Abstract
Rhes (RASD2) is a thyroid hormone–induced gene that regulates striatal motor activity and promotes neurodegeneration in Huntington disease (HD) and tauopathy. Rhes moves and transports the HD protein, polyglutamine-expanded huntingtin (mHTT), via tunneling nanotube (TNT)–like membranous protrusions between cultured neurons. However, similar intercellular Rhes transportation in the intact brain was unknown. Here, we report that Rhes induces TNT-like protrusions in the striatal medium spiny neurons (MSNs) and transported between dopamine-1 receptor (D1R)–MSNs and D2R-MSNs of intact striatum and organotypic brain slices. Notably, mHTT is robustly transported within the striatum and from the striatum to the cortical areas in the brain, and Rhes deletion diminishes such transport. Moreover, Rhes moves to the cortical regions following restricted expression in the MSNs of the striatum. Thus, Rhes is a first striatum-enriched protein demonstrated to move and transport mHTT between neurons and brain regions, providing new insights into interneuronal protein transport in the brain.
Rhes enables unprecedented physical freight of neurodegenerative disease protein between the brain regions.
INTRODUCTION
The brain’s striatum plays a critical role in motor, cognitive, and psychiatric functions, and its dysregulation results in neurological and neurodegenerative illnesses, such as Huntington disease (HD). The most common cell types in the striatum are the medium-sized spiny neurons (MSNs), and during postnatal brain development, the MSNs show the highest expression of Rhes (RASD2), a thyroid hormone–induced gene that regulates striatal motor activity (1, 2). Rhes is enriched in the synaptic fractions, associates with membranes via the farnesylation domain, and mediates dopamine-related behaviors and G protein–coupled receptor and protein kinase A signaling (1, 3). In response to psychostimulants, such as amphetamines or cocaine, Rhes also forms protein-protein complexes and alters proteomics in the striatum while also inhibiting locomotor activities (4, 5). Rhes also regulates analgesia, tolerance, and dependency behaviors related to opioids (6).
In addition to its N-terminal guanosine triphosphatase (GTPase) domain, Rhes also has a C-terminal SUMO (small ubiquitin-like modifier) E3-like domain that promotes SUMOylation of diverse substrates including mHTT and promotes its solubility and toxicity in cell and animal HD models (7–10). Rhes interacts with mammalian target of rapamycin (mTOR) kinase and promotes l-dopa–induced dyskinesia in Parkinson disease (11). Apart from its role in striatal diseases, Rhes is also linked to tau pathology, and its mislocalization in human neurons is considered a hallmark of tauopathies (12, 13). These results indicate that Rhes orchestrates neuronal abnormalities associated with neurodegenerative diseases, but the precise mechanisms of its action remain largely unknown. However, given the known importance of neuronal communication in brain function, cell-to-cell communication is likely to be important.
One intriguing new mode of cell communication occurs via transient and fragile membranous protrusions, such as tunneling nanotubes (TNTs) or cytonemes (henceforth TNT-like processes) (14, 15). TNT-like processes are distinct from neurites or filopodia in cell culture in shape, size, length, and strength. Currently, there are no cellular markers that distinguish TNT-like protrusion from neurites or filopodia. However, they are mostly F-actin positive, characteristically do not adhere to the substratum, extend up to 100 μm in length, physically connect two cells, and are vulnerable to common fixation methods. These TNT-like processes are readily observed in various cell types in culture and in vivo and can be induced in cancer tissues (16). TNT-like processes can transport organelles, such as endosomes, lysosomes, and mitochondria, to neighboring cells (17, 18). Drosophila cytonemes regulate morphogenetic signals during development and can function as glutamatergic synapses (19). The emerging evidence indicates that TNT-like protrusions may play a critical role in key organism development and growth, but their roles and regulation in the brain remain unclear (20).
We serendipitously discovered that Rhes promotes the formation of TNT-like membranous protrusions that transport cargoes such as endosomes and lysosomes (20–22). Rhes itself is transported via the TNT-like protrusions and can interact with lysosomes and mitochondria in the neighboring cells (20–22). Notably, in HD, Rhes causes a several-fold enhancement of the cell-to-cell transport of mHTT, indicating a cell-nonautonomous role of Rhes in HD. Although the exact mechanisms by which cargoes of Rhes-mediated TNTs, such as mHTT, enter the acceptor cell are unknown, our live-cell time-lapse imaging studies have revealed the involvement of endocytic-like delivery mechanisms (20–22).
These data, taken together, indicate that the Rhes moves from cell to cell via the TNT-like membranous protrusions, but whether this movement and mHTT transport can occur in the intact brain is not known. Here, we address this question by developing new reporter tools and Cre recombinase transgenic (Tg) mouse models and by live-cell imaging in organotypic brain slices and mouse brain that express MSN-specific reporter Rhes.
RESULTS
Rhes promotes protrusions and moves from D2R to D1R-MSN in corticostriatal organotypic brain slices
We showed that Rhes travels between cells in the TNT-like protrusions in primary neurons and cell lines (20–22). However, whether Rhes can induce TNT in MSNs of the striatum, where it is highly expressed, is not known. Here, we determined using live-cell imaging whether Rhes can induce TNT-like protrusion in primary MSNs and in a cytoarchitecturally intact three-dimensional (3D) brain model. Most neuronal cell types, except for neuronal PC12 cells (7, 23, 24), do not express Rhes or express it only at very low levels in culture conditions, indicating that structural and functional integrity of the striatum may be necessary to ensure Rhes expression in vivo. So, we established Flex (or “flip-excision”) genetic switch (Cre-On) replication-deficient adeno-associated virus (AAVs) with PHP.eB serotype, the most efficient vector that transduces neurons (25), encoding either enhanced green fluorescent protein (EGFP) or EGFP-Rhes. This approach restricts the reporter signal to Cre-expressing neurons; therefore, only if the reporter can move from neuron to neuron will the signal be found in non–Cre-expressing neurons. We used EGFP as a reporter for Rhes because of its photostability, which allows live-cell monitoring of the dynamic membranous protrusions.
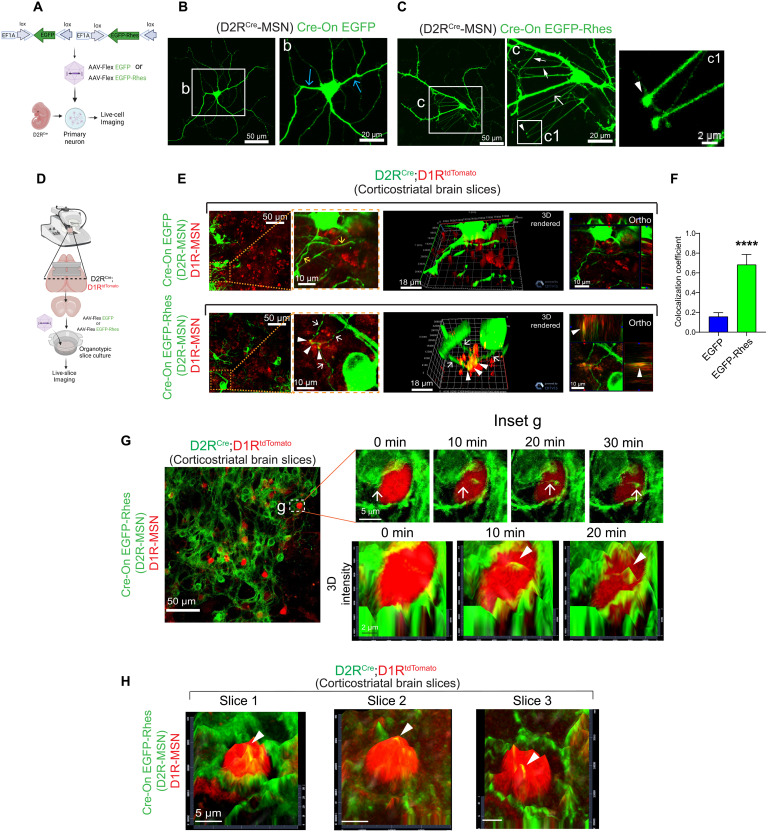
First, we determined whether Rhes can induce TNT-like protrusions in primary dopamine-2 receptor (D2R)–MSNs prepared from D2RCre mice and infected with AAV Cre-On EGFP or AAV Cre-On EGFP-Rhes (Fig. 1A). We found that D2RCre-MSNs with AAV Cre-On EGFP expression showed typical neurites characteristic of morphologically crooked and bifurcated structures emanating from the cell body (Fig. 1B, inset b, blue arrow). In contrast, Cre-On EGFP-Rhes induced the formation of long (50 to 100 μm), straight, EGFP-Rhes–positive TNT-like protrusions emanating from cell body (open arrow), as well as from the neurites (closed arrow) of D2R MSNs (Fig. 1C, inset c, arrow). These Rhes-induced TNTs also contained characteristic round vesicle-like puncta at the surface (Fig. 1C, inset c1, arrowhead), consistent with prior observations in striatal neuronal cell line (20–22).
Fig. 1. Rhes promotes TNT-like protrusions in MSNs.
