ABSTRACT
Some cardiometabolic risk factors such as dyslipidemia and insulin resistance are known to be associated with low gut microbiota richness. A link between gut microbiota richness and the diversity of consumed dietary fibers (DF) has also been reported. We introduced a larger diversity of consumed DF by using a daily consumed bread in subjects at cardiometabolic risk and assessed the impacts on the composition and functions of gut microbiota as well as on cardiometabolic profile. Thirty-nine subjects at cardiometabolic risk were included in a double-blind, randomized, cross-over, twice 8-week study, and consumed daily 150 g of standard bread or enriched with a 7-dietary fiber mixture (5.55 g and 16.05 g of fibers, respectively). Before and after intervention, stool samples were collected for gut microbiota analysis from species determination down to gene-level abundance using shotgun metagenomics, and cardiometabolic profile was assessed. Multi-fiber bread consumption significantly decreased Bacteroides vulgatus, whereas it increased Parabacteroides distasonis, Fusicatenibacter saccharivorans, an unclassified Acutalibacteraceae and an unclassified Eisenbergiella (q < 0.1). The fraction of gut microbiota carrying the gene coding for five families/subfamilies of glycoside hydrolases (CAZymes) were also increased and negatively correlated with peaks and total/incremental area under curve (tAUC/iAUC) of postprandial glycemia and insulinemia. Compared to control bread, multi-fiber bread decreased total cholesterol (−0.42 mM; q < 0.01), LDL cholesterol (−0.36 mM; q < 0.01), insulin (−2.77 mIU/l; q < 0.05), and HOMA (−0.78; q < 0.05). In conclusion, increasing the diversity of DF in a daily consumed product modifies gut microbiota composition and function and could be a relevant nutritional tool to improve cardiometabolic profile.
KEYWORDS: Dietary fiber diversity, cardiometabolic profile, gut microbiota, Parabacteroides distasonis, Eisenbergiella sp., glycoside hydrolases
Introduction
Cardiometabolic diseases (CMD) are among the leading worldwide causes of death. Indeed, ischemic heart disease, stroke, and diabetes accounted for about 16.8 million deaths in 2019. Adiposity, dyslipidemia, and insulin resistance are known to increase the risk of developing CMD.1 Studies focusing on gut microbiota analysis have shown that such deleterious phenotypes are associated with low gut microbiota gene richness.2 Furthermore, studies comparing geographically distinct populations, both close and distant, have shown that an increased consumption of fruits and vegetables, thus higher quantity in dietary fiber intake, is associated with higher gut microbiota diversity.3–7 In overweight and obese subjects initially presenting a low gene count, a short-term energy-restricted diet including an increased quantity of dietary fibers, partly restored gut microbiota richness in parallel to improvement of the cardiometabolic profile.8 Taken together, this supports a key role of diet, particularly dietary fiber, in gut microbiota modulation and potentially in cardiometabolic risk factor management;9 especially since average dietary fiber intake is less than the recommended 25–30 g/day.10,11
Interestingly, a link between gut microbiota richness and consumed vegetable diversity has also been shown,12 suggesting that beyond quantity, the diversity of dietary fiber intake may also affect gut microbiota richness. Moreover, in a pig model, in the context of a high-fat diet, an improved control of lipid metabolism has been reported following supplementation with a mixture of three dietary fibers.13 Dietary fibers are mainly composed of resistant starch and non-starch polysaccharides (NSPs) from plant cell walls, particularly found in legumes, vegetables, and cereals. Other types of carbohydrates such as oligosaccharides, gums, and sugar-alcohols also reach the colon after escaping digestion in the small intestine. Fiber NSPs are complex polymers of cellulose, hemicellulose (xylan, xyloglucan, mannans, mixed-linkage glucans …) and pectins composed of a central sugar backbone, some branched with various side-chains by different glycosidic linkage patterns to form large, diverse molecules. All these polysaccharides are embedded in a complex matrix and represent various substrates for the gut microbiota, and particularly promote fibrolytic and glycolytic microbes that are genetically equipped with a panel of enzymes, substrate-binding proteins, and transporters to effectively hydrolyze these complex polymers.14 The human gut microbiota harbors 80 to 160 families of carbohydrate active enzymes (CAZymes) in healthy humans from different countries, compared to the 4 enzyme families of the human genome, which only break down starch, trehalose, and sugars in human milk.15,16 Thus, the more complex the structure of carbohydrates, the more the enzyme systems must be diversified to allow bacteria to completely break down complex fibers and convert sugar units into energy for its maintenance and multiple-host benefits.17 In turn, fiber breakdown delivers oligosaccharides that may feed the overall microbial community organized in a trophic network and promote a more global functional impact. The diversity and characteristics of fibers are thus thought to play a key role in this selective or global modulation of commensal bacteria and the host benefits.18
In this cross-over study, we thus aimed at increasing the quantity but also the diversity of dietary fiber intake using a daily consumed product such as sourdough bread and evaluated the induced effects on gut microbiota composition and function as well as on cardiometabolic risk profile. For this purpose, we defined a mixture of 7 dietary fibers of different molecular structures, with various physiochemical properties (both soluble and insoluble). We hypothesized that this particular mixture of dietary fibers would promote efficient fiber degradation by engaging a panel of enzymes with complementary modes of action. Due to the nature and metabolism of fibers, an impact on the cardiometabolic profile is also expected in parallel with the modification of the gut microbiota composition and functions.
Results
Participants characteristics
Forty-five subjects with cardiometabolic risk profile were included and randomized: 39 completed the cross-over study (17 men, 22 women) and were analyzed (Figure 1). Ninety-two percent were dyslipidemic (high triglycerides (TG) and/or low high-density lipoprotein cholesterol (HDL-C) and/or high low-density lipoprotein cholesterol (LDL-C) and/or high total cholesterol (TC). They had neither fasting hyperglycemia nor diabetes nor elevated hs-CRP (high-sensitivity C-Reactive Protein) nor hypertension (Table 1).
Figure 1.
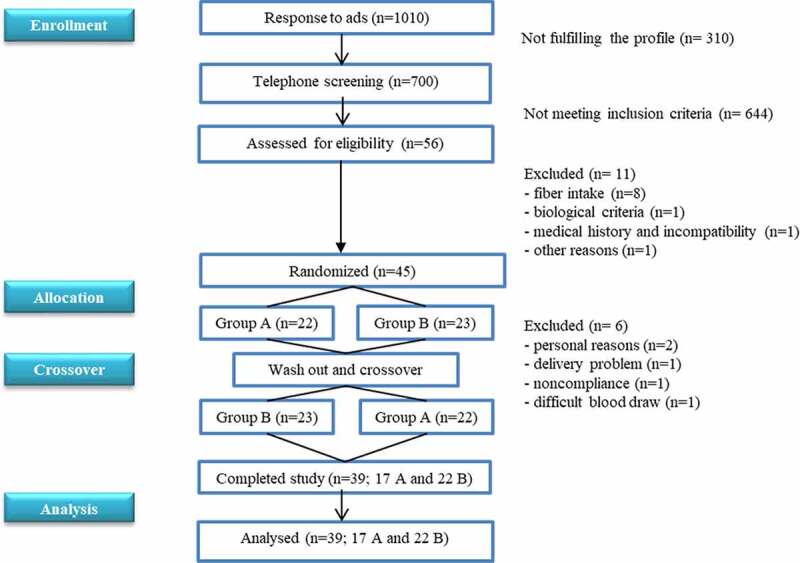
Consort flow diagram of participants.
