Abstract
Somatostatin (SS) is a peptide hormone with diverse physiological roles. By investigating a deep-water clade of fish-hunting cone snails, we show that predator-prey evolution has generated a diverse set of SS analogs, each optimized to elicit specific systemic physiological effects in prey. The increased metabolic stability, distinct SS receptor activation profiles, and chemical diversity of the venom analogs make them suitable leads for therapeutic application, including pain, cancer, and endocrine disorders. Our findings not only establish the existence of SS-like peptides in animal venoms but also serve as a model for the synergy gained from combining molecular phylogenetics and behavioral observations to optimize the discovery of natural products with biomedical potential.
Venomous cone snails with a previously unknown hunting strategy use weaponized hormones to catch fish.
INTRODUCTION
Cone snails comprise a large family of venomous marine predators (family Conidae). The complete lack of offensive mechanical weaponry in this diverse molluscan lineage has been compensated for by the evolution of highly sophisticated pharmacological strategies for prey capture (1). Because of their stability, chemical diversity, and target selectivity, cone snail toxins (conotoxins) have been developed as biomedical tools, drug leads, and one approved drug (2–4) and have elucidated previously unknown signaling pathways in health and disease (5, 6).
The ~1000 living species of cone snails can be broadly divided into those that hunt fish, gastropod mollusks, or marine worms (primarily polychaetes) (1). Because many of the molecular targets expressed in fish also play analogous important physiological roles in humans, fish-hunting Conus species have been the focus of many cone snail venom research programs. Eight lineages of fish hunters exist, comprising approximately 150 extant species (7). A phylogenetic reconstruction of representatives of these lineages is shown in Fig. 1A. Within these, diverse predation strategies have evolved for capturing fish prey.
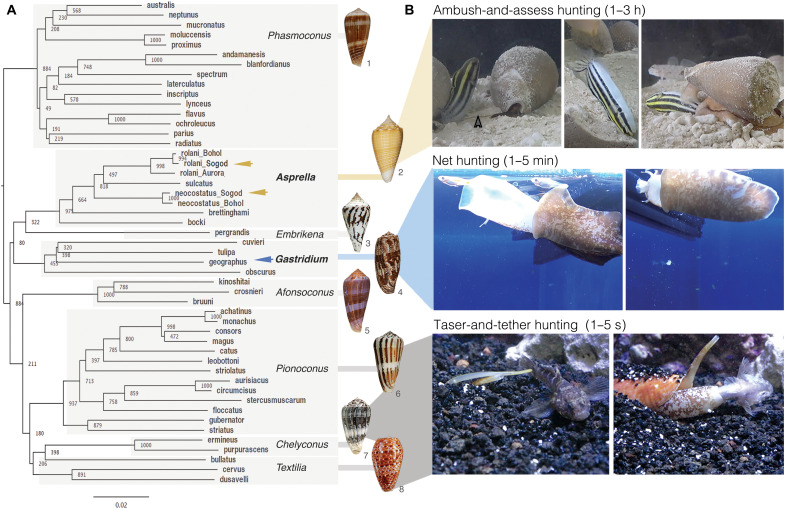
Fig. 1. Evolution of distinct predation strategies in different clades of fish-hunting cone snails.
(A) Phylogenetic tree of the eight recognized clades of fish hunters. Bootstrap values and shell images of neotypes are shown (1: Conus radiatus, 2: Conus sulcatus, 3: Conus pergrandis, 4: Conus geographus, 5: Conus kinoshitai, 6: Conus magus, 7: Conus ermineus, 8: Conus bullatus). Arrows show the species used in this work, with color corresponding to the proposed hunting strategy shown in (B). (B) Still images of three distinct predation strategies filmed for three different species of fish hunters [from top to bottom: C. neocostatus (Asprella clade, gray arrow marks proboscis loaded with radular tooth), C. geographus (Gastridium clade), and C. bullatus (Textilia clade)]. Images were extracted from supporting movie files (movies S1 to S3). Video/photo credit: Dylan Taylor and Baldomero M. Olivera, University of Utah.
The most widespread and best studied fish-hunting strategy is called taser-and-tether (8). First described in 1956, it is characterized by a rapid chemical electrocution of prey that is facilitated by the action of diverse ion channel modulators (Fig. 1B and movie S1) (8). The underlying molecular mechanisms of taser-and-tether toxins have been well characterized and have provided a rich set of pharmacological tools and drug leads, including an approved therapeutic for the treatment of chronic pain (3).
A second predation strategy, net hunting, has been observed in only two species, Conus geographus and Conus tulipa from the Gastridium clade. Behavioral observations dating back to the early 1970s showed that the net-hunting strategy is characterized by a pre-capture phase during which venom components released into the water cause fish to become both sensory deprived and disoriented, thereby facilitating prey capture (1, 9). The mixture of compounds that elicits this response was called the nirvana cabal because it makes fish appear as if under the influence of narcotic drugs (1). We provide a video recording showing that the net-hunting strategy enables simultaneous capture of multiple fish (Fig. 1 and movie S2). Nirvana-cabal toxins have proven biomedical applications, including a diagnostic tool for an autoimmune disorder (10) and a family of insulins that have inspired the design of fast-acting drug leads for diabetes (11, 12). C. geographus venom has also provided one of the most widely used pharmacological tools in molecular neuroscience for studying synaptic transmission, the calcium channel blocker ω-conotoxin GVIA (3).
In this article, we document the discovery that a largely unexplored, deep-water lineage of fish-hunting cone snails of the Asprella clade uses a markedly different predation strategy that we call “ambush-and-assess.” In contrast to the rapid and efficient capture previously described, direct observation of the Asprella envenomation behavior demonstrates that it takes 1 to 3 hours from the first strike to engulfment of prey. This unexpected observation led us to investigate venom components that potentially facilitate the idiosyncratic behavior observed. By combining the behavioral observations of prey capture by the cone snail with behavioral changes elicited by the action of individual venom components in mice, a previously uncharacterized class of analogs of the vertebrate hormone somatostatin (SS) was discovered.
RESULTS AND DISCUSSION
Deep-water cone snail species of the Asprella clade: Observations of fish-prey envenomation
Phylogenetic reconstruction using the mitochondrial cytochrome c oxidase subunit 1 (COI) marker gene highlights the existence of eight lineages of fish hunters in the genus Conus (Fig. 1A) (7). Of these, the Asprella clade is the least well known (1). Unlike the majority of cone snail species that inhabit shallow waters, Asprella species live well offshore, typically at depths of 60 to 250 m (13). This has hampered behavioral observation and venom analysis. To overcome this limitation, we established contacts with fishermen experienced in collecting marine gastropods from deep waters off the town of Sogod on Cebu Island in the Philippines. After a series of field expeditions, we collected enough specimens of two species of Asprella, Conus rolani, and Conus neocostatus for behavioral recordings and bioactivity-guided toxin discovery.
Three captive individuals of C. neocostatus were successfully documented catching different species of fish (movies S3 to S5; see table S1 for timeline of events). In all three predation episodes, C. neocostatus struck its prey with a rapid venom sting followed by immediate withdrawal (<1 s). Following envenomation, the fish initially appeared normal and continued to swim for 5 to 15 min. The snail waited during this time and did not react to any other fish that may have approached. In two of the three cases, the cone snail approached the fish again and carried out a second strike (movies S4 and S5). In all three cases, once the fish was deemed to be dead or completely incapacitated, the snail approached the fish from behind and consumed it whole. The timeline from first strike to the quiescent fish being engulfed ranged from 1 to 3.5 hours. Thus, in marked contrast to prey capture by fish-hunting cone snails of other phylogenetic lineages, prey capture by C. neocostatus is extremely slow.
We hypothesize that this seemingly inefficient predation behavior enables capture of aggressive fish that need to be approached from a distance and cannot be tethered without risk of retaliation. In line with this, we have documented fish that aggressively attack cone snails from other lineages, as they are extending their proboscis in preparation to envenomate their prey (movie S6). This behavior is also consistent with the lack of a pronounced barb on the radular tooth of Asprella species that is used by taser-and-tether hunters to hook onto their prey (1, 14–16). Killing by potentially dangerous prey has been proposed as one of the most important factors of natural selection on predatory organisms (17). Notably, the observed predation of C. neocostatus bears resemblance to the strike-and-release strategy used by rattlesnakes and other viperids of the family Viperidae, and by some of the deadliest members of the family Elapidae, including cobras (17). These snakes ambush rodent prey with an extremely rapid envenomating strike followed by a quick release of prey (18). The envenomated rodent then wanders off before ultimately succumbing to the deadly sting. Envenomation without capture provides the advantage of minimal contact with dangerous prey but adds the risk of potentially losing the prey and the challenge of relocating the envenomated animal. After envenomation, even when multiple fish were available, C. neocostatus tracks the specific individual that it struck, likely indicating that the snail can detect chemosensory cues released by the envenomated prey as has been reported for rattlesnakes (19). Although the C. neocostatus predation was only observed in captivity in a relatively small aquarium, chemosensory signaling is a common phenomenon in the marine environment where chemosensory information can be detected over longer distances than light and even sound (20, 21). Marine animals that use chemosensory cues include predators that track prey, with some fish prey synthesizing specific compounds to avoid detection (22). While we did not observe any other predatory behaviors by C. neocostatus, the ambush-and-assess hunting strategy recorded here may not be the only predation strategy used by these snails in the wild.
Anecdotal evidence suggests that ambush-and-assess hunting has also evolved in other cone snail lineages. Conus flavus, a fish-hunting species of the Phasmoconus clade, reportedly envenomates and captures fish without tethering (1). However, in contrast to C. neocostatus, envenomation by C. flavus leads to relatively faster immobilization. The extremely slow onset of action observed here suggests that the venom of Asprella snails may contain toxins that target G protein–coupled receptors (GPCRs) expressed in the somatosensory or neuroendocrine systems of prey. To specifically identify these toxins, we performed bioactivity-guided assays with an emphasis on compounds that elicited behavioral changes reminiscent of the slow-onset state of hypoactivity observed in fish after C. neocostatus envenomation.
Asprella venom elicits a slow-onset hypoactivity in mice: Identification of a previously unknown bioactive peptide
Venom was extracted from venom glands of C. rolani, a closely related Asprella species that is collected in the deepest water habitats (200 to 1250 m) (13). Unlike C. neocostatus, C. rolani can be abundantly collected, making the accumulation of larger quantities of venom for activity testing possible. However, C. rolani does not survive in captivity, and its predation behavior could therefore not be assessed. Nevertheless, given the close relatedness of these two species (13), the documented use of several of the same toxins (13), and similarities in the radular tooth morphology of species of the Asprella clade (14–16), we hypothesize that C. rolani and other Asprella species may also be ambush-and-assess hunters.
