Abstract
Vibrio penaeicida (family Vibrionaceae ) is an important bacterial pathogen that affects Japanese shrimp aquaculture. Only two whole-genome sequences of V. penaeicida are publicly available, which has hampered our understanding of the pathogenesis of shrimp vibriosis caused by this bacterium. To gain insight into the genetic features, evolution and pathogenicity of V. penaeicida, we sequenced five V. penaeicida strains (IFO 15640T, IFO 15641, IFO 15642, TUMSAT-OK1 and TUMSAT-OK2) and performed comparative genomic analyses. Virulence factors and mobile genetic elements were detected. Furthermore, average nucleotide identities (ANIs), clusters of orthologous groups and phylogenetic relationships were evaluated. The V. penaeicida genome consists of two circular chromosomes. Chromosome I sizes ranged from 4.1 to 4.3 Mb, the GC content ranged from 43.9 to 44.1 %, and the number of predicted protein-coding sequences (CDSs) ranged from 3620 to 3782. Chromosome II sizes ranged from 2.2 to 2.4 Mb, the GC content ranged from 43.5 to 43.8 %, and the number of predicted CDSs ranged from 1992 to 2273. All strains except IFO 15641 harboured one plasmid, having sizes that ranged from 150 to 285 kb. All five genomes had typical virulence factors, including adherence, anti-phagocytosis, flagella-related proteins and toxins (repeats-in-toxin and thermolabile haemolysin). The genomes also contained factors responsible for iron uptake and the type II, IV and VI secretion systems. The genome of strain TUMSAT-OK2 tended to encode more prophage regions than the other strains, whereas the genome of strain IFO 15640T had the highest number of regions encoding genomic islands. For comparative genome analysis, we used V. penaeicida (strain CAIM 285T) as a reference strain. ANIs between strain CAIM 285T and the five V. penaeicida strains were >95 %, which indicated that these strains belong to the same species. Orthology cluster analysis showed that strains TUMSAT-OK1 and TUMSAT-OK2 had the greatest number of shared gene clusters, followed by strains CAIM 285T and IFO 15640T. These strains were also the most closely related to each other in a phylogenetic analysis. This study presents the first comparative genome analysis of V. penaeicida and these results will be useful for understanding the pathogenesis of this bacterium.
Keywords: Vibrio penaeicida, comparative genomics, genome sequencing, virulence factors, mobile genetic elements
Data Summary
The translated CDSs of 105 (102 Vibrio and three Aliivibrio species) reference genomes were retrieved from the NCBI RefSeq database. The corresponding accession numbers and other details of CDSs are available in Table S1 (available in the online version of this article). Genome assemblies of four Vibrio strains and two plasmid sequences were retrieved from the NCBI database. The corresponding accession numbers and URLs are available in Table S2.
Impact Statement.
Vibrio species are ubiquitous in aquatic ecosystems, causing various infections in fish, crustaceans and shellfish. V. penaeicida is a member of the genus Vibrio that was first isolated from diseased shrimp in 1982 in Yamaguchi and Kumamoto Prefectures, Japan. In this study, we investigated the genomes of five V. penaeicida strains using whole-genome sequencing and comparative genome analysis. Our genomic analysis identified common genomic species features, multiple virulence factors and various mobile genetic elements. Furthermore, novel plasmids were identified and were found to be contained in tetracycline resistance genes. The results of this study provide valuable information to understand the genetic features, pathogenicity, evolution and phylogenetic distinctness of V. penaeicida .
Introduction
The global shrimp trade represents about 18 % of the total world fish trade in terms of value, and the global farmed shrimp market continues to grow faster than that of any other aquaculture species, with most shrimp being produced in Asia [1]. The kuruma shrimp (Marsupenaeus japonicus) is one of the most economically important reared shrimp species in Japan [2]. However, in shrimp aquaculture, microbial infections remain a major problem, with bacterial infections accounting for 20 % of total losses [1].
Vibriosis is a serious bacterial disease in shrimp aquaculture [3]. Vibrio penaeicida was first described in 1995 by Ishimaru et al. as a Gram-negative, facultatively anaerobic, motile by a single polar flagellum, and slightly curved rod-shaped bacterium [4], and was first isolated by Takahashi et al. in 1982 as the causative agent of vibriosis affecting cultured kuruma shrimp in Japan [5]. V. penaeicida has also been responsible for mortality outbreaks of Penaeus stylirostris in New Caledonia [6].
Shrimp with vibriosis are characterized by cloudiness of the abdominal muscle, especially in the sixth segment, and brown spots in the lymphoid organs and gills [5]. The pathogenicity of vibrios is due to a large number of virulence factors, including proteases, haemolysins, siderophores, cytotoxins, quorum sensing (QS), phage, biofilm formation [7–9] and flagella, which are essential for motility [10]. Among the various virulence factors, haemolysin was reported as one of the major virulence factors among Vibrio species, such as the thermolabile haemolysin (TLH) that was detected in V. parahaemolyticus [11] and V. harveyi [12]. QS is a process through which bacteria can communicate by extracellular signalling molecules called autoinducers [13] and gives bacteria the ability to control the secretion of virulence factors. For example, QS is known as the most defined virulence regulatory mechanism in V. harveyi [14] and V. cholera [15]. The type VI secretion system (T6SS), another important virulence factor, has been associated with the pathogenesis of Escherichia coli [16], V. cholerae [17], and acute hepatopancreatic necrosis disease (AHPND)-causing V. parahaemolyticus [18]. Furthermore, type IV pili, a virulence factor related to adherence, play important roles in host–cell interactions, gliding motility, DNA uptake, twitching motility and signal transduction [19].
The genomes of Vibrio species typically possess two chromosomes that are shaped by recombination and horizontal gene transfer (HGT) [20]. HGT has an important role in bacterial pathogenesis by dissemination of genes encoding virulence and antibiotic resistance through mobile genetic elements (MGEs) such as integrating conjugative elements (ICEs), genomic islands (GIs), plasmids and bacteriophages [21]. These MGEs have been identified in vibrios and have been reported to play a significant role in the evolution of Vibrio species [22]. Recently, whole genome sequencing (WGS) approaches towards microbes have provided high-quality sequences to examine virulence-associated genes [23], metabolism, drug resistance, host–pathogen interactions and host–environment reactions [24]. Currently, only two whole-genome sequences of V. penaeicida that were isolated from kuruma shrimp are available in the GenBank database. In addition, only a few studies on the pathogenesis of V. penaeicida in penaeid shrimps are available. Previous studies identified and proposed different V. penaeicida extracellular products as putative virulence factors to shrimp; for example, V. penaeicida strain AM101 secreted an extracellular thermo-labile cytotoxin, which produced 100 % mortality when it was injected into juvenile Litopenaeus stylirostris [25], and a cysteine protease-like exotoxin, which produced high mortality when injected into juvenile Litopenaeus vannamei [26]. Therefore, the present study aimed to characterize the genomes of five V. penaeicida strains using a comparative genomics approach to better understand their pathogenesis.
