Abstract
Computational drug design relies on the calculation of binding strength between two biological counterparts especially a chemical compound, i.e. a ligand, and a protein. Predicting the affinity of protein-ligand binding with reasonable accuracy is crucial for drug discovery, and enables the optimization of compounds to achieve better interaction with their target protein. In this paper, we propose a data-driven framework named DeepAtom to accurately predict the protein-ligand binding affinity. With 3D Convolutional Neural Network (3D-CNN) architecture, DeepAtom could automatically extract binding related atomic interaction patterns from the voxelized complex structure. Compared with the other CNN based approaches, our light-weight model design effectively improves the model representational capacity, even with the limited available training data. We carried out validation experiments on the PDBbind v.2016 benchmark and the independent Astex Diverse Set. We demonstrate that the less feature engineering dependent DeepAtom approach consistently outperforms the other baseline scoring methods. We also compile and propose a new benchmark dataset to further improve the model performances. With the new dataset as training input, DeepAtom achieves Pearson’s R=0.83 and RMSE=1.23 pK units on the PDBbind v.2016 core set. The promising results demonstrate that DeepAtom models can be potentially adopted in computational drug development protocols such as molecular docking and virtual screening.
Keywords: binding affinity prediction, deep learning, efficient 3D-CNN, benchmarking
I. INTRODUCTION
Binding of a molecule to a protein may start a biological process. This includes the activation or inhibition of an enzyme’s activity, and a drug molecule affecting its target protein. The binding is quantified by how strong the chemical compound, a.k.a. a ligand, binds to its counterpart protein; this quantity is called binding affinity. Modeling of biological processes and computational drug design heavily rely on calculating this binding strength. As a practical example, we may have a target protein whose relevance to a disease has been experimentally confirmed. We would like to rank the different poses of a single ligand or a library of ligands when binding to this target protein. Molecular docking and Virtual Screening (VS) take advantage of the binding affinity score to achieve this discrimination. In other words, VS assigns a score to each binding ligand, indicating how strong it binds to the target protein. To get the overall picture of how binding affinity prediction enables VS, the reader is referred to [8], [16], [55]. In the past, VS has successfully assisted the design of drugs to treat a wide range of diseases, such as type 2 diabetes, malaria, and hepatitis B [38], [40], [41].
Current approaches for quantifying the binding affinity can be categorized as physics-based, empirical, knowledge-based and descriptor-based scoring functions [34]. In spite of their merits, the conventional techniques assume a predetermined functional form which is additive. Furthermore, they need domain knowledge to extract features and formulate the scoring functions. For example, the semi-empirical force field in AutoDock [39] and empirical scoring function in X-Score [53] belong to this category. As instance, X-Score takes average of three scoring functions HPScore, HMScore, and HSScore, differing by the terms which describe the hydrophobic interactions. Each of these scoring functions comes in the form of a linear combination of the terms [52]. These conventional techniques rely on experts’ insight on which phenomena and interactions make substantial contribution to binding affinity.
Only in the past decade the machine learning algorithms have been used to score the protein-ligand binding strength in a data-driven manner. Das et al. encoded molecular shapes and property distributions on protein and ligand surfaces as the input signatures to Support Vector Machine (SVM) models [11]. Ballester and coworkers gathered data about the frequency of occurrence of each possible protein-ligand atom pair up to a distance threshold. By binning these counts, they trained their Random Forest model implicitly on short-range, middle-range, and long-range interactions. The RF-Score model is still among the top performer models in this field [2], [3], [29], [30], [56]. Durrant and McCammon developed a series of neural network-based methods for binding affinity prediction. The input features were a combination of the count of short-range ligand-protein atom pairs, electrostatics sum over these atom pairs, different ligand atom types, and a ligand’s number of rotatable bonds [13]–[15]. The de Azevedo group implemented Taba, a tool to analyze the binding affinity based on representing a protein-ligand complex as a massspring system. Their program calculates average distances for different pairs of ligand-protein atom pairs up to different distance cutoff values. Taba then takes advantage of supervised machine learning to compute the weights for contribution of these interaction types. They report improved results compared to AutoDock 4 and Vina [4], [10]. The same research group also developed SAnDReS, a computational tool for statistical analysis of docking results and development of scoring functions [5], [6], [57]. Despite their merits, most classical machine learning models described above heavily rely on biological feature engineering to extract descriptors or fingerprints; it is still based on expert knowledge and therefore biased.
Deep learning models, which belong to descriptor-based category, aim to minimize the bias due to domain knowledge. To describe the interactions between a protein and its ligand, atom-centered and grid-based methods are the most widely used techniques. Schietgat et al. developed a 3D neighborhood kernel, which took the atom spatial distances into consideration, to describe the structure of proteins and ligands [45]. More recently Gomes and coworkers [17] represented the structures of proteins and ligands as a combination of neighbor lists and atom types, to later be used in a deep network. In their case, they described the whole protein and ligand structures using their atom-centered scheme.
By contrast, grid-based approach usually limits the representation of protein and ligand interactions to a grid box defined around a protein’s binding site, and different atom information is encoded in different channels of the 3D grid. This representation puts the voxels, not the atoms, at the center of focus, and calculates the contribution of adjacent atoms to the center of each voxel. Research has been carried out for both classification and regression applications based on protein-ligand binding affinity. Wallach et al. [50] and Ragoza et al. [42] developed CNN scoring functions to classify compound poses as binders or non-binders. Jiménez et al. [25] and Marta et al. [48] designed similar deep learning models to predict the binding affinity, based on the rasterized protein-ligand input structures.
In addition to modeling approach, data reliability is a major issue for binding affinity prediction. Although a few thousands of labeled protein-ligand complexes are available, their binding affinity are reported as different experimental measures, including Kd (dissociation constant, which defines how strong a ligand binds to a protein), Ka (association constant, which is the reciprocal of Kd), Ki (inhibitor constant, a measure of binding strength when an inhibitor binds to its target), and IC50 (half-maximal inhibitory concentration), in decreasing order of reliability for the purpose of binding affinity prediction. A relevant thermodynamic property is the Gibbs free energy of binding for protein-ligand complexes. It is related to the dissociation constant as ΔG = RT lnKd where R is the gas constant and T is temperature in Kelvin. The Gibbs free energy of binding can be used to estimate the binding of ligands to the binding sites of proteins [7], [12]. It should be noted that if we indiscriminately feed all data with different types of binding affinity to a machine learning model during training phase, it will potentially introduce label noise or even incorrect labels different from ground truth. The machine learning model may then suffer from the inaccurate supervision.
The model performance is limited by the training data. The impact of including low quality data on model’s prediction power has been under debate. One the one hand, it has been reported that the Random Forest (RF) model can get a boost from being trained on an expanded dataset which also includes low quality data; the authors then recommended not to limit training on a relatively small set of high quality samples [31]. On the other hand, another group of researchers reported that expanding the training set made no significant change in their model performance [25]. We previously showed that including low quality samples improved the generalization power of our models [43].
More recently, deep learning models have exhibited their powerful superiority in a broad range of bioinformatics tasks, such as gene mutations impact prediction [49], protein folding [32] and drug discovery [46]. By stacking many well-designed neural network layers, the final model is capable of extracting useful features from raw data form and approximating highly complex functions [28]. Many advanced deep learning algorithms are developed based on convolutional neural networks (CNNs). The impressive performance of CNNs is mainly because they can effectively take advantage of spatially-local correlation in the data. Similarly, protein-ligand 3D structure naturally has such characteristics; biochemical interactions between atoms occur locally. CNNs hopefully can hierarchically compose such local spatial interactions into abstract high-dimensional global features contributing to the binding score.
