strains. The prophage-related CDSs were represented in yellow, while the insert sequence 1 (IS 1) in emerald green, the transposons in pink, the phage recombination associated CDSs in dark green, the other unnamed mobile elements in orange and the gene clusters in blue.
Four GIs related to cross-species transmission were identified in the 55 S . algae genomes. Two conjugative transfer-related GIs detected in strain 2NE11 shared 84.47 and 95.90% sequence identity with Halomonas meridiana Eplume2 and Alcanivorax N3-2A respectively. Two GIs detected in strains RQs-106 and CCU101 shared 93.35 and 97.15% sequence identity with Aeromonas hydrophila OnP3.1 and Marinobacter hydrocarbonoclasticus VT8 respectively (Table S7). The prophage-related element, playing the role of transposase, was detected in strain 18064-CSB-B-B isolated from poultry stool. Interestingly, this prophage-related element was found to be very similar to those in E. coli AR435 and Salmonella SL131 by blast homology comparison (Fig. 5c). Large quantities of insert sequences (IS) and transposases were predicted and annotated in the upstream and downstream sequences of these mobile genetic elements (MGEs), which may contribute to the genetic diversity of the accessory genome.
Prediction of virulence-associated genes
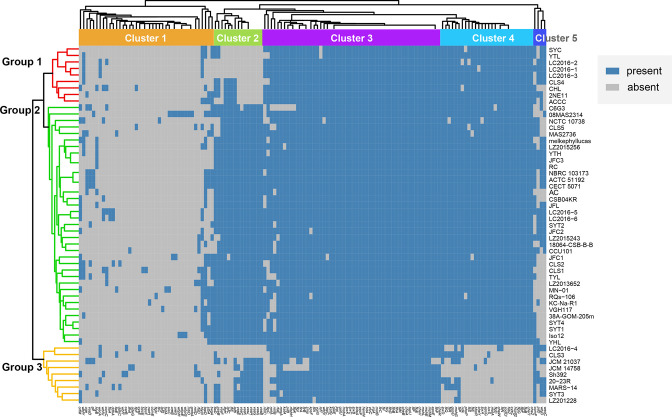
In the VFDB database, due to the lack of a virulence factor library related to Shewanella genus, we used Vibrio species as a reference for the virulence gene analysis because the phylogenetic relationship of Vibrio was closed to Shewanella . A putative virulence-associated genes pool showed great diversity, consisting of 19 classes, 59 virulence factors and 142 potential virulence genes (Table 1). The virulence-associated genes can be divided into five clusters, and the 55 S . algae strains were classified into three groups (Fig. 6). The results showed that 55 to 107 known or putative virulence genes were identified in the genome of each strain. The strains of different groups manifested specific virulence gene patterns. Most strains carried potential virulence genes related to secretion system, iron uptake and adherence, but rarely carried genes of Cluster 1, which could encode toxin, endotoxin, antiphagocytosis, acid resistance, biofilm formation and nutritional virulence. Strains from Group 1 mainly contained genes of Cluster 4 and 5, while strains from Group 2 mainly contained genes of Cluster 2, 4 and 5. Nearly all strains in Group 2 carried virulence-related genes in Cluster 3 with the main functions of secretion system, adherence and glycosylation system. The gene vasF related to VAS type VI secretion system was specific for strains in Group 2. Although strains from Group 3 mainly carried genes of Cluster 2 and 5, most strains in this group lacked a portion of potential virulence gene in Cluster 4 related to flagella, two component system, EPS type II secretion system, autoinducer-related genes and VAS effector proteins. On the whole, strains in Group 3 contained relatively the lowest number of virulence-associated genes, while strains in Group 2 contained the highest number.
Table 1.
