Abstract
Embryogenesis has long been known for its robustness to environmental factors. Although developmental tuning of embryogenesis to the environment experienced by the parent may be beneficial, little is understood on whether and how developmental patterns proactively change. Here, we show that Caenorhabditis elegans undergoes alternative embryogenesis in response to maternal gut microbes. Harmful microbes result in altered endodermal cell divisions; morphological changes, including left-right asymmetric development; double association between intestinal and primordial germ cells; and partial rescue of fecundity. The miR-35 microRNA family, which is controlled by systemic endogenous RNA interference and targets the β-transducin repeat-containing protein/cell division cycle 25 (CDC25) pathway, transmits intergenerational information to regulate cell divisions and reproduction. Our findings challenge the widespread assumption that C. elegans has an invariant cell lineage that consists of a fixed cell number and provide insights into how organisms optimize embryogenesis to adapt to environmental changes through epigenetic control.
Embryos alter their developmental pattern based on inherited microRNAs to adapt to gut microbes experienced by their mother.
INTRODUCTION
The process of embryogenesis has long been known to be extraordinarily invariable, or “canalized” (1), against extrinsic perturbations in many model systems (2–4). In viviparous species, such as mammals, embryogenesis proceeds while directly receiving substances from the mother, and it can often respond to the maternal environment, as seen in embryonic diapause upon starvation (5). However, because embryogenesis in most organisms occurs in isolation from the external environment, it is considered difficult for the embryo to directly perceive environmental factors in many cases, with few exceptions such as temperature (6). Therefore, little has been investigated about the extent to which the embryo alters its developmental patterns to adapt to various environmental changes.
The development of the nematode Caenorhabditis elegans is particularly renowned for its robustness; it is widely assumed to have exactly 959 somatic cells (1033 in males), of which 558 (560 in males) are produced during embryogenesis (4). Such highly invariable development allows for the assessment of developmental changes induced by environmental changes, genetic variations, and stochastic noises, making it useful for the study of developmental robustness and plasticity. C. elegans has been reported to alter its development in the presence of environmental stressors. When the environment is harsh for its growth during the first larval stage, it forms a dauer larva, an alternative third larval stage specialized for survival (7). Stresses such as high temperature and starvation also cause cell fate changes in vulval cells (8) and seam cells (9), although these changes are presently considered to be developmental “errors” (8, 9) rather than adaptive developmental tuning. The ability of C. elegans to change its developmental patterns during embryogenesis has not been explored.
Many studies have reported that parental experiences of various environmental factors affect the progeny in terms of metabolism, behavior, and stress resistance, and environmental cues converge onto chromatin modifications and small RNAs, such as short interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs), in the germ line to generate and transmit epigenetic information (10, 11). siRNAs are amplified via an RNA-directed RNA polymerase chain reaction, and in some animals, piRNAs are also amplified via a “ping-pong” cycle, allowing them to function as heritable epigenetic memory to transmit information across generations (10, 11). However, without a known amplification mechanism, it is unclear whether microRNAs (miRNAs) can serve as heritable epigenetic memories without being diluted. Therefore, much attention has been directed toward examining how miRNAs may trigger long-lasting chromatin modifications [e.g., (12)].
Gut microbes have attracted a great deal of attention in the past decade owing to their widespread impact on health and disease risk (13). Recent studies in mice have shown that depletion of the maternal gut microbiota by antibiotic treatment or germ-free rearing leads to deficiencies in various maternal metabolites, which can cause abnormal neurodevelopment (14) and metabolic disorders (15) in progeny. Although it is unclear whether these microbe-free conditions can reflect the natural environment in which the animal lives, these studies raise the possibility that changes in symbiotic gut microbes can influence the embryonic development of progeny.
RESULTS
Developmental plasticity in C. elegans embryogenesis
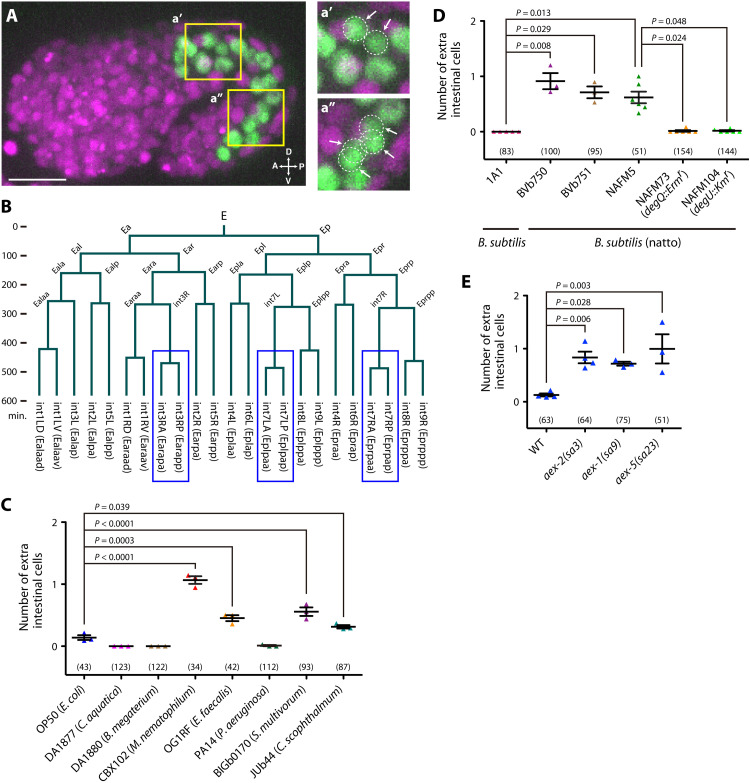
To precisely characterize the effects of environmental factors on embryogenesis, we traced the developmental behaviors of almost all cells in C. elegans embryos using long-term, single-cell level, whole-embryo imaging under various environmental conditions (see Materials and Methods). We found that when C. elegans mothers had experienced mildly harmful microbes, such as Microbacterium nematophilum (16) CBX102 and Enterococcus faecalis (17) OG1RF, the number of intestinal nuclei in their embryos was increased (Fig. 1, A to C). The extra cell nuclei were generated by cell division but not by binucleation of cells because they were surrounded by the cell membrane (40 of 40 nuclei; fig. S1A).
Fig. 1. Maternal gut microbes affect specific embryonic cell divisions in progeny.
(A) Embryo whose mother experienced CBX102. Green, intestinal nuclei (elt-2::GFP); magenta, all cell nuclei (mCherry::Histone). Magnifications of the boxed regions (yellow) are shown in the right. White dotted circles and arrows, extra cells. Scale bar, 10 μm. A, anterior; D, dorsal; P, posterior; V, ventral. (B) The intestinal cell lineage of the embryo shown in (A), acquired at ~1-min time resolution. The vertical axis is time, and a horizontal line indicates a cell division. Blue, extra divisions. (C) Numbers of extra cells in progeny whose mothers experienced various bacterial strains (shown along the x axis). Each dot represents the mean from one trial. Analysis of variance (ANOVA) with Dunnett’s posttest [F(7,16) = 83.95, P < 0.0001]. (D) Numbers of extra cells in progeny whose mothers experienced B. subtilis (1A1), B. subtilis (natto) (BVb750, BVb751, and NAFM5), or γPGA-nonproducing B. subtilis (natto) mutants (NAFM73 and NAFM104). Kruskal-Wallis and Dunn’s posttest (P = 0.0002). (E) Numbers of extra cells of defecation-defective mutants whose mothers experienced OP50. ANOVA with Dunnett’s posttest [F(3,10) = 8.675, P = 0.003]. WT, wild type. (C to E) Each dot represents the mean from one trial. Bars represent means ± SEM of independent trials. The total numbers of progeny scored in all trials are shown in parentheses.