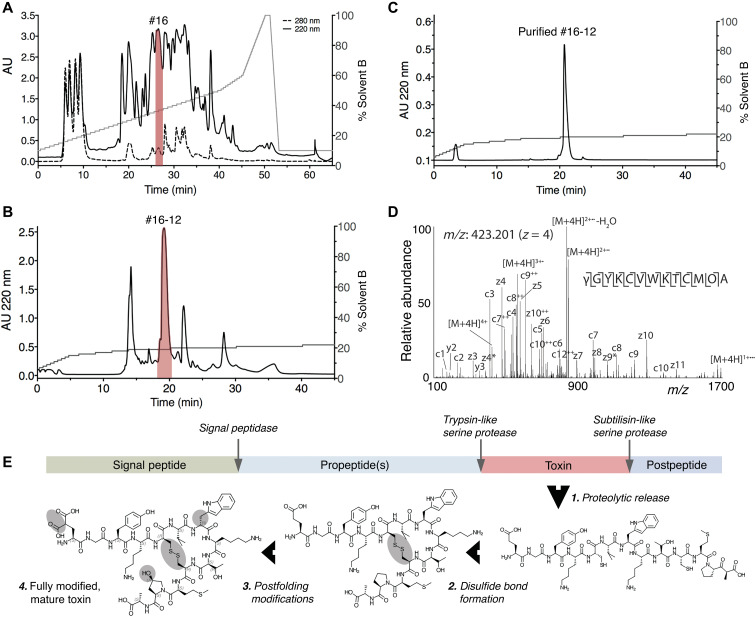
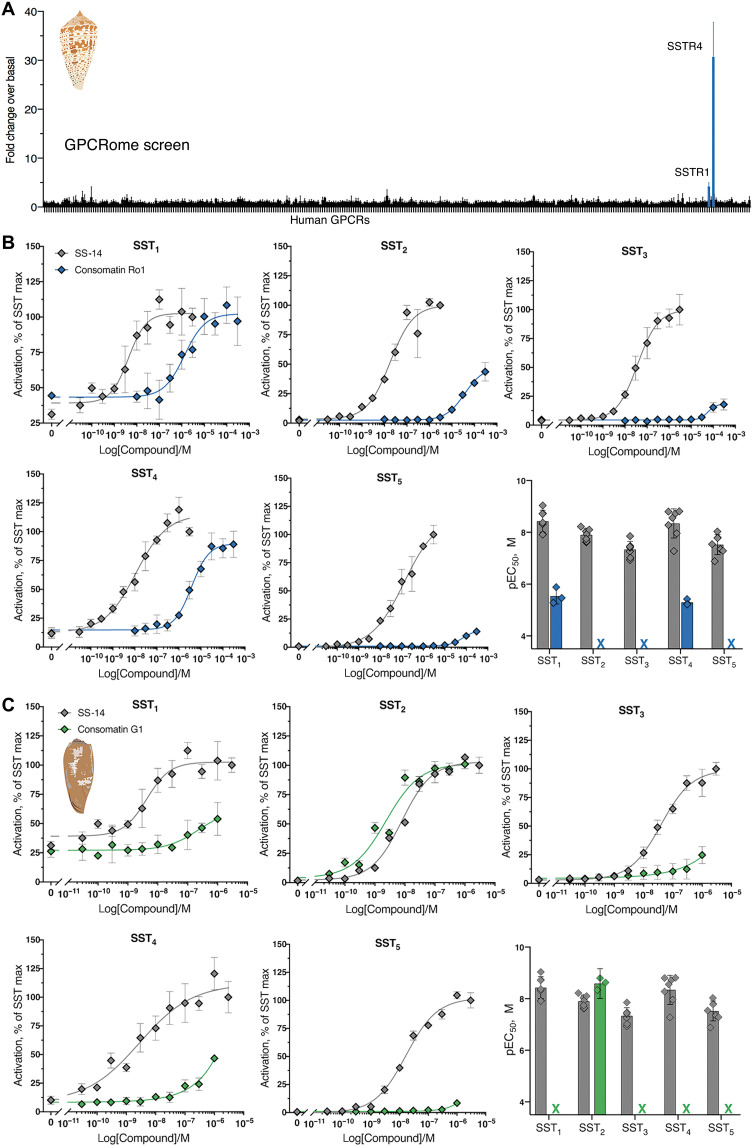
Because of their well-established use in toxinology and their closer relatedness to humans, mice were used to assay venom activity instead of fish. Activity testing of venom fractionated by reversed-phase chromatography revealed the presence of a fraction that elicited a phenotypic response upon intracranial injection in mice that was very similar to what was observed in fish: slow-onset loss of balance and a state of hypoactivity and sedation that lasted for >3 hours (fraction #16; Fig. 2A and table S2). This venom fraction was further fractionated, and the bioactivity of individual components was assessed by injecting them in mice. Subfraction #16-12 (Fig. 2, B and C) most closely recapitulated the initial effect observed in fraction 16 (table S2). The active component of subfraction #16-12 was characterized using a combination of Edman degradation, mass spectrometry (MS), and transcriptome sequencing. As determined by MS, the compound that elicited the bioactivity was a small peptide with a monoisotopic mass of 1573.65 (M+1H)1+ (fig. S1). This peptide is hereafter referred to as Consomatin Ro1.
Fig. 2. Identification of Consomatin Ro1.
(A) Reversed-phase chromatography of crude venom of the Asprella snail C. rolani and (B) subfractionation and (C) purification of Consomatin Ro1 from subfraction #16-12 (shown in red). (D) ETD MS/MS spectrum of the quadruple-charged ion of the venom peptide after reduction and alkylation with 2-methylaziridine acquired on the Orbitrap Lumos Tribrid with 30,000 resolution (at 400 m/z) and three microscans. N-terminal fragment ions (c-type ions) are indicated by ⌉, and C-terminal fragment ions (z,y-type ions) are indicated by ⌊. Doubly charged ions are indicated with ++, and z ions resulting from cleavage at cysteine and loss of the cysteine side chain are indicated with *. [M+4H]2+•• and [M+4H]1+••• indicate charge-reduced species. Because of space limitations, not all different charge states of already labeled peptide bond cleavages are indicated in the figure artwork. The mass accuracy for all fragment ions is better than 10 ppm. γ, γ-carboxyglutamate; O, hydroxyproline; w, d-tryptophan. (E) Organization of the prepropeptide of Consomatin Ro1 identified by transcriptome sequencing depicting posttranslational intermediates and processing events. AU, absorbance units.
Biochemical characterization and bioactivity of Consomatin Ro1
Tandem MS sequencing of purified Consomatin Ro1 and database searches of MS spectra against the venom gland transcriptome of C. rolani revealed the peptide’s amino acid sequence and posttranslational modifications (Fig. 2D). In addition to a disulfide bond between Cys5 and Cys10, the 13-residue peptide contains an N-terminal γ-carboxylated Glu (abbreviated as γ) and a hydroxylated Pro (abbreviated as O) in position 12 (Fig. 2, D and E). Furthermore, when sequenced by Edman degradation, Trp7 gave a lower-than-expected signal, indicating that this amino acid was potentially modified to dextrorotary (d)–Trp. To investigate the chirality of Trp7, we synthesized two analogs of Consomatin Ro1 containing either the l-Trp7 or the d-Trp7 enantiomer and compared their reversed-phase elution profile and in vivo activity to that of the native venom peptide. Only the d-Trp7 analog coeluted with the venom peptide (fig. S2). In addition, Consomatin Ro1 (d-Trp7) exhibited the same bioactivity as the purified venom peptide: Mice behaved normally for the first few minutes after intracranial injections but became progressively less active and ultimately motionless. These effects were dose dependent, with the lowest dose needed to elicit these effects being 0.52 to 0.94 mg/kg body weight (table S2). At 4 to 5 hours after injection, mice recovered, even from the highest dose tested (5.4 mg/kg). The l-Trp7 analog did not show any bioactivity when injected in mice. Whether this was caused by a lower in vivo stability of this analog was not further investigated.
Transcriptome sequencing of the C. rolani venom gland identified the full-length transcript encoding Consomatin Ro1. The translated Consomatin Ro1 precursor sequence belongs to the C-gene superfamily and shares the canonical sequence organization of most cone snail toxins, consisting of an N-terminal signal sequence for translocation into the endoplasmic reticulum and secretory pathway, followed by the propeptide region, the mature toxin, and a short postpeptide sequence (Fig. 2E and fig. S3). Consomatin Ro1 appears to be released from the prepropeptide by trypsin- and subtilisin-like serine proteases. The biological function of the pre- and postpeptide regions of the Consomatin Ro1 precursor remains to be determined but, given the high degree of modifications, may include recognition sites for modifying enzymes.
Consomatin Ro1 is a mimetic of the vertebrate hormone SS
The slow onset of action of Consomatin Ro1 in mice suggested that this peptide may not be a classical ion channel modulator but might instead target a GPCR. Comparative sequence mining of Consomatin Ro1 against an in-house database of known GPCR ligands revealed the toxin’s similarity to SS, an agonist of the G protein–coupled SS receptor family (Fig. 3).
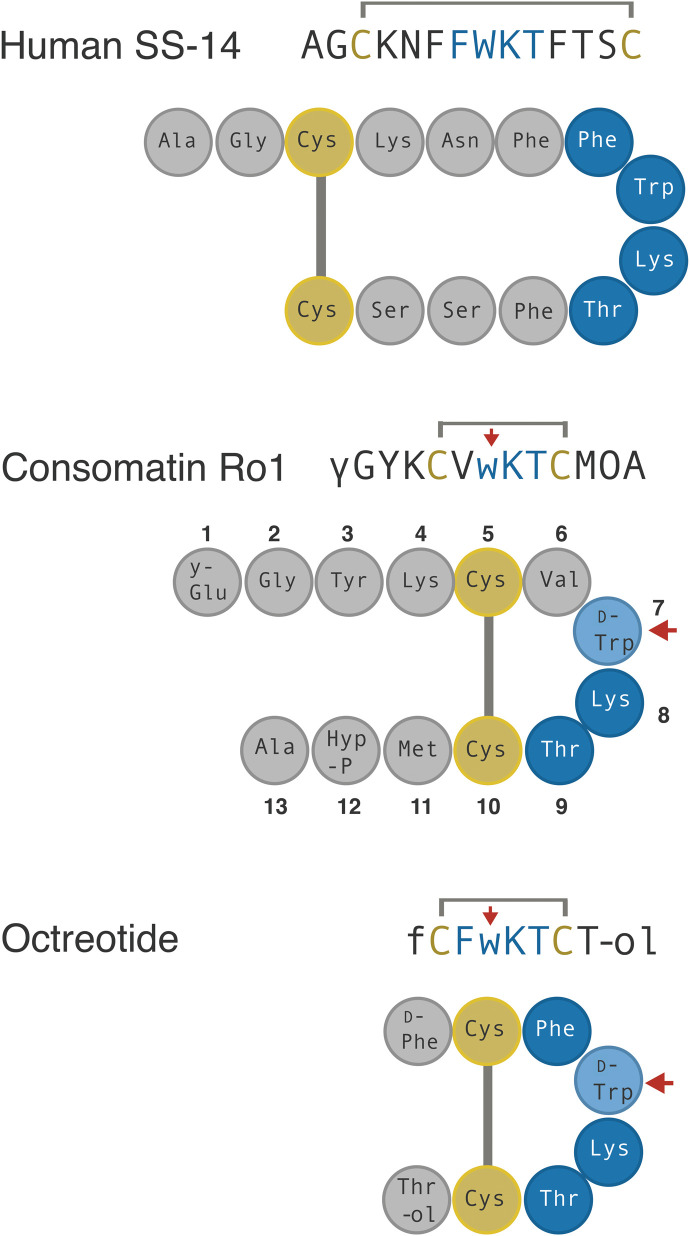
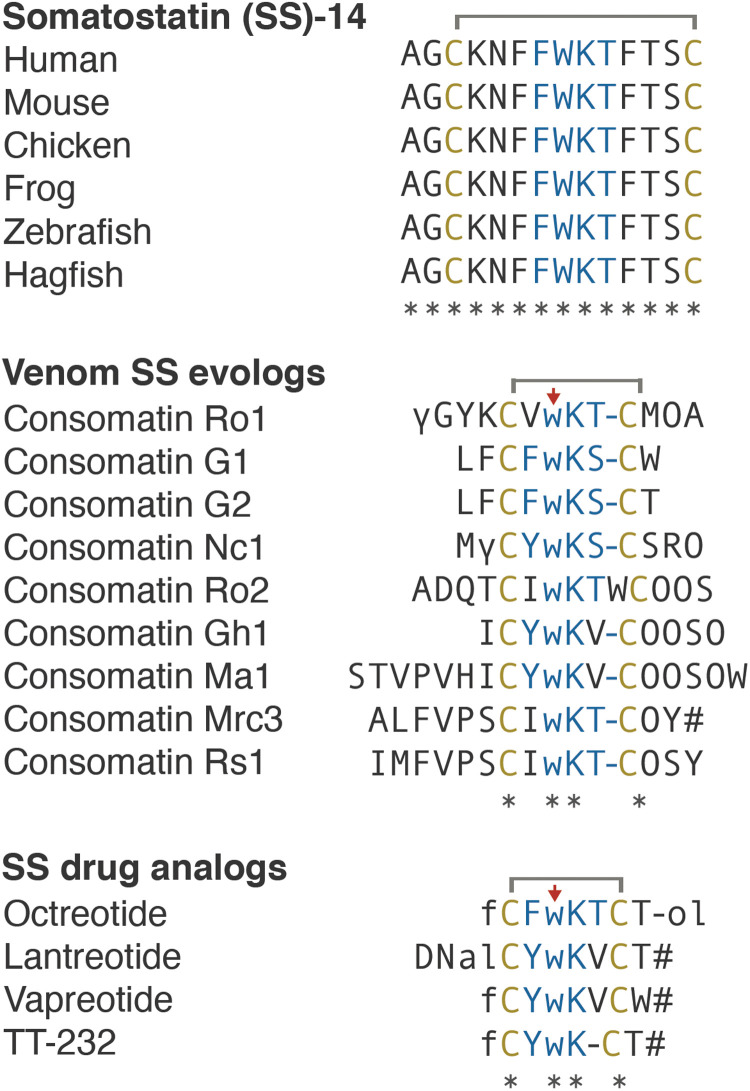
Fig. 3. Sequences and schematic representation of human SS, Consomatin Ro1, and the SS drug analog octreotide.