Methods
Bacterial strains
Strains TUMSAT-OK1 and TUMSAT-OK2 were isolated from two mass mortality events involving M. japonicus in a commercial shrimp farm in Okinawa, in 2019. The shrimp did not show any unambiguously diagnostic clinical signs at the macroscopic level (as is often the case for any shrimp infectious diseases). We therefore sought to isolate possibly pathogenic bacteria from the moribund shrimp by inoculating shrimp stomach contents onto heart infusion agar plates. The colonies grown on the plates were uniform, indicating that the moribund shrimp were infected by a single type of bacterium, which was subsequently identified as V. penaeicida by 16S rRNA gene sequencing. We isolated nine phenotypically indistinguishable isolates, from which TUMSAT-OK1 and TUMSAT-OK2 were randomly selected to represent each mass mortality event and were used in WGS. For comparative genomic analyses, we purchased three historical V. penaeicida strains, IFO 15640T, IFO 15641 and IFO 15642, from the NITE Biological Resource Center (NBRC) (Table 1). The three strains were the only strains available in public culture collections. Starting with glycerol stocks, all V. penaeicida strains were cultured on heart infusion agar plates supplemented with 2.5 % (w/v) NaCl and were incubated at 25 °C overnight. For DNA extraction, a full loopful of grown colonies was selected, cultured in heart infusion broth (Gibco) supplemented with 2.5 % (w/v) NaCl and incubated at 25 °C overnight with shaking.
Table 1.
Genome assembly statistics and annotation information of V. penaeicida strains
|
Strain |
GeneBank accession no. |
Genome Size (bp) |
GC (%) |
no. of coding sequences |
no. of rRNAs |
no. of tRNAs |
Origin or source of strain and year of isolation |
|---|---|---|---|---|---|---|---|
|
IFO 15640T | |||||||
|
Chromosome I |
4,134,604 |
44.12 |
3620 |
31 |
102 |
Kuruma shrimp, Kagoshima Prefecture, purchased from NBRC, 1989 |
|
|
Chromosome II |
2,363,338 |
43.8 |
2076 |
7 |
|||
|
Plasmid |
285,012 |
41.83 |
294 |
||||
|
IFO 15641 | |||||||
|
Chromosome I |
4,300,323 |
44.01 |
3782 |
28 |
98 |
Kuruma shrimp, Yamaguchi Prefecture, purchased from NBRC, 1986 |
|
|
Chromosome II |
2,273,513 |
43.81 |
1992 |
7 |
|||
|
IFO 15642 | |||||||
|
Chromosome I |
4,212,274 |
43.92 |
3708 |
25 |
94 |
Kuruma shrimp, Hiroshima Prefecture, purchased from NBRC |
|
|
Chromosome II |
2,363, 514 |
43.8 |
2071 |
7 |
|||
|
Plasmid |
240,687 |
41.76 |
260 |
||||
|
TUMSAT-OK1 | |||||||
|
Chromosome I |
4,213,929 |
43.94 |
3711 |
31 |
102 |
Kuruma shrimp, stomach, Okinawa Prefecture, 2019 |
|
|
Chromosome II |
2,489,285 |
43.59 |
2270 |
7 |
|||
|
Plasmid |
150,136 |
43.33 |
185 |
||||
|
TUMSAT-OK2 | |||||||
|
Chromosome I |
4,213,527 |
43.94 |
3711 |
31 |
102 |
Kuruma shrimp, stomach, Okinawa Prefecture, 2019 |
|
|
Chromosome II |
2,489,259 |
43.59 |
2273 |
7 |
|||
|
Plasmid |
150,127 |
43.33 |
187 |
||||
Whole-genome sequencing using Illumina and Nanopore sequencing
For Illumina sequencing, we extracted the bacterial genomic DNA using the standard cetyltrimethylammonium bromide method. The DNA paired-end libraries were prepared using a Nextera XT library preparation kit (Illumina), following the manufacturer’s protocol and sequenced using the MiSeq Illumina platform and MiSeq reagent kit v.2 (300 cycles; Illumina).
For Nanopore sequencing, genomic DNA was extracted using NucleoBond AXG100 columns and the NucleoBond buffer set III (Macherey-Nagel). Long-read libraries were prepared with the ligation sequencing kit (SQK-LSK109; Oxford Nanopore Technologies) and sequenced using R9.4.1 flow cells on a GridION platform. The fast5files were base-called using Guppy v.4.0.1 with the settings configdna_r9.4.1_450bps_hac and qscore_filtering.
Genome assembly
We combined Illumina short reads with Nanopore long reads to produce hybrid complete genome assemblies of V. penaeicida genomes. The raw Illumina sequencing data were quality assessed using fastp v.0.20.2 [27] with default settings. The Nanopore reads were de novo assembled using Flye v.2.7 [28] with the settings nano-raw and genome-size 7M. The Illumina reads and Nanopore reads were aligned to the assemblies using minimap2 v.2.17 [29] with the settings ax sr for Illumina reads and ax map-ont for Nanopore reads, and the resulting BAM files were used to improve the accuracy of assemblies by HyPo v.1.0.2 [30] with the settings s 6 m and c 50. Assembly quality was evaluated using Quast v.5.0.2 [31] with default options, and assembly completeness was assessed using Benchmarking Universal Single-Copy Orthologs (BUSCO) v.4.1.4 [32]. The circular topology of the chromosomes and plasmids was confirmed using Bandage v.0.8.1 [33].
Genome annotation
The genome assemblies were annotated on the Rapid Annotations using Subsystems Technology (RAST) server v.2.0 (http://rast.nmpdr.org/) [34–36] and Prokka v.1.13.3 [37, 38] with the settings force, rfam, kingdom bacteria, gram neg, genus Vibrio , usegenus. Prokka predicted the protein-coding sequences (CDSs), tRNAs and rRNAs found in the V. penaeicida genomes and RAST was used for further annotation of all predicted prophages and GIs. We used the virulence factor database (VFDB) for prediction of virulence factors (http://www.mgc.ac.cn/VFs/main.htm) [39]. The antimicrobial resistance (AMR) gene family and resistance mechanisms were identified by the Resistance Gene Identifier (RGI) (https://card.mcmaster.ca/analyze/rgi), which is based on the comprehensive antibiotic resistance database (CARD) [40].
Mobile genetic elements
Putative GIs and prophages were predicted using IslandViewer4 (http://www.pathogenomics.sfu.ca) [41] and PHAge Search Tool Enhanced Release (PHASTER) (https://phaster.ca/) [42]. Furthermore, ICEs were identified by using the ICEfinder web-based tool in ICEberg v.2.0 (https://db-mml.sjtu.edu.cn/ICEfinder/ICEfinder.html) [43].
Comparative whole-genome analysis
Average nucleotide identity (ANI)
ANI between the genomes was measured using the JSpeciesWS server (http://jspecies.ribohost.com/jspeciesws/) [44] based on blast+ (ANIb). We adopted a cutoff value of >95 % to delineate species boundaries.
Phylogenetic tree reconstruction
Two phylogenetic tree analysis methods for V. penaeicida were used. A phylogenetic orthology inference method was used to describe the genetic relatedness between V. penaeicida and other vibrios. We downloaded the translated CDSs of 105 (102 Vibrio and three Aliivibrio species) reference genomes from the NCBI RefSeq database (accessed 9 December 2021; Table S1). The protein sequences were clustered by OrthoFinder2 v.2.5.1 [45], yielding 687 single-copy orthologues unanimously conserved among all species. The protein sequences were aligned by MAFFT v.7.490 [46], and the multiple sequence alignments were used for maximum-likelihood phylogenetic analysis using IQ-TREE2 v.2.1.4-beta [47]. Another whole-genome proteome-based phylogenetic tree was reconstructed to describe the genetic relatedness among V. penaeicida strains using the Type Strain Genome Server (TYGS) (https://tygs.dsmz.de) [48, 49]. The genomes of four Vibrio strains were retrieved from the NCBI database (Table S2) and all sequences were submitted to the TYGS server in fasta format. The phylogenetic tree was built using FastME v.2.1.6.1 [50] from whole proteome-based Genome blast Distance Phylogeny (GBDP), and the tree was rooted at the midpoint [51] and visualized by iTOL v.5 (https://itol.embl.de/) [52].
