ABSTRACT
Circular RNAs (circRNAs) are non-coding RNAs that have attracted considerable attention in recent years. Owing to their distinct circular structure, circRNAs are stable in cells. Autophagy is a catabolic process that helps in the degradation and recycling of harmful or inessential biological macromolecules in cells and enables cells to adapt to stress and changes in the internal and external environments. Evidence has shown that circRNAs influence the course of a disease by regulating autophagy, which indicates that autophagy is involved in the onset and development of various diseases and can affect drug resistance (for example, it affects cisplatin resistance in tumors). In this review, we summarized the role of circRNAs in autophagy and their influence on disease onset and progression as well as drug resistance. The review will expand our understanding of tumors as well as cardiovascular and neurological diseases and also suggest novel therapeutic strategies.
Abbreviations: ACR: autophagy-related circRNA; ADSCs: adipogenic mesenchymal stem cells; AMPK: AMP-activated protein kinase; ATG: autophagy related; BCL2: BCL2 apoptosis regulator; BECN1: beclin 1; ceRNA: competing endogenous RNA; circRNA: circular RNA; CMA: chaperone-mediated autophagy; EPCs: endothelial progenitor cells; LE/MVBs: late endosomes/multivesicular bodies; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MTOR: mechanistic target of rapamycin kinase; NSCLC: non-small cell lung cancer; PDLSCs: periodontal ligament stem cells; PE: phosphatidylethanolamine; PtdIns: phosphatidylinositol; PtdIns3K: phosphatidylinositol 3-kinase; PtdIns3P: phosphatidylinositol-3-phosphate 1,2-dipalmitoyl; PTEN: phosphatase and tensin homolog; RBPs: RNA-binding proteins; SiO2: silicon dioxide; TFEB: transcription factor EB; ULK: unc-51 like autophagy activating kinase 1
KEYWORDS: Autophagy, cancer, cardiovascular disease, circRNAs, neurological disease
Introduction
Circular RNAs (circRNAs), the recently identified forms of non-coding RNAs, are formed by the reverse splicing of the 5ʹ and 3ʹ ends [1]. Owing to the lack of the 5ʹ cap and 3ʹ poly(A) tail, circRNAs are stable, are undigestible by RNases [2], and are usually expressed in the tissues and body fluids. circRNAs are not only used as diagnostic markers for diseases [3] but are also involved in the development of several types of tumors [4] and in diseases such as, cardiovascular disorders [5], metabolic diseases [6], and neurodegenerative disorders [7]. Previous studies [8,9] have shown that circRNAs not only directly affect the expression of the key proteins involved in disease development by binding RNA or proteins but also indirectly regulate cell proliferation or apoptosis by influencing autophagy, which plays an important role in disease development.
Autophagy, a process by which cells phagocytose and digest themselves, is classified into macroautophagy, microautophagy, and chaperone-mediated autophagy. This review focuses on macroautophagy. Under normal physiological conditions, cells undergo basal levels of autophagic activity. However, when stimulated by metabolic changes [10,11], oxidative stress [12], endoplasmic reticulum stress [13], mechanical damage [14,15], or protein aggregation [16,17], cells respond by swallowing and digesting aged or damaged organelles, or misfolded or accumulated proteins, to maintain homeostasis. Thus, autophagy not only helps to recover amino acids and other large molecules for protein and ATP synthesis but also degrades cellular waste, which is conducive to cell stability [18]. During the onset, development, and treatment of diseases, changes occur in the internal and external environment of cells, which stimulate autophagy to enable the cells to adapt to these changes. Autophagy is involved in the onset and development of several tumors [19–28] and diseases including cardiovascular [29,30], neurodegenerative [31,32], and metabolic diseases [33,34], as well as immune disorders [35] and in drug resistance [36]. Autophagy plays different roles in disease development, and therefore, its activation exerts various effects on cells since it promotes [37] or inhibits [38] apoptosis and regulates cellular processes that play key roles in disease progression.
Herein, we discussed and summarized the influence of circRNAs on autophagy in tumors and diseases including cardiovascular and neurological diseases. A deeper understanding of the mechanisms underlying the roles played by circRNAs in autophagy is required to elucidate the mechanisms by which autophagy affects the course of numerous diseases. It is also essential to study disease pathogenesis/pathophysiology since it provides a theoretical basis for explaining the mechanisms underlying drug resistance in cancer therapy and for identifying novel therapeutic targets to improve treatment outcomes.
Circular RNAs (circRNAs)
circRNAs were first identified in RNA viruses in 1979 [39] and were originally hypothesized to be a by-product of splicing. circRNAs demonstrate a circular structure without poly(A) tail and exhibit important biological functions. They can be stably present in cells and have specific expression during development, thereby making them a suitable molecular target against various diseases [40]. Although circRNAs are involved in normal physiological activities, they also influence the onset and development of diseases.
circRNAs are classified into the following four categories based on their origin: exon circRNAs, intron circRNAs, exon and intron combination circRNAs [41], and circRNAs encoded by viral genomes, such as EB virus-encoding circRNAs [42]. circRNAs are formed primarily through two processes, namely intron pairing-derived cyclization and lasso-derived cyclization. The former process includes introns on both sides of the circRNAs; the exon contains the reverse complementary sequences, which completes the double-stranded RNA at the shear site, which, through alternative splicing, results in the formation of different circRNAs with or without introns. Alu and GU are common complementary sequences [43,44] (Figure 1A). In the latter process, which occurs in the precursor mRNA, the connection of the downstream 5ʹ end of the exon to the upstream 3ʹ end is catalyzed to form a lasso structure, which is then spliced to form circRNAs [43] (Figure 1B). RNA-binding proteins (RBPs) regulate circRNA formation, during which RBPs bind to introns on both sides of the pre-mRNA that forms circRNA and then shear each other to form circRNAs. For example, QKI regulates circRNA production in cardiomyocytes [45], and ILF3/NF90 and the NF110 splice variant regulate circRNA production during viral infection [46] (Figure 1C).
Figure 1.
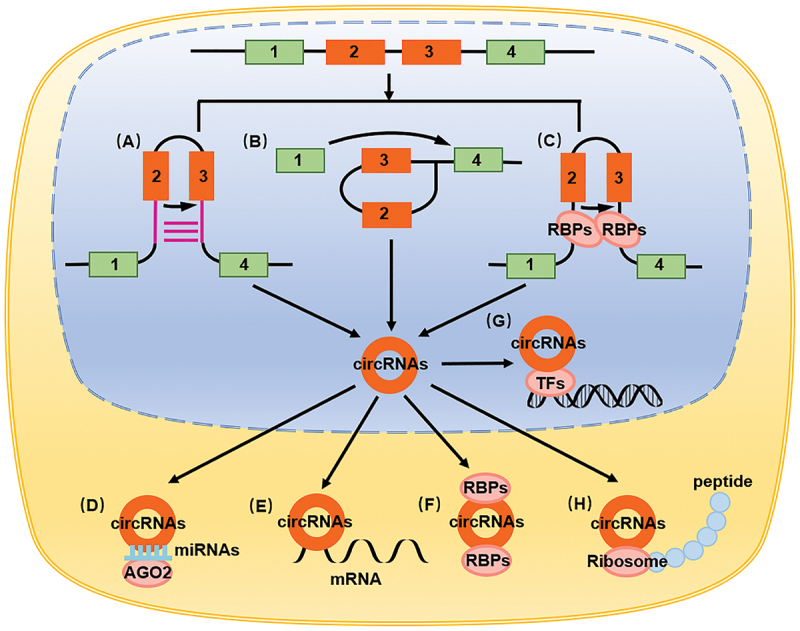
The biological characteristics and functions of circRNAs. (A) Intron pairing-derived cyclization. (B) Lasso-derived cyclization. (C) RNA binding proteins regulate circRNAs formation. (D) circRNAs adsorption of miRNA as a molecular sponge, playing the role of ceRNA. (E) circRNAs direct binding to mRNA. (F) circRNAs binding to proteins. (G) circRNAs binding to transcription factors. (H) circRNAs encoding small peptides.
CircRNAs play their roles mainly through the following ways: circRNAs combining with RNA, as molecular sponges, circRNAs adsorbing miRNA, acting as competing endogenous RNA (ceRNA), thereby affecting the function of miRNA in targeting mRNA in the occurrence and development of cancer [8,47–49], cardiovascular disorders [50], neurological disorders [51], metabolic disorders [52], and immunological disorders [53] (Figure 1D). circRNAs can also directly bind to mRNA and regulate its function. In intestinal stem cells, circPan3 promotes IL13RA1/IL-13Rα1 expression by binding to Il13ra1 mRNA, thereby stabilizing it [54] (Figure 1E). circRNAs can also bind to proteins, including RNA-binding proteins, and affect their functions. For example, the combination of circAGO2 and ELAVL1/HuR promotes the accumulation of untranslated regions at the 3ʹ end of target genes and inhibits the function of AGO2 [55] (Figure 1F). circRNAs can also bind to transcription factors and regulate its effect on target genes, thereby influencing the transcription process. The combination of circAmotl1 and STAT3 promotes its expression, while its combination with nuclear translocation on the Dnmt3a promoter region promotes its translocation [56] (Figure 1G). circRNAs reportedly demonstrate a coding function; hence, they can bind to ribosomes and encode small peptides. circZNF609 encodes small peptides in muscle cells to promote muscle cell proliferation [57], while circFBXW7 encodes small peptides FBXW7-185aa to inhibit tumor growth in glioblastoma [58] (Figure 1H).
