Abstract
Clostridioides difficile is a pathogen often associated with hospital-acquired infection or antimicrobial-induced disease; however, increasing evidence indicates infections can result from community or environmental sources. Most genomic sequencing of C. difficile has focused on clinical strains, although evidence is growing that C. difficile spores are widespread in soil and water in the environment. In this study, we sequenced 38 genomes collected from soil and water isolates in Flagstaff (AZ, USA) and Slovenia in an effort targeted towards environmental surveillance of C. difficile . At the average nucleotide identity (ANI) level, the genomes were divergent to C. difficile at a threshold consistent with different species. A phylogenetic analysis of these divergent genomes together with Clostridioides genomes available in public repositories confirmed the presence of three previously described, cryptic Clostridioide s species and added two additional clades. One of the cryptic species (C-III) was almost entirely composed of Arizona and Slovenia genomes, and contained distinct sub-groups from each region (evidenced by SNP and gene-content differences). A comparative genomics analysis identified multiple unique coding sequences per clade, which can serve as markers for subsequent environmental surveys of these cryptic species. Homologues to the C. difficile toxin genes, tcdA and tcdB, were found in cryptic species genomes, although they were not part of the typical pathogenicity locus observed in C. difficile , and in silico PCR suggested that some would not amplify with widely used PCR diagnostic tests. We also identified gene homologues in the binary toxin cluster, including some present on phage and, for what is believed to be the first time, on a plasmid. All isolates were obtained from environmental samples, so the function and disease potential of these toxin homologues is currently unknown. Enzymatic profiles of a subset of cryptic isolates (n=5) demonstrated differences, suggesting that these isolates contain substantial metabolic diversity. Antimicrobial resistance (AMR) was observed across a subset of isolates (n=4), suggesting that AMR mechanisms are intrinsic to the genus, perhaps originating from a shared environmental origin. This study greatly expands our understanding of the genomic diversity of Clostridioides . These results have implications for C. difficile One Health research, for more sensitive C. difficile diagnostics, as well as for understanding the evolutionary history of C. difficile and the development of pathogenesis.
Keywords: Clostridioides difficile, cryptic species, genomics, toxin
Data Summary
Genomic sequence data generated as part of this study have been deposited at the National Center for Biotechnology Information (NCBI) under BioProject accession numbers PRJNA438482 and PRJNA662350, with specific accession numbers as shown in Table S1. Public genomic data were downloaded from NCBI databases (Table S2).
Impact Statement.
Clostridioides difficile is a pathogen often associated with severe diarrhoea that can be difficult to treat and can cause potentially fatal infections in humans. Understanding the diversity and ecology of C. difficile and closely related bacteria is important from a public-health perspective, as this knowledge could aid in identifying sources of human infections and could improve diagnostics for detecting C. difficile . In this study, we used comparative genomic analyses to characterize bacterial isolates collected from environmental samples from distant geographical locations. The isolates are closely related to C. difficile but can be considered multiple, different species, and some isolates contain homologues of C. difficile toxin genes. This study contributes to our understanding of the diversity of Clostridioides and can provide a framework for studying pathogenic members of the genus.
Introduction
Clostridioides difficile is an anaerobic, spore-forming pathogen that can cause potentially fatal infections in humans. Although C. difficile has been associated with nosocomial infections and the human gut, the organism has been isolated from a variety of sample types and environments including non-human animals, water (including wastewater treatment facilities), household environments, and soil from urban or rural settings [1–3]. These strains are genetically similar to those isolated from humans in clinical settings, suggesting that the same strains can inhabit multiple reservoirs and that the environment is a source of human infections.
The use of whole-genome sequencing is changing our view of C. difficile composition, evolution and differentiation. In 2016, divergent types of C. difficile were observed in environmental samples based on multilocus sequence typing (MLST) analysis and ribotyping [4]. In 2014, the cryptic clade C-I was identified, although at the time only non-toxigenic strains were described [5]. In 2018, a core-genome MLST system identified the presence of two additional cryptic lineages (C-II and C-III) [6]. A recent study used average nucleotide identity (ANI) to demonstrate that each of these lineages is distinct from each other and C. difficile , representing unique genomospecies [7]. The function, geographical distribution and complete genomic complement of genomes from these lineages are understudied.
Divergent lineages were also reported to contain unique toxin-encoding regions. Three toxins are produced by C. difficile toxigenic strains in various combinations. Toxins A and B, encoded by tcdA and tcdB, respectively [8], are accepted as the main virulence factors [9], whereas the role of the binary toxin CDT has not been fully determined [10, 11]. Toxins A and B are encoded by genes in the PaLoc region, which in nontoxigenic strains is replaced by a section of DNA 115 bp in length; in divergent strains, large insertions have been observed instead of the PaLoc [4, 5, 12]. Although tcdB+ strains that lack tcdA are fully virulent in animal models [13], strains that lack tcdB can cause disease [13] but appear to be attenuated [14]. There have been numerous examples of tcdB+/tcdA− C. difficile isolates [15]. Although tcdB−/tcdA+ isolates of C. difficile have not been reported, this profile has been found in related cryptic isolates [16].
Identification of C. difficile in the clinic typically includes the detection of either the tcdB or the tcdA genes [17, 18], the 16S rRNA gene [19] and/or the tpi gene (triose phosphate isomerase) [20] through PCR or the detection of toxins A and B and/or glutamate dehydrogenase (GDH) with enzyme immunoassays [21, 22]. C. difficile have been isolated from the environment through culture-based methods using selective media [23], as the pathogen is expected to be a minor component of the microbial community. Isolates are characterized with diverse methods including colony morphology [24], MALDI-TOF [4] and molecular methods such as PFGE [25] or ribotyping [4, 26]. Following enrichment and/or isolation, many workflows focus on the detection of tcdA [27] and tcdB [28] to identify C. difficile . However, C. difficile sampling that focuses on detection of toxin genes could miss the diversity of Clostridioides that don’t possess these genes or have nucleotide differences that could evade current molecular assays. Currently, genomic characterization of environmental C. difficile isolates is less common than for clinical isolates [7]. For clinical and environmental detection of C. difficile , understanding the genomic diversity of the pathogen, as well as genetic near neighbours, is crucial for ensuring appropriately sensitive and specific assays. To better understand the genomic landscape of divergent Clostridioides lineages, we describe a comparative genomics analysis of cryptic Clostridioides isolates collected from environmental samples in two diverse geographical locations: Flagstaff, (AZ, USA) and Slovenia (Europe).
Methods
Sample collection, bacterial cultivation, DNA extraction and sample screening
The sampling and isolation of strains from Slovenia has been previously described [4, 29]. To survey C. difficile throughout Flagstaff, AZ, USA, soil (n=210) and water (n=50) samples were collected at various locations (Fig. S1, Table S1, available with the online version of this article). Two rounds of sampling were conducted between 2017 and 2019. For the first round of sampling, 50 soil and 50 water samples were collected in 2017. Soil samples were collected from the top 2 inches (5 cm) of soil using a disposable plastic spoon and stored in a quart-sized plastic bag at 4 °C until processing. For water samples, a maximum of 150 ml water was collected from each location. The water samples were filtered using a Combisart three branch manifold (Sartorius) with 250 ml sterile funnels containing Microsart filters (cellulose nitrate, 47 mm diameter, 0.2 µM pore size) and a Microsart EJet pump. Sterile Minisart syringe filters (25 mm, 0.2 µm PTFE) were attached to each branch on the apparatus for sterile venting. The filter was stored at 4 °C until processing. Two grams of soil or one-half of a cellulose nitrate membrane filter was incubated in Cdiff banana broth (Hardy Diagnostics) at 36 °C for 72 h. A 200 µl aliquot of enrichment culture was plated onto taurocholate-cefoxitin-cycloserine-fructose agar (TCCFA) and incubated for 24 h at 36 °C in a vinyl type C anaerobic chamber (Coy Laboratory Products). A 20 % (v/v) glycerol stock was created from this population for future work, and DNA was extracted from the population using a Qiagen DNeasy blood and tissue kit, and screened with Taqman PCR assays designed to detect C. difficile and the presence of the tcdB gene [30]. The C. difficile Taqman PCR assay includes primers targeting a unique C. difficile marker (CD630_24840) used previously for detection of C. difficile in dog faecal samples [30]. Samples with positive Taqman PCR assay results for the C. difficile marker were streaked for isolation on brain heart infusion agar supplemented with 0.03 % l-cysteine (BHIS) and incubated anaerobically for 24 h at 36 °C. Lawns were created for each isolate and DNA was extracted using a Qiagen DNeasy blood and tissue kit for whole-genome sequencing. For the second round of sampling in Flagstaff (160 soil samples collected in 2018 and 2019), approximately 3 g soil collected from the top 1 inch (2.5 cm) of sampling sites was added to Cdiff banana broth and incubated at 37 °C for 72 h. From these enrichments, glycerol stocks were created and DNA was extracted with a MoBio Powersoil DNA extraction kit. Samples with positive results for the C. difficile Taqman PCR assay were streaked for isolation on TCCFA; colonies were picked and streaked onto BHIS, and DNA was subsequently extracted with a Qiagen DNeasy blood and tissue kit for whole-genome sequencing.