Prediction of clusters of orthologous groups (COGs)
COGs are clusters of genes in different species that evolve from a common ancestral gene through speciation events [53]. These genes can be used to assess the evolutionary history of given genes from common ancestors. The online program OrthoVenn with default parameters E-value 1e-2 and inflation value 1.5 (https://orthovenn2.bioinfotoolkits.net/task/create) [54] was used to compare and annotate the COGs, and perform Gene Ontology (GO) enrichment analysis for the assembled genomes of the studied Vibrio strains.
Whole-genome comparative visualization
Visualization of genome comparisons was conducted to determine the genotypic differences between Vibrio strains using blast Ring Image Generator (BRIG) [55]
Results and discussion
General genomic characteristics of V. penaeicida strains
In this study, we investigated the genomic features of five V. penaeicida strains isolated from different shrimp farms in Japan to gain more insight into the pathogenesis and evolution of this important shrimp pathogen. We sequenced a total of five V. penaeicida strains using Illumina and Nanopore platforms, yielding chromosome-level hybrid assemblies. The final assemblies of each strain had two circular DNA chromosomes and four strains had one circular plasmid. BUSCO analysis yielded 99.6 % of BUSCO completeness, indicating our genome assemblies were of high quality. Chromosome I (ChrI) sizes ranged from 4.1 to 4.3 Mb, the GC content ranged from 43.9 to 44.1 %, and the number of predicted CDSs ranged from 3620 to 3782. ChrII sizes ranged from 2.2 to 2.4 Mb, the GC content ranged from 43.5 to 43.8 %, and the number of predicted CDSs ranged from 1992 to 2273. This study confirmed that the V. penaeicida genome possessed two circular DNA chromosomes, which is consistent with the results from other Vibrio species [56]; the sizes of both chromosomes were relatively constant among the V. penaeicida genomes. Strain TUMSAT-OK1 had the largest genome, whereas strain IFO 15640T harboured the largest plasmid among the V. penaeicida strains analysed in this study. The majority of recognizable genes for essential cell functions (e.g. DNA replication, RNA metabolism, biosynthetic pathways and membrane transport) and pathogenicity (e.g. iron acquisition, chemotaxis and motility, endotoxin, and immune evasion) are located on ChrI. In contrast, ChrII contains a larger percentage (45–49 %) of hypothetical genes compared with ChrI (30–32 %), as is the case in other vibrios [57, 58].
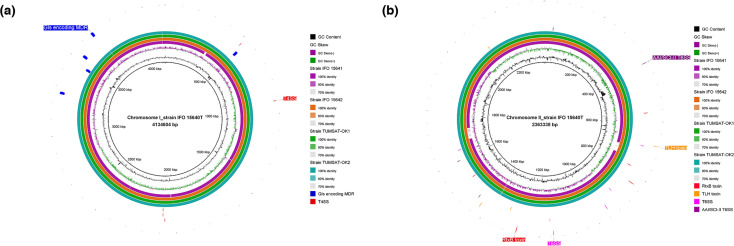
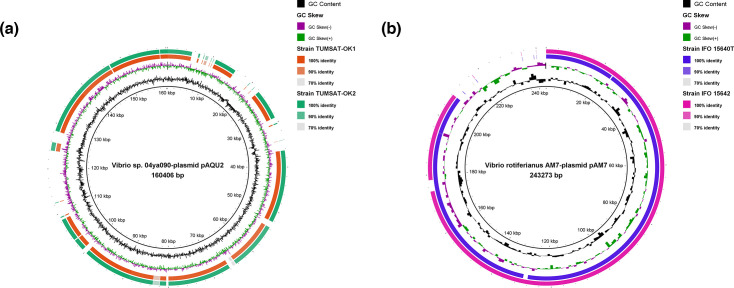
Plasmid sizes ranged from 150, 127 to 285, 012 bp, the GC content ranged from 41.7 to 43.3 %, and the predicted CDSs ranged from 185 to 294. Details of the general genomic features are summarized in Table 1. The plasmid sequences were subjected to NCBI blast searches against the NCBI non-redundant nucleotide database and showed high similarities to Vibrio sp. 04Ya090-plasmid pAQU2 and V. rotiferianus AM7-plasmid pAM7. Comparison of whole genomes and plasmids of the five V. penaeicida strains was visualized using BRIG (Figs. 1 and 2).
Fig. 1.
Genome-wide comparison of five V. penaeicida strains. Chromosome I (a) and chromosome II (b) of strain IFO 15640T used as central references. From inner to outer rings, the first ring indicates the reference genome, the second and third rings are the GC content and GC skew, and the fourth to seventh rings indicate the other V. penaeicida genomes in this study. The clockwise-arrows in the remaining rings indicate the presence, absence and location of virulence factors and genomic islands (GIs) of interest among the five V. penaeicida genomes. The virulence factors include: RtxB, repeats-in-toxin B; TLH, thermolabile haemolysin; T4SS and T6SS, type IV and VI secretion systems; and AAI/SCI-II, T6SS-3 gene cluster in enteroaggregative Escherichia coli . The GIs include GIs encoding MDR (multidrug resistance). The figure was produced using the blast Ring Image Generator (BRIG).
Fig. 2.
Circular comparison of V. penaeicida plasmids mapped against respective reference plasmids. (a) Alignments of plasmids from TUMSAT-OK1 and TUMSAT-OK2 against Vibrio sp. 04Ya090 plasmid pAQU2, and (b) alignments of plasmids from IFO 15640T and IFO 15642 against V. rotiferianus AM7 plasmid pAM7. The different colours refer to the different plasmids, GC skew and GC content, and are listed in the key. The figure was generated using the blast Ring Image Generator (BRIG).
ANI has been considered the best alternative [59, 60] to DNA–DNA hybridization (gold standard) for species delineation at the genomic level [59]. We used ANI to determine the species boundaries and measure the genetic distance between V. penaeicida genomes. ANI values were calculated using the five V. penaeicida strains and the reference strain ( V. penaeicida CAIM 285T). All ANI values were higher than the threshold value of 95 % for bacterial delineation [59], which indicated that these strains belong to the same species. Based on ANI values, strain IFO 15640T (99.99 %) was the most closely related to strain CAIM 285T, followed by strain IFO 15641 (99.91 %) and strain IFO 15642 (99.89 %). With an ANI value of 99.6 %, strains TUMSAT-OK1 and TUMSAT-OK2 were more distantly related to strain CAIM 285T. The absence of certain genomic regions in strain IFO 15641 (Fig. 1), despite its higher ANI than strain IFO 15642, can be explained by the large plasmid missing in strain IFO15641 as well as the chromosomal GIs and/or prophages. Also, strain IFO15641 had a larger ChrI than strain IFO15640T, seemingly inflated by unique GIs.