Cysteines and amino acids of the core SS receptor binding motif are shown in yellow and blue, respectively. Modifications: γ, γ-carboxyglutamate; w, d-tryptophan (marked by red arrow); O, hydroxyproline; f, d-phenylalanine; #, amidation; -ol, alcohol.
SS is a highly conserved vertebrate peptide hormone that, in humans, is predominantly secreted in the brain, pancreas, and gastrointestinal tract where it functions as an inhibitor of secretion and cell proliferation (23). SS was first found as the main inhibitor of growth hormone (GH) release from the anterior pituitary gland and has since been associated with many additional biological functions, including the inhibition of pancreatic hormone secretion, neuronal signaling, pain, and inflammation (24). SS belongs to a larger family of related peptide hormones that includes SS-14, SS-28, and cortistatin. All three peptides bind to and activate the five subtypes of the SS receptor (SST1–5) (25). Notably, the venom peptide Consomatin Ro1 shares a disulfide bond and three of the four core residues known to be important for SST1–5 activation (Trp7-Lys8-Thr9; numbering according to the sequence of Consomatin Ro1 throughout the manuscript) (Fig. 3) (25).
To the best of our knowledge, Consomatin Ro1 represents the first SS-like peptide to be isolated from venom. In addition, while SS is ubiquitously found in vertebrates, including fish (26), and has recently also been reported in invertebrate deuterostomes (27), the presence of a peptide with such a high degree of sequence similarity to SS in a snail is unexpected because the existence of SS-like peptides in protostomes is still a matter of controversy (26, 28).
In addition to SS, Consomatin Ro1 shares some sequence similarity with the peptide hormone urotensin-II (UII). UII activates the urotensin receptor (UTR), another class A GPCR, and functions as one of the most potent vasoconstrictors in humans (29). However, UII lacks several residues important for SS receptor activation and has low activity at SST1–5 (29).
Consomatin Ro1 is an evolutionarily optimized stable analog of SS
Given its important roles in human physiology, SS has been investigated as a drug lead for various diseases, including hypersecretory tumors, diabetes, cancer, pain, and inflammation (25). Early attempts to develop SS as a drug proved unsuccessful because of the hormone’s very short in vivo half-life and lack of receptor subtype selectivity (30). These limitations fueled efforts to design metabolically stable, subtype-selective SS analogs (31). The three main features that are commonly shared between these analogs are a modified N terminus, a shortened cyclic core, and the incorporation of d-Trp within the conserved Phe-Trp-Lys-Thr receptor binding motif (Fig. 3). These modifications stabilize a characteristic pharmacophoric β-turn motif, provide increased resistance to proteolytic degradation, and confer subtype selectivity (31). For example, the in vivo half-life of octreotide (trade name Sandostatin), an SS drug analog approved for the treatment of GH-producing tumors, is 90 min following intravenous infusion compared to 1 to 3 min for native human SS (32, 33).
Remarkably, not only does the venom peptide Consomatin Ro1 display all three of the drug-like features introduced into minimized therapeutic SS analogs: Even the disulfide bond and d-Trp are located at the same positions (Fig. 3). Consistent with this, the in vitro half-life of Consomatin Ro1 when incubated with human plasma at 37°C is >158 hours compared to 5.5 hours for native human SS (fig. S4). While incorporation of d-enantiomers is a common strategy used for enhancing the metabolic stability of compounds in drug design, in nature, d–amino acid–containing peptides and proteins are extremely rare, particularly in metazoans (34). To the best of our knowledge, no other natural product has ever been reported to share a d-enantiomer with a synthetic peptide drug.
The x-ray structure of Consomatin Ro1 closely aligns with that of the SS drug analog octreotide
To compare Consomatin Ro1 to SS and therapeutic SS drug analogs, we determined the x-ray structure of this toxin at 1.95-Å resolution. Crystallographic data and refinement statistics are provided in table S3. The structure of Consomatin Ro1 is characterized by two β strands and a β turn in the cyclic core (residues Phe6–d-Trp7–Lys8–Thr9) between the Cys5-to-Cys10 disulfide linkage (Fig. 4A). The crystal of Consomatin Ro1 contained six molecules per asymmetric unit, arranged in an antiparallel β sheet, wrapped around a large polyethylene glycol (PEG) molecule (figs. S5 and S6). All six copies adopt essentially the same conformation and overlap closely, with root mean square deviation (RMSD) = 0.27 to 0.50 Å in pairwise overlaps of 10 Cα positions (residues 3 to 12). The conformation of the cyclic core region is very similar in all copies, characterized by a side-by-side stacking arrangement of d-Trp7 and Lys8, known to be important for SS receptor binding and activation (figs. S5 and S6) (25). The d-Trp7–Lys8 pair of side chains is oriented slightly differently in copy d of the six molecules in asymmetric unit as a result of crystal-packing interactions (fig. S5).
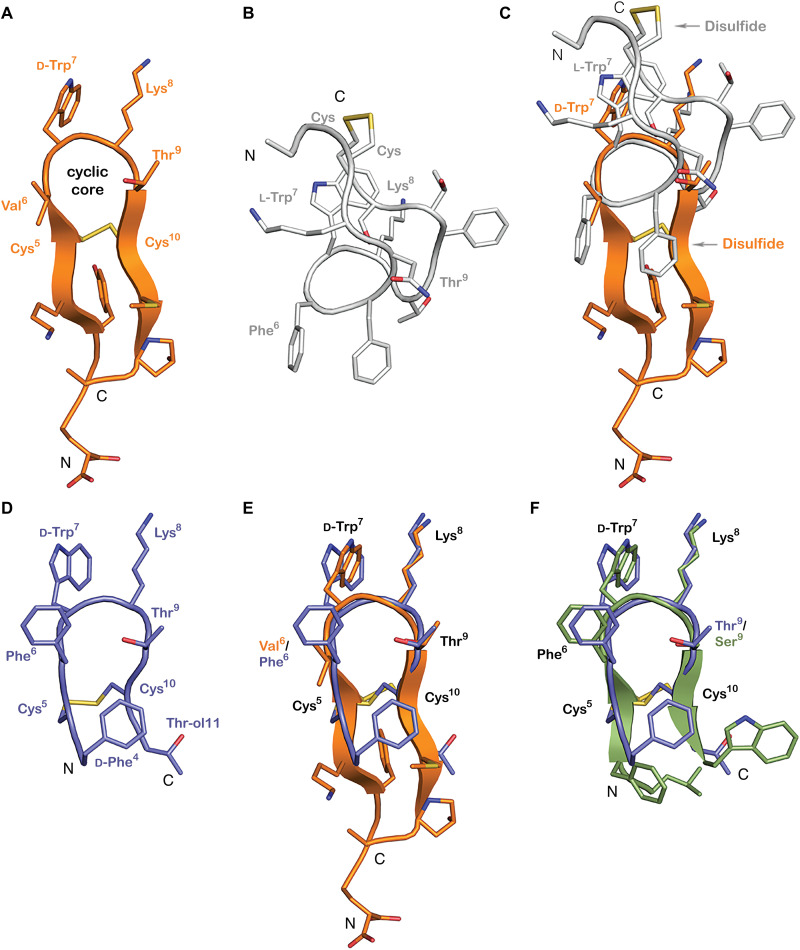
Fig. 4. Consomatin Ro1 and G1 are structural mimetics of the SS drug analog octreotide.
(A) X-ray structure of Consomatin Ro1 at 1.95-Å resolution (PDB ID: 7SMU). (B) Nuclear magnetic resonance (NMR) solution structure of human SS-14 obtained during heparin-induced fibril formation (PDB ID: 2MI1). (C) Alignment of the structure of Consomatin Ro1 (orange) with that of SS-14 (gray). (D) NMR solution structure of octreotide (PDB ID: 1SOC). (E) Alignment of the structure of Consomatin Ro1 (orange) with that of octreotide (purple) showing nearly identical backbone conformation and orientation of d-Trp7, Lys8, Thr9, and the disulfide bond, but differences in the amino acid composition and side-chain arrangements of Val/Phe6 and of residues outside the cyclic core. (F) Alignment of a homology model of Consomatin G1 (green), based on the structure of Ro1, with that of octreotide (purple), suggesting that the molecules share strong structural similarities. Numbering of residues according to that of Consomatin Ro1.
Despite the sequence similarities between Consomatin Ro1 and human SS [Protein Data Bank (PDB) ID: 2MI1], the backbone conformations and side-chain orientations of the two peptides are quite distinct (Fig. 4, B and C). The apex of the turn of human SS “cups” in the opposite direction of Consomatin Ro1, making it impossible to overlap the structure on more than the four residues of the apex of the β turn. Residues Trp7 and Lys8 of human SS (numbering taken from Consomatin Ro1) occupy similar positions, although they are in different rotamers from Consomatin Ro1 (Fig. 4C).
In contrast, Consomatin Ro1 displays notable similarities with the solution structure of the SS drug analog octreotide (PDB ID: 1SOC; Fig. 4D). The backbone of Consomatin Ro1 aligns closely with octreotide, and their d-Trp7–Lys8–Thr9 residues adopt nearly identical conformations as in octreotide (Fig. 4E). The most notable structural differences are seen in the side-chain orientation of Phe6/Val6 within the cyclic core and between residues outside of the cyclic core motif (Fig. 4E). Thus, the venom peptide Consomatin Ro1 represents a nature-derived, structural mimetic of octreotide and possibly other SS drug analogs for which structural data are not available.
Octreotide was developed as a stable drug agonist of SST2, a receptor target for the treatment of neuroendocrine disorders such as acromegaly and gastroenteropancreatic neuroendocrine tumors (35). Correspondingly, octreotide preferentially activated SST2 over SST3 and SST5 and displayed little to no activity on SST1 or SST4 (35). These observations on the subtype selectivity of octreotide and other SS analogs prompted us to investigate the activity of the Consomatin Ro1 venom peptide at the five human SS receptors and related GPCRs.
