Abstract
Background:
The Prostate Imaging Reporting and Data System (PI-RADS) provides guidelines for risk stratification of lesions detected on multiparametric MRI (mpMRI) of the prostate but suffers from high intra/interreader variability.
Purpose:
To develop an artificial intelligence (AI) solution for PI-RADS classification and compare its performance with an expert radiologist using targeted biopsy results.
Study Type:
Retrospective study including data from our institution and the publicly available ProstateX dataset.
Population:
In all, 687 patients who underwent mpMRI of the prostate and had one or more detectable lesions (PI-RADS score >1) according to PI-RADSv2.
Field Strength/Sequence:
T2-weighted, diffusion-weighted imaging (DWI; five evenly spaced b values between b = 0–750 s/mm2) for apparent diffusion coefficient (ADC) mapping, high b-value DWI (b = 1500 or 2000 s/mm2), and dynamic contrast-enhanced T1-weighted series were obtained at 3.0T.
Assessment:
PI-RADS lesions were segmented by a radiologist. Bounding boxes around the T2/ADC/high-b value segmentations were stacked and saved as JPEGs. These images were used to train a convolutional neural network (CNN). The PI-RADS scores obtained by the CNN were compared with radiologist scores. The cancer detection rate was measured from a subset of patients who underwent biopsy.
Statistical Tests:
Agreement between the AI and the radiologist-driven PI-RADS scores was assessed using a kappa score, and differences between categorical variables were assessed with a Wald test.
Results:
For the 1034 detection lesions, the kappa score for the AI system vs. the expert radiologist was moderate, at 0.40. However, there was no significant difference in the rates of detection of clinically significant cancer for any PI-RADS score in 86 patients undergoing targeted biopsy (P = 0.4–0.6).
Data Conclusion:
We developed an AI system for assignment of a PI-RADS score on segmented lesions on mpMRI with moderate agreement with an expert radiologist and a similar ability to detect clinically significant cancer.
Level of Evidence:
4
Technical Efficacy Stage:
2204
PROSTATE CANCER is the most common non-cutaneous cancer in men, with an estimated 175,000 new diagnoses in 2019 in the United States.1 The traditional method of diagnosis for prostate cancer is an ultrasound-guided template sampling of the prostate in men deemed at risk for prostate cancer based on elevated serum prostate-specific antigen (PSA) or positive digital rectal exam screening tests.2 Multiparametric magnetic resonance imaging (mpMRI), including diffusion-weighted imaging, dynamic contrast-enhanced MRI, of the prostate has allowed for enhanced visualization of lesions and improved biopsy targeting.3 Utilizing MRI/ultrasound fusion-guided biopsy techniques, sampling of MRI-defined lesions suspicious for prostate cancer has been shown to improve the detection of clinically significant prostate cancer.4,5
The second version of the Prostate Imaging and Reporting Data System (PI-RADSv2) attempts to standardize the acquisition, interpretation, and reporting of mpMRI6 in order to improve cancer detection at prostate MRI. The PI-RADSv2 documentation provides a detailed description for assignment of detected lesions into categories with increasing risk of detection of clinically significant cancer as the score increases.7 PI-RADSv2 has been shown to be useful in refining the risk of detection of clinically significant prostate cancer on biopsy compared to clinical data alone8; very recently, PI-RADSv2 has been revised to its v2.1 version9 and clinical studies are under way to show its potential improvements in detecting clinically significant prostate cancer. However, the performance of PI-RADS has been hindered by poor interreader and intrareader agreement. Interreader agreement has been reported as less than 50%, and intrareader agreement is reportedly in the range of 60–74%.10 This variation results in substantial discrepancies in diagnostic performance of targeted biopsies, with clinically significant cancer detection rates from MR-guided biopsies varying by as much as 40% for PI-RADS 5 lesions.11 Therefore, in order to make PI-RADS categorization more uniform, we sought to develop a deep-learning artificial intelligence (AI) system to classify radiologist-identified lesions into PI-RADSv2 risk categories. Here we aim to describe this system and compare its performance with an expert radiologist using targeted biopsy results.