The classification of virulence related factors of S. algae species against VFDB
|
Cluster |
VF-class |
Virulence factor |
Functions of productions |
|---|---|---|---|
|
C1 |
Acid resistance |
Urease |
Protein biosynthesis |
|
C1 |
Adherence |
Accessory colonization factor, Mannose-sensitive hemagglutinin4, Type IV pilus, Flagella4, Flp type IV pili, LPS O-antigen, Tap type IV pili, tad locus |
Flagellum assembly, pilus assembly, protein secretion, signals transduction, translocation, hydrolase |
|
C1 |
Anaerobic respiration |
Nitrate reductase |
Catalytic activity |
|
C1 |
Antiphagocytosis |
Capsular polysaccharide |
LPS biosynthesis and metabolism, catalytic activity |
|
C1 |
Biofilm formation |
AdeFGH efflux pump/transport autoinducer |
Catalytic activity |
|
C1 |
Chemotaxis and motility |
Flagella |
Flagellum biogenesis, motor activity, pathogenesis |
|
C1 |
Efflux pump |
AcrAB, MtrCDE |
Efflux |
|
C1 |
Toxin |
Hemolysin III, Phytotoxin phaseolotoxin, Endotoxin |
Signalling receptor, protein kinase activity |
|
C1 |
Immune evasion |
Capsule, LPS, Exopolysaccharide |
Metabolism, stationary phase survival, catalytic activity, biosynthesis |
|
C1 |
Nutritional virulence |
Biotin metabolism, Cysteine acquisition, Pyrimidine biosynthesis |
Biotin metabolism, chemotaxis signal transduction, signals transmission |
|
C2 |
Adherence |
Accessory colonization factor, Mannose-sensitive hemagglutinin4, Type IV pilus, Flagella4, Flp type IV pili, LPS O-antigen, Tap type IV pili, tad locus |
Flagellum assembly, pilus assembly, protein secretion, signals transduction, translocation, hydrolase |
|
C2 |
Chemotaxis and motility |
Flagella |
Flagellum biogenesis, motor activity, pathogenesis |
|
C2 |
Secretion system |
EPS type II secretion system, VAS type VI secretion system3, T4SS effectors |
Protein secretion and transport, virulence, chaperone |
|
C2 |
Glycosylation system |
O linked flagellar glycosylation |
Catalytic activity |
|
C3 |
Adherence |
Accessory colonization factor, Mannose-sensitive hemagglutinin4, Type IV pilus, Flagella4, Flp type IV pili, LPS O-antigen, Tap type IV pili, tad locus |
Flagellum assembly, pilus assembly, protein secretion, signals transduction, translocation, hydrolase |
|
C3 |
Antiphagocytosis |
Capsular polysaccharide |
LPS biosynthesis and metabolism, catalytic activity |
|
C3 |
Chemotaxis and motility |
Flagella |
Flagellum biogenesis, motor activity, pathogenesis |
|
C3 |
Immune evasion |
Capsule, LPS, Exopolysaccharide |
Metabolism, stationary phase survival, catalytic activity, biosynthesis |
|
C3 |
Iron uptake |
Enterobactin receptors, Periplasmic binding protein-dependent ABC transport systems, Cytochrome c maturation locus, Haemophilus iron transport locus, biosynthesis |
Transcription activation, signalling receptor, transport, catalase activity |
|
C3 |
Endotoxin |
Lipooligosaccharide (LOS) (Haemophilus) |
Catalytic activity |
|
C3 |
Protease |
Zn2+ metallophrotease |
Endoprotease |
|
C3 |
Stress adaptation |
Catalase, SodCI |
Catalytic activity, destroys radicals |
|
C4 |
Adherence |
Accessory colonization factor, Mannose-sensitive hemagglutinin4, Type IV pilus, Flagella4, Flp type IV pili, LPS O-antigen, Tap type IV pili, tad locus |
Flagellum assembly, pilus assembly, protein secretion, signals transduction, translocation, hydrolase |
|
C4 |
Chemotaxis and motility |
Flagella |
Flagellum biogenesis, motor activity, pathogenesis |
|
C4 |
Endotoxin |
LOS (Haemophilus) |
Endotoxin |
|
C4 |
Immune evasion |
Capsule, LPS, Exopolysaccharide |
Metabolism, stationary phase survival, catalytic activity, biosynthesis |
|
C4 |
Phagosome arresting |
Nucleoside diphosphate kinase |
Nucleoside triphosphates synthesis |
|
C4 |
Quorum sensing |
Autoinducer-2, PhoPQ, Two component system |
Autoinducer synthesis, regulation of transcription |
|
C4 |
Regulation |
Two component system |
Signal transduction |
|
C4 |
Stress adaptation |
Catalase, SodCI |
Catalytic activity, destroys radicals |
|
C4 |
Toxin |
Hemolysin III, Phytotoxin phaseolotoxin, Endotoxin |
Signalling receptor, protein kinase activity |
|
C4 |
Secretion system |
EPS type II secretion system, VAS type VI secretion system3, T4SS effectors |
Protein secretion and transport, virulence, chaperone |
|
C5 |
Iron uptake |
Enterobactin receptors, Periplasmic binding protein-dependent ABC transport systems, Cytochrome c maturation locus, Haemophilus iron transport locus, biosynthesis |
Transcription activation, signalling receptor, transport, catalase activity |
|
C5 |
Adherence |
Accessory colonization factor, Mannose-sensitive hemagglutinin4, Type IV pilus, Flagella4, Flp type IV pili, LPS O-antigen, Tap type IV pili, tad locus |
Flagellum assembly, pilus assembly, protein secretion, signals transduction, translocation, hydrolase |
|
C5 |
Antiphagocytosis |
Capsular polysaccharide |
LPS biosynthesis and metabolism, catalytic activity |
Fig. 6.