Our goal in this paper is twofold. First, we aim to develop an end-to-end solution for accurate prediction of binding affinity which 1) gets as input the 3D structural data for the complex form of a protein and ligand, 2) requires minimum feature engineering, and 3) achieves state-of-the-art prediction performance. Second, we aim to systematically analyze the publicly available protein-ligand binding affinity data and propose a new benchmark for more reliable model learning.
Herein, we propose the framework DeepAtom to accurately predict the protein-ligand binding affinity. The 3D structure of a protein-ligand complex is first rasterized into a 3D grid box, which is centralized on the ligand center in the protein binding site. Each voxel has several input channels, embedding the different raw information of atoms located around the voxel. Thus each voxel aggregates the information from its surrounding atoms to the corresponding channels, using the algorithm first described in [24], which we call PCMax algorithm throughout the text. A light-weight 3D-CNN model is then developed to hierarchically extract useful atom interaction features supervised by the binding affinity labels. As a data-driven approach, it effectively avoids a priori functional form assumptions. More importantly, our efficient architecture design significantly improves the model representational and generalization capacity even trained with the limited available data of protein-ligand complex structures, in this case a few thousand complexes. It also effectively reduces the amount of both computation and memory cost introduced by 3D convolution operations and speeds up the training process.
We present comprehensive experiments on the standard benchmark test set, called PDBbind v.2016 core set [51] and an additional test set, called Astex Diverse Set [18]. Randomly initialized and evaluated for 5 times, DeepAtom consistently outperforms the baseline state-of-the-art models studied here. In order to further improve the model performance, we also critically study the publicly available complex data and propose a new benchmark dataset. It further improves the DeepAtom performance, with potential benefits to the future research in the binding affinity scoring field. It has the capacity to be plugged into computational drug development protocols such as molecular docking and virtual screening. With this aim, we provide the benchmark dataset in the supplement.
II. MATERIALS AND METHODS
A. Input Featurization and Processing
1). Protein-ligand Complex:
The standard datasets, such as PDBbind and Binding MOAD, include the structures of protein and ligand in their bound form, a.k.a. their complex, deposited in a single PDB file. The strength of ligand binding to protein has been determined for each structure using experimental techniques such as isothermal titration calorimetry (ITC) and spectroscopic shift assays [27]. This binding affinity data is used as the ground truth labels for these protein-ligand complexes.
2). Grid Size & Resolution:
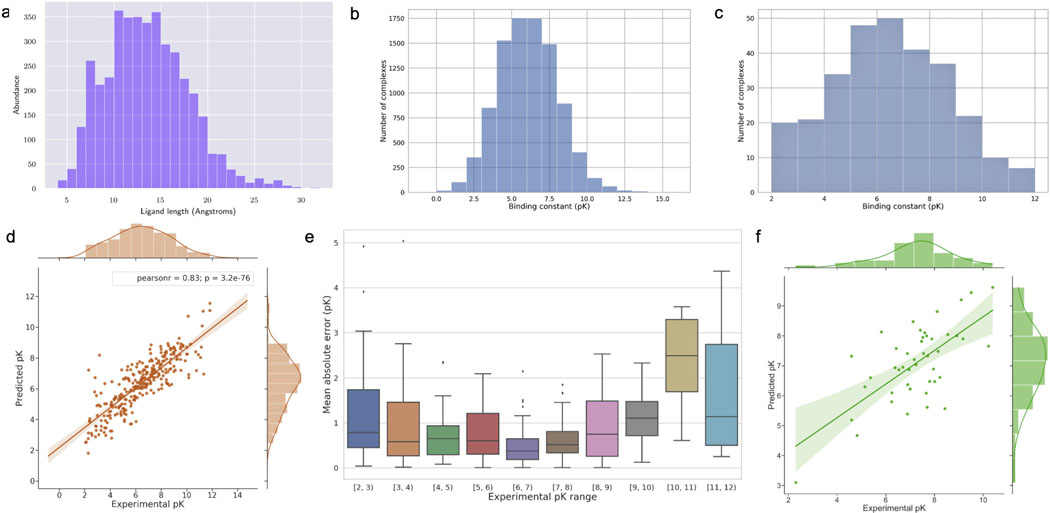
We calculate the distribution of end-to-end distances for all ligands in the PDBbind v.2016 refined and core datasets. This gives us clues to define a box size of 32 Å, which is the same as the end-to-end distance for the longest ligand in these two datasets, so there is no need to filter out any. If the dimension of the grid box was defined smaller than the length of a ligand, the terminal sides of the ligand might fall outside the box and the model would lose data relevant to those moieties. Even if the initial orientation of such a ligand allowed it to fit inside the grid box, later data augmentation would most certainly generate input structures with data loss. The distribution of ligand lengths in the PDBbind v.2016 refined and core subsets is illustrated in Fig. 3a.
Fig. 3:
a. Ligand length distribution in the PDBbind v.2016 refined and core sets. b. Binding data distribution of the training set in our proposed benchmark. c. Binding data distribution of the core set. d. DeepAtom prediction results for the core set. e. The distribution of MAE between DeepAtom prediction and target complexes with different binding ranges. f. DeepAtom prediction results for the Astex Diverse Set.
The van der Waals radius of the 9 major heavy atoms (C, N, O, P, S, F, Cl, Br, I) used in our study is greater than 1.4 Å. As a simplified view, an atom’s rvdw can be assumed as a measure of its size; it is defined as half of the internuclear separation of two non-bonded atoms of the same element on their closest possible approach. A grid resolution larger than 2 × rvdw cannot differentiate two atoms from each other. On the other hand, a finer resolution brings about much higher computational cost. As a trade-off between accuracy and efficiency, we set the grid resolution as 1.0 Å.
3). Features / Atom Types:
A difference between deep learning in computer vision and structural biology applications is the use of RGB channels as a standard in the former where the model can directly consume them. However, in our case, there is no consensus on the channels for representing protein and ligand structures. We used 11 Arpeggio atom types, based on the potential interactions each atom may get involved in [26]; they include features such as Hydrogen bond acceptor and donor, positive or negative, hydrophobic, and aromatic atom types. These properties are similar to pharmacophoric features in medicinal chemistry [54]. The protein and ligand atoms are described by 11 Arpeggio atom types and an excluded volume feature, where discrimination is made between protein and ligand atoms. This resulted in (11 + 1) × 2 = 24 features.