Table 1.
Subjects’ anthropometric, metabolic characteristics and habitual dietary fiber intake at baseline (n = 39)
| |
female |
male |
||||
|---|---|---|---|---|---|---|
| n = 22 | n = 17 | |||||
| Age (years) | 43.0 | ± | 10.6 | 41.6 | ± | 12.9 |
| Weight (kg) | 75.7 | ± | 8.9 | 93.9 | ± | 10.8 |
| Height (m) | 1.6 | ± | 0.1 | 1.8 | ± | 0.1 |
| BMI (kg/m2) | 28.4 | ± | 2.6 | 29.0 | ± | 3.1 |
| Waist circumference (cm) | 97.1 | ± | 6.6 | 103.9 | ± | 7.9 |
| Hip circumference (cm) | 110.0 | ± | 6.0 | 108.4 | ± | 6.7 |
| Blood biomarkers | ||||||
| TG (mmol/L) | 1.2 | ± | 0.4 | 1.3 | ± | 0.6 |
| TC (mmol/L) | 5.8 | ± | 1.0 | 5.2 | ± | 0.7 |
| HDL-C (mM) | 1.4 | ± | 0.3 | 1.1 | ± | 0.2 |
| LDL-C (mM) | 3.9 | ± | 0.9 | 3.5 | ± | 0.6 |
| Glucose (mmol/L) | 4.7 | ± | 0.3 | 5.1 | ± | 0.5 |
| hs-CRP (mg/L) | 3.0 | ± | 3.1 | 3.7 | ± | 5.6 |
| HbA1c % | 5.2 | ± | 0.2 | 5.3 | ± | 0.4 |
| Blood pressure (mmHg) | ||||||
| Systolic pressure | 120.0 | ± | 13.2 | 125.1 | ± | 12.4 |
| Diastolic pressure |
71.9 |
± |
11.0 |
78.9 |
± |
9.2 |
| Habitual dietary fiber intake | ||||||
| Quantity (g/d) | 17.45 | ± | 1.6 | 16.3 | ± | 2.3 |
| Diversity (food group/d) | 3 | ± | 0.1 | 3 | ± | 0.1 |
Data are expressed as mean ± SD. BMI: body mass index; TG: triglyceride; TC: total cholesterol, HDL-C: high-density lipoprotein cholesterol, LDL-C: low-density lipoprotein cholesterol; hs-CRP: high sensitivity C reactive protein; HbA1c: glycated hemoglobin.
Participants dietary intake and compliance
The mean compliance to the dietary interventions (control (CTL) and multi-fiber (MF) bread consumption) was high (99%). Dietary records showed that subjects did not modify their energy and macronutrient intakes throughout the study. Likewise, the daily dietary fiber intake (without including consumed bread) in quantity and diversity remained low (<20 g/day, <3 fiber-rich food group/day) and was not modified throughout the study (Table S1). Consumed fiber-rich food items were mainly from starchy food, vegetables, and fruits (Figure S1).
Impact of dietary interventions on gut microbiota composition
Only the multi-fiber bread significantly modified gut microbiota composition
The two-month consumption of MF bread altered gut microbiota composition by modifying the relative abundance of specific gut bacterial species. Bacteroides vulgatus msp_0069 significantly decreased (cliff’s delta (CD), effect size: −0.27, small), whereas Parabacteroides distasonis msp_0012 (CD: 0.72, large), Fusicatenibacter saccharivorans msp_0154 (CD: 0.49, large), the unclassified Acutalibacteraceae msp_0291 (CD: 0.47, medium), and the unclassified Eisenbergiella msp_0125 (CD: 0.46, medium) significantly increased (q ≤ 0.1) (Figure 2). CTL bread did not significantly alter gut microbiota composition. Consistently, when assessing intra-individual changes, of species relative abundance, based on Bray-Curtis dissimilarity index before and after each bread consumption, we observed that compared to control bread, Bray-Curtis dissimilarity index of gut microbiota was larger with multi-fiber bread consumption 0.38 ± 0.08 vs 0.34 ± 0.10 (p = .02) (Figure S2).
Figure 2.
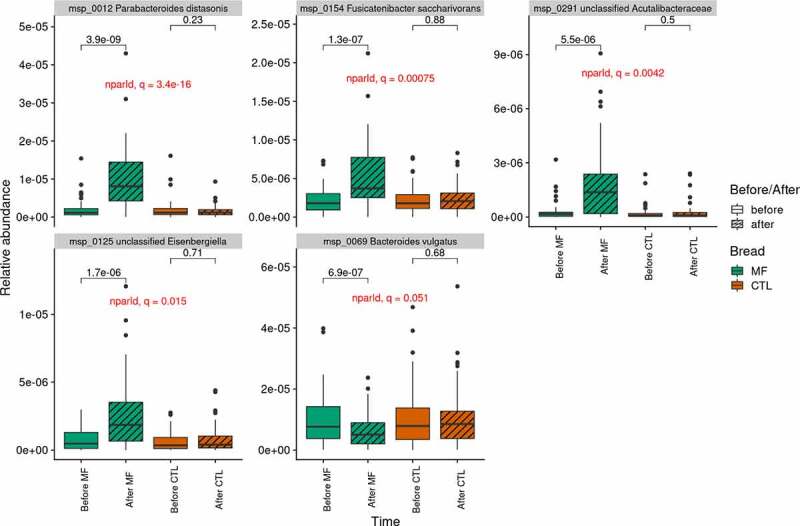
Abundance of the MSP significantly impacted by multi-fiber bread (n=39).
Increased MSP (panel 1-5); decreased MSP (panel 6). MSP: Metagenomic Species; MF: multi-fiber bread; CTL: control bread. P-values from Wilcoxon signed-rank test are displayed.
The washout period was appropriate. When assessing intra-individuals Bray-Curtis dissimilarity index between the different visits in the group that consumed MF bread and the group that consumed CTL bread during the period 1, we actually showed that there was no difference concerning the dissimilarity between V2 and V5 in the two groups (p = .75) although the dissimilarity between V2 – V4 was different (p = .06). Moreover, the species, which were increased or decreased in abundance with the multi-fiber bread showed no significant difference between V2 (baseline) and V5 (after wash-out) (Figure S3).
All along the dietary intervention, microbiota richness in terms of gene count and MSP (Metagenomic Species) count did not significantly differ (CD: 0.07 and 0.04 negligible respectively) (Figure 3).
Figure 3.
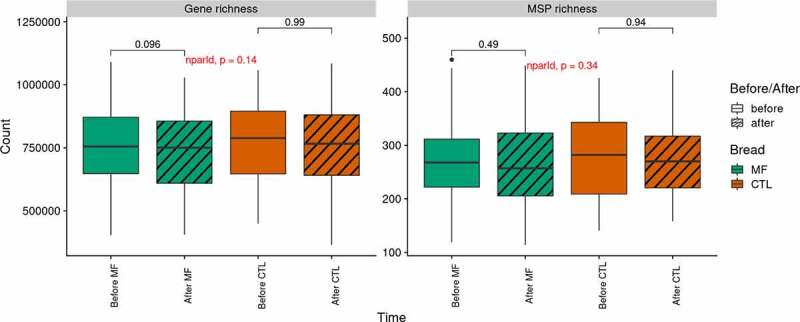
Gut microbiota richness throughout the interventions (n=39).