The C. elegans intestine consists of nine intestinal rings (int1 to int9) arranged in an anterior-to-posterior sequence. The first int ring (int1) contains four cells, and the other int rings (int2 to int9) each contain two left and right cells (intXL and intXR) (18). All 20 intestinal cells are generated from the endoderm founder cell termed “E” during embryogenesis (4, 18). Our imaging and subsequent cell tracking in embryos whose mothers had experienced CBX102 showed that in most cases, the extra divisions induced by CBX102 occurred at either int7L (Eplpa) or int7R (Eprpa), or both, increasing the number of intestinal cells to 21 or 22 (fig. S1B). A small fraction of the extra divisions (8 of 66) occurred at int3R (Earap) (fig. S1B), increasing the number of intestinal cells up to 23 (Fig. 1B). These extra cell divisions occurred almost simultaneously with, or slightly after, the increase in the intestinal cell number from 16 to 20 (Fig. 1B and fig. S1C). In the int7 divisions, there is a left-right asymmetric tendency for int7R to divide along the anterior-posterior axis more often than int7L (fig. S1D). Binucleation, which occurs in all int3 to int7 cells and some int8 and int9 cells during the postembryonic development (18), appeared to occur also in extra cells (18 of 18 cells; fig. S1E).
Changes in cell numbers caused by CBX102 appeared to be specific to the endodermal lineage. We manually traced the entire cell lineage in three embryos with CBX102-induced extra division in the E lineage up to the 500- to 550-cell stage. Other than the extra divisions in the E lineage, all cell divisions and programmed cell deaths up to that stage were normal when compared to Sulston’s reference lineage (fig. S2).
Extra cells were also induced by Sphingobacterium multivorum BIGb0170 and Chryseobacterium scophthalmum JUb44 (Fig. 1C), both of which are part of the natural microbiota of C. elegans that colonize the gut and retard the growth of C. elegans (19), implying that the developmental changes also occur in the natural habitat of C. elegans. Escherichia coli OP50, the standard food for C. elegans in laboratory culture but reported to be weakly toxic (17, 20), had a slight but detectable activity to induce extra cells (Fig. 1C). Sulston et al. (4) noticed that an extra cell was “occasionally” produced in the intestine when they described the entire embryonic cell lineage. Our results suggest that their observation is an environmentally induced phenomenon by OP50 rather than intrinsic variability due to less robust control of the lineage. In contrast, no extra cells were observed on the benign bacteria Comamonas aquatica (21) DA1877 and Bacillus subtilis (20) 1A1 (Fig. 1, C and D).
How do microbes induce the developmental changes? Given the diversity of the microbial species, we reasoned that it is more likely to be a physiological state of the worm than common chemicals from the microbes. Highly pathogenic Pseudomonas aeruginosa (17) PA14 and the hard-to-eat bacterium Bacillus megaterium (21) DA1880 did not induce extra cells (Fig. 1C), suggesting that general health, developmental speed, or nutrient deficiency in the mother is not a critical determinant. We noticed that all the microbes that induced extra cells are described as viscous (22, 23) and/or cause “constipation,” “distention,” or “lumen bloating” in the C. elegans intestine (fig. S3A) (16, 24–26). It has recently been suggested that bloating of the intestinal lumen is a “danger signal” that signifies microbial colonization (24). We examined defecation-defective mutants that are known to exhibit intestinal bloating, namely, aex-1, aex-2, and aex-5 (27, 28), and found that their intestinal cell numbers increased on OP50 (Fig. 1E). Furthermore, restricting the entry of microbes into the gut tube (pharynx, intestine, and rectum) in mothers suppressed extra cells induced by CBX102, and this suppression was unaffected when worms were crossed with males with normal feeding (fig. S3, B to D), suggesting that the microbe acts in the maternal gut. We next exposed parental worms to B. subtilis (natto). B. subtilis (natto) is classified as the same species as the model organism B. subtilis but is characterized by the production of a hard-to-digest biofilm composed of the biopolymer γ-poly-dl-glutamic acid (γPGA), a major constituent of the sticky strings of the Japanese food Natto and an important virulence factor for anthrax (29, 30). B. subtilis (natto) strains strongly induced extra cells in C. elegans embryos (Fig. 1D). B. subtilis (natto) mutants defective in γPGA synthesis (29) did not cause extra cells (Fig. 1D), indicating that γPGA is required for the induction, although we cannot rule out the possibility that the mutations may affect additional aspects of the bacteria. B. subtilis (natto) also slowed the growth of worms in a γPGA-dependent manner (fig. S3E). Together, these results suggest that the maldigestion of maternal gut microbes that are highly proliferative and/or produce a viscous biofilm is an important factor for this developmental plasticity.
Consequences of alternative embryogenesis
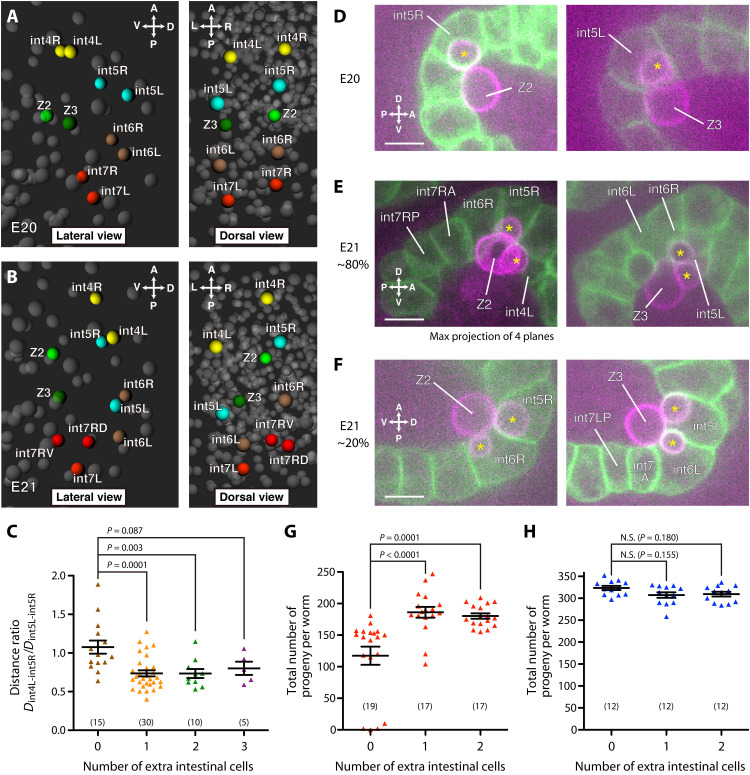
To better understand the effect of maternal gut microbes, we further examined the embryonic development and fitness of the progeny that had extra cells. First, extra cells are accompanied by changes in the symmetry of intestinal development. In embryos that had extra cells, the right-side intestinal cells near the mid-intestine moved anteriorly, resulting in a left-right asymmetric cell arrangement (Fig. 2, A and B, fig. S4A, and movies S1 and S2). For example, int5R was paired with int4L (Fig. 2, B and C) instead of the normal int5L, and int5R was even slightly anterior to int4L in ~60% of embryos (fig. S4B). This left-right asymmetric cell arrangement seemed to be resolved by the inclusion of int3R in the int2 ring (fig. S4C). The left-right asymmetry in the directions of the extra int7 division axis (fig. S1D) appears to be consistent with the left-right asymmetric cell arrangement.