The heatmap of the virulence genes in 55 S . algae strains. On the left, the S. algae strains were divided into three groups (Group 1, Group 2 and Group 3) according to the clustering situation. On the top was the clusters of virulence-related genes, which were marked as Cluster 1 to Cluster 5, respectively. The blue colour indicated the presence of genes while the grey colour indicated the absence.
S. algae as a reservoir and a vehicle of potential antimicrobial resistance
We found genes associated with the resistance to eight antibiotics in the S. algae genomes (Table 2). The genes resisted to β-lactams (bla OXA, bla CTX, bla CMY, bla TEM, bla VEB), aminoglycosides (armA, aph(3'')-Ib, aph(6)-Id, ant(2'')-Ia, aadA2), quinolones (qnrA), phenicols (cml, catA1, floR), macrolides (erm), sulfonamides (dfr, sul), tetracyclines (tet), lincosamides (vga) were detected by BLAST+ against the database. The bla OXA gene was located in all strains with the major genotype of bla OXA-SHE (81.8%). Thirty-six strains carried qnr with the most common genotype of qnrA3. The environmental strain KC-Na-R1 carried the highest number of antimicrobial resistance genes, which indicated a capacity to resist multiple antibiotics. The genetic environments of identified resistance genes had been analysed. Twenty-two strains had mobile genetic elements (MGEs) in the upstream and downstream 10 kb sequences of resistance genes. The resistance genes possessed by strain KC-Na-R1 had the most abundant MGEs in its genetic environment. The detailed information was shown in the Table S8.
Table 2.
|
Category |
Genotype |
no. of strains |
Positive rate (%) |
|---|---|---|---|
|
β-lactam |
bla OXA-SHE, bla OXA-10, bla OXA-55, bla CTX-M-15, bla CMY-2, bla TEM-2, bla VEB-1 |
55 |
100 |
|
Quinolones |
qnrA1, qnrA3, qnrA5, qnrA7 |
36 |
65.5 |
|
Aminoglycosides |
armA, aph(3'')-Ib, aph(6)-Id, ant(2'')-Ia, aadA1, aadA2 |
12 |
21.8 |
|
Sulfonamides |
dfrA12, dfrA27, sul1, sul2 |
12 |
21.8 |
|
Amide alcohols |
cmlA1, catA1, floR |
8 |
14.5 |
|
Lincosamides |
vga(A) |
1 |
1.8 |
|
Tetracyclines |
tet(A) |
1 |
1.8 |
|
Macrolides |
erm(42) |
1 |
1.8 |
Antibiotic resistance screening of S. algae strains
The antimicrobial susceptibility of 12 clinical strains was shown in Table S9. All strains were resistant to cefazolin, but sensitive to minocycline, amikacin, cefepime, meropenem, doxycycline, minocycline, gentamicin, kanamycin, streptomycin. Also, these strains were sensitive to four kinds of aminoglycosides (including gentamicin, amikacin, kanamycin, and streptomycin). Six strains isolated from patients of Laizhou, China in 2015 were multi-drug resistant (insensitive to three or more types of antibiotics).
Discussion
Our study firstly provided a finer-scale comparison of S. algae at the intra-species level by sequencing 12 new S. algae strains and performed pan-, core- and accessory-genome analysis together with another 43 S . algae strains available in GenBank.
Pan-genome analysis explored the gene pool, unique genes and functional information underlying bacterial diversification. The number of accessory genes was 5.2 times that of core genes, which demonstrated that S. algae has great diversity in terms of genomic characteristics. The phylogenetic tree based on core genome demonstrated that the 55 S . algae strains were clustered into three separate clades, with most strains in China in clade 1 and 2. Compared with the report by Fang et al. [14], the phylogeny of S. algae in our study was consistent with the topological structure and evolutionary relationship of the phylogenetic tree constructed by MLSA method. When comparing the type strain of S. algae with those of other phylogenetically closed Shewanella species based on genome alignments, we observed that there were a large number of rearrangement and duplication events within different Shewanella species. The COG function annotation showed that the unique genes of S. algae were mainly related to signal transduction and transcription mechanisms, which may also indicate that accessory genes, especially the unique genes acquired from different environments, could be important for S. algae strains to increase their survival ability (Fig. S2).