4). Occupancy Type:
This hyper-parameter defines how each atom impacts its surrounding environment. In our work, each atom can affect its neighbor voxels up to double of its van der Waals radius rvdw through a pair correlation potential. We use the Atom-to-voxel PCMax algorithm, described in [24], where each atom makes a continuous contribution n(r) to its neighbor voxels as defined by Eq. 1. At the center of a voxel, only the maximum effect from contributing atoms is kept.
| (1) |
B. Network Architecture
To extract the atomic interaction information from the voxelized protein-ligand complex data, a straightforward approach is to extend 2D-CNNs to 3D by using 3D convolution kernels. One channel of the output feature map at the location (i, j, k) is computed by the standard convolution as follows:
| (2) |
where h represents the input with M channels, represents one filter weight, and S, T, R are side length of the filter. However, 3D convolution itself will massively inflate the amount of trainable network parameters, due to the increase in the input and kernel dimensions. Specifically, if N is the number of output channels, one standard convolution layer will introduce S · T · R · M · N parameters. More importantly, in order to improve the learning ability and achieve higher prediction accuracy, a general trend has been to make the model deeper and more complicated [47], [19], [23]. This in turn necessitates a higher requirement for a large scale high-quality training dataset. By contrast, for the affinity prediction problem, only a few thousands of protein-ligand complexes with experimentally determined binding affinity data are available. This issue discourages the use of network architectures with too many trainable parameters, because overfitting is likely to occur when the network has high complexity whereas a relatively small data set is available for training. Indeed, another 3D CNN-based affinity prediction work [42] also encountered the overfitting issue. After empirically optimizing the model depth and width, they ultimately reduced the network to only three convolutional layers. Similarly, Pafnucy [48] was developed as a 3D CNN model with three convolutional and three dense layers.
Our model is inspired by the light-weight network architectures, and aims to achieve the best trade-off between the prediction accuracy and the model complexity in terms of learnable parameters. A series of related network structures have been recently proposed, such as Xception [9], MobileNet v1 [20] and v2 [44], ShuffleNet v1 [58] and v2 [35], and CondenseNet [22]. Based on the practical guidelines for efficient CNN architecture design and the corresponding ShuffleNet units described in [35], we propose a novel light-weight 3D CNN model, which can be effectively trained with deeper layers by the limited training samples. It improves the prediction performance by a large margin, but does not significantly increase model complexity.
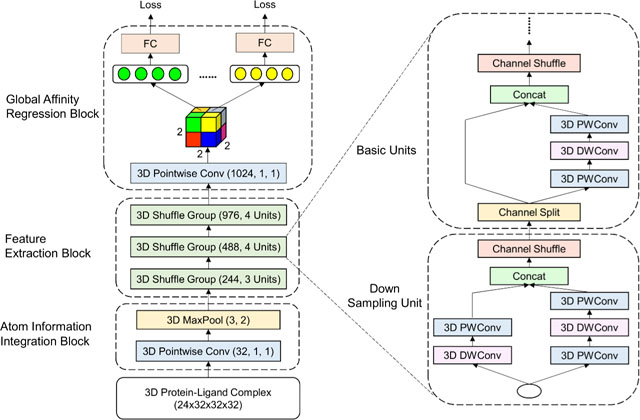
Specifically, as shown in Fig. 2 our model consists of three building blocks, namely atom information integration block, stacked feature extraction block, and global affinity regression block.
Fig. 2: Network architecture.
Each Conv layer is specified by its number of channels, kernel size and stride. The 3D MaxPool layer has kernel size 3 and stride 2. For the 3D Shuffle Groups, the numbers in parentheses denote the number of output channels and repeat time of the unit. Only the first unit has down sampling layer, where the DWConv layer has kernel size 3 and stride 2. In the remaining units, DWConv with kernel size 3 and stride 1, as well as PWConv with kernel size 1 and stride 1 are utilized. Eight losses are calculated based on the shared weights FC layer output. Two dropout layers are appended before the last 3D Pointwise Conv and FC layers respectively.
In the atom information integration block, a pointwise (PW, 1 × 1 × 1) convolution layer defined in Eq. 3 with non-linear activation function is first utilized to fuse the atom information across different channels.
| (3) |
This cascaded cross-channel parametric pooling structure brings about an improvement compared to the empirical scoring functions. For instance, AutoDock software’s scoring function is composed of a linear combination of interaction types, such as Hydrogen bonding and electrostatic interactions [39]. The pointwise convolution layer in our model is followed by a 3D max pooling layer to increase the translational invariance of the network and reduce the input dimension. The output of this block has the grid size of 16 × 16 × 16.
The feature extraction block consists of multiple consecutive 3D shuffle units, and according to the number of channels in their outputs, they are categorized into three groups. At the beginning of the unit, a channel split operator equally splits the input of feature channels into two branches. Data in one branch is sequentially processed by a pointwise convolution, a 3 × 3 × 3 depthwise (DW) convolution and an additional pointwise convolution. All three layers have the same number of input and output channels N. Depthwise convolutional layer performs the spatial convolution independently over every channel of an input:
| (4) |
where ⊙ denotes the element-wise product. Although depthwise convolution does not combine different input channels, the two neighbor regular pointwise convolutions effectively fuse the information across the channels. The other branch is kept as identity until it is concatenated with the output from the first branch. This identity branch can be regarded as an effective feature reuse design, which strengthens feature propagation and reduces the number of parameters. This strategy is inspired by ResNet model where the shortcut connections enable the model circumvent the vanishing/exploding gradient problem [19]. Within a basic unit, the depthwise and pointwise convolutions respectively introduce and parameters. Therefore, using a basic unit to replace the standard convolution, we obtain the parameter reduction as follows:
| (5) |
DeepAtom uses 3 × 3 × 3 depthwise convolutions and the number of channels are set as 244, 488, and 976. Therefore, with the efficient model design, we can easily obtain more than 20 times parameters reduction, which enable us to stack deeper layers to improve the model learning capacity.
At the end of the units, the channel shuffle operation is applied to enable the information flow across the two branches. Particularly, the channel shuffle operation first divides the feature map in each branch into several subgroups, then mixes the branches with different subgroups, as inspired by [58]. When the spatial down sampling is applied, the channel split operator in the shuffle unit is removed, and the number of output channels is doubled. In each group, only the first shuffle unit has the down sampling layer, and the remaining units keep the input dimension.
After stacking three shuffle groups, the original 3D input data is down sampled to a 1024 × 2 × 2 × 2 4D tensor (3 grids along x, y, z axes and 1024 channels). The global affinity regression block first slices the tensor into 2×2×2 = 8 vectors with dimension 1024. Based on the prior shuffle groups, the receptive field of each vector covers the entire raw volume, so we set up the affinity prediction task for each vector. A shared weights fully connected (FC) layer consumes each vector to construct regression loss, and it enables us to train the top layers more thoroughly and further avoid overfitting. In testing phase, outputs from the multiple hidden vectors are averaged right before the FC layer to stabilize the prediction.
In the architecture, we adopt the leaky rectified linear unit as the activation function. A batch normalization layer is appended after each convolution operation to speed up the training. The mean squared error is set as our affinity regression loss for model learning.
1). Training:
The model is updated by Adam algorithm with default parameters for momentum scheduling (β1 = 0.9, β2=0.999). Training the model from scratch, we set the initial learning rate as 0.001 and the weight decay to 4 × 10−5. Our model is implemented using PyTorch (version 0.4). With batch size of 256, the model is trained for around 60 epochs on 2 Nvidia P40 GPU cards.
2). Data Augmentation:
The publicly available biological datasets contain only thousands of complexes with reliable experimental binding affinity value. Directly training on these insufficient samples easily makes the deep learning model suffer from the overfitting problem. Data augmentation is proved as an effective approach to enhance the deep learning model performance. In our experiments, each of the original samples gets randomly translated and rotated. To ensure the ligands stay inside the grid box, the center of grid box is limited to move up to 0.5 Å in an arbitrary direction. Enabling such transformations significantly improves the training and model capacity. In order to reduce the variance, the augmented samples of each protein-ligand complex are averaged during the prediction phase.