Material and Methods
Datasets
Images were acquired from three independent cohorts: the first two from our institution and the third from an open-source dataset. The purpose of including three distinct datasets was to expose the neural network to diverse data in order to reduce overfitting to one center or one specific patient population (ie, only those undergoing surgery). The first cohort (Cohort 1) included all patients undergoing mpMRI prior to radical prostatectomy between January 2015 and November 2018 as part of an Institutional Review Board (IRB)-approved protocol (Clinical Trials.gov Identifier: NCT02594202). This time period was selected based on the implementation of PI-RADSv2 scoring at our institution. The second cohort (Cohort 2) included all patients enrolled in an institutional protocol for patients undergoing multiparametric prostate MRI for evaluation of known or suspicious prostate cancer between study initiation (February 2018) and when study enrollment was halted (November 2018) (Clinical Trials.gov Identifier: NCT03354416). For both cohorts, per IRB requirements, informed consent was obtained from participants. The inclusion criterion was having a prostate MRI with a prospective PIRADS scoring 2 or greater. The exclusion criterion was having prior treatment (n = 11 patients) who had received prior androgen deprivation therapy and were excluded from these cohorts.
Both cohorts were scanned using a Phillips Achieva 3.0T MRI (Achieva 3.0-T-TX; Philips Healthcare, Best, the Netherlands) using an endorectal coil (BPX-30; Medrad, Pittsburgh, PA) or phased array surface coils (Philips Healthcare). A T2-weighted turbo-spin-echo acquisition was obtained in the axial, sagittal, and coronal planes. Diffusion-weighted imaging for production of apparent diffusion coefficient (ADC) maps using a monoexponential decay model and a separate high b-value (b = 1500 or b = 2000) sequences. Dynamic contrast-enhanced (DCE) sequences were obtained by administering gadoterate meglutamine (Dotarem, Bloomington, IN) through a peripheral vein at a dose of 0.1 mm/kg of body weight and a rate of 3 mL/s. All pulse sequence image acquisition parameters are detailed in Table S1 in the Supplemental Material from our prior publication.12 The pathologic outcome was assessed in patients who underwent an MRI-transrectal ultrasonography (MRI-TRUS) fusion-guided biopsy at our institution within a 6-month period after the acquisition of the MRI. Each lesion was biopsied twice—one was obtained in the axial plane and a second was obtained in the sagittal plane.
A third cohort was obtained from the publicly available PROSTATEx training dataset, which is described in detail on the challenge website (https://www.aapm.org/GrandChallenge/PROSTATEx-2/default.asp). Briefly, this dataset features multiparametric prostate MRIs included in two challenges for prostate cancer detection and risk stratification.13 The PROSTATEx dataset also had T2-weighted, diffusion-weighted series. No endorectal coil was utilized in this dataset. The imaging acquisition parameters are included as Table S1 in the Supplemental Material.
PI-RADS Classification
Lesion detection and PI-RADS scoring for all three cohorts was performed by a single radiologist with more than 10 years of experience in the interpretation of prostate MRIs (>1200 MRIs/year). All assessments of PI-RADs scores were performed prospectively as part of the clinical workflow prior to the beginning of this study and were not altered during this study. The radiologist was blinded to pathologic outcomes of all lesions during the labeling process. Segmentation of all detected lesions was performed by the same radiologist using software developed for research purposes (pseg, iCAD, Nashua, NH), and ground truth scoring of the lesions was performed according to PI-RADSv2 guidelines. PI-RADSV2.1 was not utilized for this research since it became available after the study was conducted. The boundaries drawn on multiple slices of the lesions were determined using visual inspection of all available MRI sequences and saved with reference coordinates on the T2-weighted axial sequence in MIPAV VOI format. In total, there were 1034 lesions detected in 687 patients.
Data Processing
The overall data processing scheme is demonstrated in Fig. 1. Due to differences in acquisition parameters and spatial resolution in T2 and diffusion sequences (ie, slice thickness difference between T2 and ADC), the ADC and high-b series were aligned using a 3D affine geometric transformation and resampled to the matrix size (ie, spatial resolution) of T2 series (affine3D, MatLab, MathWorks, Natick, MA). Each slice was then normalized to a minimum of 0 and a maximum of 255. The minimum and maximum values of the polygon segmentations in both x and y dimensions were parsed and used to determine a bounding box around the lesion on each slice. The bounding boxes were then padded by 10 voxels to ensure the immediate surrounding tissue was contained within the bounding box. The coordinates for the padded bounding box were used to select a patch from the T2-weighted image, ADC map, and high-b-value series, which were then saved as a three-channel JPEG image.