Autophagy
Autophagy was first observed in the circulatory system in 1963 [59]. With technological advances, autophagy pathways and mechanisms of autophagy have been documented well [60]. Autophagy is divided into three categories based on the process through which it occurs, namely macroautophagy (commonly known as autophagy) [61], microautophagy, and chaperone-mediated autophagy (CMA). Microautophagy refers to the direct phagocytosis of cytoplasmic substances into endosomes [62], which requires HSPA8/Hsc70 to bind to the target protein and deliver it to late endosomes/multivesicular bodies (LE/MVBs), where the target protein is eventually digested [63]. CMA refers to the process in which an unfolded or misfolded protein with a specific amino acid sequence is recognized and bound by a chaperone and translocated into the lysosome for digestion [64]. This also requires HSPA8 to bind to the target protein, be recognized by LAMP2A on the lysosome, and finally be absorbed and digested by the lysosome [63].
Research has elucidated the specific steps involving autophagy and the associated signaling pathways. The process of autophagy involves the following four steps: 1) After autophagy induction by external stimuli, the cells form a flat, double-layered membranous structure in the cytoplasm, which acts as the precursor of autophagy; 2) The autophagy precursor continues to extend at both ends, encapsulating the cellular components directed for digestion including the cytoplasm, organelles, and proteins; 3) These cellular components are then phagocytosed inside the membranous structure, thereby forming a sealed autophagosome. Once formed, the autophagosome interacts directly with the lysosomes or fuses with the phagocytic vesicle, gulp vesicle, or endosome through intracellular endocytosis and then combines with the lysosome; 4) Following the degradation of the cargo by digestive enzymes present in the lysosome, the amino acids and fatty acids are released into the cytoplasm for cell reuse, and the digested residues are either expelled out of the cell or remain in the cytoplasm for excretion [65–68].
The intracellular signaling pathways regulating autophagy can either inhibit or activate autophagy. When the cellular homeostasis is maintained and when the internal and external environments of the cell are stable, the cell is then subjected to inhibitory autophagy signals. These signals activate MTOR (mechanistic target of rapamycin kinase) complex 1 (MTORC1), which accelerates the phosphorylation of ULK1 (unc-51 like autophagy activating kinase 1) at Ser757, inhibits the key initiation complex of autophagy formation, and thereby inhibits autophagy in the cell [69,70]. Conversely, when the cell is stimulated by an extracellular autophagy activation signal, the activity of intracellular MTORC1 is inhibited, the inhibitory phosphorylation of ULK1 is blocked, and the ULK complex composed of ULK1-ATG13-RB1CC1/FIP200-ATG101 is activated [71]. The activated ULK complex initiates the activity of the class III phosphatidylinositol 3-kinase (PtdIns3K) complex [72], including BECN1 and PIK3C3/VPS34 [73], which convert phosphatidylinositol (PtdIns) to phosphatidylinositol-3-phosphate 1,2-dipalmitoyl (PtdIns3P) [74], thus promoting the formation of the autophagosome precursor, the phagophore. Subsequently, two ubiquitin-like proteins are generated in the cell; the first is LC3-II. Intracellular MAP1LC3/LC3 (microtubule associated protein 1 light chain 3) is processed by the ATG4 protease to form the soluble cytosolic LC3-I, which is conjugated by ATG3 and ATG7 to phosphatidylethanolamine (PE) to form the membrane-associated LC3-II (LC3-PE) [75]. Intracellular ATG12 first interacts with ATG7, forming the ATG12-ATG7 complex. Subsequently, it interacts with ATG10 and ATG5 and forms the ATG12–ATG5 complex [76] and then interacts with ATG16L1 and forms the ATG12–ATG5-ATG16L1 complex [61,77], which functions as an E3 enzyme for LC3-II formation. The two complexes, LC3-II and ATG12–ATG5-ATG16L1, jointly promote the formation of the autophagosome [78].
Cells regulate autophagy by influencing the formation of key complexes such as the ULK1 and class III PtdIns3K complexes. The autophagy inhibition signal activates PI3K-AKT and MTORC1 to inhibit autophagy [79]. The BCL2 anti-apoptotic protein inhibits autophagy by inhibiting the activation of BECN1, which is part of the class III PtdIns3K complexes [80]. Upon activation, the RAS-RAF-MAP2K1/Mek-MAPK/Erk signaling pathway inhibits autophagy by activating MTORC1 [81] and promoting the expression of BCL2 [82,83]. In contrast, MAPK/p38 inhibits autophagy by inhibiting the activation of ULK1 [84]. Upon activation, the WNT-CTNNB1/β-catenin signaling pathway indirectly inhibits autophagy by inhibiting the expression of LC3-II and BECN1 [85]. Cytoplasmic TP53/p53 indirectly inhibits autophagy by inhibiting the activation of AMP-activated protein kinase (AMPK) [86]. The AMPK signaling pathway activation via autophagy signals can directly promote the phosphorylation of ULK1 at the Ser317 and Ser777 sites to promote autophagy [70,71]. In contrast, the AMPK signaling pathway can also indirectly promote the activation of the ULK complex by inhibiting MTORC1 activity through the phosphorylation of TSC2 and RPTOR [71,87,88], thereby promoting autophagy. PTEN (phosphatase and tensin homolog) can inhibit the PI3K-AKT-MTORC1 signaling pathway and ultimately promote autophagy by inhibiting the phosphorylation of AKT-MTORC1 [89]. The RAS-RAF-MAP2K1-MAPK signaling pathway directly promotes LC3-II expression and activates autophagy [90]. Activation of the MAPK/p38 signaling pathway promotes autophagy by upregulating the expression of BECN1 and ATG7 [91]. Transcription factor TP53 and TFEB (transcription factor EB) promote the transcription and synthesis of autophagy-associated proteins and promote autophagy [92].
In summary, the autophagy-associated pathways are complex and diverse. circRNAs affect autophagy by affecting the key molecules involved in the process, thereby regulating the biological functions of cells (Figure 2).
Figure 2.
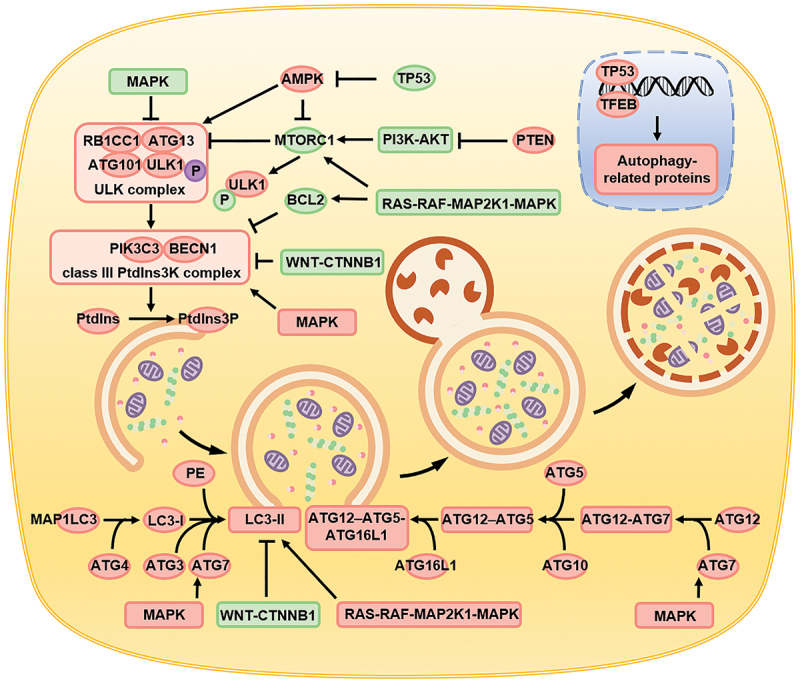
Schematic diagram depicting the process of autophagy, the biomolecules involved, and the signaling pathways regulated. Autophagy is primarily divided into four steps, namely the formation of autophagy precursors, the encapsulation of the cell components directed for digestion by the autophagosomes, the interaction of the autophagosomes with lysosomes to form the autolysosomes, and ultimately digestion and degradation of the autolysosomes. Signaling pathways such as AMPK, PI3K-AKT-MTORC1, MAPK, WNT-CTNNB1 regulate autophagy by affecting key proteins like the ULK complex, class III PItd3nsK complex, ATG protein family, and LC3-II.