C. Dataset Preparation
1). PDBbind Dataset:
PDBbind is the standard dataset for developing models to predict the binding affinity of protein-ligand complexes [51]; it has three subsets, namely core, refined, and general. The general subset includes complexes with relatively lower quality; it contains lower resolution structures, and the experimental affinities for some structures are reported as IC50 values. The refined dataset is a higher quality subset of the general dataset. It includes complex structures with better resolution and excludes any complex with IC50 data only; IC50 is a less preferred experimental measure of binding affinity due to its dependence on the concentration of both protein and ligand. More specifically, the refined subset excludes NMR-resolved structures; it contains complexes with resolution better than 2.5 Å and R-factor values lower than 0.250. It also filters out complexes where the ligand has any missing fragments or the protein binding site has any missing backbone or side chain fragments. In addition, it rejects complexes with extreme binding affinity values (Kd or Ki > 10 mM; Kd or Ki < 1 pM). To avoid complicated cases, the ternary complexes are also removed, such as when a substrate binds in vicinity of a cofactor [33]. In total, PDBbind v.2016 refined dataset includes 4057 complexes. The core dataset is a subset of refined data, clustered with a threshold of 90% sequence similarity; five representative complexes are picked for each of the 58 protein family clusters in order to cover the affinity range better. This results in 290 complexes in the core subset, which serves as the standard test set to evaluate the scoring approaches. We further split the rest of 3767 non-overlapping complexes between the refined and core into two disjoint subsets: (i) 10% of complexes (377) are randomly selected and used as the validation set, (ii) the rest (3390 complexes) are used for training, which is named as “training set-1”.
2). Proposed Benchmark Training Set:
In order to compile an improved benchmark dataset, we use PDBbind data as well as a complementary source of protein-ligand binding affinity data, namely Binding MOAD [1], [21]. In order to incorporate the recently updated complexes, we start from PDBbind v.2018 dataset, and extract all complexes with either Kd, Ka, or Ki data from general and refined subsets. It is worth noting that we exclude the complexes shared with the core subset to prevent the data leakage. We follow the same steps with Binding MOAD data. A few filtration steps are also necessary: first if a complex has reported Kd/Ka data in one database and Ki in the other, we keep the Kd/Ka data only. Second, complexes with a peptide as their ligand are discarded. We do not filter the complexes based on their structure resolution nor perform any clustering on them in terms of protein sequence or structure; clustering is typically done to later reduce the dataset into representative samples. The limited availability of the experimental affinity data discourages further removal of samples, although the dataset is biased towards some structures, e.g. the congeneric series. These are the 3D structures in Protein Data Bank where they share the same protein, complexed with different ligands.
In total, the final benchmark dataset contains 10 383 complexes. Please note that in contrast to NMR structures which contain multiple 3D models, a PDB file from X-ray crystallography contains a single 3D structure only. Our proposed benchmark dataset includes almost exclusively X-ray structures, with only one structure existing in each PDB file. Merely 63 complexes come from NMR experiments. We get only the first model from these PDB files.
As mentioned earlier, there is a debate over how the inclusion of lower quality data in training affects the model performance. Different groups have reported controversial results [25], [31]. Our previous work demonstrated an improvement in model performance when we did not try to limit the dataset based on its quality [43]. Therefore, we do not filter based on the structure resolution nor complexes with Ki data. Also, we do not perform any clustering on the proposed benchmark dataset in terms of protein sequence or structure.
Compared with the refined subset of PDBbind, this dataset almost doubles the number of samples with Kd/Ka/Ki data and is expected to improve the performance of binding affinity scoring techniques. The full list of the proposed benchmark dataset for model training is provided in the Supplementary Table1. The pKd/pKa/pKi value for each complex is reported to make it easier for other researchers to use the proposed dataset. The binding score of the complexes in this dataset ranges from −0.15 to 15.22 in pk units, and the score distribution is shown in Supplementary Fig. S1.
To train the scoring approaches, we split the proposed benchmark dataset into two subsets: (i) we randomly select 1000 samples from non-overlapping complexes between PDBbind v.2016 refined and core sets. (ii) the rest (9383 complexes) are for training, named as “training set-2”.
3). Astex Diverse Set:
This dataset was developed in 2007. It includes 85 protein-ligand complexes filtered to be of interest specifically to pharmaceutical and agrochemical industries [18]. Among these 85 complexes, 64 of them include binding affinity data. To avoid data leakage, we remove 19 complexes which are shared between Astex and our training set-2.
D. Other Methods for Comparison
In Section III, we compare DeepAtom with three state-of-the-art and open-source scoring approaches: Pafnucy model [48], RF-Score [3], and X-Score [53]. For Pafnucy and RF-Score, we use the open-source codes provided by the authors, and use their suggested hyper-parameters to re-train models on the same datasets as DeepAtom. For X-Score, we take the results from paper [25], where the authors used the publicly available binaries to make predictions on the same PDBbind v.2016 core set.
III. RESULTS & DISCUSSION
In this section, we describe the training and benchmarking protein-ligand complex data for DeepAtom. The evaluation details are presented along with discussion of the results.
A. Evaluation Metrics
To comprehensively evaluate the model performance, we use Root Mean Square Error (RMSE) and Mean Absolute Error (MAE) to measure the prediction error, and use Pearson correlation coefficient (R) and standard deviation (SD) in regression to measure the linear correlation between prediction and the experimental value. The SD is defined in Eq. 6.
| (6) |
where xi and yi respectively represent the predicted and experimental values of binding affinity for the ith complex; a and b are the intercept and the slope of the regression line, respectively.
B. Model Comparison with “Training Set-1”
We first train DeepAtom, RF-Score and Pafnucy on the “training set-1” with 3390 complexes described in Section II-C1, and evaluate them on the PDBbind v.2016 core set, which is unseen to the model during its training and validation. Each approach is randomly initialized and evaluated for 5 times. The mean and the standard deviation (in the parentheses) of the four evaluation metrics are presented in Table I for testing, and Supplemental Table S1 for validation. Learning with the very limited samples, DeepAtom outperforms the similar 3D CNN-based Pafnucy by a large margin, which demonstrates that our light-weight architecture design enables effective training with the deep layers and significantly improves its learning and generalization capacity. Our improved results are also likely to come from using a moderate level of chemical details in the features. As shown by Ballester et al. in their paper [3], the incorporation of sophisticated features such as Sybyl atom types indeed reduces the model performance, compared to moderate elements atom types. Our previous research confirms this too [43]. In addition to their atom types, Pafnucy adds more complicated features such as atomic partial charges and atom hybridization, somewhat similar to Sybyl atom types. The charge contributions might be inferred in our model from the Arpeggio atom types where the features also indicate the potential type of electrostatic interactions. We believe the inclusion of more sophisticated features might have indeed lowered the performance of Pafnucy model.
TABLE I:
Results on PDBbind v.2016 core set with “training set-1”. In each table cell, mean value over five runs is reported as well as the standard deviation in parentheses.
| RMSE | MAE | SD | R | |
|---|---|---|---|---|
| DeepAtom | 1.318 (0.212) | 1.039 (0.016) | 1.286 (0.015) | 0.807 (0.005) |
| RF-Score | 1.403 (0.002) | 1.134 (0.003) | 1.293 (0.002) | 0.803 (0.001) |
| Pafnucy | 1.553 (0.031) | 1.261 (0.027) | 1.521 (0.037) | 0.722 (0.017) |
On the other hand, DeepAtom achieves the comparable performance with the conventional machine learning method RF-Score, although as a practical guideline, training a supervised deep learning model generally requires larger datasets. It suggested that our model has greater potential to provide more accurate prediction, given enough training data.