Fig. 2. Extra cells coincide with left-right asymmetric development, increasing the primordial germ cell–intestine association.
(A and B) Three-dimensional reconstruction of centers of cell nuclei around the mid-intestine of 1.5-fold stage embryos. An embryo without extra cell (A) (E20) and one with an extra cell (B) (E21), whose mothers experienced CBX102. Left: lateral view. Right: dorsal view. Yellow, int4L/R cells; cyan, int5L/R cells; brown, int6L/R cells; red, int7L/R cells and int7R daughters; yellow-green, Z2; green, Z3; gray, other cells. (C) Ratios of the distance between int4L and int5R (Dint4L-int5R) to that between int5L and int5R (Dint5L-int5R) in embryos whose mothers experienced CBX102. ANOVA with Dunnett’s posttest [F(3,56) = 7.147, P = 0.0004]. The numbers of embryos are indicated in parentheses. (D to F) Embryo without extra cell (D) and embryos with one extra cell (E and F), whose mothers experienced CBX102. Green, intestinal cell membranes; magenta, PGC membranes. Single focal plane images are shown unless otherwise indicated. Left: Z2 and its lobes (asterisks). Right: Z3 and its lobes (asterisks). Scale bars, 10 μm. (G and H) Numbers of total progeny from worms grown on mixture of 10% CBX102 and 90% OP50 (G) and 100% OP50 (H). Intestinal cell numbers were determined in F1 worms born to mothers that had experienced CBX102, and numbers of F2 progeny of individually cultured F1 worms were counted. Kruskal-Wallis and Dunn’s posttest [(G) P < 0.0001; (H) P = 0.1356]. N.S., not significant. Each dot represents one F1 worm. The numbers of F1 worms tested are indicated in parentheses. (C, G, and H) Bars represent means ± SEM.
Second, embryos with extra cells increase the physical association between the endoderm and primordial germ cells (PGCs), a process that is widely conserved in invertebrate and vertebrate animals (31–34). From the 88-cell stage to the L1 larval stage, C. elegans has two PGCs (4), namely, Z2 and Z3. In embryos without extra cells whose mothers experienced CBX102, Z2 and Z3 took on an hourglass shape and each inserted a lobe into int5R and int5L, respectively (Fig. 2D), as reported in canonical embryogenesis (4, 18, 32). In embryos with extra cells, PGCs formed additional lobes, which associate with other mid-intestinal cells (29 of 30 embryos; Fig. 2, E and F). The identity of the partner intestinal cells appeared to be correlated with the degree of left-right asymmetry of the intestine (Fig. 2, E and F, and fig. S5A). The doubling of lobes seems to be enabled by the positional changes in the intestinal cells (Fig. 2B and fig. S5A). The PGC lobes are digested by the partner intestinal cells during late embryogenesis, possibly allowing for clearance of unwanted materials (32). Consistent with the doubling of lobes, the size of PGCs at the L1 stage after lobe removal was reduced in the presence of extra cells (fig. S5B).
Third, worms with extra cells show partial rescue of fecundity when CBX102 is present; they produced >50% more progeny than worms without extra cells (Fig. 2G and fig. S6A). In contrast, on OP50, they did not produce more progeny (Fig. 2H and fig. S6B). Their developmental speed on CBX102 was comparable to that of worms without the extra cells (fig. S6C), implying that the reproductive fitness was not due to better general health caused by the increased number of intestinal cells. Although the extent to which altered PGC morphology contributes to fecundity is unknown, the three observations suggest that maternal gut microbes lead to an alternative developmental program with extra cells and reproductive fitness.
Epigenetic control of alternative development
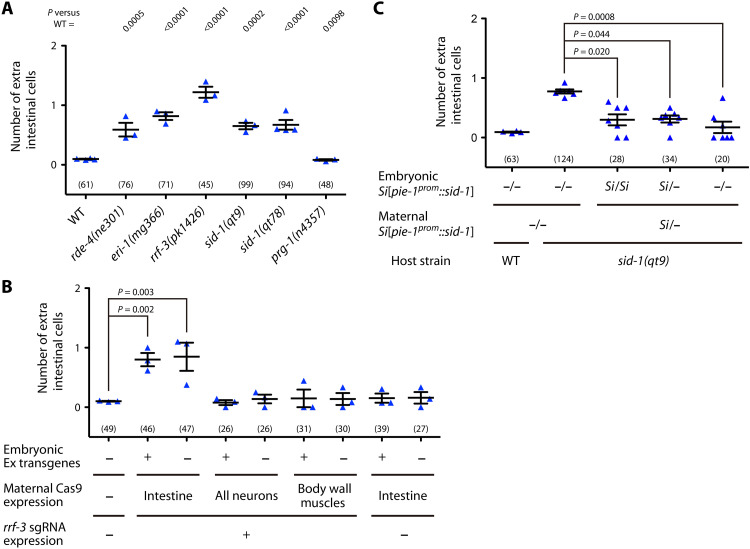
The C. elegans embryo is isolated from the external environment by the eggshell, and thus, information on microbes is highly likely to be transmitted through the maternal germ line. To understand how the information is processed in the mother and the embryo, we investigated the developmental phenotype of mutants of genes implicated in stress responses. We found that endogenous RNA interference (endo-RNAi), RNAi caused by endogenous siRNAs (35, 36), is involved in this plasticity. Mutants of rde-4, eri-1, and rrf-3, all of which are components of the enhanced RNAi (ERI) complex required for endo-RNAi biogenesis (37, 38), showed increases in intestinal cells on OP50 (Fig. 3A), suggesting that endo-RNAi acts to prevent the extra divisions. This result may be consistent with an increased intestinal cell nuclei number caused by RNAi of eri-5 (39), which is another ERI component (37, 38). In contrast, mutants of prg-1, a PIWI Argonaute involved in the piRNA pathway (40, 41), did not show an increase in intestinal cells (Fig. 3A). In C. elegans, intercellular transport of double-stranded RNAs (dsRNAs) leads to the propagation of RNAi from cell to cell (systemic RNAi), including transmission from soma to germ line (42). Systemic RNAi requires the systemic RNA interference deficiency–1 (SID-1) dsRNA importer (43–46), the worm ortholog of mammalian SID-1 transmembrane family member 1 (SIDT1) and SIDT2, and SID-1–dependent intercellular propagation has been suggested to function in endo-RNAi (47, 48). Mutants of sid-1 also showed increases in intestinal cells (Fig. 3A).
Fig. 3. Maternal endogenous RNAi and SID-1 regulate extra cells.
(A) Numbers of extra cells of RNAi and prg-1 mutants whose mothers experienced OP50. ANOVA with Dunnett’s posttest [F(6,16) = 32.93, P < 0.0001]. (B) Cas9 was maternally expressed in a tissue-specific manner with (“+”) or without (“−”) rrf-3 single-guide RNAs (sgRNAs) from extrachromosomal transgenic arrays (Ex), and numbers of extra cells were counted in array-positive (+) and array-negative (−) (stochastic array loss) embryos. All mothers experienced OP50. ANOVA with Dunnett’s posttest [F(8,18) = 6.908, P = 0.0003]. (C) The sid-1 complementary DNA (cDNA) was expressed specifically in the germ line from a single-copy–inserted (Si) transgene, and numbers of extra cells in progeny segregated from mothers heterozygous for the transgene (Si/−) or from mothers lacking the transgene (−/−) were counted. All mothers experienced OP50. Kruskal-Wallis and Dunn’s posttest (P = 0.002). (A to C) Bars represent means ± SEM of independent trials. Each dot represents the mean from one trial. The total numbers of progeny scored in all trials are shown in parentheses.