Gene and MSP richness (left and right panel, respectively) evolution per bread. MSP: Metagenomic Species; MF: multi-fiber bread; CTL: control bread. P-values from Wilcoxon signed-rank test are displayed.
Impact of dietary interventions on gut microbiota functions
Only the multi-fiber bread significantly modified gut microbiota functions
The bioconversion potential of dietary fibers according to the microbial CAZymes patterns was assessed as the fraction of gut microbiota carrying a given CAZyme, i.e. the sum of species (MSP) with at least one gene annotated with this CAZyme (see Methods). Out of the 200 examined CAZyme families, MF bread impacted the relative abundance of CAZymes from eight different families and sub-families, among which six were significantly increased and two significantly decreased (absolute CD ranging from 0.23, small to 0.58, large, q < 0.1), while remaining stable with control bread (Figure 4). The increased CAZymes belong to glycoside hydrolase (GH) families (GH30_2, GH43_8, GH43_28, GH43_33, and GH76) and glycosyl-transferase (GT) family (GT17), while the decreased CAZymes belong to polysaccharide lyase (PL) family (PL26) and GH13_29 subfamily. Among the increased putative GHs, two GH30_2 proteins shared 51% and 52% aminoacid (aa) identity with a characterized β-D-xylosidase from rumen bacterium (ADO20355), respectively. One putative GH43_8 protein has 37% aa identity with a β-D-galactofuranosidase from Bacteroides salyersiae (EIY66405) and a GH43_33 sequence shared 39% aa identity with an α-L-arabinofuranosidase from Halothermothrix orenii H168 (ACL70803). Two GH76 putative proteins presented 31% and 38% aa identity with an endo-α-1,6-mannanase from Bacillus circulans (BAA75632), respectively. No characterized protein sequence was yet available for similarity search to the GH43_28 subfamily. Consistently, all the five increased putative GH families were found in P. distasonis msp_0012, two of them were also found in F. saccharivorans msp_0154 (GH43_28) and in the unclassified Eisenbergiella msp_0215 (GH76), respectively. Furthermore, the protein sequences of GH30_2, GH43_8, GH43_28, GH43_33, and GH76 were identical to those of several strains of P. distasonis already annotated for CAZymes (http://www.cazy.org/bP.html). Homologous proteins were also found in numerous P. distasonis sequences available in the databases (not shown).
Figure 4.
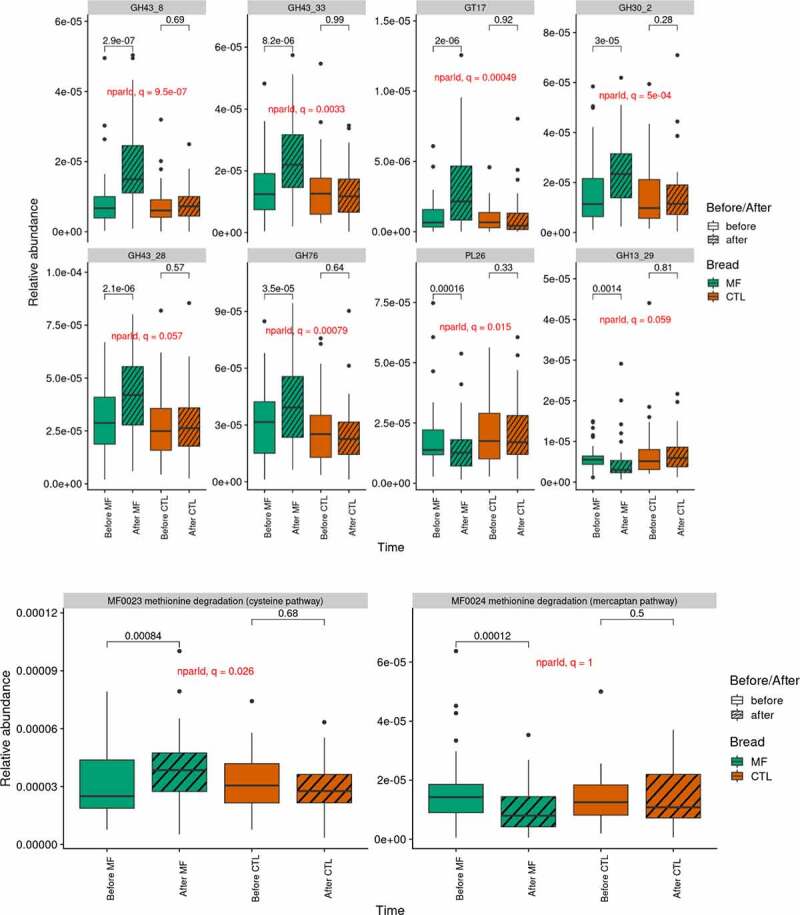
Gut microbiota function (n=39)
Gene abundance of CAZymes (A) and GMM (B) impacted by multi-fiber bread. GMM: gut metabolic modules; MF: multi-fiber bread; CTL: control bread. P-values from Wilcoxon signed-rank test are displayed. GH: glycoside hydrolase; GT: glycosyl transferase; PL: polysaccharide lyase.
Regarding gut-metabolic modules (GMM), only the cysteine pathway of methionine degradation (methionine = > L-homocysteine) changed significantly during the intervention and was increased with MF bread consumption (CD: 0.32, small; p = .026) (Figure 4). Interestingly, the mercaptan pathway of methionine degradation (methionine = > methanethiol) showed the opposite evolution with similar effect size, non-significantly after multiple test, though (p = .04, q = 1). Short-chain fatty acids-related GMM were not modulated during the intervention.
Impact of dietary interventions on metabolic parameters
The multi-fiber bread significantly improve lipid profile and insulin sensitivity
Compared to CTL bread, MF bread significantly decreased total cholesterol (TC: −0.42 mM), low-density lipoprotein cholesterol (LDL-C:-0.36 mM), insulin (−2.77 mUI/l) and homeostasis model assessment (HOMA: −0.78) (q < 0.05) (Figure 5). There was no significant impact of MF bread on anthropometric and inflammatory parameters as well as on the postprandial metabolism of glucose in response to a standardized test meal challenge (Table 2).
Figure 5.
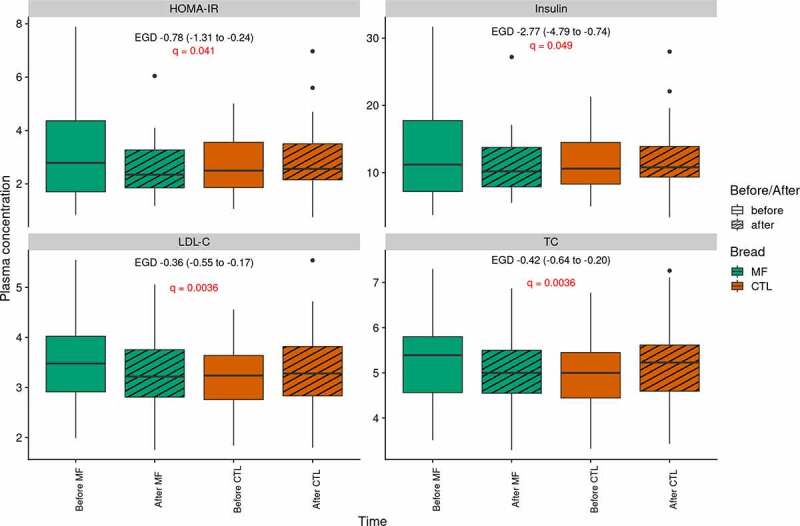
Effects of multi-fiber bread compared to control bread on lipid profile and insulin sensitivity (n=39).