Consomatin Ro1 preferentially activates two of the five subtypes of the human SS receptors
Consomatin Ro1 was screened against a panel of 318 human GPCRs, including all five subtypes of the SS receptor, using the PRESTO-Tango β-arrestin recruitment assay (36). When tested at 10 μM, Consomatin Ro1 activated SST4 and, to a lesser extent, SST1. At this concentration, the peptide did not activate any of the other 313 GPCRs tested, including the UTR and ∂-, κ-, and m-opioid receptors (DOR, KOR, and MOR) (Fig. 5A). Subsequent concentration-response studies of Consomatin Ro1 at the five human SSTs confirmed preferential activation of SST1/SST4 with little or no activity at SST2, SST3, and SST5 (Fig. 5B). The median effective concentration (EC50) value of Consomatin Ro1 is 2.9 μM [pEC50 ± 95% confidence interval (CI95) = 5.5 ± 0.8] at SST1 and 5.1 μM (pEC50 ± CI95 = 5.3 ± 0.3) at SST4 (Fig. 5B and table S4; calculated from three independent experiments each carried out in triplicate). EC50 values at SST2, SST3, and SST5 could not be determined using a maximum concentration of 300 μM. For comparison, the EC50 of human SS-14 is between 1 and 97 nM for the five SSTs (Fig. 5 and table S4), which correlates well with previously reported receptor binding data (37). Consomatin Ro1, though having much lower potency than human SS, displays subtype preference toward SST1/SST4. While not tested here, it is conceivable that the toxin is more potent at one or more of the eight homologous receptors expressed in fish, which share ~60% sequence identity with human SS receptors (26).
Fig. 5. Consomatin Ro1 and G1 selectively activate the human SS receptors.
(A) PRESTO-Tango screen of Consomatin Ro1 (10 μM) at 318 human GPCRs (x axis) identified SST4 and SST1 as the molecular target. Plotted values represent means ± SD of four technical replicates. (B and C) Representative concentration-response curves at the five human SS receptor subtypes comparing (B) Consomatin Ro1 or (C) Consomatin G1 with human SS-14 using the β-arrestin recruitment assay (error bars represent SD of three technical replicates), as well as bar graphs showing the respective pEC50 values (error bars represent CI95 of three to seven independent repeats). RLU, relative luminescence units. Illustration credit: Paula Flórez Salcedo, University of Utah.
A single injection of Consomatin Ro1 provides antinociception and antihyperalgesia in two mouse models of acute pain
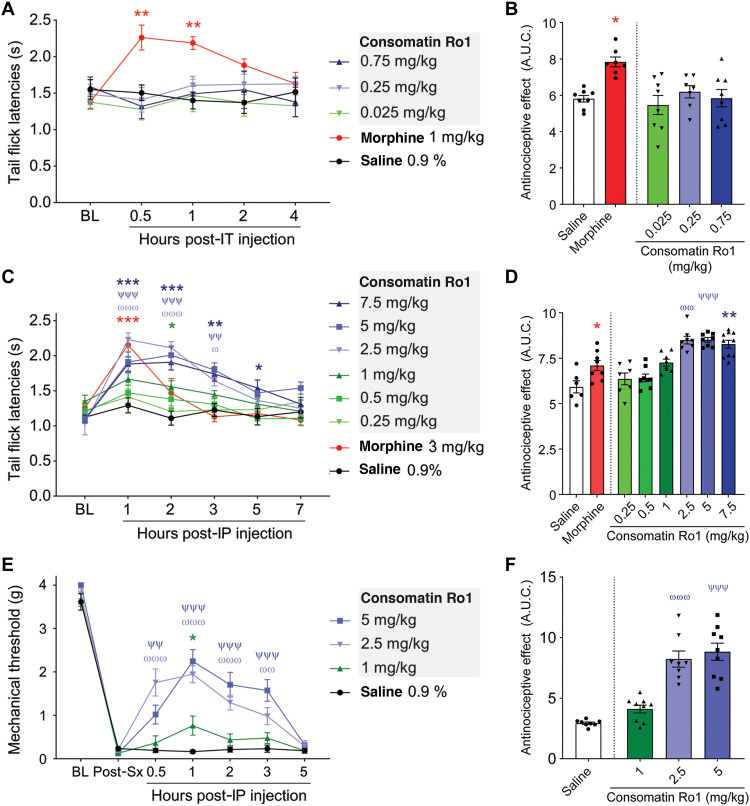
In addition to its inhibitory role in hormone secretion and cell proliferation, SS is expressed in sensory and sympathetic neurons where it can reduce nociception and inflammation (38). For example, the SS peptide mimetic TT-232, which was initially developed for its endocrine antitumor activity (39), was later shown to provide analgesia in animal models of acute and neuropathic pain (40). The two subtypes that were associated with these effects were SST1 and SST4, which are both activated by Consomatin Ro1 at low micromolar concentration. These observations led us to investigate the potential analgesic properties of the venom peptide using two mouse models of pain.
Antinociceptive effects were first evaluated through measurement of tail withdrawal latencies during noxious heat exposure using the tail flick test (41). To assess whether the toxin’s effects were mediated via receptors expressed in the central or peripheral nervous system, Consomatin Ro1 was either injected intrathecally or intraperitoneally before measurements. Mice injected intrathecally with multiple doses of the venom peptide (0.025 to 0.75 mg/kg) did not show any significant increase in tail flick latencies compared to saline (Fig. 6, A and B). Morphine (1 mg/kg, intrathecally) injection (positive control) resulted in robust antinociception confirming the sensitivity of the assay (Fig. 6, A and B). However, when Consomatin Ro1 was injected via the intraperitoneal route, dose-dependent antinociception was observed, with the maximal effect observed at 2.5 mg/kg, which lasted up to 3 hours (Fig. 6, C and D). Higher doses (5 and 7.5 mg/kg) did not result in any further antinociception. The observed effect was similar in magnitude to that of morphine at 3 mg/kg (Fig. 6, C and D). It should be noted that the highest intrathecal dose tested (0.75 mg/kg) that demonstrated no efficacy was only one-third of the Emax of the intraperitoneal dose. By contrast, typically 1/10th to 1/100th of a systemic dose is effective when delivered intrathecally, if a drug is acting in the spinal circuit (42, 43). Because Consomatin Ro1 is a peptide, it is highly unlikely that intraperitoneal dosing resulted in crossing of sufficient amount of the drug across the blood-brain barrier to induce an effect in the central nervous system (CNS). Together, these findings suggest that the analgesia-inducing site of action of Consomatin Ro1 is outside the CNS.
Fig. 6. Consomatin Ro1 provides analgesia in two mouse models of acute pain.
(A and B) Sensitivity to thermal noxious stimuli was evaluated by dipping mouse tails into hot water (52°C) and recording tail flick latencies (TFLs). (A) Mouse TFLs after a single intrathecal injection of saline, morphine, or multiple doses of Consomatin Ro1. TFLs were captured over a 4-hour period [n = 7 to 8 per group, two-way analysis of variance (ANOVA) with Dunnett’s post hoc test]. (B) Area under the curve between baseline (BL) and 4-hour condition, presenting global TFLs when animals received intrathecal (IT) injections (Kruskal-Wallis followed by Dunn’s post hoc test). (C) Mouse TFLs after a single intraperitoneal (IP) injection of saline, morphine, or multiple doses of Consomatin Ro1. TFLs were captured over a 7-hour period (n = 6 to 10 per group, two-way ANOVA with Dunnett’s post hoc test). (D) Area under the curve (A.U.C.) between baseline and 7-hour condition, presenting global TFLs when animals received intraperitoneal injections (Kruskal-Wallis followed by Dunn’s post hoc test). (E) Analgesic effect of Consomatin Ro1 on acute postsurgical pain. Plantar incision surgery intraperitoneal injections. Mechanical sensitivity was assessed using von Frey filaments (n = 8 to 9 per group, two-way ANOVA with Dunnett’s post hoc test). (F) Area under the curve between baseline and 5-hour condition, presenting mechanical withdrawal thresholds when animals received intraperitoneal injections (Kruskal-Wallis followed by Dunn’s post hoc test). Results correspond to means ± SEM. */ω/ψP < 0.05, **/ωω/ψψP < 0.01, and ***/ωωω/ψψψP < 0.001. Red asterisks illustrate significant difference between morphine and saline condition, blue asterisks: 7.5 mg versus saline, ψ: 5 mg versus saline, ω: 2.5 mg versus saline, green asterisks: 1 mg/kg versus saline.
We next evaluated the peptide’s efficacy in a mouse model of acute postsurgical pain (44). Following plantar incision surgery on the hind paw, mechanical sensitivity was determined by measuring the withdrawal response of the hind paw with a series of calibrated filaments (von Frey). Twenty-four hours after surgery, mice developed mechanical allodynia, which did not change after intraperitoneal injection of saline solution (control). However, a single injection of Consomatin Ro1 reversed mechanical hypersensitivity in a dose-dependent manner, with the effect lasting up to 3 hours (Fig. 6, E and F). The maximal effect was observed at 2.5 mg/kg, similar to that observed in the tail flick assay. Similar effective intraperitoneal doses have recently been reported for the small-molecule SST4 agonist J-2156 in a rat model of breast cancer–induced bone pain (45). Together, these data demonstrate that Consomatin Ro1 represents a new lead for the potential development of an analgesic that acts via opioid receptor–independent pathways.
In the natural setting, inhibition of pain sensing pathways in prey may help inhibit the escape (fight-or-flight) response of fish following envenomation (46), thereby facilitating prey capture by slow-moving Asprella snails. Notably, intramuscular injection of 10 nmol Consomatin Ro1 in goldfish (n = 4, body weight 1.11 ± 0.08 g) or zebrafish (n = 3, age 3 to 5 months) did not result in any obvious phenotype. Similarly, intramuscular injections of 5 nmol of SS in goldfish did not show any changes in the behavior in goldfish. However, when applied to cultured neurons from the zebrafish larva CNS, both Consomatin Ro1 and SS induced a reproducible block of the potassium chloride–evoked calcium signals in a subset of cells (fig. S7). These findings suggest that Consomatin Ro1 alone does not produce a visible phenotype but may inhibit or reduce the escape response triggered by the action of other toxins in the venom cocktail. Alternatively, Consomatin Ro1 may only produce a visible phenotype when injected in combination with other toxins in an additive or synergistic manner. More comprehensive studies are needed to elucidate the effect of Consomatin Ro1 and other toxins from C. rolani on fish behavior.
Consomatin Ro1 defines a previously uncharacterized class of venom evologs
The discovery of Consomatin Ro1 led us to search for other SS-like peptides in cone snail venom. To specifically identify candidates that likely activate the SS receptor, we limited our search to sequences that shared at least three of the four core residues important for receptor activation (Phe6-Trp7-Lys8-Thr9) or that had conservative substitutions at these positions. Sequence discovery was performed using the venom gland transcriptomes of Asprella snails and by assembling and mining the transcriptomes of 37 additional cone snail species available in the National Center for Biotechnology Information, DNA Data Bank of Japan, and China National GeneBank sequence repositories (table S5). Searching a total of 602,377 assembled transcripts from these venom gland datasets led to the identification of 18 additional consomatins, 2 from the Asprella snails C. rolani and C. neocostatus, 2 from the net hunter C. geographus, and 14 from Africonus and Varioconus, two lineages of worm-hunting cone snails endemic to West Africa (Fig. 7 and fig. S3) (47). One of the C. geographus sequences was previously reported as “G042 Contulakin precursor conopeptide” (GenBank ID BAO65575), but its sequence similarity to SS was not recognized or stated (48). The name Contulakin was given on the basis of the first C-superfamily toxin identified in C. geographus, Contulakin-G, a neurotensin-like peptide that activates the human neurotensin receptor (49).
Fig. 7. Comparative sequence alignments of vertebrate SS, selected Consomatin evologs, and SS drug analogs.
Identical amino acids are denoted by an asterisk (*). Mature toxins are shown with posttranslational modifications based on processing and modification of Consomatin Ro1 (modifications and color codes as shown in Fig. 3). See fig. S3 for precursor sequences of all consomatin sequences reported in this study.