(A) AAV Flex Cre-On viral vector design and infection into the D1RCre (Drd1aCre) mouse primary MSNs. (B and C) Confocal images of D2RCre-MSN expressing AAV Cre-On GFP (B) or AAV Cre-On GFP-Rhes (C). Inset b: Blue arrow indicates neurites. Inset c: White open arrow indicates TNT-like protrusions from cell body. Closed arrow indicates TNT-like protrusion from the neurites. Arrowhead indicates GFP-Rhes–positive vesicle-like blub. (D) Organotypic slice culture from D2RCre;D1RtdTomato mice infected with viral particles. (E) Confocal live-slice images and insets of organotypic brain slices transduced with Cre-On AAV viral particles. Yellow and white arrows show neuronal processes (green). The arrowhead shows GFP-Rhes puncta (red). 3D rendering and orthogonal (Ortho, single plane) display show EGFP-Rhes puncta in D1RtdTomato MSN (arrowhead). (F) Pearson’s coefficient for colocalization (n = 12, D1RtdTomato neurons from three slices); data are means ± SEM; Student’s t test, ****P < 0.0001. (G) Confocal time-lapse imaging of brain slices. Inset g (top) shows EGFP-Rhes–positive TNT-like protrusions (0 to 30 min) connecting D2RCre-MSN (green) to D1RtdTomato-MSN (red). Inset g (bottom) shows 3D intensity of movement of Rhes (arrowhead) at different time points. (H) 3D intensity from three different brain slice experiments.
Other characteristic features distinguishing TNT-like structures from neurites or filopodia in culture are their sensitivity to commonly used fixation and ability to stretch above the substratum (26). We found that Rhes-induced TNTs in D2R-MSN are disintegrated by fixative, paraformaldehyde (PFA) (fig. S1, A and B, dotted region, arrow), while certain protrusions that are most likely neurites remained intact [fig. S1, A and B, dotted region, arrowhead, compare DIC (Differential Interference Contrast) image]. PFA treatment also destroyed Rhes-induced TNT-like protrusions (arrow) while sparing filopodia-like structures (arrowhead) in the striatal neuronal cell line (fig. S1, C and D). We also found that Rhes-positive TNT-like protrusions are distinguishable from Rhes-positive neurites (fig. S1E). As shown in two examples, we found that Rhes-positive neurites are attached to the substratum (fig. S1, E and F, insets e and f, arrowhead). In contrast, Rhes-positive TNT-like protrusions hover above the substratum and connect to nearby cell body (fig. S1E, inset e, arrow) or neuronal processes (fig. S1F, inset f, arrow).
Together, these results demonstrate that Rhes induces characteristic TNT-like structures that are distinct from neurites or filopodia in the primary MSNs. The data also show that these structures are highly fragile, further emphasizing the necessity of extracting dynamic information about TNT-like structures from live-cell imaging and reporter tools.
We next determined whether TNT-like structures are formed by Rhes in vivo. Two distinct types of MSNs, expressing either D1R or D2R, have been well characterized in the striatum. Approaches involving in situ anatomical studies and Tg reporter mice have now established that less than 5% of the MSNs coexpress D1R and D2R, and these double-positive MSNs are distributed uniformly throughout the striatum (27, 28).
We used the organotypic corticostriatal brain slices from postnatal 4- to 8-day-old pups from D2RCre;D1RtdTomato mice and transduced the slices with AAV Cre-On EGFP or Cre-On EGFP-Rhes particles (Fig. 1D). As expected, the D2R neurons (Cre+) showed EGFP or EGFP-Rhes signals, marked by green fluorescence. Only a few EGFP-only–positive protrusions (yellow arrow) were observed in the Cre-On EGFP cultures, but with no clear localization with D1RtdTomato (red) neurons (Fig. 1E, top). By contrast, we observed multiple EGFP-Rhes–positive protrusions (white arrow) interacting and colocalizing (arrowhead) with D1RtdTomato (red) neurons (Fig. 1E, bottom). A 3D rendering and orthogonal projections further confirmed a clear colocalization of EGFP-Rhes protrusions with D1R-MSNs (Fig. 1, D, arrowhead, and F). Unlike their appearance in dissociated cultures (Fig. 1B), the EGFP-Rhes–positive membranous protrusions in slices appeared crooked, similar to those seen in mouse corneal explants of Tg mice expressing Cx3cr1GFP, CD11ceYFP, or major histocompatibility complex (MHC) class IIGFP (29–31). Because there are no immunohistochemical or molecular markers that can reliably discriminate TNTs from other processes, we designate these Rhes-positive structures in slices as membranous protrusions. These results indicate that Rhes induces membranous protrusions in a complex interconnected 3D organotypic brain slices.
We also imaged EGFP-Rhes–positive protrusions from D2RCre-MSNs traversing through the D1RtdTomato-MSNs by time-lapse confocal imaging of organotypic corticostriatal brain slices (Fig. 1G, inset g). We found a time-dependent induction (0 to 30 min) of EGFP-Rhes–positive protrusions from D2RCre-MSNs, which associates with the D1RtdTomato-MSNs (Fig. 1G, inset g, arrow, top). Using 3D intensity, we showed that these protrusions merge with the D1RtdTomato-MSN surface (Fig. 1G, inset g, arrowhead, bottom). This was consistently observed in three independent slice preparations, indicating potential Rhes delivery into D1R-MSN (Fig. 1H, arrowhead). Slices that express Cre-On EGFP alone did not show any obvious merging of EGFP protrusions with D1R-MSNs (fig. S2). These results showed that Rhes can induce membranous protrusions and connect with the neighboring neurons in an intact 3D brain architecture.
Rhes moves between D1R-MSN and D2R-MSN in the striatum
We next investigated the possible in vivo Rhes transport between MSNs by crossing D1RCre/+ mice with D2REGFP/+ mice. We tagged Rhes with TurboRFP, because of its intracellular stability, which allows red fluorescent protein (RFP) to be measured over a longer period of time. We confirmed that the AAV reporter construct expresses RFP-Rhes in a Cre-dependent manner and forms TNT-like protrusions (arrow) containing vesicular puncta-like bulb in their edge (arrowhead) in striatal neuronal cells in culture (fig. S3, A and B).
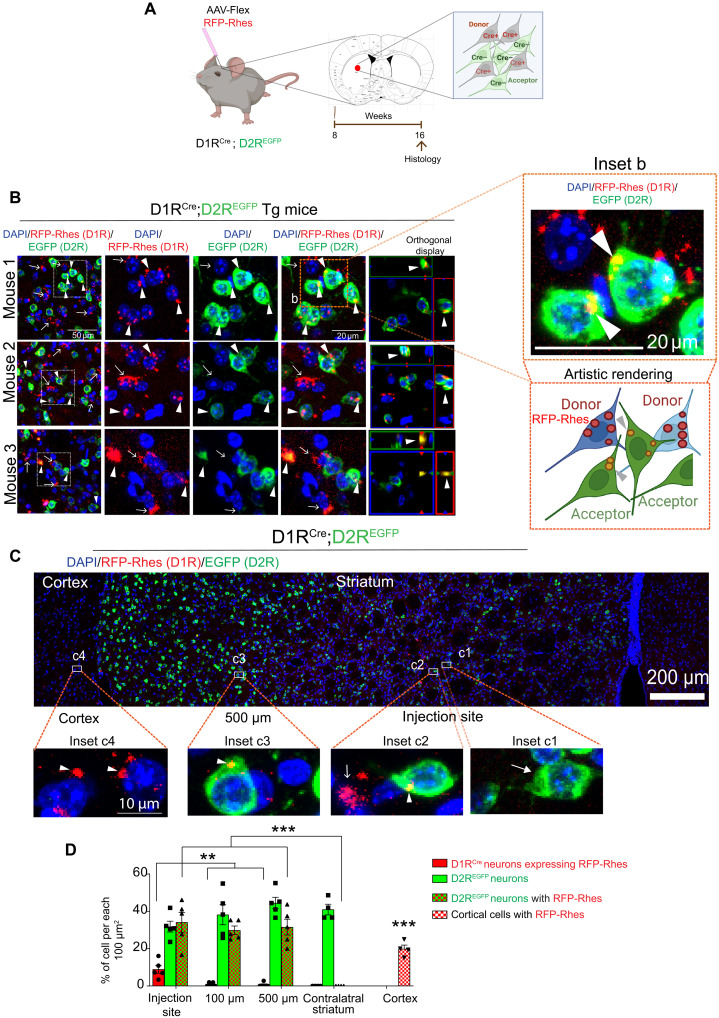
We then injected Cre-On AAV-RFP-Rhes or AAV-RFP (control) particles into the striatum in one hemisphere of 2-month-old D1RCre;D2REGFP Tg mice brain, where D1R-MSN (Cre+) is considered as donor and D2R-EGFP (Cre−) as acceptor (Fig. 2A). At 8 weeks after injection, the brain was perfused, and sections were processed for confocal imaging with 4′,6-diamidino-2-phenylindole (DAPI) as a nuclear marker. RFP-Rhes (red, Fig. 2B, arrow) signals in the D1RCre-MSN and EGFP (green) signals in D2REGFP-MSN were detected in the striatum (Fig. 2B). In addition, we found a robust RFP-Rhes signal associated with D2R-EGFP+ neurons in the striatum, and this was consistent in three different mice (Fig. 2B, arrowhead). The RFP-Rhes signals were colocalized with EGFP neurons (Fig. 2B, orthogonal display, arrowhead) and were found in the perinuclear cytoplasm of D2R MSNs (Fig. 2B, inset b, arrowhead). Using artistic rendering, we depicted the transported RFP-Rhes from D1R-MSN donor to D2R-MSN acceptor (Fig. 2B, inset b, bottom).
Fig. 2. Rhes moves from D1R-MSN to D2R-MSN in vivo.