Sequencing, genome assembly and MLST
All DNA was sequenced on the Illumina MiSeq platform; for a subset of genomes, sequencing was also performed on the Nanopore MinION platform. DNA extracted from Flagstaff isolates was prepared for multiplexed, paired-end sequencing with a 500 bp insert using standard PCR library amplification (KAPA Biosystems). DNA extracted from Slovenian isolates was prepared using the Nextera XT DNA library preparation kit (Illumina). Genomes were sequenced on the Illumina MiSeq platform. Illumina data was adapter trimmed with BBDuk v38.86 (https://sourceforge.net/projects/bbmap/), assembled with Spades v3.10.0 or v3.14.0 [31], polished with Pilon v1.22 [32] and contigs >500 nt were annotated with Prokka v1.14.6 [33]. For MinION sequencing, samples were streaked for isolation from glycerol stocks using BHIS and incubated anaerobically at 37 °C for 24 h; high molecular mass DNA was extracted using the Quick-DNA HMW MagBead kit (Zymo). DNA was assessed for quality using a standard genomic 50 kb Fragment Analyzer kit (Agilent) to ensure mean DNA fragments >60 000 kb. Libraries were prepared using a SQK-LSK109 1D ligation gDNA kit with the native barcoding gDNA kit (Oxford Nanopore). Libraries were loaded onto a R9/R9.4 flow cell and MinION sequencing was performed for 60 h using MinKNOW software. Guppy v3.22 was used for basecalling using the 9.4.1_450bps_hac workflow, and reads were trimmed with Porechop v0.2.4 (https://github.com/rrwick/Porechop). Unicycler v0.4.8 [34] was used to generate hybrid assemblies where Illumina and MinION data were available. MLST profiles of sequenced genomes were determined by stringMLST v0.6.3 (-k 45) [35].
Publicly available genomic data
Genome assemblies matching the search term ‘Clostridioides’ were downloaded from GenBank on 20 October 2020 (n=2520). Two of these genome assemblies were discarded due to anomalous G+C content (the excluded genomes had G+C content >44 mol%, the G+C content of included genomes ranged from 28.04–32.78 mol%). Paired-end Illumina reads for isolates identified as belonging to cryptic Clostridioides lineages by Knight et al. [7] were downloaded from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA). To identify additional genomes belonging to cryptic Clostridioides lineages, the SRA was also searched for ‘Clostridioides difficile’ on 20 October 2020 and reads released after 1 January 2020 were downloaded (this set of reads includes data released after the 2020 Knight et al. publication [7]). MLST profiles were assigned to paired-end Illumina data with stringMLST v0.6.3 (-k 45) [35]. Reads with MLST profiles known to be associated with C. difficile clades 1–5 were discarded. The remaining reads were assembled as described above and assemblies with anomalous genome size or G+C content were discarded (the assembly size and G+C content of included genomes are listed in Table S2). Genome assemblies were then compared with mash v2.1 [36] and only genomes associated with cryptic Clostridioides lineages closely related to C. difficile were retained. The final data set included 2518 GenBank assemblies and 28 genomes downloaded from the SRA (Table S2).
ANI
After an initial screening of genomes with mash, a set of 101 reference genomes was chosen from the entire data set (all genomes in Table S1 and 63 genomes in Table S2); this smaller set of genomes included all genomes associated with cryptic Clostridioides lineages and representative genomes from each C. difficile clade (1–5) (Tables S1 and S2). These genomes were compared with pyani (v0.2.10 – https://github.com/widdowquinn/pyani) [37], using the MUMmer v3.23 [38] alignment option. An ANI (ANIm value) of 95 % was used to delineate species/lineage boundaries, which is consistent with published criteria [39].
Core-genome SNP phylogeny
A core-genome SNP analysis was performed for the set of 101 reference genomes. All genomes were aligned against the completed C. difficile genome CD630 (GenBank accession no. NC_009089) using NUCmer v3.1 [40] and SNPs were called with nasp v1.2 [41]. Any SNP that fell within a duplicated region in the CD630 genome, based on a self-alignment with NUCmer, was filtered from downstream analyses. A phylogeny was inferred from a concatenation of SNPs with iq-tree v2.0.3 [42], using the TN+F+ASC+R3 substitution model [43].
A core-genome SNP analysis was also performed on C-III genomes within nasp by aligning Illumina sequencing reads to the completed assembly for ES-W-0016-02 (GenBank accession numbers CP061361 and CP061362) with bwa-mem v0.7.17 [44], and calling SNPs with the UnifiedGenotyper method in gatk v3.4-46 [45, 46]. SNPs were filtered from downstream analyses if the depth of coverage was less than 5x, the proportion of base calls was less than 0.9, the SNP was identified in a duplicated region of the reference genome, or if the SNP was in close proximity to another SNP call (within 5 positions), which may indicate a low-quality SNP call [47]. A phylogeny was generated with iq-tree as described above (substitution model TVM+F+R3, SNPs queried from C. difficile CD630 were used to root the phylogeny).
Comparative genomics and pan-genomics
To determine the sizes of the pan and core genomes of the novel clades, ‘.gff’ files produced by Prokka were processed by Panaroo v1.2.3 [48] using default settings. Coding region sequences (CDSs) were considered to be part of the core genome if they were conserved in >99 % of genomes surveyed. CDSs unique to each clade were identified with the Large-scale blast Score Ratio (ls-bsr) pipeline v1.2.2 [49]. CDSs for a representative genome in each clade were screened against all genome assemblies (n=2584) using the blastn alignment option. A CDS was considered a lineage-specific marker if the CDS had a blast score ratio (BSR) [50] value ≥0.8 for all in-group genome assemblies and a BSR value <0.4 for all out-group genome assemblies. Unique regions were translated with Biopython and the clusters of orthologous groups (COG) annotation was determined by eggNOG-Mapper v2.0.1–14 [51].
In addition, the protein sequences for genes associated with toxins in C. difficile were screened against the reference genome set (n=101) with tblastn v2.10.0+ [52] in conjunction with ls-bsr. The resulting matrix was converted into a binary presence (‘1’) or absence (‘0’) matrix based on a BSR threshold of 0.5; this is equivalent to 50 % identity over 100 % of the protein length. The resulting heatmap was visualized with the Interactive Tree of Life [53]. The gene order of the PaLoc and the CdtLoc in selected genomes was visualized by genoPlotR [54].
Plasmid and phage screen
Previously published sequences from C. difficile plasmids [55] and phage [56] were downloaded from GenBank; additional extrachromosomal regions were downloaded based on the study by Hornung et al. [57]. Circular contigs assembled in this study that were not associated with the chromosome were added to these other sequences and a dendrogram was created with MashPy, a pipeline that clusters pairwise mash distances (https://gist.github.com/jasonsahl/24c7cb0fb78b4769521752193a43b219).
Enzymatic profiles of isolates from new clades
Enzymatic profiles for five isolates representing newly identified clades were evaluated using the API 20A system (bioMérieux). Five anaerobic isolates were grown overnight on BHIS at 37 °C within an anaerobic chamber. After 24 h, isolates were suspended in Mueller Hinton broth (Millipore Sigma) to a 1 McFarland turbidity standard. Using this suspension, bacterial lawns were created on Brucella blood agar with haemin and vitamin K media (Hardy Diagnostics). Isolates were grown on blood media overnight at 37 °C for 24 h under anaerobic conditions. Samples were suspended in inoculation medium API 20A (Anaerobe) from API system (bioMérieux) to a 3 McFarland turbidity standard and then added into microtubes according to the manufacturer’s instructions. The API 20A strip consisted of 20 microtubes with dehydrated substrates. All test strips were incubated at 37 °C with humidity overnight and tests were scored after 24 h. These reactions were scored according to the manufacturers 'Reading Table'. The following tests were determined: gelatine (GEL), urea (URE), aesculin hydrolysis (ESC), indole synthesis (IND) and the fermentation ability of 16 carbohydrates – d-glucose (GLU), d-mannitol (MAN), d-lactose (LAC), d-sucrose (SAC), d-maltose (MAL), salicin (SAL), d-xylose (XYL), l-arabinose (ARA), glycerol (GLY), d-cellobiose (CEL), d-mannose (MNE), d-melezitose (MLZ), d-raffinose (RAF), d-sorbitol (SOR), l-rhamnose (RHA) and d-trehalose (TRE). Additionally, a catalase reaction in which the addition of hydrogen peroxide (3%) was added to culture and the presence (positive result) or absence (negative result) of bubbles was recorded.