circRNAs regulate autophagy in diseases
Cancer
circRNAs activating autophagy
Various circRNAs promote autophagy in a diverse range of tumors. Studies have shown that the expression of circDNMT1 increases in patients with breast cancer. circDNMT1 binds to the TP53 protein in the cytoplasm and leads to its migration to the nucleus [93]. The accumulation of TP53 in the nucleus promotes the expression of autophagy-related genes, thereby inducing autophagy in breast cancer cell. circDNMT1 also combines with HNRNPD/AUF1 and promotes its entry into the nucleus, thus enhancing the stability of the DNMT1 mRNA and upregulating DNMT1 expression. A high expression of DNMT1 in the nucleus inhibits the transcription of TP53 gene. The combination not only promotes autophagy in breast cancer cells via TP53-related mechanisms, but also regulates the process through HNRNPD [93]. The expression of circCDYL is often upregulated and associated with poor prognosis in breast cancer patients. circCDYL acts as a molecular sponge and adsorbs MIR1275, thus inhibiting its function; this, in turn, leads to the upregulation of intracellular ATG7 and ULK1 expression, promoting autophagy, which ultimately triggers the proliferation of breast cancer cells [94]. In patients with breast cancer resistant to paclitaxel, circ0006528 expression is increased, while that of CDK8 is upregulated due to the inhibition of MIR1299 expression, which indirectly promotes the expression of LC3-II, thereby inducing autophagy in breast cancer cells, enhancing proliferation and migration capacity, and causing resistance to paclitaxel in patients [95]. circABCB10 is also highly expressed in patients with paclitaxel resistance. As a molecular sponge, circABCB10 adsorbs LET7A-5p to function as ceRNA, thereby promoting the expression of DUSP7 and ultimately promoting the expression of LC3-II and autophagy in breast cancer cells and causing drug resistance [96]. Patients with epithelial ovarian cancer show increased expression of circMUC16, which has been found to be associated with poor prognosis. Conversely, circMUC16 adsorbs MIR199A-5p to mitigate the inhibition of BECN1 and RUNX1. BECN1 overexpression can promote autophagy, while RUNX1 overexpression can, through negative feedback loop, promote circMUC16 production. Meanwhile, circMUC16 can directly bind ATG13, which constitutes the ULK complex, to promote its expression. Together, the two pathways promote autophagy in ovarian cancer cells, and cancer cell proliferation and migration [9]. In patients with cervical cancer, the expression of circMTO1 is upregulated. As a molecular sponge, circMTO1 can adsorb MIR6893 and play the role of ceRNA. This indirectly promotes the expression of BECN1, thereby promoting autophagy, eventually leading to enhanced invasion and migration of the cervical cancer cells and drug resistance [97]. Upregulation of hsa_circ_0023404 expression in cervical cancer cells promotes the expression of BECN1, thereby promoting autophagy, inhibiting apoptosis, and imparting resistance to cisplatin [98]. During apatinib treatment of gastric cancer, the expression of circRACGAP1 is upregulated. Adsorption of MIR3657 promotes the expression of ATG7, enhances autophagy in gastric cancer cells, inhibits apoptosis, and induces apatinib resistance. Blockade of the expression of circRACGAP1 may improve the therapeutic effect of apatinib [99]. In liver cancer cells treated with sorafenib, the upregulated circIARS directly combines with ALKBH5 and inhibits its function, which dissociates BCL2 and BECN1 and promotes autophagy [100]. In patients with HCC resistant to cisplatin, circPVT1 expression is upregulated to promote YAP1 expression by targeting MIR30A-5p, indirectly promoting LC3-II expression, and ultimately leading to autophagy and cisplatin resistance [101]. In patients with non-small cell lung cancer (NSCLC), the expression of hsa_circ_0085131 is upregulated to adsorb MIR654-5p, as a molecular sponge, to promote the expression of ATG7, and this ultimately results in autophagy, leading to cisplatin resistance [102]. Papillary thyroid cancer is characterized by the increased expression of circEIF6, which inhibits MIR144-3p, thereby indirectly promoting the expression of LC3-II, and ultimately leading to enhanced autophagy and cisplatin resistance [103]. In renal clear cell carcinoma, the expression of hsa_circ_0035483 is upregulated, which promotes the expression of LC3-II following targeted inhibition of MIR335, which promotes autophagy and imparts resistance to gemcitabine [104]. In imatinib-resistant chronic myeloid leukemia patients, the expression of circ0009910 is upregulated in the serum. circ0009910 binds MIR34A-5p and inhibits MIR34A-5p binding to ULK1 mRNA, thereby increasing the expression of ULK1. circ0009910 also promotes the phosphorylation of ULK1, and expression of BECN1 and LC3-II, ultimately promoting autophagy to inhibit apoptosis and provide resistance to imatinib [105]. In patients with adriamycin-resistant acute myeloid leukemia, the expression of circPAN3 is increased in the serum, which in turn, promotes the phosphorylation and activation of AMPK, and inhibits MTOR phosphorylation to promote autophagy. It also directly promotes the expression of BECN1 and LC3-II, thereby further promoting autophagy, inhibiting apoptosis, and causing adriamycin resistance [106]. In patients with radiotherapy-resistant prostate cancer, circCCNB2 expression is upregulated; additionally, LC3-II and BECN1 expression are also indirectly upregulated by MIR30B-5p-KIF18A to promote autophagy and treatment resistance [107].
circRNAs inhibiting autophagy
circRNAs not only influence tumor development by promoting autophagy, but also regulate disease progression by inhibiting autophagy. In patients with lung cancer, circHIPK3 acts as a molecular sponge and adsorbs MIR124-3p. It plays the role of a ceRNA and promotes the expression of IL6R and STAT3, both of which promote the phosphorylation of STAT3. Phosphorylated STAT3 inhibits the phosphorylation of PRKAA and activates the AMPK signaling pathway, thereby inhibiting autophagy and promoting the invasion and migration of lung cancer cells [108]. In patients with gastric cancer, the expression of circNRIP1 increases and is associated with poor prognosis. circNRIP1 competes with MIR149-5p to activate the AKT-MTOR signaling pathway, thus inhibiting autophagy in gastric cancer cells, and promoting cell invasion, metastasis, and proliferation [109]. circ0032821 is also highly expressed in patients with gastric cancer, where it promotes the malignant phenotype by inhibiting autophagy, which may be associated with the activation of MAPK/Erk signaling pathway [110]. circKHDC4/KIAA0907 is expressed at low levels in patients with gastric cancer; the adsorption of MIR452-5p promotes the expression of KAT6B, and indirectly inhibits the expression of LC3-II and BECN1, and ultimately autophagy, thereby promoting apoptosis [111]. circCUL2 is also expressed at low levels in patients with gastric cancer, and acts as a molecular sponge to adsorb MIR142-3p in cisplatin-resistant cell lines to promote the expression of ROCK2, and indirectly inhibits the expression of LC3-II and BECN1, and ultimately autophagy, thereby promoting cisplatin resistance in gastric cancer cells [112]. The low expression level of circMCTP2 in patients with cisplatin-resistant gastric cancer promotes MTMR3 expression by targeting MIR99A-5p, and indirectly inhibits BECN1 and LC3-II expression, and ultimately autophagy [113]. In esophageal squamous cell carcinoma, CDR1-AS/ciRS-7 promotes the expression of EGFR following adsorption of MIR1299, which acts as a molecular sponge. EGFR activates the AKT-MTOR signaling pathway, which inhibits autophagy, and affects the invasion and proliferation of esophageal cancer cells [114]. In patients with glioma, the drug matrine downregulates the expression of circ104075, which inhibits autophagy by activating the AKT-MTOR signaling pathway [115]. circ104075 also promotes the WNT-CTNNB1 signaling pathway [116] and inhibits autophagy by promoting the expression of BCL9. Therefore, matrine promotes cell autophagy and apoptosis, thus making it a candidate drug for treating glioblastoma patients [115]. Matrine downregulates the expression of circ0027345 in liver cancer cells. circ0027345 indirectly inhibits the expression of LC3-II and BECN1 via MIR345-5p-HOXD3, inhibits the onset of autophagy, improves autophagy in liver cancer cells subjected to matrine treatment, and inhibits the growth, migration, and invasion of liver cancer cells [117]. In patients with cervical cancer, increased expression of hsa_circ_0000515 was observed, which is correlated with adverse recovery. hsa_circ_0000515 competitively combines with MIR326 and promotes the expression of ELK1, inhibits the expression of BECN1 and LC3-II, inhibits autophagy, and promotes the proliferation and invasion of cervical cancer cells [118]. circ0000285 is also highly expressed in cervical cancer. By inhibiting the expression of MIR197-3p, ELK1 expression is increased, thereby inhibiting autophagy, promoting cell proliferation, and inhibiting apoptosis [119]. The expression of circCRIM1 is upregulated in osteosarcoma, and adsorption of MIR432-5p, as a molecular sponge, promotes the expression of HDAC4, which indirectly inhibits the expression of LC3-II and BECN1, inhibits autophagy, and promotes the invasion, metastasis, and proliferation of osteosarcoma cells [120]. circKMT2D expression is downregulated in H2O2-treated osteosarcoma cells; circKMT2D inhibits the expression of MIR210 and BECN1, and ultimately autophagy [121] (Figure 3).
Figure 3.
