Abstract
The general stress response (GSR) in Listeria monocytogenes plays a critical role in the survival of this pathogen in the host gastrointestinal tract. The GSR is regulated by the alternative sigma factor B (σB), whose role in protection against acid stress is well established. Here, we investigated the involvement of the stressosome, a sensory hub, in transducing low pH signals to induce the GSR. Mild acid shock (15 min at pH 5.0) activated σB and conferred protection against a subsequent lethal pH challenge. A mutant strain where the stressosome subunit RsbR1 was solely present retained the ability to induce σB activity at pH 5.0. The role of stressosome phosphorylation in signal transduction was investigated by mutating the putative phosphorylation sites in the core stressosome proteins RsbR1 (rsbR1-T175A, -T209A, -T241A) and RsbS (rsbS-S56A), or the stressosome kinase RsbT (rsbT-N49A). The rsbS S56A and rsbT N49A mutations abolished the response to low pH. The rsbR1-T209A and rsbR1-T241A mutants displayed constitutive σB activity. Mild acid shock upregulates invasion genes inlAB and stimulates epithelial cell invasion, effects that were abolished in mutants with an inactive or overactive stressosome. Overall, the results show that the stressosome is required for acid-induced activation of σB in L. monocytogenes. Furthermore, they show that RsbR1 can function independently of its paralogues and signal transduction requires RsbT-mediated phosphorylation of RsbS on S56 and RsbR1 on T209 but not T175. These insights shed light on the mechanisms of signal transduction that activate the GSR in L. monocytogenes in response to acidic environments, and highlight the role this sensory process in the early stages of the infectious cycle.
Author summary
The stress sensing hub known as the stressosome, found in many bacterial and archaeal lineages, plays a crucial role in both stress tolerance and virulence in the food-borne pathogen Listeria monocytogenes. However, the mechanisms that lead to its activation and the subsequent activation of the general stress response have remained elusive. In this study, we examined the signal transduction mechanisms that operate in the stressosome in response to acid stress. We found that only one of the five putative sensory proteins present in L. monocytogenes, RsbR1, was required for effective transduction of acid tress signals. We further found that phosphorylation of RsbS and RsbR1, mediated by the RsbT kinase, is essential for signal transduction. Failure to phosphorylate RsbS on Serine 56 completely abolished acid sensing by the stressosome, which prevented the development of adaptive acid tolerance. The acid-induced activation of internalin gene expression was also abolished in mutants with defective stressosome signalling, suggesting a role for the stressosome in the invasion of host cells. Together the data provide new insights into the mechanisms that activate the stressosome in response to acid stress and highlight the role this sensory hub plays in virulence.
Introduction
The firmicute Listeria monocytogenes is a stress-tolerant Gram-positive bacterium that causes listeriosis, a food-borne infection that poses a particular risk for immunocompromised individuals and for pregnant women that is often associated with a high mortality rates (ranging from 20 to 30%) [1]. The ability of L. monocytogenes to survive and proliferate in a broad range of harsh environments, including those found in food processing facilities and the human gastrointestinal (GI), tract makes it a particular problem for producers of ready-to-eat foods [2–4]. Following the entry into the host, L. monocytogenes can withstand an array of potentially lethal conditions in the GI tract, such as the extreme acid pH of the stomach, high osmotic pressure and high concentration of bile salts in the duodenum (reviewed in [5,6]. L. monocytogenes employs the alternative sigma factor B (σB) to upregulate a regulon of approximately 300 genes, which collectively confer an array of homeostatic and protective mechanisms that allow survival under severe stress conditions (reviewed in [7,8]. L. monocytogenes relies on σB to enhance survival in the GI tract and to establish infection [9]. A ΔsigB mutant exhibits attenuated virulence [10–12]. This has been partly attributed to the σB-dependent expression of the internalins A and B, encoded by inlA and inlB, respectively. These surface-expressed invasins promote internalization of L. monocytogenes into epithelial cells expressing the E-cadherin and C-Met receptors, respectively [13–15]. Interestingly, exposure of L. monocytogenes to acid shock increases inlA and inlB expression and invasiveness in epithelial cells [14,16,17]. Additionally, loss of sigB is associated with a marked reduction in survival at extreme low pH [18]. The σB regulon is upregulated by mild stress conditions [19–24]. Although important for L. monocytogenes acid tolerance, ΔsigB mutants are still able to develop further resistance towards lethal acid pH when pre-exposed to mild acidic stress (pH 5.0) for one hour [25–27]. This phenomenon is known as the adaptive acid tolerance response (ATR) [28,29]. While it is well established that σB plays a critical role in acid tolerance its role in the ATR is poorly understood. Genes known to be involved in acid tolerance are upregulated in a σB-dependent manner following a mild acid treatment [23,30–32] but the mechanisms of signal transduction involved have not been described yet.
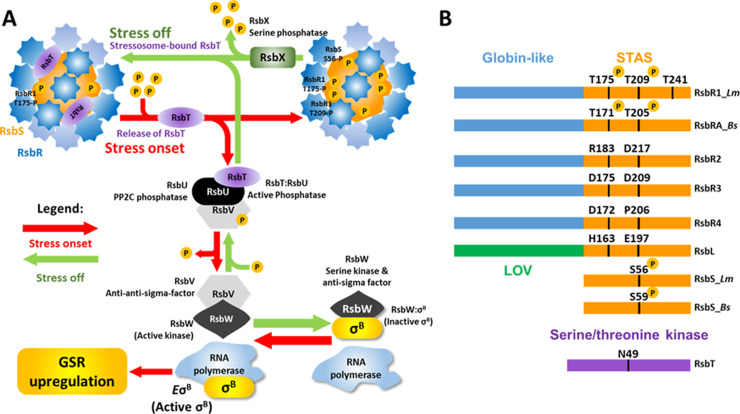
The regulatory pathway of σB activation in L. monocytogenes was initially inferred from Bacillus subtilis due to the high degree of conservation between the sigB operons of these two genera, composed of rsbR, rsbS, rsbT, rsbU, rsbV, rsbW, sigB and rsbX [33,34]. These operons encode components of a signal transduction cascade responsible for triggering the dissociation of σB from RsbW (anti-sigma factor) during the onset of stress, allowing the formation of the holoenzyme RNA polymerase: σB (EσB) and consequently the upregulation of the σB regulon [35, 36] (Fig 1A). At the top of the signal transduction pathway is a high molecular weight protein complex (~1.8 MDa) called the stressosome that serves as a sensory hub, integrating environmental signals into the σB pathway (Fig 1A) [37–39]. Genes encoding this macromolecular complex are found in a phylogenetically diverse range of species in both bacteria and archaea [40]. The stressosome functions as an environmental and nutritional stress sensor in L. monocytogenes [37,41–43]. The L. monocytogenes stressosome is composed of RsbR1, an orthologue of B. subtilis RsbRA, and its paralogues, RsbR2 (Lmo0161), RsbL (Lmo0799), RsbR3 (Lmo1642) and potentially RsbR4 (Lmo1842), although the latter has not yet formally been shown to be part of the stressosome complex. Interestingly, the N-terminal domains of the RsbR paralogues of L. monocytogenes and B. subtilis share little amino acid conservation, suggesting that the sensory roles of these proteins may differ between species. The only exception are the blue light sensors RsbL and the B. subtilis homologue YtvA, which are known to interact with the light-excitable flavin mononucleotide through their Light-Oxygen-Voltage (LOV)-containing N-terminal domains [44,45]. These proteins combine with the core protein RsbS and the serine-threonine kinase RsbT to form a pseudo-icosahedral complex with a 2:1:1 ratio (RsbR1:RsbS:RsbT) [46] (Fig 1A). The current model suggests that the N-terminal domains of RsbR1, which form protruding turrets, are responsible for sensing environmental stresses. Following signal detection it is thought that a structural rearrangement propagates into the stressosome core leading to the activation of kinase RsbT, which phosphorylates RsbR1 at threonine 209 (T209) and RsbS at serine 56 (S56), promoting the release of RsbT from the stressosome core and signal cascade initiation [39,46]. Thus far, this model does not take into account interactions among the RsbR1 paralogues, although a recent study suggests that they may act negatively on the stressosome assembly [42]. It is currently unknown if RsbR1 and its paralogues are able to form heterodimers within the stressosome, although in B. subtilis YtvA dimers (the homologue of RsbL) can form heterotetramers with RsbRA dimers [47]. In the present study, we sought to clarify the role of RsbR1 in acid sensing in a genetic background where the other paralogues were absent, to determine whether signal transduction could be mediated solely by RsbR1.
Fig 1. Model depicting the stressosome-mediated regulation of σB and alignment of stressosome proteins.
(A) The putative sensory proteins RsbR1 and its paralogues RsbR2, RsbR3, RsbL, the scaffold protein RsbS and the serine-threonine kinase RsbT form the stressosome. Environmental/nutritional stress is hypothesized to be sensed by one or more RsbR1 paralogue via its N-terminal non-haem binding globin domains forming the turrets at the stressosome surface (blue). The stressosome core (orange) is composed of the STAS domains of both RsbS and RsbR1 C-termini. Stress triggers T209 phosphorylation in RsbR1 and S56 in RsbS, freeing RsbT (purple) from the stressosome core. The phosphatase RsbU interacts with the free RsbT, activating its phosphatase activity towards RsbV. Once dephosphorylated, the serine kinase and anti-sigma factor RsbW, which has higher affinity with the non-phosphorylated RsbV, releases σB and phosphorylates RsbV once more. σB is then able to interact with the RNApol forming the holoenzyme EσB enabling the upregulation of the general stress regulon. The phosphatase RsbX is responsible for the deactivation of the stressosome by dephosphorylating its residues and enabling the re-sequestration of RsbT and the inactivation of the signal cascade. The model shown incorporates the current available data from studies on both Listeria and Bacillus (see text for citations). (B) Domain organization of L. monocytogenes RsbR1_Lm and its paralogues, RsbS_Lm and RsbT and B. subtilis RsbRA_Bs and RsbS_Bs. The N-terminal domains of RsbR1 and paralogues fold into non-haem binding globin-like domains that form the stressosome turrets, while the blue-light sensor RsbL folds into an LOV domain. The C-termini of RsbR1 and paralogues fold into STAS domains forming the stressosome core along with RsbS. The serine-threonine kinase RsbT responsible for the stressosome phosphorylation requires the ATP-Mg2+ chelating residue N49 for its kinase activity.
In the stressosome core the RsbR1/RsbRA C-terminal domain and RsbS, fold into Sulphate Transporter and Anti-Sigma factor antagonist (STAS) domains [48,49]. Several residues in the STAS domains of B. subtilis RsbRA (T171 and T205) and RsbS (S59) are known to be phosphorylated and they play a critical role in the stressosome activation [50,51]. Similarly, L. monocytogenes RsbR1 also possesses putative phosphorylation sites at positions T175, T209 and T241, and S56 in RsbS (Fig 1B). However, these threonine residues are absent in the remaining paralogues of RsbR1 and so far, only RsbR1-T175 phosphorylation has been confirmed [42]. The RsbT kinase is responsible for phosphorylation of all residues in the RsbR1 and RsbS and the inactivation or deletion of this protein results on the impairment of the σB activation in both L. monocytogenes and B. subtilis [41,42,52–54]. Once the stress has dissipated or the bacterium has adequately responded to it, the stressosome is restored to the ground state by dephosphorylation of the core proteins, through the action of the serine phosphatase RsbX [52,55–58].
In this study, we examined the role of the L. monocytogenes stressosome in sensing mild acid stress and in activating the general stress response and conferring adaptive acid resistance. We constructed an array of mutations, where either the paralogues of RsbR1 were deleted, the kinase activity of RsbT was inactivated, or the putative phosphorylation sites in the stressosome core were substituted with alanine. Our results revealed that RsbR1 has the ability to activate σB in response to mild acid stress when the other four paralogues were absent. The phosphorylation sites in RsbR1 (T175 and T209) modulate the intensity of σB activity in the absence of stress, but have no role in the stressosome’s low pH sensing. Furthermore, the RsbS S56 phosphorylation site and the kinase activity of RsbT are crucial for σB activation at low pH. The extent of σB activation in response to mild acid stress correlates closely with the survival under lethal acidic conditions (pH 2.5). We also show that the stressosome is required for the induction of the internalins inlA and inlB under conditions of low pH stress and for the modulation of L. monocytogenes internalization into epithelial cells. Together, our results point to a central role for the stressosome in acid stress sensing, the development of acid resistance and in the regulation of host cell invasion, through the activation and modulation of σB activity. These findings point to the stressosome as a possible target for new antimicrobials to control this pathogen in food, food-processing environments and in the gastrointestinal tract.