Currently, the pathogenic mechanism of S. algae is not clear. Wu et al. found homologous genes of hlyA, hlyB, hlyD and tolC in the genome of S. algae STY3, which encoded hemolysin and transport functions, and speculated that the hemolytic activity may be an important virulence factor of S. algae [31]. Gallacher et al. confirmed that S. algae begins to produce tetrodotoxin after 12 h of culture [32]. Also, TTX was detected in S. algae strains isolated from food poisoning specimens by Wang et al., which suggests the possibility of TTX as a pathogenic substance of S. algae [33]. Alazea M. Tamez et al. have discovered and proposed candidate virulence factors of S. algae , including hemolysin, the type VI secretion system (T6SS), microbial collagenase, DNase, type IV pili, curli, twin-arginine translocation system, ClpP and urease [34].
In order to further explore the pathogenic characteristics of S. algae , 142 potential virulence-associated genes were extracted against the VFDB online database, showing a rich diversity of virulence-associated factors. There were significant differences in the virulence genes carried by strains in different groups (Fig. 6). Thirty-four core virulence-associated genes were found in all 55 strains. The presence of hlyB was only found in eight strains, and the complete RTX family that encodes a hemolysin failed to be predicted, which may explain why the function of hemolysis was irregular and difficult to be detected by Janda et al. [5]. The prokaryotic hlyB gene product is a member of a superfamily of ATP-binding transport proteins [35], located in the inner membrane of Escherichia coli and dedicated to the secretion of the toxin (alpha-hemolysin) HlyA in E. coli [36]. Therefore, hemolysin may be one of the pathogenic substances of S. algae . In addition, the common genes related to type IV pili, which are regulated by external stimuli for directed movement, provided a fresh perspective on pathogenicity [37]. We found that gene vasF, which was related to VAS type VI secretion system (T6SS), was specific for S. algae strains in group 2 (Fig. 6). T6SS was involved in a variety of cellular processes by secreting effector proteins into the extracellular milieu. Takahiko Ishikawa et al. demonstrated that the T6SS of wild-type V. cholerae O1 strains was functional and that its expression was controlled by specific environments in a pathogen-adaptive fashion [38], which indicated the S. algae strains in group 2 may have great adaptability to the environment.
Different bacteria in the gut flora could compete with other species through contact-dependent mechanisms, such as the T6SS. For example, Salmonella Typhimurium could utilize the T6SS to inject effector proteins into target cells to enhance its intestinal colonization ability [39]. Vibrio cholerae could use its T6SS to kill the symbiotic bacteria to promote colonization and enhance its pathogenicity, revealing that the absence of symbiotic bacteria may weaken the severity of diseases caused by V. cholerae, and V. cholerae enhances its pathogenicity through competition with symbiotic bacteria [40]. In this study, most clinical strains did not carry the vas gene, which means that clinical strains may not be very competitive for colonization since S. algae was just considered as an opportunistic pathogen. Considering S. algae is widely distributed in the oceans and has a potential to carry vas genes for colonization, it is necessary to strengthen S. algae environmental monitoring. Almost all strains contain katA gene, the product of which can decompose hydrogen peroxide, protect cells from the toxic effects of hydrogen peroxide, and enhance the bacterial colonization ability in the host [41]. Most of the S. algae strains also contained a stress-regulating gene sodCI. This gene encodes superoxide dismutase, which can destroy free radicals produced by host cells so as to eradicate bacteria, and enhance the bacterial resistance [42]. In our study, the genomes of 45 strains contained rmlA gene, which was related to capsular polysaccharide. It has been demonstrated that the mutations of the rmlA gene reduced biofilm-forming capacity of Stenotrophomonas maltophilia [43]. Our study showed that the ratio of rmlA gene in environmental S. algae strains was higher than that in its clinical counterparts, which revealed new insights into the environmental adaptability of S. algae . Two strains (strains 08MAS2314 and JFC1) contained urease-related genes ureB and ureG. Fu et al. demonstrated that the products of ureB and ureG genes are important virulence factors of Vibrio harveyi and enhance the pathogenicity to fish [44]. Also, Mehta et al. confirmed ureG gene could encode urease, which was essential for the colonization of Helicobacter pylori in stomach [45]. Most Brucella spp. also show strong urease activity. The products encoded by ureB and ureG are involved in protecting Brucella spp. from gastric acid and thus contribute to the pathogenicity [46]. Therefore, these two genes may also play an important role in the pathogenic process of S. algae . O-antigen, a virulence factor of Gram-negative bacteria, is the outermost structure of lipopolysaccharide. O-antigen is important for environmental adaptability because it is the main target of bacteriophages and the host immune system. O-antigen can induce a strong immune response in the host [47]. In our study, 19 strains carry the O-antigen synthesis-related genes. At the current stage, we failed to effectively detect the relatedness between virulence-associated genes and the sources of S. algae strains, suggesting that there may be minimal difference between clinical and environmental stains in terms of virulence-associated genes. Thus, we inferred that the pathogenicity of S. algae is affected by many factors such as complex interaction between the host and the environment, which is in accordance with the report of Janda et al. [1].