Antimicrobial-resistance (AMR) profiles of select isolates
Four cryptic Clostridioides isolates representing two cryptic lineages were screened for AMR against the following drugs known to induce C. difficile infection (CDI): ceftriaxone, ceftazidime, clindamycin, ciprofloxacin, imipenem, cefotaxime, cefepime, erythromycin, moxifloxacin and levofloxacin. Minimum inhibitory concentrations (MICs) were determined across these ten antimicrobials using ETESTs (bioMérieux) as per the Clinical and Laboratory Standards Institute (CLSI) 2020 guidelines, where available; for ciprofloxacin, no breakpoint was listed and the breakpoint for moxifloxacin was used, as has been done previously [58]. Samples were streaked for isolation from glycerol stocks using BHIS and incubated anaerobically at 37 °C for 24 h. An initial inoculum was prepared by making a Mueller Hinton broth suspension of isolated colonies from the overnight growth on BHIS plates. The bacterial suspension was adjusted to achieve turbidity equivalent to a 1.0 McFarland turbidity standard (Remel). Bacterial lawns were created from this suspension using a sterile swab and Brucella blood agar with haemin and vitamin K (Hardy Diagnostics) according to standard protocols. ETESTs were applied on plates following a 10 min dry time and plates were incubated at 37 °C for 48 h, after which MICs were scored according to the manufacturer’s instructions. All MIC testing was performed in biological duplicate.
In silico predicted AMR profiling
The set of 101 reference genomes were screened with the Comprehensive Antimicrobial Resistance Database (CARD) [59] genes with blastn and ls-bsr. Any CARD region with a BSR ≥0.75 was considered to be conserved. Protein sequences of interest (GyrA and GyrB) were identified with tblastn v2.9.0+ [52], aligned with muscle v3.8.31 [60] and examined for mutations associated with AMR listed in the CARD database [59].
16S rRNA gene and rpoB gene analyses
16S rRNA gene sequences were extracted from Prokka annotation files for the reference set genomes (n=101). For genome assemblies with multiple 16S copies, a single sequence was selected for each genome (the longest clostridial 16S gene sequence for each genome). No 16S rRNA gene sequence was retrieved for two genomes in the reference set. Sequences were aligned with muscle v3.8.31 [60] and a maximum-likelihood phylogeny was inferred with iq-tree v1.6.12 [61] using the selected best fit substitution model (HKY+F+R2) [43]. The V4 region of the 16S rRNA gene commonly targeted in microbiome research (515F, 806R) [62] was extracted from sequences and analysed as described above (amplicon sequences were predicted for 89 genomes and included in this analysis, substitution model JC). rpoB gene sequences were also extracted from Prokka annotation files and aligned with muscle, followed by generation of a maximum-likelihood phylogeny with iq-tree (substitution model TIM2+F+I+G4). To identify fragments of the rpoB gene that would be a good target for clade differentiation, the rpoB gene was split into 300 bp fragments and processed with Phylomark v1.6 [63] in order to find the fragment that best separated the clades; a phylogeny was inferred from the resulting alignment with FastTree2 v2.1.10 [64].
In silico PCR
Primers and probes associated with different targets (Table S3) were screened against the reference genome set (n=101) with usearch (v11.0.667_i86linux32) [60] using the search_pcr and search_oligodb commands with default settings. The primers screened include those that target the signature previously determined to be unique to C. difficile [30], two sets of primers targeting four toxins [65, 66] and a primer set that we developed for more inclusive tcdB detection (Table S3).
Results
Sample collection, isolation and genome sequencing
A total of 260 samples were collected from soil and water around Flagstaff (AZ, USA) (Fig. S1, Table S1), in an effort targeted towards C. difficile surveillance. Positive results for a Taqman PCR assay targeting C. difficile [30] were observed for approximately 50 % of the 260 environmental samples, and isolates from these samples were whole-genome sequenced. Twenty-one of these sequenced isolates either had a multilocus sequence type (ST) associated with cryptic Clostridioides species or had a novel ST and were included in this study. The 21 isolates originated from 19 samples, as 2 samples included isolates with two multilocus STs associated with cryptic clades. Additional studies focusing on C. difficile isolated from the Flagstaff sampling effort are in progress. Slovenian isolates with STs associated with cryptic clades were collected from soil (n=14), puddle water (n=1) [4] and dandelion (n=2) [29], resulting in 17 genomes.
Sequenced genome clade designation
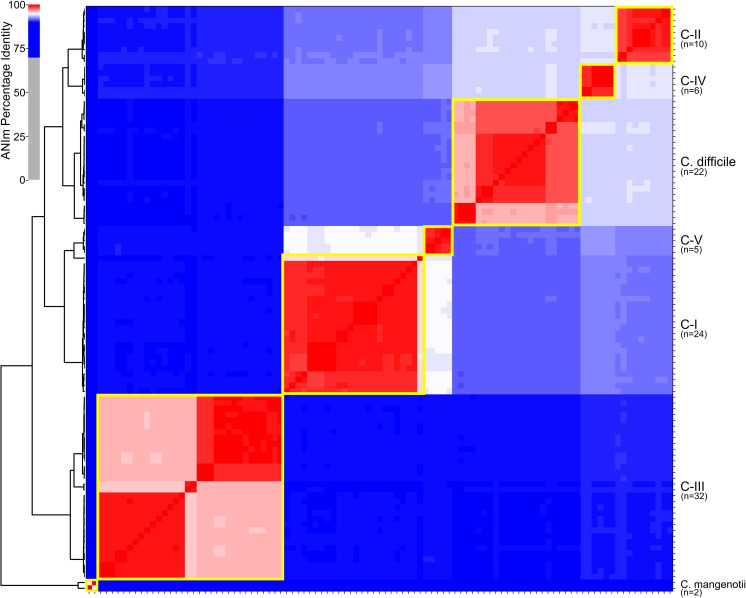
Following sequencing and genome assembly, an initial screen was performed using mash to verify assignment of genomes into cryptic clades. After this initial screen, a set of 101 genomes that included all cryptic species genomes and representative genomes from C. difficile were analysed with pyani (Table S4). The results demonstrate the presence of five defined clades (C-I–C-V, within each clade ANI values are >95 %) that are distinct from C. difficile (Fig. 1). The pairwise pyani distances (Table S4) between clades demonstrates that each clade represents a separate species based on published thresholds [39], confirming recently published results [7]. Isolate genomes from Flagstaff fall within two of these novel clades, including one that has not been described previously (clade C-IV). One of these novel clades (C-III) primarily includes genomes from environmental isolates collected in rural eastern Slovenia [4] and Flagstaff (AZ, USA). Genomes from Slovenia isolates fall into clades C-I, C-II and C-III.
Fig. 1.
Heat map and clustering of genomes based upon pairwise ANI values (ANIm value computed with pyani [37]). Yellow boxes indicate species/lineage boundaries of 95 % ANI.
Core-genome phylogeny and toxin screen
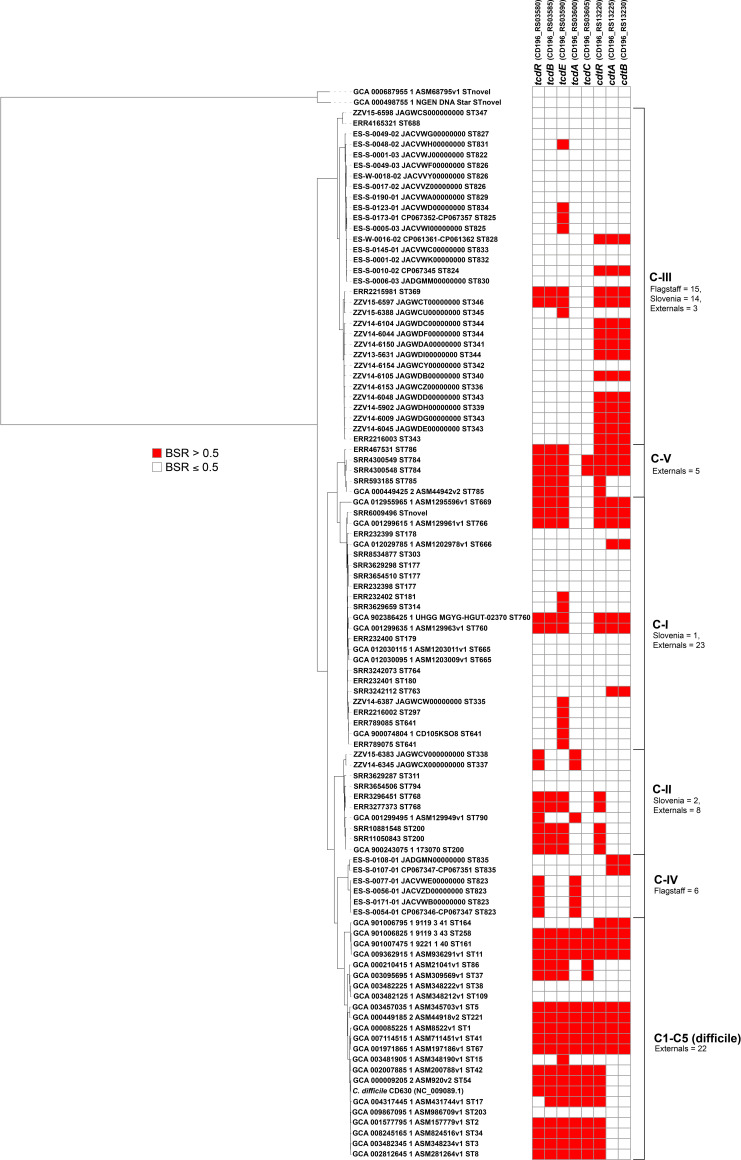
A core-genome phylogeny rooted with Clostridium. mangenotii (Fig. 2) demonstrated a similar topology to the pyani distance cluster. C. difficile toxin sequences were screened against the reference genome set and the presence/absence of each toxin (based on BSR value cut-off of 0.5, see Methods) was correlated with each genome (Fig. 2). The results demonstrate that there are homologues to tcdB in clades C-I, C-II, C-III and C-V. There were also homologues to tcdA in clades C-II and C-IV. There were homologues to binary toxins (cdtA, cdtB) in all cryptic clades except for C-II.