Results
The transcription of lmo0596 is induced by acid in σB-dependent manner
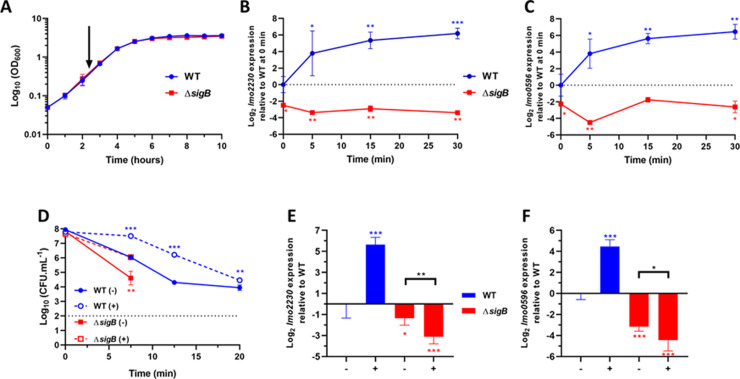
The σB regulon in L. monocytogenes is composed of a large number of genes that are heterogeneously expressed in response to the stress encountered [9]. The expression of the gene lmo2230, which encodes a putative arsenate reductase, is well established as a reliable σB-reporter [24,42,59], however we sought to use an additional σB-reporter gene in this study. The gene lmo0596 was a potential target, as it possesses a σB promoter and had been shown in other studies to belong to the σB regulon [9,60,61]. Although, Lmo0596 has no predicted function, it is homologous to Escherichia coli HdeD, a transmembrane protein that plays a role in acid tolerance [62–65]. In vitro transcription experiments showed that lmo0596 transcription is σB-dependent and PrfA-independent in L. monocytogenes [66], while in vivo studies showed it to be highly σB-dependent [59,60]. In addition, acid treatment at pH 5.0 upregulates lmo0596 within the first 15 min post stress onset [17]. To validate lmo0596 as a σB-reporter, the L. monocytogenes wild type strain EGD-e and an isogenic ΔsigB mutant strain were grown to mid log-phase (OD600 = 0.4) (Fig 2A), and the transcription of lmo0596 and lmo2230 were then measured following acid treatment (pH 5.0). The transcript levels of both lmo0596 and lmo2230 increased after the acid treatment in the wild type strain, plateauing at 15 min, while transcript levels remained unchanged in the ΔsigB strain (Figs 2B and 2C and S1A). The expression of lmo0596 also increases as the pH decreases, reaching its maximum at pH ~5.0–5.5 (S1B Fig). The presence of a putative PrfA box in the regulatory region upstream from lmo0596 prompted us to examine whether PrfA might also influence its transcription. Northern blots used to measure lmo0596 transcription in wild type and ΔprfA backgrounds showed that PrfA does not influence lmo0596 expression under the conditions tested (S1A Fig). Overall, these results showed that lmo0596 transcription was induced by mild acid treatment in a σB-dependent manner, making it suitable to use as a reporter of σB activity.
Fig 2. σB-dependent transcription of lmo2230 and lmo0596 is upregulated rapidly under mild acid stress conditions.
(A) Growth curves obtained from Log10 (OD600) measurements of wild type and ΔsigB strains in BHI at 37°C. The arrow points to OD600 = 0.4, the mid-log phase of L. monocytogenes in BHI. Time-lapse expression measurements of the genes (B) lmo2230 and (C) lmo0596 in wild type and ΔsigB strains grown to OD600 = 0.4 and subsequently treated at a mild pH 5.0 (HCl 5 M) at 0 min. Samples were taken at 0, 5, 15 and 30 min post treatment. Results were converted to Log2 relative to the wild type strain at time 0 min. (D) Mid-log phase cultures untreated at pH 7.0 (-) or treat at pH 5.0 (+) for 15 min and subsequently challenged in acidified BHI (pH 2.5). The dashed line represents the detection threshold. Samples were taken at 0, 7.5, 12.5 and 20 min. Survival data is expressed as Log10 (CFU.mL-1). Gene expression of lmo2230 (E) and lmo0596 (F) in wild type and ΔsigB strains grown to OD600 = 0.4. Samples were taken 15 min after untreated (-) or treated (+) at pH 5.0. Gene expression was obtained from RT-qPCR and converted to Log2 gene expression relative to the wild type strain at time 0 min or untreated. Statistical analysis was performed using a paired Student’s t-test relative to the wild type untreated after 15 min, for RT-qPCR data, and untreated wild type at time 0 min, for acid challenge data (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
σB activation in response to sub-lethal acid stress was measured following exposure of mid log-phase cultures to pH 5.0. To determine whether a sub-lethal acid exposure for 15 min influenced acid resistance, a survival experiment was performed at pH 2.5 to follow the resistance of bacteria that were either unadapted or pre-adapted at pH 5.0 for 15 min. There was a significant increase in acid resistance in the pre-adapted parental strain EGD-e (Fig 2D). As expected, the sigB deletion mutant was highly acid sensitive with no detectable survivors evident by 12.5 min post acid challenge (Fig 2D). At 15 min post adaptation there was a significant increase in the transcription of both lmo2230 and lmo0596 relative to the unadapted cultures (4.47–5.65 log2-fold increase) in the wild type (Fig 2E and 2F). Together these data show that acid-induced σB activity can be measured at the transcriptional level and the role of σB in the acid resistant phenotype is evident under these conditions.
RsbR1 paralogues are dispensable for acid signal transduction and acid adaptation
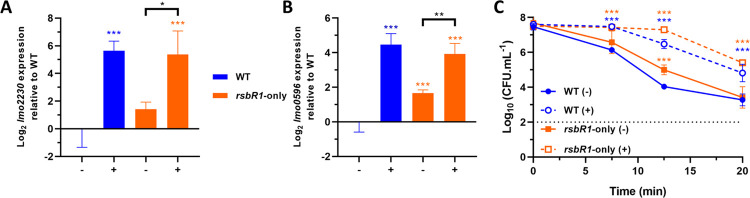
To investigate the role of the putative sensory component of the stressosome RsbR1 in transducing the acid signals into the σB activation pathway, we constructed a mutant strain where each of the three RsbR1 paralogues, RsbR2, RsbR3, and RsbR4 were deleted and the light sensing paralogue RsbL was inactivated by a mutation that produces a light-blind variant (C56A; [67]). The resulting genotype: ΔrsbR2 ΔrsbR3 ΔrsbR4 rsbL-C56A [42], is hereafter designated “rsbR1-only” for simplicity. When this mutant strain was subjected to the same acid exposure described above (pH 5.0), the transcription of the two σB reporter genes, lmo2230 and lmo0596, was found to be strongly induced (Fig 3A and 3B) and there was a significant increase in acid resistance at pH 2.5, similar to that observed in the wild type (Fig 3C). The possibility that residual signal transduction activity was present in the mutated rsbL-C56A gene in the RsbR1-only strain was tested by measuring σB activation by sub-lethal acid exposure in a mutant background where all four paralogues were deleted (ΔrsbR2 ΔrsbR3 ΔrsbR4 ΔrsbL) (Table 1). The behaviour of this strain was essentially identical to the RsbR1-only strain in terms of induced σB activity (S2A and S2B Fig) and acid resistance (S2C Fig). Thus, a strain lacking all known RsbR1 paralogues retains the ability to transduce acid signals via the stressosome and activate σB in response to acid adaptation at pH 5.0.
Fig 3. RsbR1 can act independently of its paralogues to transduce the low pH signals to activate σB and enhance acid tolerance.
Expression of σB-dependent genes (A) lmo2230 and (B) lmo0596, obtained from mid-log phase cultures of wild type and rsbR1-only strains untreated (-) and treated (+) for 15 min in pH 5.0 at 37°C. Gene expression is expressed as Log2 relative gene expression. (C) Acid challenge of mid-log phase cultures treated in pH 5.0 for 15 min at 37°C, as previously described in Fig 2D. Survival data is expressed as Log10 (CFU.mL-1). The rsbR1-only genotype consists on the following mutations, ΔrsbR2; ΔrsbR3; ΔrsbR4; rsbL (C56A). Statistical analysis was performed using a paired Student’s t-test relative to the wild type untreated after 15 min, for RT-qPCR data, and untreated wild type at time 0 min, for acid challenge data (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
Table 1. Plasmids and strains used in this study.
| Plasmid or strain | Reference, Source |
| Plasmids | |
| pMAD; ori pE194ts; PclpB::bgaB; Eryr; Ampr | [68] |
| pEX-K168::rsbS S56A; ori pUC; Kanr | Eurofins Genomics |
| pMAD::rsbR1 T175A; Eryr; Ampr | [42] |
| pMAD::rsbR1 T209A; Eryr; Ampr | This study |
| pMAD::rsbS S56A; Eryr; Ampr | This study |
| pMAD::rsbR1 T241A; Eryr; Ampr | This study |
| pMAD::rsbT N49A; Eryr; Ampr | [42] |
| pMAD::ΔrsbL; Eryr; Ampr | [69] |
| pIMK3; ori p15A; IPTG-controlled gene expression Phelp::lacOid; Kanr | [70] |
| pDNG7 (pIMK3-Phelp::lacOid::rsbR1 rsbS rsbT); Kanr a | This study |
| Strains/Mutants | |
| Escherichia coli One Shot TOP10 | Invitrogen |
| Listeria monocytogenes EGD-e WT | K. Boor |
| L. monocytogenes EGD-e ΔsigB | [59] |
| L. monocytogenes EGD-e ΔprfA | [9] |
| L. monocytogenes EGD-e (rsbL C56A) | [67] |
| L. monocytogenes EGD-e (rsbL C56A ΔrsbR2 ΔrsbR3 ΔrsbR4) | [42] |
| L. monocytogenes EGD-e (ΔrsbL ΔrsbR2 ΔrsbR3 ΔrsbR4) | This study |
| L. monocytogenes EGD-e (rsbL C56A ΔrsbR2 ΔrsbR3 ΔrsbR4 rsbR1 T175A) | This study |
| L. monocytogenes EGD-e (rsbL C56A ΔrsbR2 ΔrsbR3 ΔrsbR4 rsbR1 T209A) | This study |
| L. monocytogenes EGD-e (rsbL C56A ΔrsbR2 ΔrsbR3 ΔrsbR4 rsbR1 T241A) | This study |
| L. monocytogenes EGD-e (rsbL C56A ΔrsbR2 ΔrsbR3 ΔrsbR4, rsbS S56A) | This study |
| L. monocytogenes EGD-e (rsbR1 T175A) | [42] |
| L. monocytogenes EGD-e (rsbR1 T209A) | This study |
| L. monocytogenes EGD-e (rsbR1 T241A) | This study |
| L. monocytogenes EGD-e (rsbS S56A) | This study |
| L. monocytogenes EGD-e (rsbT N49A) | [42] |
| L. monocytogenes EGD-e; rsbS- (formally C12:C14); Eryr b | [69] |
| Complemented mutant strains | |
| L. monocytogenes EGD-e + pDNG7; Kanr | This study |
| L. monocytogenes EGD-e ΔsigB + pDNG7;Kanr | This study |
| L. monocytogenes EGD-e rsbL C56A ΔrsbR2 ΔrsbR3 ΔrsbR4 + pDNG7; Kanr | This study |
| L. monocytogenes EGD-e rsbR1 T175A + pDNG7; Kanr | This study |
| L. monocytogenes EGD-e rsbR1 T209A + pDNG7; Kanr | This study |
| L. monocytogenes EGD-e rsbR1 T241A + pDNG7; Kanr | This study |
| L. monocytogenes EGD-e rsbS S56A + pDNG7; Kanr | This study |
| L. monocytogenes EGD-e rsbT N49A + pDNG7; Kanr | This study |
apDNG7 was constructed by cloning the open reading frames of rsbR1, rsbS and rsbT upstream of the IPTG inducible promoter Phelp::lacOid of the vector pIMK3.
bStrain harbouring a frameshift allele in rsbS and a transposon insertion between sreB and lmo0596. The transposon sequence contains the erythromycin resistance gene (ermC). This strain was renamed as rsbS-.