It is increasingly agreed that S. algae is a dominant human pathogen in the genus Shewanella when people are exposed to marine niches containing this pathogen through occupational or recreational activities. Documented illnesses linked to S. algae included skin and soft tissue infections, bacteremia, and otitis media. Generally, S. algae mostly infect people with an impaired immune system [2, 48, 49]. Also, we found no clear distinction in (potential) virulence determinants between environmental and clinical isolates of S. algae , which suggests that most S. algae strains may only cause opportunistic infections. Accordingly, we could reasonably speculate that the infection of S. algae is a preliminary condition rather than a decisive factor for human disease, and the pathogenic process is the result of the synergistic effect of multiple factors.
GIs harbouring a cluster of genes, are defined as probable horizontal origins in bacterial or archaeal genomes [27]. As one of the major drivers of genome evolution, GI can enhance the fitness of bacteria within a niche. There were 167 GIs and 65 phage-related elements detected in our study. Four GIs were associated with cross-species horizontal transfer. The GIs detected in strain 2NE11 were highly homologous to those in Halomonas meridiana Eplume2 and Alcanivorax N3-2A. Also, two GIs, which were detected in strains RQs-106 and CCU101 respectively, were highly homologous to those in Aeromonas hydrophila ONP3.1 and Marinobacter hydrocarbonoclasticus VT8 respectively. The sequences of prophage-related elements detected in strain 18064-CSB-B-B were 100 % similar to those in E. coli AR435 and Salmonella SL131. The GIs carried a suite of virulence genes and mobile elements, and the prophages contained genes encoding transposase and integrase. The horizontal gene transfer across species breaks down the boundaries of kinship and makes gene flow more complex. Therefore, we speculated that the cross-species gene transfer caused by GIs or prophage-related elements contributes to the evolution and independent acquisitions of virulence factors of S. algae . In addition, strains 2NE11, VGH117 and RQs-106 harboured GIs that encode several acyltransferases. Acyltransferases are enzymes which transfer acyl groups to specific targets and may be an important factor regulating the production of virulence factors, motility and biofilm formation.
The CRISPR-Cas system is a prokaryotic adaptive immune system which has evolved to get rid of foreign invading genes from viruses. The system is found in many bacteria, showing great complexity and diversity [50]. In our study, 23 strains harboured CRISPR-Cas system with CAS-TypeIE and CAS-TypeIF as the main Cas types. It is worth pointing out that the strain C6G3 has an incomplete CRISPR-Cas system, which means that this strain may have a defect in its natural immune defenses process.
Related literature reported that the aquatic bacterium Shewanella was a reservoir for MCR-4 mobile colistin resistance [51], and S. algae was the source of plasmid-mediated QnrA determinants [52]. A variety of drug-resistant genetic elements were identified in some Shewanella spp. with environmental or clinical origins, highlighting that the genus is probably a vehicle and a reservoir of antibiotic resistance genes [9, 10, 53–55]. These genes showed resistance to different antibiotic classes, including β-lactams, quinolones, aminoglycosides, macrolides and carbapenems. In this study, all 55 S . algae strains were found to carry antibiotic resistance genes, among which the carriage rate of bla OXA gene was 100 %, and that of qnr gene followed. It is worth noting that the plasmids of strains 18064-CSB-B-B and CCU101 were proved to carry RTFs, which contribute to the spread of antibiotic-resistance genes. Over the past few years, various investigations have extensively reported drug-resistant (XDR) Shewanella species from environmental and human infection samples, like XDR S. xiamenensis [10]. Among the 55 S. algae strains in our study, 67.3 % contained genes associated with resistance to more than one class of antibiotics (strain KC-NA-R1 carried the most number: 14). In addition, clinical strains have a higher frequency of carrying resistance genes of aminoglycosides (21.9%) compared with environmental strains (8.7 %), which may reveal an early warning for clinical surveillance and treatment of S. algae infection, since aminoglycosides are more often preferred in the treatment of severe infection caused by aerobic Gram-negative bacteria.
It is worth noting that, the drug-resistant phenotype of the 55 strains in our study did not match the drug-resistant genes carried in their genomes. Several reasons may account for this phenomenon. First, the genomes of most strains were not complete so the drug-resistant genes may be interrupted. Also, although the antimicrobial resistant phenotypes were determined by the corresponding genotypes, there was no absolute consistency between them [56, 57]. Jianhua Yin et al. discovered that the blaA gene was strongly induced by ampicillin at high (50 µg ml−1), but not at low levels (2.5 µg ml−1) [58]. These reasons may explain part of the mismatch that the 12 clinical strains in this study had a high frequency of carrying the resistance genes of aminoglycosides but were all sensitive to four tested aminoglycoside drugs.