The mature toxin regions of additional consomatins described here were predicted using the same proteolytic cleavage sites and posttranslational modifications as those that appear to be involved in the processing of Consomatin Ro1. Sequence identity of these predicted mature toxins to human SS ranges from 14% for Consomatin Ma1 to 38% for Consomatin Ro1 (Fig. 7 and fig. S3). Notably, while SSs are highly conserved peptide hormones in vertebrates, sequences of consomatins are hypervariable with the exception of a conserved cysteine framework and Trp7-Lys8 motif (see sequence alignment in Fig. 7). The same pattern of variable and conserved residues can be observed for SS drug analogs, further emphasizing that consomatins represent an unexplored class of drug-like ligands of the SSTs or related receptors. To distinguish the venom peptides from the native prey hormones and synthetic hormone analogs, we propose the name “evologs” for receptor ligands evolved by an organism to mimic the native ligand in another animal. Because of their streamlined role in inducing specific physiological changes in the other animal, evologs have distinctive biochemical and functional properties that make them ideal biomedical tools and drug leads.
Astonishingly, the sequences of the two consomatin evologs identified in the fish hunter C. geographus, Consomatins G1 and G2, are nearly identical to the drug octreotide (80 to 90% overall sequence similarity; Fig. 7). An alignment of a structural model of Consomatin G1 with the structure of octreotide underscores their close similarities (Fig. 4F). Consistent with this observation, synthetic Consomatin G1 is a highly potent and selective activator of the human SST2 (EC50 = 2.6 nM, pEC50 ± CI95 = 8.6 ± 0.6; Fig. 5C and table S4). The phenylalanine residue preceding the first cysteine was observed to be important for SST2 potency during the development of octreotide (50). Previous work had identified a minimal “core” FWKT sequence from SS, which, when inserted between two cysteines, was sufficient to retain inhibition of glucagon and GH release (51), functions that are largely attributed to SST2 activation (25). These core differences between Ro1 and G1 could to some degree explain the different subtype selectivity and potency profiles of these two peptides. Further studies are needed to fully elucidate the role of the core motif and the residues outside of the core. Given that SST2 is an important pharmacological target for the treatment of acromegaly and neuroendocrine tumors (35), Consomatin G1 has potential as a therapeutic for the treatment of these disorders.
In addition to its role in inhibiting GH secretion from the pituitary gland, SST2 is expressed in the pancreas where its activation suppresses the release of insulin and glucagon (52). Considering that Consomatin G1 was identified in the venom gland transcriptome of C. geographus, a species that uses fast-acting insulins to induce hypoglycemia in its fish prey (11), we hypothesize that Consomatin G1 may have evolved to modulate pancreatic hormone secretion, thereby exacerbating a hypoglycemic phenotype.
We note that the true chemical identity of Consomatin G1 and other consomatins identified from venom gland transcriptome data may differ from our predictions, particularly with respect to the nature of posttranslational modifications and proteolytic N- and C-terminal cleavage sites. Thus, future studies on extracted venom are needed to elucidate the nature of the mature peptides deriving from these SS-like sequences. However, although the exact chemical identity and biological role of Consomatin G1 in the venom of C. geographus are yet to be experimentally determined, the discovery of additional SS-like peptides capable of activating the human SSTs at nanomolar potency establishes the evolution and wider use of SS evologs in cone snails and the presence of SS-like peptides in invertebrate deuterostomes in general. Furthermore, the presence of a large number of consomatins in several worm-hunting species suggests the existence of an SST-like receptor in marine worms with similar ligand binding sites to the vertebrate SSTs. Identifying such receptor-ligand system in the future will likely provide fundamental insight into the evolution of the SS signaling system.
Bioinformatic transcriptome mining revealed that several of the 39 analyzed cone snail species express C-superfamily sequences that, while not meeting our criteria for being SS-like, share a pair of cysteines and two of the four cyclic core residues with SS and related peptide hormones (Trp7-Lys8). Two of these sequences, Contulakin-Lt1 and Contulakin-Lt2, from the venom gland transcriptome of Conus litteratus [UniProt ID Q2I2P1 and Q2I2P2 (53)], were previously reported to have sequence similarity to an invertebrate UII-like peptide (54). Their biological activity was not assessed. Whether the C. litteratus toxins and other C-superfamily sequences with lower sequence similarity to SS evolved to target the SSTs or another receptor family has not been addressed but likely represents an untapped opportunity for the identification of additional GPCR ligands with interesting biological activities.
Two different conotoxins have previously been described as antagonists of SST3, τ-conotoxin LiC32 from Conus lividus and τ-conotoxin CnVA from Conus consors (55). While these conotoxins do not share any sequence or structural similarity with consomatins and exhibit a different mode of action (antagonist versus agonist), their existence further highlights the potential of cone snail venoms for the discovery of GPCR modulators.
In conclusion, we show that through predator-prey evolution, venomous snails have mastered SS drug design to likely induce diverse physiological endpoints in prey. With hundreds of cone snail species yet to be sequenced, we anticipate the future discovery of many more consomatins with diverse receptor activation profiles that exert distinctive systemic effects. Together with our recent discovery of fast-acting venom insulins that has led to the design of a new drug lead for the treatment of diabetes (12, 56), this study provides a powerful example of the evolution of optimized, drug-like evologs of human hormones in venomous predators.
MATERIALS AND METHODS
Specimen collection
C. rolani and C. neocostatus snails were collected from the waters of Sogod, Cebu, Philippines by tangle nets at depths of around 150 to 300 m. Collection of the snails was done under the gratuitous permits (GP-0053-11, GP-0063-12, and GP-0084-15) issued by the Bureau of Fisheries and Aquatic Resources to the University of the Philippines Marine Science Institute. Specimen identification was initially performed by morphological examination and later verified by sequence analysis of the COI gene as previously described (57). For DNA/RNA analysis, venom glands were dissected and stored in RNAlater (Thermo Fisher Scientific) until analysis. For venom extraction, venom glands were dissected and stored at −80°C until analysis.
Phylogenetic analysis
Phylogenetic analysis was performed using the COI barcode sequence for each species. Sequences were aligned, and the neighbor-joining trees method was used to generate the phylogenetic tree shown in Fig. 1 using T-Coffee with default options (58). The tree was visualized using Tree Dyn 198.3 (59).
Extraction and fractionation of C. rolani venom
Frozen venom glands of C. rolani were thawed in 30% acetonitrile (CH3CN) with 0.2% trifluoroacetic acid (TFA). Venom glands were cut with fine scissors and then homogenized. The mixture was centrifuged at 15,000 rcf for 10 min as previously described (60). The supernatant was fractionated by reversed-phase high-performance liquid chromatography (RP-HPLC) on a Vydac preparative C18 column. Aliquots of the fractions and fraction pools were dried for bioactivity testing. Active HPLC fractions were further subfractionated using an analytical C18 column (Vydac 238TP; 5 μm, 250 mm × 4.60 mm). Gradient elution was done at a flow rate of 1 ml/min with increasing solvent B concentration (90% CH3CN in 0.1% TFA) in solvent A (nanopure water acidified with 0.1% TFA) at 0.1% solvent B/min. Absorbance was measured at 220 and 280 nm.
Mass determination and sequencing of Consomatin Ro1
The mass of the active fraction was determined using electrospray ionization mass spectrometry (ESI MS) at the Salk Institute for Biological Sciences Mass Spectrometry Core Facility. Before sequencing, an aliquot of the fraction was reduced using 26 μl of 50 mM dithiothreitol (Sigma-Aldrich) and incubated at 65°C for 30 min. The reduced peptide was alkylated with 35 μl of 1.5 M iodoacetamide (Sigma-Aldrich) and incubated in the dark at room temperature for 15 min. All solutions were buffered with tris (pH 7–8). The reduced and alkylated peptide was run in the HPLC from 10 to 25% solvent B (90% CH3CN in 0.1% TFA) in 45 min. The amino acid sequence of the peptide was determined by automated Edman degradation at the Protein Facility of the Iowa State University and by tandem MS sequencing.
For tandem MS, an aliquot of the sample was dried in SpeedVac and subsequently reduced and alkylated in vapor using 1% 2-methylaziridine and 2% trimethylphosphine in 50% acetonitrile and 100 mM ammonium bicarbonate (pH 8.4) for 60 min at room temperature. Alkylation vapor was removed, and the sample was reconstituted in 0.5% acetic acid. Aliquots (1 μg) of the now reduced and alkylated sample were loaded onto a 75 μm by 50 cm PepMap EasySpray column using the autosampler of an Easy nLC-1000 nano-HPLC coupled to an Orbitrap Fusion Lumos Tribrid mass spectrometer. The sample was eluted using a flow rate of 0.2 μl/min with a gradient of 0 to 100% B [solvent A = 0.5% acetic acid, solvent B = 90% acetonitrile in 0.5% acetic acid (v/v)] in 90 min, a spray voltage of 2.0 kV, and a 3-s cycle time. MS1 scans were acquired at 120,000 resolution [at 400 mass/charge ratio (m/z)], 50-ms maximum injection time. MS2 was acquired on precursors that carry 4 to 20 charges using the following settings: three microscans, 2 m/z isolation window, target value of 5 × 104 ions, and 200-ms maximum injection time. Each precursor was subjected to electron transfer dissociation (ETD) and HCD fragmentation using the following conditions: 30,000 resolution (at 400 m/z), ETD using 60-ms ion reaction time, and higher energy collision dissociation (HCD) using 32% normalized collision energy. Raw files were searched using search engine Byonic (Protein Metrics) allowing for a 10-ppm (parts per million) mass tolerance and common conotoxin post-translational modifications (PTMs). The peptide sequence was then manually verified.
Transcriptome sequencing of the C. rolani and C. neocostatus venom gland
Total RNA was extracted from the venom glands of C. rolani and C. neocostatus using the Direct-zol RNA Extraction Kit (Zymo Research), with on-column deoxyribonuclease treatment, according to the manufacturer’s instructions. Complementary DNA library preparation and sequencing were performed by the University of Utah High-Throughput Genomics Core Facility as previously described (56). Briefly, 125-cycle paired-end sequencing was performed on an Illumina HiSeq2000 instrument at an 80% standard cluster density. Adapter trimming of demultiplexed raw reads was performed using fqtrim (v0.9.4), followed by quality trimming and filtering using prinseq-lite (61). Error correction was performed using the BBnorm ecc tool, part of the BBtools package (open-source software, Joint Genome Institute). Trimmed and error-corrected reads were assembled using Trinity (version 2.2.1) (62) with a k-mer length of 31 and a minimum k-mer coverage of 10. Assembled transcripts were annotated using a blastx search (63) (E value setting of 1 × 10−3) against a combined database derived from UniProt and an in-house cone snail venom transcript library.
Peptide synthesis, folding, and purification
Peptide synthesis
Consomatin Ro1 was synthesized using an Apex 396 automated peptide synthesizer (AAPPTec, Louisville, KY) applying standard solid-phase Fmoc (9-fluorenylmethyloxy-carbonyl) protocols. Peptide was constructed on preloaded Fmoc-Ala-Wang resin (substitution: 0.38 mmol/g; Peptides International, Louisville, KY). All standard amino acids were purchased from AAPPTec except for the following: N-α-Fmoc-O-t-butyl-l-trans-4-hydroxyproline (Hyp) was purchased from EMD Millipore (Billerica, MA), Fmoc-γ-carboxy-l-glutamic acid γ,γ-di-t-butyl ester (Gla) was purchased from Advanced ChemTech (Louisville, KY), and Nα-Fmoc-N(in)-Boc-d-tryptophan (d-Trp) was purchased from Chem-Impex International Inc. (Wood Dale, IL). Side-chain protection for the amino acids was as follows: Cys, trityl (Trt); Gla, t-butyl ester (OtBu); Lys and d-Trp, tert-butyloxycarbonyl (Boc); Hyp, Thr, and Tyr, tert-butyl ether (tBu). A 10-fold excess of standard amino acids was used except for Fmoc-Gla(OtBu)2, Fmoc-Hyp(tBu), and Fmoc-D-Trp(Boc)-OH, for which fivefold excess was used. The coupling activation was achieved with one equivalent of 0.4 M benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate (PyBOP) and two equivalents of 2 M N,N-diisopropylethyl amine in N-methyl-2 pyrrolidone as the solvent. Each coupling reaction was conducted for 60 min, except for the special amino acids for which the reaction was conducted for 90 min. Fmoc deprotection was carried out for 20 min with 20% piperidine in dimethylformamide.