Role of stressosome phosphorylation in acid adaptation
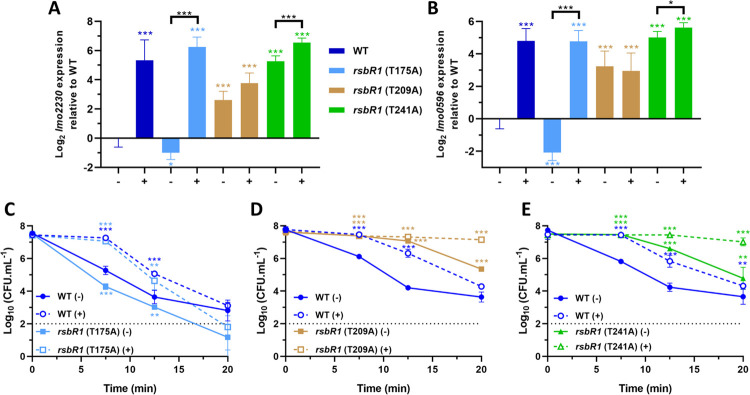
A comparison of the STAS C-terminal domains of RsbR1 with the homologue RsbRA from B. subtilis reveals conserved threonine residues at positions 175, 209 and 241, respectively, of the L. monocytogenes protein, one of which (T175) is phosphorylated in vivo in both organisms ([42,50]; Fig 4A and 4B). To investigate the contribution of these phosphorylation sites in acid signal transduction these residues were substituted with alanine in the RsbR1-only background (Table 1) and the respective mutants subjected to the acid adaptation regime at pH 5.0. The rsbR1-T175A substitution did not prevent acid induction of σB activity (Fig 4A and 4B). In contrast the rsbR1-T209A substitution essentially abolished the acid induced transcription of the reporter genes and resulted in a significantly higher baseline transcription level compared to unadapted wild type bacteria (2.61–3.25 log2-fold higher; Fig 4A and 4B, respectively). Similarly, the rsbR1-T241A mutant also exhibited increased baseline transcription, however the expression of the reporter genes was further increased by acid in this mutant. When the effects of these substitutions on acid resistance at pH 2.5 were measured, the rsbR1-T175A strain behaved similar to the wild type, showing an adaptive response to lethal acid challenge (Fig 4C). There was a small but significant decrease (~ 1 log10 CFU.ml-1 between 7.5 and 20 min) in the acid resistance of the unadapted rsbR1-T175A mutant strain and this is correlated with a small reduction (~ 1.5 log2-fold; p = 0.016) in the transcription of the two reporter genes in the unadapted conditions compared to the wild type. The rsbR1-T209A and rsbR1-T241A mutants showed a marked increase (p < 0.001) in acid resistance compared to the wild type even in the unadapted culture (Fig 4D and 4E) and this correlated with the increased σB activity recorded by the reporter gene transcript levels (Fig 4A and 4B). We also examined lmo2230 and lmo0596 expression in genetic backgrounds with or without the RsbR1 paralogues when each of the three putative phosphorylation sites was separately mutated. Their expression pattern was not significantly influenced by the presence or absence of the RsbR1 paralogues (S3 and S4 Figs).
Fig 4. RsbR1 residues T209 and T241 are required for normal acid-mediated activation of σB and development of acid tolerance.
Expression of σB-dependent genes (A) lmo2230 and (B) lmo0596, obtained from mid-log phase cultures of wild type and rsbR1 (T175A), rsbR1 (T209A) and rsbR1 (T241A) strains untreated (-) and treated (+) for 15 min in pH 5.0 at 37°C. Gene expression is expressed as Log2 relative gene expression. Acid challenge of mid-log phase of (C) rsbR1 (T175A), (D) rsbR1 (T209A) and (E) rsbR1 (T241A) cultures treated in pH 5.0 for 15 min at 37°C, as previously described in Fig 2D. Survival data is expressed as Log10 (CFU.mL-1). Statistical analysis was performed using a paired Student’s t-test relative to the wild type untreated after 15 min, for RT-qPCR data, and untreated wild type at time 0 min, for acid challenge data (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
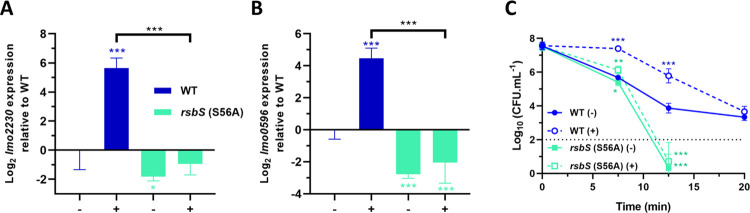
The T209 residue of RsbR1 aligns with a conserved serine (S56) at the equivalent position in the STAS domain of L. monocytogenes RsbS (Fig 1B) and this RsbS residue is known to be important for signal transduction in the B. subtilis stressosome [51]. We constructed a chromosomal allele of rsbS in the wild type background resulting in an RsbS S56A substitution (Table 1). The activation of σB during acid adaptation at pH 5.0 was completely abolished in this background, as indicated by the reporter genes. The transcription of these genes was also significantly lower (p = 0.0457 and <0.005 for lmo2230 and lmo0596, respectively) than the wild type under unadapted conditions (Fig 5A and 5B). Adaptation at pH 5.0 to a lethal acid challenge (pH 2.5) was abolished in the rsbS-S56A mutant and the unadapted strain was also markedly more sensitive to acid that the wild type, showing no detectable survivors after 20 min (Fig 5C). Thus, the increased acid sensitivity of the rsbS-S56A mutant correlates with an inability to induce σB activity during the adaptation at pH 5.0, highlighting the importance of the S56 residue in the transduction of acid signals through the stressosome.
Fig 5. RsbS residue S56 is essential for the transduction of the low pH signal and activation of σB.
Expression of σB-dependent genes (A) lmo2230 and (B) lmo0596, obtained from mid-log phase cultures of wild type and rsbS (S56A) strains untreated (-) and treated (+) for 15 min in pH 5.0 at 37°C. Gene expression is represented as Log2 relative gene expression. (C) Acid challenge of mid-log phase cultures treated in pH 5.0 for 15 min at 37°C, as previously described in Fig 2D. Survival data is expressed as Log10 (CFU.mL-1). Statistical analysis was performed using a paired Student’s t-test relative to the wild type untreated after 15 min, for RT-qPCR data, and untreated wild type at time 0 min, for acid challenge data (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
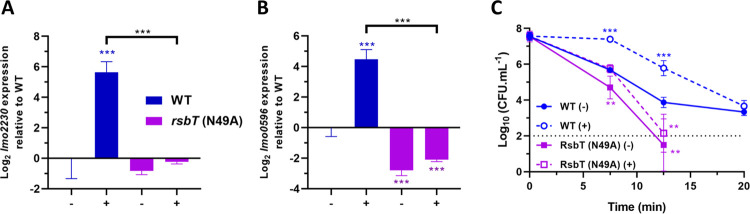
To determine the role of stressosome phosphorylation in acid stress signal transduction, we used a strain where the kinase activity of RsbT was inactivated through mutation. The rsbT-N49A mutation was recently shown to abolish the kinase activity of RsbT [42]. The acid induction of the two σB reporter genes at pH 5.0 was essentially eliminated in the rsbT-N49A background (Fig 6A and 6B) and this correlated with substantial decrease in acid resistance at pH 2.5 (Fig 6C).
Fig 6. The kinase activity of RsbT is essential for acid sensing by the stressosome.
Expression of σB-dependent genes (A) lmo2230 and (B) lmo0596, obtained from mid-log phase cultures of wild type and rsbT (N49A) strains untreated (-) and treated (+) for 15 min in pH 5.0 at 37°C. Gene expression is represented as Log2 relative gene expression. (C) Acid challenge of mid-log phase cultures treated in pH 5.0 for 15 min at 37°C, as previously described in Fig 2D. Survival data is expressed as Log10 (CFU.mL-1). Statistical analysis was performed using a paired Student’s t-test relative to the wild type untreated after 15 min, for RT-qPCR data, and untreated wild type at time 0 min, for acid challenge data (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
To confirm that the mutations introduced into rsbR1, rsbS and rsbT were responsible for the observed effects on acid survival, we provided a functional copy in trans. The single mutant strains were transformed with an IPTG inducible vector (pDNG7) bearing the parental rsbR1-rsbS-rsbT open reading frames (Table 1), and then grown to stationary phase, where σB is highly active and σB-defective strains exhibit a pronounced sensitivity in extreme acidic pH [18,59]. Stationary phase cultures were challenged in BHI at pH 2.5 at 37°C. The wild type strain showed further increase in acid tolerance when IPTG was added to the medium (S6A and S6B Fig). Interestingly, the acid tolerance of the rsbR1 T209A and T241A strains, that had elevated baseline σB activity, showed no further increase, suggesting that the complementation with native stressosome is unable to outcompete the mutated RsbR1 and attenuate σB activity. Finally, the complementation of the stressosome inactive strains, rsbS S56A and rsbT N49A, successfully restored their phenotypes but not for the ΔsigB strain (S6A–S6C Fig), showing that the provision of native stressosome in trans can restore the signal transduction in these strains.
To investigate the effects of the rsbR1 (T175A, T209A and T241A), rsbS (S56A) and rsbT (N49A) mutations on stressosome phosphorylation following sub-lethal acid treatment, we determined the phosphorylation patterns of RsbR1 and RsbS using the Phos-Tag electrophoresis system. A single RsbR1 band, probably corresponding to the unphosphorylated isoform, was observed in the rsbT-N49A kinase mutant. In contrast, two bands with reduced electrophoretic mobility were apparent in the wild type, presumed to correspond to different phosphorylated isoforms of higher molecular weight (Figs 7A and S5). The rsbR1-T175A and rsbR1-T209A mutations each resulted in the loss of one of the upper bands, suggesting that each band might represent a different singly phosphorylated isoform. Since we detected only two singly phosphorylated isoforms of RsbR1 on the Phos-Tag Western blots (Fig 7A) it appears that phosphorylation of T175 and T209 may be mutually exclusive, at least under the conditions we investigated. This raises the possibility that the temporal sequence of the phosphorylation and dephosphorylation reactions might contribute to regulatory control of RsbT release from the stressosome. The acid adaptation at pH 5.0 had minimal effects on the phosphorylation patterns detected, although there was some evidence in the wild type, rsbR1-T175A background that T209 was slightly more phosphorylated following acid exposure (Fig 7A and 7B). Finally, the phosphorylation patterns of RsbR1 were not affected in the rsbR1-T241A mutant compared to the wild type, suggesting that T241 is unlikely to be phosphorylated in L. monocytogenes (Fig 7A).
Fig 7. Phosphorylation of RbsR1 on residues T175, T209 and RsbS on residue S56 requires the kinase RsbT.
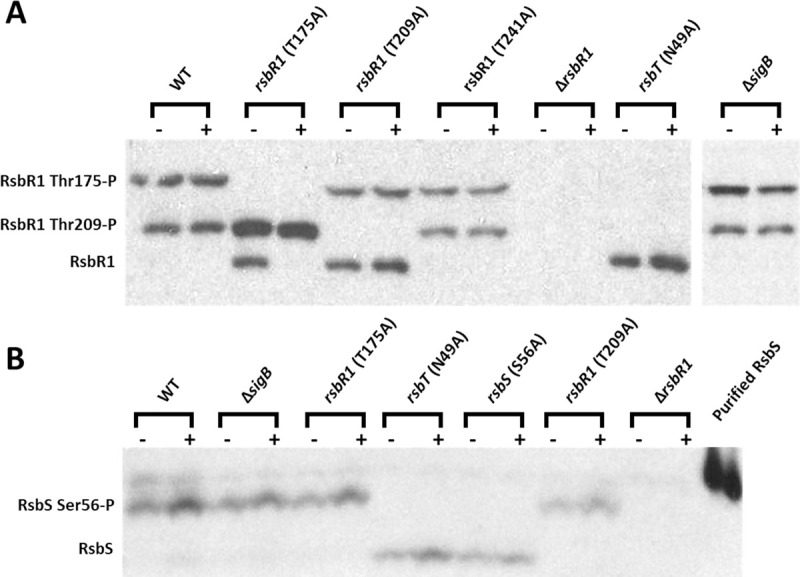
Western blots probed with (A) anti-RsbR1 and (B) anti-RsbS antibodies. Wild type and all mutant strains were grown to mid-log phase at 37°C in BHI and then untreated (-) or treated (+) at pH 5.0 for 15 min. Total protein extracted was separated in Phos-tag gels. The lower bands correspond to non-phosphorylated isoforms of RsbR1 or RsbS in the respective blots. The middle bands in anti-RsbR1 blots correspond to mono-phosphorylated RsbR1 at the T209 and the higher band corresponds to mono-phosphorylated RsbR1 at T175. The higher band on the RsbS blot corresponds to RsbS phosphorylated on S56. The ΔrsbR1 mutant extracts were included as negative controls since RsbR1 and RsbS are not expressed in this strain.