Plasma concentrations of clinical variables impacted by multi-fiber bread. MF: multi-fiber bread; CTL: control bread. HOMA IR: homeostasic model assessment of insulin resistance; TC: total cholesterol; LDL-C: low-density lipoprotein cholesterol. EGD: estimated group difference. P-values from mixed linear model for repeated measures are displayed.
Table 2.
Effects of multi-fiber bread compared to control bread on metabolic parameters (n = 39)
| |
Fiber-enriched bread |
Control Bread |
Effect of multi-fiber bread |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | Post-Intervention | Baseline | Post-Intervention | EGD | 95% IC | |||||||||||
| Anthropometric parameters | ||||||||||||||||
| Weight (kg) | 84.65 | ± | 13.7 | 84.79 | ± | 13.35 | 84.59 | ± | 13.3 | 84.67 | ± | 13.43 | 0.06 | (−0.65 | to | 0.78) |
| BMI (kg/m2) | 29.02 | ± | 2.76 | 29.1 | ± | 2.70 | 29.03 | ± | 2.81 | 29.05 | ± | 2.72 | 0.06 | (−0.19 | to | 0.30) |
| Fat mass (kg) | 32.86 | ± | 8.11 | 32.7 | ± | 8.04 | 32.83 | ± | 8.19 | 32.83 | ± | 8.06 | 0.11 | (−0.48 | to | 0.69) |
| Waist circumference (cm) | 100.23 | ± | 8.37 | 100.28 | ± | 8.44 | 99.74 | ± | 8.24 | 100.17 | ± | 8.02 | −0.38 | (−1.64 | to | 0.87) |
| Height circumference (cm) | 109.91 | ± | 5.87 | 110.05 | ± | 5.99 | 109.79 | ± | 5.88 | 110.28 | ± | 6.21 | −0.35 | (−1.15 | to | 0.46) |
| Fasting metabolic parameters | ||||||||||||||||
| Glucose (mM) | 5.35 | ± | 0.49 | 5.22 | ± | 0.40 | 5.31 | ± | 0.48 | 5.29 | ± | 0.40 | −0.11 | (−0.26 | to | 0.04) |
| NEFA (µM) | 446.1 | ± | 159.15 | 438.28 | ± | 163.74 | 494.1 | ± | 161.99 | 484.87 | ± | 221.14 | 1.41 | (−90.6 | to | 93.4) |
| Insulin (mIU/l) | 13.41 | ± | 7.63 | 10.94 | ± | 4.37 | 11.34 | ± | 4.18 | 11.96 | ± | 4.89 | −2.77* | (−4.79 | to | −0.74) |
| HOMA | 3.23 | ± | 1.92 | 2.54 | ± | 1.01 | 2.69 | ± | 1.04 | 2.83 | ± | 1.23 | −0.78* | (−1.31 | to | −0.24) |
| TG (mM) | 1.32 | ± | 0.51 | 1.26 | ± | 0.43 | 1.29 | ± | 0.59 | 1.28 | ± | 0.49 | −0.05 | (−0.23 | to | 0.14) |
| Total cholesterol (mM) | 5.29 | ± | 0.92 | 5.03 | ± | 0.86 | 4.97 | ± | 0.78 | 5.13 | ± | 0.85 | −0.42** | (−0.64 | to | −0.20) |
| HDL cholesterol (mM) | 1.24 | ± | 0.27 | 1.19 | ± | 0.30 | 1.19 | ± | 0.26 | 1.20 | ± | 0.31 | −0.05 | (−0.11 | to | 0.00) |
| LDL cholesterol (mM) | 3.46 | ± | 0.80 | 3.27 | ± | 0.72 | 3.20 | ± | 0.66 | 3.36 | ± | 0.77 | −0.36** | (−0.55 | to | −0.17) |
| CRPus (mg/L) | 3.36 | ± | 6.47 | 3.41 | ± | 4.68 | 4.00 | ± | 5.69 | 2.93 | ± | 4.28 | 0.28 | (−0.22 | to | 0.79) |
| RMR (kcal) | 1490.27 | ± | 330.33 | 1699.97 | ± | 332.94 | 1717.04 | ± | 359.58 | 1700.98 | ± | 334 | 0.01 | (−0.02 | to | 0.05) |
| CD14 (µg/mL) | 1.53 | ± | 0.42 | 1.45 | ± | 0.42 | 1.57 | ± | 0.42 | 1.48 | ± | 0.32 | 0.01 | (−0.19 | to | 0.21) |
| LBP (µg/mL) | 16.06 | ± | 6.94 | 16.33 | ± | 5.80 | 16.20 | ± | 6.51 | 15.81 | ± | 7.49 | 0.67 | (−2.27 | to | 3.61) |
| Ratio LBP/CD14 | 10.81 | ± | 4.69 | 12.69 | ± | 8.75 | 10.55 | ± | 4.38 | 10.96 | ± | 5.3 | 1.67 | (−1.26 | to | 4.61) |
| Postprandial metabolic parameters | ||||||||||||||||
| Glucose tAUC (mM*min) | 1031.3 | ± | 93.8 | 1024.47 | ± | 73.46 | 1048.32 | ± | 96.24 | 1043.61 | ± | 122.98 | −2.12 | (−40.78 | to | 36.54) |
| Glucose iAUC (mM*min) | 147.16 | ± | 118.39 | 151.14 | ± | 103.46 | 197.89 | ± | 132.42 | 192.77 | ± | 151.53 | 9.10 | (−41.02 | to | 59.23) |
| Glucose peak (mM) | 7.26 | ± | 1.04 | 6.99 | ± | 0.76 | 7.25 | ± | 1.09 | 7.18 | ± | 1.05 | −0.2 | (−0.62 | to | 0.23) |
| Insulin tAUC (mUI/l*min) | 7676 | ± | 2951.96 | 7568.86 | ± | 3181.22 | 7707.86 | ± | 3098.25 | 7694.25 | ± | 3568.1 | 24.18 | (−1090 | to | 1139) |
| Insulin iAUC (mUI/l*min) | 5827.22 | ± | 2804.61 | 5837.54 | ± | 2941.38 | 5991.59 | ± | 2901.58 | 5996.27 | ± | 3252.8 | 112 | (−1085 | to | 1308) |
| Insulin peak (mIU/l) | 90.28 | ± | 51.08 | 87.39 | ± | 63.01 | 86.27 | ± | 41.12 | 88.26 | ± | 57.99 | −6.44 | (−28.88 | to | 15.99) |
Data are expressed as mean ± SD. Effects of multi-fiber bread were analyzed using linear mixed model for repeated measures with heterogeneous Toeplitz or autoregressive as covariance structure. MF: multi-fiber bread; CTL: control bread; BMI: body mass index; RMR: resting metabolic rate; HOMA IR: homeostasic model assessment of insulin resistance; TC: total cholesterol; HDL cholesterol: high-density lipoprotein cholesterol; LDL-C: low-density lipoprotein cholesterol, TG: triglyceride; NEFA: non-esterified fatty acid; hs-CRP: high sensitivity C-reactive protein. EGD: estimated group difference. Adjusted p value *< 0.05 ** <0.01.