(A) AAV-Flex in vivo model. (B) Confocal images of brain sections from D1RCre;D2REGFP mice injected with Cre-On RFP-Rhes in the striatum. Arrow indicates expression of RFP-Rhes in D1RCre(+) neurons. Arrowhead indicates RFP-Rhes expression in D2REGFP Cre(−) neurons. Arrowhead in the orthogonal display shows that RFP-Rhes puncta are colocalized with D2REGFP neuron. Artistic rendering of inset b is shown. (C) Horizontal reconstruction of confocal images of D1RCre;D2REGFP mice injected with Cre-On RFP-Rhes in the striatum. At the injected site, inset c1 shows D2REGFP neuron (closed arrow), and inset c2 shows D1RCre neurons expressing RFP-Rhes (open arrow) and D2REGFP neurons with RFP-Rhes (arrowhead). Inset c3 shows RFP-Rhes (arrowhead) in D2REGFP neurons, 500 μm away from the injected site, and inset c4 shows RFP-Rhes in the ipsilateral cortical cells. (D) Bar graph shows quantification of the % of indicated neurons from the injected site, and 100 and 500 μm away from the injection in the striatum, ipsilateral cortex [as in (C)], and contralateral striatum (n = 5 per injection, 3 male, 2 female). Data are means ± SEM. **P < 0.01 and ***P < 0.001, two-way ANOVA and Bonferroni post hoc test.
We also determined the numbers of D1R (RFP-Rhes), D2R (EGFP), and D2R neurons containing RFP-Rhes in 100-μm2–sized striatal areas at the injection site, 100 and 500 μm away from the injection site and in the cortical areas of the brain sections of the D1RCre;D2REGFP Tg mice (Fig. 2, C, insets c1 to c3, and D).
At the injection site, we found 30% of D2REGFP only neurons (Fig. 2, C, inset c1, closed arrow, and D) and 10% of D1RCre MSNs expressing RFP-Rhes (Fig. 2, C, inset c2, open arrow, and D). We also observed that at the injected site, ~30% of D2REGFP MSNs were positive for one or more RFP-Rhes puncta (Fig. 2, C, inset c2, arrowhead, and D). Even at 100 and 500 μm away from the injection site, 30% of D2REGFP neurons remained positive for RFP-Rhes (Fig. 2, C, inset c3, arrowhead, and D), indicating that Rhes transits from D1R-MSNs to D2R-MSNs in the striatum. No similar colocalization was observed in the contralateral side of the striatum, indicating that Rhes is unable to move from ipsilateral striatum to contralateral striatum (Fig. 2D). We also observed strong signals for RFP-Rhes in the cortex of D1RCre;D2REGFP mice (Fig. 2, C, inset c4, arrowhead, and D), indicating Rhes movement from striatum to cortex. Together, these results confirmed that Rhes can move from D1R-MSNs to D2R-MSNs within the striatum as well as from D1R-MSNs to the cortex in the mice brain.
Rhes promotes mHTT spreading in the brain
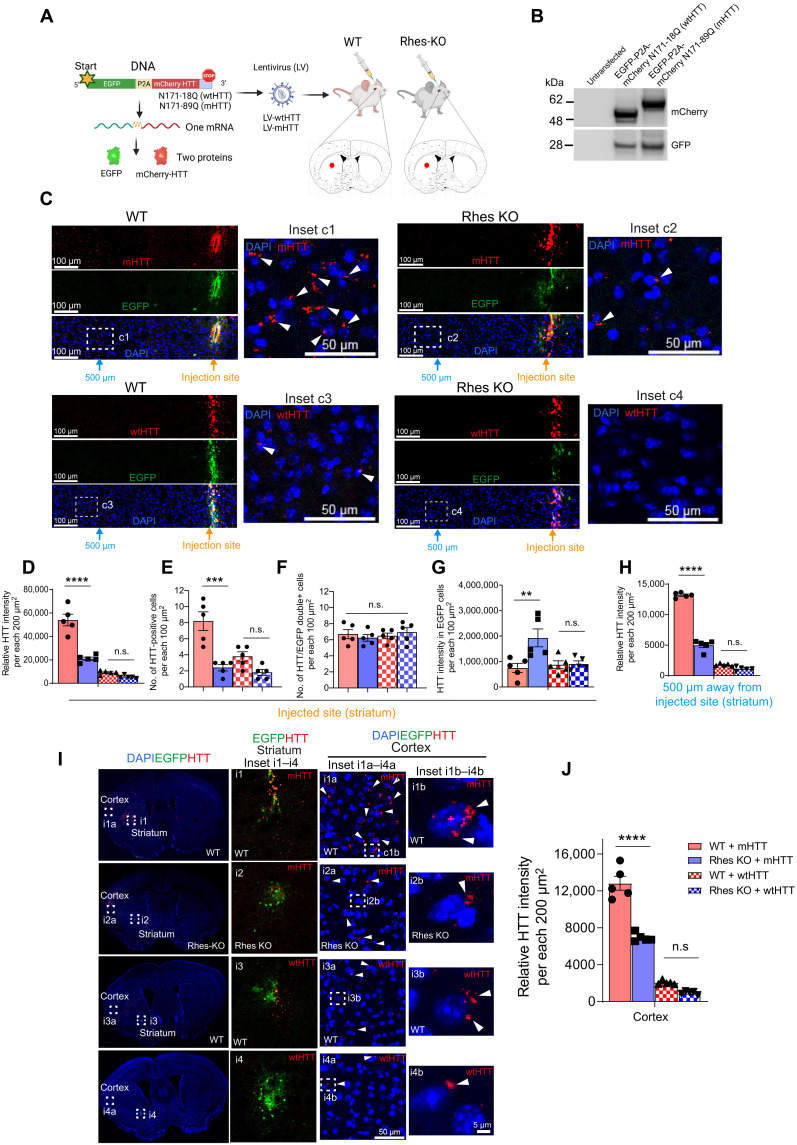
We previously showed that Rhes is a major driver of the cell-to-cell transportation of mHTT in immortalized striatal cells (22). Our finding that Rhes is transported between neurons in vivo prompted us to investigate whether mHTT can also be transported by endogenous Rhes. We prepared lentiviral (LV) bicistronic vectors expressing EGFP and mCherry-tagged HTT containing N-terminal 171 amino acids with 18Q (LV-wtHTT) or 89Q (LV-mHTT) poly-glutamine (Q) repeats, separated by the P2A sequence (Fig. 3A) (32). In this scenario, EGFP and mCherry-HTT will be expressed as two separate proteins in an infected cell, thereby allowing the investigation of HTT transport by monitoring the mCherry fluorescence. We first validated the LV vectors and confirmed similar expression levels of two separate proteins by Western blotting in striatal neuronal cell line (Fig. 3B). We then stereotaxically injected LV-wtHTT or LV-mHTT unilaterally into the striatum of 2-month-old wild-type (WT) and Rhes knockout (KO) mice. After 2 months, the brains were fixed and processed for EGFP and mCherry fluorescence detection by confocal microscopy. As expected, we observed EGFP (green) and mCherry (HTT, red) coexpression at the injection site of the striatum (Fig. 3C). We also detected the mCherry-mHTT expression alone in the 200-μm2 area around the injection site, indicating mHTT movement within the striatum (Fig. 3, C, inset c1, arrowhead, and D); however, the mCherry-HTT signal intensity was markedly reduced in the Rhes KO mice (Fig. 3, C, inset c2, arrowhead, and D). Furthermore, the number of mCherry-mHTT alone–positive cells was also substantially diminished in the striatal injected site of the Rhes KO mice compared to WT mice (Fig. 3E), but the total number of cells positive for both EGFP/mCherry was unaltered between the genotypes (Fig. 3F). This result indicates that LV infection occurs similarly between WT and Rhes KO striatum. We found a notable increase in the mCherry-mHTT intensity in the EGFP-positive cells in the striatum of Rhes KO mice compared to WT (Fig. 3G). Such differences were not observed in the mCherry-wtHTT intensity between the genotypes (Fig. 3G), indicating that mHTT particularly accumulates and spreads less in the Rhes KO striatum.
Fig. 3. Rhes promotes cell-to-cell transport of mHTT in vivo.
(A) Graphical representation of LV vectors and injection. (B) Western blotting of striatal neuronal cells. (C) Confocal image of a brain section of EGFP (green) and wtHTT or mHTT (mCherry, red) at the injection site and 500 μm away from it. Insets c1 to c4 show an enlarged portion. Arrowhead, perinuclear mHTT. Cell nuclei, DAPI (blue). (D to H) Quantification of the intensity of HTT (D), number of HTT-positive cells (E), the total number of HTT/EGFP double-positive cells (F), HTT intensity in the EGFP-positive cells (G), and mHTT intensity 500 μm away from the injection site (H). (I) Confocal images of brain sections from WT and Rhes KO mice showing the expression of EGFP (green) and wtHTT or mHTT (mCherry, red) and cell nuclei stained with DAPI (blue). Insets i1 to i4 show magnified images from the striatum, and insets i1a to i4a and i1b to i4b show magnified images from the cortex. Arrowhead indicates mHTT or wtHTT in the cortex. (J) Quantification of the intensity of HTT in 200-μm2 area in the ipsilateral cortex (n = 5 per injection, 3 male, 2 female). Data are means ± SEM. ****P < 0.0001; ***P < 0.001; **P < 0.01; not significant, one-way ANOVA and Tukey’s multiple comparisons test.
We also detected the mCherry-mHTT expression alone 500 μm away from the injection site in the WT mice of the striatum, which is markedly diminished in Rhes KO (Fig. 3H). Moreover, little wtHTT was transported within the striatum compared to mHTT, and no significant differences were detected between WT and Rhes KO mice (Fig. 3, C, insets c3 and c4, and D to H).
Besides striatum (Fig. 3I, insets i1 to i4), a strong mCherry-mHTT expression was observed in the cortex of the WT mice, which was also much diminished in the Rhes KO mice cortex (Fig. 3, I, insets i1a to i4a, arrowhead, and J). When compared to mHTT, only few wtHTT puncta were seen in WT, and it was not significantly different than Rhes KO cortex (Fig. 3, I, arrowhead, and J). Most mHTT and wtHTT puncta were distributed perinuclearly in the cortical cells of WT and Rhes KO brain (Fig. 3I, insets i1b to i4b, arrowhead). These observations indicate that the mHTT protein is transported between neurons and to cortical areas and that Rhes is a major driver for such transport.