Anti-RsbS blotting confirmed the disappearance of the phosphorylated isoform (upper band) of RsbS in the rsbS-S56A and rsbT-N49A backgrounds (Fig 7B). The specificity of the anti-RsbS antibodies was confirmed using a ΔrsbR1 mutant that is polar and abolishes RsbS expression [42]. Overall these data confirmed the loss of phosphorylation of RsbR1 and RsbS in the site directed mutant strains, thereby supporting the conclusion that phosphorylation of RsbR1 (at residue T209) and RsbS (at residue S56) is necessary for transducing acid stress signals through the stressosome.
Acid-induced virulence gene expression requires functional stressosome signal transduction
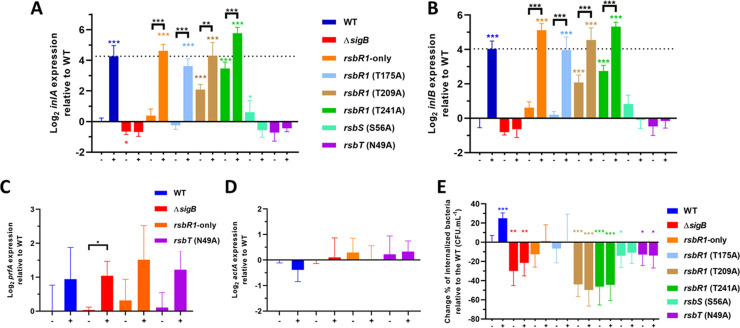
There are several lines of evidence that σB plays an important role in transcription during the gastrointestinal stage of the infectious cycle [9,71,72]. We investigated whether the invasion genes inlA and inlB, required for penetration across the intestinal barrier, and known to be partly under σB control [13,14], might be induced in response to the low pH signals found in the GI tract. Using the same acid adaptation regime described above (pH 5.0 for 15 min) we found that both inlA and inlB were strongly induced (~ 4 log2-fold) in the wild type but not in a sigB mutant (Fig 8A and 8B). Since both genes belong to one operon, the shared pattern of expression was not surprising. Next the role of the stressosome in this acid induced regulation was investigated. A strain carrying RsbR1 but none of its four paralogues (RsbR1-only) showed a very similar pattern of inlAB transcription to the wild type. Mutations abolishing the kinase activity of RsbT (rsbT-N49A) or the phosphorylation of RsbS (rsbS-S56A) eliminated the acid-induced transcription of the inlAB operon (Fig 8A and 8B). Mutations in rsbR1 produced a less pronounced effect on inlAB induction by acid, with the rsbR1-T175A mutant showing a very similar response to the wild type and the rsbR1-T209A mutant showing a significantly higher level of inlAB transcription in the untreated condition (Fig 8A and 8B). Together these data strongly suggest that RsbR1 is sufficient to sense and transduce acid stress signals into the stressosome and show that this is necessary for acid induced regulation of the virulence invasion genes inlA and inlB.
Fig 8. Acid sensing via the stressosome triggers upregulation of the inlAB invasion operon and influences Caco-2 cell invasion without affecting PrfA activity.
(A) inlA, (B) inlB, (C) prfA and (D) actA expression was obtained from mid-log phase cultures of strains used in this study. Cultures were untreated (-) and treated (+) in pH 5.0 for 15 min at 37°C. (E) Caco-2 invasion experiments performed on untreated (-) and treated (+) cultures in pH 5.0 for 15 min. L. monocytogenes wild type and mutant strains were in contact with the Caco-2 epithelial cells for 1 h and extracellular bacteria were eliminated with extensive washing and the supplementation of 50 μg.mL-1 of gentamicin to fresh tissue culture medium. Quantification of internalized L. monocytogenes was obtained from lysed Caco-2 cells 1 h after the gentamicin treatment. The untreated wild type strain was arbitrary converted to 0% and all samples were normalized to percentage of CFU.mL-1 relative to the untreated wild type. Statistical analysis was performed using a paired Student’s t-test relative to the wild type untreated after 15 min (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
The possibility that induction of inlAB by acid could involve the virulence gene regulator PrfA was examined by measuring the transcription of prfA and the PrfA-dependent actA under the same experimental conditions. The prfA transcript increased slightly (~ 1 log2-fold) following acid treatment, as has been reported by others [17]. This increase was evident in a mutant lacking σB and all the strains carrying RsbR1, RsbS and RsbT mutations (Fig 8C) and was therefore independent of σB and stressosome function. This small increase in prfA transcript levels did not affect PrfA activity since the levels of the highly PrfA-dependent (but not σB-dependent) transcript actA were not affected by acid treatment (Fig 8C and 8D). Together these data confirm that the involvement of σB in the acid-induced transcription of inlAB was direct and did not involve an indirect effect on PrfA.
Finally, we investigated whether the acid-induced activation of σB via the stressosome affected host cell invasion. Compared to the untreated wild type bacteria, the acid-exposed wild type bacteria displayed an increase in Caco-2 cell uptake of approximately 25% in the invasion of Caco-2 epithelial cells (Fig 8E). Mutants lacking sigB or carrying any of the stressosome alleles all showed a reduced invasion under the untreated conditions and the presence of acid failed to stimulate invasion. Surprisingly, even those strains that showed increased inlAB transcription (RsbR1 T209A, RsbR1 T241A) showed a reduced invasion rate compared to the wild type parental strain (Fig 8E). It is possible that post-transcriptional control of inlAB expression influenced the invasion rate, but further investigation of this phenomenon was beyond the scope of this study. Overall, the data showed that mutations that blocked acid-induced σB activation also affected the acid stimulated increase in host cell invasion.
Discussion
In this study, we sought to understand the signal transduction mechanisms that allow the stressosome of L. monocytogenes to transduce acid stress signals leading to activation of σB and the GSR. While it has been known for over two decades that σB plays a critical role in acid stress tolerance in this pathogen [18,73] nothing was known about how, or indeed whether, acid stress influenced σB activity. Here we show that the L. monocytogenes stressosome is responsible for the rapid transduction of the low pH signals. Specifically RsbR1 in conjunction with RsbS and RsbT, independently of the other four RsbR1 paralogues, are sufficient to initiate the downstream signal transduction events leading to σB activation. The kinase activity of RsbT and phosphorylation of RsbS on residue S56 are essential for transduction of the acid stress signal. The activation of σB by mild acidic stress (pH 5) results in the upregulation of σB-dependent genes and produces an increase in acid resistance to lethal acid pH challenge.
Although the overall composition of the L. monocytogenes stressosome, which comprises RsbR1 and its paralogues (RsbL, RsbR2, RsbR3), RsbS and RsbT, is now fairly well understood [37,42,46], the stoichiometry of individual stressosomes and heterogeneity of stressosome composition within cells and populations is still unknown. The outward facing turrets on the stressosome, derived from the N-terminal domains of RsbR1/RsbRA and paralogues, have been suggested to be the most likely sensory domains, although the nature of the signals detected has in most cases been elusive. As postulated by Murray and colleagues, the non-haem globin domains, found in most RsbR1/RsbRA-like proteins, may transiently interact with small signal molecules [74], but evidence for these has not been forthcoming. There may be some redundancy in the sensory capacity of RsbR1/RsbRA-like proteins since studies on the B. subtilis stressosome have shown that different RsbR paralogues can respond to ethanol and oxidative stress [51,75,76]. Here we sought to establish whether RsbR1 was capable of sensing and transducing acid stress signals via the stressosome. While it was not possible to delete rsbR1 without affecting the expression of the downstream genes (rsbS, rsbT, rsbU; [42]) it was possible to construct a strain lacking functional versions of all four paralogues (RsbL, RsbR2, RsbR3, RsbR4) and this mutant displayed a near-wild type activation of σB in response to acid stress (Fig 3A and 3B). The results show that RsbR1 is capable of sensing and transducing acid stress, independently of its paralogues, although the precise nature of the signals detected will require further research.
From studies on the non-pathogenic bacterium B. subtilis we have learned that the stressosome is a dynamic protein complex whose activation depends on phosphorylation of residues in RsbRA and RsbS [50,53], although acid stress has not been investigated as a stressor in this organism. In L. monocytogenes, RsbR1 is phosphorylated on residues T175 [77] and T209, but the phosphorylation sites are not conserved in the RsbR1 paralogues (Fig 1B). In this study, we confirm the phosphorylation of RsbR1 T209 and show that loss of this phosphorylation leads to derepressed σB activity under no-stress conditions and loss of responsiveness to acid. The finding that a substitution of RsbR1 residue T241 with alanine produces a similar phenotype provides evidence that this residue plays an important role is modulating signal transduction activity, although it does not appear to be phosphorylated in L. monocytogenes (Fig 7A).
Although a detailed mechanistic model accounting for signal transduction through the stressosome is not possible at this time, we propose a new working model to aid further experimental work on the system. The outward-facing dimeric N-terminal turret domains of RsbR1 most likely function in signal detection [78], although the nature of that signal remains to be determined. The acid stress signal could either be a change in the local proton concentration or some secondary consequence of acidification. The signal is likely to produce a conformation change of the N-terminal domains that propagates into the alpha-helical linker domains and into the STAS domains, resulting in the release of RsbT and consequently activation of σB via the downstream partner switching module [8,39]. We propose that the release of RsbT from the stressosome likely requires the phosphorylation of RsbS on S56, since acid sensing is abolished in the RsbS S56A variant and in the kinase-inactive mutant (Figs 5 and 7B). In this regard it is noteworthy that substitutions to negatively charged residues in B. subtilis RsbS (S59D) and Moorella thermoacetica MtS (S58D; MtS is a homologue of RsbS) abolish the interaction with the cognate kinase [79,80]. Furthermore, these mutations lock the stressosome in an active state, resulting in constitutive σB activity, which is deleterious for the bacteria [53,80,81].
The role of RsbR1 phosphorylation on residues T175 and T209 in modulating the response to acid is less clear but we speculate that they may influence the propagation of the signal that results in phosphorylation of RsbS. Altering T209 to alanine increased the background σB activity in the absence of acid stress and essentially abolished the responsiveness of the system to acid, while the T175A substitution did not affect acid sensing (Fig 4). The finding that the phosphorylation state of RsbR1 and RsbS doesn’t change much in response to osmotic [42] or acid stress (Fig 7A and 7B) is apparently in contradiction with the finding that phosphorylation is necessary for signal transduction, as evidenced by the absence of signal transduction in a kinase deficient mutant (Fig 6; [42]). One possibility we have not ruled out is that the method we used to determine the phosphorylation state of RsbR1 and RsbS is not sufficiently robust to exclude the possibility of phosphorylation during the protein extraction procedure (which includes a boiling step). Indeed, a previous study demonstrated that temperature stress rapidly stimulates the activation of σB in L. monocytogenes [82]. Thus, the boiling step used in the protein extract step could inadvertently lead to phosphorylation of the RsbR1 and RsbS, making it difficult to observe the effects of acid stress on RsbR1/S phosphorylation. Phosphomimetic substitutions that add negatively charged residues to the phosphorylatable sites in RsbR1 and RsbS will help to further clarify this issue.
While the phosphorylation undoubtedly plays a pivotal role in the activation of the stressosome, the precise mechanisms that result in release of RsbT are unknown at present. It has been suggested that conformational changes may occur at the STAS domains of RsbR1 and RsbS, prompting the release of RsbT from the stressosome core [39]. A recent study investigating the L. monocytogenes stressosome structure found T241 residing in RsbR1 α3 helix, described as “flexible loop”, which is located in close proximity to the T209 phosphorylation site [46]. Moreover, a recent Cryo-EM structural analysis of the phosphorylated L. innocua stressosome suggests the possibility of hydrogen bond formation between T209-PO4 and residue T241 (Christine Ziegler, personal communication). Given that amino acid substitutions to alanine at both of T209 and T241 lead to constitutive σB activity (Figs 4A and 4B and S3C–S3F), we speculate that the phosphorylation of T209 and its subsequent interaction with T241 likely determines the relative position of this flexible loop within the stressosome. We speculate that the loop’s position may influence the magnitude of the signal transduced by the stressosome, perhaps by affecting the propagation of the signal from one protomer to another. Further structural and genetic studies will be needed to fully elucidate the role phosphorylation in signal transduction. The role of the putative phosphatase RsbX in resetting the sensing–ready state of the stressosome has recently been suggested [56] but further work will be needed to elucidate the details of precisely how it interacts with the stressosome.