Peptide cleavage and purification
Consomatin Ro1 was cleaved from 103 mg of resin by a 1.5-hour treatment with 1 ml of Reagent K (TFA/water/phenol/thioanisole/1,2-ethanedithiol 82.5/5/5/5/2.5 by volume). Next, the cleavage mixture was filtered and precipitated with 10 ml of cold methyl-tert-butyl ether (MTBE). The crude peptide was then precipitated by centrifugation at 7000 rcf for 6 min and washed once with 10 ml of cold MTBE. The crude peptide was purified by RP-HPLC using a semi-preparative C18 Vydac column (250 mm × 10 mm, 5-μm particle size; 218TP510) eluted with a linear gradient ranging from 10 to 40% solvent B in 30 min at a flow rate of 4 ml/min. The HPLC solvents were 0.1% (v/v) TFA in water (solvent A) and 0.1% TFA (v/v) in 90% aqueous acetonitrile (v/v) (solvent B). The eluent was monitored by measuring absorbance at 220/280 nm. Purity of the peptide was assessed by analytical C18 Vydac RP-HPLC (218TP54; 250 mm × 4.6 mm, 5-μm particle size) using the same gradient as described above with a flow rate of 1 ml/min. The peptide was quantified by ultraviolet absorbance at 280 nm, using an extinction coefficient (ε) value of 6990 M−1 cm−1. Of 103 mg of the cleaved resin, 8.3 mg of the linear peptide was prepared.
Oxidative folding of Consomatin Ro1
Two hundred nanomoles of linear Consomatin Ro1 was resuspended in 1 ml of 0.01% TFA solution and added to a solution containing 5 ml of 0.2 M tris-HCl (pH 7.5) plus 0.2 mM EDTA, 0.5 ml of 20 mM reduced and 0.5 ml of 20 mM oxidized glutathione, and 3 ml of water. Final peptide concentration in the folding mixture was 20 μM. The folding reaction was conducted for 2 hours and quenched with formic acid to a final concentration of 8%. The quenched reaction mixture was then separated by RP-HPLC using a semipreparative C18 column and a linear gradient ranging from 10 to 40% of solvent B in 30 min with a flow rate of 4 ml/min. The eluent was monitored by absorbance at 220/280 nm. Purity of the folded peptide was assessed by an analytical C18 Vydac RP-HPLC using the gradient described above, with a flow rate of 1 ml/min. Pure fully folded Consomatin Ro1 was quantified by absorbance at 280 nm as described for the linear peptide, which was later verified by amino acid analysis performed by the University of Utah DNA/Peptide Facility. The final Consomatin Ro1 (0.71 mg) was obtained from 1.89 mg of the linear peptide with 98% purity. The molecular mass of Consomatin Ro1 was confirmed by ESI MS (calculated monoisotopic MH+1: 1573.64, determined monoisotopic MH+1: 1573.64) at the University of Utah Mass Spectrometry and Proteomics Core Facility. An additional mass of 1595.63 Da was also identified, which corresponded to sodium adduct: [M+Na]+ ion.
Consomatin G1 was custom-synthesized by GenScript. The peptide’s mass and purity (>95%) were verified by RP-HPLC and MS.
Coelution of native and synthetic peptides
The native and synthetic Consomatin Ro1 were separately loaded on an analytical C18 column (Vydac 238TP; 5 μm, 250 mm × 74.60 mm) and profiled using a linear gradient from 19 to 21% solvent B in 20 min. A 1:2 mixture of the native and synthetic Consomatin Ro1 was also applied on the same column using the same gradient and conditions.
Determination of the crystal structure of Consomatin Ro1
Lyophilized Consomatin Ro1 was dissolved in water at 40 mg/ml and filtered with a 0.2-μm filter. This solution was diluted with water for crystallization trials. For crystal screening, wild-type (WT) Consomatin Ro1 (at 20 mg/ml) was mixed in a 1:1 ratio with commercial crystallization solutions in sitting drop vapor diffusion crystal trays. Consomatin Ro1 crystallized after 11 days at 21°C in SaltRx (Hampton Research), condition D10 [4.0 M sodium nitrate and 0.1 M sodium acetate trihydrate (pH 4.6)]. A WT crystal was prepared for data collection by suspending the crystal in a small nylon loop, briefly immersing in fresh mother liquor for 20 s, and cryocooling by plunging in liquid nitrogen. A platinum heavy atom–soaked crystal was prepared by soaking for 5 min in 20 μl of fresh mother liquor containing 0.1 mM K2PtCl4. The crystal was then cryocooled by plunging in liquid nitrogen (without back soaking).
X-ray diffraction data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) with the crystal maintained at 100 K. WT data were collected with 1.7711-Å x-rays in an effort to optimize sulfur anomalous scattering. Data were integrated and scaled using XDS (64) and AIMLESS (table S3) (65). A fluorescence scan of a platinum-soaked crystal indicated the presence of platinum, and the x-ray energy was chosen on the basis of this scan to optimize anomalous scattering. The resulting platinum derivative data were sufficient to determine initial phases by single-wavelength anomalous diffraction analysis using the program Autosol (66) in the PHENIX software package (66). The resulting electron density map was readily interpretable, and an initial model was built using the program Coot (67). The initial model comprised most of six copies of Consomatin Ro1 and was partially refined (68) against the platinum derivative dataset. The model was then used to calculate model-based phases, and model building and refinement (using CCP4 program Refmac5) (69, 70) were continued using the WT dataset. The model was initially refined to Rcryst = 0.275 and Rfree = 0.302 with good geometry. The model was evaluated using MolProbity (71) and judged to be of good quality; however, a large region of unexplained density was visible in the FoFc electron density map. The density strongly resembled that of a PEG molecule, although PEG was not a component of the crystallization solution and was not thought to be present in the protein solution. Models of several PEG molecules were tested by model building and refinement until the best fit to the density was obtained with a molecule of composition C33O17H67 (predicted molecular weight ~ 750 Da). The model was improved by including the PEG molecule and was refined to Rcryst = 0.239 and Rfree = 0.283 with good geometry. The structural homology model of the venom peptide Consomatin G1 was predicted by changing the side chains of Ro1 to those of Consomatin G1 in Coot (67), based on the sequence alignment (Fig. 7).
Plasma stability assays
The in vitro stability of Consomatin Ro1 was investigated in human plasma (mixed gender) and compared to human SS-14. Peptides were incubated in 500 μl of plasma at 37°C with a final concentration of 3 μM. Samples (40 μl) were taken at 0-, 5-, 15-, 30-, 60-, 120-, 240-, 480-, and 1440-min time points. A twofold volume of cold acetonitrile with 0.1% formic acid was added to terminate the incubation. Samples were applied on a C18 column (Waters CSH; 2.1 mm × 50 mm, 1.7-μm particle size) with a flow rate of 0.650 ml/min and a gradient profile of 2 to 20% solvent B in 3.5 min and then 20 to 98% solvent B in 0.5 min and run on a Thermo Vanquish Horizon UHPLC. The HPLC solvents were 0.1% formic acid in water (solvent A) and acetonitrile (solvent B). MS data were acquired using a Thermo Q-Exactive Focus Orbitrap mass spectrometer at 35,000 resolution (full width at half maximum at m/z 200) for full scan and 17,500 resolution for MS2 in data-dependent acquisition mode. Parent compound disappearance, based on relative LC-MS peak area (0 min = 100%), was used to calculate half-lives. Propanthelin bromide (1 μM) was used as a control, and two replicates for each compound were analyzed.
Initial GPCRome screening
Functional data were provided by the National Institute of Mental Health’s Psychoactive Drug Screening Program (NIMH PDSP), contract no. HHSN-271-2013-00017-C. The NIMH PDSP is directed by B. L. Roth at the University of North Carolina at Chapel Hill and J. Driscoll at NIMH, Bethesda, MD, USA. Consomatin Ro1 was tested on a panel of GPCRs using the PRESTO-Tango system (36), which uses HTLA cells with a tTA-dependent luciferase reporter and a β-arrestin 2 tobacco etch virus protease fusion gene. HTLA cells were plated in 384-well plates overnight before transfection of the receptor constructs (318 target receptors). Assay plates with the compounds were incubated overnight at 37°C. The compounds and the media were removed, and Bright-Glo reagent (Promega) was added to determine luciferase activity. Results were presented in fold of the average basal; normal activity is 0.5- to 2.0-fold of basal level (~4 relative luminescence units). Dopamine receptor DRD2 with 100 nM quinpirole was used as an assay control. Follow-up assays were done on the SS receptors.
β-Arrestin recruitment assay (PRESTO-Tango)
HTLA cells (a gift from B. L. Roth) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) (Biowest), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Thermo Fisher Scientific) (HTLA medium), with hygromycin B (100 μg/ml; Thermo Fisher Scientific) and puromycin (2 μg/ml; Thermo Fisher Scientific) added. On day 1, 3 million cells were seeded in T25 flasks in 5 ml of HTLA medium and incubated overnight. On day 2, medium was changed, and cells were transfected using PolyFect (Qiagen) according to the manufacturer’s protocol. On day 3, the cells were resuspended in DMEM supplemented with 1% defined FBS (Thermo Fisher Scientific) (assay medium), and 15,000 cells in 40 μl per well were seeded in poly-d-lysine–coated white clear-bottom 384-well plates (Corning) and incubated overnight. On day 4, medium was changed, and ligands were diluted to appropriate concentrations (5× final concentration) in Hanks’ balanced salt solution (HBSS) (Thermo Fisher Scientific) supplemented with 20 mM Hepes (Sigma-Aldrich), 1 mM CaCl2, and 1 mM MgCl2, pH adjusted to 7.4 with 10 M NaOH (assay buffer), which was supplemented with 0.1% bovine serum albumin (ligand buffer). Ten microliters per well was added, and cells were incubated overnight. On day 5, the medium and compounds were removed from the cells, and 20 μl per well of a 1:20 dilution of Bright-Glo (Promega) in assay buffer supplemented with 0.01% Pluronic F68 (Thermo Fisher Scientific) was added to the cells. Plates were incubated for 20 min in the dark at room temperature, and luminescence was measured on Molecular Devices SpectraMax iD5 (Molecular Devices) with each well integrated for 1 s.
IACUC approval
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the Universities of Utah and Arizona. Protocol numbers are as follows: #14-08018 (approved period, 20 August 2014 to 19 August 2017), #17-07020 (approved period, 1 August 2017 to 31 July 2020), and #15-578 (approved period, 3 July 2018 to 3 July 2021).
Intracranial mouse bioassay
Swiss Webster mice (12 to 22 days old) were injected intracranially with the peptide dissolved in 15 μl of normal saline solution (NSS; 0.9% NaCl). Mouse behaviors in response to stimuli such as prodding were observed for at least an hour. Behavioral differences between the treated and control groups were recorded.
Intraperitoneal administration of Consomatin Ro1 in mouse
For intraperitoneal injection, the injection site (left lower abdominal quadrant) was wiped with 2% chlorhexidine. Mice were manually restrained, abdomen side up, with cranial end pointed down. With an angle of 15° to 20°, the abdominal cavity was punctured to inject either 200 μl of saline solution or 0.25, 0.5, 1, 2.5, 5, or 7.5 mg/kg of Consomatin Ro1. A solution of morphine sulfate (3 mg/kg) was injected as positive control. Before injection, aspiration was attempted to ensure that an abdominal viscus had not been penetrated.