Rhes moves from the striatum to the cortical areas of the brain
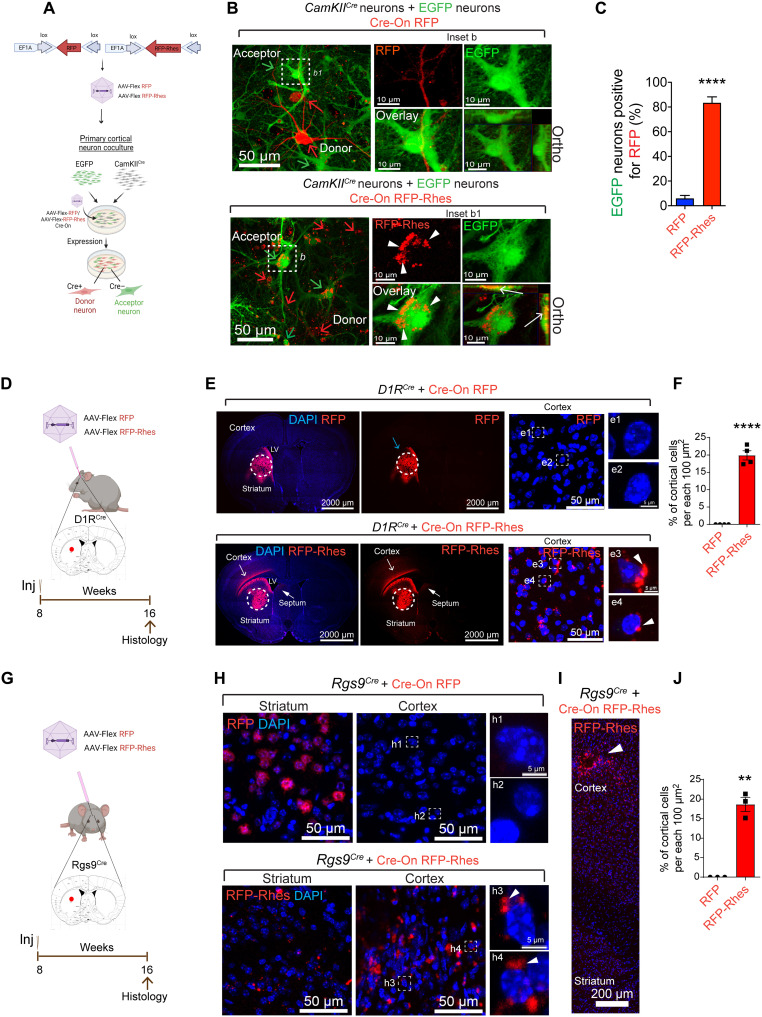
The above data suggested that Rhes can move to the cortical region, indicating propensity for extrastriatal migration. First, we examined whether Rhes might be transferred between cortical neurons in vitro. We cocultured cortical neurons of EGFP Tg mice and CamKIICre Tg mice and infected them with AAV Cre-On RFP or AAV Cre-On RFP-Rhes viral particles (Fig. 4A). Three days after the infection, we observed two distinct neuronal populations expressing Cre-On RFP in CamKIICre neurons (donor) and EGFP neurons (acceptor) without any apparent overlap (Fig. 4B). Little or no RFP signal was found in EGFP neurons (Fig. 4B, inset b). A robust transport of Cre-On RFP-Rhes from CamKIICre (donor) neuron was found in the perinuclear regions of the EGFP (Cre−) cortical acceptor neurons (Fig. 4B, inset b1, arrowhead). Orthogonal rendering further confirmed that RFP-Rhes was colocalized with the EGFP neurons (Fig. 4B, inset b1, arrow). Overall, RFP-Rhes, but not RFP alone, is transported efficiently between cortical neurons (Fig. 4C).
Fig. 4. Rhes moves from striatum to cortex.
(A) AAV-Flex and coculture model. (B) Confocal images. Red arrow, CamKIICre neurons (Cre+, donor) expressing Cre-On RFP alone or RFP-Rhes. Green arrow, EGFP neurons (Cre−, acceptor). Inset b1, arrowhead, and arrow (orthogonal display), Cre-On RFP-Rhes in the EGFP neurons. (C) Quantification of EGFP neurons positive for Cre-On RFP (n = 29) and RFP-Rhes (n = 41). Data are means ± SEM; Student’s t test, ****P < 0.0001. (D to J) AAV-Flex injection into D1RCre [Drd1aCre, (D)] or Rgs-9Cre striatum (G). Coronal brain section of the D1RCre mice (E) or Rgs-9Cre (H) injected with Cre-On RFP or RFP-Rhes. DAPI (nuclei, blue). RFP in the striatum (blue arrow). RFP-Rhes in the cortical region (open arrow) and septal regions (closed arrow). Insets e1 to e4 and h1 to h4 show the high-magnification cortex. Insets e3 and e4 (E) and h3 and h4 (H) show perinuclear Rhes in cortical cells (arrowhead). A vertical reconstruction of RFP-Rhes (arrowhead) in the Rgs-9Cre cortex (I). Arrowhead shows RFP-Rhes in the cortex. Quantification of cortical cells in the cortex of D1RCre mice [n = 4 per injection, 2 male, 2 female, (F)] or Rgs9Cre [n = 3 per injection, all male, (J)] positive for RFP or RFP-Rhes. Data are means ± SEM; Student’s t test, **P < 0.01 and ****P < 0.0001.
Next, we set out to determine further whether the intraregional transport of Rhes can occur from the striatum to cortex using D1RCre mice (Fig. 4D). We stereotaxically injected AAV Cre-On RFP (control) or Cre-On RFP-Rhes particles unilaterally into the adult mouse striatum of Drd1aCre/+ Tg mice (Fig. 4D), in which D1R-positive MSNs are primarily restricted to the striatum, as confirmed in Drd1aEGFP mice (fig. S4A). At ~8 weeks after injection, the Drd1aCre/+ Tg mouse brains were perfused, and sections were analyzed for RFP and RFP-Rhes signal by confocal microscopy. As expected, we found Cre-On RFP expression highly restricted to the striatum (Fig. 4E, top, blue arrow). By contrast, the Cre-On RFP-Rhes signals were found in the striatum, but strong signals were also observed in the cortical areas (layers V and VI) of the brain (open arrow), with some signal also evident in the septum region (closed arrow) (Fig. 4E, bottom). The RFP-Rhes signal, but not the RFP signal alone, was consistently found as perinuclear cytoplasmic structures in the cortical cells of the Drd1aCre/+ mouse cortex (Fig. 4, E, insets e1 to e4, arrowhead, and F).
We further confirmed the cortical transport of Rhes in Rgs9Cre mice (Fig. 4G) that primarily expresses Cre in the striatal MSNs (33, 34). As expected, striatal injection of Cre-On RFP in the Rgs9Cre mouse striatum resulted in RFP expression effectively restricted to the striatum (fig. S4B and Fig. 4H, top). By contrast, striatal injection of Cre-On RFP-Rhes resulted in RFP-Rhes signals in the striatum and also cortical areas of the Rgs9Cre mice (Fig. 4, H, bottom, and I and J). Signal from RFP-Rhes, but not RFP alone, was observed in the perinuclear cytoplasm of the cortical cells (Fig. 4, H, insets h1 to h4, arrowhead, and J), consistent with the observations in D1RCre mice (Fig. 4E). Together, these results indicated that Rhes protein can transit efficiently in vitro between primary cortical neurons and in vivo from the striatum to the cortex.
DISCUSSION
In this report, we provide compelling evidence that the brain-enriched protein Rhes can move from neuron to neuron in dissociated neuron preparations, in brain slices, and in intact mouse brains. Previous work has shown that prion proteins, as well as misfolded proteins, such as α-synuclein, amyloid, and tau, can move from neuron to neuron, although the mechanisms are not fully understood (35, 36). The Arc protein, which shows virus-like properties, can be transported between neurons via extracellular vesicles/exosomes (37, 38). Rhes does not share homology with prions or with Arc protein, nor is it present in exosomes (fig. S5). The findings presented here broaden the transport mechanisms for structurally unrelated proteins between neurons.
Our data suggest that neuron-to-neuron Rhes transport involves TNT-like mechanisms. Rhes is expressed in PC12 cells (7, 23, 24), in which TNTs were first reported (39). We confirmed that the depletion of endogenous Rhes in the PC12 cells diminishes TNTs, indicating that Rhes is required for TNT formation or maintenance (fig. S6). Rhes, in addition to engineering actin-based TNT-like membranous protrusions, travels by this route and is delivered to the acceptor cells (20–22). Clues by which this occurs emerged from time-lapse cell imaging studies showing that the TNT-like protrusions touch the acceptor cells and deliver vesicle-like puncta in a process that resembles endocytosis-like mechanisms of cargo uptake (20, 22). The live-cell imaging of brain slices further demonstrated that the protrusions and their related delivery mechanisms are likely operational in vivo (Fig. 1) and that Rhes transport from neuron to neuron in the adult brain (Fig. 2) can occur by a protrusion-based mechanism (20, 22).
Anatomical and electrophysiological studies have shown that D1R and D2R MSNs are interconnected by collateral axons (40–43). Our present work showed that Rhes moves between MSNs, adding a new dimension to neuronal communication in the striatum. Beyond the MSNs, the interneurons of the striatum are also positive for EGFP positivity in D2REGFP mice (44). Therefore, the possibility that Rhes might be transported to other cell types, such as interneurons in the striatum, cannot be ruled out.
Neuron-to-neuron communication via synapses and neuron to glia via extracellular fluid is necessary for neuronal functioning. Rhes-mediated TNT-like communication may provide a network of direct routes connecting the cell body and the synapses of neurons in the brain. Rhes is highly abundant in the neuronal synaptic fraction (4), where it may bridge synapses via the tiny TNT-like protrusions to transport proteins or vesicular cargoes depending upon the functional and metabolic demands of neurons and their disease state.