Multiple studies have shown that pre-treating L. monocytogenes with mild acidic pH increases its invasion towards Caco-2 cells [15–17] and this correlates well with increased inlA expression under these conditions [15,17,23]. Our study shows that the σB-dependent induction of the inlAB operon under mild acidic conditions is dependent on the stressosome’s sensory capacity (Fig 8A and 8B). In addition, we showed that the enhancement of L. monocytogenes EGD-e invasion is critically dependent on the normal function of stressosome sensing properties and a proper σB upregulation, as either lower or higher baseline activity abrogates L. monocytogenes capacity to invade Caco-2 cells (Fig 8E). Interestingly, the high baseline σB activity produced in the RsbR1 T209A and T241A strains correlated with a decrease in invasion, even though transcription of the inlAB operon was increased in this genetic background. This result demonstrates clearly that host cell invasion does not solely depend on the transcription of invasion genes, and further studies will be required to determine if this effect is due to some unknown post-transcriptional effect on inlAB expression. Indeed post-transcriptional control of inlA expression has been reported before [83]. Overall, our results support the idea that σB activity during the early stages of the infection cycle plays crucial role in priming the pathogen for host cell invasion and that the stressosome participates by helping to integrate pH-related signals from the environment of GI.
Overall, this study has established that the stressosome plays an active role in sensing and responding to acid stress by modulating the expression of the GSR. The signal transduction mechanism requires the kinase activity of RsbT and the phosphorylation of S56 on RsbS. In RsbR1, both T209 and T241 constrain σB activation in no-stress conditions and are necessary for a normal response to acid stress, while T175 appears to be dispensable for acid sensing. This signal transduction pathway has a demonstrable role in the development of acid resistance, likely influencing the survival of this pathogen in the food chain, food-processing environments and in the acidic conditions encountered within the host gastrointestinal tract. This pathway likely influences the progression of the infectious cycle during listeriosis through the impact on the expression of virulence factors that influence host cell invasion in the GI tract.
Material and methods
Bacterial strains, plasmids, and primers
L. monocytogenes EGD-e (serovar 1/2a) and E. coli TOP10 strains, plasmids and primers used in this study are listed in Table 1 and Table 2. Strains were grown in BHI broth or agar (LabM, UK) at 37°C with constant shaking at 150 rpm.min-1 at initial neutral pH ~ 7.4, unless otherwise specified. Cells were grown for 16 h until stationary phase was reached or further diluted in fresh BHI to an initial OD600 = 0.05 and further allowed to grow until mid-log phase (OD600 = 0.4). The following antibiotics were added to the media to a final concentration when required: erythromycin (Ery) at 5 μg.ml-1 for L. monocytogenes strains, ampicillin (Amp) at 100 μg.ml-1 for E. coli strains and kanamycin (Km) 50 μg.ml-1 for both microorganisms.
Table 2. Primers used in this study.
| Primer sequence (5’-3’) | Target |
|---|---|
| GTCGACGATCCCATTATGCTTTGGC | pIMK3_F |
| AAACTGCAGATGTATAAAGATTTTGCAAACTTCATCCGT | rsbRST_PstI_V2_Fa |
| CTAGGTCGACTTACCGAACCCATTTCGTTGT | rsbRST_SalI_V2_Ra |
| GTCCACCTCGGTTGGCCAAT | rsbR1 (T175A)_F |
| GGCACGCTCCGCATCTATC | rsbR1 (T175A)_R |
| AAGAAAAACCTGAAGATGATGC | rsbR1 (T209A)_F |
| TACCATAGCGTCTACAACAG | rsbR1 (T209A)_R |
| AGAAATCGCGCAAACAATC | rsbS (S56A)_F |
| GATGAAAGCGTCGATAAAATCG | rsbS (S56A)_R |
| TGGGAAAAGACAGTTTCTATCCA | rsbR1 (T241A)_F |
| GTTTACAATAGCTTGAGCG | rsbR1 (T241A)_R |
| CACCATGTATAAAGATTTTGCAAACTTCATCCG | rsbR1_up_flank_F |
| GATTGAATTTCGTCATCTTCCG | lmo0799_flank_F |
| CTCGCCATGTGTTTCTTTTTTC | lmo0799_flank_R |
| RT-qPCR | |
| TGGGGAGCAAACAGGATTAG | 16S_F |
| TAAGGTTCTTCGCGTTGCTT | 16S_R |
| CATATTCGAAGTGCCATTGC | lmo2230_F |
| CTGAACTAGGTGAATAAGACAAAC | lmo2230_R |
| GGGTACTAGCTGACGGAATTTTATC | lmo0596_F |
| CCCACATACCGAAAAGTAATACGAG | lmo0596_R |
| AGCTCACGAGTATTAGCGAGA | prfA_F |
| GCCTGCTCGCTAATGACTTC | prfA_R |
| CCTGCCACAAAACCACAAGA | actA_F |
| CTTCAATGCCAGCAGAACGA | actA_R |
| AAAGAACCAAAGGCACCAAC | inlA_F |
| ATTCCCGCCGTTATTTGTTG | inlA_R |
| GGCGCTAAACACGTGAATAA | inlB_F |
| GGCGCTGACATAACGAGT | inlB_R |
| Northern blots | |
| GTTTCTGGCTGGGTACTAGC | lmo0596_NB_F |
| GCCAATTGTACGCATAATACC | lmo0596_NB_R |
aUnderlined bases represent the restriction sequences used.
Construction of genetically modified L. monocytogenes
The codon 56 TCA (Serine) in rsbS gene was changed to GCT (Alanine). In both cases, silent mutations were added to the two adjunct codons in order to discern mutant from WT codons by PCR during mutagenesis while taking in consideration the codon frequencies in L. monocytogenes EGD-e strain (rsbS WT -ATTGATTCATTTATT- to rsbS S56A -ATCGACGCTTTCATC-). The mutagenic sequences, each with a total length of 612 bp, including SalI and BamHI restriction sites in each edges, were artificially synthetized in the vector pEX-K168::rsbS (S56A) (Eurofins Genomics, Germany). The mutagenic sequences were subsequently cloned into shuttle vector pMAD, resulting in the pMAD::rsbS (S56A) vector. L. monocytogenes electrocompetent cells were created as previously described [70]. The electrocompetent L. monocytogenes WT strain was transformed with pMAD::rsbS (S56A). The quadruple mutant strain (lmo0799 (C56A); Δlmo0161; Δlmo1642; Δlmo1842) was further separately transformed with pMAD::Δlmo0799, pMAD::rsbR1 (T175A), pMAD::rsbR1 (T209A) and pMAD::rsbS (S56A). Electroporated cells were plated in BHI supplemented with Ery. Chromosomal integration and subsequent excision was achieved through a two-step recombination as previously described [67]. Colony PCR was made with the primers rsbR1_upflank_F paired with either rsbR1 (T175A)_R, rsbR1 (T209A)_R, rsbR1 (T241A)_R and rsbS (S56A)_R, were used to identify the chromosomal mutation in the respective genes rsbR1, rsbS and rsbT, while the deletion of lmo0799 was verified with primers lmo0799_flank_F and lmo0799_flank_R. All genomic DNA from each mutant strains were whole genome sequence to confirm the chromosomal mutation and identify additional secondary mutations.
Growth kinetics
Stationary-phase cells of the L. monocytogenes EGD-e WT and ΔsigB were diluted and adjusted to an initial OD600 of 0.05 and grown in fresh BHI at 37°C. OD600 measurements were taken every hour for 10 h. Three biological replicates were made.
Acid shock treatment in L. monocytogenes
Stationary phase cultures grown at 37°C were diluted to an initial OD600 of 0.05 in fresh BHI and allowed to grow at 37°C until mid-log phase was reached (OD600 = 0.4). Cultures were either acidified or not to pH 5.0 with HCl 5 M and further incubated for 15 min at 37°C. Cultures were diluted in 1:10 of BHI previously acidified to pH 2.5 with HCl 5 M. Samples were taken at 0, 7.5, 12.5 and 20 min, serial diluted from 10−2 to 10−7 in PBS and plated in BHI agar. Plates were then incubated at 37°C for 24 hours and colonies were counted. At least three biological replicates were made.
RNA extraction and RT-qPCR
Cultures of L. monocytogenes EGD-e and its isogenic mutants were grown until mid-log phase and subjected to acid treatment for 15 min as previous described. Cultures were diluted in RNAlater (Sigma, Germany) at a 1:5 ratio to stop the transcription. Total RNA was extracted using an RNeasy minikit (Qiagen, Netherlands) according to the manufacturer’s recommendations. Cells were disrupted by bead beating twice in FastPrep-24 at a speed of 6 m.s-1 for 40 s. DNA was digested with Turbo DNA-free (Invitrogen, UK) according to the manufacturer’s recommendations. The RNA integrity was verified by electrophoresis in 0.7% (w/v) agarose gels. Synthetises of cDNA was performed with SuperScript III First-Strand Synthesis System (Invitrogen) according to the manufacturer’s recommendations. cDNA was quantified using NanoDrop 2000c (ThermoFisher Scientific, UK) and diluted to a final concentration of 7 ng.ml-1. RT-qPCR was performed using a QuantiTect SYBR Green PCR kit (Qiagen) and primers for the target genes (Table 2). Primer efficiency for 16S, lmo2230, lmo0596, inlA, inlB, prfA and actA were previously tested using cDNA. Samples were analysed on LightCycler 480 system (Roche) with the following parameters: 95°C for 15 min; 45 cycles of 15 s at 95°C, 15 s at 53°C, and 30 s at 72°C; a melting curve drawn for 5 s at 95°C and 1 min at 55°C, followed by increases of 0.11°C.s-1 until 95°C was reached; and cooling for 30 s at 40°C. Cycle quantification values were calculated by using LightCycler 480 software version 1.5.1 (Roche, Switzerland) and the Pfaffl relative expression formula [84, 85]. The expression of 16S rRNA was used as a reference gene. Results are expressed as Log2-fold relative expression ratios normalized against the expression of L. monocytogenes WT strain in the absence of stress. At least three independent biological replicates were performed.
Time-lapse of σB response towards mild acidic pH
Stationary cultures of L. monocytogenes WT and ΔsigB were grown at 37°C were diluted to an initial OD600 of 0.05 in fresh BHI and allowed to grow at 37°C until mid-log phase was reached (OD600 = 0.4). Cultures were then acidified until pH 5.0 with HCl 5 M and further incubated up to 30 min. Samples were taken at 0, 5, 15 and 30 min post acidification and RT-qPCR were performed. At least three biological replicates were made.
Caco-2 cell invasion assay
Caco-2 cell line human colon adenocarcinoma (Merck, UK) were cultivated in DMEM (Sigma) supplemented with 10% (v/v) FBS (Sigma), 1x MEM non-essential Amino acids (Gibco, UK) and Pen-Strep (Merck) at 37°C, in 5% CO2 and at 90–95% humidity. 5 x 104 cells of non-differentiated Caco-2 (passages 12 to 15) were seeded 48 h prior infection in a 24 well-plate. L. monocytogenes strains were separately grown until mid-log phase (OD600 = 0.4) and acid shocked at pH 5.0 for 15 min at 37°C and washed twice in DPBS and adjusted to a multiplicity of infection of 100 in DMEM without antibiotics. Caco-2 cells were covered with DMEM containing L. monocytogenes strains. The bacteria were in contact with the eukaryotic cells for 1 h and subsequently washed twice with PBS and covered with 500 μL of DMEM supplemented with 50 μg.mL-1 of gentamicin for 1 h. Subsequently, the wells were washed twice with DPBS and the Caco-2 cells were lysed with 1 mL of cold PBS supplemented with 0.1% (v/v) Triton X-100. Intracellular bacteria was determined by plating serial diluted lysates in BHI and incubated for 24 h at 37°C. Results were calculated with the difference between the CFU.mL-1 counts of the untreated wild type strain and each respective mutant.
Identification of phosphorylated isoforms of RsbR1 and RsbS
Cultures and protein extraction were performed as previously described by others [42] with some minor changes. L. monocytogenes WT and its isogenic mutants were acid shocked cultures before being boiled for 20 s at 100°C to stop the metabolism before harvesting the cells by centrifugation. Total protein standardization was confirmed by SDS-PAGE (S5 Fig).