FIGURE 1:
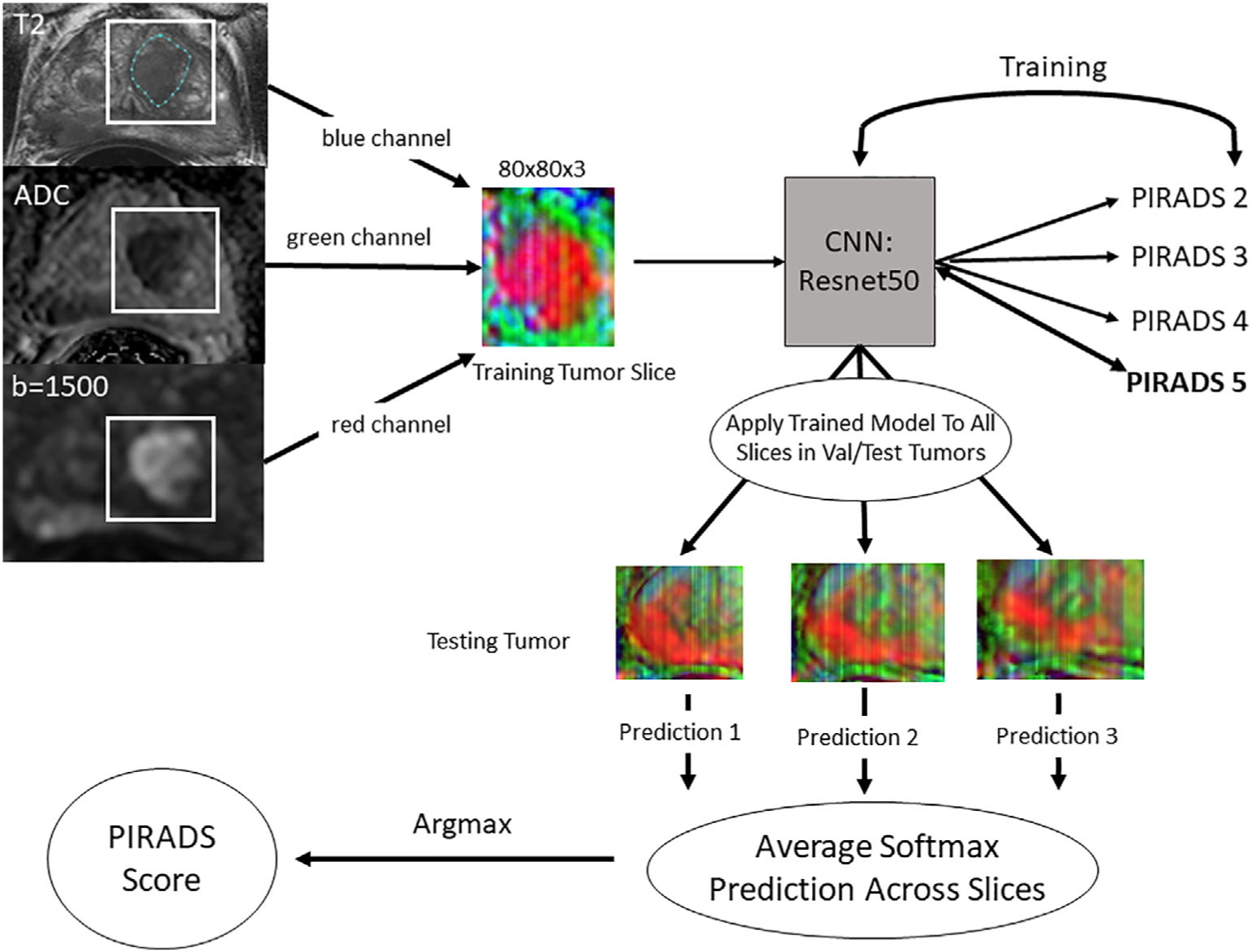
Workflow for data processing, model training, and per-lesion model application. The lesion was segmented on the T2-weighted axial series while viewing the corresponding ADC map and the high-b-value images. All three series were aligned so they were in the same physical space. The lesion segmentation was used to determine the maximum and minimum x and y values, then a bounding box was drawn around the lesion with 10-voxel padding. These cropped images from each series were placed into a three-channel array and saved as JPEGs. All images were resized to 80 × 80. The lesions were then split into 70/20/10 train/validation/test sets and slices were extracted from the training and validation datasets for model training. A Resnet 34 convolutional neural network (CNN) was trained using these slices. The fully trained model was then applied on a slice-by-slice basis for each lesion in the validation and test datasets and the predictions were averaged. The largest average score was then considered the PI-RADS score.