Previous studies have reported the cell-to-cell transport of mHTT in human HD patient striatum, as well as in mouse and Drosophila HD models (45–50). Extracellular vesicles and endocytosis are suspected to be involved in the mHTT delivery and uptake (51); however, the details of the molecular mechanism that drives mHTT transport between neurons in the brain are unclear. Notably, before disease onset, the striatal atrophy and loss of corticostriatal white matter occur with succeeding cell death in the cortex in the later stage of the disease (52, 53). Thus, a stereotypical pattern of neurodegeneration and neuron loss occurs in HD (54, 55), but whether it involves mHTT spreading is unknown. Consistent with earlier work (56), we showed that mHTT can be transported by TNT-like protrusions and that Rhes, which strongly interacts with mHTT (7), accelerates this process several fold (22). In this report, we confirm that mHTT spreading occurs in WT mice; however, the spreading is markedly diminished in the Rhes-KO brain (Fig. 3), indicating that Rhes is a physiological mediator of mHTT transport in vivo. Although Rhes is absent in the exosomes in the WT mice (fig. S5), we cannot exclude that exosomes or other concomitant mechanisms participate in Rhes-mediated interneuronal mHTT transport in the HD brain.
Several studies have indicated that mHTT has prion-like properties that may contribute to its cell-to-cell transmission (57). Previous work showed that TNTs may serve as a route for the transport of prion-like protein between cultured cells (58). An interesting possibility is that Rhes may increase the transmission of prion-like mHTT in vivo via the TNT-like processes. Recent work has shown that tau pathology and spreading can occur via TNTs, and Rhes is a critical determinant of tau pathology (12, 13, 59). The spread of tau pathology can occur from the striatum, and tau is also associated with HD pathology (60–62). We found that Rhes interacts with striatal low-density lipoprotein receptor–related protein 1 (LRP1) (4), which is involved in tau uptake and spread (63). Thus, Rhes may have a binary function in neurodegenerative disease pathology by promoting transport of both misfolded tau and mHTT from the striatum.
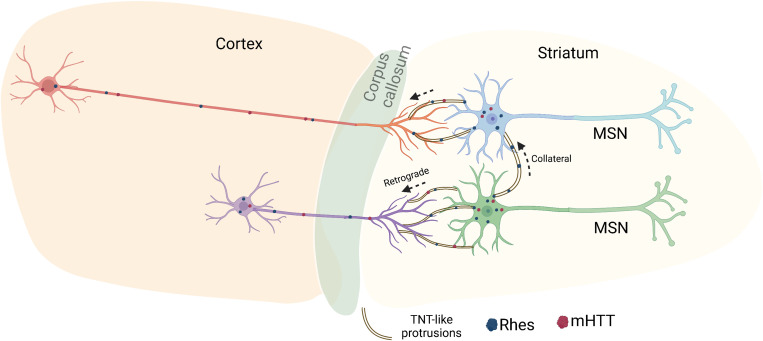
One intriguing question is how Rhes moves from the striatum to the cortex (Fig. 4). Our data support the model that Rhes promotes the formation of TNT-like processes from the MSN to connect to the axonal projections arising from the cortex (corticostriatal fibers), thereby allowing retrograde transport of Rhes (Fig. 5). Retrograde transport of proteins, and particularly the plant protein horseradish peroxidase (HRP), is well documented. HRP is transported in membrane-bound vesicles within the axons of ganglion cells in the optic tectum of the chick and rat sciatic nerves (64, 65). HRP injected into the striatum was later found in various regions of the cortex in monkeys (66). Similar to HRP (65), Rhes is found localized with lysosomes in the acceptor cells (22), indicating that HRP and Rhes might share similar destination mechanisms. On lysosomes, Rhes might influence mTORC1 signaling and autophagy (24). Besides lysosomes, Rhes is also localized to endosomes and damaged mitochondria in the acceptor cells (21, 22). Therefore, we predict that conserved intercellular retrograde transport and docking mechanisms are involved in transporting Rhes from the TNT-like protrusions of the striatal MSNs to the cortex (Fig. 5).
Fig. 5. Neuron-to-neuron Rhes transport model.
Our data indicate that Rhes moves between MSNs in the striatum as well as to the cortical areas. Live-cell imaging data from primary neurons and organotypic slice data indicate that TNT-like membranous protrusions are the key routes Rhes contacts the neighboring neurons. Thus, we predict that Rhes transports and facilitates mHTT movements in vivo potentially via the direct physical contact of neurons via membranous protrusions. Both collateral contact between MSNs and corticostriatal contacts of MSN to the cortical projections may occur via TNT-like membranous protrusions.
Our in vitro studies also indicated an involvement of Rhes in cell-to-cell transport of receptors, such as D1R, D2R, and histamine-3 receptor, but not of TMEM214, an endoplasmic reticulum–associated transmembrane protein (fig. S7), suggesting that this specific transport of cargoes by Rhes may modulate cell-autonomous signaling between neurons in an unprecedented manner. Future work should clarify the cellular and molecular mechanisms by which Rhes moves and transports cargoes between neurons and their physiological role in the brain.
In summary, our results demonstrate that Rhes transits and transports mHTT in the brain most likely involving protrusion-based communication routes between neurons and interconnecting neural pathways. The findings presented here indicate the potential of developing new therapeutic approaches for interfering with Rhes-mediated mHTT spreading in the brain to slow or prevent HD.
MATERIALS AND METHODS
Animals
Surgery and stereotaxic injections were made in adult animals (8 weeks old). Animals were housed in groups of three to five on a 12-hour light/12-hour dark cycle, and food and water were ad libitum provided. We strictly adhered to 3Rs (Replacement, Reduction, and Refinement), and all protocols were approved by the Institutional Animal Care and Use Committee at The Scripps Research Institute, Florida.
For Rhes_Flex Cre-On expression, we used previously well-characterized Cre or reporter Tg mice. D1RCre mice were obtained from MMRRC [030989-UCD-Hemi-F; B6.FVB(Cg)-Tg(Drd1-Cre)EY262Gsat/Mmucd]. D2RCre mice were purchased from MMRRC [032108-UCD-Hemi-F; B6.FVB(Cg)-Tg(Drd2-Cre)ER44Gsat/Mmucd]. D2REGFP mice were purchased from MMRRC (Mutant Mouse Resource & Research Center) [000230-UNC-Hemi-M; Tg(Drd2-EGFP)S118Gsat/Mmnc]. CamKIICre Tg mice [005359 B6.Cg-Tg(Camk2a-cre)T29-1Stl/J, homozygous], D1RTd-Tomato mice [016204; B6.Cg-Tg(Drd1a-tdTomato)6Calak/J, Hemi-M], and EGFP Tg mice [006567; C57BL/6-Tg(CAG-EGFP)131Osb/LeySopJ, homozygous] were obtained from The Jackson Laboratory. RGS9Cre mice were produced as described before (33). Double Tg mice (D1RTd-Tomato/D2RCre and D1RCre/D2REGFP) were obtained by crossing male D1RTd-Tomato with female D2RCre and female D1RCre with male D2REGFP, respectively. Female mice with the Cre transgenes were used for breeding, according to the suggestions from the company. Heterozygous condition for the Cre transgene was used for all the experiments, with the exception of RGS9-Cre, which was used in (male) homozygous form.
Construct details
Cre-On RFP and Cre-On Turbo RFP-Rhes was cloned in pAAV (FLEX Cre-On) vector under EF1A promoter (VectorBuilder). Similarly, Cre-On EGFP and Cre-On EGFP-Rhes were cloned in pAAV (FLEX Cre-On) under CAG promoter (VectorBuilder). Human RASD2 (NM_014310.3) sequences were used for Rhes expression. The details of vector and cloning can be found in fig. S8. The vectors were packaged in AAV-PHP.eB serotype and ultrapurified using VectorBuilder service (www.vectorbuilder.com). The mCherry-HTT N171 18Q or 89Q was cloned in third-generation LV vector for bicistronic expression of EGFP and the gene of interest (Addgene, 24129) and transfected in human embryonic kidney (HEK) 293T cells along with packaging plasmids. After 24 hours of transfection, the medium was changed and cells were incubated for additional 48 hours. Supernatant was collected and virus was purified, and the titer was determined by infecting HEK 293T cells by a serial dilution. pEGFP-N-Drd1 plasmid was a gift from K. Mykytyn (Addgene, 104358), GFP-DRD2 plasmid was a gift from J.-M. Arrang (Addgene, 24099), pH3R-mCherry-N1 plasmid was a gift from D. Gadella (Addgene, 84327), and Tmem214 pmRFP-N2 plasmid was a gift from E. Schirmer (Addgene, 62047). Scrambled guide RNA (gRNA) and rat-specific gRNA CRISPR constructs were custom-made from VectorBuilder. The institutional review board compliance and regulation were strictly followed while handling the AAVs and lentivirus.
Stereotaxic striatal injections
Intrastriatal surgery for AAV/lentivirus infusion was carried out using the stereotaxic coordinates as described in our previous studies (8). Briefly, adult (8 to 10 weeks old) male and female mice were injected with the virus according to the designed experiment. For surgery, the mice were anesthetized through the constant delivery of isoflurane in oxygen while mounted in a stereotaxic frame (David Kopf Instruments). Unilateral injection was made into the striatum at the following coordinates: ML (mediolateral) = ±1.6, AP (anteroposterior) = +1.0; DV (dorsoventral) = −3.6 from bregma. Viruses were injected in 0.5-μl volume. The following viruses were used: AAV RFP-Rhes (Flex) Cre-On, 1.24 × 1012 gc (genome copies)/ml; AAV RFP (Flex) Cre-On, 2.39 × 1012 gc/ml; AAV GFP-Rhes (Flex) Cre-On, 1.28 × 1012 gc/ml; AAV GFP (Flex) Cre-On, 2.12 × 1012 gc/ml; lentivirus GFP-P2A-mCherry HTT N171-18Q, 1.0 × 1012 gc/ml; lentivirus GFP-P2A-mCherry HTT N171-89Q, 1.0 × 1012 gc/ml.