Supplementary Data
Funding information
This work was supported by the National Science and Technology Infrastructure of China (National Pathogen Resource Center-NPRC-32), the National Science and Technology Fundamental Resources Investigation Program of China (2021FY100904) and Youth Fund of Liaoning Provincial Department of Education (QL202005).
Author contributions
Conceptualization, Q.W. and D.W.; designed the work, Z.H. and K.Y.; performed the experiments, Z.H., K.Y., S.F. and Y.X.; collected samples and isolated strains, Z.H., K.Y. and D.W.; analysed the data, Z.H., K.Y. and S.F.; wrote the manuscript, Z.H., K.Y., S.F., Y.X. and D.W. All authors contributed to the article and approved the submitted version.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This study used strains obtained from clinical and environmental sources and the Institutional Human Ethics Committee of the Institute did not require the study to be reviewed or approved because the strains were from laboratory stock. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements. The data were analysed anonymously and reported.
Footnotes
Abbreviations: ANI, average nucleotide identity; AST, antimicrobial susceptibility testing; CARD, comprehensive antibiotic research database; COG, Clusters of Orthologous Groups; GIs, genomic Islands; IS, insert sequence; MDR, multidrug-resistant; MGEs, mobile genetic elements; MIC, minimal inhibitory concentration; MLSA, multilocus sequence analysis; SNP, single nucleotide polymorphism; T6SS, type VI secretion system; VFDB, virulence factor database.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Nine supplementary tables and two supplementary figures are available with the online version of this article.
References
- 1.Janda JM, Abbott SL. The genus Shewanella: from the briny depths below to human pathogen. Crit Rev Microbiol. 2014;40:293–312. doi: 10.3109/1040841X.2012.726209. [DOI] [PubMed] [Google Scholar]
- 2.Hau HH, Gralnick JA. Ecology and biotechnology of the genus Shewanella . Annu Rev Microbiol. 2007;61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- 3.Inohana Y, Katsuya S, Koga R, Kouzuma A, Watanabe K. Shewanella algae relatives capable of generating electricity from acetate contribute to coastal-sediment microbial fuel cells treating complex organic matter. Microbes Environ. 2020;35 doi: 10.1264/jsme2.ME19161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aigle A, Bonin P, Fernandez-Nunez N, Loriod B, Guasco S, et al. The nature of the electron acceptor (MnIV/NO3) triggers the differential expression of genes associated with stress and ammonium limitation responses in Shewanella algae C6G3. FEMS Microbiol Lett. 2018;365 doi: 10.1093/femsle/fny068. [DOI] [PubMed] [Google Scholar]
- 5.Khashe S, Janda JM. Biochemical and pathogenic properties of Shewanella alga and Shewanella putrefaciens . J Clin Microbiol. 1998;36:783–787. doi: 10.1128/JCM.36.3.783-787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nozue H, Hayashi T, Hashimoto Y, Ezaki T, Hamasaki K, et al. Isolation and characterization of Shewanella alga from human clinical specimens and emendation of the description of S. alga Simidu et al., 1990, 335. Int J Syst Bacteriol. 1992;42:628–634. doi: 10.1099/00207713-42-4-628. [DOI] [PubMed] [Google Scholar]
- 7.Hong J, Steen C, Wong E, Keong B. Shewanella: an important, emerging and lethal pathogen in a patient with recurrent presentations of cholangitis. BMJ Case Rep. 2020;13:12. doi: 10.1136/bcr-2020-237655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer MJ, Stone-Garza KK, Croom D, Andreoli C, Woodson P, et al. Shewanella algae Infections in United States naval special warfare trainees. Open Forum Infect Dis. 2019;6:fz442. doi: 10.1093/ofid/ofz442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao JY, Zhao SM, Mu XD, Xiao Z. Genetic characterization of plasmid-mediated quinolone resistance gene qnrS2 in Pseudoalteromonas and Shewanella isolates from seawater. FEMS Microbiol Lett. 2017;364 doi: 10.1093/femsle/fnw295. [DOI] [PubMed] [Google Scholar]
- 10.Yousfi K, Bekal S, Usongo V, Touati A. Current trends of human infections and antibiotic resistance of the genus Shewanella . Eur J Clin Microbiol Infect Dis. 2017;36:1353–1362. doi: 10.1007/s10096-017-2962-3. [DOI] [PubMed] [Google Scholar]
- 11.da Costa WF, Giambiagi-deMarval M, Laport MS. Shewanella harboring antimicrobial and copper resistance genes in sea urchins (Paracentrotus lividus) from the Crozon peninsula (Brittany, France) Infect Genet Evol. 2020;85:104437. doi: 10.1016/j.meegid.2020.104437. [DOI] [PubMed] [Google Scholar]