Virulence factors and AMR profiles
Identification of virulence factors is essential for estimating the pathogenicity of a given bacterium because these factors are directly responsible for pathogenic bacteria infecting and damaging the host [61]. VFDB revealed putative virulence factors in the V. penaeicida genomes, including genes involved in: adherence; anti-phagocytosis; chemotaxis and motility; iron uptake and acquisition; QS; type II, IV, and VI secretion system proteins; and endotoxins and toxins [repeats-in-toxin (RTX) and TLH]. These putative virulence factors are well known in the genus Vibrio [62], and many of them were closely related to those found in other Vibrio species such as Vibrio cholerae O1 biovar El Tor str. N16961, V. cholerae O395, V. parahaemolyticus RIMD 2210633, V. vulnificus CMCP6 and V. vulnificus YJ016. They were also closely related to those found in other bacterial taxa such as Aeromonas , Acinetobacter , Escherichia coli , Pseudomonas , Legionella , Coxiella , Shigella , Mycobacterium , Yersinia , Bacillus and Haemophilus (Table 2).
Table 2.
Potential virulence factor profiles of the five V. penaeicida strains predicted using the virulence factor database (VFDB)
|
Classification |
Virulence factors |
Related genes |
Location |
|---|---|---|---|
|
Adherence |
Accessory colonization factor |
acfB |
Chromosome II |
|
Mannose-sensitive HA |
mshA–N |
Chromosome I |
|
|
Type IV pilus |
pilA, pilB, pilC, pilD |
Chromosome I |
|
|
Flp type IV pili ( Aeromonas )† |
flpC, flpF, flpH |
Chromosome I |
|
|
Tad locus ( Haemophilus )‡ |
tadA |
Chromosome II |
|
|
Hsp60 ( Legionella )§ |
htpB |
Chromosome II |
|
|
Antiphagocytosis |
Capsular polysaccharide |
wbfB, wbfU, wbfY, wbfV/wcvB |
Chromosome I |
|
cpsA, cpsC |
Chromosome II |
||
|
Chemotaxis and motility |
Flagella |
cheA, cheB, cheR, cheV, cheW, cheY, cheZ, filM, flaA, flaC, flaD, flaE, flgA, flgB, flgC, flgD, flgE, flgF–N, flhA, flhB, flhF, flhG, fliA, fliD, fliE, fliF, fliG, fliH, fliL, fliJ, fliL, fliN, fliO, flip, fliQ, fliR, fliS, flrA, flrB, flrc, motA, motB, motX, motY |
Chromosome I |
|
cheA |
Chromosome II |
||
|
Iron uptake |
Enterobactin receptors |
vctA |
Chromosome II |
|
Periplasmic binding protein dependent |
vctC, vctD, vctG, viuC, viuD, viuG, viuP |
Chromosome II |
|
|
Acinetobactin ( Acinetobacter ) |
basG |
Chromosome I |
|
|
Pyochelin ( Pseudomonas ) |
pchB |
Chromosome I |
|
|
Iron/magnesium transport ( Escherichia coli ) |
sitA, sitB, sitC, sitD |
Chromosome II |
|
|
Vibriobactin biosynthesis |
vibA, vibB, vibC, vibE, vibF |
Chromosome II |
|
|
Vibriobactin utilization |
viuA, viuB |
Chromosome II |
|
|
Quorum sensing |
Cholerae autoinducer-1 |
cqsA |
Chromosome II |
|
Cholerae autoinducer-2 |
luxS |
Chromosome I |
|
|
Secretion system |
EPS type II secretion system¶ |
epsC, epsE, epsF, epsG, epsH, epsI, epsJ, epsK, epsL, epsM, epsN, gspD |
Chromosome I |
|
VAS effector protein |
hcp-2, vgrG-2 |
Chromosome II |
|
|
VAS T6SS** |
vasA, vasB, vasC, vasD, vasE, vasF, vasG, vasH, vasJ, vasK |
Chromosome II |
|
|
vasK |
Chromosome I |
||
|
Hcp secretion island1 encoded T6SS ( Pseudomonas )†† |
clpV1 |
Chromosome I |
|
|
T4SS effectors ( Coxiella )‡‡ |
Chromosome I |
||
|
AAI/SCI-II T6SS ( Escherichia coli )§§ |
aaiL |
Chromosome II* |
|
|
T6SS ( Aeromonas ) |
Chromosome II |
||
|
Toxin |
Repeats-in-toxin (RTX) |
rtxB |
Chromosome II |
|
Thermolabile haemolysin (TLH) |
tlh |
Chromosome II |
|
|
Cell surface component |
Trehalose-recycling ABC transporter ( Mycobacterium ) |
sugC |
Chromosome I Chromosome II |
|
Endotoxin |
LOS ( Haemophilus )¶¶ |
kdkA, IgtF, lpxA, lpxD, lpxK, msbA, waaQ |
Chromosome I |
|
Immune evasion |
LPS (glycosylation) (S higella )*** |
gtrB |
Chromosome I |
|
Iron acquisition |
(Bacillibactin) ( Bacillus ) |
entE |
Chromosome I |
|
O-Ag ( Yersinia ) |
wcaG |
Chromosome I |
|
|
Regulation |
Two-component system ( Acinetobacter ) |
bfmR |
Chromosome I |
*Missing from TUMSAT-OK1.
†Flp, fimbrial low-molecular weight protein.
‡Tad, tight adherence.
§Hsp, heat shock protein.
¶Eps, extracellular protein secretion.
**T6SS, type VI secretion system.
††Hcp, haemolysin co-regulated protein.
§§AAI/SCI-II, T6SS-3 gene cluster in enteroaggregative Escherichia coli.
¶¶LOS, lipooligosaccharide.
***LPS, lipopolysaccharide.
The putative virulence factors included virulence-related genes shared by other Vibrio spcies, including the tlh gene, which encodes thermolabile haemolysin toxin with phospholipase activity [63]. The V. penaeicida tlh gene was present on ChrI, and this was also the case in V. cholerae [64] and V. harveyi 345 [65]. tlh is also considered a species-specific marker for V. parahaemolyticus [66, 67]. RTX are toxins with cytolytic activity that are produced by a wide range of pathogenic Gram-negative bacteria and are transported from the cytoplasm to the surface of the cell by transport proteins encoded by rtxB and rtxD genes [68]. In the current study, the rtxB gene was detected in ChrII of V. penaeicida . Similarly, previous studies have shown that the rtxB gene was detected in V. alginolyticus, V. metschnikovii, V. anguillarum and V. fujianensis [69]. Moreover, The V. penaeicida genomes harboured the pilA, B, C and D genes encoding the components of the type IV pilus, which mediates bacterial motility on solid surfaces, host–cell adhesion, bacteriophage adsorption, microcolony formation and transformation [70]. This result is in accordance with that of a recent study, which identified a type IV pilus in the genome of V. parahaemolyticus causing disease in Penaeus vannamei [71]. QS gives bacteria the ability to control several critical processes, including virulence factor secretion, antibiotic production, biofilm formation, motility, bioluminescence, development of genetic competence and sporulation [72]. In this study, QS-related genes, such as CqsA and LuxS, genes of cholera autoinducer 1 and 2, respectively, were distributed in both chromosomes of the V. penaeicida genome. CqsA and LuxS genes have also been recently reported in V. parahaemolyticus associated with disease outbreak in shrimp [73]. Extracellular protein secretion (EPS) type II secretion system is mediated by the genes epsC–epsN, which were reported to be essential for secretion of the cholera toxin (the main virulence factor of V. cholerae ) [74], and were located on ChrI of the V. penaeicida . Furthermore, other secretion systems, including types IV and VI (T4SS and T6SS), were also predicted. The T4SS and T6SS are capable of delivering the virulence factors into adjacent eukaryotic cells, but only T6SSs have been reported to inject lethal toxins into prokaryotic cells [75]. The T4SS and T6SS components were detected on ChrI and ChrII, respectively. T6SS-3 gene cluster (also called aai or sci-2), which has been identified in the genome of enteroaggregative Escherichia coli (strain 17-2) [76, 77], was found in the genomes of four V. penaeicida strains. In summary, our results demonstrate that V. penaeicida shares many of the virulence factors with other Vibrio species, although the possibility remains that as-yet-unknown virulence factors exist, which may be annotated as hypothetical proteins in our current genome annotation scheme.