C. Model Comparison with “Training Set-2”
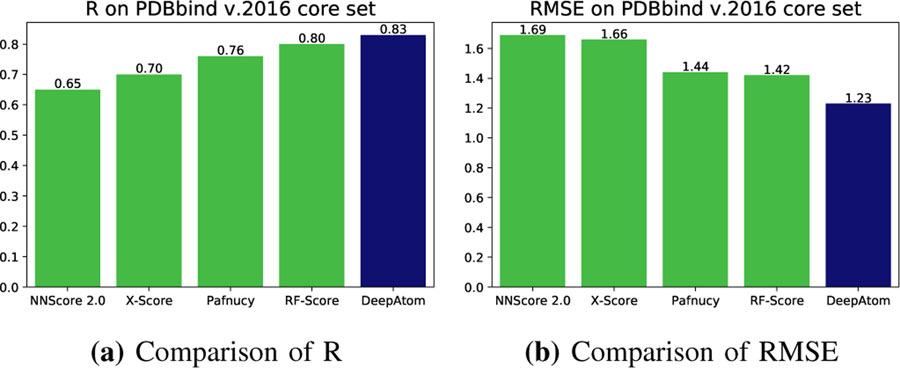
Next, we use our proposed “training set-2” to re-train DeepAtom, RF-Score and Pafnucy models, as well as the NNScore 2.0 model [13]–[15]. Then we evaluate them on the PDBbind v.2016 core set. Similarly, 5 different runs are conducted for the first three scoring methods to stabilize the results. For NNScore 2.0 model, 20 neural networks are trained on the input complexes and the final prediction takes average over all outputs. Fig. 4 shows the comparison of results in terms of mean value of the R and RMSE, also including the X-Score prediction results. Table II presents the detailed results of the mentioned methods. As shown, DeepAtom outperforms all the other approaches across all four measurements by a large margin. It achieves the best Pearson correlation coefficient of 0.83 and RMSE of 1.23 in pK units, compared with RF-Score results: R = 0.80 and RMSE = 1.42, Pafnucy results: R = 0.76 and RMSE = 1.44, and NNScore results: R = 0.65 and RMSE = 1.69. To the best of our knowledge, DeepAtom achieves the state-of-the-art performance on this well-known benchmark. The corresponding validation results are shown in the Supplementary Table S2.
Fig. 4:
Comparison of scoring methods on PDBbind v.2016 core set.
TABLE II:
Results on PDBbind v.2016 core with “training set-2”.
| RMSE | MAE | SD | R | |
|---|---|---|---|---|
| DeepAtom | 1.232 (0.011) | 0.904 (0.019) | 1.222 (0.011) | 0.831 (0.003) |
| RF-Score | 1.419 (0.002) | 1.124 (0.001) | 1.304 (0.002) | 0.801 (0.000) |
| Pafnucy | 1.443 (0.021) | 1.164 (0.019) | 1.424 (0.022) | 0.761 (0.008) |
| NNScore 2.0 | 1.692 | 1.323 | 1.656 | 0.648 |
Fig. 3d shows the correlation between the prediction results of one DeepAtom model and the experimental binding affinity data. DeepAtom gives the highly correlated prediction on the PDBbind v.2016 core set. To further investigate the model performance on different ranges of the binding data, we visualize the binding affinity distribution of the training set (Fig. 3b) in the proposed benchmark, PDBbind v.2016 core set (Fig. 3c) and the corresponding DeepAtom RMSE value within each pK unit range (Fig. 3e). From Fig. 3b and 3c, we observe that the binding scores of our training samples are intensively located in the middle range (from 3 to 9 pK units) which is highly similar to the core set. Fig. 3e shows that DeepAtom obtains better prediction results with lower MAE values in this middle range, compared to the less frequent binding scores. For a data-driven approach, the distribution of training data plays a crucial role in its performance. Because the number of training samples falling in the middle range is much larger than the samples with marginal affinity values, DeepAtom naturally performs better for the complexes in the middle range during the testing stage. It also suggests that diversifying the training samples is promising for DeepAtom to provide more accurate and reliable predictions. Furthermore because the compounds of interest during computational drug design and lead optimization usually lie in this middle range, these graphs give us more confidence in model predictions in real-world applications.
We also compare DeepAtom with the RF-Score, Pafnucy, and NNScore models on the independent Astex Diverse Set. Table III shows that DeepAtom again significantly outperforms the others over all the measurements. The prediction results averaged over 7 DeepAtom models are illustrated in Fig. 3f.
TABLE III:
Results on Astex Diverse Set with “training set-2”.
| RMSE | MAE | SD | R | |
|---|---|---|---|---|
| DeepAtom | 1.199 (0.061) | 0.913 (0.047) | 1.152 (0.037) | 0.651 (0.028) |
| RF-Score | 1.228 (0.007) | 0.946 (0.011) | 1.218 (0.007) | 0.598 (0.006) |
| Pafnucy | 1.368 (0.120) | 1.095 (0.117) | 1.300 (0.074) | 0.509 (0.081) |
| NNScore 2.0 | 1.509 | 1.142 | 1.310 | 0.510 |
D. Evaluation of the Proposed Benchmark Dataset
Comparison of Tables I and II reveals that training on our proposed benchmark dataset results in a significant improvement of the model performances, especially for the deep leaning based approaches. For example, DeepAtom increases the R from 0.81 to 0.83 and decreases the RMSE from 1.32 to 1.23. The difference between the two tables confirms the effectiveness of our proposed new benchmark dataset, where the model trained on our new dataset provides more accurate predictions. Although the dataset contains some complexes with low resolution structure, such low-quality data does not introduce obvious label noise. On the contrary, this extended dataset provides more reliable complex data which can effectively improve the generalization power of binding affinity prediction algorithms.
It is worth noting that our proposed benchmark dataset extends the standard refined set by including the complexes from PDBbind general subset and Binding MOAD database only when the experimental affinity data is either Kd, Ka, or Ki. While Kd and Ki as equilibrium constants may be compared if derived from multiple binding assays, dependence of IC50 values on experimental settings discourages its comparison across different assays [33].
E. Hyper-parameter Optimization
Although DeepAtom is an end-to-end data-driven approach for binding affinity prediction, some hyper-parameters are inevitably introduced especially in the input featurization process. To finalize the optimal data representation of protein-ligand complex for DeepAtom prediction, we implement systematic optimization experiments over the related hyper-parameters. In all of following comparison experiments, our deep learning models are trained on the “training set-1” with 3390 complexes, and evaluated on the corresponding validation set with 377 complexes; the prediction performance is measured by Pearson’s R and RMSE.