Association between gut microbiota composition and function, and metabolic parameters
We correlated the delta (after – before multi-fiber bread consumption) of impacted metagenomics features with those of clinical variables related to cholesterol and glucose metabolisms (Figure 6). We showed that a group of features enriched after multi-fiber bread consumption, P. distasonis, an unclassified Eisenbergiella, methionine degradation, and a set of GHs (GH30_2, GH43_8, GH43_28, GH43_33, GH76) were negatively correlated with the peaks and total/incremental area under curve (tAUC/iAUC) of postprandial glycemia and insulin, in response to a standardized test meal challenge.
Figure 6.

Correlation between metagenomic features and clinical variables (n=39).
Heatmap of Spearman’s correlation coefficient between deltas (after – before multi-fiber bread) of metagenomics features impacted by MF bread (in rows) and deltas of clinical variables related to cholesterol and glucose metabolisms (in columns). The right side bar stands for the metagenomics features Cliff’s Delta (red: decreased with MF, green: increased with MF). Black dots stand for correlations that are significant after Benjamini-Hochberg correction (q ≤ 0.1). Empty circles stand for correlations that are significant regarding non-corrected p-values (p ≤ 0.05). P. distasonis and the modules/CAZymes it carries are denoted with ‘*’ at the end of their name. CD: Cliff’s Delta; GH: glycoside hydrolase; GT: glycosyl-transferase; PL: polysaccharide lyase.
Discussion
To our knowledge, this study investigated for the first time the impact of increasing the diversity of the dietary fiber intake, beyond the quantity, on the composition and the functions of the gut microbiota as well as on cardiometabolic profiles using a daily consumed product such as bread. We demonstrated that in subjects at cardiometabolic risk, such interventions alter gut microbiota composition and function by modifying the relative abundance of specific gut bacterial species, CAZymes and GMM, accompanied by a significant improvement of cholesterol and insulin sensitivity parameters.
In this study, we showed that in a population of subjects at cardiometabolic risk of whom 92% were dyslipidemic, 2 months of consumption of multi-fiber bread significantly decreased total cholesterol (−0.42 mM) and LDL cholesterol (−0.36 mM) compared to 2 months of consumption of standard bread. Epidemiological studies have shown an inverse association between dietary fiber intake and cholesterol levels.19 Dietary intervention trials (2 to 16 weeks), based on different fiber-enriched diets with a dose varying from 5 to 23 g/d and conducted in dyslipidemic subjects, consistently showed a significant mean decrease of total and LDL cholesterol between 0.1 and 0.3 mM and 0.1 to 0.2 mM, respectively.20–22 Such decrease of LDL cholesterol is important in terms of reducing cardiometabolic risks, as 1 mM LDL cholesterol reduction was shown to be associated with a 19% lower risk of coronary mortality23 and a much greater decrease of 52% CM risk in case of early intervention.24
Literature suggests that the cholesterol-lowering effect of dietary fiber is due to different factors. First, the transformation of dietary cholesterol to coprostanol by coprostanoligenic bacteria including Bacteroides sp. D8 closely related to B. vulgatus, based on its morphological and biochemical characteristics.25 However, we reported here a decrease of B. vulgatus not supporting a major impact of this mechanism on the cholesterol-lowering effect of the mixture of fibers. Then, fibers are also known to affect bile acids. In the colon, bacteria can modify bile acids by hydrolyzing them, resulting in cholesterol-lowering. Bacteria capable of such reactions are those with bile salt hydrolase activity including bacteria belonging to Bacteroides, Parabacteroides and Clostridium genera26,27 and particularly P.distasonis.28,29 Since P.distasonis was significantly increased after multi-fiber bread, this mechanism could have been involved in the cholesterol-lowering effect of the mixture of fibers.
Interestingly, multi-fiber bread consumption significantly improved insulin sensitivity parameters. Epidemiological studies have reported a positive relation between dietary fiber intake and insulin sensitivity.30,31 A clinical study conducted in overweight and at cardiometabolic risk subjects has reported an improvement of insulin sensitivity estimated by HOMA following diet incorporating resistant starch, with an average HOMA decrease of 10.4%.32 Other studies all involving at cardiometabolic risk subjects, supported that wholegrain, oat-based products, β-glucan extract, and refined-grain products had no impact on insulin sensitivity.24–26 Here, we demonstrated a much larger 21%-decrease of HOMA and thus insulin resistance.
In this study, we hypothesized that using the dietary fiber mixture instead of a single one would promote gut microbiota diversity and functions since it would stimulate the metabolism of a larger number of ecological niches and generate a larger variety of by-products usable by the entire microbial ecosystem. However, we did not detect any change in gut microbiota richness in our study. Previously, Cotillard et al. showed that in overweight and obese subjects, a short-term energy-restricted diet including an increased quantity of dietary fibers partly restored the gut microbiota richness, but only in the subgroup initially presenting a markedly lower gene count (LGC) (compared to a higher gene count subgroup (HGC). Due to differing sequencing technologies used in this study and that of Cotillard et al., it was not possible to robustly compare gut microbiota richness between the subjects from the two studies. However, it has been reported that the body mass index (BMI) negatively correlates with gut microbiota richness.33,34 As the subjects included in this study presented a lower BMI compared to those from Cotillard et al. (28.9 ± 2.8 vs 33.2 ± 0.6), their gut microbiota richness may be higher, which could explain why we did not impact this parameter. Consistently, when comparing our cohort to a nationality- and sequencing technology-matched overweight cohort34 (our cohort < BMI: 31.4 ± 3.0 < Cotillard et al. cohort), richness was 17% higher than for this cohort (data not shown). Baseline microbiota richness could therefore be an important inclusion criterion to consider for such studies. Although we managed to increase both the quantity and the diversity of dietary fiber intake of included subjects, modifying one component of the diet without energy restriction or change in macronutrient composition may not have been sufficient to significantly alter overall microbiota richness in this population. However, we observed a significant decrease in abundance of B. vulgatus and a significant increase of P. distasonis, F. saccharivorans, an unclassified Acutalibacteriaceae, and an unclassified Eisenbergiella after 2 months of multi-fiber bread consumption. Multi-fiber bread nutritional interventions thus more specifically targeted certain taxa. Previous studies have also evidenced an increase in the level of P. distasonis after a dietary intervention using either single fiber supplementation35,36 or mixtures.37
Most if not all, glycoside hydrolases families increased with multi-fiber bread consumption have been found in P. distasonis, emphasizing the role this species may play in fiber supplementation. Among them, GH43_8/28/33 and GH30_2 are thought to be debranching enzymes aiding in the degradation of NSPs such as arabinoxylan from hemicelluloses, and galactomannan found in gums and pectin, although very few information is available in the literature regarding these subfamilies (from zero to two characterized members). Furthermore, GH43 are among the most abundant CAZymes found in the human gut microbiome and probably access to a wide range of complex carbohydrates.38 All together, these enzymes families are consistent with the nature of plant cell wall polysaccharides or gums provided by the cereals of multi-fiber bread and locust bean gum, respectively. GH76 are putative α-1,6-endomannanases dedicated to the degradation of mannans from yeast glycoproteins. Both multi-fiber and control breads are sourdough bread, but the presence of various types of fibers in the multi-fiber bread during the fermentation process with yeasts appeared to enhance bacteria bearing enzymes that hydrolyze yeast cell wall mannans. By regularly consuming yeast-leavened bread and other fermented products, Bacteroidetes in the gut microbiota have evolved the capacity to metabolize these glycans, in particular Bacteroides and Parabacteroides.39 Multi-fiber bread could therefore have enhanced this metabolic function through the increase of P. distasonis. Concerning the decreased families, GH13_29 includes α-glucosidase and α-phosphotrehalases related to the hydrolysis of starch and trehalose. Enzymes encoding-genes from this subfamily are rather observed in the genome of Enterobacteriacea, and lactic acid bacteria40,41 but many other GH13 enzymes dedicated to the disruption of alpha glycoside linkages are found in P. distasonis according to the CAZy database, PL26 is thought to degrade rhamnogalacturonan from pectins but only one fungal enzyme from this family has been characterized so far,42 therefore other functional characteristics may exist. Nevertheless, numerous PL26 encoding genes have been found in Bacteroides species including B. vulgatus43 which is also significantly decreased with MF bread consumption. B. vulgatus has been reported to be associated with alteration of insulin resistance.44 Thus, its observed decrease could partly explain the improvement of insulin sensitivity. Moreover, we showed a significant increase of Parabacteroides distasonis whose involvement in type 2 bile acid production and farnesoid X receptor (FXR) pathway activation resulting in insulin sensitivity improvement has recently been shown.45 Moreover, the methionine degradation to L-homocysteine was the metagenomics features that displayed the highest number of significant correlations with the postprandial glycemia and insulinemia parameters. Both methionine and homocysteine have been related to insulin resistance.46–51 Hence, the modulation of methionine metabolism through the increase of P. distasonis could be a possible mechanism to explain the decrease of insulin observed in this study.