Intrathecal administration of Consomatin Ro1 by lumbar puncture in mouse
For single administration, mice were sedated with 2% isoflurane in O2 anesthesia delivered at 2 liters/min before the injection. Increasing doses (0.025 to 0.75 mg/kg) of Consomatin Ro1 or morphine (1 mg/kg) were administered by lumbar puncture as described by Hylden and Wilcox (72). Briefly, mice were lightly restrained by the pelvic girdle in one hand (shaved, skin wiped with 2% chlorhexidine), while the syringe was held in the other hand at an angle of about 20° above the vertebral column. A 21-gauge needle was inserted into the tissue to one side of the lumbar 5 (L5) or L6 spinous process so that it slipped into the groove between the spinous and transverse processes. The needle was then moved carefully forward to the intervertebral space as the angle of the syringe was decreased to about 10°. The tip of the needle was inserted so that approximately 0.5 cm was within the vertebral column. The solution was then injected in a volume of 5 μl, and the needle was rotated on withdrawal.
Measurement of thermal sensory thresholds: Tail flick test in mouse
Antinociceptive effect was assessed through measurement of tail withdrawal latencies using the tail flick test. A hot water chamber was set at 52°C. Adult mice were restrained in a cylinder, and the distal two-thirds of the tail was dipped into hot water. To record tail flick latencies, a chronometer was started when the mouse tail was dipped into water and stopped as soon as the mouse withdrew its tail. Results correspond to the mean time when animals start to flick their tail out of the water. A cutoff time is set at 10 s before manual removal of the tail to prevent tissue damage.
Paw incision model of postsurgical pain in mouse
To analyze acute postsurgical pain, animals were subjected to an incisional surgery based on the procedure previously described by Brennan et al. (44). A 0.5-cm long incision was made through skin and fascia of the plantar surface of the left hind paw. The plantaris muscle was then elevated and longitudinally incised, leaving the muscle origin and insertion intact. After hemostasis with gentle pressure, the skin was closed with two mattress sutures of 5-0 nylon on a curved needle. Sham animals underwent anesthesia, and their left hind paw was scrubbed with Betadine and ethanol (70%). Only incision was performed, without damaging the muscle.
Measurement of tactile sensory thresholds: Von Frey test
The assessment of tactile sensory thresholds was determined by measuring the withdrawal response to probing the plantar surface of the hind paw with a series of calibrated filaments (von Frey) 24 hours after surgery. Each filament was applied perpendicularly to the plantar surface of the paw of a mouse held in suspended wire mesh cages. The “up and down” method was used to identify the mechanical force required for a paw withdrawal. Data were analyzed with the nonparametric method of Dixon, as described by Chaplan et al. (73). Results are expressed as the mean withdrawal threshold that induces a paw withdrawal response in 50% of animals.
Fish bioassays
Consomatin Ro1 (10 nmol) or human SS (5 nmol) was dissolved in 10 μl of NSS and injected into the muscle under the dorsal fin of a goldfish (Carassius auratus) (0.85 to 2.5 g body weight) or zebrafish (Danio rerio, wild type, age 3 to 5 months). Swimming and breathing behaviors were observed by eye and compared to fish injected with 10 μl of NSS (control).
Calcium imaging of zebrafish neurons
Primary neuronal cultures were obtained from the whole brain of zebrafish larvae (6 to 9 days after fertilization). Cells were dissociated with a combination of enzymatic and mechanical trituration. Briefly, the dissociated brain was transferred to HBSS containing 0.25% (w/v) trypsin and incubated at 37°C for 12 min. Cells were gently washed several times with minimum essential medium (MEM) (Thermo Fisher Scientific) to stop the enzymatic activity. After the last wash, mechanical trituration was performed with a series of three fire-polished pipettes of different diameters. The solution was filtered using a 70-μm cell strainer and centrifuged at 50 rcf for 5 min. The supernatant was removed, and cells were resuspended in MEM plus B27 supplement (Thermo Fisher Scientific) until the cell pellet was fully dissolved. The dissociated neurons were plated on poly-d-lysine–coated plates (Corning) and incubated overnight at room temperature in Neurobasal-A (Thermo Fisher Scientific) plus B27 supplement. Glucose was used to adjust to an osmolarity of 296 mOsm. One hour before calcium imaging experiments, cells were loaded with 3.5 μM Fura2-AM dye (Sigma-Aldrich) and incubated at room temperature for 1 hour. Imaging was performed using ×10 magnification, an intermittent excitation light of 340 and 380 nm was used, and excitation was monitored at 510 nm. A 12-bit camera was used to capture one image every 2 s. The calcium transients were elicited by application of 30 mM KCl (~15 s) followed by six washes with extracellular solution [134 mM NaCl, 3 mM KCl, 2 mM CaCl2, 1.2 mM MgCl2, 10 mM glucose, and 10 mM Hepes (pH 7.8)]. Consomatin Ro1 and human SS-14 were applied for 4 min in extracellular solution at 5 μM and 100 nM, respectively. NIS-Elements were used to acquire the data that were further processed with CellProfiler v.3.1.9 (https://cellprofiler.org/). Custom-built scripts in Python 3.7.2 and R were used to generate the calcium imaging traces shown in fig. S7.
Acknowledgments
We thank J. Erchegyi, C. Miller, and our friend and colleague J. Rivier (deceased, 2019) for peptide synthesis; K. Chase for assistance with transcriptome assemblies; P. Huynh and S. D. Robinson for initial behavioral recording; N. Barghi and A. O. Lluisma for help with initial sequence analysis; and N. Saguil for assistance with specimen collection under the Department of Agriculture–Bureau of Fisheries and Aquatic Resources–issued gratuitous permit nos. 0053-11, 0063-12, and 0084-15. We also thank the NIMH Psychoactive Drug Screening Program for initial screening of Consomatin Ro1; the High-Throughput Genomics Core Facility, the DNA Sequencing Core Facility, the DNA/Peptide Facility, and the Mass Spectrometry and Proteomics Core Facility at the University of Utah (MS equipment was obtained through a Shared Instrumentation Grant 1 S10 OD018210 01A1); the Iowa State University for Edman sequencing; and W. Low at the Salk Institute for initial mass spectrometric analysis. The mass spectrometric experiments were, in part, supported by a shared instrumentation grant from the NIH, 1S10OD010582-01A1, for the purchase of an Orbitrap Fusion Lumos Tribrid mass spectrometer. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the Department of Energy’s Office of Biological and Environmental Research and by the NIH, National Institute of General Medical Sciences (P41GM103393).
Funding: This research was funded by a Department of Defense grant (W81XWH-17-1-0413 to B.M.O.), a Villum Young Investigator Grant (19063 to H.S.-H.), Department of Science and Technology–Philippine Council for Health Research and Development grant FP 140015 (to G.P.C.), a USAID/Philippines through the Science, Technology, Research, and Innovation for Development (STRIDE) Scholarship Program grant (to I.B.L.R.), the Benning Society (to C.P.H.), and NIH grants (NINDS K08NS1042727 and R01NS116694 to A.P.).
Author contributions: I.B.L.R. and H.S.-H. discovered Consomatin Ro1. I.B.L.R. performed bioactivity testing in mice. I.B.L.R. and J.S.I. carried out venom extractions, fractionations, and compound purifications. J.G. synthesized peptides. M.W. performed phylogenetic analysis. D.T. maintained cone snails and performed behavioral observations and recording. I.B.L.R., H.S.-H., and W.R. performed venom extractions, derivatization, and mass spectrometric sequencing. B.U. led mass spectrometric sequencing. I.B.L.R. and W.E.B.-Y. performed and analyzed GPCR assays. W.E.B.-Y. and H.B.-O. led GPCR assays. I.B.L.R. crystallized proteins. F.G.W. performed crystallographic analyses. C.P.H. oversaw x-ray crystallography. L.F.M. performed pain assays. A.P. led pain assays. P.F.S. performed fish experiments. H.S.-H. performed transcriptomics analyses. G.P.C., B.M.O., and H.S.-H. led the research. H.S.-H. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. In addition, structural data are available in the PDB (7SMU). Sequences of consomatins identified here are available in GenBank under the following accession numbers: Consomatin Ro1: MT409178, Consomatin Ro2: MW390882, Consomatin Nc1: MW390884, Consomatin G1: MW390883, and Consomatin G2 (annotated as G042 Contulakin precursor conopeptide): BAO65575. Sequencing reads for transcriptome studies are available in the GenBank SRA under accession numbers SAMN22417933 and SAMN22417934. The venoms of C. rolani and C. neocostatus analyzed in this study can be provided by G.P.C. and B.M.O. pending scientific review and a completed material transfer agreement. Requests for these venoms should be submitted to olivera@biology.utah.edu.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S5
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S6
REFERENCES AND NOTES
- 1.Olivera B. M., Seger J., Horvath M. P., Fedosov A. E., Prey-capture strategies of fish-hunting cone snails: Behavior, neurobiology and evolution. Brain Behav. Evol. 86, 58–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King G. F., Venoms as a platform for human drugs: Translating toxins into therapeutics. Expert. Opin. Biol. Ther. 11, 1469–1484 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Safavi-Hemami H., Brogan S. E., Olivera B. M., Pain therapeutics from cone snail venoms: From ziconotide to novel non-opioid pathways. J. Proteome 190, 12–20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yarotskyy V., Elmslie K. S., Omega-conotoxin GVIA alters gating charge movement of N-type (CaV2.2) calcium channels. J. Neurophysiol. 101, 332–340 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Miljanich G. P., Ziconotide: Neuronal calcium channel blocker for treating severe chronic pain. Curr. Med. Chem. 11, 3029–3040 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Richter K., Sagawe S., Hecker A., Küllmar M., Askevold I., Damm J., Heldmann S., Pöhlmann M., Ruhrmann S., Sander M., Schlüter K. D., Wilker S., König I. R., Kummer W., Padberg W., Hone A. J., McIntosh J. M., Zakrzewicz A. T., Koch C., Grau V., C-reactive protein stimulates nicotinic acetylcholine receptors to control ATP-mediated monocytic inflammasome activation. Front. Immunol. 9, 1604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aman J. W., Imperial J. S., Ueberheide B., Zhang M. M., Aguilar M., Taylor D., Watkins M., Yoshikami D., Showers-Corneli P., Safavi-Hemami H., Biggs J., Teichert R. W., Olivera B. M., Insights into the origins of fish hunting in venomous cone snails from studies of Conus tessulatus. Proc. Natl. Acad. Sci. U.S.A. 112, 5087–5092 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olivera B. M., ConusVenom peptides: Reflections from the biology of clades and species. Annu. Rev. Ecol. Syst. 33, 25–47 (2002). [Google Scholar]
- 9.Johnson C. R., Stablum W., Observations on the feeding behavior of Conus geographus (Gastropoda: Toxoglossa). Pac. Sci. 25, 109–111 (1971). [Google Scholar]
- 10.Motomura M., Johnston I., Lang B., Vincent A., Newsom-Davis J., An improved diagnostic assay for Lambert-Eaton myasthenic syndrome. J. Neurol. Neurosurg. Psychiatry 58, 85–87 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safavi-Hemami H., Gajewiak J., Karanth S., Robinson S. D., Ueberheide B., Douglass A. D., Schlegel A., Imperial J. S., Watkins M., Bandyopadhyay P. K., Yandell M., Li Q., Purcell A. W., Norton R. S., Ellgaard L., Olivera B. M., Specialized insulin is used for chemical warfare by fish-hunting cone snails. Proc. Natl. Acad. Sci. U.S.A. 112, 1743–1748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong X., Menting J. G., Disotuar M. M., Smith N. A., Delaine C. A., Ghabash G., Agrawal R., Wang X., He X., Fisher S. J., MacRaild C. A., Norton R. S., Gajewiak J., Forbes B. E., Smith B. J., Safavi-Hemami H., Olivera B., Lawrence M. C., Chou D. H. C., A structurally minimized yet fully active insulin based on cone-snail venom insulin principles. Nat. Struct. Mol. Biol. 27, 615–624 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olivera B., Watkins M., Puillandre N., Tenorio M. J., Hidden diversity in the Asprella clade: Description of Conus (Asprella) neocostatus sp. nov. (Gastropoda, Conidae). Xenophora Taxonomy 33, 22–29 (2021). [Google Scholar]