Northern-blots probed for lmo0596
Total RNA was isolated from L. monocytogenes WT, ΔsigB, ΔprfA and rsbS- (transposon mutant C14:C12), containing a frameshift in rsbS open reading frame, as previously described by others [86] with some modifications. WT and mutants strains were allowed to grow until late-log phase (OD600 = 0.8) and acid shocked before transcription was stopped. The DNA probe for lmo0596 was generated as previously described [69].
Validation of lmo0596 as a σB-dependent gene under mild low pH stress
Late-log phase cultures (OD600 = 0.8) of L. monocytogenes WT, ΔsigB and Δlmo0596 were acid shocked at pH 5.0, while WT and ΔsigB strains were treated with a range of pH’s (non-acidified pH 7.0, 6.0, 5.5, 5.0 and 4.5). Total RNA extraction, cDNA synthesis and RT-qPCR were performed as described above.
Genetic complementation of L. monocytogenes mutant strains, acid tolerance and Congo Red phenotyping
Genetic complementation was achieved by amplifying rsbR1-rsbS-rsbT open reading frames using the primers rsbRST_PstI_V2_F and rsbRST_SalI_V2_R. The obtained amplicon and the IPTG inducible vector, pIMK3, were separately digested using PstI and SalI and ligated afterwards. The resulting plasmid, pDNG7, was transformed into E. coli TOP10 competent cells. The pDNG7 vector was transformed into electrocompetent L. monocytogenes strains and plated into BHI supplemented with Kanamycin 50 μg.ml-1. Complementation was verified using the primers pIMK3_F and rsbRST_SalI_V2_R on the obtained colonies. Obtained complemented strains were grown at 37°C for 16 h in BHI supplemented or not with 1 mM of IPTG (Sigma) until stationary phase was reached. Strains were challenged in BHI (pH 2.5) acidified with HCl 5 M and samples were taken at 0 and 30 min post challenge, serial diluted in PBS and plated in BHI agar plates. 10 μL of stationary phase cultures of the complemented strains were plated on BHI agar plates supplemented with Congo Red (Sigma) 25 μg.mL-1 and IPTG 1 mM. Plates were incubated at 37°C then transferred to 30°C for 3 days. At least two biological replicates were made.
Whole-genome sequencing and Single Nucleotide Polymorphism (SNP) analysis
The gDNA of all mutants strains constructed in this study was extracted using DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer recommendations. The obtained genomic material was analysed via Illumina sequencing by MicrobesNG (Birmingham, UK) and the resulting trimmed reads were analysed using Breseq software [87] to identify additional SNP in the mutant strains chromosome. The nucleotide sequence of L. monocytogenes EGD-e chromosome (NCBI RefSeq accession no. NC_003210.1) was used as reference in this analysis.
Statistical analysis
All statistical analysis, Student’s t-test were performed using GraphPad Prism 8.
Supporting information
Cultures of wild type, ΔsigB, ΔprfA and rsbS- (formerly C12:C14) strains were grown to late-log phase (OD600 = 0.8) and untreated (-) and treated (+) samples in pH 5.0 or a range of pH (7.0 to 4.5) for 15 min. Total RNA was extracted as described in material and methods. rsbS- mutant consists on a transposon strain which also carries a frameshift in rsbS resulting in the premature stop codon and a polar effect on RsbT translation (57). (A) Northern blots probed for lmo0596. (B) RT-qPCR results for lmo0596 expression in wild type and ΔsigB strains in BHI adjusted to a pH range (7, 6, 5.5, 5 and 4.5). Gene expression obtained by RT-qPCR is expressed as Log2 relative gene expression. Statistical analysis was performed using a paired student t test relative to the untreated wild type after 15 min (NS = not significant; *, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
(TIF)
Expression of σB-dependent genes (A) lmo2230 and (B) lmo0596, obtained from mid-log phase cultures of rsbR1-only (rsbL C56A) and rsbR1-only (ΔrsbL) strains untreated (-) and treated (+) for 15 min in pH 5.0 at 37°C. (C) Acid challenge of mid-log phase cultures treated in pH 5.0 for 15 min at 37°C, as previously described in Fig 2D. Survival data is expressed as Log10 (CFU.mL-1). The rsbR1-only (rsbL C56A) genotype ΔrsbR2 ΔrsbR3 ΔrsbR4 rsbL (C56A) and the genotype of rsbR1-only (ΔrsbL) consists on ΔrsbR2 ΔrsbR3 ΔrsbR4 ΔrsbL. Statistical analysis was performed using a paired student t test relative to the untreated rsbR1-only (rsbL C56A) after 15 min, for RT-qPCR data and untreated wild type at time 0 min, for acid challenge data. (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
(TIF)
Expression of σB-dependent genes (A, C, E, G) lmo2230 and (B, D, F, H) lmo0596, obtained from mid-log phase cultures of rsbR1 (T175A), rsbR1 (T209A), rsbR1 (T241A), rsbS (S56A) and their respective rsbR1-only backgrounds. These strains were untreated (-) and treated (+) for 15 min in pH 5.0 at 37°C. The rsbR1-only backgrounds consist on the following genotype ΔrsbR2; ΔrsbR3; ΔrsbR4; rsbL (C56A). Statistical analysis was performed using a paired student t test relative to the untreated wild type strain after 15 min (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
(TIF)
Acid challenge of mid-log phase cultures of (A) rsbR1 (T175A), (B) rsbR1 (T209A), (C) rsbR1 (T241A), (D) rsbS (S56A) and their respective rsbR1-only backgrounds, treated in BHI acidified to pH 5.0 for 15 min at 37°C. Survival data is expressed as Log10 (CFU.mL-1). The rsbR1-only backgrounds consist in ΔrsbR2; ΔrsbR3; ΔrsbR4; rsbL (C56A). The dashed line represents the detection threshold. Samples were taken at 0, 7.5, 12.5 and 20 min. Survival data is expressed as Log10 (CFU.mL-1). Statistical analysis was performed using a paired student t test relative to the respective untreated parental strain after at time 0 min (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
(TIF)
SDS-PAGE gels of normalized total protein extracts used for the Phos-tag and western-blots made in this study. Total protein extractions were obtain from mid-log phase cultures untreated (-) and treated (+) in pH 5.0 for 15 min at 37°C. Each protein sample was obtained from ~7 x 107 cells. Total protein was separated in 12% acrylamide/bis-acrylamide gels and subsequently stained with Coomassie dye.
(TIF)
Mutant strains were genetically complemented with pDNG7, which contains the native rsbR rsbS rsbT open reading frame under the control of a leaky IPTG inducible promoter (Phelp::lacOid). (A) Complemented (+) and non-complemented (-) strains were grown to stationary phase without IPTG in BHI at 37°C and then challenged in BHI acidified to pH 2.5. (B) Complemented strains were grown with (+) and without IPTG (-) and subsequently challenged to pH 2.5. Results are expressed in Log10 (CFU.mL-1). (C) Stationary phase cultures were plated in BHI agar containing 25 μg.mL-1 of Congo Red and 1 mM of IPTG and further incubated at 37°C for 24 h and transferred to 30°C for more 3 days.
(TIF)
Acknowledgments
We are grateful to Christine Ziegler (University of Regensburg) and her colleagues for helpful discussions and for sharing results with us prior to their publication. We thank members of the PATHSENSE consortium for helpful discussions and suggestions during the course of this research.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors DNG, TT, DG, AB, JJ, FGP, COP were supported by funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 721456 (https://ec.europa.eu/research/mariecurieactions/). The author MGP was funded by grant no. PGC2018-096364-B-I00 from the Spanish Ministry of Science and Innovation (https://www.ciencia.gob.es/en/Ministerio/mision-y-organizacion.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. Listeriosis [Internet]. 2018 [cited 2021 Jun 2]. Available from: https://www.who.int/news-room/fact-sheets/detail/listeriosis
- 2.Barbuddhe SB, Chakraborty T. Listeria as an Enteroinvasive Gastrointestinal Pathogen. In: Sasakawa C, editor. Molecular Mechanisms of Bacterial Infection via the Gut [Internet]. Berlin, Heidelberg: Springer; 2009. [cited 2021 Jun 2]. p. 173–95. (Current Topics in Microbiology and Immunology). Available from: 10.1007/978-3-642-01846-6_6 [DOI] [PubMed] [Google Scholar]
- 3.Martínez-Suárez JV, Ortiz S, López-Alonso V. Potential Impact of the Resistance to Quaternary Ammonium Disinfectants on the Persistence of Listeria monocytogenes in Food Processing Environments. Front Microbiol. 2016;7. doi: 10.3389/fmicb.2016.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.NicAogáin K, O’Byrne CP. The Role of Stress and Stress Adaptations in Determining the Fate of the Bacterial Pathogen Listeria monocytogenes in the Food Chain. Front Microbiol [Internet]. 2016. Nov 23 [cited 2020 Aug 10];7. Available from: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01865/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis ML, Ricke SC, Donaldson JR. Establishment of Listeria monocytogenes in the Gastrointestinal Tract. Microorganisms. 2019. Mar;7(3):75. doi: 10.3390/microorganisms7030075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandhi M, Chikindas ML. Listeria: A foodborne pathogen that knows how to survive. International Journal of Food Microbiology. 2007. Jan 1;113(1):1–15. doi: 10.1016/j.ijfoodmicro.2006.07.008 [DOI] [PubMed] [Google Scholar]
- 7.Arcari T, Feger M-L, Guerreiro DN, Wu J, O’Byrne CP. Comparative Review of the Responses of Listeria monocytogenes and Escherichia coli to Low pH Stress. Genes. 2020. Nov;11(11):1330. doi: 10.3390/genes11111330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerreiro DN, Arcari T, O’Byrne CP. The σB-Mediated General Stress Response of Listeria monocytogenes: Life and Death Decision Making in a Pathogen. Front Microbiol [Internet]. 2020. [cited 2020 Aug 17];11. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01505/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009. Jun;459(7249):950–6. doi: 10.1038/nature08080 [DOI] [PubMed] [Google Scholar]
- 10.Chaturongakul S, Raengpradub S, Palmer ME, Bergholz TM, Orsi RH, Hu Y, et al. Transcriptomic and Phenotypic Analyses Identify Coregulated, Overlapping Regulons among PrfA, CtsR, HrcA, and the Alternative Sigma Factors σB, σC, σH, and σL in Listeria monocytogenes. Appl Environ Microbiol. 2011. Jan;77(1):187–200. doi: 10.1128/AEM.00952-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garner MR, Njaa BL, Wiedmann M, Boor KJ. Sigma B Contributes to Listeria monocytogenes Gastrointestinal Infection but Not to Systemic Spread in the Guinea Pig Infection Model. Infect Immun. 2006. Feb;74(2):876–86. doi: 10.1128/IAI.74.2.876-886.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver HF, Orsi RH, Wiedmann M, Boor KJ. Listeria monocytogenes σB Has a Small Core Regulon and a Conserved Role in Virulence but Makes Differential Contributions to Stress Tolerance across a Diverse Collection of Strains. Appl Environ Microbiol. 2010. Jul;76(13):4216–32. doi: 10.1128/AEM.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, Boor KJ, Marquis H. Listeria monocytogenes σB Contributes to Invasion of Human Intestinal Epithelial Cells. Infect Immun. 2004. Dec;72(12):7374–8. doi: 10.1128/IAI.72.12.7374-7378.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H, Marquis H, Boor KJ. σB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology (Reading). 2005. Oct;151(Pt 10):3215–22. doi: 10.1099/mic.0.28070-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werbrouck H, Vermeulen A, Van Coillie E, Messens W, Herman L, Devlieghere F, et al. Influence of acid stress on survival, expression of virulence genes and invasion capacity into Caco-2 cells of Listeria monocytogenes strains of different origins. International Journal of Food Microbiology. 2009. Aug 31;134(1):140–6. doi: 10.1016/j.ijfoodmicro.2009.03.022 [DOI] [PubMed] [Google Scholar]
- 16.Conte MP, Petrone G, Di Biase AM, Ammendolia MG, Superti F, Seganti L. Acid tolerance in Listeria monocytogenes influences invasiveness of enterocyte-like cells and macrophage-like cells. Microb Pathog. 2000. Sep;29(3):137–44. doi: 10.1006/mpat.2000.0379 [DOI] [PubMed] [Google Scholar]