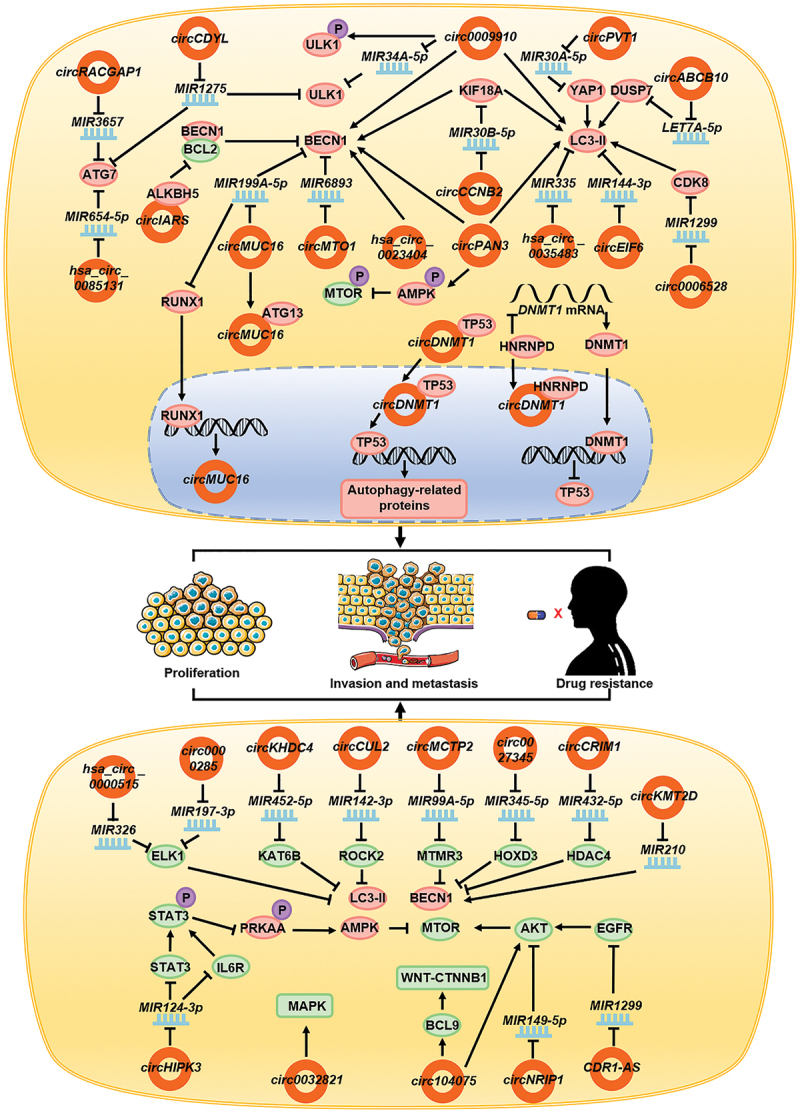
circRNAs affect tumor development and drug resistance by regulating autophagy. In tumor cells, circDNMT1, circCDYL, circ0006528, circABCB10, circMUC16, circMTO1, hsa_circ_0023404, circRACGAP1, circIARS, circPVT1, hsa_circ_0085131, circEIF6, hsa_circ_0035483, circ0009910, circPAN3, and circCCNB2 promote autophagy by promoting the functions of key proteins involved in the autophagy process, whereas circHIPK3, circNRIP1, circ0032821, circKHDC4, circCUL2, circMCTP2, CDR1-AS, circ104075, circ0027345, has_circ_0000515, circ0000285, circCRIM1, and circKMT2D inhibit autophagy by inhibiting the functions of key proteins in the autophagy process. Eventually circRNAs lead to the enhancement of tumor cell invasion, metastasis, proliferation, and drug resistance in patients.
Cardiovascular diseases
circRNAs also influence the course of cardiovascular diseases by regulating autophagy. During myocardial hypoxia/reoxygenation, the expression of circ101237 gradually increases with time and promotes autophagy. The molecular mechanism involves circ101237, as a molecular sponge, which adsorbs Let7a-5p, thereby inhibiting its function and increasing the expression of IGF2BP3 [122]. In turn, IGF2BP3 combines with the 5ʹ UTR of Igf2 mRNA and promotes the expression of IGF2 [123], which phosphorylates and activates the MAPK/Erk signaling pathway [124]. This promotes the expression of LC3-II, thereby leading to promotion of autophagy and inhibition of apoptosis in cardiomyocytes [122]. In H2O2-stimulated cardiomyocytes exhibiting oxidative damage, circHipk2, as a molecular sponge, promoted the expression of ATG101 through Mir485-5p, promoted autophagy, and accelerated H2O2-induced cardiomyocyte apoptosis [125]. In mouse myocardial infarction model, circPan3 expression upregulation promoted FOXO3 expression by inhibiting Mir221; FOXO3, as a transcription factor, bound to the promoter region of Atg7, to promote its expression, and ultimately autophagy [126].
The expression of circZnf292 is upregulated in ischemic heart disease. circZnf292 activates the WNT-CTNNB1 and MTOR signaling pathways by inhibiting the expression of BNIP3, thereby leading to the inhibition of autophagy and reducing damage in cardiomyocytes [127]. During cardiac ischemia-reperfusion injury, the expression of autophagy-related circRNAs (ACRs) is downregulated. Acr directly combines with DNMT3B and inhibits the DNA methylation of Pink1 promoter, thereby promoting the expression of PINK1. In turn, PINK1 promotes the phosphorylation of RIPOR2/FAM65B, which inhibits autophagy in myocardial cells, inhibits apoptosis, reduces the myocardial infarction area, and plays a protective role in myocardial ischemia-reperfusion injury [128]. circPan3 is also downregulated in myocardial ischemia-reperfusion injury, which promotes the expression of PINK1 by adsorbing Mir421, thereby inhibiting the expression of autophagy-associated proteins, and eventually the onset of autophagy, and reducing cell apoptosis [129] (Figure 4).
Figure 4.
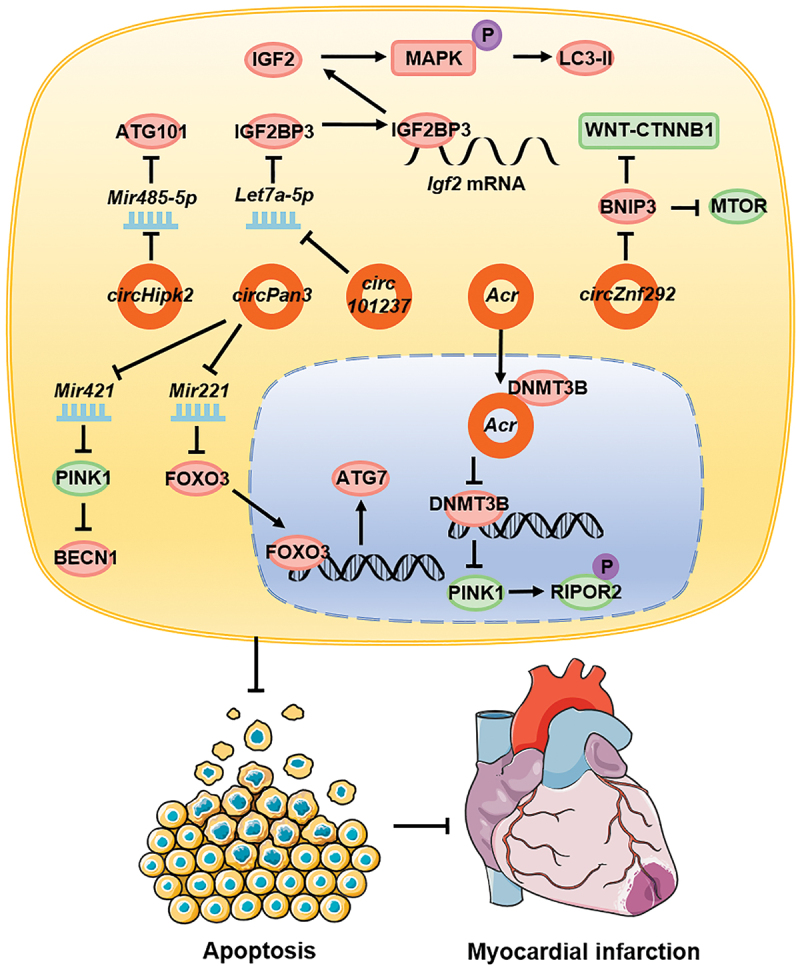
circRNAs affect cardiovascular diseases by regulating autophagy. In cardiovascular diseases, circ101237, circHipk2, and circPan3 promote autophagy whereas circZnf292, Acr, and circPan3 inhibit autophagy, and this ultimately leads to reduced cardiomyocyte apoptosis and a reduced myocardial necrosis area.
Neurological disorders
circRNAs activating autophagy
Autophagy plays an important role in neurological diseases. During cerebral ischemia-reperfusion injury, circ016719 expression is upregulated in neurons, and this promotes autophagy. circ016719 promotes the expression of MAP2K6 through competitive binding of Mir29c. MAP2K6, as a key molecule upstream of the MAPK/p38 signaling pathway [130], promotes the expression of BECN1, thereby promoting autophagy and apoptosis in neurons [131]. In astrocytes, circHectd1 is upregulated, which combines with Mir142 and inhibits its function. Moreover, it promotes the expression of TIPARP, which upregulates the expression of LC3-II and activates autophagy, thus enabling the astrocytes to function during stroke [132]. During cerebral ischemia injury, circAkap7 derived from exosomes can promote the expression of ATG12, by inhibiting Mir155-5p, can promote autophagy, and can improve the ischemic injury caused by oxidative stress [133]. circLrp1b expression is upregulated in patients with traumatic brain injury, promoting DRAM2 expression by inhibiting Mir27a-3p function, indirectly promoting ATG5 expression, and ultimately promoting the expression of autophagy-associated molecular markers, autophagy activation, and inflammatory processes [134]. In patients with Parkinson disease, circDLGAP4 acts as a molecular sponge and competes with MIR134-5p to inhibit its function. It promotes the expression of BECN1 and LC3-II, thereby promoting autophagy, inhibiting apoptosis, and reducing mitochondrial damage [135]. During astrocyte activation, the high expression of circHipk2 inhibits Mir124-2hg, increases SIGMAR1 expression, and promotes the expression of LC3-II, thereby promoting autophagy and inhibiting the inflammatory response of astrocytes [136]. In the neuropathic pain model, the expression of Cdr1-as/ciRS-7 is upregulated in the dorsal horn of the spinal cord and associated with pain progression. Following the adsorption of Mir135a-5p as a molecular sponge, Cdr1-as promotes its function and increases the expression of BECN1 and LC3-II, thereby promoting autophagy and regulating inflammation [137]. In a mice model of Alzheimer disease [138,139], the expression of circNf1-419 was found to be upregulated. Following interaction with DNM1 (dynamin 1) and AP2B1, circNf1-419 activated the AMPK signaling pathway and ULK1, either directly or indirectly, by inhibiting MTORC1, which in turn activated autophagy in astrocytes and mitigated Alzheimer symptoms in mice [138,139].
circRNAs inhibiting autophagy
Stimulation of craniocerebral injury by H2O2 leads to the downregulation of circHipk3 expression. circHipk3 promotes the expression of Mir455, which enhances the expression of NFE2L2/NRF2, activates the MAPK/Erk signaling pathway, inhibits the expression of BECN1, and suppresses autophagy. Furthermore, during a craniocerebral injury, circHipk3 affects neuronal cell apoptosis [140]. During peripheral nerve injury, the expression of circ2837 is downregulated. circ2837 acts as a molecular sponge and binds to Mir34a and inhibits its function. Thus, circ2837 inhibits the expression of LC3-II, thereby inhibiting autophagy in the nerve cells and playing a neuroprotective role [141]. In a diabetic peripheral neuropathy study [142], stimulation of Schwann cells with high-glucose downregulated the expression of circRNAs associated with autophagy. Acr activated the PI3K-AKT-MTORC1 signaling pathway by interacting with Mir145-3p. Therefore, stimulation by high glucose concentration leads to autophagy, ultimately slowing cell death [142] (Figure 5).