Fig. 2.
A core-genome, maximum-likelihood SNP phylogeny of 101 reference genomes rooted with C. mangenotii (top of phylogeny) demonstrating a similar topology to the clustering of genomes with ANI values. The presence or absence of C. difficile toxins (tcdR, tcdB, tcdE, tcdA, tcdC, cdtR, cdtA, cdtB) is displayed in the heat map to the right of the phylogeny. Toxin presence or absence was determined with a BSR cut-off value of 0.5 (see Methods). The numbers of genomes from Flagstaff (this study), Slovenia (this study) or downloaded from the NCBI (labelled as external) in each species/lineage are listed next to the heat map.
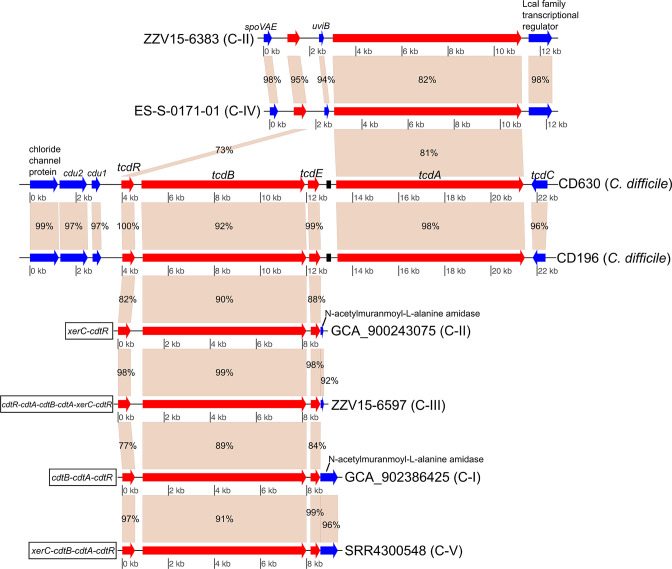
For cryptic clade genome assemblies in which homologues to tcdB were observed (groups C-I, C-II, C-III and C-V), no homologue to tcdA was identified. However, homologous flanking genes were observed in many of the cryptic species genome assemblies containing tcdB homologues (Fig. 3), suggesting common integration sites not associated with the traditional C. difficile PaLoc. Genes associated with the binary toxin locus were present adjacent to the tcdB gene cluster, which has been described elsewhere [7, 67]. In cryptic clade genomes with homologues to tcdA (C-II and C-IV), the tcdA genes were also not part of the traditional PaLoc gene cluster. These novel tcdA clusters contained similar proteins in the same gene order despite the isolates originating from regions separated by large geographical distances. One of these isolates was collected from the USA and the other isolate was collected from Slovenia; another C-II genome (accession no. GCF_001299495.1) from isolate RA09-70, collected in Paris, France, had the identical gene structure to these two genomes. In these clusters, tcdR was upstream of the tcdA homologue, although another coding region (uviB) was found directly adjacent to the tcdA homologue.
Fig. 3.
Gene map of tcdB and tcdA regions in genomes from various species/lineages containing C. difficile toxin homologues. Toxin gene homologues (tcdR, tcdB, tcdE, tcdA) are displayed in red, and flanking genes are displayed in blue. tcdB homologues were observed in genomes in groups C-I, C-II, C-III and C-V, and tcdA homologues were observed in genomes in groups C-II and C-IV. No cryptic lineage genomes contained both tcdB and tcdA homologues. The figure was generated with genoPlotR [54]. Numbers on genes indicate pairwise blast identities.
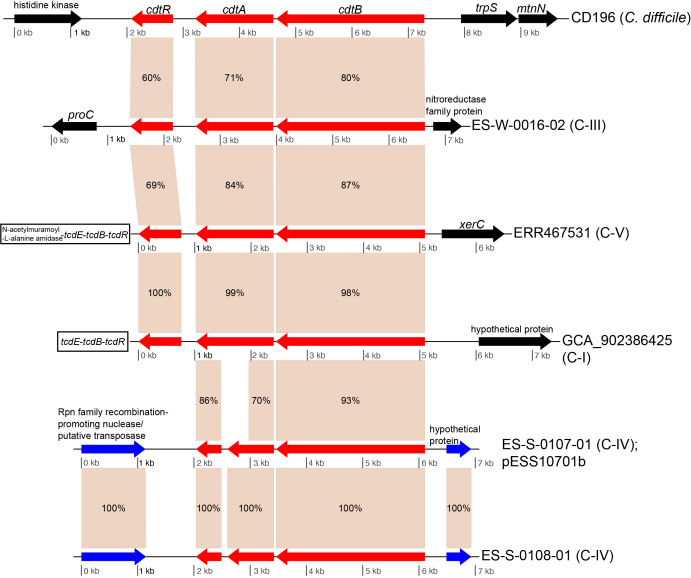
For the binary toxin cluster, CdtLoc, homologous genes were observed across isolates from clades C-I, C-III, C-IV and C-V (Fig. 4). Two of the genomes in clade C-IV contained homologues for cdtA and cdtB, but were missing cdtR. An investigation of the gene structure of these genomes demonstrated that the cdtA homologue was split between two predicted coding regions. The entire locus was identified on a plasmid in isolate ES-S-0107-01, suggesting mobile transport and explaining the lack of the locus in other genomes from the clade. ES-S-108-01 was isolated from a sample taken on the same day as ES-S-0107-01, but from a different location, and demonstrated the same gene structure as ES-S-0107-01. Although we only have a draft genome assembly for isolate ES-S-0108-01, the similarity in gene content to ES-S-107-01, as well as the coverage of the pESS10701b plasmid (Fig. S2), suggests that the binary toxin homologues also are found on an extrachromosomal element in this isolate.
Fig. 4.
Gene map of binary toxin gene regions in genomes from various species. Binary toxin homologues are displayed in red. Binary toxin gene homologues were observed in isolates from clades C-I, C-III, C-IV and C-V. The figure was generated with genoPlotR [54]. Numbers on genes indicate pairwise blast identities. Blue arrows indicate homologous genes outside of the toxin cluster while black boxes indicate genes with no homolog in other gene clusters.
Pan-genomics of novel clades
Panaroo was used to calculate the pan and core genome for each novel clade (Table 1). Unique sequences were identified for most lineages with ls-bsr (Tables 1 and S5), which is consistent with previous work [7]. For each clade, the pan-genome size is much larger than the core-genome size, suggesting that the pan-genomes are being driven by the acquisition of exogenous genes in the environment, which is seen in some other pathogenic bacteria such as Burkholderia pseudomallei [68].
Table 1.