Model Training and Implementation
The lesion-level data from the three datasets were combined into a single dataset, which was then split randomly using a random number generator into three datasets on the patient level—training (70%, N = 482), validation (20%, N = 137), and test (10%, N = 68). For the training dataset, if a tumor segmentation spanned more than three slices, the first and last slices were discarded in the training dataset to allow for training on the most representative part of the tumor. All images were resized to 80 × 80 prior to training. A four-class (PI-RADS 2–5) convolutional neural network (CNN) was trained using the fastai library (https://github.com/fastai). A Resnet 34 architecture14 with weights initialized from a model pretrained on ImageNet15 was utilized for training with only the densely connected layers adjusted. The learning rate was set at 8.0 × 10−3. The batch size was 256. A label-smoothing loss function was utilized.16 Standard data augmentation with vertical flips, rotation up to 15°, warping up to 0.05, and lighting changes of 0.05 were utilized. An additional data augmentation strategy that inserts parts of one image into another called mixup was utilized.17 The model was trained on an NVIDIA Titan RTX GPU for a planned 50 epochs with the best model saved during the training process. The trained model was then applied to each slice in a lesion in the test and validation datasets and the softmax outputs for each PI-RADSv2 class were recorded. Softmax values were averaged across all MRI slices and the highest value was chosen as the per-lesion PI-RADS score.
Assessment of Interreader Agreement and Upgrading/Downgrading
In order to determine the interreader agreement from patients within this cohort, a random subsample of 50 patients was taken from the publicly available ProstateX dataset. A second body radiologist with a cumulative experience of 15 years, blinded to the purpose of the re-review, was shown the T2, ADC, and high b-value slices from the midpoint in the lesion without the lesion marked and was asked to assess the PI-RADS score of the lesion within the representative slice. Upgrading of the PI-RADS score was defined as an AI-generated PI-RADS score that was higher than the radiologist PI-RADS score. Downgrading was defined as an AI-generated PI-RADS score that was lower than the radiologist PI-RADS score. The cancer detection rate was defined as the rate of detection of any prostate cancer. Clinically significant cancer detection was defined as Gleason Grade 3 + 4 or higher.
Statistical Analysis
The performance of the AI PI-RADS model was assessed in two ways. First, the agreement of the deep-learning PI-RADS classification and the radiologist’s PI-RADS classification was calculated using a Cohen’s kappa and a weighted kappa score. The kappa values were interpreted relative to kappa scores in the literature. A total of 2000 bootstrap samples were then performed on the lesion level by random sampling to generate a 95% confidence interval. The second metric of performance was assessment of the relationship between radiologists-assessed PI-RADS score and the deep-learning model’s predicted PI-RADS scores on biopsy outcome. This was assessed using accuracy, defined as the percent of AI-generated PI-RADS scored that matched the radiologist-generated PI-RADS scores. We also assessed the percentage of the Al-generated score falling within one PI-RADS score of the radiologist-generated score. A Wald test was utilized to test for differences in cancer detection rates between radiologist-assigned PI-RADS score vs. the Al-assigned PI-RADS score, estimated from 2000 bootstrap samples on the lesion level with P < 0.05 considered statistically significant.
Code Availability
All code utilized for the development of the deep-learning model is available at https://github.com/NIH-MIP/semiautomated_PIRADS. A downloadable demo notebook including the trained model is available upon request.
Results
Cohort Characteristics
Available clinical characteristics are listed in Table 1. There were a total of 4130 slices with 345 PI-RADS 2 slices (8%), 994 PI-RADS 3 slices (24%), 1141 PI-RADS 4 slices (28%), and 1650 PI-RADS 5 slices (40%). Table 2 demonstrates the breakdown of patients and slices stratified by PI-RADS score for the training, validation, and test set.
TABLE 1.
Clinical Characteristics
| Cohort 1 (125 patients) | Cohort 2 (269 patients) | ProstateX (293 patients) | |
|---|---|---|---|
| Age (median, range) | 67 (46–81) | 68 (48–89) | 66 (48–83) |
| Weight, kg (median, range) | 85 (60–132) | 83 (31–146) | — |
| Whole prostate size, cc (median, range) | 38 (10–255) | 60 (21–265) | 54 (17–259) |
| Transition zone size, cc (median, range) | 17 (5–104) | 34 (3–217) | 31 (2–171) |
| Number lesions ≥PI-RADS2 | 221 | 414 | 399 |
| Endorectal coil use (N, %) | 95 (76) | 96 (36) | 0 (0) |
| Location | |||
| Right side (N, %) | 90 (40) | 151 (36) | 166 (41) |
| Left side (N, %) | 86 (39) | 217 (52) | 183 (46) |
| Midline (N, %) | 47 (21) | 47 (11) | 53 (13) |
| Zone | |||
| PZ (N, %) | 153 (69) | 272 (66) | 168 (42) |
| TZ (N, %) | 67 (30) | 134 (32) | 233 (58) |
| CZ (N, %) | 1 (<1) | 8 (2) | 1 (<1) |
| PZ + TZ (N, %) | 2 (<1) | 1 (<1) | 0 (0) |
| PI-RADS category | |||
| 2 (N, %) | 11 (5) | 67 (16) | 35 (9) |
| 3 (N, %) | 41 (18) | 122 (29) | 155 (39) |
| 4 (N, %) | 99 (44) | 169 (41) | 82 (20) |
| 5 (N, %) | 72 (32) | 57 (14) | 130 (32) |
TABLE 2.