Figure 5.
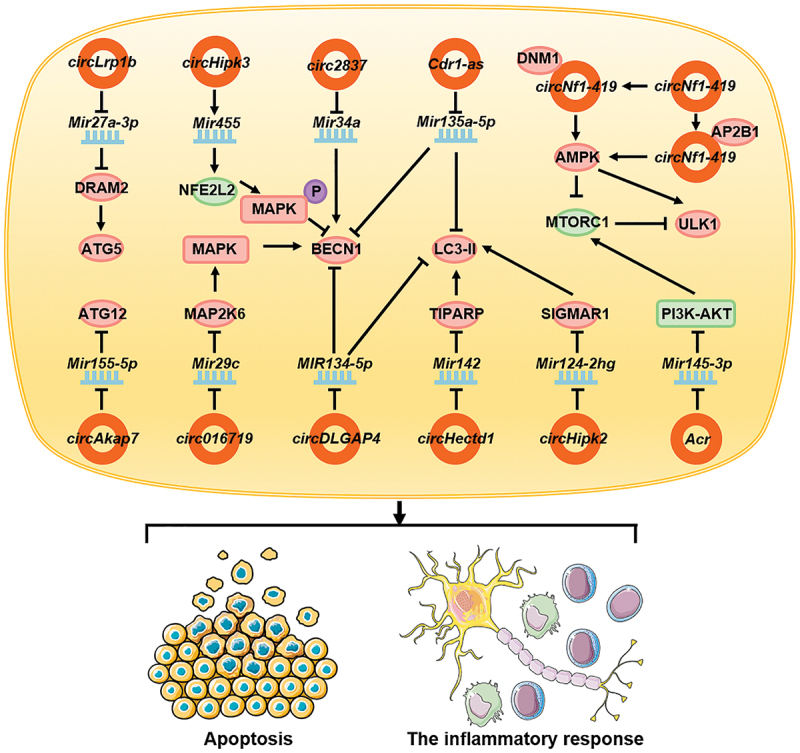
circRNAs affect neuronal diseases by regulating autophagy. In neurological diseases, circ016719, circHectd1, circAkap7, circLrp1b, circDLGAP4, circHipk2, Cdr1-as, and circNf1-419 promote autophagy in neuronal cells or glial cells by promoting the function of autophagy-related proteins; in contrast, circHipk3, circ2837, and Acr inhibit autophagy by inhibiting the functions of autophagy-related proteins. They ultimately affect the apoptosis of nerve cells and the inflammatory response of the nervous system.
Miscellaneous roles
The somatic inflammatory cells of patients with tubal inflammation show reduced expression of circEIF3K. Following binding with MIR139-5p, circEIF3K inhibits the functions of MIR139-5p, the phosphorylation of MAPK/Erk, and the expression of BCL2, which promotes the autophagy of inflammatory cells, thereby inhibiting cell vitality and promoting apoptosis [143]. circCDK8 is highly expressed in periodontal ligament stem cells (PDLSCs) during hypoxia, which promotes the expression of ATG5, thereby directly promoting autophagy [144]. Autologous adipogenic mesenchymal stem cells (ADSCs) can secrete exosomes containing mmu_circ_0000250, act on endothelial progenitor cells (EPCs), promote SIRT1 expression through the adsorption of Mir128-3p, promote autophagy, inhibit cell apoptosis, and contribute to the healing of diabetic wounds [145]. Exosomes secreted by ADSCs also contain mmu_circ_0000623, which can activate cell autophagy through Mir125-ATG4D, thereby inhibiting liver fibrosis [146]. Hsa_circ_0026827 is highly expressed in human dental pulp stem cells and can promote BECN1 expression through MIR188-3p, autophagy, and osteoblast differentiation [147]. In a study investigating the effects of cadmium poisoning on the body, CdCl2 induced the high expression of hsa_circ_0040768 in HepG2 cells, thereby increasing the expression of MAP1LC3B, promoting autophagy, and inducing HepG2 cell apoptosis [148].
Induction by silicon dioxide (SiO2) inhibits the expression of circHECTD1 in the lungs in a concentration-dependent manner. circHECTD1 inhibits the expression of LC3-II by inhibiting the expression of HECTD1, thereby inhibiting autophagy, invasion, migration, and activation in lung fibroblasts. This leads to increased pulmonary fibrosis under the action of SiO2 [149]. However, in patients with silicosis, the expression of circ012091 is downregulated. in lung fibroblasts. circ012091 inhibits the expression of the autophagy-associated protein LC3-II by inhibiting the expression of PPP1R13B, a major pro-apoptotic protein of the TP53 family, thus inhibiting autophagy in the cells. The occurrence of apoptosis and endoplasmic reticulum stress inhibits the proliferation and migration of lung fibroblasts, which ultimately leads to the aggravation of pulmonary fibrosis in silicosis patients [150]. The expression of circGATAD2A is upregulated following infection with influenza virus. circGATAD2A increases viral replication by inhibiting autophagy [151]. During inflammatory bowel disease, circPabpn1 interacts with ELAVL1/HuR and inhibits its binding to Atg16l1 mRNA, resulting in the decreased stability of Atg16l1 mRNA and reduced expression of ATG16L1, thereby inhibiting autophagy in intestinal epithelial cells [152] (Figure 6).
Figure 6.
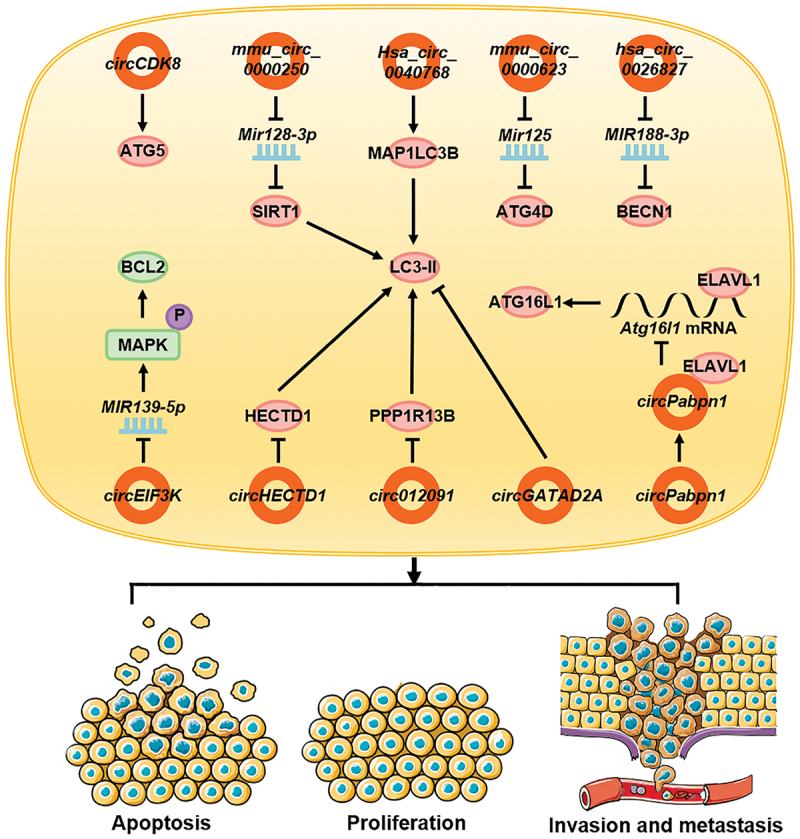
circRNAs affect other diseases by regulating autophagy. In inflammatory cells, circEIF3K promotes autophagy and affects apoptosis. In stem cells, circCDK8, mmu_circ_0000250, mmu_circ_0000623, and hsa_circ_0026827 promote autophagy and affect its function. In cadmium-exposed cells, hsa_circ_0040768 promotes autophagy and apoptosis whereas in fibroblasts, intestinal epithelial cells, and virus-infected cells, circHECTD1, circ012091, circGATAD2A, and circPabpn1 inhibit autophagy and cause cell proliferation, invasion, and migration.