Conditional knockout of rrf-3 using somatic CRISPR-Cas9 (49) in the maternal intestine, but not in maternal neurons or body wall muscles, induced extra cells on OP50 (Fig. 3B), making the intestine a candidate as the source of endo-RNAi. Furthermore, the sid-1 mutant phenotype was rescued by germline-specific expression of sid-1 driven from a single-copy transgene (Si[pie-1prom::sid-1]) (Fig. 3C), implying a role for soma-to-germline transmission of dsRNAs. The rescue effect was observed in transgene-free embryos (−/−) born to heterozygous mothers (Si/−) (Fig. 3C), and sid-1 mutants crossed with wild-type males also showed an increase in extra cells (fig. S7A), indicating that the maternal action of sid-1 is important. Last, mutations in rde-4, rrf-3, or sid-1 did not enhance the extra cells induced by CBX102 (fig. S7B), which is consistent with the notion that systemic endo-RNAi is the major response pathway to the microbe.
Next, we focused on miRNAs of the miR-35 family (miR-35–42, hereafter referred to as miR-35fam) as potential epigenetic regulators, because their expression is highly enriched in the germ line (50) and they positively regulate intestinal cell numbers (51). In mir-35–41(nDf50) and mir-35–41(gk262) deletion mutants (51, 52), both of which lack a gene cluster consisting of seven of the eight family members, CBX102-induced extra cells were completely suppressed (Fig. 4A). Furthermore, introduction of nEx1187, a multicopy mir-35 expression array (52, 53), induced extra cells on OP50 (Fig. 4B). This induction of extra cells was observed in array-negative embryos born to array-positive mothers (Fig. 4B), indicating that increased maternal mir-35 expression is sufficient for the regulation. Notably, nEx1187 also led to an improved fecundity on harmful microbes. We cultured mothers with the nEx1187 array on OP50 and isolated array-negative embryos with or without extra cells. When they were raised on CBX102, worms with extra cells produced more progeny (Fig. 4C and fig. S8A). Reverse transcription quantitative polymerase chain reaction (RT-qPCR) showed that mature miR-35-3p and miR-40-3p, which are miRNAs expressed from the mir-35–41 cluster, were increased in the germ line when worms had experienced CBX102 or B. subtilis (natto) BVb750 (Fig. 4D). The miRNAs were not detected in the mir-35–41(nDf50) deletion mutants, indicating the specificity of our qPCR (Fig. 4D). The introduction of nEx1187 also increased the miR-35-3p expression in the maternal germ line (fig. S8B), which may be due to leaky expression from the array that evades germline silencing. Mutants of the endo-RNAi pathway and the nEx1187 strain showed normal defecation (fig. S8, C and D), showing that these genes act in parallel or downstream of intestinal stress. Together, these results show that the expression level of miR-35fam in the maternal germ line increases in response to the microbes and that miR-35fam up-regulation activates the alternative developmental program including extra cells and fitness.
Fig. 4. The miR-35 microRNA family is involved in alternative embryogenesis.
(A) Numbers of extra cells of mir-35–41 mutants whose mothers were exposed to OP50 or CBX102. Kruskal-Wallis and Dunn’s posttest (P = 0.001). (B) mir-35(+) was expressed from an extrachromosomal transgenic array in the wild-type, lin-23(zb11), and cdc-25.2(g52) backgrounds, and numbers of extra cells were counted in array-positive (+) and array-negative (−) (stochastic array loss) embryos. ANOVA with Dunnett’s posttest [F(7,23) = 51.48, P < 0.0001]. (A and B) Each dot represents the mean from one trial. The total numbers of progeny scored in all trials are shown in parentheses. (C) Numbers of total progeny from worms grown on 10% CBX102. Intestinal cell numbers were determined in F1 embryos born to P0 mothers that had had the mir-35(+) array, and numbers of F2 progeny of individually cultured F1 worms without the array were counted. Kruskal-Wallis and Dunn’s posttest (P = 0.0004). Each dot represents one F1 worm. The numbers of F1 worms tested are shown in parentheses. (D) RNA was purified from dissected gonads, and expression levels of mature miRNAs were quantified by RT-qPCR. ANOVA with Dunnett’s posttest. N.D., not detected. (E) Quantification of the expression of mir-35–41prom::GFP in wild-type embryos whose mothers experienced OP50 or CBX102. (F and G) Quantification of the mir-35–41prom::GFP expression in embryos whose mothers experienced CBX102 (F) or OP50 (G). (E and F) Two-tailed Mann-Whitney test. (G) Kruskal-Wallis and Dunn’s posttest (P = 0.0001). (E to G) The numbers of embryos are indicated in parentheses. (A to G) Bars represent means ± SEM. (H) A model for the mechanism of the epigenetic regulation of the alternative embryonic development.
Next, we examined zygotic expression of miR-35fam using wwIs8[mir-35–41prom::GFP], a transcriptional reporter driven by the promoter of the mir-35–41 cluster (54), which is silenced in the maternal germ line but active in embryos. The expression of the reporter was elevated in embryos whose mothers had experienced CBX102 (Fig. 4E and fig. S9A). Furthermore, the reporter expression was reduced in the mir-35–41(nDf50) mutant background (Fig. 4F). Thus, similar to the positive feedback–based epigenetic inheritance of chromatin structure (55), the long-lasting response transmitted from mother to mid-stage embryo may be enabled by an autoregulatory feedback loop of miR-35fam, where miR-35fam positively controls its own expression. The reporter expression was increased in mutants of rrf-3 and sid-1 (Fig. 4G), implying that the expression of miR-35fam is repressed by RNAi. Last, the mir-35–41(nDf50) mutation completely suppressed the extra cells in RNAi mutants (fig. S9B), further suggesting that miR-35fam acts downstream of the RNAi pathway.
An experimentally verified functional miR-35fam–binding site (51) is embedded in the 3′ untranslated region (3′UTR) of lin-23 (fig. S9C), the C. elegans ortholog of the mammalian F-box proteins β-transducin repeat-containing protein–1 (β-TrCP-1) and β-TrCP-2. LIN-23 negatively regulates the intestinal cell number by degrading cell division cycle 25 (CDC-25), a family of cell cycle–activating phosphatases, such as CDC-25.2 (56). To investigate whether miR-35fam regulates extra cells via the repression of lin-23, we used CRISPR-Cas9 to generate lin-23(zb11), a short deletion allele in which the miR-35fam–binding site in lin-23 3′UTR is disrupted (fig. S9C). The lin-23(zb11) mutation and mir-35–41(nDf50) increased the level of lin-23 mRNA (fig. S9D). While the lin-23(zb11) mutation did not lead to any gross phenotype, it, as well as a hypomorphic mutation in cdc-25.2, suppressed the nEx1187-induced extra cells (Fig. 4B), suggesting that the LIN-23/CDC-25 pathway acts downstream of miR-35fam (Fig. 4H), although the incomplete suppression of extra cells in lin-23(zb11) (Fig. 4B) suggests the potential involvement of miR-35fam target(s) other than lin-23.