Lesion and Slice Quantification of Training, Validation Test
| PI-RADS score | Training (70%) (482 patients) | Validation (20%) (137 patients) | Test (10%) (68 patients) | Total (687 patients) | ||||
|---|---|---|---|---|---|---|---|---|
| Lesion (N,%) | Slices (N,%) | Lesion (N,%) | Slices (N,%) | Lesion (N,%) | Slices (N,%) | Lesion (N,%) | Slices (N,%) | |
| 2 | 78 (11) | 229 (8) | 25 (12) | 80 (9) | 10 (11) | 36 (9) | 113 (11) | 345 (8) |
| 3 | 238 (33) | 735 (26) | 51 (24) | 165 (19) | 26 (27) | 94 (23) | 315 (30) | 994 (24) |
| 4 | 238 (33) | 777 (27) | 79 (37) | 259 (29) | 30 (32) | 105 (26) | 347 (34) | 1141 (28) |
| 5 | 173 (24) | 1094 (39) | 57 (27) | 381 (43) | 29 (31) | 175 (43) | 259 (25) | 1650 (40) |
| Total | 727 | 2835 | 212 | 885 | 95 | 410 | 1034 | 4130 |
Model Training and Validation
On a slice-by-slice basis, the accuracy of the AI-generated PI-RADS score compared with the radiologist PI-RADS score in the validation set was 58%. The fully trained model was then applied to all slices in each lesion in the validation and test sets and a lesion-based assessment was rendered. The agreement between the radiologist PI-RADS score and the AI-based PI-RADS classification is shown in Table 3. When the test set and validation set results were combined, there was agreement in 58% of lesions. Agreement was lowest for PI-RADS 2 lesions (6%) and highest for PI-RADS 5 lesions (80%). The kappa score was 0.40 (0.32–0.48), P < 0.001, reflecting moderate agreement. The weighted kappa score was 0.43. The upgrading and downgrading rates can be seen in Table 4—there were nearly twice as many tumors that were upgraded (85/307, 28%) compared with tumors that were downgraded (45/307, 15%). This is particularly notable in the PI-RADS 3 category, where there was a 44% rate of upgrading compared with a 4% rate of downgrading. When the evaluation was performed + one PI-RADS score, the correct classification rate was achieved in 86%.
TABLE 3.
Agreement Between AI-Generated PI-RADS Score and Radiologist-Generated PI-RADS Score for 307 Lesions in 205 Patients in the Validation and Test Sets
| Radiologist PI-RADS score | Validation | Test | Validation + test | |||
|---|---|---|---|---|---|---|
| Correct validation | Validation ± 1 | Correct test | Test ± 1 | Correct overall | Overall ± 1 | |
| 2 | 0/25 (0%) | 9/25 (36%) | 2/10 (20%) | 6/10 (60%) | 2/35 (6%) | 15/35 (43%) |
| 3 | 29/51 (57%) | 45/51 (88%) | 11/26 (42%) | 24/26 (92%) | 40/77 (52%) | 69/77 (90%) |
| 4 | 51/79 (65%) | 74/79 (94%) | 15/30 (50%) | 29/30 (97%) | 66/109 (52%) | 103/109 (95%) |
| 5 | 44/57 (77%) | 50/57 (88%) | 25/29 (86%) | 27/29 (93%) | 69/86 (80%) | 77/86 (90%) |
| Overall | 124/212 (38%) | 178/212 (84%) | 53/95 (56%) | 86/95 (91%) | 177/307 (58%) | 264/307 (86%) |
TABLE 4.