Conclusions and future prospects
The structural characteristics of circRNAs enable them to stably exist in cells, and not only serve as a potential marker for early diagnosis but are also vital for disease progression. Autophagy plays an important role in the onset and development of diseases. circRNAs play important roles in the regulation of autophagy, by primarily affecting the process in two ways. It either affects the expression of key proteins involved in autophagy or affects the activation or inhibition of signaling pathways regulating autophagy. Most studies have focused on the effect of circRNAs on the expression of autophagy marker proteins such as LC3-II or SQSTM1/p62. However, the mechanisms by which circRNAs affect protein expression have not been examined. circRNAs have several functions; however, studies have primarily focused on the ceRNA model. circRNAs act as molecular sponges to adsorb miRNAs, thereby hindering their inhibitory function against target downstream mRNA. Additionally, circRNAs function through various pathways. With advancements in research, the different modes of regulation can be identified, thereby enriching the existing knowledge on the autophagy regulatory network of circRNAs. Expression levels of several circRNAs are altered during disease development, but the mechanism remains unknown. Targeting proteins produced by circRNAs may be a potential treatment strategy against various diseases. Further understanding of the influence of circRNAs on disease progression through autophagy can help to design targeted therapies.
Autophagy, a method of regulating cell metabolism, which also enables their functioning, can promote apoptosis and plays diverse roles in various diseases. Therefore, studies on autophagy should focus on its association with disease development and prognosis. The regulatory mechanisms of autophagy are remarkably complex; moreover, the same signaling pathways may function differently under different cellular metabolic subsystems and extracellular microenvironments. Further studies are warranted to better understand the regulation and function of these pathways under different circumstances, which may also be associated with the metabolic state of the cells. Therefore, studies on autophagy should not only focus on the changes in autophagy-related proteins, but also focus on the changes in cellular metabolism and the regulation of autophagy by circRNAs. Despite growing research on circRNAs and their role in autophagy, the mechanism underlying circRNA-mediated regulation of autophagy in the pathogenesis of tumor, cardiovascular disease, and neurodegeneration remains to be elucidated. Whether these circRNAs can be used as targets for early diagnosis and post-treatment analysis remains to be determined. There are few studies on drugs targeting circRNAs and autophagy-associated proteins and signaling pathways as novel treatment strategies against the above-mentioned diseases. Using various experimental models, these unanswered questions will be addressed in the future.
Supplementary Material
Funding Statement
This work was supported in part by grants from the National Natural Science Foundation of China [81803025, 81872278, 81972776, 82072374, 82003243, 82002239 and 82073135], the Overseas Expertise Introduction Project for Discipline Innovation [111 Project, No. 111-2-12], the Natural Science Foundation of Hunan Province [2019JJ50354, and 2020JJ4766], Central South University Graduate Research and Innovation Project [2020zzts232], and Mittal Student Innovation Program [GCX2020321Y].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Wang Y, Mo Y, Gong Z, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pandey PR, Rout PK, Das A, et al. RPAD (RNase R treatment, polyadenylation, and poly(A)+ RNA depletion) method to isolate highly pure circular RNA. Methods. 2019;155:41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fan CM, Wang JP, Tang YY, et al. circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci. 2019;110:2180–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].He R, Liu P, Xie X, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fan X, Weng X, Zhao Y, et al. Circular RNAs in cardiovascular disease: an overview. Biomed Res Int. 2017;2017:5135781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fang Y, Wang X, Li W, et al. Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int J Mol Med. 2018;42:1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lu D, Xu AD.. Mini review: circular RNAs as potential clinical biomarkers for disorders in the central nervous system. Front Genet. 2016;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gan X, Zhu H, Jiang X, et al. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer. 2020;19:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kuma A, Mizushima N.. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin Cell Dev Biol. 2010;21:683–690. [DOI] [PubMed] [Google Scholar]
- [11].Fan C, Zhang S, Gong Z, et al. Emerging role of metabolic reprogramming in tumor immune evasion and immunotherapy. Sci China Life Sci. 2020;64:534-547. [DOI] [PubMed] [Google Scholar]
- [12].Marino ML, Fais S, Djavaheri-Mergny M, et al. Proton pump inhibition induces autophagy as a survival mechanism following oxidative stress in human melanoma cells. Cell Death Dis. 2010;1:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kouroku Y, Fujita E, Tanida I, et al. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. [DOI] [PubMed] [Google Scholar]
- [14].Zhang X, Jing Y, Qin C, et al. Mechanical stress regulates autophagic flux to affect apoptosis after spinal cord injury. J Cell Mol Med. 2020;24:12765-12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li D, Lu Z, Xu Z, et al. Spironolactone promotes autophagy via inhibiting PI3K/AKT/mTOR signalling pathway and reduce adhesive capacity damage in podocytes under mechanical stress. Biosci Rep. 2016;36:e00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fu Y, Sun X, Lu B. HIPK3 modulates autophagy and HTT protein levels in neuronal and mouse models of Huntington disease. Autophagy. 2018;14:169–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Catanese A, Olde Heuvel F, Mulaw M, et al. Retinoic acid worsens ATG10-dependent autophagy impairment in TBK1-mutant hiPSC-derived motoneurons through SQSTM1/p62 accumulation. Autophagy. 2019;15:1719–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004. ;6:463–477. [DOI] [PubMed] [Google Scholar]
- [19].Sato K, Tsuchihara K, Fujii S, et al. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res. 2007;67:9677–9684. [DOI] [PubMed] [Google Scholar]
- [20].Nassour J, Radford R, Correia A, et al. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature. 2019;565:659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lu Y, Liu LL, Liu SS, et al. Celecoxib suppresses autophagy and enhances cytotoxicity of imatinib in imatinib-resistant chronic myeloid leukemia cells. J Transl Med. 2016;14:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Thomas S, Sharma N, Golden EB, et al. Preferential killing of triple-negative breast cancer cells in vitro and in vivo when pharmacological aggravators of endoplasmic reticulum stress are combined with autophagy inhibitors. Cancer Lett. 2012;325:63–71. [DOI] [PubMed] [Google Scholar]
- [23].Zhu L, Li L, Zhang Q, et al. NOS1 S-nitrosylates PTEN and inhibits autophagy in nasopharyngeal carcinoma cells. Cell Death Discov. 2017;3:17011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu G, Pei F, Yang F, et al. Role of autophagy and apoptosis in non-small-cell lung cancer. Int J Mol Sci. 2017;18:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tang L, Wei F, Wu Y, et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J Exp Clin Cancer Res. 2018;37:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang YA, Li XL, Mo YZ, et al. Effects of tumor metabolic microenvironment on regulatory T cells. Mol Cancer. 2018;17:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jiang X, Wang J, Deng X, et al. The role of microenvironment in tumor angiogenesis. J Exp Clin Cancer Res. 2020;39:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wei X, Chen Y, Jiang X, et al. Mechanisms of vasculogenic mimicry in hypoxic tumor microenvironments. Mol Cancer. 2021;20:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang X, Guo Z, Ding Z, et al. Inflammation, autophagy, and apoptosis after myocardial infarction. J Am Heart Assoc. 2018;7:e008024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu CY, Zhang YH, Li RB, et al. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun. 2018;9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zare-Shahabadi A, Masliah E, Johnson GV, et al. Autophagy in Alzheimer’s disease. Rev Neurosci. 2015;26:385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rivero-Ríos P, Madero-Pérez J, Fernández B, et al. Targeting the autophagy/lysosomal degradation pathway in Parkinson’s disease. Curr Neuropharmacol. 2016;14:238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kim J, Lim YM, Lee MS. The role of autophagy in systemic metabolism and human-type diabetes. Mol Cells. 2018;41:11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wei F, Wang D, Wei J, et al. Metabolic crosstalk in the tumor microenvironment regulates antitumor immunosuppression and immunotherapy resisitance. Cell Mol Life Sci. 2021;78:173–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yin H, Wu H, Chen Y, et al. The therapeutic and pathogenic role of autophagy in autoimmune diseases. Front Immunol. 2018;9:1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang C, Yang Y, Sun L, et al. Baicalin reverses radioresistance in nasopharyngeal carcinoma by downregulating autophagy. Cancer Cell Int. 2020;20:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Xu F, Yan W, Cheng Y. Pou4f3 gene mutation promotes autophagy and apoptosis of cochlear hair cells in cisplatin-induced deafness mice. Arch Biochem Biophys. 2020;680:108224. [DOI] [PubMed] [Google Scholar]