DISCUSSION
We propose here that maternal exposure to harmful gut microbes can lead to an alternative embryogenesis program with adaptive advantage and that the alternative program is epigenetically controlled by the endo-RNAi pathway and the miR-35 miRNA family (Fig. 4H). These findings challenge the widespread assumption that C. elegans has an invariant cell lineage. Our results suggest that the maldigestion of maternal gut microbes, which causes intestinal distention and poses a great survival risk to worms (16, 24–26), triggers the developmental plasticity. However, because of our current technological limitations, we cannot exclude a potential involvement of a different chemical or physical action(s) of the microbes. The function of the endo-RNAi pathway in the intestine raises the possibility of it serving as a mechanism to monitor and broadcast organ health. However, it is not yet clear what siRNAs are involved, how they are regulated by the presence of specific gut microbes, or what their effectors that serve as the systemic signal to the germ line are. The miR-35fam regulates the lineage by targeting the cell cycle regulator β-TrCP/LIN-23, but it is less clear how miR-35fam affects germline development and fecundity of the progeny.
Our results suggest that intergenerationally inherited miR-35fam responses induce specific endodermal cell divisions and adaptive advantages by controlling cell cycle regulators. Assuming that intergenerationally induced miRNA responses can be an entity of epigenetic memory by a simple transcriptional feedback loop, they may greatly expand the current repertoire of intergenerational epigenetic controls that allow sequence-specific gene regulation. While the cell cycle regulators LIN-23 and CDC-25 are ubiquitously expressed in the embryo to exert pleiotropic functions, small changes in their activities result in organ-specific abnormalities in endodermal cell divisions (57, 58). Our results are consistent with these known phenotypes and the notion that miRNAs tune the expression of their targets within a range (59, 60). The cells that divide excessively in response to maternal gut microbes (fig. S1B) are sisters of the cells that divide when the number of endodermal cells increases from 16 to 20 (Fig. 1B). Such a lineage relationship may be key to creating the specificity of the developmental changes.
The interactions between animals and microbes in their environments are complex. Some are beneficial, such as maternal gut microbes in mice providing essential metabolites for normal embryonic development (14, 15). In contrast, many studies, including a report that germ-free mice can live 1.5 times longer than normal (61), have suggested that animals are adversely affected by chronic exposure to some indigenous microbes. Our results show that such stress can induce developmental tuning in animals with mosaic (determinate) development. Given the obvious plasticity of regulative development and the ability of the microbiome to affect systemic signaling (13), our study raises the question of whether such exposure could induce adaptive tuning of embryogenesis in humans and other complex animals.
In most animals, including humans, the development of the egg (and the embryo in viviparous animals) occurs near the intestine. This proximity makes the mother-to-embryo nutrition transport efficient, but it also has the disadvantage that the effects of harmful gut microbes can easily reach the next generation. For example, in humans, there is a great risk of Listeria monocytogenes vertically infecting the fetus in utero from the intestine, causing miscarriage and neonatal death (62). Future research may further reveal the mechanisms that protect the next generation of cells from harmful gut microbes through developmental changes.
MATERIALS AND METHODS
Strains and culture
Standard methods were used to construct and culture C. elegans strains (63–66). Worms were cultured on nematode growth medium (NGM) plates containing nystatin (50 U/ml; Sigma-Aldrich) at 20° to 21°C. Bristol N2 was used as the wild type. The strains used in this study are listed in table S1.
Embryo mounting and fluorescence microscopy
Preparation and mounting of embryos were performed as previously described (67–69) with modifications. Briefly, ~20 gravid adult worms were picked to ~20 μl of Boyd buffer [5 mM Hepes (pH 7.2), NaCl (3.5 g/liter), KCl (2.4 g/liter), Na2HPO4 (0.4 g/liter), CaCl2 (0.2 g/liter), MgCl2 (0.2 g/liter), and 0.2% glucose] and washed three times with Boyd buffer. Worms were then cut with a 25-gauge needle (BD PrecisionGlide), and early embryos at the 2- to 30-cell stage were collected with an eyelash. Embryos were transferred with a glass needle and mounted in ~1.5 μl of Boyd buffer containing ~50 polystyrene-based microspheres (20 μm in diameter, Polysciences) between a 24 mm by 50 mm glass slide and an 18 mm by 18 mm glass coverslip, which was sealed at the edges with melted Vaseline.
Images of live embryos were acquired with an Olympus UPlanSAPO 60× silicone oil objective [numerical aperture (NA), 1.30] or an Olympus UPlanSAPO 40× silicone oil objective (NA, 1.25). The microscopes used were (i) a spinning-disk confocal microscope (Quorum Technologies Inc.) consisting of a Zeiss Axio Observer Z1, a Yokogawa CSU-X1 spinning-disk unit, and two Hamamatsu C9100-13 EM-CCD cameras and (ii) an instant structured illumination microscope (Visitech iSIM) consisting of an Olympus IX73 and a Hamamatsu Flash 4.0v2 scientific complementary metal-oxide-semiconductor (sCMOS) camera. Total fluorescence of the mir-35–41::GFP (green fluorescent protein) reporter (Fig. 4, E to G) was measured in ~30-cell stage embryos using Fiji software (National Institutes of Health).
Cell lineage analyses
For cell lineage analyses, embryos were imaged with 1-μm z-steps across 29 μm and with 75-s intervals. Positions of cell nuclei and timing of cell divisions were determined with StarryNite and AceTree software (68–70). Directions of cell divisions and distance between cells were calculated from the central coordinates of the cell nuclei. Three-dimensional reconstruction of cell nuclei positions in embryos was performed by WormGUIDES software (69, 71).
Exposure of worms to a microbial lawn
For preculture of microbial strains, a freshly streaked colony was inoculated and grown at 30°C in LB broth with the exceptions of OG1RF, which was cultured in brain heart infusion broth (Difco), and JUb44 and BIGb0170, which were cultured at 25°C. After 24 hours, 100 μl of preculture was spread onto a 60-mm NGM plate and then incubated at 20° to 21°C for 24 hours.
NGM plates for culturing parental worms were seeded with OP50 and incubated at 20° to 21°C for 2 days. The plates were stored at 4°C for 2 to 20 days until the day of use. Parental worms were cultivated on the OP50-seeded plates until L4 larval to young adult stages. The parental worms were collected and washed five times with M9 buffer [KH2PO4 (3 g/liter), Na2HPO4 (6 g/liter), NaCl (5 g/liter), 1 mM MgSO4, and 0.03% gelatin], and then ~50 worms were transferred to a plate seeded with each indicated microbial strain. After 24 hours of incubation at 20° to 21°C, their embryos were collected and mounted as described above. Ivermectin was first dissolved in dimethyl sulfoxide at a concentration of 10 μg/ml and added to melted NGM agar to give a final concentration of 1 ng/ml before pouring.
Quantification of intestinal cell numbers
Numbers of intestinal cells were counted in 1.5- to 3-fold stage embryos using the fluorescence of stIs10453[elt-2prom::elt-2::gfp] or the combination of rrIs1[elt-2prom::nls::gfp] and ujIs113[pie-1prom::mCherry::H2B, nhr-2prom::mCherry::his-24]. Because the expression of rrIs1 was variable and occasionally undetectable in some cells, the additional fluorescence of ujIs113 was required to accurately count intestinal cells. The distribution of the number of intestinal cells in each trial is shown in data S1.