- 17.Neuhaus K, Satorhelyi P, Schauer K, Scherer S, Fuchs TM. Acid shock of Listeria monocytogenes at low environmental temperatures induces prfA, epithelial cell invasion, and lethality towards Caenorhabditis elegans. BMC Genomics. 2013. Apr;14:285. doi: 10.1186/1471-2164-14-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiedmann M, Arvik TJ, Hurley RJ, Boor KJ. General Stress Transcription Factor ςB and Its Role in Acid Tolerance and Virulence ofListeria monocytogenes. Journal of Bacteriology. 1998. Jul 15;180(14):3650–6. doi: 10.1128/JB.180.14.3650-3656.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowman JP, Chang KJL, Pinfold T, Ross T. Transcriptomic and Phenotypic Responses of Listeria monocytogenes Strains Possessing Different Growth Efficiencies under Acidic Conditions. Applied and Environmental Microbiology. 2010;76(14):4836–50. doi: 10.1128/AEM.00315-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaturongakul S, Boor KJ. σB Activation under Environmental and Energy Stress Conditions in Listeria monocytogenes [Internet]. Applied and Environmental Microbiology. 2006. [cited 2021 Jun 8]. Available from: https://journals.asm.org/doi/abs/10.1128/AEM.03058-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorey A, Lee B-H, Rotter B, O’Byrne CP. Blue Light Sensing in Listeria monocytogenes Is Temperature-Dependent and the Transcriptional Response to It Is Predominantly SigB-Dependent. Frontiers in Microbiology. 2019;10:2497. doi: 10.3389/fmicb.2019.02497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivy RA, Wiedmann M, Boor KJ. Listeria monocytogenes Grown at 7°C Shows Reduced Acid Survival and an Altered Transcriptional Response to Acid Shock Compared to L. monocytogenes Grown at 37°C. Appl Environ Microbiol. 2012. Jun 1;78(11):3824–36. doi: 10.1128/AEM.00051-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sue D, Fink D, Wiedmann M, Boor KJY 2004. σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology. 2004;150(11):3843–55. doi: 10.1099/mic.0.27257-0 [DOI] [PubMed] [Google Scholar]
- 24.Utratna M, Shaw I, Starr E, O’Byrne CP. Rapid, Transient, and Proportional Activation of σB in Response to Osmotic Stress in Listeria monocytogenes. Applied and Environmental Microbiology. 2011. Nov 1;77(21):7841–5. doi: 10.1128/AEM.05732-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreira A O ’Byrne CP, Boor KJ. Role of σB in Heat, Ethanol, Acid, and Oxidative Stress Resistance and during Carbon Starvation in Listeria monocytogenes. Appl Environ Microbiol. 2001. Oct 1;67(10):4454–7. doi: 10.1128/AEM.67.10.4454-4457.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreira A, Sue D, O’Byrne CP, Boor KJ. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Applied and Environmental Microbiology. 2003;69(5):2692–8. doi: 10.1128/AEM.69.5.2692-2698.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Driscoll B, Gahan C, Hill C. Adaptive acid tolerance response in Listeria monocytogenes: isolation of an acid-tolerant mutant which demonstrates increased virulence [Internet]. Applied and Environmental Microbiology. 1996. [cited 2021 Jun 8]. Available from: https://journals.asm.org/doi/abs/10.1128/aem.62.5.1693-1698.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis MJ, Coote PJ, O’Byrne CPY 1996. Acid tolerance in Listeria monocytogenes: the adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology. 1996;142(10):2975–82. [DOI] [PubMed] [Google Scholar]
- 29.Kroll RG, Patchett RA. Induced acid tolerance in Listeria monocytogenes. Letters in Applied Microbiology. 1992;14(5):224–7. [Google Scholar]
- 30.He K, Xin Y, Shan Y, Zhang X, Song H, Fang W. Phosphorylation residue T175 in RsbR protein is required for efficient induction of sigma B factor and survival of Listeria monocytogenes under acidic stress. J Zhejiang Univ Sci B. 2019. Aug 1;20(8):660–9. doi: 10.1631/jzus.B1800551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Utratna M. The development of reporters to measure the general stress response in Listeria monocytogenes: population and single cell studies on the activation of σB [Doctoral Thesis]. National University of Ireland, Galway; 2012. [Google Scholar]
- 32.Wemekamp-Kamphuis HH, Wouters JA, Leeuw PPLA de, Hain T, Chakraborty T, Abee T. Identification of Sigma Factor σB-Controlled Genes and Their Impact on Acid Stress, High Hydrostatic Pressure, and Freeze Survival in Listeria monocytogenes EGD-e. Applied and Environmental Microbiology. 2004. Jun;70(6):3457–66. doi: 10.1128/AEM.70.6.3457-3466.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferreira A, Gray M, Wiedmann M, Boor KJ. Comparative Genomic Analysis of the sigB Operon in Listeria monocytogenes and in Other Gram-Positive Bacteria. Curr Microbiol. 2004. Jan 1;48(1):39–46. doi: 10.1007/s00284-003-4020-x [DOI] [PubMed] [Google Scholar]
- 34.Wise AA, Price CW. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor sigma B in response to environmental signals. J Bacteriol. 1995. Jan;177(1):123–33. doi: 10.1128/jb.177.1.123-133.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson AK, Haldenwang WG. Characterization of a regulatory network that controls sigma B expression in Bacillus subtilis. J Bacteriol. 1992. Feb;174(3):749–57. doi: 10.1128/jb.174.3.749-757.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boylan SA, Rutherford A, Thomas SM, Price CW. Activation of Bacillus subtilis transcription factor sigma B by a regulatory pathway responsive to stationary-phase signals. J Bacteriol. 1992. Jun;174(11):3695–706. doi: 10.1128/jb.174.11.3695-3706.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Impens F, Rolhion N, Radoshevich L, Bécavin C, Duval M, Mellin J, et al. N-terminomics identifies Prli42 as a membrane miniprotein conserved in Firmicutes and critical for stressosome activation in Listeria monocytogenes. Nature Microbiology. 2017. Feb 13;2(5):1–12. doi: 10.1038/nmicrobiol.2017.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marles-Wright J, Lewis RJ. The stressosome: molecular architecture of a signalling hub. Biochemical Society Transactions. 2010. Jul 26;38(4):928–33. doi: 10.1042/BST0380928 [DOI] [PubMed] [Google Scholar]
- 39.Pané-Farré J, Quin MB, Lewis RJ, Marles-Wright J. Structure and Function of the Stressosome Signalling Hub. In: Harris JR, Marles-Wright J, editors. Macromolecular Protein Complexes: Structure and Function [Internet]. Cham: Springer International Publishing; 2017. [cited 2021 Jun 3]. p. 1–41. (Subcellular Biochemistry). Available from: 10.1007/978-3-319-46503-6_1 [DOI] [PubMed] [Google Scholar]
- 40.Pané-Farré J, Lewis RJ, Stülke J. The RsbRST Stress Module in Bacteria: A Signalling System That May Interact with Different Output Modules. MIP. 2005;9(2):65–76. doi: 10.1159/000088837 [DOI] [PubMed] [Google Scholar]
- 41.Chaturongakul S, Boor KJ. RsbT and RsbV Contribute to σB-Dependent Survival under Environmental, Energy, and Intracellular Stress Conditions in Listeria monocytogenes. Appl Environ Microbiol. 2004. Sep;70(9):5349–56. doi: 10.1128/AEM.70.9.5349-5356.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dessaux C, Guerreiro DN, Pucciarelli MG, O’Byrne CP, García-del Portillo F. Impact of osmotic stress on the phosphorylation and subcellular location of Listeria monocytogenes stressosome proteins. Sci Rep. 2020. Nov 30;10(1):20837. doi: 10.1038/s41598-020-77738-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez L, Reeves A, Haldenwang W. Stressosomes Formed in Bacillus subtilis from the RsbR Protein of Listeria monocytogenes Allow σB Activation following Exposure to either Physical or Nutritional Stress. J Bacteriol. 2010. Dec;192(23):6279–86. doi: 10.1128/JB.00467-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Losi A, Polverini E, Quest B, Gärtner W. First Evidence for Phototropin-Related Blue-Light Receptors in Prokaryotes. Biophysical Journal. 2002. May 1;82(5):2627–34. doi: 10.1016/S0006-3495(02)75604-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ondrusch N, Kreft J. Blue and Red Light Modulates SigB-Dependent Gene Transcription, Swimming Motility and Invasiveness in Listeria monocytogenes. PLOS ONE. 2011. Jan 11;6(1):e16151. doi: 10.1371/journal.pone.0016151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams AH, Redzej A, Rolhion N, Costa TRD, Rifflet A, Waksman G, et al. The cryo-electron microscopy supramolecular structure of the bacterial stressosome unveils its mechanism of activation. Nat Commun. 2019. Jul 8;10(1):3005. doi: 10.1038/s41467-019-10782-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi S, Nakasone Y, Hellingwerf KJ, Terazima M. Photoreaction Dynamics of a Full-Length Protein YtvA and Intermolecular Interaction with RsbRA. Biochemistry. 2020. Dec 22;59(50):4703–10. doi: 10.1021/acs.biochem.0c00888 [DOI] [PubMed] [Google Scholar]
- 48.Chen C-C, Lewis RJ, Harris R, Yudkin MD, Delumeau O. A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Molecular Microbiology. 2003;49(6):1657–69. doi: 10.1046/j.1365-2958.2003.03663.x [DOI] [PubMed] [Google Scholar]
- 49.Sharma AK, Rigby AC, Alper SL. STAS Domain Structure and Function. CPB. 2011;28(3):407–22. doi: 10.1159/000335104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaidenko TA, Yang X, Lee YM, Price CW. Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the environmental stress signaling pathway of Bacillus subtilis 11Edited by M. Gottesman. Journal of Molecular Biology. 1999. Apr 23;288(1):29–39. doi: 10.1006/jmbi.1999.2665 [DOI] [PubMed] [Google Scholar]
- 51.Kim T-J, Gaidenko TA, Price CW. A Multicomponent Protein Complex Mediates Environmental Stress Signaling in Bacillus subtilis. Journal of Molecular Biology. 2004. Jul 30;341(1):135–50. doi: 10.1016/j.jmb.2004.05.043 [DOI] [PubMed] [Google Scholar]
- 52.Eymann C, Schulz S, Gronau K, Becher D, Hecker M, Price CW. In vivo phosphorylation patterns of key stressosome proteins define a second feedback loop that limits activation of Bacillus subtilis σB. Molecular Microbiology. 2011;80(3):798–810. doi: 10.1111/j.1365-2958.2011.07609.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim T-J, Gaidenko TA, Price CW. In Vivo Phosphorylation of Partner Switching Regulators Correlates with Stress Transmission in the Environmental Signaling Pathway of Bacillus subtilis. J Bacteriol. 2004. Sep;186(18):6124–32. doi: 10.1128/JB.186.18.6124-6132.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woodbury RL, Luo T, Grant L, Haldenwang WG. Mutational Analysis of RsbT, an Activator of the Bacillus subtilis Stress Response Transcription Factor, σB. J Bacteriol. 2004. May;186(9):2789–97. doi: 10.1128/JB.186.9.2789-2797.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen C-C, Yudkin MD, Delumeau O. Phosphorylation and RsbX-Dependent Dephosphorylation of RsbR in the RsbR-RsbS Complex of Bacillus subtilis. J Bacteriol. 2004. Oct;186(20):6830–6. doi: 10.1128/JB.186.20.6830-6836.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oliveira AH, Tiensuu T, Guerreiro DN, Tükenmez H, Dessaux C, García-del Portillo F, et al. Listeria monocytogenes requires the RsbX protein to prevent SigB-activation under non-stressed conditions. Journal of Bacteriology. 2021;JB.00486-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voelker U, Luo T, Smirnova N, Haldenwang W. Stress activation of Bacillus subtilis sigma B can occur in the absence of the sigma B negative regulator RsbX. J Bacteriol. 1997. Mar;179(6):1980–4. doi: 10.1128/jb.179.6.1980-1984.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia Y, Xin Y, Li X, Fang W. To Modulate Survival under Secondary Stress Conditions, Listeria monocytogenes 10403S Employs RsbX To Downregulate σB Activity in the Poststress Recovery Stage or Stationary Phase. Applied and Environmental Microbiology. 2016. Feb 15;82(4):1126–35. doi: 10.1128/AEM.03218-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guerreiro DN, Wu J, Dessaux C, Oliveira AH, Tiensuu T, Gudynaite D, et al. Mild Stress Conditions during Laboratory Culture Promote the Proliferation of Mutations That Negatively Affect Sigma B Activity in Listeria monocytogenes. J Bacteriol. 2020. Apr 9;202(9). doi: 10.1128/JB.00751-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ollinger J, Wiedmann M, Boor KJ. σB- and PrfA-Dependent Transcription of Genes Previously Classified as Putative Constituents of the Listeria monocytogenes PrfA Regulon. Foodborne Pathogens and Disease. 2008. Jun 1;5(3):281–93. doi: 10.1089/fpd.2008.0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wurtzel O, Sesto N, Karunker I, Edelheit S, Bécavin C, Archambaud C, et al. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Molecular Systems Biology. 2012. Jan 1;8(1):583. doi: 10.1038/msb.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arnold CN, McElhanon J, Lee A, Leonhart R, Siegele DA. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. Journal of Bacteriology. 2001;183(7):2178–86. doi: 10.1128/JB.183.7.2178-2186.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Masuda N, Church GM. Regulatory network of acid resistance genes in Escherichia coli. Molecular Microbiology. 2003;48(3):699–712. doi: 10.1046/j.1365-2958.2003.03477.x [DOI] [PubMed] [Google Scholar]