- [38].Zhu HL, Xu XF, Shi XT, et al. Activation of autophagy inhibits cadmium-triggered apoptosis in human placental trophoblasts and mouse placenta. Environ Pollut. 2019;254:112991. [DOI] [PubMed] [Google Scholar]
- [39].Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. [DOI] [PubMed] [Google Scholar]
- [41].Zhang H, Shen Y, Li Z, et al. The biogenesis and biological functions of circular RNAs and their molecular diagnostic values in cancers. J Clin Lab Anal. 2020;34:e23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Liu Q, Shuai M, Xia Y. Knockdown of EBV-encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma cell proliferation and metastasis through sponging multiple miRNAs. Cancer Manag Res. 2019;11:8023–8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. [DOI] [PubMed] [Google Scholar]
- [45].Gupta SK, Garg A, Bar C, et al. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ Res. 2018;122:246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Li X, Liu C-X, Xue W, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. 2017;67:214–227.e7. [DOI] [PubMed] [Google Scholar]
- [47].Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. [DOI] [PubMed] [Google Scholar]
- [48].Tang L, Xiong W, Zhang L, et al. circSETD3 regulates MAPRE1 through miR-615-5p and miR-1538 sponges to promote migration and invasion in nasopharyngeal carcinoma. Oncogene. 2021;40:307–321. [DOI] [PubMed] [Google Scholar]
- [49].Fan C, Qu H, Xiong F, et al. CircARHGAP12 promotes nasopharyngeal carcinoma migration and invasion via ezrin-mediated cytoskeletal remodeling. Cancer Lett. 2021;496:41–56. [DOI] [PubMed] [Google Scholar]
- [50].Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. [DOI] [PubMed] [Google Scholar]
- [51].Xie BS, Wang YQ, Lin Y, et al. Circular RNA expression profiles alter significantly after traumatic brain injury in rats. J Neurotrauma. 2018;35:1659–1666. [DOI] [PubMed] [Google Scholar]
- [52].Zhu K, Hu X, Chen H, et al. Downregulation of circRNA DMNT3B contributes to diabetic retinal vascular dysfunction through targeting miR-20b-5p and BAMBI. EBioMedicine. 2019;49:341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhang C, Wang X, Chen Y, et al. The down-regulation of hsa_circ_0012919, the sponge for miR-125a-3p, contributes to DNA methylation of CD11a and CD70 in CD4+ T cells of systemic lupus erythematous. Clin Sci (Lond). 2018;132:2285–2298. [DOI] [PubMed] [Google Scholar]
- [54].Zhu P, Zhu X, Wu J, et al. IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat Immunol. 2019;20:183–194. [DOI] [PubMed] [Google Scholar]
- [55].Chen Y, Yang F, Fang E, et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yang ZG, Awan FM, Du WW, et al. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol Ther. 2017;25:2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Radewa J. Observations on autophagocytosis phenomena in the blood. Z Rheumaforsch. 1963;22:36–46. [PubMed] [Google Scholar]
- [60].Lekli I, Ray D, Mukherjee S, et al. Co-ordinated autophagy with resveratrol and γ-tocotrienol confers synergetic cardioprotection. J Cell Mol Med. 2010;14:2506–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;2:1542–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2014;7:673–682. [DOI] [PubMed] [Google Scholar]
- [63].Tekirdag K, Cuervo AM. Chaperone-mediated autophagy and endosomal microautophagy: joint by a chaperone. J Biol Chem. 2018;293:5414–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Massey A, Kiffin R, Cuervo AM. Pathophysiology of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2420–2434. [DOI] [PubMed] [Google Scholar]
- [65].Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chiu HC, Richart S, Lin FY, et al. The interplay of reovirus with autophagy. Biomed Res Int. 2014;2014:483657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sergin I, Razani B. Self-eating in the plaque: what macrophage autophagy reveals about atherosclerosis. Trends Endocrinol Metab. 2014;25:225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Stead ER, Castillo-Quan JI, Miguel VEM, et al. Agephagy - adapting autophagy for health during aging. Front Cell Dev Biol. 2019;7:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Jung CH, Ro SH, Cao J, et al. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Wong PM, Puente C, Ganley IG, et al. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Russell RC, Tian Y, Yuan H, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–364. [DOI] [PubMed] [Google Scholar]
- [75].Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. [DOI] [PubMed] [Google Scholar]
- [77].Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. [DOI] [PubMed] [Google Scholar]
- [79].Wu Y, Liu X, Qin Z, et al. Low-frequency ultrasound enhances chemotherapy sensitivity and induces autophagy in PTX-resistant PC-3 cells via the endoplasmic reticulum stress-mediated PI3K/Akt/mTOR signaling pathway. Onco Targets Ther. 2018;11:5621–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Maiuri MC, Le Toumelin G, Criollo A, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Carriere A, Romeo Y, Acosta-Jaquez HA, et al. ERK1/2 phosphorylate raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1). J Biol Chem. 2011;286:567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Holmberg J, Durbeej M. Laminin-211 in skeletal muscle function. Cell Adh Migr. 2013;7:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Li D, Wei Y, Xu S, et al. A systematic review and meta-analysis of bidirectional effect of arsenic on ERK signaling pathway. Mol Med Rep. 2018;17:4422–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].He Y, She H, Zhang T, et al. p38 MAPK inhibits autophagy and promotes microglial inflammatory responses by phosphorylating ULK1. J Cell Biol. 2018;217:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Lin R, Feng J, Dong S, et al. Regulation of autophagy of prostate cancer cells by beta-catenin signaling. Cell Physiol Biochem. 2015;35:926–932. [DOI] [PubMed] [Google Scholar]
- [86].Tasdemir E, Maiuri MC, Galluzzi L, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Gwinn DM, Shackelford DB, Egan DF, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Jia J, Abudu YP, Claude-Taupin A, et al. Galectins control mTOR in response to endomembrane damage. Mol Cell. 2018;70:120–135.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Czarny P, Pawlowska E, Bialkowska-Warzecha J, et al. Autophagy in DNA damage response. Int J Mol Sci. 2015;16:2641–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Kim JH, Hong SK, Wu PK, et al. Raf/MEK/ERK can regulate cellular levels of LC3B and SQSTM1/p62 at expression levels. Exp Cell Res. 2014;327:340–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].McClung JM, Judge AR, Powers SK, et al. p38 MAPK links oxidative stress to autophagy-related gene expression in cachectic muscle wasting. Am J Physiol Cell Physiol. 2010;298:C542–C549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Crighton D, Wilkinson S, O’Prey J, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. [DOI] [PubMed] [Google Scholar]
- [93].Du WW, Yang W, Li X, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829–5842. [DOI] [PubMed] [Google Scholar]
- [94].Liang G, Ling Y, Mehrpour M, et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer. 2020;19:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Liu G, Zhang Z, Song Q, et al. Circ_0006528 contributes to paclitaxel resistance of breast cancer cells by regulating miR-1299/CDK8 axis. Onco Targets Ther. 2020;13:9497–9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Yang W, Gong P, Yang Y, et al. Circ-ABCB10 contributes to paclitaxel resistance in breast cancer through Let-7a-5p/DUSP7 axis. Cancer Manag Res. 2020;12:2327–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Chen M, Ai G, Zhou J, et al. circMTO1 promotes tumorigenesis and chemoresistance of cervical cancer via regulating miR-6893. Biomed Pharmacother. 2019;117:109064. [DOI] [PubMed] [Google Scholar]
- [98].Guo J, Chen M, Ai G, et al. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed Pharmacother. 2019;115:108957. [DOI] [PubMed] [Google Scholar]
- [99].Ma L, Wang Z, Xie M, et al. Silencing of circRACGAP1 sensitizes gastric cancer cells to apatinib via modulating autophagy by targeting miR-3657 and ATG7. Cell Death Dis. 2020;11:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Liu Z, Wang Q, Wang X, et al. Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov. 2020;6:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Yao W, Guo P, Mu Q, et al. Exosome-derived Circ-PVT1 contributes to cisplatin resistance by regulating autophagy, invasion, and apoptosis via miR-30a-5p/YAP1 axis in gastric cancer cells. Cancer Biother Radiopharm. 2020. DOI: 10.1089/cbr.2020.3578. [DOI] [PubMed] [Google Scholar]
- [102].Kong R. Circular RNA hsa_circ_0085131 is involved in cisplatin-resistance of non-small-cell lung cancer cells by regulating autophagy. Cell Biol Int. 2020;44:1945–1956. [DOI] [PubMed] [Google Scholar]
- [103].Liu F, Zhang J, Qin L, et al. Circular RNA EIF6 (Hsa_circ_0060060) sponges miR-144-3p to promote the cisplatin-resistance of human thyroid carcinoma cells by autophagy regulation. Aging (Albany NY). 2018;10:3806–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Yan L, Liu G, Cao H, et al. Hsa_circ_0035483 sponges hsa-miR-335 to promote the gemcitabine-resistance of human renal cancer cells by autophagy regulation. Biochem Biophys Res Commun. 2019;519:172–178. [DOI] [PubMed] [Google Scholar]
- [105].Cao HX, Miao CF, Sang LN, et al. Circ_0009910 promotes imatinib resistance through ULK1-induced autophagy by sponging miR-34a-5p in chronic myeloid leukemia. Life Sci. 2020;243:117255. [DOI] [PubMed] [Google Scholar]
- [106].Shang J, Chen WM, Liu S, et al. CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk Res. 2019;85:106198. [DOI] [PubMed] [Google Scholar]