Intestinal cell numbers in cross progeny (figs. S3C and S7A) were counted as follows. him-8(−) males containing the stIs10453[elt-2prom::elt-2::gfp] marker and him-8(+) hermaphrodites lacking any fluorescent marker were raised until mid-L4 stage on OP50-seeded plates. The males and hermaphrodites were separately collected, washed five times with M9 buffer, and transferred to NGM plates on which CBX102 or OP50 was grown for 18 hours. After 6 hours of separated preexposure, 50 males and 10 hermaphrodites were picked to a new NGM plate on which CBX102 or OP50 was grown for 24 hours. After 24 hours of incubation at 20° to 21°C, embryos were collected from the hermaphrodites and mounted as described above. Intestinal cells were observed only in the cross progeny, which carried the stIs10453 marker.
Brood size, growth, and defecation assays
L4 and young adult worms (P0) grown on OP50 were collected and washed five times with M9 buffer, and then ~50 worms were transferred to a plate seeded with CBX102 (Fig. 2, G and H, and fig. S6) or with OP50 (Fig. 4C and fig. S8A). After 30 hours, 1.5-fold embryos (F1) were directly picked into ~50 μl of M9 buffer dropped on a 24 mm by 50 mm glass slide, and their ELT-2::GFP fluorescence was observed with the spinning-disk confocal microscope described above. Embryos without extra intestinal cell (E20), embryos with one extra intestinal cell produced by either int7L or int7R divisions (E21), and embryos with two extra intestinal cells produced by both int7L and int7R divisions (E22) were isolated. The F1 embryos were individually washed with 1% sodium hypochlorite to avoid microbial contamination and then plated onto NGM plates seeded with OP50 or onto NGM plates seeded with a mixture of stationary-phase cultures of CBX102 and OP50 mixed in a volumetric ratio of 1:9 (10% CBX102 plates).
For brood size assays, the isolated F1 embryos were grown until young adults, and all the F2 hatchlings were killed and counted every 24 hours until the F1 worms no longer produced progeny. For growth assays on CBX102 (fig. S6C), the isolated F1 embryos were allowed to develop at 20°C on 10% CBX102 plates, and their developmental stages were determined after 72 and 80 hours.
For growth assays on the E. coli and B. subtilis strains (fig. S3E), early embryos were prepared by hypochlorite treatment of gravid hermaphrodites raised on OP50 and were transferred onto a new plate seeded with an indicated E. coli or B. subtilis strain. The embryos were allowed to develop at 20°C for 65 hours, and their developmental stages were determined. Defecation cycle length, the duration between the first muscular contraction of one defecation and the first muscular contraction of the next defecation, was measured in 1-day-old young adult worms at 20°C under a dissecting microscope.
Isolation of gonadal miRNAs and RT-qPCR
For isolation of gonadal miRNAs, young adults were transferred into egg buffer [25 mM Hepes (pH 7.2), 118 mM NaCl, 48 mM KCl, 2 mM CaCl2, and 2 mM MgCl2] and washed five times with egg buffer immediately before dissection. The gonads were isolated from 100 worms by cutting the worms behind the pharyngeal bulb and in front of the spermatheca with a 25-gauge needle. The isolated gonads were placed into 300 μl of TRIzol on ice. Sixty microliters of chloroform was added to it, and after centrifugation, the aqueous phase was transferred into a new tube. After adding 1.5 volumes of ethanol, the samples were loaded into a miRNeasy column (Qiagen) and purified according to the supplier’s instructions. The purified samples were reverse-transcribed using the miScript reverse transcriptase and HiSpec buffer (Qiagen).
For isolation of mRNAs, approximately 20 1-day-old gravid adults were placed into 300 μl of TRIzol and frozen. After thawing, total RNA was purified as described above with an RNeasy column (Qiagen). The purified samples were reverse-transcribed using Quantitect reverse transcription (Qiagen).
For qPCR, the QuantiTect SYBR Green PCR kit (Qiagen) was used on the QuantStudio 6 Flex real-time PCR system (Applied Biosystems). Primers specific for miR-2-5p, miR-47-3p, miR-35-3p, mir-40-3p, miR-42-3p, lin-23, and act-1 were obtained from the miScript Primer Assay kit and the Quantitect Primer Assay kit (Qiagen). miR-47-3p (72) and act-1 were used as the endogenous control genes, and the results were analyzed using the comparative Ct method.
Germline transformation
Expression constructs were injected at 10 to 50 ng/μl. pG-unc-122prom::mCherry and pCFJ90 (myo-2prom::mCherry, a gift from E. Jorgensen, Addgene plasmid #19327) were used as injection markers. In each case, the total concentration of injected DNA was 100 ng/μl.
The zb11 allele of lin-23 was generated using the CRISPR-Cas9 system (66). The single-guide RNA (sgRNA) expression construct containing the target sequence 5′-GGTTTGGTTGATTTCTGCAC-3′ was obtained using a PCR-fusion technique, and the PCR product was injected at 15 ng/μl together with pDD162 (50 ng/μl; eft-3prom::Cas9, a gift from B. Goldstein, Addgene plasmid #47549). After injection, F1 animals were screened for a deletion by single-worm PCR using the primers 5′-TATGGATCACCTGGGCGGAG-3′ and 5′-GAGGAAAAGTTGGGAAGGGGA-3′. The resulting lin-23(zb11) mutant line was backcrossed twice to the original strain before use. The pie-1prom::sid-1(cDNA)::pie-1 3′UTR transgene in the pCFJ150-pie-1prom-sid-1(cDNA)-pie-1 3′UTR plasmid (see below) was integrated as a single copy into the oxTi185 site on chromosome I of the EG8078 strain using MosSCI (65).
Transgene constructions
The construction of pDEST-Cas9, pENTR-ges-1prom, pENTR-myo-3prom, pENTR-rimb-1prom, and pG-unc-122prom::mCherry was described previously (73–75). pG-ges-1prom-Cas9, pG-rimb-1prom-Cas9, and pG-myo-3prom-Cas9 were made by LR recombination reactions between the pENTR plasmids and pDEST-Cas9. For pCFJ150-pie-1prom-sid-1(cDNA)-pie-1 3′UTR, the sid-1 complementary DNA (cDNA) was amplified from the genomic DNA of the XE1375 strain (76) with the primers 5′-gtgtAGGCCTaaaaATGATTCGTGTTTATTTGATAATTTTAATGCA-3′ and 5′-gtgtGCTAGCCTAGAAAATGTTAATCGAAGTTTTGCGT-3′ and inserted into the NaeI–NheI sites of pCFJ150-mCherry(dpiRNA)::ANI-1(AHPH) (a gift from H.-C. Lee, Addgene plasmid #107939). Three expression constructs of U6prom::rrf-3 sgRNAs were amplified by PCR from pDD162 using the primers 5′-GTATTGTGTTCGTTGAGTGACC-3′, 5′-TGGCTTAACTATGCGGCATC-3′, 5′-gGAAATGTTGATTGACTCTGgttttagagctagaaatagc-3′, 5′-CAGAGTCAATCAACATTTCcaagacatctcgcaataggag-3′, 5′-gAGAAAACGAAGCACAGAGGgttttagagctagaaatagc-3′, 5′-CCTCTGTGCTTCGTTTTCTcaagacatctcgcaataggag-3′, 5′-gCTCAAATCTCGCATACGAGgttttagagctagaaatagc-3′, and 5′-CTCGTATGCGAGATTTGAGcaagacatctcgcaataggag-3′ (target sequences were GGAAATGTTGATTGACTCTG, GAGAAAACGAAGCACAGAGG, and GCTCAAATCTCGCATACGAG) and were cointroduced. Further details of the expression constructs will be provided upon request.