Tetracycline resistance is a common phenotypic characteristic of Vibrio species that has been reported in environmental samples and Asian aquaculture sectors [78–80]. CARD analysis identified genes that mediate tetracycline resistance, including tetB and tetR on the plasmids harboured by strains IFO 15640Tand IFO 15642. Similarly, a previous study identified the tetB gene-encoded plasmid in V. parahaemolyticus strains associated with AHPND from shrimp [81].
Identification of MGEs
The PHASTER web server was used to identify and annotate putative prophage sequences in the V. penaeicida genomes. The predicted prophages are summarized in Table 3 and the detailed features are listed in Table S3. The putative prophages PHAGE_Bacill_vB_BtS_BMBtp14 and PHAGE_Salmon_118970_sal3 were only found in TUMSAT-OK2, whereas PHAGE_Vibrio_12B12, PHAGE_Entero_mEp235 and PHAGE_Escher_ArgO145 were found in all five strains. We detected a total of seven intact prophage sequences in the V. penaeicida chromosomes; PHAGE_Vibrio_12B12 belongs to the family Myoviridae, whereas PHAGE_Escher_ArgO145 and PHAGE_Shigel_POCJ13 belong to the family Podoviridae as per the Virus-Host Database [82]. In other studies, prophages belonging to the family Podoviridae were detected in V. parahaemolyticus [83] and Vibrio alginolyticus [84]. The majority of CDSs of the predicted prophages encoded unknown hypothetical proteins and it is unclear whether the prophages contribute to the virulence of this bacterium.
Table 3.
Predicted prophages in the genomes of the V. penaeicida strains
+, One copy existed; ++, two copies existed; –, absent.
|
Predicted prophage |
IFO 15640T |
IFO 15641 |
IFO 15642 |
TUMSAT-OK1 |
TUMSAT-OK2 |
|---|---|---|---|---|---|
|
PHAGE_Vibrio_12B12 |
+ |
++ |
+ |
++ |
++ |
|
PHAGE_Sulfit_pCB2047_A |
+ |
− |
− |
+ |
− |
|
PHAGE_Entero_mEp235 |
+ |
+ |
+ |
+ |
+ |
|
PHAGE_Escher_ArgO145 |
+ |
+ |
+ |
+ |
++ |
|
PHAGE_Sulfit_pCB2047_C |
− |
+ |
+ |
− |
+ |
|
PHAGE_Pseudo_phi2 |
− |
+ |
− |
+ |
+ |
|
PHAGE_Escher_TL_2011b |
− |
− |
+ |
+ |
− |
|
PHAGE_Shigel_SfIV |
− |
− |
− |
+ |
+ |
|
PHAGE_Shigel_POCJ13 |
− |
− |
− |
+ |
− |
|
PHAGE_Bacill_vB_BtS_BMBtp14 |
− |
− |
− |
− |
+ |
|
PHAGE_Salmon_118970_sal3 |
− |
− |
− |
− |
+ |
GIs are typically recognized as large segments of genomic DNA that range in size from 10 to 200 kb, and GIs smaller than 10 kb are known as genomic islets [85]. In total, we detected 86 GIs in the V. penaeicida genomes using IslandViewer4. A total of 104 transposase genes were predicted, the majority of which were classified into the IS_3 family. The putative GIs and their features are listed in Table S4. Strain IFO 15640T had the highest number of regions encoding GIs, suggesting that this strain has experienced numerous HGT events mediated by GIs. GIs encoding multidrug resistance were identified in the V. penaeicida genomes. V. penaeicida had another large putative GI that did not contain any virulence factors but contained genes for essential cell functions. In contrast to V. penaeicida, large GIs encoding virulence factors have been found in V. vulnificus and V. parahaemolyticus [86].
ICEs are one of the MGEs that can integrate into the host chromosome and shape the behaviour of bacterial communities [87]. ICE-finder predicted one putative T4SS-type ICE of 86,243 bp in ChrI of strain IFO 15641, indicating this ICE may facilitate transfer of the T4SS, which is involved in the pathogenicity of most Gram-negative bacteria [88]. Moreover, three ICEs were detected in the plasmids of strains IFO 15642, TUMSAT-OK1 and TUMSAT-OK2, which harboured essential genes for the ability of plasmids to perform conjugative transfer [e.g. transfer (tra), T4SS, type IV coupling protein and relaxase genes] [89]. This result indicated that these plasmids are conjugative. A T4SS-mediated ICE was also detected on ChrI of strain IFO 15641. Our data indicated that various MGEs, such as plasmids, prophages, GIs and T4SS-type ICE, were detected in the five V. penaeicida genomes; this suggests strongly that V. penaeicida is capable of acquiring new traits, including virulence and antibiotic resistance genes via HGT by these MGEs which have a strong influence on the genomic evolution of vibrios [22].
Phylogenetic analysis
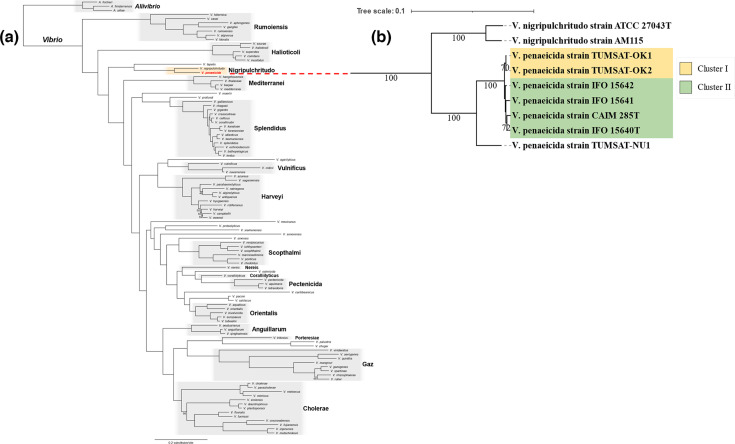
Two phylogenetic trees were reconstructed to identify the species closely related to V. penaeicida and to describe the genetic relatedness among V. penaeicida strains. Phylogenetic analysis (Fig. 3a) was based on sequences coding for 687 single-copy conserved genes found in the 105 species (102 Vibrio and three Aliivibrio species), which was reconstructed using the maximum-likelihood method. The resulting phylogenetic tree showed that the 102 Vibrio species were divided into 16 clades and V. penaeicida belongs to the clade of Nigripulchritudo. This clade was located close to the clades Halioticoli and Mediterranei, but distant from the clades Rumoiensis, Splendidus, Vulnificus and Harveyi.