Pan-genomic information for cryptic species genomes
|
Clade |
No. of genomes |
Core genome |
Pan-genome |
Unique regions |
|---|---|---|---|---|
|
C-I |
24 |
3039 |
6369 |
5 |
|
C-II |
10 |
2891 |
5148 |
0 |
|
C-III |
32 |
3014 |
8135 |
53 |
|
C-IV |
6 |
3073 |
4390 |
40 |
|
C-V |
5 |
3626 |
5133 |
27 |
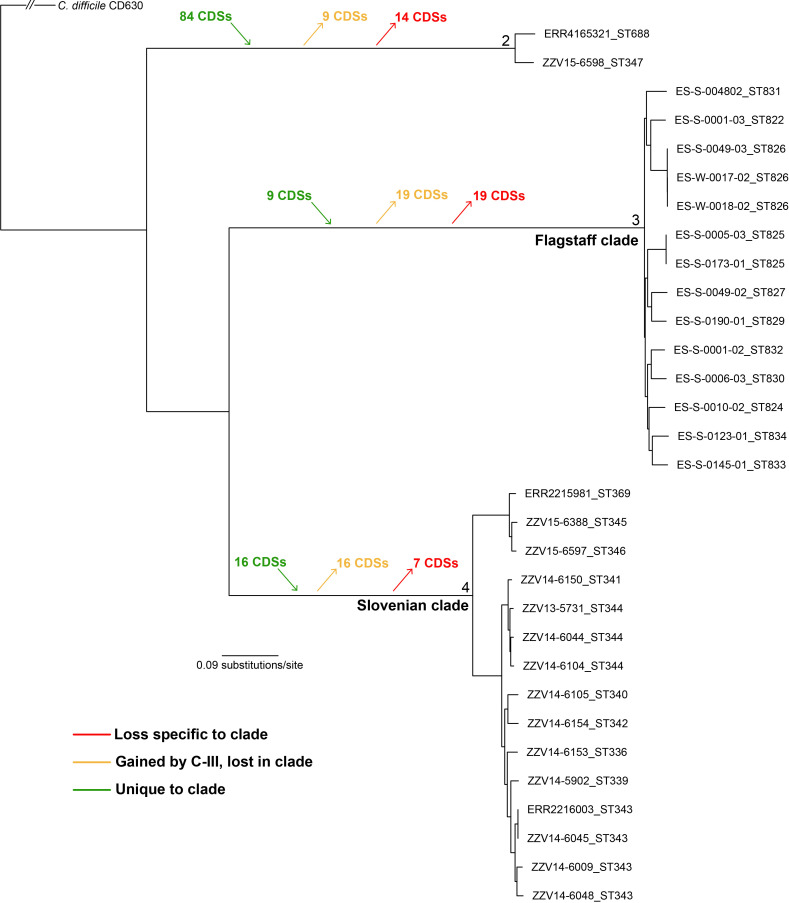
A comparative pan-genomics analysis was also performed between Flagstaff and Slovenian genomes in clade C-III, which fell into two distinct clades (differentiated by thousands of SNPs) in a core-genome SNP phylogeny (Fig. 5). The results demonstrate the presence of 9 unique coding regions to the Flagstaff clade, 16 unique coding regions to the large Slovenian clade, and 84 regions unique to a small clade including a genome from Slovenia and a genome from an isolate collected from septic arthritis (ERR4165321) (Fig. 5, Table S6). Other regions were identified that were highly conserved in Clostridioides genomes, but lost in all C-III genomes, as well as regions that were only conserved in C-III genomes, but were then lost in sub-clades (Fig. 5, Table S6).
Fig. 5.
A core-genome, maximum-likelihood SNP phylogeny of C-III genomes displaying coding regions conserved or lost among lineages. Unique coding regions are present in all three major lineages of C-III, based on an analysis with ls-bsr (see Methods). The phylogeny was rooted by querying SNPs in C. difficile CD630.
Completed representative genomes from novel clades
Finished genomes were generated for five isolates from groups C-III and C-IV. For three of the isolates, plasmids were identified (Table 2). For four of the strains, phage sequences were identified from both Myoviridae and Siphoviridae, based on a cluster dendrogram with previously published and annotated phage (Fig. S3). All assignments of phage or plasmid were based on sequence similarity to these previously characterized elements. Reads from all genomes sequenced in this study were mapped against all extrachromosomal elements and the breadth of coverage was calculated (Fig. S2). The phiSemix9P1 phage (KX905163.1) was present in multiple clade C-III genomes from Slovenia; this phage contains genes from the binary toxin cluster, including cdtR, cdtA and cdtB.
Table 2.
Finished genome assembly information
|
Genome |
Accession no. |
Element type |
Element length (bp) |
|---|---|---|---|
|
ES-W-0016-02 |
Chromosome |
4 189 111 |
|
|
|
Plasmid pESW1602 |
305 867 |
|
|
|
Phage pCD1602_4 |
132 519 |
|
|
ES-S-0010-02 |
Chromosome |
4 259 110 |
|
|
ES-S-0173-01 |
Chromosome |
4 100 889 |
|
|
|
Plasmid pESS17301a |
136 427 |
|
|
|
Plasmid pESS17301c |
47 667 |
|
|
|
Plasmid pESS17301d |
11 693 |
|
|
|
Plasmid pESS17301e |
4119 |
|
|
|
Phage ES-S-0173-01 |
113 019 |
|
|
ES-S-0054-01 |
Chromosome |
3 925 358 |
|
|
|
Phage pCD5401_3 |
34 243 |
|
|
ES-S-0107-01 |
Chromosome |
3 896 990 |
|
|
|
Plasmid pESS10701b |
105 438 |
|
|
|
Plasmid pESS10701c |
64 238 |
|
|
|
Plasmid pESS10701d |
18 539 |
|
|
|
Phage ES-S-0107-01 |
132 924 |
Biochemical characterization
Five Flagstaff isolates were processed with the API 20A kit (bioMérieux) in order to identify enzymatic differences between isolates and to compare the isolates to published results for C. difficile . According to the manufacturer’s documentation, at least some C. difficile isolates have produced positive results for 12 of the included reactions; many of these reactions are not commonly reported as positive for C. difficile (see the manufacturer’s documentation). Positive results were observed for at least one of the tested isolates for all reactions except MLZ (Table 3). Only one isolate, ES-W-0016-02, produced positive results for reactions not reported for C. difficile in the manufacturer’s documentation; for this isolate, positive reactions were observed for LAC, SAC, MAL, ARA and catalase production (CAT). The results indicate that differentiating these cryptic lineages based solely on this assay may be difficult; however, the results also demonstrate that there is variation within and between groups (Table 3), suggesting that these isolates are capable of different metabolic roles.
Table 3.
Biochemical characterization of Flagstaff isolates
Grey highlighting indicates reactions that have been observed to be positive in at least some C. difficile isolates according to the manufacturer.
|
Sample |
Clade |
IND |
URE |
GLU |
MAN |
LAC |
SAC |
MAL |
SAL |
XYL |
ARA |
GEL |
ESC |
GLY |
CEL |
MNE |
MLZ |
RAF |
SOR |
RHA |
TRE |
CAT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
H2O |
na |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
|
ES-S-0010-02 |
C-III |
− |
− |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
− |
+ |
− |
− |
− |
− |
− |
− |
|
ES-W-0016-02 |
C-III |
− |
− |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
− |
+ |
+ |
− |
− |
+ |
+ |
+ |
+ |
|
ES-S-0173-01 |
C-III |
− |
− |
+ |
+ |
− |
− |
− |
+ |
− |
− |
+ |
+ |
− |
− |
+ |
− |
− |
− |
− |
− |
− |
|
ES-S-0054-01 |
C-IV |
− |
− |
+ |
+ |
− |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
+ |
− |
− |
+ |
− |
+ |
− |
|
ES-S-0107-01 |
C-IV |
− |
− |
+ |
+ |
− |
− |
− |
+ |
+ |
− |
− |
+ |
− |
+ |
+ |
− |
− |
− |
− |
+ |
− |
na, Not applicable; IND, indole synthesis; URE, urea; GLU, D-glucose; MAN, D-mannitol; LAC, D-lactose; SAC, D-sucrose; MAL, D-maltose; SAL, salicin; XYL, D-xylose; ARA, L-arabinose; GEL, gelatine; ESC, aesculin hydrolysis; GLY, glycerol; CEL, D-cellobiose; MNE, D-mannose; MLZ, D-melezitose; RAF, D-raffinose; SOR, D-sorbitol; RHA, L-rhamnose; TRE, D-trehalose.
AMR profiles
Four isolates from cryptic groups C-III and C-IV were tested for resistance to 10 antimicrobials associated with induction of CDI (Table 4). The results demonstrate reduced susceptibility or resistance to the tested cephalosporin antibiotics (ceftriaxone, ceftazidime, cefotaxime, cefepime). The tested isolates also show resistance to the fluoroquinolones ciprofloxacin and levofloxacin (Table 4). A screen of genes from the CARD database identified only a single gene (3003835 cdeA) that was observed in cryptic species genomes (BSR values >0.75). A screen for mutations in the gyrA and gyrB genes associated with fluoroquinolone resistance (mutations listed in the CARD database) indicated the presence of three gyrB mutations for the cryptic group isolates (Table S7). While the presence of the cdeA gene and these gyrB mutations may play a role in resistance to fluoroquinolones. the genomic features conferring resistance to antimicrobial drug families are complex and not well understood, and resistance to ciprofloxacin seems to be common among C. difficile with various genomic features [69]. Therefore, the observed resistance to the tested antimicrobials may be due to intrinsic resistance or as yet undescribed acquired genomic features.
Table 4.