Upgrading and Downgrading of PI-RADS Score by Dataset
| PI-RADS score | Validation | Test | Validation + test | |||
|---|---|---|---|---|---|---|
| Upgraded | Downgraded | Upgraded | Downgraded | Upgraded | Downgraded | |
| 2 | 25/25 (100%) | 0/25 (0%) | 8/10 (80%) | 0/10 (0%) | 33/35 (94%) | 0/35 (0%) |
| 3 | 21/51 (41%) | 1/51 (2%) | 13/26 (40%) | 2/26 (8%) | 34/77 (44%) | 3/77 (4%) |
| 4 | 11/79 (14%) | 17/79 (22%) | 7/30 (23%) | 8/30 (27%) | 18/109 (17%) | 25/109 (23%) |
| 5 | 0/57 (0%) | 13/57 (23%) | 0/29 (0%) | 4/29 (14%) | 0/86 (0%) | 17/86 (20%) |
| Overall | 57/212 (27%) | 31/212 (15%) | 28/95 (30%) | 14/95 (15%) | 85/307 (28%) | 45/307 (15%) |
Interreader Agreement
Two radiologists agreed on the PI-RADS score in 25/50 cases (50%). The two readers agreed within 1 PI-RADS score in 38/50 cases (76%). The kappa score for agreement was 0.340 (95% confidence interval [CI] 0.17–0.51). Of those 25 cases classified as incorrect, 16/50 were upgraded (32%), while 9/50 were downgraded (18%).
Correlation With Pathology Results
Of the 307 lesions in the validation/test set, 188 were derived from in-house patients and the remainder were derived from prostateX patients. Of the lesions derived from patients studied at our institution, 86 lesions (46%) underwent an MRI/ultrasound fusion-guided biopsy within 6 months following MRI acquisition. The association between the radiologist- and Al-assigned PI-RADS scores and pathologic outcome of the targeted biopsies was determined. For the radiologist-assigned PI-RADS score, the rates of biopsy-positive clinically significant prostate cancer (Gleason 7 or above) was 0%, 50%, 40%, and 79% for PI-RADS 2, 3, 4, and 5, respectively. For the AI-derived PI-RADS scores, the rates of biopsy-positive clinically significant prostate cancer were 0%, 40%, 39%, and 85% for PI-RADS 2, 3, 4, and 5, respectively. There were no statistically significant differences between the AI-assigned and radiologist-assigned PI-RADS scores in the rates of clinically significant prostate cancer (P = 0.59 for PI-RADS 3, P = 0.36 for PI-RADS 4, and P = 0.47 for PI-RADS 5) (Table 5).
TABLE 5.
Model Performance: Cancer Detection Rate for 86 Patients Who Underwent Prostate Biopsy
| PI-RADS score | Radiologist | Deep-learning model | ||||
|---|---|---|---|---|---|---|
| N | Cancer detection (%) | Clinically significant cancer detection (%) | N | Cancer detection (%) | Clinically significant cancer detection (%) | |
| 2 | 4 | 50 | 0 | 1 | 0 | 0 |
| 3 | 14 | 57 | 50 | 15 | 53 | 40 |
| 4 | 40 | 60 | 40 | 44 | 61 | 39 |
| 5 | 28 | 89 | 79 | 26 | 92 | 85 |
Discussion
Multiparametric MRI has been shown to be useful in detecting tumors within the prostate by combining anatomic (T2-weighted, T2W) and quantitative (diffusion-weighted and dynamic contrast-enhanced) sequences.18 MRI provides anatomic localization of the tumor, which allows for the specific targeting of lesions during prostate biopsy.4 The addition of MRI has increased the detection of clinically significant cancer, although the magnitude of this increase varies.4,5 PIRADSv2 (and now PI-RADS v2.1) serves as a unifying and standardized scoring system across institutions to assess the risk of clinically significant prostate cancer given specific lesion morphology and appearance.7 However, the assignment of a PI-RADS score relies on individual assessments of qualitative attributes of lesions such as the determination of which T2W MRI lesions are heterogeneous (PI-RADS 3) vs. homogenous (PI-RADS 4). The subjective nature of these assessments leaves room for disagreement among clinicians and prior studies19,20have demonstrated substantial interreader and intrareader variability in PI-RADS classification, similar to what we experienced in the 50-patient experiment in our current study.10 This variability results in difficulty with consistently mapping PI-RADS scores to the risk of prostate cancer.11 In a prior multicenter study, experienced radiologists were shown the same multiparametric prostate MRI study 1 month apart and were asked to assign a PI-RADS score to detected lesions. Interestingly, these radiologists disagreed with their own diagnosed 15–40% of cases when asked to read the same scan a month later.10 Similar findings have been reported in additional studies.20,21 Considering that PI-RADS scores of MRI-detected lesions are routinely used to determine if a biopsy is to be performed, such inconsistencies hinder widespread use of MRI-derived information in prostate cancer care. An AI model, properly trained with accurate annotation, has the potential to produce a consistent PI-RADS score, which should elevate the intraobserver agreement,22 improving the confidence of clinicians in the results of MRI.