Tissue fixation for imaging
Tissue was minimally processed to avoid loss of endogenous fluorescent signal. Briefly, 8 weeks after the stereotaxic injection, mice were anesthetized and perfused with cold saline solution (10 ml, 1.5 ml/min rate perfusion), followed by 4% PFA (10 ml, 1.5 ml/min rate perfusion). Mouse brains were collected and postfixed overnight in 4% PFA, cryoprotected in a sucrose/phosphate-buffered saline (PBS) gradient at 4°C (10, 20, and 30%), and then embedded in Tissue-Tek OCT (optimal cutting temperature) compound (Sakura). Coronal sections (20 μm) were collected on Superfrost/Plus slides, counterstained with DAPI, and mounted using Fluoromount-G mounting medium (Thermo Fisher Scientific). Images were obtained with a Zeiss LSM 880 microscope and processed using ZEN software (Zeiss).
Organotypic cultures
Organotypic corticostriatal slices were prepared from Tg postnatal 4- to 8-day-old mouse pups of both sexes from D2RCre+/−;D1RtdTomato+/− mice, as described in our earlier work (67). Briefly, the animals were quickly decapitated, the brain was removed, and the cerebellum and frontal hemisphere were cut. Corticostriatal coronal slices (350 μm thick) were obtained using a vibratome (Leica). Slices were collected and kept in dissection medium [minimum essential medium (MEM) (M7278, Millipore Sigma), 1% GlutaMAX (Thermo Fisher Scientific), and 1% penicillin/streptomycin (Thermo Fisher Scientific)]. Single slice was placed on interface-style Millicell culture inserts (PICM0RG50, 30 μm, hydrophilic PTFE (polytetrafluoroethylene), 0.4-μm pore size) in six-well culture plates containing 1 ml of sterile slice culture medium [50% MEM, 25% Basal Medium Eagle (B1522, Millipore Sigma), 25% heat-inactivated horse serum (Thermo Fisher Scientific), 0.6% glucose (G8769, Millipore Sigma), 2 mM GlutaMAX, and 1% antibiotic-antimycotic (15240096, Thermo Fisher Scientific)]. Brain slices were incubated at 37°C in 5% CO2. Two days later, 800 μl of the culture medium was removed and replaced with 800 μl of the culture medium. On day 5, the culture medium was replaced with low-serum Neurobasal-N1 medium (94.5% Neurobasal plus medium, 0.5% heat-inactivated horse serum, 1× N1 supplement, 2 mM glutamine, 0.6% glucose, and 1% antibiotic-antimycotic). The culture medium (Neurobasal-N1 medium) was exchanged every 3 to 4 days. On day 7, in vitro slice cultures were infected with 0.5 μl of Cre-On GFP or Cre-On GFP-Rhes using a droplet method (67). For live imaging (7 to 10 days after infection), slices were cut out of the insert (still attached to membrane), transferred into glass-bottom dishes (D11140H, Matsunami Glass) with a drop of imaging medium (A14291DJ, Thermo Fisher Scientific), and imaged using a Zeiss LSM 880 microscope, and the videos were analyzed using ZEN software.
Primary neuron culture and coculture experiments
Primary neuron culture was performed as described by Sharma and Subramaniam (22). Briefly, striata of postnatal day 1 D2RCre mice were removed and digested at 37°C for 15 min in a final concentration of 0.25% papain and resuspended in neuronal plating medium (Neurobasal-A medium, Thermo Fisher Scientific), with 5% fetal bovine serum (FBS), 0.5 mM GlutaMAX, and 1% penicillin-streptomycin. Tissues were dissociated by trituration with a pipette. Further, cells were plated in 35-mm glass-bottom dishes (D11140H, Matsunami) coated with poly-d-lysine (100 μg/ml) at the density of 2 × 105 cells per dish. Dishes were maintained in a 37°C, 5% CO2 incubator. After the cells adhered (1 to 3 hours after plating), plating media were replaced with growth media (Neurobasal-A medium, 2% B27, 0.5 mM GlutaMAX, and 1% penicillin-streptomycin). Neurons were infected with AAV Cre-On EGFP or Cre-On EGFP-Rhes at DIV (day in vitro) 10 [multiplicity of infection (MOI), 10]. Confocal mages were acquired after 48 hours in live-cell imaging solution as described earlier (22). For fixation experiments, similar strategy was used for primary neuron culture as above. Live-cell imaging was acquired from primary neurons or striatal neuronal cells expressing GFP-Rhes. Later, cells were fixed with 1% PFA for 10 s and images were captured from the same field to assess the effect of PFA fixation on TNT stability. For coculture experiments, primary cortical neurons were prepared from CamKIICre mice and EGFP Tg mice. Cells were plated in 1:1 ratio and cultured and infected using AAV Cre-On RFP or Cre-On RFP-Rhes (MOI 10) as mentioned above. The percentage of EGFP neurons positive for RFP signal for RFP Cre-On and RFP-Rhes Cre-On groups was quantified. PC12 cells stably expressing scramble or rat-Rasd2 gRNA were grown as described before (24).
Confocal imaging
Fixed tissue and organotypic cultures were imaged using a Zeiss LSM 880 confocal system. Whole coronal reconstructions were acquired using a 20× objective, with a Z-stack of three planes. High-magnification images and partial corticostriatal reconstructions were acquired using a 63× objective, and optical zoom was adjusted according to the field of interest. Live imaging of organotypic slices was acquired using a 40× objective with a 0.5 optical zoom. Primary neurons and striatal neuronal cells transfected with reporter Rhes and reporter cargoes were live-cell imaged as described before (22).
Quantification
Confocal images were used to quantify the protein expression using ImageJ software. The average of two to three sections from each mouse was used for group analysis, and the average of two to three different areas from each region (injection site, at 100 and 500 μm from the injection site, contralateral striatum) was analyzed. The proportion of D1, D2, and cortical cells with Rhes, as well as D2 alone without Rhes, after injection was made by quantifying each kind of neuron in 100-μm2 areas from each region. The total number of nuclei stained with DAPI was considered as 100% for each 100 μm2 analyzed. Similarly, ipsilateral cortical area intensity of RFP or RFP-Rhes was determined in D1RCre and Rgs9Cre mice brain sections in 100-μm2 area.
Transportation of the mCherry-HTT (N171-18Q) protein or the mCherry-mHTT (N171-89Q) protein was measured using ImageJ software by quantifying the relative intensity for the HTT or mHTT expression (mCherry, red channel). The average of two areas (200 μm2) from each region (injection site, 500 μm from the injection site and cortex) was analyzed for group results (site injection was identified by the EGFP expression). Quantification of the number of EGFP/mCherry double-positive cells in the striatal area of the WT and Rhes-KO mice was carried out in three to five regions of interest (ROIs) per mice in 100-μm2 injected striatal area. mCherry-HTT/mHTT intensity in EGFP-positive cells in 100-μm2 injected striatal area was carried out in three to five ROIs per mice (n = 5 mice, mixed sex).
Exosome isolation
The exosome isolation was performed using a protocol described previously (68). Briefly, the striatum tissue was dissected out, weighed, and transferred to a 50-ml tube containing collagenase (75 U/ml) in PBS (total volume of PBS was 6 ml). The tissue was then incubated in a shaking water bath at 37°C for a total of 30 min. The tube was mixed three times during incubation by gentle inversion (after 10-min incubation). The collagenase was diluted by adding ice-cold PBS (up to 44 ml to make the total volume of 50 ml). Once the tissue was settled down, around 48 ml of PBS was discarded. The washing step was repeated one more time to further dilute out the collagenase. Protease and phosphatase inhibitors (Millipore Sigma) were added to a final concentration of 1× in the remaining 2 ml of PBS. Tissue was introduced to gentle pipetting (10 strokes). The dissociated tissue was spun at 300g for 5 min at 4°C, the pellet was collected for input, and the supernatant was transferred to a fresh tube and spun at 2000g for 10 min at 4°C. The pellet was discarded, and the supernatant was spun at 10,000g for 30 min at 4°C. The supernatant was overlaid on sucrose cushion (10, 20, 30, 40, and 50%). The gradient was spun for 3 hours at 180,000g (average) at 4°C (SW41 Beckman ultracentrifuge). After the spin, the top of the gradient was removed and discarded, and the 10% fraction (F1) was designated number 1. Fractions 2 (20%), 3 (30%) 4 (40%), and 5 (50%) were subsequently collected, and the protein was precipitated via the methanol-chloroform method (21). Exosome isolation was confirmed by flotillin antibody in Western blot.
Western blotting
Striatal neuronal cells (STHdhQ7/Q7) grown in growth medium containing Dulbecco’s modified Eagle’s medium with high glucose (Thermo Fisher Scientific) with 10% FBS, transfected with indicated plasmids, or infected with AAVs and lysed in radioimmunoprecipitation assay buffer and equal proteins were loaded onto SDS–polyacrylamide gel electrophoresis and transferred as described before (4). Primary antibodies against Rhes (1:500; FabGennix, #RHES-101AP), GFP (1:5000; Cell Signaling Technology, #2956), mCherry (1:5000; Novus Biologicals, #NBP2-25157), and flotillin (1:1000; Cell Signaling Technology, #18634) and secondary antibodies conjugated to HRP (1:10,000; The Jackson Laboratory) were used.