AMR profiles (ETESTs) for Flagstaff isolates. Units are in microgram/milliliter (μg/ml)
|
Antimicrobial |
Antimicrobial class |
ES-W-0016-02 (C-III) |
ES-S-0010-02 (C-III) |
ES-S-0173-01 (C-III) |
ES-S-0054-01 (C-IV) |
Replicate |
R breakpoint |
S breakpoint |
Reference |
|---|---|---|---|---|---|---|---|---|---|
|
Ceftriaxone (CRO) |
Cephalosporin |
32 |
48 |
48 |
12 |
Rep1 |
≥64 |
≤16 |
CLSI 2020 |
|
32 |
48 |
48 |
12 |
Rep2 |
|
|
|
||
|
Ceftazidime (CAZ) |
Cephalosporin |
>256 |
>256 |
24 |
32 |
Rep1 |
≥64 |
≤16 |
CLSI 2020 |
|
>256 |
>256 |
24 |
32 |
Rep2 |
|
|
|
||
|
Cefotaxime (CXT) |
Cephalosporin |
>32 |
>32 |
>32 |
>32 |
Rep1 |
≥64 |
≤16 |
CLSI 2020 |
|
>32 |
>32 |
>32 |
>32 |
Rep2 |
|
|
|
||
|
Cefepime (FEP) |
Cephalosporin |
>256 |
>256 |
>256 |
>256 |
Rep1 |
≥16 |
≤2 |
CLSI 2020* |
|
>256 |
>256 |
>256 |
>256 |
Rep2 |
|
|
|
||
|
Clindamycin (CLI) |
Macrolide |
8 |
12 |
2 |
16 |
Rep1 |
≥8 |
≤2 |
CLSI 2020 |
|
8 |
12 |
2 |
16 |
Rep2 |
|
|
|
||
|
Erythromycin (ERY) |
Macrolide |
1 |
1 |
1 |
0.25 |
Rep1 |
≥8 |
≤2 |
EUCAST 2014 |
|
1 |
1 |
1 |
0.25 |
Rep2 |
|
|
|
||
|
Imipenem (IMP) |
Carbapenem |
8 |
6 |
6 |
4 |
Rep1 |
≥16 |
≤4 |
CLSI 2014 |
|
8 |
6 |
6 |
4 |
Rep2 |
|
|
|
||
|
Ciprofloxacin (CIP) |
Fluoroquinolone |
16 |
>32 |
>32 |
>32 |
Rep1 |
≥8 |
≤2 |
CLSI 2020† |
|
16 |
>32 |
>32 |
>32 |
Rep2 |
|
|
|
||
|
Moxifloxacin (MXF) |
Fluoroquinolone |
1.5 |
1.5 |
1.5 |
2 |
Rep1 |
≥8 |
≤2 |
CLSI 2020 |
|
1.5 |
1.5 |
1.5 |
2 |
Rep2 |
|
|
|
||
|
Levofloxacin (LVX) |
Fluoroquinolone |
8 |
8 |
8 |
16 |
Rep1 |
≥2 |
≤0.5 |
CLSI 2020* |
|
8 |
8 |
8 |
16 |
Rep2 |
|
|
|
||
|
Susceptible |
|
||||||||
|
Intermediate |
|
||||||||
|
Resistant |
|
*Breakpoint for Enterobacteriaceae used.
†Breakpoint for moxifloxacin used.
EUCAST, European Committee on Antimicrobial Susceptibility Testing.
16S rRNA and rpoB analysis of Clostridioides
To determine the ability of single marker genes to resolve Clostridioides groups, regions associated with the 16S rRNA gene (Fig. S4) and rpoB (Fig. S5) were extracted from genome assemblies in the reference genome set (n=101). The 16S rRNA gene provided some resolution between Clostridioides clades (Fig. S4a), although the clustering was not fully representative of the core-genome phylogeny. When the 16S rRNA V4 region was extracted from genomes in the reference set, few SNP differences were identified between genomes (Fig. S4b) and most C. difficile and cryptic species genomes were indistinguishable. It should be noted that the quality of 16S rRNA gene sequences in some genome assemblies may be poor, which could impact the performance of this locus at differentiating species. The full-length rpoB gene, which is 3717 bp in length, provided much better separation between groups than the 16S rRNA gene (Fig. S5a). Phylomark identified several 300 bp candidate regions of the rpoB gene that were amenable to Illumina sequencing and provided consistent clade separation (Fig. S5b); the region used to infer the phylogeny in Fig. S5(b) was associated with positions 750 to 1050 in the rpoB gene (accession no. CD630_00660).
In silico PCR
To assess the potential performance of PCR assays for detecting C. difficile and toxin genes, primers were screened against genome assemblies in silico (Table S8). PCR primers for the previously characterized marker unique to C. difficile were predicted to amplify across all C. difficile as well as all cryptic clades, and amplification for C. mangenotii was not predicted. The probe for this assay was not predicted to bind for C-II genomes and one C-V genome (Table S8). This assay detects not only C. difficile , but also closely related cryptic lineages. Assays published previously to detect tcdA, tcdB, cdtA and cdtB returned variable results. An assay designed to amplify tcdA in C. difficile [65] was not predicted to amplify tcdA sequences for some C. difficile and cryptic clade genomes for which ls-bsr results indicate the presence of tcdA homologues. A second assay targeting tcdA [66] was predicted to amplify tcdA sequences in all C. difficile genomes identified as containing tcdA genes plus two C. difficile genomes that ls-bsr did not identify as tcdA-positive (these two genomes contain truncated tcdA sequences); this assay was predicted to amplify tcdA homologues in C-II genomes but not tcdA homologues in C-IV genomes. Several PCR assays targeting tcdB were tested in silico. ls-bsr results indicated 34 reference set genomes contained tcdB sequences (17 C . difficile genomes and 17 cryptic clade genomes). In silico amplification prediction indicated that one assay designed to amplify tcdB in C. difficile [65] may fail to detect some tcdB homologues in cryptic species. Two other assays targeting tcdB ([66], this study) were predicted to provide more broad amplification of tcdB sequences in cryptic clades. Predicted amplification of these two primer sets agree with ls-bsr results except amplification with the primer set generated as part of this study is not predicted for one genome (ERR2215981_ST369, clade C-III) for which ls-bsr results indicate the presence of a tcdB homologue. Also, this primer set is predicted to amplify a partial tcdB gene sequence present in a genome for which ls-bsr results indicate the absence of the gene. It should be noted that predicted in silico PCR amplification and in vitro amplification results may differ in cases where a few nucleotide mismatches are present.
Discussion
C. difficile is an important human pathogen associated with potentially serious disease manifestation. Most C. difficile studies have focused on the presence of the pathogen in the hospital, especially due to concerns of hospital-acquired infection [70] and antibiotic-associated CDI [71]. However, recent work has identified C. difficile spores widely distributed in the environment [4], suggesting a largely understudied reservoir of C. difficile . Because of the oxygenated environments where spores have been found and the strictly anaerobic nature of the pathogen [72], most believe that C. difficile spores represent a dormant state until a new host is found [73]. Additional surveillance for C. difficile in anaerobic environments (e.g. wetlands) may uncover an unknown ecological function besides mammalian infection, although the pathogen is likely adapted to animal hosts.
In this study, we surveyed soil and water in Flagstaff (AZ, USA), to detect C. difficile spores; we combined our results with a study focused on the environmental detection of C. difficile in Slovenia [4]. Following enrichment, a qPCR (quantitative PCR) assay that targets a chromosomal marker thought to be specific to C. difficile was employed for Flagstaff samples; the chromosomal marker was determined to be specific to C. difficile based on the analysis of a smaller number of genomes before the identification of the cryptic species [30]. Slovenian strains were isolated without enrichment using chromID C. difficile selective agar (bioMérieux). Isolates were identified by typical colony morphology, followed by MALDI-TOF identification and further molecular confirmation with 16S rRNA gene sequencing as described previously [4]. Though different isolation and identification methods were used at each location, whole-genome sequencing followed by an analysis based on ANI demonstrated that some genomes from each location were highly divergent from known C. difficile genomes. By including all Clostridioides genomes from GenBank, as well divergent genomes collected from environmental sites in Flagstaff and Slovenia, the identification of five divergent clades were identified that were distinct from C. difficile .
Pairwise ANI values confirmed that three clades (C-I–C-III) represent potentially novel species, based on an ANI threshold of 95 %; this result was recently published [7], although it doesn’t include the full complement of genomes analysed here. In this study, we publish genomes from an additional clade (C-IV), representing a close, extant near-neighbour species of C. difficile . We also demonstrate the presence of an additional novel clade (C-V) that shows pairwise ANI values less than 95 % to the next closest genome, a threshold used previously to delineate bacterial species [39]. Phenotypic information was obtained for isolates from two near-neighbour genomospecies, including information on enzymatic activity and AMR. Observed differences in phenotype, in addition to genomic ANI differences, suggests that these clades in fact represent novel, cryptic species that expand the known genomically characterized diversity of Clostridioides .
In clade C-III, substantial genomic differences were observed between genomes collected from the USA and Slovenia (differentiated by thousands of core-genome SNPs; see Fig. 5). These isolates were identified by a relatively limited sampling effort, suggesting that enormous, untapped diversity of cryptic Clostridioides likely exists in soil and water worldwide. In this study, we identified genomic signatures that are unique to four of the five clades (C-I, C-III, C-IV, C-V), suggesting that targeted assays could be designed to probe for isolates in these clades. However, there may be other Clostridioides isolates in the environment that also contain these signatures, which could further expand the diversity of the genus.