This study describes a deep-learning-based image classification AI system that assigns a PI-RADS score to a lesion detected and segmented by a radiologist. This system was trained on a slice-by-slice basis, then applied across all slices in the validation and test sets in order to obtain a PI-RADS score for each lesion. The agreement of the AI system with the expert radiologist was only in the moderate range (kappa = 0.40); however, this is within the range of previously reported inter- and intrareader agreements, which have been reported as low as 0.24 among multiple radiologists.10 The AI system performed better on PI-RADS 5 lesions than other PI-RADS scores. When the AI system predicted the PI-RADS score incorrectly, it often was within one PI-RADS score range of the expert radiologist’s PI-RADS score, leading to an 86% correct classification rate within 1 PI-RADS score error range.
Ultimately, the most important function of the PI-RADS score is to provide a consistent mapping between a lesion detected at MRI and clinically significant cancer on biopsy. While PI-RADS has been shown to have sensitivity (0.89) and specificity (0.73) for overall cancer detection,23 the rates of detection of clinically significant cancer have not been as impressive, specifically in lesions with a PI-RADS 4 score, indicating that a “clinically significant cancer is likely to be present.”24 In our study, we validated the results of Mehralivand et al25 on a different patient cohort, demonstrating a detection rate of clinically significant cancer in the minority (22.1%) of PI-RADS 4 lesions. Although the pathologic outcomes included in this study were based on TRUS/MRI fusion-guided targeted biopsy, which may miss or undersample the lesions, a notable finding from this study was that the rates of detection of clinically significant cancer within each PI-RADS category was similar between radiologist-assigned PI-RADS scores and AI-assigned PI-RADS scores. The discrepancy between the lack of agreement on PI-RADS scores overall and the presence of agreement in biopsy outcomes is likely the result of the ability of the system to categorize PI-RADS close to the correct score. Because the rates of clinically significant cancer are within 10% between a PI-RADS 3 vs. 4,24 this type of misclassification may not make a difference in relation to biopsy outcome. Providing a consistent method of classifying MRI findings to predict clinically significant cancer may allow treating physicians to have more confidence in making biopsy and treatment decisions using the PI-RADS scoring system. Provided an AI algorithm has been properly validated, the reproducible nature of AI-based PIRADS prediction may provide consistent correlation with pathological outcome.
Limitations
Our study has limitations. First, this AI system requires manual lesion segmentation, which makes the assumption that the lesion can be both accurately detected and delineated. Second, this study relied on a retrospective cohort of patients from two different institutions, and, therefore, patient selection was subject to unknown biases. The AI system has a possible tendency to upgrade PI-RADS categories of some lesions, which could potentially alter the need for biopsy. Additionally, we utilized PIRADSv2 in our study instead of PIRADSv2.1, which was just released while our study was still being conducted at the training and validation phases. Finally, the available pathologic outcomes of this study were based on targeted biopsy, not whole-mount pathology. Half of the patients in the validation/test sets did not undergo a prostate biopsy after the MRI, introducing a potential source of bias in the determination of biopsy outcomes. Including a cohort where all patients underwent radical prostatectomy may potentially bias the lesion population towards a relatively higher Gleason grade cohort.
Conclusion
We described an AI model that can assign a PI-RADS score to a lesion that is identified and segmented on a multiparametric prostate MRI. The AI system’s agreement with an expert radiologist was similar to intra- and interreader agreement in prior studies. There was no difference in detection of clinically significant prostate cancer within PI-RADS scores between the AI and radiologist PI-RADS classifications. An AI-based classification system may improve consistency in the PI-RADS classification system.
Supplementary Material
Acknowledgments
This project was funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This project was supported in part by the Intramural Research Program of the NIH.