Statistical analysis
Data are presented as means ± SEM as indicated. Variance was found to be similar between the groups tested. Statistical analysis was performed with a Student’s t test or one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test or two-way ANOVA followed by Bonferroni post hoc test. Significance was set at P < 0.05. All statistical tests were performed using Prism 9.0 (GraphPad software).
Acknowledgments
We would like to thank D. Fitzpatrick, R. Davis, and B. Xu for critical reading of the manuscript and for constructive suggestions. We thank M. Benilous for administrative help and the members of the laboratory for continuous support and collaborative atmosphere. We would like to thank members at the Max Planck Institute of Neuroscience, Jupiter, FL microscopy core for help and expertise. We also thank BioRender application tools for model preparation as described in the article.
Funding: This research was supported by NIH/NINDS R01-NS087019-01A1, NIH/NINDS R01-NS094577-01A1, a grant from Cure for Huntington Disease Research Initiative (CHDI) foundation, and Scripps bridge funding.
Author contributions: S.S. conceptualized the project and co-designed experiments with U.N.R.-J., who performed mouse experiments, immunohistochemistry, immunostaining, confocal, colocalization analysis, primary neuron, and slice culture and analyzed the data. M.S. designed P2A constructs, generated lentivirus, and performed primary neuron, cell culture, and exosome experiments. N.S. co-designed and prepared brain slice experiments and in vivo injections as well as Western blotting. Y.L. generated the Rgs9Cre mice. S.B. interpreted the data and provided intellectual contribution. S.S. wrote the manuscript with inputs from coauthors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
REFERENCES AND NOTES
- 1.Vargiu P., Morte B., Manzano J., Perez J., de Abajo R., Gregor Sutcliffe J., Bernal J., Thyroid hormone regulation of rhes, a novel Ras homolog gene expressed in the striatum. Brain Res. Mol. Brain Res. 94, 1–8 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Falk J. D., Vargiu P., Foye P. E., Usui H., Perez J., Danielson P. E., Lerner D. L., Bernal J., Sutcliffe J. G., Rhes: A striatal-specific Ras homolog related to Dexras1. J. Neurosci. Res. 57, 782–788 (1999). [PubMed] [Google Scholar]
- 3.Napolitano F., D’Angelo L., de Girolamo P., Avallone L., de Lange P., Usiello A., The thyroid hormone-target gene rhes a novel crossroad for neurological and psychiatric disorders: New insights from animal models. Neuroscience 384, 419–428 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Shahani N., Swarnkar S., Giovinazzo V., Morgenweck J., Bohn L. M., Scharager-Tapia C., Pascal B., Martinez-Acedo P., Khare K., Subramaniam S., RasGRP1 promotes amphetamine-induced motor behavior through a Rhes interaction network (“Rhesactome”) in the striatum. Sci. Signal. 9, ra111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Napolitano F., De Rosa A., Russo R., Di Maio A., Garofalo M., Federici M., Migliarini S., Ledonne A., Rizzo F. R., Avallone L., Nuzzo T., Biagini T., Pasqualetti M., Mercuri N. B., Mazza T., Chambery A., Usiello A., The striatal-enriched protein Rhes is a critical modulator of cocaine-induced molecular and behavioral responses. Sci. Rep. 9, 15294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee F. A., Baiamonte B. A., Spano D., Lahoste G. J., Soignier R. D., Harrison L. M., Mice lacking rhes show altered morphine analgesia, tolerance, and dependence. Neurosci. Lett. 489, 182–186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subramaniam S., Sixt K. M., Barrow R., Snyder S. H., Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science 324, 1327–1330 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swarnkar S., Chen Y., Pryor W. M., Shahani N., Page D. T., Subramaniam S., Ectopic expression of the striatal-enriched GTPase Rhes elicits cerebellar degeneration and an ataxia phenotype in Huntington’s disease. Neurobiol. Dis. 82, 66–77 (2015). [DOI] [PubMed] [Google Scholar]
- 9.O. Rivera, M. Sharma, N. Shahani, U. N. Ramírez-Jarquín, G. Crynen, P. Karunadharma, F. McManus, T. Pierre, S. Subramaniam, Rhes, a striatal enriched protein, regulates post-translational small-ubiquitin-like-modifier (SUMO) modification of nuclear proteins and alters gene expression. bioRxiv 2020.06.18.160044 [Preprint]. 19 June 2020. 10.1101/2020.06.18.160044. [DOI] [PMC free article] [PubMed]
- 10.Subramaniam S., Mealer R. G., Sixt K. M., Barrow R. K., Usiello A., Snyder S. H., Rhes, a physiologic regulator of sumoylation, enhances cross-sumoylation between the basic sumoylation enzymes E1 and Ubc9. J. Biol. Chem. 285, 20428–20432 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramaniam S., Napolitano F., Mealer R. G., Kim S., Errico F., Barrow R., Shahani N., Tyagi R., Snyder S. H., Usiello A., Rhes, a striatal-enriched small G protein, mediates mTOR signaling and l-DOPA–induced dyskinesia. Nat. Neurosci. 15, 191–193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrenberg A. J., Leng K., Letourneau K. N., Hernandez I., Lew C., Seeley W. W., Spina S., Miller B., Heinsen H., Kampmann M., Kosik K. S., Grinberg L. T., Patterns of neuronal Rhes as a novel hallmark of tauopathies. Acta Neuropathol. 141, 651–666 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez I., Luna G., Rauch J. N., Reis S. A., Giroux M., Karch C. M., Boctor D., Sibih Y. E., Storm N. J., Diaz A., Kaushik S., Zekanowski C., Kang A. A., Hinman C. R., Cerovac V., Guzman E., Zhou H., Haggarty S. J., Goate A. M., Fisher S. K., Cuervo A. M., Kosik K. S., A farnesyltransferase inhibitor activates lysosomes and reduces tau pathology in mice with tauopathy. Sci. Transl. Med. 11, eaat3005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornberg T. B., A path to pattern. Curr. Top. Dev. Biol. 116, 551–567 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Zurzolo C., Tunneling nanotubes: Reshaping connectivity. Curr. Opin. Cell Biol. 71, 139–147 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Sahu P., Jena S. R., Samanta L., Tunneling nanotubes: A versatile target for cancer therapy. Curr. Cancer Drug Targets 18, 514–521 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Lim Y. S., Tang B. L., Intercellular organelle trafficking by membranous nanotube connections: A possible new role in cellular rejuvenation? Cell Commun. Adhes. 19, 39–44 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Pinto G., Saenz-de-Santa-Maria I., Chastagner P., Perthame E., Delmas C., Toulas C., Moyal-Jonathan-Cohen E., Brou C., Zurzolo C., Patient-derived glioblastoma stem cells transfer mitochondria through tunneling nanotubes in tumor organoids. Biochem. J. 478, 21–39 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang H., Liu S., Kornberg T. B., Glutamate signaling at cytoneme synapses. Science 363, 948–955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramaniam S., Rhes tunnels: A radical new way of communication in the brain’s striatum? Bioessays 42, e1900231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma M., Jarquin U. N. R., Rivera O., Kazantzis M., Eshraghi M., Shahani N., Sharma V., Tapia R., Subramaniam S., Rhes, a striatal-enriched protein, promotes mitophagy via Nix. Proc. Natl. Acad. Sci. U.S.A. 116, 23760–23771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma M., Subramaniam S., Rhes travels from cell to cell and transports Huntington disease protein via TNT-like protrusion. J. Cell Biol. 218, 1972–1993 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vargiu P., De Abajo R., Garcia-Ranea J. A., Valencia A., Santisteban P., Crespo P., Bernal J., The small GTP-binding protein, Rhes, regulates signal transduction from G protein-coupled receptors. Oncogene 23, 559–568 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Mealer R. G., Murray A. J., Shahani N., Subramaniam S., Snyder S. H., Rhes, a striatal-selective protein implicated in Huntington disease, binds beclin-1 and activates autophagy. J. Biol. Chem. 289, 3547–3554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dayton R. D., Grames M. S., Klein R. L., More expansive gene transfer to the rat CNS: AAV PHP.EB vector dose-response and comparison to AAV PHP.B. Gene Ther. 25, 392–400 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Dubois F., Benard M., Jean-Jacques B., Schapman D., Roberge H., Lebon A., Goux D., Monterroso B., Elie N., Komuro H., Bazille C., Levallet J., Bergot E., Levallet G., Galas L., Investigating tunneling nanotubes in cancer cells: Guidelines for structural and functional studies through cell imaging. Biomed. Res. Int. 2020, 2701345 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thibault D., Loustalot F., Fortin G. M., Bourque M. J., Trudeau L. E., Evaluation of D1 and D2 dopamine receptor segregation in the developing striatum using BAC transgenic mice. PLOS ONE 8, e67219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren K., Guo B., Dai C., Yao H., Sun T., Liu X., Bai Z., Wang W., Wu S., Striatal distribution and cytoarchitecture of dopamine receptor subtype 1 and 2: Evidence from double-labeling transgenic mice. Front. Neural Circuits 11, 57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salas-Vidal E., Lomeli H., Imaging filopodia dynamics in the mouse blastocyst. Dev. Biol. 265, 75–89 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Chinnery H. R., Pearlman E., McMenamin P. G., Cutting edge: Membrane nanotubes in vivo: A feature of MHC class II+ cells in the mouse cornea. J. Immunol. 180, 5779–5783 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seyed-Razavi Y., Hickey M. J., Kuffova L., McMenamin P. G., Chinnery H. R., Membrane nanotubes in myeloid cells in the adult mouse cornea represent a novel mode of immune cell interaction. Immunol. Cell Biol. 91, 89–95 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Liu Z., Chen O., Wall J. B. J., Zheng M., Zhou Y., Wang L., Vaseghi H. R., Qian L., Liu J., Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 7, 2193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao G. Y., Li Y., Xu B., Ablation of TrkB expression in RGS9-2 cells leads to hyperphagic obesity. Mol. Metab. 