- [107].Cai F, Li J, Zhang J, et al. Knockdown of Circ_CCNB2 sensitizes prostate cancer to radiation through repressing autophagy by the miR-30b-5p/KIF18A axis. Cancer Biother Radiopharm. 2020. DOI: 10.1089/cbr.2019.3538. [DOI] [PubMed] [Google Scholar]
- [108].Chen X, Mao R, Su W, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha signaling in STK11 mutant lung cancer. Autophagy. 2020;16:659-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Jiang Y, Zhang Y, Chu F, et al. Circ_0032821 acts as an oncogene in cell proliferation, metastasis and autophagy in human gastric cancer cells in vitro and in vivo through activating MEK1/ERK1/2 signaling pathway. Cancer Cell Int. 2020;20:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Zhu L, Wang C, Lin S, et al. CircKIAA0907 retards cell growth, cell cycle, and autophagy of gastric cancer in vitro and inhibits tumorigenesis in vivo via the miR-452-5p/KAT6B axis. Med Sci Monit. 2020;26:e924160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Peng L, Sang H, Wei S, et al. circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol Cancer. 2020;19:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Sun G, Li Z, He Z, et al. Circular RNA MCTP2 inhibits cisplatin resistance in gastric cancer by miR-99a-5p-mediated induction of MTMR3 expression. J Exp Clin Cancer Res. 2020;39:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Meng L, Liu S, Ding P, et al. Circular RNA ciRS-7 inhibits autophagy of ESCC cells by functioning as miR-1299 sponge to target EGFR signaling. J Cell Biochem. 2020;121:1039–1049. [DOI] [PubMed] [Google Scholar]
- [115].Chi G, Xu D, Zhang B, et al. Matrine induces apoptosis and autophagy of glioma cell line U251 by regulation of circRNA-104075/BCL-9. Chem Biol Interact. 2019;308:198–205. [DOI] [PubMed] [Google Scholar]
- [116].Xu Y, Yang Z, Yuan H, et al. PCDH10 inhibits cell proliferation of multiple myeloma via the negative regulation of the Wnt/beta-catenin/BCL-9 signaling pathway. Oncol Rep. 2015;34:747–754. [DOI] [PubMed] [Google Scholar]
- [117].Lin S, Zhuang J, Zhu L, et al. Matrine inhibits cell growth, migration, invasion and promotes autophagy in hepatocellular carcinoma by regulation of circ_0027345/miR-345-5p/HOXD3 axis. Cancer Cell Int. 2020;20:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Tang Q, Chen Z, Zhao L. Circular RNA hsa_circ_0000515 acts as a miR-326 sponge to promote cervical cancer progression through up-regulation of ELK1. Aging (Albany NY). 2019;11:9982–9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Zhang W, Zhang S. Downregulation of circRNA_0000285 suppresses cervical cancer development by regulating miR197-3p-ELK1 axis. Cancer Manag Res. 2020;12:8663–8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Liu J, Feng G, Li Z, et al. Knockdown of CircCRIM1 inhibits HDAC4 to impede osteosarcoma proliferation, migration, and invasion and facilitate autophagy by targeting miR-432-5p. Cancer Manag Res. 2020;12:10199–10210. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [121].Zhang J, Chou X, Zhuang M, et al. circKMT2D contributes to H2O2-attenuated osteosarcoma progression via the miR-210/autophagy pathway. Exp Ther Med. 2020;20:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Gan J, Yuan J, Liu Y, et al. Circular RNA_101237 mediates anoxia/reoxygenation injury by targeting let‑7a‑5p/IGF2BP3 in cardiomyocytes. Int J Mol Med. 2020;45:451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Lederer M, Bley N, Schleifer C, et al. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol. 2014;29:3–12. [DOI] [PubMed] [Google Scholar]
- [124].Panebianco F, Kelly LM, Liu P, et al. THADA fusion is a mechanism of IGF2BP3 activation and IGF1R signaling in thyroid cancer. Proc Natl Acad Sci U S A. 2017;114:2307–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Zhou J, Li L, Hu H, et al. Circ-HIPK2 accelerates cell apoptosis and autophagy in myocardial oxidative injury by sponging miR-485-5p and targeting ATG101. J Cardiovasc Pharmacol. 2020;76:427–436. [DOI] [PubMed] [Google Scholar]
- [126].Li F, Long TY, Bi SS, et al. circPAN3 exerts a profibrotic role via sponging miR-221 through FoxO3/ATG7-activated autophagy in a rat model of myocardial infarction. Life Sci. 2020;257:118015. [DOI] [PubMed] [Google Scholar]
- [127].Ren Q, Li H, Wang X. The circular RNA ZNF292 alleviates OGD-induced injury in H9c2 cells via targeting BNIP3. Cell Cycle. 2019;18:3365–3377. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [128].Zhou LY, Zhai M, Huang Y, et al. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/FAM65B pathway. Cell Death Differ. 2019;26:1299–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Zhang CL, Long TY, Bi SS, et al. CircPAN3 ameliorates myocardial ischaemia/reperfusion injury by targeting miR-421/Pink1 axis-mediated autophagy suppression. Lab Invest. 2020;101:89–103. [DOI] [PubMed] [Google Scholar]
- [130].Zhu X, Rottkamp CA, Hartzler A, et al. Activation of MKK6, an upstream activator of p38, in Alzheimer’s disease. J Neurochem. 2001;79:311-318. [DOI] [PubMed] [Google Scholar]
- [131].Tang C, Ou J, Kou L, et al. Circ_016719 plays a critical role in neuron cell apoptosis induced by I/R via targeting miR-29c/Map2k6. Mol Cell Probes. 2020;49:101478. [DOI] [PubMed] [Google Scholar]
- [132].Han B, Zhang Y, Zhang Y, et al. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. 2018;14:1164–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Xu L, Ji H, Jiang Y, et al. Exosomes derived from CircAkap7-modified adipose-derived mesenchymal stem cells protect against cerebral ischemic injury. Front Cell Dev Biol. 2020;8:569977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Li H, Lu C, Yao W, et al. Dexmedetomidine inhibits inflammatory response and autophagy through the circLrp1b/miR-27a-3p/Dram2 pathway in a rat model of traumatic brain injury. Aging (Albany NY). 2020;12:21687–21705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Feng Z, Zhang L, Wang S, et al. Circular RNA circDLGAP4 exerts neuroprotective effects via modulating miR-134-5p/CREB pathway in Parkinson’s disease. Biochem Biophys Res Commun. 2020;522:388–394. [DOI] [PubMed] [Google Scholar]
- [136].Huang R, Zhang Y, Han B, et al. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy. 2017;13:1722–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Cai W, Zhang Y, Su Z. ciRS-7 targeting miR-135a-5p promotes neuropathic pain in CCI rats via inflammation and autophagy. Gene. 2020;736:144386. [DOI] [PubMed] [Google Scholar]
- [138].Diling C, Yinrui G, Longkai Q, et al. Circular RNA NF1-419 enhances autophagy to ameliorate senile dementia by binding dynamin-1 and adaptor protein 2 B1 in AD-like mice. Aging (Albany NY). 2019;11:12002–12031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [139].Diling C, Longkai Q, Yinrui G, et al. CircNF1-419 improves the gut microbiome structure and function in AD-like mice. Aging (Albany NY). 2020;12(1):260–287. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [140].Chen X, Chen J, Jin S, et al. The circular RNA HIPK3 reduces hydrogen peroxide‐induced injury of PC‐12 cells by upregulation of miR‐455. J Cell Biochem. 2020. DOI: 10.1002/jcb.29660. [DOI] [PubMed] [Google Scholar]
- [141].Zhou ZB, Niu YL, Huang GX, et al. Silencing of circRNA.2837 plays a protective role in sciatic nerve injury by sponging the miR-34 family via regulating neuronal autophagy. Mol Ther Nucleic Acids. 2018;12:718–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Liu Y, Chen X, Yao J, et al. Circular RNA ACR relieves high glucose-aroused RSC96 cell apoptosis and autophagy via declining microRNA-145-3p. J Cell Biochem. 2019. DOI: 10.1002/jcb.29568. [DOI] [PubMed] [Google Scholar]
- [143].Yao L, Xu B, Li X. Neisseria gonorrhoeae-induced salpingitis is targeted by circular RNA EIF3K via miR-139-5p and regulating MAPK/NF-kappaB signaling pathway to promotes apoptosis and autophagy bacterial cells. Microb Pathog. 2020;142:104051. [DOI] [PubMed] [Google Scholar]
- [144].Zheng J, Zhu X, He Y, et al. CircCDK8 regulates osteogenic differentiation and apoptosis of PDLSCs by inducing ER stress/autophagy during hypoxia. Ann N Y Acad Sci. 2020;1485:56–70. [DOI] [PubMed] [Google Scholar]
- [145].Shi R, Jin Y, Hu W, et al. Exosomes derived from mmu_circ_0000250-modified adipose-derived mesenchymal stem cells promote wound healing in diabetic mice by inducing miR-128-3p/SIRT1-mediated autophagy. Am J Physiol Cell Physiol. 2020;318:C848–C856. [DOI] [PubMed] [Google Scholar]
- [146].Zhu M, Liu X, Li W, et al. Exosomes derived from mmu_circ_0000623-modified ADSCs prevent liver fibrosis via activating autophagy. Hum Exp Toxicol. 2020;39:1619–1627. [DOI] [PubMed] [Google Scholar]
- [147].Ji F, Zhu L, Pan J, et al. hsa_circ_0026827 promotes osteoblast differentiation of human dental pulp stem cells through the Beclin1 and RUNX1 signaling pathways by sponging miR-188-3p. Front Cell Dev Biol. 2020;8:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Hao R, Li F, Song X, et al. Caffeic acid phenethyl ester against cadmium induced toxicity mediated by CircRNA modulates autophagy in HepG2 cells. Ecotoxicol Environ Saf. 2020;197:110610. [DOI] [PubMed] [Google Scholar]
- [149].Chu H, Wang W, Luo W, et al. CircHECTD1 mediates pulmonary fibroblast activation via HECTD1. Ther Adv Chronic Dis. 2019;10:2040622319891558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Cheng Y, Luo W, Li Z, et al. CircRNA-012091/PPP1R13B-mediated lung fibrotic response in silicosis via endoplasmic reticulum stress and autophagy. Am J Respir Cell Mol Biol. 2019;61:380–391. [DOI] [PubMed] [Google Scholar]
- [151].Yu T, Ding Y, Zhang Y, et al. Circular RNA GATAD2A promotes H1N1 replication through inhibiting autophagy. Vet Microbiol. 2019;231:238–245. [DOI] [PubMed] [Google Scholar]
- [152].Li -X-X, Xiao L, Chung HK, et al. Interaction between HuR and circPABPN1 modulates autophagy in the intestinal epithelium by altering ATG16L1 translation. Mol Cell Biol. 2020;40:e00492-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