It is well known that studying the impact of dietary interventions on human microbiota has some limitations, such as a limited number of participants like in our study, and the fact that studies normally provide fecal samples reflecting the microbiota of the distal colon, but do not allow access to the microbiota of the actual site of food fermentation (cecum and proximal colon).52 Beyond the effects on gut microbiota functions mediated by the modulation of the microbiome, we would expect observing effects on the metabolome.
In conclusion, increasing the diversity of dietary fibers in a daily consumed product modified gut microbiota composition and promoted GH families involved in the degradation of plant polysaccharides and locust bean gum found in the seven selected dietary fibers. The parallel improvement of lipid and insulin sensitivity parameters suggests that such intervention could be a relevant nutritional approach to improve cardiometabolic profile and further prevent cardiometabolic risk.
Materials and methods
Study participants
Forty-five subjects with CMR profile were included in this study between November 2017 and July 2018. Inclusion criteria included age: 18–70 years old, body mass index (BMI): 25–35 kg/m2, waist circumference >80 cm for women and >96 cm for men, daily bread consumption <200 g/day, low dietary fiber intake <20 g/day, stable weight and moderate physical activity, no known gastrointestinal disease, no previous bariatric surgery, no use of antibiotics or other drugs interfering with microbiota composition in the 3 months prior and during the study.
Study design
This study was a single-center, randomized, double-blind crossover trial conducted at the Human Nutrition Research Center of Rhône-Alpes (CRNH-RA) and carried out in accordance with the Second Declaration of Helsinki and French Jardé’s law. It was reported and registered on http://www.clinicaltrials.gov (NCT03875898). All participants received and signed informed consent approved by the Scientific Ethics Committee of Bordeaux Sud-Ouest and Outre-Mer III.
After a 1-week run-in period (150 g/day of standard sourdough bread (control CTL)), subjects received two 8-weeks dietary interventions in a random order according to randomization. The two dietary interventions were: 1) 150 g/d of sourdough bread enriched with a mixture of 7 selected dietary fibers (multi-fiber; MF) and 2) 150 g/d of CTL. Group A started with MF, whereas group B started with CTL. A 4 to 6-weeks washout period and another run-in separated the two interventions (Figure S4). CTL and MF bread were structurally matched (color, size, and texture), although had different dietary fiber content (Table S2). 150 g of MF bread contained 16.05 g of 7 selected fibers with a 1:1 ratio of insoluble to soluble fiber.
Every week, baked and frozen breads were directly delivered to the subjects in individual sealed pack of 3 × 50 g and stored in their freezer until consumption. Compliance was checked by the number of returned empty packs.
During the intervention periods, subjects were asked to maintain the same diet in terms of caloric and macronutrients content and were ordered to fill in a 3-day dietary record. Data from this 3-day dietary record were processed using NUTRILOG© which allowed to estimate energy and macronutrient intake. In parallel, the subjects also completed a questionnaire concerning 128 fiber-rich food items to evaluate their dietary fiber intake in terms of diversity.12 A fiber-rich food group (breakfast and biscuits, starchy food, dried vegetables, vegetables, fruits, nuts, chocolate, and probiotics) is made of different items (for example, apple is an item belonging to fruits). For each subject, fiber-rich food group was taken into account when he/she ate at least one item/day of this group on 2 days out of three.
At the beginning and the end of each nutritional intervention period, metabolic assessment days were scheduled to evaluate CM risk factors (V2, V4, V5, and V7).
During the 3 days before metabolic assessment days, subjects were ordered to collect a stool sample at ambient temperature using a specific kit containing RNAlater® as stabilizing solution, following the International Human Microbiome Standards (IHMS, SOP 05) (http://www.human-microbiome.org/index.php?id=Sop&num=005) and send it to the French National Research Institute of Agriculture, food and Environment (INRAE) MetaGénoPolis (mgps.eu) for analysis. Intermediate visits with dietician were also scheduled to ensure a good compliance (V3 and V6).
On metabolic assessment days, subjects arrived at CRNH-RA after an 8 h overnight fast following the ingestion of a standard low dietary fiber evening meal (one serving of lean meat or fish, rice, a dairy product and fruit compote). Body weight, fat mass percentage, height, and waist circumference were measured using standardized methodologies with a calibrated weighing scale, a Bodystat Quadscan 4000 (BQ4000; Bodystat Ltd. Douglas, UK) stadiometer and non-elastic tape, respectively. BMI was calculated as weight/height2. The resting metabolic rate (RMR) was measured by indirect calorimetry using a QUARK calorimeter (Cosmed, Rome, Italy). Subjects were served a breakfast at T0 (the study product (multi-fiber or control bread), hot drink, jam, and butter) and a standardized challenge test meal at T240. Fasting and postprandial blood (T0, T15 T30 T45 T60 T90 T120 T180 T210 T240 T255 T270 T300 T330 T360 T390 T420) were collected using an antecubital vein catheter.
Biochemical blood analyses
Collected blood was centrifuged immediately for 10 min at 4,500 rpm. Plasma was stored at −20°C until the assays were conducted.
Glycemia was measured by spectrophotometry according to Architect Abbott Hexokinase method; C-reactive protein (CRP) by immunoturbidimetry; Non-esterified Fatty Acid (NEFA) by colorimetric technic; total cholesterol (TC), HDL-cholesterol (HDL-C) and triglyceride (TG) by spectrometry using Architect Module Chimie Abbott method; insulin by radio immunoassay (RIA) according to RIA CisBio IBA method, LBP and sCD14 by sandwich ELISA according to CliniSciences instructions. LDL-cholesterol was calculated using Friedwald formula and HOMA as plasma glucose (mmol/L) × plasma insulin (mUI/L)/22.5.