- 12.Zago V, Veschetti L, Patuzzo C, Malerba G, Lleo MM. Resistome, mobilome and virulome analysis of Shewanella algae and Vibrio spp. strains isolated in italian aquaculture centers. Microorganisms. 2020;8:572. doi: 10.3390/microorganisms8040572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu W, Miller S, Chiu CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol. 2019;14:319–338. doi: 10.1146/annurev-pathmechdis-012418-012751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang Y, Wang Y, Liu Z, Dai H, Cai H, et al. Multilocus sequence analysis, a rapid and accurate tool for taxonomic classification, evolutionary relationship determination, and population biology studies of the genus Shewanella . Appl Environ Microbiol. 2019;85:e03126-03118. doi: 10.1128/AEM.03126-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorell K, Meier-Kolthoff JP, Sjöling Å, Martín-Rodríguez AJ. The genus Shewanella: from the briny depths below to human pathogen. Front Microbiol. 2019;10:1861. doi: 10.3389/fmicb.2019.01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown J, Pirrung M, McCue LA. FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics. 2017;33:3137–3139. doi: 10.1093/bioinformatics/btx373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honaas LA, Wafula EK, Wickett NJ, Der JP, Zhang Y, et al. Selecting superior de novo transcriptome assemblies: lessons learned by leveraging the best plant genome. PLoS One. 2016;11:e0146062. doi: 10.1371/journal.pone.0146062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee I, Ouk Kim Y, Park SC, Chun J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 21.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 22.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 26.Tonkin-Hill G, Lees JA, Bentley SD, Frost SDW, Corander J. Fast hierarchical Bayesian analysis of population structure. Nucleic Acids Res. 2019;47:5539–5549. doi: 10.1093/nar/gkz361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertelli C, Laird MR, Williams KP, Simon Fraser University Research Computing Group. Lau BY, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.CLSI Performance Standards for Antimicrobial Susceptibility Testing. 28th edn. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2018. [Google Scholar]
- 31.Wu Z-Y, Ho S-P, Cheng J-F, Tung K-C, Hong Y-K, et al. Whole-genome characterization of Shewanella algae strain SYT3 isolated from seawater reveals insight into hemolysis. Future Microbiol. 2018;13:1709–1717. doi: 10.2217/fmb-2018-0267. [DOI] [PubMed] [Google Scholar]
- 32.Gallacher S, Birkbeck TH. A tissue culture assay for direct detection of sodium channel blocking toxins in bacterial culture supernates. FEMS Microbiol Lett. 1992;71:101–107. doi: 10.1016/0378-1097(92)90549-4. [DOI] [PubMed] [Google Scholar]
- 33.Wang D, Wang Y, Huang H, Lin J, Xiao D, et al. Identification of tetrodotoxin-producing Shewanella spp. from feces of food poisoning patients and food samples. Gut Pathog. 2013;5:15. doi: 10.1186/1757-4749-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamez AM, McLaughlin RW, Li J, Wan X, Zheng J. Searching for putative virulence factors in the genomes of Shewanella indica and Shewanella algae . Arch Microbiol. 2021;203:683–692. doi: 10.1007/s00203-020-02060-1. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Y, Ojeda-May P, Nagaraju M, Kim B, Pu J. Mapping free energy pathways for ATP Hydrolysis in the E. coli ABC Transporter HlyB by the String Method. Molecules. 2018;23:10. doi: 10.3390/molecules23102652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cross MA, Koronakis V, Stanley PL, Hughes C. HlyB-dependent secretion of hemolysin by uropathogenic Escherichia coli requires conserved sequences flanking the chromosomal hly determinant. J Bacteriol. 1990;172:1217–1224. doi: 10.1128/jb.172.3.1217-1224.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyd JM, Dacanay A, Knickle LC, Touhami A, Brown LL, et al. Contribution of type IV pili to the virulence of Aeromonas salmonicida subsp. salmonicida in atlantic salmon (Salmo salar L.) Infect Immun. 2008;76:1445–1455. doi: 10.1128/IAI.01019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishikawa T, Sabharwal D, Bröms J, Milton DL, Sjöstedt A, et al. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect Immun. 2012;80:575–584. doi: 10.1128/IAI.