2, 491–497 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang M. T., Yokoi F., Yin H. H., Lovinger D. M., Wang Y., Li Y., Disrupted motor learning and long-term synaptic plasticity in mice lacking NMDAR1 in the striatum. Proc. Natl. Acad. Sci. U.S.A. 103, 15254–15259 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis A. A., Leyns C. E. G., Holtzman D. M., Intercellular spread of protein aggregates in neurodegenerative disease. Annu. Rev. Cell Dev. Biol. 34, 545–568 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng C., Trojanowski J. Q., Lee V. M., Protein transmission in neurodegenerative disease. Nat. Rev. Neurol. 16, 199–212 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashley J., Cordy B., Lucia D., Fradkin L. G., Budnik V., Thomson T., Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell 172, 262–274.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastuzyn E. D., Day C. E., Kearns R. B., Kyrke-Smith M., Taibi A. V., McCormick J., Yoder N., Belnap D. M., Erlendsson S., Morado D. R., Briggs J. A. G., Feschotte C., Shepherd J. D., The neuronal gene arc encodes a repurposed retrotransposon Gag protein that mediates intercellular RNA transfer. Cell 173, 275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rustom A., Saffrich R., Markovic I., Walther P., Gerdes H. H., Nanotubular highways for intercellular organelle transport. Science 303, 1007–1010 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Bolam J. P., Somogyi P., Takagi H., Fodor I., Smith A. D., Localization of substance P-like immunoreactivity in neurons and nerve terminals in the neostriatum of the rat: A correlated light and electron microscopic study. J. Neurocytol. 12, 325–344 (1983). [DOI] [PubMed] [Google Scholar]
- 41.Yung K. K., Smith A. D., Levey A. I., Bolam J. P., Synaptic connections between spiny neurons of the direct and indirect pathways in the neostriatum of the rat: Evidence from dopamine receptor and neuropeptide immunostaining. Eur. J. Neurosci. 8, 861–869 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Taverna S., Ilijic E., Surmeier D. J., Recurrent collateral connections of striatal medium spiny neurons are disrupted in models of Parkinson’s disease. J. Neurosci. 28, 5504–5512 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei W., Ding S., Zhou F. M., Dopaminergic treatment weakens medium spiny neuron collateral inhibition in the parkinsonian striatum. J. Neurophysiol. 117, 987–999 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertran-Gonzalez J., Bosch C., Maroteaux M., Matamales M., Herve D., Valjent E., Girault J. A., Opposing patterns of signaling activation in dopamine D1 and D2 receptor-expressing striatal neurons in response to cocaine and haloperidol. J. Neurosci. 28, 5671–5685 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cicchetti F., Lacroix S., Cisbani G., Vallieres N., Saint-Pierre M., St-Amour I., Tolouei R., Skepper J. N., Hauser R. A., Mantovani D., Barker R. A., Freeman T. B., Mutant huntingtin is present in neuronal grafts in Huntington disease patients. Ann. Neurol. 76, 31–42 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Pecho-Vrieseling E., Rieker C., Fuchs S., Bleckmann D., Esposito M. S., Botta P., Goldstein C., Bernhard M., Galimberti I., Muller M., Luthi A., Arber S., Bouwmeester T., van der Putten H., Di Giorgio F. P., Transneuronal propagation of mutant huntingtin contributes to non-cell autonomous pathology in neurons. Nat. Neurosci. 17, 1064–1072 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Jeon I., Cicchetti F., Cisbani G., Lee S., Li E., Bae J., Lee N., Li L., Im W., Kim M., Kim H. S., Oh S. H., Kim T. A., Ko J. J., Aube B., Oueslati A., Kim Y. J., Song J., Human-to-mouse prion-like propagation of mutant huntingtin protein. Acta Neuropathol. 132, 577–592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Babcock D. T., Ganetzky B., Transcellular spreading of huntingtin aggregates in the Drosophila brain. Proc. Natl. Acad. Sci. U.S.A. 112, E5427–E5433 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pearce M. M. P., Spartz E. J., Hong W., Luo L., Kopito R. R., Prion-like transmission of neuronal huntingtin aggregates to phagocytic glia in the Drosophila brain. Nat. Commun. 6, 6768 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Donnelly K. M., DeLorenzo O. R., Zaya A. D., Pisano G. E., Thu W. M., Luo L., Kopito R. R., Panning Pearce M. M., Phagocytic glia are obligatory intermediates in transmission of mutant huntingtin aggregates across neuronal synapses. eLife 9, e58499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuang X., Nunn K., Jiang J., Castellano P., Hardikar U., Horgan A., Kong J., Tan Z., Dai W., Structural insight into transmissive mutant huntingtin species by correlative light and electron microscopy and cryo-electron tomography. Biochem. Biophys. Res. Commun. 560, 99–104 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McColgan P., Seunarine K. K., Razi A., Cole J. H., Gregory S., Durr A., Roos R. A., Stout J. C., Landwehrmeyer B., Scahill R. I., Clark C. A., Rees G., Tabrizi S. J.; Track-HD Investigators , Selective vulnerability of rich club brain regions is an organizational principle of structural connectivity loss in Huntington’s disease. Brain 138, 3327–3344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rub U., Seidel K., Heinsen H., Vonsattel J. P., den Dunnen W. F., Korf H. W., Huntington’s disease (HD): The neuropathology of a multisystem neurodegenerative disorder of the human brain. Brain Pathol. 26, 726–740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McColgan P., Helbling S., Vaculciakova L., Pine K., Wagstyl K., Attar F. M., Edwards L., Papoutsi M., Wei Y., Van den Heuvel M. P., Tabrizi S. J., Rees G., Weiskopf N., Relating quantitative 7T MRI across cortical depths to cytoarchitectonics, gene expression and connectomics. Hum. Brain Mapp. 42, 4996–5009 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vonsattel J. P., Myers R. H., Stevens T. J., Ferrante R. J., Bird E. D., Richardson E. P. Jr., Neuropathological classification of Huntington’s disease. J. Neuropathol. Exp. Neurol. 44, 559–577 (1985). [DOI] [PubMed] [Google Scholar]
- 56.Costanzo M., Abounit S., Marzo L., Danckaert A., Chamoun Z., Roux P., Zurzolo C., Transfer of polyglutamine aggregates in neuronal cells occurs in tunneling nanotubes. J. Cell Sci. 126, 3678–3685 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Alpaugh M., Denis H. L., Cicchetti F., Prion-like properties of the mutant huntingtin protein in living organisms: The evidence and the relevance. Mol. Psychiatry (2021). [DOI] [PubMed] [Google Scholar]
- 58.Gousset K., Schiff E., Langevin C., Marijanovic Z., Caputo A., Browman D. T., Chenouard N., de Chaumont F., Martino A., Enninga J., Olivo-Marin J. C., Mannel D., Zurzolo C., Prions hijack tunnelling nanotubes for intercellular spread. Nat. Cell Biol. 11, 328–336 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Tardivel M., Begard S., Bousset L., Dujardin S., Coens A., Melki R., Buee L., Colin M., Tunneling nanotube (TNT)-mediated neuron-to neuron transfer of pathological Tau protein assemblies. Acta Neuropathol. Commun. 4, 117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zareba-Paslawska J., Patra K., Kluzer L., Revesz T., Svenningsson P., Tau isoform-driven CBD pathology transmission in oligodendrocytes in humanized tau mice. Front. Neurol. 11, 589471 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vuono R., Winder-Rhodes S., de Silva R., Cisbani G., Drouin-Ouellet J.; REGISTRY Investigators of the European Huntington’s Disease Network, Spillantini M. G., Cicchetti F., Barker R. A., The role of tau in the pathological process and clinical expression of Huntington’s disease. Brain 138, 1907–1918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schulze Westhoff M., Osmanovic A., Meissner C., Heck J., Mahmoudi N., Hendrich C., Berding G., Seifert J., Bleich S., Frieling H., Kruger T., Groh A., An unusual presentation of Huntington’s disease. Clin. Case Rep. 9, e04547 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rauch J. N., Luna G., Guzman E., Audouard M., Challis C., Sibih Y. E., Leshuk C., Hernandez I., Wegmann S., Hyman B. T., Gradinaru V., Kampmann M., Kosik K. S., LRP1 is a master regulator of tau uptake and spread. Nature 580, 381–385 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.LaVail J. H., LaVail M. M., Retrograde axonal transport in the central nervous system. Science 176, 1416–1417 (1972). [DOI] [PubMed] [Google Scholar]
- 65.Feher E., Electron microscopic study of retrograde axonal transport of horseradish peroxidase. Int. Rev. Cytol. 90, 1–29 (1984). [DOI] [PubMed] [Google Scholar]
- 66.Jones E. G., Coulter J. D., Burton H., Porter R., Cells of origin and terminal distribution of corticostriatal fibers arising in the sensory-motor cortex of monkeys. J. Comp. Neurol. 173, 53–80 (1977). [DOI] [PubMed] [Google Scholar]
- 67.Shahani N., Subramaniam S., Wolf T., Tackenberg C., Brandt R., Tau aggregation and progressive neuronal degeneration in the absence of changes in spine density and morphology after targeted expression of Alzheimer’s disease-relevant tau constructs in organotypic hippocampal slices. J. Neurosci. 26, 6103–6114 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vella L. J., Scicluna B. J., Cheng L., Bawden E. G., Masters C. L., Ang C. S., Willamson N., McLean C., Barnham K. J., Hill A. F., A rigorous method to enrich for exosomes from brain tissue. J. Extracell. Vesicles 6, 1348885 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8