Microbiota analysis
DNA extraction of stool samples and shotgun sequencing
DNA extraction from aliquots of fecal samples was performed following the IHMS SOP 07 V2 (http://www.human-microbiome.org/index.php?id=Sop&num=005). DNA was quantitated using Qubit Fluorometric Quantitation (ThermoFisher Scientific, Waltham, US) and qualified using DNA size profiling on a Fragment Analyzer (Agilent Technologies, Santa Clara, US). Three µg of high molecular weight DNA (>10 kbp) was used to build the library. Shearing of DNA into fragments of approximately 150 bp was performed using an ultrasonicator (Covaris, Woburn, US) and DNA fragment library construction was performed using the Ion Plus Fragment Library and Ion Xpress Barcode Adapters Kits (ThermoFisher Scientific, Waltham, US). Purified and amplified DNA fragment libraries were sequenced using the Ion Proton Sequencer (ThermoFisher Scientific, Waltham, US), with a minimum of 20 million high-quality reads of 150 bp (in average) generated per library.
Microbial gene count table
To create the gene count table, the METEOR software was used: first, reads were filtered for low-quality by AlienTrimmer (https://forgemia.inra.fr/metagenopolis/meteor).8 Reads that aligned to the human genome (identity > 95%) were also discarded. Remaining reads were trimmed to 80 bases and mapped to the Integrated Gut Catalog 2 (IGC2),53,54 comprising 10.4 million of genes, using Bowtie2.55 First, the unique mapped reads (reads mapped to a unique gene in the catalog) were attributed to their corresponding genes. Second, the shared reads (reads that mapped with the same alignment score to multiple genes in the catalog) were attributed according to the ratio of their unique mapping counts of the captured genes. The resulting count table was further processed using the R package MetaOMineR v1.31.2 It was downsized to 14 million mapped reads to take into account differences in sequencing depth and in mapping rate across samples. Then the downsized matrix was normalized for gene length and transformed into a frequency matrix (fragments per kilobase of exon model per million reads mapped) (FPKM) normalization). Gene count was computed as the number of genes detected (i.e., whose abundance is strictly positive) in a given sample after downsizing.
Metagenomic Species (MSP) profiles
The IGC2 catalog was organized into 1990 Metagenomic Species (MSP), clusters of minimum 100 genes, using MSPminer54 MSP taxonomy was assigned with the Genome Taxonomy Database.56 Relative abundance of an MSP was computed as the mean abundance of its 100 ‘marker’ genes (that is, the genes that correlate the most altogether). If less than 10% of ‘marker’ genes were seen in a sample, the abundance of the MSP was set to 0. Relative abundances at higher taxonomical ranks were computed as the sum of the MSP that belong to a given taxa. MSP count was assessed as the number of MSP present in a sample (that is, whose abundance is strictly positive).
Microbiome functional potential
Genes from the IGC2 catalog were mapped with diamond57 onto KEGG orthologs (KO) from the KEGG database (version 8.9).58 Each gene was assigned to the best-ranked KO among hits with e-value <10–5 and a bit score >60. For a given sample, we assessed the abundance of a KEGG module or a Gut-Metabolic Modules (GMMs)59 with the following procedure:
We restricted the gene content of each MSP detected in the sample to its genes also detected in the sample, plus the core genes of the MSP;
We computed the fraction of the module present in the restricted set of genes for the pair sample/MSP;
We considered that an MSP carried a module in a given sample if the computed fraction was above 90%;
We summed the abundance of all MSPs that carried the module for a given sample.
The IGC2 was also annotated on the Carbohydrate-Active enZYmes Database (CAZy, November 2018) (http://www.cazy.org/60).54 The abundance of each CAZYme group (glycoside hydrolases (GH), glycosyl-transferases (GT), polysaccharide lyases (PL), and carbohydrate esterases (CE)) was computed using the same process as KEGG modules and GMM.
The abundance of each CAZyme group (glycoside hydrolases (GH), glycosyl-transferases (GT), polysaccharide lyases (PL), and carbohydrate esterases (CE)) was computed using the same process as KEGG modules and GMM.
Statistics
The primary outcome of this study was an increase of 20% of gut microbiota richness which corresponds to 76 000 genes according to the distribution of gut microbiota richness of two cohorts of obese and overweight subjects.2,8 An increase of 76 000 genes allowed the individuals to move from Low Gene Count (LGC) status to High Gene Count (HGC) status. LCG and HGC individuals significantly differed on metabolic profile, HGC individuals presenting a better metabolic profile. Assessing the impacts on cardiometabolic profile was actually the secondary outcome of this study.
According to these data, to achieve a significance of p < .05 and a power (type II error) of 95%, a size of 40 subjects was required.
All statistical analyses and graphs were performed with R software v3.6.0 and SAS software 9.4 TS Level 1M6.61
To assess if a given metagenomic feature was impacted by the multi-fiber bread, we used the R package nparLD62 to perform non-parametric analysis of the longitudinal data. P-values from the interaction of the factors “bread” and “visit” were corrected for multiple test with the Benjamini–Hochberg procedure. A feature was considered significant when corrected p-values ≤0.1. Finally, Wilcoxon signed-rank test were performed on each bread separately (i.e., before/after multi-fiber bread, and before/after control bread) to assess which bread impacted the feature. Effect size was computed using the Cliff’s Delta with the R package effsize.63 This measure gives an information similar to log-fold change, but is comprised between −1 and 1. (0: no effect; +1 or −1: large effect). The magnitude of the effect size d is assessed as negligible if |d| <0.147, small if |d| <0.33, medium if |d| <0.474, and large otherwise.64
For clinical variables, the effect of multi-fiber bread was evaluated by calculating the estimated group difference, i.e., the difference between the changes induced by each of the two breads. A linear mixed model for repeated measures, with Toeplitz or autoregressive structure (AR) as covariance structure, was used to determine whether the difference between the changes induced by each of the two breads was statistically significant. Bread, time, period, and sequence were included as fixed variables. In order to account for variability between subjects and to adjust for any nonspecific differences, subjects were included as random effects. When the normality of the model residuals was not assumed, a logarithmic transformation of the data has been done.
Correlations between variables were performed using Spearman’s correlations. All p-values were adjusted for multiple testing with the Benjamini Hochberg Procedure. Unless stated otherwise, a corrected p-value is considered significant if inferior to 0.1; a non-adjusted p-value is significant if inferior to 0.05.
Funding Statement
This study was funded by Bridor and in part by Metagenopolis grant Agence Nationale de la Recherche ANR-11-DPBS-0001. JD acknowledges funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Grant agreement ERC-2017-AdG No. 788191 - Homo.symbiosus). We thank all the subjects who participated in this study. We thank Bernard Henrissat who annotated the IGC2 with CAZy.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The raw sequencing data are deposited into the European Nucleotide Archive (ENA) of EBI (https://www.ebi.ac.uk/ena/browser/home) under PRJEB48663. The study protocol and the datasets generated during and/or analyzed during the current study, including deidentified participant data will be available from the corresponding author on reasonable request.