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A. 2016;113:E5044–51. doi: 10.1073/pnas.1608858113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fast D, Kostiuk B, Foley E, Pukatzki S. Commensal pathogen competition impacts host viability. Proc Natl Acad Sci U S A. 2018;115:7099–7104. doi: 10.1073/pnas.1802165115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bandyopadhyay P, Steinman HM. Catalase-peroxidases of Legionella pneumophila: cloning of the katA gene and studies of KatA function. J Bacteriol. 2000;182:6679–6686. doi: 10.1128/JB.182.23.6679-6686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnakumar R, Kim B, Mollo EA, Imlay JA, Slauch JM. Structural properties of periplasmic SodCI that correlate with virulence in Salmonella enterica serovar Typhimurium. J Bacteriol. 2007;189:4343–4352. doi: 10.1128/JB.00010-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhuo C, Zhao Q, Xiao S. The impact of spgM, rpfF, rmlA gene distribution on biofilm formation in Stenotrophomonas maltophilia . PLoS One. 2014;9:e108409. doi: 10.1371/journal.pone.0108409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu S, Ni P, Yang Q, Hu H, Wang Q, et al. Delineating the key virulence factors and intraspecies divergence of Vibrio harveyi via whole-genome sequencing. Can J Microbiol. 2021;67:231–248. doi: 10.1139/cjm-2020-0079. [DOI] [PubMed] [Google Scholar]
- 45.Mehta N, Benoit S, Maier RJ. Roles of conserved nucleotide-binding domains in accessory proteins, HypB and UreG, in the maturation of nickel-enzymes required for efficient Helicobacter pylori colonization. Microb Pathog. 2003;35:229–234. doi: 10.1016/s0882-4010(03)00151-7. [DOI] [PubMed] [Google Scholar]
- 46.Sangari FJ, Seoane A, Rodríguez MC, Agüero J, García Lobo JM. Characterization of the urease operon of Brucella abortus and assessment of its role in virulence of the bacterium. Infect Immun. 2007;75:774–780. doi: 10.1128/IAI.01244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lerouge I, Vanderleyden J. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol Rev. 2002;26:17–47. doi: 10.1111/j.1574-6976.2002.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 48.Goyal R, Kaur N, Thakur R. Human soft tissue infection by the emerging pathogen Shewanella algae . J Infect Dev Ctries. 2011;5:310–312. doi: 10.3855/jidc.1436. [DOI] [PubMed] [Google Scholar]
- 49.Takata T, Chikumi H, Morishita S, Hamada S, Hoi S, et al. Shewanella algae bacteremia in an end-stage renal disease patient: a case report and review of the literature. Intern Med. 2017;56:729–732. doi: 10.2169/internalmedicine.56.7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H, Wei W, Huang M, Umar Z, Feng Y. Definition of a Family of Nonmobile Colistin Resistance (NMCR-1) Determinants Suggests Aquatic Reservoirs for MCR-4. Adv Sci (Weinh) 2019;6:1900038. doi: 10.1002/advs.201900038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother. 2005;56:463–469. doi: 10.1093/jac/dki245. [DOI] [PubMed] [Google Scholar]
- 53.Almuzara M, Montaña S, Lazzaro T, Uong S, Parmeciano Di Noto G, et al. Genetic analysis of a PER-2-producing Shewanella sp. strain harbouring a variety of mobile genetic elements and antibiotic resistance determinants. J Glob Antimicrob Resist. 2017;11:81–86. doi: 10.1016/j.jgar.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Tacão M, Araújo S, Vendas M, Alves A, Henriques I. Shewanella species as the origin of bla OXA-48 genes: insights into gene diversity, associated phenotypes and possible transfer mechanisms. Int J Antimicrob Agents. 2018;51:340–348. doi: 10.1016/j.ijantimicag.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 55.Potron A, Poirel L, Nordmann P. Origin of OXA-181, an emerging carbapenem-hydrolyzing oxacillinase, as a chromosomal gene in Shewanella xiamenensis . Antimicrob Agents Chemother. 2011;55:4405–4407. doi: 10.1128/AAC.00681-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pornsukarom S, van Vliet AHM, Thakur S. Whole genome sequencing analysis of multiple Salmonella serovars provides insights into phylogenetic relatedness, antimicrobial resistance, and virulence markers across humans, food animals and agriculture environmental sources. BMC Genomics. 2018;19:801. doi: 10.1186/s12864-018-5137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ho J, Jelfs P, Sintchencko V. Phenotypically occult multidrug-resistant Mycobacterium tuberculosis: dilemmas in diagnosis and treatment. J Antimicrob Chemother. 2013;68:2915–2920. doi: 10.1093/jac/dkt284. [DOI] [PubMed] [Google Scholar]
- 58.Yin JH, Sun YY, Mao YT, Jin M, Gao HC. PBP1a/LpoA but not PBP1b/LpoB are involved in regulation of the major β-lactamase gene blaA in Shewanella oneidensis . Antimicrob Agents Chemother. 2015;59:3357–3364. doi: 10.1128/AAC.04669-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.