1). Feature/Atom Types:
We consider three different descriptors with 11, 24, and 60 features. The first descriptor characterizes both the protein and ligand atoms with the same 11 Arpeoggio atom types. The second descriptor is described in Section II-A3. The third descriptor includes 40 ANOLEA atom types to describe protein atoms and 9 heavy-atom element types to describe ligand atoms, in addition to the 11 Arpeggio atom types. The ANOLEA atom types describe each protein atom based on its bond connectivity, chemical nature, and whether it belongs to side-chain or backbone of the amino acid [36] [37]. This provides a fine grained representation of protein atoms with more information about the local bonded neighbors of each atom than pharmacophoric features. For the rest of controlled variables, we use the simple binary scheme to represent the occupancy types, and set the resolution of 3D grid box as 1.0 Å. From Table IV, we can see that when both protein and ligand atoms are treated the same (the first descriptor), a lower performance is obtained; training the models on PDBbind dataset needs extracting the structures of free protein and free ligand from the complex, assuming that the conformational change upon ligand binding is negligible. Therefore, binding affinity prediction relies on the inter-molecular interactions between protein and ligand atoms, while the intra-molecular energies are cancelled out. In this case, ignoring the distinction between protein and ligand atoms makes it difficult for the network to recognize these crucial protein-ligand inter-molecular interactions.
TABLE IV:
Validation performance with various feature/atom types.
| Num of Features | Resolution | Occupancy | RMSE | R |
|---|---|---|---|---|
| 11 | 1.485 | 0.706 | ||
| 24 | 1.0 | Binary | 1.360 | 0.737 |
| 60 | 1.359 | 0.737 |
2). Resolution:
High-resolution rasterized data can adequately capture the fine-grained features and changes in the local spatial regions. However, it will cause excessive memory usage and heavy computational cost. Thus, there exists a trade-off between prediction performance and computational efficiency. Based on our analysis, we pick the resolution as 1.0 Å and 0.5 Å, both of which are less than the smallest 2 × rvdw value of 1.4 Å for the 9 major heavy atoms. Table V shows that with an increase in resolution, DeepAtom prediction performance improves. However, this slight improvement comes with a large increase in the computational cost, especially when the more demanding occupancy strategy such as PCMax is utilized later; therefore we select 1.0 Å as the optimal resolution value.
TABLE V:
Validation performance with different resolutions.
| Num of Features | Resolution | Occupancy | RMSE | R |
|---|---|---|---|---|
| 24 | 0.5 | Binary | 1.357 | 0.739 |
| 1.0 | 1.360 | 0.737 |
3). Occupancy Type:
Occupancy type describes how each atom impacts its surrounding environment. Several different strategies have been proposed, such as binary, Gaussian [42] and PCMax [24]. The binary occupancy discretizes an atom’s impact over the voxel. For example, if the distance between an atom and a voxel center is shorter than the atom’s van der Waals radius, the corresponding voxel channel will be activated as 1, otherwise deactivated as 0. In contrast, the Gaussian and PCMax approaches can represent an atom’s impact by a continuous numerical value, which can contain richer information. The impact can also decay smoothly when the distance increases. We compare the binary and PCMax occupancy types, on the basis of the optimal 24 feature/atom types and 1.0 Å grid resolution, where the cutoff distance for binary and PCMax strategies is set to rvdw and 2×rvdw, respectively. Table VI shows that DeepAtom with PCMax occupancy type achieves better performance. Considering the similarity between Gaussian and PCMax algorithms, we expect them yield comparable results.
TABLE VI:
Validation performance with different occupancy types.
| Num of Features | Resolution | Occupancy | RMSE | Pearson’s R |
|---|---|---|---|---|
| 24 | 1.0 | Binary | 1.360 | 0.737 |
| PCMax | 1.348 | 0.741 |
4). Averaging at the Testing Time:
As an effective strategy, data augmentation is also used to improve the DeepAtom performance. In addition to augmenting data for training, we also run the trained model on multiple augmented versions of test data and average the results to reduce the prediction variance. We evaluate multiple test data versions, including 1, 12 and 24, where the value 1 means only the original test set is used without the averaging operation. We observe that increasing the test set versions can favorably reduce the variance of predictions and further improve the performance.
IV. CONCLUSION
In this paper, we proposed the framework DeepAtom to accurately predict the protein-ligand binding affinity. An efficient 3D-CNN architecture is proposed to effectively improve the model learning capacity with limited available complexes data for training. In a purely data-driven manner without a priori functional form assumptions, DeepAtom outperforms the studied baseline state-of-the-art deep learning, machine learning and conventional scoring techniques. We also proposed a new benchmark dataset to further improve the model performance. The promising results on independent challenging datasets demonstrated DeepAtom can be potentially adopted in computational drug development protocols such as molecular docking and virtual screening.
Supplementary Material
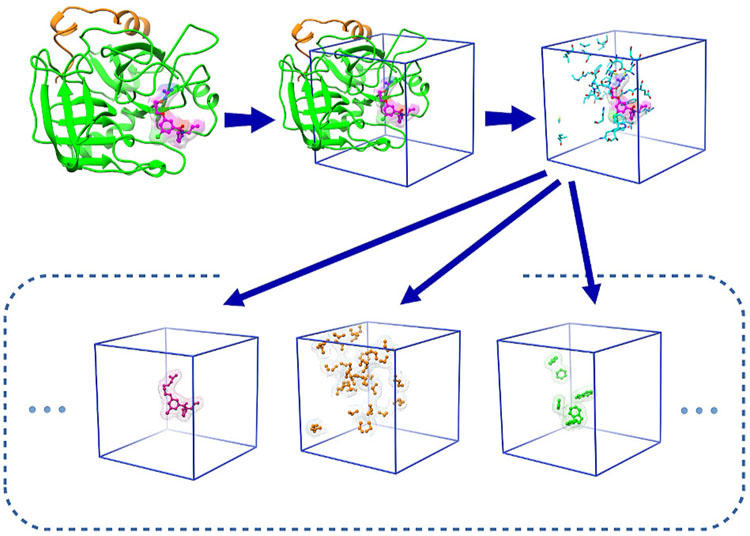
Fig. 1: Local box featurization (3D data representation).
The grid box encompasses the area around the binding site, centered on the ligand. Each channel includes only a specific feature, e.g. from left to right, the three channels shown are the excluded volume channel for the ligand as well as the hydrophobic and aromatic channels for protein. Each sample is described in terms of 24 channels in total.
V. ACKNOWLEDGMENTS
This work is partially supported by National Science Foundation (CNS-1842407) and National Institutes of Health (R01GM110240; R01NS088437; R01CA212403). We thank Yaxia Yuan for helpful discussions and comments that improved the manuscript. We also thank the anonymous reviewers for helpful suggestions and comments that helped improve the paper.
Biography

Mohammad A. Rezaei received his B.Sc. degree in Physics from AmirKabir University of Technology, Tehran Polytechnic, Tehran, Iran, in 2005 and his Master’s degree in Biophysics from the Ohio State University, Columbus, Ohio, in 2016. He received his Ph.D. in Computational Chemistry from the University of Florida in Gainesville, Florida in 2019. Currently he is a postdoc fellow in Simulation & Modeling Sciences, Pfizer R&D in Cambridge, Massachusetts. His current research interests include computational drug discovery, structure- and ligand-based drug design, cheminformatics, machine learning, deep learning, and generative models in chemistry.

Yanjun Li received the B.E. degree from the University of Electronic Science and Technology of China (UESTC), Chengdu, China, in 2012, and the dual M.S. degrees from UESTC, and Waseda University, Japan, in 2015. He is currently pursuing a Ph.D. degree with the Department of Computer and Information Science and Engineering, University of Florida, Gainesville, FL, USA. His current research interests include machine/deep learning, drug discovery, and smart healthcare.