Statistical analysis
Statistical analyses were performed with a statistic package (Prism v.9.0.2, GraphPad software). Error bars indicate SEM. Statistical comparisons were performed with two-tailed one-way analysis of variance (ANOVA) with Dunnett’s posttest or Tukey’s posttest, two-tailed Kruskal-Wallis with Dunn’s posttest, two-tailed t test, or two-tailed Mann-Whitney test. All experiments were repeated three to seven times with similar results. Raw data that support the findings of this study are available on Figshare with the DOI 10.6084/m9.figshare.18133622.v2.
Acknowledgments
We thank the Caenorhabditis Genetics Center (funded by the NIH Office of Research Infrastructure Programs P40 OD010440), NBRP (NIG, Japan)—B. subtilis, NBRP (Tokyo Women’s Medical University, Japan)—C. elegans, the Bacillus Genetic Stock Center, J. Nance, and D. A. Garsin for strains.
Funding: This work was supported by HFSP long-term fellowship LT000938/2017 (to H.O.), NIH grant R01GM097576 (to Z.B.), and MSK Cancer Center Support/Core P30CA008748.
Author contributions: Conceptualization: H.O. and Z.B. Methodology: H.O. and Z.B. Investigation: H.O. Visualization: H.O. Funding acquisition: H.O. and Z.B. Project administration: H.O. and Z.B. Supervision: Z.B. Writing—original draft: H.O. Writing—review and editing: H.O. and Z.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Raw data that support the findings of this study are available on Figshare with the DOI 10.6084/m9.figshare.18133622.v2. Requests for all strains and plasmids generated in this study should be submitted to H.O. and Z.B. The codes for StarryNite and AceTree used in this study are available at https://github.com/zhirongbaolab/StarryNite and https://github.com/zhirongbaolab/AceTree, respectively, and Figshare with the DOI 10.6084/m9.figshare.18133622.v2.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Table S1
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
Data S1
REFERENCES AND NOTES
- 1.Waddington C. H., Canalization of development and the inheritance of acquired characters. Nature 150, 563–565 (1942). [Google Scholar]
- 2.Eldar A., Dorfman R., Weiss D., Ashe H., Shilo B., Barkai N., Robustness of the BMP morphogen gradient in Drosophila embryonic patterning. Nature 419, 304–308 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Spemann H., Entwicklungsphysiologische Studien am Tritonei III. Arch. f. Entw. Mech. 16, 551–631 (1903). [Google Scholar]
- 4.Sulston J. E., Schierenberg E., White J. G., Thomson J. N., The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119 (1983). [DOI] [PubMed] [Google Scholar]
- 5.Renfree M. B., Fenelon J. C., The enigma of embryonic diapause. Development 144, 3199–3210 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Gilbert S. F., Ecological developmental biology: Developmental biology meets the real world. Dev. Biol. 233, 1–12 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Cassada R. C., Russell R. L., The dauer larva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 46, 326–342 (1975). [DOI] [PubMed] [Google Scholar]
- 8.Braendle C., Félix M.-A., Plasticity and errors of a robust developmental system in different environments. Dev. Cell 15, 714–724 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Hintze M., Koneru S. L., Gilbert S. P. R., Katsanos D., Lambert J., Barkoulas M., A cell fate switch in the Caenorhabditis elegans seam cell lineage occurs through modulation of the Wnt asymmetry pathway in response to temperature increase. Genetics 214, 927–939 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavalli G., Heard E., Advances in epigenetics link genetics to the environment and disease. Nature 571, 489–499 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Duempelmann L., Skribbe M., Bühler M., Small RNAs in the transgenerational inheritance of epigenetic information. Trends Genet. 36, 203–214 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Khraiwesh B., Arif M. A., Seumel G. I., Ossowski S., Weigel D., Reski R., Frank W., Transcriptional control of gene expression by microRNAs. Cell 140, 111–122 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Sommer F., Bäckhed F., The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Vuong H. E., Pronovost G. N., Williams D. W., Coley E. J. L., Siegler E. L., Qiu A., Kazantsev M., Wilson C. J., Rendon T., Hsiao E. Y., The maternal microbiome modulates fetal neurodevelopment in mice. Nature 586, 281–286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura I., Miyamoto J., Ohue-Kitano R., Watanabe K., Yamada T., Onuki M., Aoki R., Isobe Y., Kashihara D., Inoue D., Inaba A., Takamura Y., Taira S., Kumaki S., Watanabe M., Ito M., Nakagawa F., Irie J., Kakuta H., Shinohara M., Iwatsuki K., Tsujimoto G., Ohno H., Arita M., Itoh H., Hase K., Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science 367, eaaw8429 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Hodgkin J., Kuwabara P. E., Corneliussen B., A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr. Biol. 10, 1615–1618 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Darby C., Interactions with microbial pathogens. WormBook 1–15 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maduro M. F., Gut development in C. elegans. Semin. Cell Dev. Biol. 66, 3–11 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Dirksen P., Assié A., Zimmermann J., Zhang F., Tietje A., Marsh S. A., Félix M., Shapira M., Kaleta C., Schulenburg H., Samuel B. S., CeMbio—The Caenorhabditis elegans microbiome resource. G3 10, 3025–3039 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garsin D. A., Villanueva J. M., Begun J., Kim D. H., Sifri C. D., Calderwood S. B., Ruvkun G., Ausubel F. M., Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300, 1921 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Avery L., Shtonda B. B., Food transport in the C. elegans pharynx. J. Exp. Biol. 206, 2441–2457 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powell J. R., Ausubel F. M., Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. Methods Mol. Biol. 415, 403–427 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Avery L., You Y.-J., C. elegans feeding. WormBook 1–23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh J., Aballay A., Microbial colonization activates an immune fight-and-flight response via neuroendocrine signaling. Dev. Cell 49, 89–99.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuen G. J., Ausubel F. M., Both live and dead Enterococci activate Caenorhabditis elegans host defense via immune and stress pathways. Virulence 9, 683–699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garsin D. A., Sifri C. D., Mylonakis E., Qin X., Singh K. V., Murray B. E., Calderwood S. B., Ausubel F. M., A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. U.S.A. 98, 10892–10897 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahoney T. R., Luo S., Round E. K., Brauner M., Gottschalk A., Thomas J. H., Nonet M. L., Intestinal signaling to GABAergic neurons regulates a rhythmic behavior in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 105, 16350–16355 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doi M., Iwasaki K., Regulation of retrograde signaling at neuromuscular junctions by the novel C2 domain protein AEX-1. Neuron 33, 249–259 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Do T.-H., Suzuki Y., Abe N., Kaneko J., Itoh Y., Kimura K., Mutations suppressing the loss of DegQ function in Bacillus subtilis (natto) poly-γ-glutamate synthesis. Appl. Environ. Microbiol. 