Fig. 3.
Phylogenetic tree analysis. (a) Phylogenetic tree of 102 Vibrio species, and three Aliivibrio species used as the out-group. A total of 687 single-copy conserved genes (237 491 amino acids) were used to build a maximum-likelihood phylogenetic tree by IQ-TREE v2.1.4-beta (substitution model: LG+F+I+G4 [91], 1000 UFBoot [92] replicates). The UFBoot support value was 100 % unless indicated beside the corresponding node. Species clades within the genus Vibrio were defined based on Sawabe et al. [93], with minor modifications to reconcile paraphyly. (b) Phylogenetic tree of V. penaeicida strains based on whole-genome proteome data and two strains of V. nigripulchritudo used as the out-group. Numbers at each node indicate GBDP pseudo-bootstrap support values from 100 replications.
Additional phylogenetic analysis (Fig. 3b) was conducted based on whole-genome proteome data of V. penaeicida genomes. The resulting tree split the V. penaeicida strains into two clusters: cluster I consisted of strains TUMSAT-OK1 and TUMSAT-OK2, while cluster II consisted of strains IFO 15640T, IFO 15641, IFO 15642 and CAIM 285T. Similar to the ANI results, CAIM 285T and IFO 15640T were the closest relatives, whereas strains TUMSAT-OK1 and TUMSAT-OK2 were most closely related to each other and most distant from the other strains. Our results showed that ANI is an effective tool for accurately measuring the genetic distance between genomes. TUMSAT-NU1 was distantly related to all other V. penaeicida strains with substantial sequence divergence, suggesting that there may be unexplored divergent V. penaeicida strains circulating in aquaculture environments. Therefore, characterization of the genetic diversity will be important for tracking the emergence of novel pathogenic strains.
Orthologous gene analysis
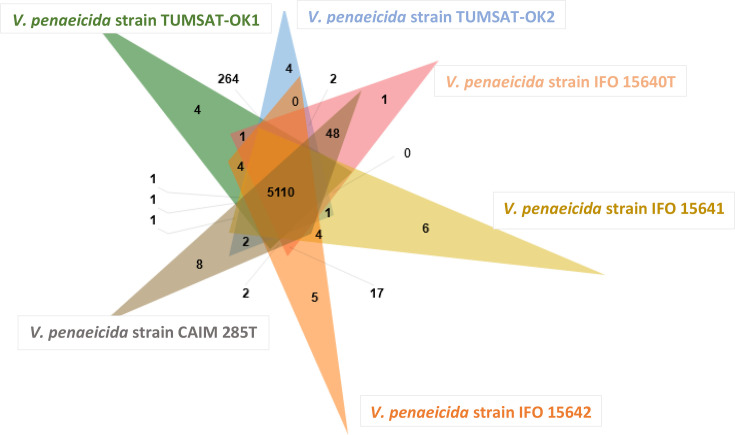
Orthologous genes usually retain functions similar to those of their ancestral genes [90]. COG analysis identified a total of 5110 orthologous clusters that were shared among the six V. penaeicida strains (Table S5). Strains TUMSAT-OK1 and TUMSAT-OK2 had the highest number of shared gene clusters (n=264) (Table S6), followed by strains CAIM 285T and IFO 15640T (n=48) (Table S7), reflecting their close relationship to each other in the phylogenetic analysis, compared with the other V. penaeicida strains. A total of 28 strain-specific clusters were found among these strains. V. penaeicida strain CAIM 285T had the highest number of unique gene clusters (n=8), of which one gene cluster was annotated as transketolase activity GO:0004802, whereas V. penaeicida IFO 15640T had the lowest number of unique gene clusters (n=1), which was annotated as transposition GO:0032196 (Fig. 4). GO enrichment analysis revealed that the following GO terms were enriched among the shared gene clusters: DNA restriction-modification system GO:0009307, transposition GO:0032196, conjugation GO:0000746, nitrate assimilation GO:0042128 and transposition, DNA-mediated GO:0006313.
Fig. 4.
Venn diagram showing the distribution of shared and unique orthologous gene clusters among six V. penaeicida strains as visualized by OrthoVenn2. A total of 5110 shared clusters of orthologous groups were identified in these six strains.
In summary, this study presents a comparative genomic analysis of five V. penaeicida strains and identified common features of these strains. The information regarding virulence factors (adherence, anti-phagocytosis, motility, iron uptake and acquisition, QS, secretion systems, and toxins), which was obtained via the VFDB, will enhance our understanding of V. penaeicida pathogenesis and will guide vaccine development. In addition, various MGEs, such as plasmids, prophages, GIs and a T4SS-type ICE, were identified, indicating that this bacterium is capable of acquiring new genetic information through HGT, which has a significant role in bacterial evolution and affects pathogenesis. Our study provides valuable information for understanding the genetic features, pathogenicity, phylogeny and evolution of V. penaeicida .
Supplementary Data
Funding information
This work was supported in part by the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT scholarship).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMR, antimicrobial resistance; ANI, average nucleotide identity; bp, base pair; BRIG, BLAST Ring Image Generator; BUSCO, Benchmarking Universal Single-Copy Orthologs; CARD, comprehensive antibiotic resistance database; CDSs, protein-coding sequences; Chr, chromosome; COGs, clusters of orthologous groups; EPS, extracellular protein secretion; Flp, fimbrial low-molecular weight protein; GBDP, Genome BLAST Distance Phylogeny; GI, genomic island; GO, Gene Ontology; Hcp, haemolysin co-regulated protein; HGT, horizontal gene transfer; Hsp, heat shock protein; ICE, integrating conjugative element; LOS, lipooligosaccharide; LPS, lipopolysaccharide; MGE, mobile genetic element; NBRC, NITE Biological Resource Center; PHASTER, PHAge Search Tool Enhanced Release; QS, quorum sensing; RAST, Rapid Annotations using Subsystems Technology; RGI, Resistance Gene Identifier; RTX, repeats-in-toxin; T, type strain; Tad, tight adherence; TLH, thermolabile haemolysin; tra, transfer; T4SS, type IV secretion system; T6SS, type VI secretion system; TYGS, Type Strain Genome Server; VFDB, virulence factor database; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Seven supplementary tables are available with the online version of this article.