Dapeng Wu (S’98–M’04–SM’06–F’13) is a professor at the Department of Electrical and Computer Engineering, University of Florida, Gainesville, FL. His research interests are in the areas of machine learning, networking, communications, signal processing, computer vision, smart grid, and information and network security. He received University of Florida Term Professorship Award in 2017, University of Florida Research Foundation Professorship Award in 2009, AFOSR Young Investigator Program (YIP) Award in 2009, ONR Young Investigator Program (YIP) Award in 2008, NSF CAREER award in 2007, the IEEE Circuits and Systems for Video Technology (CSVT) Transactions Best Paper Award for Year 2001, and the Best Paper Awards in IEEE GLOBECOM 2011 and International Conference on Quality of Service in Heterogeneous Wired/Wireless Networks (QShine) 2006. Currently, he serves as Editor in Chief of IEEE Transactions on Network Science and Engineering. He is an IEEE Fellow.

Xiaolin (Andy) Li is a Partner of Tongdun Technology, heading the AI Institute and Cognization Lab. He was a professor at University of Florida. As the founding center director, he founded the first national center on deep learning: NSF Center for Big Learning with colleagues at University of Florida, Carnegie Mellon University, University of Oregon, and UMKC, funded by National Science Foundation and dozens of industry members. His research interests include deep learning, federated learning, cloud computing, precision medicine, financial technology, and security & privacy. He received a PhD in computer engineering from Rutgers University.

Chenglong Li was born in Susong, China in 1965. He received his B.Sc. in Chemistry and M.Sc. in Physical Chemistry in Beijing University in 1985 and 1988, respectively. He received his Ph.D. in Biophysics in Cornell University in 2000. He is now the Nicholas Bodor Professor for Drug Discovery at the University of Florida; a professor of Medicinal Chemistry, Biochemistry and Biophysics. He is the Director of NIH/NIGMS Chemistry-Biology Interface Predoctoral Training Program at UF; and Associate Director of the Center for Natural Products, Drug Discovery and Development (CNPD3). His main research interest is molecular recognition from electronic to organismal levels with combined computational and experimental approaches.
Footnotes
REFERENCES
- [1].Ahmed A, Smith RD, Clark JJ, Dunbar JBJ, and Carlson HA. Recent improvements to binding moad: a resource for protein-ligand binding affinities and structures. Nucleic Acids Research, 43(D1):D465–D469, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ballester PJ and Mitchell JB. A machine learning approach to predicting protein-ligand binding affinity with applications to molecular docking. Bioinformatics, 26(9):1169–1175, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ballester PJ, Schreyer A, and Blundell TL. Does a more precise chemical description of protein-ligand complexes lead to more accurate prediction of binding affinity? Journal of chemical information and modeling, 54(3):944–955, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bitencourt-Ferreira G, da Silva AD, and de Azevedo WF Jr. Application of machine learning techniques to predict binding affinity for drug targets. a study of cyclin-dependent kinase 2. Current medicinal chemistry, 2019. [DOI] [PubMed] [Google Scholar]
- [5].Bitencourt-Ferreira G and de Azevedo WF Jr. Sandres: a computational tool for docking. In Methods in Molecular Biology, volume 2053, pages 51–65, 2019. [DOI] [PubMed] [Google Scholar]
- [6].Bitencourt-Ferreira G, Rizzotto C, and de Azevedo WF Jr. Machine learning-based scoring functions. development and applications with sandres. Current medicinal chemistry, 2020. [DOI] [PubMed] [Google Scholar]
- [7].Borea PA, Varani K, Gessi S, Gilli P, and Dalpiaz A. Receptor binding thermodynamics as a tool for linking drug efficacy and affinity. Il Farmaco, 53(4):249–254, 1998. [DOI] [PubMed] [Google Scholar]
- [8].C Braga R, M Alves V, C Silva A, N Nascimento M, C Silva F, M Liao L, and H Andrade C. Virtual screening strategies in medicinal chemistry: the state of the art and current challenges. Current topics in medicinal chemistry, 14(16):1899–1912, 2014. [DOI] [PubMed] [Google Scholar]
- [9].Chollet F. Xception: Deep learning with depthwise separable convolutions. In Computer Vision and Pattern Recognition, 2017. [Google Scholar]
- [10].da Silva AD, Bitencourt-Ferreira G, and de Azevedo WF Jr. Taba: a tool to analyze the binding affinity. Journal of computational chemistry, 41(1):69–73, 2020. [DOI] [PubMed] [Google Scholar]
- [11].Das S, Krein MP, and Breneman CM. Binding affinity prediction with property encoded shape distribution signatures. Journal of chemical information and modeling, 50(2):298–308, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Du X, Li Y, Xia Y-L, Ai S-M, Liang J, Sang P, Ji X-L, and Liu S-Q. Insights into protein–ligand interactions: Mechanisms, models, and methods. International journal of molecular sciences, 17(2):144, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Durrant JD, Friedman AJ, Rogers KE, and McCammon JA. Comparing neural-network scoring functions and the state of the art: applications to common library screening. Journal of chemical information and modeling, 53(7):1726–1735, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Durrant JD and McCammon JA. Nnscore: a neural-network-based scoring function for the characterization of protein-ligand complexes. Journal of chemical information and modeling, 50(10):1865–1871, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Durrant JD and McCammon JA. Nnscore 2.0: a neural network receptor-ligand scoring function. Journal of chemical information and modeling, 51(11):2897–2903, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Evanthia L, George S, K VD, and Zoe C. Structure-based virtual screening for drug discovery: principles, applications and recent advances. Current Topics in Medicinal Chemistry, 14(16):1923–1938, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gomes J, Ramsundar B, Feinberg EN, and Pande VS. Atomic convolutional networks for predicting protein-ligand binding affinity. arXiv:1703.10603, 2017. [Google Scholar]
- [18].Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WT, Mortenson PN, and Murray CW. Diverse, high-quality test set for the validation of protein- ligand docking performance. Journal of medicinal chemistry, 50(4):726–741, 2007. [DOI] [PubMed] [Google Scholar]
- [19].He K, Zhang X, Ren S, and Sun J. Deep residual learning for image recognition. In Computer Vision and Pattern Recognition, 2016. [Google Scholar]
- [20].Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, Andreetto M, and Adam H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv:1704.04861, 2017. [Google Scholar]
- [21].Hu L, Benson ML, Smith RD, Lerner MG, and Carlson HA. Binding moad (mother of all databases). Proteins, 60(3):333–340, 2005. [DOI] [PubMed] [Google Scholar]
- [22].Huang G, Liu S, van der Maaten L, and Weinberger KQ. Condensenet: An efficient densenet using learned group convolutions. In Computer Vision and Pattern Recognition, 2018. [Google Scholar]
- [23].Huang G, Liu Z, Van Der Maaten L, and Weinberger KQ. Densely connected convolutional networks. In Computer Vision and Pattern Recognition, 2017. [Google Scholar]
- [24].Jiménez J, Doerr S, Martínez-Rosell G, Rose A, and De Fabritiis G. Deepsite: protein-binding site predictor using 3d-convolutional neural networks. Bioinformatics, 33(19):3036–3042, 2017. [DOI] [PubMed] [Google Scholar]