An in silico screen of toxin genes demonstrated that there were homologues of tcdB, tcdA, cdtA and cdtB in many near-neighbour genomes (>0.5 BSR value, see Methods). For clade C-III genomes sequenced in this study, many genomes were positive for binary toxin homologues (Fig. 2). Binary toxins in C. difficile have been associated with increased disease severity [10], and while the role of binary toxins in CDI is still under investigation, binary toxins may impact disease by suppressing the host immune response [74] or increasing adhesion to host cells [75]. The role of the binary toxin homologues in environmental C-III genomes is unknown, but if divergence estimates of this lineage compared to C. difficile are accurate, these genes are potentially millions of years old [7] and have an unknown ecological function. In a C-IV genome, the binary toxin homologues were located on a plasmid, although no homologue to cdtR was identified. Although intact binary toxin genes have previously been identified on a phage [76], this is the first example of binary toxin genes being located on a plasmid. A screen of all reads from this study demonstrated that the phiSemix9P1 phage, which contains a binary toxin gene locus, was present in multiple (n=8) C-III clade genomes from Slovenia, suggesting that the presence of binary toxin genes on mobile elements is much more widespread than previously thought.
Homologues to tcdB were identified in multiple cryptic clades, although they demonstrated substantial amino acid differences to tcdB in C. difficile (Fig. 2). Some clades demonstrated a broad conservation of these genes, whereas other clades demonstrated within-clade diversity, suggesting horizontal acquisition rather than vertical inheritance and divergence. Substantial variability in tcdB and horizontal transfer combined with module rearrangement was also described for previously recognized (clade 1–5) C . difficile strains [15]. Previously, it was suggested that differences in tcdB from cryptic species could result in PCR-based assay failures due to nucleotide mismatches [7]. An in silico screen indicated that published assays potentially amplify some cryptic tcdB and tcdA variants (Table S8); however, the diversity of these toxin genes must be considered when attempting to detect clinical infections with these cryptic species if PCR-based assays are used for pathogen detection. The in silico PCR requires close primer/probe matches, and an amplicon could perhaps still be generated if single nucleotide mismatches were present. For example, the ‘unique’ C. difficile marker used in this study may not be expected to identify some cryptic species using in silico PCR (Table S8) due to probe mismatches, directly conflicting with our in vitro observations.
Clade C-III genomes contain a tcdA homologue that demonstrates significant amino acid differences to tcdA in C. difficile (Fig. 2). Although tcdA+/tcdB− C-III isolates were all from soil, one C-II tcdA+/tcdB− isolate was associated with antibiotic-induced disease [16]. Future work, potentially using animal models of CDI, will determine the disease potential of these strains.
Previously, the 16S rRNA gene has been used as a method for speciation between closely related species within a genus [77]. In this study, we extracted 16S rRNA genes from all Clostridioides genomes and inferred a phylogeny. However, the phylogeny failed to distinguish between Clostridioides genomes (Fig. S4a), suggesting that a different marker is required. When just the V4 region of the 16S rRNA gene was extracted, almost no resolution was identified (Fig. S4b). In an environmental survey based on sequence data from this region, V4 amplicons would erroneously identify the presence of C. difficile if cryptic species were present instead. The rpoB gene has been applied to identify bacterial species [78], and we determined that the rpoB gene did a much better job of species differentiation (Fig. S5a), although the longer gene (>3 kb) would be more difficult to sequence and analyse using either Sanger or Illumina sequencing. We did identify a region of the rpoB gene that is of a reasonable size for amplicon sequencing on the Illumina platform and generally groups the cryptic species into the correct clades, although the deeper branching order is not consistent with the core-genome phylogeny. Primers could target this region for deep sequencing of environmental samples in the search for previously described cryptic species or potentially new and undiscovered clades.
The addition of these genomes provides a background for designing more specific and sensitive C. difficile diagnostics. Previously, we demonstrated how the inclusion of near-neighbour genomes results in an erosion of the diagnostic signature space [79, 80], but is critical for environmental surveillance. Although the ecology of isolates from these cryptic species is largely unknown, an investigation into unique genes may provide insight into the function of these isolates. As the cultures were grown in anaerobic conditions, but were collected from surface soils and water, an assumption is that these spores were dormant in the environment. Additional work looking at oxygen tolerance in isolates from these lineages may identify differences to C. difficile , representing a different evolutionary history and a unique ecological role.
The results of this work significantly expand the genomic landscape of Clostridioides . Although some of the genomes from cryptic species were previously published, this study adds significant genomic diversity to the genus. Divergent strains are frequently associated with atypical variants of all four toxin genes (tcdB, tcdA, cdtA, cdtB), which are inserted in different chromosomal regions than the PaLoc or CdtLoc or are associated with mobile genetic elements (e.g. plasmids). The isolates collected, sequenced and analysed in this study were from just two locations and of limited geographical and temporal diversity. A targeted survey of the environment from multiple states in the USA and multiple countries in Europe and Asia is expected to expand our understanding of these cryptic species, perhaps adding additional context into the evolution and diversification of a globally distributed human pathogen.
Supplementary Data
Funding information
This research was funded in part by the Flinn Foundation (http://www.flinn.org/), grants Flinn 2234 and Flinn 2062. The funders had no role in study design, data collection and analysis, decision to publish nor preparation of the manuscript
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMR, antimicrobial resistance; ANI, average nucleotide identity; ARA, L-arabinose; BSR, BLAST score ratio; CDI, Clostridioides difficile infection; CDS, coding region sequence; CLSI, Clinical and Laboratory Standards Institute; IND, indole synthesis; LAC, D-lactose; MAL, D-maltose; MIC, minimum inhibitory concentration; MLST, multilocus sequence typing; NCBI, National Center for Biotechnology Information; SAC, D-sucrose; SRA, Sequence Read Archive; ST, sequence type.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Eight supplementary tables and five supplementary figures are available with the online version of this article.
References
- 1.Janezic S, Smrke J, Rupnik M. Isolation of Clostridioides difficile from different outdoor sites in the domestic environment. Anaerobe. 2020;62:102183. doi: 10.1016/j.anaerobe.2020.102183. [DOI] [PubMed] [Google Scholar]
- 2.Lim SC, Knight DR, Riley TV. Clostridium difficile and one health. Clin Microbiol Infect. 2020;26:857–863. doi: 10.1016/j.cmi.2019.10.023. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez Diaz C, Seyboldt C, Rupnik M. Non-human C. difficile reservoirs and sources: animals, food, environment. Adv Exp Med Biol. 2018;1050:227–243. doi: 10.1007/978-3-319-72799-8_13. [DOI] [PubMed] [Google Scholar]
- 4.Janezic S, Potocnik M, Zidaric V, Rupnik M. Highly divergent Clostridium difficile strains isolated from the environment. PLoS One. 2016;11:e0167101. doi: 10.1371/journal.pone.0167101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dingle KE, Elliott B, Robinson E, Griffiths D, Eyre DW, et al. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evol. 2014;6:36–52. doi: 10.1093/gbe/evt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bletz S, Janezic S, Harmsen D, Rupnik M, Mellmann A. Defining and evaluating a core genome multilocus sequence typing scheme for genome-wide typing of Clostridium difficile . J Clin Microbiol. 2018;56:e01987-17. doi: 10.1128/JCM.01987-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knight DR, Imwattana K, Kullin B, Guerrero-Araya E, Paredes-Sabja D, et al. Major genetic discontinuity and novel toxigenic species in Clostridioides difficile taxonomy. elife. 2021;10:e64325. doi: 10.7554/eLife.64325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammond GA, Johnson JL. The toxigenic element of Clostridium difficile strain VPI 10463. Microb Pathog. 1995;19:203–213. doi: 10.1016/s0882-4010(95)90263-5. [DOI] [PubMed] [Google Scholar]