References
- 1.FAO Toward sustainability in shrimp industry. 2020. http://www.fao.org/in-action/globefish/market-reports/resource-detail/en/c/1261310/
- 2.Sano T, Fukuda H. Principal microbial diseases of mariculture in Japan. Aquaculture. 1987;67:59–69. doi: 10.1016/0044-8486(87)90008-1. [DOI] [Google Scholar]
- 3.Takahashi Y, Itami T, Maeda M, Kondo M. Bacterial and viral diseases of kuruma shrimp (Penaeus japonicus) in Japan. Fish Pathol. 1998;33:357–364. doi: 10.3147/jsfp.33.357. [DOI] [Google Scholar]
- 4.Ishimaru K, Akagawa-Matsushita M, Muroga K. Vibrio penaeicida sp. nov., a pathogen of kuruma prawns (Penaeus japonicus) Int J Syst Bacteriol. 1995;45:134–138. doi: 10.1099/00207713-45-1-134. [DOI] [Google Scholar]
- 5.Takahashi Y, Shimoyama Y, Momoyama K. Pathogenicity and characteristics of Vibrio sp. isolated from cultured kuruma prawn Penaeus japonicus bate. Nippon Suisan Gakkai Shi. 1985;51:721–730. doi: 10.2331/suisan.51.721. [DOI] [Google Scholar]
- 6.Costa R, Mermoud I, Koblavi S, Morlet B, Haffner P, et al. Isolation and characterization of bacteria associated with a Penaeus stylirostris disease (Syndrome 93) in New Caledonia. Aquaculture. 1998;164:297–309. doi: 10.1016/S0044-8486(98)00195-1. [DOI] [Google Scholar]
- 7.Aguirre-Guzman G, Mejia Ruiz H, Ascencio F. A review of extracellular virulence product of Vibrio species important in diseases of cultivated shrimp. Aquac Research. 2004;35:1395–1404. doi: 10.1111/j.1365-2109.2004.01165.x. [DOI] [Google Scholar]
- 8.Nakhamchik A, Wilde C, Rowe-Magnus DA. Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus . Appl Environ Microbiol. 2008;74:4199–4209. doi: 10.1128/AEM.00176-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rønneseth A, Castillo D, D’Alvise P, Tønnesen Ø, Haugland G, et al. Comparative assessment of Vibrio virulence in marine fish larvae. J Fish Dis. 2017;40:1373–1385. doi: 10.1111/jfd.12612. [DOI] [PubMed] [Google Scholar]
- 10.Thompson FL, Gevers D, Thompson CC, Dawyndt P, Naser S, et al. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol. 2005;71:5107–5115. doi: 10.1128/AEM.71.9.5107-5115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taniguchi H, Hirano H, Kubomura S, Higashi K, Mizuguchi Y. Comparison of the nucleotide sequences of the genes for the thermostable direct hemolysin and the thermolabile hemolysin from Vibrio parahaemolyticus . Microb Pathog. 1986;1:425–432. doi: 10.1016/0882-4010(86)90004-5. [DOI] [PubMed] [Google Scholar]
- 12.Zhang XH, Meaden PG, Austin B. Duplication of hemolysin genes in a virulent isolate of Vibrio harveyi . Appl Environ Microbiol. 2001;67:3161–3167. doi: 10.1128/AEM.67.7.3161-3167.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee S, Bassler BL. Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol. 2019;17:371–382. doi: 10.1038/s41579-019-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Defoirdt T, Boon N, Sorgeloos P, Verstraete W, Bossier P. Quorum sensing and quorum quenching in Vibrio harveyi: lessons learned from in vivo work. ISME J. 2008;2:19–26. doi: 10.1038/ismej.2007.92. [DOI] [PubMed] [Google Scholar]
- 15.Zheng J, Shin OS, Cameron DE, Mekalanos JJ. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae . Proc Natl Acad Sci U S A. 2010;107:21128–21133. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navarro-Garcia F, Ruiz-Perez F, Cataldi Á, Larzábal M. Type VI secretion system in pathogenic Escherichia coli: structure, role in virulence, and acquisition. Front Microbiol. 2019;10:1965. doi: 10.3389/fmicb.2019.01965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salomon D, Gonzalez H, Updegraff BL, Orth K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS One. 2013;8:e61086. doi: 10.1371/journal.pone.0061086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craig L, Li J. Type IV pili: paradoxes in form and function. Curr Opin Struct Biol. 2008;18:267–277. doi: 10.1016/j.sbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, et al. Vibrio spp. infections. Nat Rev Dis Primers. 2018;4:8. doi: 10.1038/s41572-018-0005-8. [DOI] [PubMed] [Google Scholar]
- 21.Sobecky PA, Hazen TH. Horizontal gene transfer and mobile genetic elements in marine systems. Methods Mol Biol. 2009;532:435–453. doi: 10.1007/978-1-60327-853-9_25. [DOI] [PubMed] [Google Scholar]
- 22.Hazen TH, Pan L, Gu JD, Sobecky PA. The contribution of mobile genetic elements to the evolution and ecology of Vibrios . FEMS Microbiol Ecol. 2010;74:485–499. doi: 10.1111/j.1574-6941.2010.00937.x. [DOI] [PubMed] [Google Scholar]
- 23.Talagrand-Reboul E, Colston SM, Graf J, Lamy B, Jumas-Bilak E. Comparative and evolutionary genomics of isolates provide insight into the pathoadaptation of Aeromonas . Genome Biol Evol. 2020;12:535–552. doi: 10.1093/gbe/evaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang CY, Yang CH, Lai CH, Huang YH, Lin JN. Comparative genomics of 86 whole-genome sequences in the six species of the Eizabethkingia genus reveals intraspecific and interspecific divergence. Sci Rep. 2019;9:19167. doi: 10.1038/s41598-019-55795-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goarant C, Herlin J, Brizard R, Marteau AL, Martin C, et al. Toxic factors of Vibrio strains pathogenic to shrimp. Dis Aquat Organ. 2000;40:101–107. doi: 10.3354/dao040101. [DOI] [PubMed] [Google Scholar]
- 26.Aguirre-Guzmán G, Ascencio F, Saulnier D. Pathogenicity of Vibrio penaeicida for white shrimp Litopenaeus vannamei: a cysteine protease-like exotoxin as a virulence factor. Dis Aquat Organ. 2005;67:201–207. doi: 10.3354/dao067201. [DOI] [PubMed] [Google Scholar]
- 27.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 29.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kundu R, Casey J, Sung WK. Hypo: super-fast & accurate polisher for long read genome assemblies. bioRxiv. 2019 [Google Scholar]
- 31.Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics. 2018;34:i142–i150. doi: 10.1093/bioinformatics/bty266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seppey M, Manni M, Zdobnov EM. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol Biol. 2019;1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- 33.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:1–15. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–14. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:1–6. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 38.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertelli C, Laird MR, Williams KP, Lau BY, et al. Simon Fraser University Research Computing Group Islandviewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:30–35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu M, Li X, Xie Y, Bi D, Sun J, et al. ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019;47:D660–D665. doi: 10.1093/nar/gky1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henz SR, Huson DH, Auch AF, Nieselt-Struwe K, Schuster SC. Whole-genome prokaryotic phylogeny. Bioinformatics. 2005;21:2329–2335. doi: 10.1093/bioinformatics/bth324. [DOI] [PubMed] [Google Scholar]
- 50.Lefort V, Desper R, Gascuel O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol. 2015;32:2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farris JS. Estimating phylogenetic trees from distance matrices. The American Naturalist. 1972;106:645–668. doi: 10.1086/282802. [DOI] [Google Scholar]
- 52.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. doi: 10.2307/2412448. [DOI] [PubMed] [Google Scholar]