- [25].Jiménez J, Skalic M, Martínez-Rosell G, and De Fabritiis G. K deep: Protein-ligand absolute binding affinity prediction via 3d-convolutional neural networks. Journal of chemical information and modeling, 58(2):287–296, 2018. [DOI] [PubMed] [Google Scholar]
- [26].Jubb HC, Higueruelo AP, Ochoa-Montaño B, Pitt WR, Ascher DB, and Blundell TL. Arpeggio: A web server for calculating and visualising interatomic interactions in protein structures. Journal of molecular biology, 429(3):365–371, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kairys V, Baranauskiene L, Kazlauskiene M, Matulis D, and Kazlauskas E. Binding affinity in drug design: experimental and computational techniques. Expert opinion on drug discovery, 14(8):755–768, 2019. [DOI] [PubMed] [Google Scholar]
- [28].LeCun Y, Bengio Y, and Hinton G. Deep learning. nature, 521(7553):436, 2015. [DOI] [PubMed] [Google Scholar]
- [29].Li H, Leung K-S, Ballester PJ, and Wong M-H. istar: a web platform for large-scale protein-ligand docking. PloS one, 9(1):e85678, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li H, Leung K-S, Wong M-H, and Ballester PJ. The impact of docking pose generation error on the prediction of binding affinity. In Computational Intelligence Methods for Bioinformatics and Biostatistics, 2015. [Google Scholar]
- [31].Li H, Leung K-S, Wong M-H, and Ballester PJ. Low-quality structural and interaction data improves binding affinity prediction via random forest. Molecules, 20(6):10947–10962, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Li Y, Kang H, Ye K, Yin S, and Li X. Foldingzero: Protein folding from scratch in hydrophobic-polar model. In Workshop on Deep Reinforcement Learning at NeurIPS, 2018. [Google Scholar]
- [33].Li Y, Liu Z, Li J, Han L, Liu J, Zhao Z, and Wang R. Comparative assessment of scoring functions on an updated benchmark: 1. compilation of the test set. Journal of chemical information and modeling, 54(6):1700–1716, 2014. [DOI] [PubMed] [Google Scholar]
- [34].Liu J and Wang R. Classification of current scoring functions. Journal of chemical information and modeling, 55(3):475–482, 2015. [DOI] [PubMed] [Google Scholar]
- [35].Ma N, Zhang X, Zheng H-T, and Sun J. Shufflenet v2: Practical guidelines for efficient cnn architecture design. In European Conference on Computer Vision, 2018. [Google Scholar]
- [36].Melo F and Feytmans E. Novel knowledge-based mean force potential at atomic level. Journal of molecular biology, 267(1):207–222, 1997. [DOI] [PubMed] [Google Scholar]
- [37].Melo F and Feytmans E. Assessing protein structures with a non-local atomic interaction energy. Journal of molecular biology, 277(5):1141–1152, 1998. [DOI] [PubMed] [Google Scholar]
- [38].Mohcine E-I, Arnaud B, Anass K, Soumaya WMP,B, and Sayeh E. Virtual screening in hepatitis b virus drug discovery: Current state-of-the-art and future perspectives. Current Medicinal Chemistry, 25(23):2709–2721, 2018. [DOI] [PubMed] [Google Scholar]
- [39].Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, and Olson AJ. Autodock4 and autodocktools4: Automated docking with selective receptor flexibility. Journal of computational chemistry, 30(16):2785–2791, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pantaleao SQ, Fujii DG, Maltarollo VG, da C Silva D, Trossini GH, Weber KC, Scott LP, and Honorio KM. The role of qsar and virtual screening studies in type 2 diabetes drug discovery. Medicinal Chemistry, 13(8):706–720, 2017. [DOI] [PubMed] [Google Scholar]
- [41].Priyanka S, Sunita T, and Imran SM. Recent progress in the identification and development of anti-malarial agents using virtual screening based approaches. Combinatorial Chemistry & High Throughput Screening, 18(3):257–268, 2015. [DOI] [PubMed] [Google Scholar]
- [42].Ragoza M, Hochuli J, Idrobo E, Sunseri J, and Koes DR. Protein-ligand scoring with convolutional neural networks. Journal of chemical information and modeling, 57(4):942–957, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rezaei MA, Li Y, Li X, and Li C. Improving the accuracy of protein-ligand binding affinity prediction by deep learning models: benchmark and model. ChemRxiv: 10.26434/chemrxiv.9866912.v1, 2019. [DOI] [Google Scholar]
- [44].Sandler M, Howard A, Zhu M, Zhmoginov A, and Chen L-C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Computer Vision and Pattern Recognition, 2018. [Google Scholar]
- [45].Schietgat L, Fannes T, and Ramon J. Predicting protein function and protein-ligand interaction with the 3d neighborhood kernel. In International Conference on Discovery Science. Springer, 2015. [Google Scholar]
- [46].Segler MH, Preuss M, and Waller MP. Planning chemical syntheses with deep neural networks and symbolic ai. Nature, 555(7698):604, 2018. [DOI] [PubMed] [Google Scholar]
- [47].Simonyan K and Zisserman A. Very deep convolutional networks for large-scale image recognition. In International Conference on Learning Representations, 2015 [Google Scholar]
- [48].Stepniewska-Dziubinska MM, Zielenkiewicz P, and Siedlecki P. Development and evaluation of a deep learning model for protein-ligand binding affinity prediction. Bioinformatics, 34(21):3666–3674, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sundaram L, Gao H, Padigepati SR, McRae JF, Li Y, Kosmicki JA, Fritzilas N, Hakenberg J, Dutta A, Shon J, et al. Predicting the clinical impact of human mutation with deep neural networks. Nature genetics, 50(8):1161, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wallach I, Dzamba M, and Heifets A. Atomnet: a deep convolutional neural network for bioactivity prediction in structure-based drug discovery. arXiv:1510.02855, 2015. [Google Scholar]
- [51].Wang R, Fang X, Lu Y, Yang C-Y, and Wang S. The pdbbind database: Methodologies and updates. Journal of Medicinal Chemistry, 48:4111–4119, 2005. [DOI] [PubMed] [Google Scholar]
- [52].Wang R, Lai L, and Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. Journal of Computer-Aided Molecular Design, 16(1):11–26, 2002. [DOI] [PubMed] [Google Scholar]
- [53].Wang R, Lu Y, and Wang S. Comparative evaluation of 11 scoring functions for molecular docking. Journal of Medicinal Chemistry, 46(12):2287–2303, 2003. [DOI] [PubMed] [Google Scholar]
- [54].Wang X, Han W, Yan X, Zhang J, Yang M, and Jiang P. Pharmacophore features for machine learning in pharmaceutical virtual screening. Molecular diversity, 2019. [DOI] [PubMed] [Google Scholar]
- [55].Wingert BM and Camacho CJ. Improving small molecule virtual screening strategies for the next generation of therapeutics. Current opinion in chemical biology, 44:87–92, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wójcikowski M, Siedlecki P, and Ballester PJ. Building machine-learning scoring functions for structure-based prediction of intermolecular binding affinity. Methods in molecular biology, 2053:1–12, 2019. [DOI] [PubMed] [Google Scholar]
- [57].Xavier MM, Heck GS, de Avila MB, Levin NMB, Pintro VO, Carvalho NL, and de Azevedo WF. Sandres a computational tool for statistical analysis of docking results and development of scoring functions. Combinatorial chemistry & high throughput screening, 19(10):801–812, 2016. [DOI] [PubMed] [Google Scholar]
- [58].Zhang X, Zhou X, Lin M, and Sun J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. In Computer Vision and Pattern Recognition, 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.