77, 8249–8258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogunleye A., Bhat A., Irorere V. U., Hill D., Williams C., Radecka I., Poly-γ-glutamic acid: Production, properties and applications. Microbiology 161, 1–17 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Molyneaux K. A., Stallock J., Schaible K., Wylie C., Time-lapse analysis of living mouse germ cell migration. Dev. Biol. 240, 488–498 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Abdu Y., Maniscalco C., Heddleston J. M., Chew T. L., Nance J., Developmentally programmed germ cell remodelling by endodermal cell cannibalism. Nat. Cell Biol. 18, 1302–1310 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Degennaro M., Hurd T. R., Siekhaus D. E., Biteau B., Jasper H., Lehmann R., Peroxiredoxin stabilization of DE-cadherin promotes primordial germ cell adhesion. Dev. Cell 20, 233–243 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takamura K., Fujimura M., Yamaguchi Y., Primordial germ cells originate from the endodermal strand cells in the ascidian Ciona intestinalis. Dev. Genes Evol. 212, 11–18 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Ambros V., Lee R. C., Lavanway A., Williams P. T., Jewell D., MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13, 807–818 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Ruby J. G., Jan C., Player C., Axtell M. J., Lee W., Nusbaum C., Ge H., Bartel D. P., Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193–1207 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Billi A. C., Fischer S. E. J., Kim J. K., Endogenous RNAi pathways in C. elegans. WormBook 1–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grishok A., Biology and mechanisms of short RNAs in Caenorhabditis elegans. Adv. Genet. 83, 1–69 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Roy S. H., Tobin D. V., Memar N., Beltz E., Holmen J., Clayton J. E., Chiu D. J., Young L. D., Green T. H., Lubin I., Liu Y., Conradt B., Saito R. M., A complex regulatory network coordinating cell cycles during C. elegans development is revealed by a genome-wide RNAi screen. G3 4, 795–804 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Batista P. J., Ruby J. G., Claycomb J. M., Chiang R., Fahlgren N., Kasschau K. D., Chaves D. A., Gu W., Vasale J. J., Duan S., Conte D. Jr., Luo S., Schroth G. P., Carrington J. C., Bartel D. P., Mello C. C., PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31, 67–78 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang G., Reinke V., A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 18, 861–867 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C., Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Winston W. M., Molodowitch C., Hunter C. P., Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Feinberg E. H., Hunter C. P., Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301, 1545–1547 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Jose A. M., Smith J. J., Hunter C. P., Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc. Natl. Acad. Sci. U.S.A. 106, 2283–2288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shih J. D., Hunter C. P., SID-1 is a dsRNA-selective dsRNA-gated channel. RNA 17, 1057–1065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kadekar P., Roy R., AMPK regulates germline stem cell quiescence and integrity through an endogenous small RNA pathway. PLOS Biol. 17, e3000309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Posner R., Toker I. A., Antonova O., Star E., Anava S., Azmon E., Hendricks M., Bracha S., Gingold H., Rechavi O., Neuronal small RNAs control behavior transgenerationally. Cell 177, 1814–1826.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen Z., Zhang X., Chai Y., Zhu Z., Yi P., Feng G., Li W., Ou G., Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev. Cell 30, 625–636 (2014). [DOI] [PubMed] [Google Scholar]
- 50.McEwen T. J., Yao Q., Yun S., Lee C.-Y., Bennett K. L., Small RNA in situ hybridization in Caenorhabditis elegans, combined with RNA-seq, identifies germline-enriched microRNAs. Dev. Biol. 418, 248–257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu M., Liu P., Zhang L., Cai Q., Gao G., Zhang W., Zhu Z., Liu D., Fan Q., mir-35 is involved in intestine cell G1/S transition and germ cell proliferation in C. elegans. Cell Res. 21, 1605–1618 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McJunkin K., Ambros V., The embryonic mir-35 family of microRNAs promotes multiple aspects of fecundity in Caenorhabditis elegans. G3 4, 1747–1754 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alvarez-Saavedra E., Horvitz H. R., Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20, 367–373 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez N. J., Ow M. C., Reece-Hoyes J. S., Barrasa M. I., Ambros V. R., Walhout A. J. M., Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 18, 2005–2015 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Margueron R., Reinberg D., Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 11, 285–296 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Son M., Kawasaki I., Oh B.-K., Shim Y.-H., LIN-23, an E3 ubiquitin ligase component, is required for the repression of CDC-25.2 activity during intestinal development in Caenorhabditis elegans. Mol. Cells 39, 834–840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kostic I., Roy R., Organ-specific cell division abnormalities caused by mutation in a general cell cycle regulator in C. elegans. Development 129, 2155–2165 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Hebeisen M., Roy R., CDC-25.1 stability is regulated by distinct domains to restrict cell division during embryogenesis in C. elegans. Development 135, 1259–1269 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P., The impact of microRNAs on protein output. Nature 455, 64–71 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N., Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Gordon H. A., Bruckner-Kardoss E., Wostmann B. S., Aging in germ-free mice: Life tables and lesions observed at natural death. J. Gerontol. 21, 380–387 (1966). [DOI] [PubMed] [Google Scholar]
- 62.Radoshevich L., Cossart P., Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 16, 32–46 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frøkjær-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9, 117–118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–1034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bao Z., Murray J. I., Mounting Caenorhabditis elegans embryos for live imaging of embryogenesis. Cold Spring Harb. Protoc. 2011, pdb.prot065599 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Bao Z., Murray J. I., Boyle T., Ooi S. L., Sandel M. J., Waterston R. H., Automated cell lineage tracing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 103, 2707–2712 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moyle M. W., Barnes K. M., Kuchroo M., Gonopolskiy A., Duncan L. H., Sengupta T., Shao L., Guo M., Santella A., Christensen R., Kumar A., Wu Y., Moon K. R., Wolf G., Krishnaswamy S., Bao Z., Shroff H., Mohler W. A., Colón-Ramos D. A., Structural and developmental principles of neuropil assembly in C. elegans. Nature 591, 99–104 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murray J. I., Bao Z., Boyle T. J., Waterston R. H., The lineaging of fluorescently-labeled Caenorhabditis elegans embryos with StarryNite and AceTree. Nat. Protoc. 1, 1468–1476 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Santella A., Catena R., Kovacevic I., Shah P., Yu Z., Marquina-Solis J., Kumar A., Wu Y., Schaff J., Colón-Ramos D., Shroff H., Mohler W. A., Bao Z., WormGUIDES: An interactive single cell developmental atlas and tool for collaborative multidimensional data exploration. BMC Bioinformatics 16, 189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kagias K., Podolska A., Pocock R., Reliable reference miRNAs for quantitative gene expression analysis of stress responses in Caenorhabditis elegans. BMC Genomics 15, 222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohno H., Sakai N., Adachi T., Iino Y., Dynamics of presynaptic diacylglycerol in a sensory neuron encode differences between past and current stimulus intensity. Cell Rep. 20, 2294–2303 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Ohno H., Yoshida M., Sato T., Kato J., Miyazato M., Kojima M., Ida T., Iino Y., Luqin-like RYamide peptides regulate food-evoked responses in C. elegans. eLife 6, e28877 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ohno H., Kato S., Naito Y., Kunitomo H., Tomioka M., Iino Y., Role of synaptic phosphatidylinositol 3-kinase in a behavioral learning response in C. elegans. Science 345, 313–317 (2014). [DOI] [PubMed] [Google Scholar]
- 76.Firnhaber C., Hammarlund M., Neuron-specific feeding RNAi in C. elegans and its use in a screen for essential genes required for GABA neuron function. PLOS Genet. 9, e1003921 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Table S1
Movies S1 and S2
Data S1