- 54.Xu L, Dong Z, Fang L, Luo Y, Wei Z, et al. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019;47:W52–W58. doi: 10.1093/nar/gkz333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:1–10. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okada K, Iida T, Kita-Tsukamoto K, Honda T. Vibrios commonly possess two chromosomes. J Bacteriol. 2005;187:752–757. doi: 10.1128/JB.187.2.752-757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae . Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naka H, Dias GM, Thompson CC, Dubay C, Thompson FL, et al. Complete genome sequence of the marine fish pathogen Vibrio anguillarum harboring the pJM1 virulence plasmid and genomic comparison with other virulent strains of V. anguillarum and V. ordalii . Infect Immun. 2011;79:2889–2900. doi: 10.1128/IAI.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Figueras MJ, Beaz-Hidalgo R, Hossain MJ, Liles MR. Taxonomic affiliation of new genomes should be verified using average nucleotide identity and multilocus phylogenetic analysis. Genome Announc. 2014;2:00927–14. doi: 10.1128/genomeA.00927-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ina-Salwany MY, Al-Saari N, Mohamad A, Mursidi F-A, Mohd-Aris A, et al. Vibriosis in fish: a review on disease development and prevention. J Aquat Anim Health. 2019;31:3–22. doi: 10.1002/aah.10045. [DOI] [PubMed] [Google Scholar]
- 62.Pérez-Reytor D, Jaña V, Pavez L, Navarrete P, García K. Accessory toxins of Vibrio pathogens and their role in epithelial disruption during infection. Front Microbiol. 2018;9:2248. doi: 10.3389/fmicb.2018.02248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shinoda S, Matsuoka H, Tsuchie T, Miyoshi S, Yamamoto S, et al. Purification and characterization of a lecithin-dependent haemolysin from Escherichia coli transformed by a Vibrio parahaemolyticus gene. J Gen Microbiol. 1991;137:2705–2711. doi: 10.1099/00221287-137-12-2705. [DOI] [PubMed] [Google Scholar]
- 64.Xu M, Wu J, Chen L. Virulence, antimicrobial and heavy metal tolerance, and genetic diversity of Vibrio cholerae recovered from commonly consumed freshwater fish. Environ Sci Pollut Res Int. 2019;26:27338–27352. doi: 10.1007/s11356-019-05287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng Y, Xu H, Su Y, Liu S, Xu L, et al. Horizontal gene transfer contributes to virulence and antibiotic resistance of Vibrio harveyi 345 based on complete genome sequence analysis. BMC Genomics. 2019;20:761. doi: 10.1186/s12864-019-6137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bej AK, Patterson DP, Brasher CW, Vickery MC, Jones DD, et al. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh . J Microbiol Methods. 1999;36:215–225. doi: 10.1016/s0167-7012(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 67.DePaola A, Nordstrom JL, Bowers JC, Wells JG, Cook DW. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl Environ Microbiol. 2003;69:1521–1526. doi: 10.1128/AEM.69.3.1521-1526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lally ET, Hill RB, Kieba IR, Korostoff J. The interaction between RTX toxins and target cells. Trends Microbiol. 1999;7:356–361. doi: 10.1016/s0966-842x(99)01530-9. [DOI] [PubMed] [Google Scholar]
- 69.Håkonsholm F, Lunestad BT, Aguirre Sánchez JR, Martinez-Urtaza J, Marathe NP, et al. Vibrios from the Norwegian marine environment: Characterization of associated antibiotic resistance and virulence genes. Microbiologyopen. 2020;9:e1093. doi: 10.1002/mbo3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, et al. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003;11:1139–1150. doi: 10.1016/s1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 71.Zhang X, Sun J, Chen F, Qi H, Chen L, et al. Phenotypic and genomic characterization of a Vibrio parahaemolyticus strain causing disease in Penaeus vannamei provides insights into its niche adaptation and pathogenic mechanism. Microb Genom. 2021;7:000549. doi: 10.1099/mgen.0.000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rocha-Estrada J, Aceves-Diez AE, Guarneros G, de la Torre M. The RNPP family of quorum-sensing proteins in Gram-positive bacteria. Appl Microbiol Biotechnol. 2010;87:913–923. doi: 10.1007/s00253-010-2651-y. [DOI] [PubMed] [Google Scholar]
- 73.Prithvisagar KS, Krishna Kumar B, Kodama T, Rai P, Iida T, et al. Whole genome analysis unveils genetic diversity and potential virulence determinants in Vibrio parahaemolyticus associated with disease outbreak among cultured Litopenaeus vannamei (Pacific white shrimp) in India. Virulence. 2021;12:1936–1949. doi: 10.1080/21505594.2021.1947448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abendroth J, Kreger AC, Hol WGJ. The dimer formed by the periplasmic domain of EpsL from the Type 2 Secretion System of Vibrio parahaemolyticus . J Struct Biol. 2009;168:313–322. doi: 10.1016/j.jsb.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hood RD, Singh P, Hsu F, Güvener T, Carl MA, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Journet L, Cascales E. The Type VI secretion system in Escherichia coli and related species. EcoSal Plus. 2016;7 doi: 10.1128/ecosalplus.ESP-0009-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brunet YR, Espinosa L, Harchouni S, Mignot T, Cascales E. Imaging type VI secretion-mediated bacterial killing. Cell Rep. 2013;3:36–41. doi: 10.1016/j.celrep.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 78.Wong HC, Liu SH, Wang TK, Lee CL, Chiou CS, et al. Characteristics of Vibrio parahaemolyticus O3:K6 from Asia. Appl Environ Microbiol. 2000;66:3981–3986. doi: 10.1128/AEM.66.9.3981-3986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mandal S, DebMandal M, Pal NK. Plasmid mediated antibiotic resistance of Vibrio cholerae O1 biotype El Tor serotype Ogawa associated with an outbreak in Kolkata, India. Asian Pacific Journal of Tropical Medicine. 2010;3:637–641. doi: 10.1016/S1995-7645(10)60154-X. [DOI] [Google Scholar]
- 80.Yu L, Zhou Y, Wang R, Lou J, Zhang L, et al. Multiple antibiotic resistance of Vibrio cholerae serogroup O139 in China from 1993 to 2009. PLoS One. 2012;7:e38633. doi: 10.1371/journal.pone.0038633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han JE, Mohney LL, Tang KFJ, Pantoja CR, Lightner DV. Plasmid mediated tetracycline resistance of Vibrio parahaemolyticus associated with acute hepatopancreatic necrosis disease (AHPND) in shrimps. Aquaculture Reports. 2015;2:17–21. doi: 10.1016/j.aqrep.2015.04.003. [DOI] [Google Scholar]
- 82.Mihara T, Nishimura Y, Shimizu Y, Nishiyama H, Yoshikawa G, et al. Linking virus genomes with host taxonomy. Viruses. 2016;8:66. doi: 10.3390/v8030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tan CW, Rukayadi Y, Hasan H, Abdul-Mutalib N-A, Jambari NN, et al. Isolation and characterization of six Vibrio parahaemolyticus lytic bacteriophages from seafood samples. Front Microbiol. 2021;12:616548. doi: 10.3389/fmicb.2021.616548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luo P, Yun L, Li Y, Tian Y, Liu Q, et al. Complete genomic sequence of the Vibrio alginolyticus bacteriophage Vp670 and characterization of the lysis-related genes, cwlQ and holA . BMC Genomics. 2018;19:741. doi: 10.1186/s12864-018-5131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 86.Klein S, Pipes S, Lovell CR. Occurrence and significance of pathogenicity and fitness islands in environmental vibrios. AMB Express. 2018;8:177. doi: 10.1186/s13568-018-0704-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wozniak RAF, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 88.Christie PJ, Whitaker N, González-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta. 2014;1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Virolle C, Goldlust K, Djermoun S, Bigot S, Lesterlin C. Plasmid transfer by conjugation in gram-negative bacteria: from the cellular to the community level. Genes (Basel) 2020;11:E1239. doi: 10.3390/genes11111239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sawabe T, Ogura Y, Matsumura Y, Feng G, Amin AR, et al. Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front Microbiol. 2013;4:414. doi: 10.3389/fmicb.2013.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.