- 14.Rolán E., Raybaudi-Massilia G., New investigation on the radular teeth of Conus (Prosobranchia: Conidae). Part I. Argonauta 8, 6–59 (1994). [Google Scholar]
- 15.J. K. Tucker, M. J. Tenorio, Systematic Classification of Recent and Fossil Conoidean Gastropods: With Keys to the Genera of Cone Shells (Conchbooks, 2009). [Google Scholar]
- 16.J. K. Tucker, M. J. Tenorio, Illustrated Catalog of Living cone Shells (MdM Publishing, Wellington, Florida, USA, 2013). [Google Scholar]
- 17.Vermeij G. J., The evolutionary interaction among species: Selection, escalation, and coevolution. Annu. Rev. Ecol. Syst. 25, 219–236 (1994). [Google Scholar]
- 18.Kardong K., Rattlesnake strike behavior: Kinematics. J. Exp. Biol. 201 ( Pt 6), 837–850 (1998). [DOI] [PubMed] [Google Scholar]
- 19.D. Chiszar, C. W. Radcliffe, K. M. Scudder, D. Duvall, in Chemical Signals in Vertebrates D. Müller-Schwarze, R. M. Silverstein, Eds. (Springer, 1983), vol. 25, pp. 1–24. [Google Scholar]
- 20.Weissburg M. J., The fluid dynamical context of chemosensory behavior. Biol. Bull. 198, 188–202 (2000). [DOI] [PubMed] [Google Scholar]
- 21.M. Weissburg, in Chemical Ecology in Aquatic Systems (Oxford Univ. Press, 2012), p. 96. [Google Scholar]
- 22.Barimo J. F., Walsh P. J., Use of urea as a chemosensory cloaking molecule by a bony fish. J. Exp. Biol. 209, 4254–4261 (2006). [DOI] [PubMed] [Google Scholar]
- 23.V. Martinez, Somatostatin, in Handbook of Biologically Active Peptides, A. J. Kastin, Ed. (Academic Press, 2013), chap. 180, pp. 1320–1329. [Google Scholar]
- 24.Viollet C., Lepousez G., Loudes C., Videau C., Simon A., Epelbaum J., Somatostatinergic systems in brain: Networks and functions. Mol. Cell. Endocrinol. 286, 75–87 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Moller L. N., Stidsen C. E., Hartmann B., Holst J. J., Somatostatin receptors. Biochim. Biophys. Acta 1616, 1–84 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Tostivint H., Ocampo Daza D., Bergqvist C. A., Quan F. B., Bougerol M., Lihrmann I., Larhammar D., Molecular evolution of GPCRs: Somatostatin/urotensin II receptors. J. Mol. Endocrinol. 52, T61–T86 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y., Yañez Guerra L. A., Egertová M., Zampronio C. G., Jones A. M., Elphick M. R., Molecular and functional characterization of somatostatin-type signalling in a deuterostome invertebrate. Open Biol. 10, 200172 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirabeau O., Joly J. S., Molecular evolution of peptidergic signaling systems in bilaterians. Proc. Natl. Acad. Sci. U.S.A. 110, E2028–E2037 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ong K. L., Lam K. S., Cheung B. M., Urotensin II: Its function in health and its role in disease. Cardiovasc. Drugs Ther. 19, 65–75 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Pless J., From somatostatin to Sandostatin: History and chemistry. Metab. Clin. Exp. 41, 5–6 (1992). [DOI] [PubMed] [Google Scholar]
- 31.Weckbecker G., Lewis I., Albert R., Schmid H. A., Hoyer D., Bruns C., Opportunities in somatostatin research: Biological, chemical and therapeutic aspects. Nat. Rev. Drug Discov. 2, 999–1017 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Martín-Gago P., Aragón E., Gomez-Caminals M., Fernández-Carneado J., Ramón R., Martin-Malpartida P., Verdaguer X., López-Ruiz P., Colás B., Cortes M. A., Ponsati B., Macias M. J., Riera A., Insights into structure-activity relationships of somatostatin analogs containing mesitylalanine. Molecules 18, 14564–14584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel Y. C., Wheatley T., In vivo and in vitro plasma disappearance and metabolism of somatostatin-28 and somatostatin-14 in the rat. Endocrinology 112, 220–225 (1983). [DOI] [PubMed] [Google Scholar]
- 34.Bai L., Sheeley S., Sweedler J. V., Analysis of endogenous d-amino acid–containing peptides in metazoa. Bioanal. Rev. 1, 7–24 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anthony L., Freda P. U., From somatostatin to octreotide LAR: Evolution of a somatostatin analogue. Curr. Med. Res. Opin. 25, 2989–2999 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kroeze W. K., Sassano M. F., Huang X. P., Lansu K., McCorvy J. D., Giguère P. M., Sciaky N., Roth B. L., PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 22, 362–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel Y. C., Srikant C. B., Subtype selectivity of peptide analogs for all five cloned human somatostatin receptors (hsstr 1-5). Endocrinology 135, 2814–2817 (1994). [DOI] [PubMed] [Google Scholar]
- 38.Pinter E., Helyes Z., Szolcsanyi J., Inhibitory effect of somatostatin on inflammation and nociception. Pharmacol. Ther. 112, 440–456 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Keri G., Erchegyi J., Horvath A., Mezo I., Idei M., Vantus T., Balogh A., Vadasz Z., Bokonyi G., Seprodi J., Teplan I., Csuka O., Tejeda M., Gaal D., Szegedi Z., Szende B., Roze C., Kalthoff H., Ullrich A., A tumor-selective somatostatin analog (TT-232) with strong in vitro and in vivo antitumor activity. Proc. Natl. Acad. Sci. U.S.A. 93, 12513–12518 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szolcsányi J., Bölcskei K., Szabó A., Pintér E., Petho G., Elekes K., Börzsei R., Almási R., Szuts T., Kéri G., Helyes Z., Analgesic effect of TT-232, a heptapeptide somatostatin analogue, in acute pain models of the rat and the mouse and in streptozotocin-induced diabetic mechanical allodynia. Eur. J. Pharmacol. 498, 103–109 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Taber R. I., Predictive value of analgesic assays in mice and rats. Adv. Biochem. Psychopharmacol. 8, 191–211 (1973). [PubMed] [Google Scholar]
- 42.Gorlin A. W., Rosenfeld D. M., Maloney J., Wie C. S., Garvey J. M., Trentman T. L., Survey of pain specialists regarding conversion of high-dose intravenous to neuraxial opioids. J. Pain Res. 9, 693–700 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen J. W., Hofer K., Cumber D. M., Wagstaff J. D., Layer R. T., McCabe R. T., Yaksh T. L., An assessment of the antinociceptive efficacy of intrathecal and epidural contulakin-G in rats and dogs. Anesth. Analg. 104, 1505, 1513 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Brennan T. J., Zahn P. K., Pogatzki-Zahn E. M., Mechanisms of incisional pain. Anesthesiol. Clin. North Am. 23, 1–20 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Shenoy P. A., Kuo A., Khan N., Gorham L., Nicholson J. R., Corradini L., Vetter I., Smith M. T., The somatostatin receptor-4 agonist J-2156 alleviates mechanical hypersensitivity in a rat model of breast cancer induced bone pain. Front. Pharmacol. 9, 495 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diochot S., Baron A., Salinas M., Douguet D., Scarzello S., Dabert-Gay A. S., Debayle D., Friend V., Alloui A., Lazdunski M., Lingueglia E., Black mamba venom peptides target acid-sensing ion channels to abolish pain. Nature 490, 552–555 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Abalde S., Tenorio M. J., Afonso C. M. L., Zardoya R., Comparative transcriptomics of the venoms of continental and insular radiations of West African cones. Proc. Biol. Sci. 287, 20200794 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dutertre S., Jin A. H., Vetter I., Hamilton B., Sunagar K., Lavergne V., Dutertre V., Fry B. G., Antunes A., Venter D. J., Alewood P. F., Lewis R. J., Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat. Commun. 5, 3521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Craig A. G., Norberg T., Griffin D., Hoeger C., Akhtar M., Schmidt K., Low W., Dykert J., Richelson E., Navarro V., Mazella J., Watkins M., Hillyard D., Imperial J., Cruz L. J., Olivera B. M., Contulakin-G, an O-glycosylated invertebrate neurotensin. J. Biol. Chem. 274, 13752–13759 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Bauer W., Briner U., Doepfner W., Haller R., Huguenin R., Marbach P., Petcher T. J., Pless J., SMS 201–995: A very potent and selective octapeptide analogue of somatostatin with prolonged action. Life Sci. 31, 1133–1140 (1982). [DOI] [PubMed] [Google Scholar]
- 51.Veber D. F., Holly F. W., Nutt R. F., Bergstrand S. J., Brady S. F., Hirschmann R., Glitzer M. S., Saperstein R., Highly active cyclic and bicyclic somatostatin analogues of reduced ring size. Nature 280, 512–514 (1979). [DOI] [PubMed] [Google Scholar]
- 52.Kailey B., van de Bunt M., Cheley S., Johnson P. R., MacDonald P. E., Gloyn A. L., Rorsman P., Braun M., SSTR2 is the functionally dominant somatostatin receptor in human pancreatic β- and α-cells. Am. J. Physiol. Endocrinol. Metab. 303, E1107–E1116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pi C., Liu J., Peng C., Liu Y., Jiang X., Zhao Y., Tang S., Wang L., Dong M., Chen S., Xu A., Diversity and evolution of conotoxins based on gene expression profiling of Conus litteratus. Genomics 88, 809–819 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Romanova E. V., Sasaki K., Alexeeva V., Vilim F. S., Jing J., Richmond T. A., Weiss K. R., Sweedler J. V., Urotensin II in invertebrates: From structure to function in Aplysia californica. PLOS ONE 7, e48764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrel C., Hocking H. G., Reynaud M., Upert G., Favreau P., Biass D., Paolini-Bertrand M., Peigneur S., Tytgat J., Gilles N., Hartley O., Boelens R., Stocklin R., Servent D., Identification, structural and pharmacological characterization of τ-CnVA, a conopeptide that selectively interacts with somatostatin sst3 receptor. Biochem. Pharmacol. 85, 1663–1671 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Ahorukomeye P., Disotuar M. M., Gajewiak J., Karanth S., Watkins M., Robinson S. D., Flórez Salcedo P., Smith N. A., Smith B. J., Schlegel A., Forbes B. E., Olivera B., Hung-Chieh Chou D., Safavi-Hemami H., Fish-hunting cone snail venoms are a rich source of minimized ligands of the vertebrate insulin receptor. eLife 8, e41574 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Q., Watkins M., Robinson S. D., Safavi-Hemami H., Yandell M., Discovery of novel conotoxin candidates using machine learning. Toxins 10, 503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Notredame C., Higgins D. G., Heringa J., T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., Claverie J.-M., Gascuel O., Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Imperial J. S., Chen P., Sporning A., Terlau H., Daly N. L., Craik D. J., Alewood P. F., Olivera B. M., Tyrosine-rich conopeptides affect voltage-gated K+ channels. J. Biol. Chem. 283, 23026–23032 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmieder R., Edwards R., Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., Couger M. B., Eccles D., Li B., Lieber M., MacManes M. D., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C. N., Henschel R., LeDuc R. D., Friedman N., Regev A., De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 64.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Sect. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., Hung L. W., Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., Coy A. M., Nicholas S. J. M., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. III, Snoeyink J., Adams P. D., Lovell S. C., Richardson J. S., Richardson D. C., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hylden J. L., Wilcox G. L., Intrathecal morphine in mice: A new technique. Eur. J. Pharmacol. 67, 313–316 (1980). [DOI] [PubMed] [Google Scholar]
- 73.Chaplan S. R., Bach F. W., Pogrel J. W., Chung J. M., Yaksh T. L., Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods 53, 55–63 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S5
Movies S1 to S6