- 9.Lyras D, O’Connor JR, Howarth PM, Sambol SP, Carter GP, et al. Toxin B is essential for virulence of Clostridium difficile . Nature. 2009;458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerding DN, Johnson S, Rupnik M, Aktories K. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes. 2014;5:15–27. doi: 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. Clostridium difficile infection. Nat Rev Dis Primers. 2016;2:16020. doi: 10.1038/nrdp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janezic S, Marín M, Martín A, Rupnik M. A new type of toxin A-negative, toxin B-positive Clostridium difficile strain lacking a complete tcdA gene. J Clin Microbiol. 2015;53:692–695. doi: 10.1128/JCM.02211-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuehne SA, Collery MM, Kelly ML, Cartman ST, Cockayne A, et al. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis. 2014;209:83–86. doi: 10.1093/infdis/jit426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, et al. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. mBio. 2015;6:e00551. doi: 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janezic S, Dingle K, Alvin J, Accetto T, Didelot X, et al. Comparative genomics of Clostridioides difficile toxinotypes identifies module-based toxin gene evolution. Microb Genom. 2020;6:000449. doi: 10.1099/mgen.0.000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monot M, Eckert C, Lemire A, Hamiot A, Dubois T, et al. Clostridium difficile: new insights into the evolution of the pathogenicity locus. Sci Rep. 2015;5:15023. doi: 10.1038/srep15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eastwood K, Else P, Charlett A, Wilcox M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol. 2009;47:3211–3217. doi: 10.1128/JCM.01082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gateau C, Couturier J, Coia J, Barbut F. How to: diagnose infection caused by Clostridium difficile . Clin Microbiol Infect. 2018;24:463–468. doi: 10.1016/j.cmi.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Mutters R, Nonnenmacher C, Susin C, Albrecht U, Kropatsch R, et al. Quantitative detection of Clostridium difficile in hospital environmental samples by real-time polymerase chain reaction. J Hosp Infect. 2009;71:43–48. doi: 10.1016/j.jhin.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Lemee L, Dhalluin A, Testelin S, Mattrat M-A, Maillard K, et al. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile . J Clin Microbiol. 2004;42:5710–5714. doi: 10.1128/JCM.42.12.5710-5714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagdasarian N, Rao K, Malani PN. Diagnosis and treatment of Clostridium difficile in adults: a systematic review. JAMA. 2015;313:398–408. doi: 10.1001/jama.2014.17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen VK, Rihn B, Heckel C, Bisseret F, Girardot R, et al. Enzyme immunoassay (ELISA) for detection of Clostridium difficile toxin B in specimens of faeces. J Med Microbiol. 1990;31:251–257. doi: 10.1099/00222615-31-4-251. [DOI] [PubMed] [Google Scholar]
- 23.Kim KH, Fekety R, Batts DH, Brown D, Cudmore M, et al. Isolation of Clostridium difficile from the environment and contacts of patients with antibiotic-associated colitis. J Infect Dis. 1981;143:42–50. doi: 10.1093/infdis/143.1.42. [DOI] [PubMed] [Google Scholar]
- 24.Weese JS, Avery BP, Rousseau J, Reid-Smith RJ. Detection and enumeration of Clostridium difficile spores in retail beef and pork. Appl Environ Microbiol. 2009;75:5009–5011. doi: 10.1128/AEM.00480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dumford DM, 3rd, Nerandzic MM, Eckstein BC, Donskey CJ. What is on that keyboard? Detecting hidden environmental reservoirs of Clostridium difficile during an outbreak associated with North American pulsed-field gel electrophoresis type 1 strains. Am J Infect Control. 2009;37:15–19. doi: 10.1016/j.ajic.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Palacios A, Staempfli HR, Duffield T, Weese JS. Clostridium difficile in retail ground meat, Canada. Emerg Infect Dis. 2007;13:485–487. doi: 10.3201/eid1303.060988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dove CH, Wang SZ, Price SB, Phelps CJ, Lyerly DM, et al. Molecular characterization of the Clostridium difficile toxin A gene. Infect Immun. 1990;58:480–488. doi: 10.1128/iai.58.2.480-488.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barroso LA, Wang SZ, Phelps CJ, Johnson JL, Wilkins TD. Nucleotide sequence of Clostridium difficile toxin B gene. Nucleic Acids Res. 1990;18:4004. doi: 10.1093/nar/18.13.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tkalec V, Janezic S, Skok B, Simonic T, Mesaric S, et al. High Clostridium difficile contamination rates of domestic and imported potatoes compared to some other vegetables in Slovenia. Food Microbiol. 2019;78:194–200. doi: 10.1016/j.fm.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 30.Stone NE, Sidak-Loftis LC, Sahl JW, Vazquez AJ, Wiggins KB, et al. More than 50% of Clostridium difficile isolates from pet dogs in Flagstaff, USA, carry toxigenic genotypes. PLoS One. 2016;11:e0164504. doi: 10.1371/journal.pone.0164504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 34.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta A, Jordan IK, Rishishwar L. stringMLST: a fast k-mer based tool for multilocus sequence typing. Bioinformatics. 2017;33:119–121. doi: 10.1093/bioinformatics/btw586. [DOI] [PubMed] [Google Scholar]
- 36.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 38.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics. 2003;10:10.3.1. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- 41.Sahl JW, Lemmer D, Travis J, Schupp JM, Gillece JD, et al. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb Genom. 2016;2:e000074. doi: 10.1099/mgen.0.000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013:1303.3997 [Google Scholar]
- 45.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bush SJ. Generalizable characteristics of false-positive bacterial variant calls. Microb Genom. 2021;7:000615. doi: 10.1099/mgen.0.000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020;21:180. doi: 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sahl JW, Caporaso JG, Rasko DA, Keim P. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ. 2014;2:e332. doi: 10.7717/peerj.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasko DA, Myers GSA, Ravel J. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics. 2005;6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gertz EM, Yu Y-K, Agarwala R, Schäffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 2006;4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 54.Guy L, Kultima JR, Andersson SGE. genoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2010;26:2334–2335. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amy J, Bulach D, Knight D, Riley T, Johanesen P, et al. Identification of large cryptic plasmids in Clostridioides (Clostridium) difficile . Plasmid. 2018;96–97:25–38. doi: 10.1016/j.plasmid.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 56.Fortier LC. Bacteriophages contribute to shaping Clostridioides (Clostridium) difficile species. Front Microbiol. 2018;9:2033. doi: 10.3389/fmicb.2018.02033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hornung BVH, Kuijper EJ, Smits WK. An in silico survey of Clostridioides difficile extrachromosomal elements. Microb Genom. 2019;5:000296. doi: 10.1099/mgen.0.000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Büchler AC, Rampini SK, Stelling S, Ledergerber B, Peter S, et al. Antibiotic susceptibility of Clostridium difficile is similar worldwide over two decades despite widespread use of broad-spectrum antibiotics: an analysis done at the University Hospital of Zurich. BMC Infect Dis. 2014;14:607. doi: 10.1186/s12879-014-0607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108 (Suppl. 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sahl JW, Matalka MN, Rasko DA. Phylomark, a tool to identify conserved phylogenetic markers from whole-genome alignments. Appl Environ Microbiol. 2012;78:4884–4892. doi: 10.1128/AEM.00929-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Terhes G, Urbán E, Sóki J, Hamid KA, Nagy E. Community-acquired Clostridium difficile diarrhea caused by binary toxin, toxin A, and toxin B gene-positive isolates in Hungary. J Clin Microbiol. 2004;42:4316–4318. doi: 10.1128/JCM.42.9.4316-4318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Persson S, Torpdahl M, Olsen KEP. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect. 2008;14:1057–1064. doi: 10.1111/j.1469-0691.2008.02092.x. [DOI] [PubMed] [Google Scholar]
- 67.Ramírez-Vargas G, López-Ureña D, Badilla A, Orozco-Aguilar J, Murillo T, et al. Novel clade C-I Clostridium difficile strains escape diagnostic tests, differ in pathogenicity potential and carry toxins on extrachromosomal elements. Sci Rep. 2018;8:13951. doi: 10.1038/s41598-018-32390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hall CM, Jaramillo S, Jimenez R, Stone NE, Centner H, et al. Burkholderia pseudomallei, the causative agent of melioidosis, is rare but ecologically established and widely dispersed in the environment in Puerto Rico. PLoS Negl Trop Dis. 2019;13:e0007727. doi: 10.1371/journal.pntd.0007727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang H, Weintraub A, Fang H, Nord CE. Antimicrobial resistance in Clostridium difficile . Int J Antimicrob Agents. 2009;34:516–522. doi: 10.1016/j.ijantimicag.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 70.Forster AJ, Taljaard M, Oake N, Wilson K, Roth V, et al. The effect of hospital-acquired infection with Clostridium difficile on length of stay in hospital. CMAJ. 2012;184:37–42. doi: 10.1503/cmaj.110543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mullish BH, Williams HR. Clostridium difficile infection and antibiotic-associated diarrhoea. Clin Med. 2018;18:237–241. doi: 10.7861/clinmedicine.18-3-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sorg JA, Dineen SS. Laboratory maintenance of Clostridium difficile . Curr Protoc Microbiol. 2009;9:9A.1. doi: 10.1002/9780471729259.mc09a01s12. [DOI] [PubMed] [Google Scholar]
- 73.Edwards AN, McBride SM. Isolating and purifying Clostridium difficile spores. Methods Mol Biol. 2016;1476:117–128. doi: 10.1007/978-1-4939-6361-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cowardin CA, Buonomo EL, Saleh MM, Wilson MG, Burgess SL, et al. The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat Microbiol. 2016;1:16108. doi: 10.1038/nmicrobiol.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aktories K, Papatheodorou P, Schwan C. Binary Clostridium difficile toxin (CDT) – a virulence factor disturbing the cytoskeleton. Anaerobe. 2018;53:21–29. doi: 10.1016/j.anaerobe.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 76.Riedel T, Wittmann J, Bunk B, Schober I, Spröer C, et al. A Clostridioides difficile bacteriophage genome encodes functional binary toxin-associated genes. J Biotechnol. 2017;250:23–28. doi: 10.1016/j.jbiotec.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 77.Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 78.Adékambi T, Drancourt M, Raoult D. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol. 2009;17:37–45. doi: 10.1016/j.tim.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 79.Sahl JW, Vazquez AJ, Hall CM, Busch JD, Tuanyok A, et al. The effects of signal erosion and core genome reduction on the identification of diagnostic markers. mBio. 2016;7:e00846-16. doi: 10.1128/mBio.00846-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Williamson CHD, Wagner DM, Keim P, Sahl JW. Developing inclusivity and exclusivity panels for testing diagnostic and detection tools targeting Burkholderia pseudomallei, the causative agent of melioidosis. J AOAC Int. 2018;101:1920–1926. doi: 10.5740/jaoacint.18-0014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.