Footnotes
Additional supporting information may be found in the online version of this article
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 2.Ankerst DP, Straubinger J, Selig K, et al. A contemporary prostate biopsy risk calculator based on multiple heterogeneous cohorts. Eur Urol 2018;74(2):197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehralivand S, Shih JH, Harmon S, et al. A grading system for the assessment of risk of extraprostatic extension of prostate cancer at multiparametric MRI. Radiology 2019;290(3):709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddiqui MM, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/-ultrasound fusion–guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA 2015;313(4):390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasivisvanathan V, Rannikko AS, Borghi M, et al. MRI-targeted or standard biopsy for prostate-cancer diagnosis. N Engl J Med 2018;378(19): 1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turkbey B, Choyke PL. PIRADS 2.0: What is new? Diagn Interv Radiol 2015;21(5):382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barentsz JO, Weinreb JC, Verma S, et al. Synopsis of the PI-RADS v2 guidelines for multiparametric prostate magnetic resonance imaging and recommendations for use. Eur Urol 2016;69(1):41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park SY, Jung DC, Oh YT, et al. Prostate cancer: PI-RADS version 2 helps preoperatively predict clinically significant cancers. Radiology 2016;280(1):108–116. [DOI] [PubMed] [Google Scholar]
- 9.Turkbey B, Rosenkrantz AB, Haider MA, et al. Prostate Imaging Reporting and Data System version 2.1: 2019 update of Prostate Imaging Reporting and Data System version 2. Eur Urol 2019;76(3): 340–351. [DOI] [PubMed] [Google Scholar]
- 10.Smith CP, Harmon SA, Barrett T, et al. Intra- and inter-reader reproducibility of PI-RADSv2: A multireader study. J Magn Reson Imaging 2019; 49(6):1694–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonn GA, Fan RE, Ghanouni P, et al. Prostate magnetic resonance imaging interpretation varies substantially across radiologists. Eur Urol Focus 2017;5(4):592–599. [DOI] [PubMed] [Google Scholar]
- 12.Greer MD, Choyke PL, Turkbey B. PI-RADSv2: How we do it. J Magn Reson Imaging 2017;46(1):11–23. [DOI] [PubMed] [Google Scholar]
- 13.Armato SG, Huisman H, Drukker K, et al. PROSTATEx challenges for computerized classification of prostate lesions from multiparametric magnetic resonance images. J Med Imaging 2018;5(04):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. arXiv:151203385 [cs]. 2015. Available from: http://arxiv.org/abs/1512.03385 [Google Scholar]
- 15.Deng D, Dong W, Socher R, Li L, Li K, Li F. ImageNet: A large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition 2009; 248–255. [Google Scholar]
- 16.He T, Zhang Z, Zhang H, Zhang Z, Xie J, Li M. Bag of tricks for image classification with convolutional neural networks. arXiv:181201187 [cs]. 2018. Available from: http://arxiv.org/abs/1812.01187 [Google Scholar]
- 17.Zhang H, Cisse M, Dauphin YN, Lopez-Paz D. mixup: Beyond empirical risk minimization. arXiv:171009412 [cs, stat]. 2018. Available from: http://arxiv.org/abs/1710.09412 [Google Scholar]
- 18.Turkbey B, Mani H, Shah V, et al. Multiparametric 3T prostate magnetic resonance imaging to detect cancer: Histopathological correlation using prostatectomy specimens processed in customized magnetic resonance imaging based molds. J Urol 2011;186(5):1818–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller BG, Shih JH, Sankineni S, et al. Prostate cancer: Interobserver agreement and accuracy with the revised prostate imaging reporting and data system at multiparametric MR imaging. Radiology 2015;277 (3):741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenkrantz AB, Ginocchio LA, Cornfeld D, et al. Interobserver reproducibility of the PI-RADS version 2 lexicon: A multicenter study of six experienced prostate radiologists. Radiology 2016;280(3):793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greer MD, Shih JH, Barrett T, et al. All over the map: An interobserver agreement study of tumor location based on the PI-RADSv2 sector map: Agreement on prostate mpMRI PI-RADS map. J Magn Reson Imaging 2018;48(2):482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greer MD, Lay N, Shih JH, et al. Computer-aided diagnosis prior to conventional interpretation of prostate mpMRI: An international multireader study. Eur Radiol 2018;28(10):4407–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woo S, Suh CH, Kim SY, Cho JY, Kim SH. Diagnostic performance of Prostate Imaging Reporting and Data System version 2 for detection of prostate cancer: A systematic review and diagnostic meta-analysis. Eur Urol 2017;72(2):177–188. [DOI] [PubMed] [Google Scholar]
- 24.Weinreb JC, Barentsz JO, Choyke PL, et al. PI-RADS prostate imaging – Reporting and data system: 2015, version 2. Eur Urol 2016;69(1):16–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehralivand S, Bednarova S, Shih Joanna H, et al. Prospective evaluation of PI-RADS™ version 2 using the International Society of Urological Pathology Prostate Cancer Grade Group System. J Urol 2017;198 (3):583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