- 64.Mates AK, Sayed AK, Foster JW. Products of the Escherichia coli Acid Fitness Island Attenuate Metabolite Stress at Extremely Low pH and Mediate a Cell Density-Dependent Acid Resistance. Journal of Bacteriology. 2007. Apr;189(7):2759–68. doi: 10.1128/JB.01490-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamanaka Y, Aizawa S-I, Yamamoto K. The hdeD Gene Represses the Expression of Flagellum Biosynthesis via LrhA in Escherichia coli K-12. Journal of Bacteriology. 2021;204(1):JB.00420–21. doi: 10.1128/JB.00420-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rauch M, Luo Q, Müller-Altrock S, Goebel W. SigB-Dependent In Vitro Transcription of prfA and Some Newly Identified Genes of Listeria monocytogenes Whose Expression Is Affected by PrfA In Vivo. Journal of Bacteriology. 2005. Jan 15;187(2):800–4. doi: 10.1128/JB.187.2.800-804.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O’Donoghue B, NicAogáin K, Bennett C, Conneely A, Tiensuu T, Johansson J, et al. Blue-Light Inhibition of Listeria monocytogenes Growth Is Mediated by Reactive Oxygen Species and Is Influenced by σB and the Blue-Light Sensor Lmo0799. Appl Environ Microbiol. 2016. Jun 13;82(13):4017–27. doi: 10.1128/AEM.00685-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arnaud M, Chastanet A, Débarbouillé M. New Vector for Efficient Allelic Replacement in Naturally Nontransformable, Low-GC-Content, Gram-Positive Bacteria. Applied and Environmental Microbiology. 2004. Nov 1;70(11):6887–91. doi: 10.1128/AEM.70.11.6887-6891.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tiensuu T, Andersson C, Rydén P, Johansson J. Cycles of light and dark co-ordinate reversible colony differentiation in Listeria monocytogenes. Molecular Microbiology. 2013;87(4):909–24. doi: 10.1111/mmi.12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Monk IR, Gahan CGM, Hill C. Tools for Functional Postgenomic Analysis of Listeria monocytogenes. Appl Environ Microbiol. 2008. Jul;74(13):3921–34. doi: 10.1128/AEM.00314-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Begley M, Sleator RD, Gahan CGM, Hill C. Contribution of Three Bile-Associated Loci, bsh, pva, and btlB, to Gastrointestinal Persistence and Bile Tolerance of Listeria monocytogenes. Infection and Immunity. 2005. Feb 1;73(2):894–904. doi: 10.1128/IAI.73.2.894-904.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watson D, Sleator RD, Casey PG, Hill C, Gahan CGM. Specific Osmolyte Transporters Mediate Bile Tolerance in Listeria monocytogenes. Infection and Immunity. 2009. Nov 1;77(11):4895–904. doi: 10.1128/IAI.00153-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abram F, Starr E, Karatzas KAG, Matlawska-Wasowska K, Boyd A, Wiedmann M, et al. Identification of Components of the Sigma B Regulon in Listeria monocytogenes That Contribute to Acid and Salt Tolerance. AEM. 2008. Nov 15;74(22):6848–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murray JW, Delumeau O, Lewis RJ. Structure of a nonheme globin in environmental stress signaling. PNAS. 2005. Nov 29;102(48):17320–5. doi: 10.1073/pnas.0506599102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cabeen MT, Russell JR, Paulsson J, Losick R. Use of a microfluidic platform to uncover basic features of energy and environmental stress responses in individual cells of Bacillus subtilis. PLOS Genetics. 2017. Jul 20;13(7):e1006901. doi: 10.1371/journal.pgen.1006901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tran V, Geraci K, Midili G, Satterwhite W, Wright R, Bonilla CY. Resilience to oxidative and nitrosative stress is mediated by the stressosome, RsbP and SigB in Bacillus subtilis. Journal of Basic Microbiology. 2019;59(8):834–45. doi: 10.1002/jobm.201900076 [DOI] [PubMed] [Google Scholar]
- 77.Dessaux C, Pucciarelli MG, Guerreiro DN, O’Byrne CP, García-del Portillo F. Activation of the Listeria monocytogenes Stressosome in the Intracellular Eukaryotic Environment. Applied and Environmental Microbiology. 2021;87(12):e00397–21. doi: 10.1128/AEM.00397-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walker JA, Rivera S, Weinert EE. Mechanism and Role of Globin-Coupled Sensor Signalling. In: Poole RK, editor. Advances in Microbial Physiology [Internet]. Academic Press; 2017. [cited 2021 Sep 22]. p. 133–69. Available from: https://www.sciencedirect.com/science/article/pii/S0065291117300231 doi: 10.1016/bs.ampbs.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quin MB, Berrisford JM, Newman JA, Baslé A, Lewis RJ, Marles-Wright J. The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling. Structure. 2012. Feb 8;20(2):350–63. doi: 10.1016/j.str.2012.01.003 [DOI] [PubMed] [Google Scholar]
- 80.Yang X, Kang CM, Brody MS, Price CW. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996. Sep 15;10(18):2265–75. doi: 10.1101/gad.10.18.2265 [DOI] [PubMed] [Google Scholar]
- 81.Kang CM, Brody MS, Akbar S, Yang X, Price CW. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor sigma(b) in response to environmental stress. J Bacteriol. 1996. Jul;178(13):3846–53. doi: 10.1128/jb.178.13.3846-3853.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guldimann C, Guariglia-Oropeza V, Harrand S, Kent D, Boor KJ, Wiedmann M. Stochastic and Differential Activation of σB and PrfA in Listeria monocytogenes at the Single Cell Level under Different Environmental Stress Conditions. Frontiers in Microbiology. 2017;8:348. doi: 10.3389/fmicb.2017.00348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stritzker J, Schoen C, Goebel W. Enhanced Synthesis of Internalin A in aro Mutants of Listeria monocytogenes Indicates Posttranscriptional Control of the inlAB mRNA. Journal of Bacteriology. 2005. Apr 15;187(8):2836–45. doi: 10.1128/JB.187.8.2836-2845.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pfaffl MW, Georgieva TM, Georgiev IP, Ontsouka E, Hageleit M, Blum JW. Real-time RT-PCR quantification of insulin-like growth factor (IGF)-1, IGF-1 receptor, IGF-2, IGF-2 receptor, insulin receptor, growth hormone receptor, IGF-binding proteins 1, 2 and 3 in the bovine species. Domestic Animal Endocrinology. 2002. Apr 1;22(2):91–102. doi: 10.1016/s0739-7240(01)00128-x [DOI] [PubMed] [Google Scholar]
- 85.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Research. 2001. May 1;29(9):e45–e45. doi: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, et al. A trans-Acting Riboswitch Controls Expression of the Virulence Regulator PrfA in Listeria monocytogenes. Cell. 2009. Nov 13;139(4):770–9. doi: 10.1016/j.cell.2009.08.046 [DOI] [PubMed] [Google Scholar]
- 87.Deatherage DE, Barrick JE. Identification of Mutations in Laboratory-Evolved Microbes from Next-Generation Sequencing Data Using breseq. In: Sun L, Shou W, editors. Engineering and Analyzing Multicellular Systems: Methods and Protocols [Internet]. New York, NY: Springer; 2014. [cited 2021 Jun 3]. p. 165–88. (Methods in Molecular Biology). Available from: 10.1007/978-1-4939-0554-6_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cultures of wild type, ΔsigB, ΔprfA and rsbS- (formerly C12:C14) strains were grown to late-log phase (OD600 = 0.8) and untreated (-) and treated (+) samples in pH 5.0 or a range of pH (7.0 to 4.5) for 15 min. Total RNA was extracted as described in material and methods. rsbS- mutant consists on a transposon strain which also carries a frameshift in rsbS resulting in the premature stop codon and a polar effect on RsbT translation (57). (A) Northern blots probed for lmo0596. (B) RT-qPCR results for lmo0596 expression in wild type and ΔsigB strains in BHI adjusted to a pH range (7, 6, 5.5, 5 and 4.5). Gene expression obtained by RT-qPCR is expressed as Log2 relative gene expression. Statistical analysis was performed using a paired student t test relative to the untreated wild type after 15 min (NS = not significant; *, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
(TIF)
Expression of σB-dependent genes (A) lmo2230 and (B) lmo0596, obtained from mid-log phase cultures of rsbR1-only (rsbL C56A) and rsbR1-only (ΔrsbL) strains untreated (-) and treated (+) for 15 min in pH 5.0 at 37°C. (C) Acid challenge of mid-log phase cultures treated in pH 5.0 for 15 min at 37°C, as previously described in Fig 2D. Survival data is expressed as Log10 (CFU.mL-1). The rsbR1-only (rsbL C56A) genotype ΔrsbR2 ΔrsbR3 ΔrsbR4 rsbL (C56A) and the genotype of rsbR1-only (ΔrsbL) consists on ΔrsbR2 ΔrsbR3 ΔrsbR4 ΔrsbL. Statistical analysis was performed using a paired student t test relative to the untreated rsbR1-only (rsbL C56A) after 15 min, for RT-qPCR data and untreated wild type at time 0 min, for acid challenge data. (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
(TIF)
Expression of σB-dependent genes (A, C, E, G) lmo2230 and (B, D, F, H) lmo0596, obtained from mid-log phase cultures of rsbR1 (T175A), rsbR1 (T209A), rsbR1 (T241A), rsbS (S56A) and their respective rsbR1-only backgrounds. These strains were untreated (-) and treated (+) for 15 min in pH 5.0 at 37°C. The rsbR1-only backgrounds consist on the following genotype ΔrsbR2; ΔrsbR3; ΔrsbR4; rsbL (C56A). Statistical analysis was performed using a paired student t test relative to the untreated wild type strain after 15 min (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
(TIF)
Acid challenge of mid-log phase cultures of (A) rsbR1 (T175A), (B) rsbR1 (T209A), (C) rsbR1 (T241A), (D) rsbS (S56A) and their respective rsbR1-only backgrounds, treated in BHI acidified to pH 5.0 for 15 min at 37°C. Survival data is expressed as Log10 (CFU.mL-1). The rsbR1-only backgrounds consist in ΔrsbR2; ΔrsbR3; ΔrsbR4; rsbL (C56A). The dashed line represents the detection threshold. Samples were taken at 0, 7.5, 12.5 and 20 min. Survival data is expressed as Log10 (CFU.mL-1). Statistical analysis was performed using a paired student t test relative to the respective untreated parental strain after at time 0 min (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.001).
(TIF)
SDS-PAGE gels of normalized total protein extracts used for the Phos-tag and western-blots made in this study. Total protein extractions were obtain from mid-log phase cultures untreated (-) and treated (+) in pH 5.0 for 15 min at 37°C. Each protein sample was obtained from ~7 x 107 cells. Total protein was separated in 12% acrylamide/bis-acrylamide gels and subsequently stained with Coomassie dye.
(TIF)
Mutant strains were genetically complemented with pDNG7, which contains the native rsbR rsbS rsbT open reading frame under the control of a leaky IPTG inducible promoter (Phelp::lacOid). (A) Complemented (+) and non-complemented (-) strains were grown to stationary phase without IPTG in BHI at 37°C and then challenged in BHI acidified to pH 2.5. (B) Complemented strains were grown with (+) and without IPTG (-) and subsequently challenged to pH 2.5. Results are expressed in Log10 (CFU.mL-1). (C) Stationary phase cultures were plated in BHI agar containing 25 μg.mL-1 of Congo Red and 1 mM of IPTG and further incubated at 37°C for 24 h and transferred to 30°C for more 3 days.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.