References
- 1.Risk factors [Internet] . World Heart Federation. Accessed 19 February 2021. https://www.world-heart-federation.org/resources/risk-factors/
- 2.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto J-M, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013. Aug 29;500(7464):541–16. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 3.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, and Lionetti P.. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010. Aug 17;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012. May 9;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014. Apr;15(5):3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcón Ó, et al. The microbiome of uncontacted Amerindians. Sci Adv. 2015. Apr 3;1(3). doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong LC, Holmes BA, Cotillard A, Habi-Rachedi F, Brazeilles R, Gougis S, Gausserès N, Cani PD, Fellahi S, and Bastard JP. Dietary patterns differently associate with inflammation and gut microbiota in overweight and obese subjects. PloS One. 2014;9(10):e109434. doi: 10.1371/journal.pone.0109434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013. Aug 29;500(7464):585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 9.Makki K, Deehan EC, Walter J, Bäckhed F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe. 2018; Jun 13;23(6):705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 10.EFSA . Scientific Opinion on Dietary Reference Values for carbohydrates and dietary fibre. EFSA J. 2010;8(3):1462. [Google Scholar]
- 11.Actualisation des repères du PNNS: révision des repères de consommations alimentaires [Internet] . Accessed 22 June 2021. https://www.anses.fr/fr/system/files/NUT2012SA0103Ra-1.pdf
- 12.Tap J, Furet J-P, Bensaada M, Philippe C, Roth H, Rabot S, Lakhdari O, Lombard V, Henrissat B, Corthier G, et al. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ Microbiol. 2015. Dec;17(12):4954–4964. doi: 10.1111/1462-2920.13006. [DOI] [PubMed] [Google Scholar]
- 13.Mohamed AB, Rémond D, Chambon C, Sayd T, Hébraud M, Capel F, Cohade B, Hafnaoui N, Béchet D, and Coudy-Gandilhon C. A mix of dietary fermentable fibers improves lipids handling by the liver of overfed minipigs. J Nutr Biochem. 2018Dec, 8(65), 72–82. [DOI] [PubMed] [Google Scholar]
- 14.Mirande C, Kadlecikova E, Matulova M, Capek P, Bernalier-Donadille A, Forano E, Béra‐Maillet C. Dietary fibre degradation and fermentation by two xylanolytic bacteria Bacteroides xylanisolvens XB1A T and Roseburia intestinalis XB6B4 from the human intestine. J Appl Microbiol. 2010. Aug;109(2):451–460. doi: 10.1111/j.1365-2672.2010.04671.x. [DOI] [PubMed] [Google Scholar]
- 15.El Kaoutari A, Armougom F, Leroy Q, Vialettes B, Million M, Raoult D, Henrissat B. Development and validation of a microarray for the investigation of the CAZymes encoded by the human gut microbiome. PloS One. 2013;8(12):e84033. doi: 10.1371/journal.pone.0084033. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Kaur K, Khatri I, Akhtar A, Subramanian S, Ramya TNC. Metagenomics analysis reveals features unique to Indian distal gut microbiota. PloS One. 2020;15(4):e0231197. doi: 10.1371/journal.pone.0231197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012; Jul 14;3(4):289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantu-Jungles TM, Hamaker BR. New View on Dietary Fiber Selection for Predictable Shifts in Gut Microbiota. mBio. 2020;11(1):e02179–19. doi: 10.1128/mBio.02179-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu K, Bowman R, Welch AA, Luben RN, Wareham N, Khaw K-T, Bingham SA. Apolipoprotein E polymorphisms, dietary fat and fibre, and serum lipids: the EPIC Norfolk study. Eur Heart J. 2007. Dec;28(23):2930–2936. doi: 10.1093/eurheartj/ehm482. [DOI] [PubMed] [Google Scholar]
- 20.Plb H, Ross AB, Kristensen M. Whole-grain and blood lipid changes in apparently healthy adults: a systematic review and meta-analysis of randomized controlled studies. Am J Clin Nutr. 2015. Sep;102(3):556–572. [DOI] [PubMed] [Google Scholar]
- 21.Zhu X, Sun X, Wang M, Zhang C, Cao Y, Mo G, Liang J, Zhu S. Quantitative assessment of the effects of beta-glucan consumption on serum lipid profile and glucose level in hypercholesterolemic subjects. Nutr Metab Cardiovasc Dis. 2015; Aug 1;25(8):714–723. doi: 10.1016/j.numecd.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Bazzano LA, Thompson AM, Tees MT, Nguyen CH, Winham DM. Non-Soy Legume Consumption Lowers Cholesterol Levels: a Meta-Analysis of Randomized Controlled Trials. Nutr Metab Cardiovasc Dis NMCD. 2011. Feb;21(2):94–103. doi: 10.1016/j.numecd.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baigent C, Keech A, Kearney PM, Blackwell L, Buck G, Pollicino C, Kirby A, Sourjina T, and Peto R. Efficacy and Safety of Cholesterol-Lowering Treatment: prospective Meta-Analysis of Data from 90,056 Participants in 14 Randomised Trials of Statins. Lancet Lond Engl. 2005. Oct 8;366(9493):1267–1278. [DOI] [PubMed] [Google Scholar]
- 24.Ference BA. Causal Effect of Lipids and Lipoproteins on Atherosclerosis: lessons from Genomic Studies. Cardiol Clin. 2018; May 1;36(2):203–211. doi: 10.1016/j.ccl.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Gerard P, Lepercq P, Leclerc M, Gavini F, Raibaud P, Juste C. Bacteroides sp. Strain D8, the First Cholesterol-Reducing Bacterium Isolated from Human Feces. Appl Environ Microbiol 2007. Oct;1(73):5742–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chand D, Avinash VS, Yadav Y, Pundle AV, Suresh CG, Ramasamy S. Molecular features of bile salt hydrolases and relevance in human health. Biochim Biophys Acta BBA - Gen Subj. 2017; Jan 1;1861(1, Part A):2981–2991. doi: 10.1016/j.bbagen.2016.09.024. [DOI] [PubMed] [Google Scholar]
- 27.Song Z, Cai Y, Lao X, Wang X, Lin X, Cui Y, Kalavagunta PK, Liao J, Jin L, Shang J, et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome. 2019. Jan 23;7(1):9. doi: 10.1186/s40168-019-0628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao L, Seaton SC, Ndousse-Fetter S, Adhikari AA, DiBenedetto N, Mina AI, Banks AS, Bry L, Devlin AS. A selective gut bacterial bile salt hydrolase alters host metabolism. eLife. 2018. Jul;17(7):e37182. doi: 10.7554/eLife.37182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ridlon JM, Devendran S, Alves JM, Doden H, Wolf PG, Pereira GV, Ly L, Volland A, Takei H, Nittono H, et al. The ‘ in vivo lifestyle’ of bile acid 7α-dehydroxylating bacteria: comparative genomics, metatranscriptomic, and bile acid metabolomics analysis of a defined microbial community in gnotobiotic mice. Gut Microbes. 2020. May 3;11(3):381–404. doi: 10.1080/19490976.2019.1618173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liese AD, Roach AK, Sparks KC, Marquart L, D’Agostino Jr RB, Mayer-Davis EJ. D’Agostino Jr RB, Mayer-Davis EJ. Whole-grain intake and insulin sensitivity: the Insulin Resistance Atherosclerosis Study. Am J Clin Nutr. 2003; Nov 1;78(5):965–971. doi: 10.1093/ajcn/78.5.965. [DOI] [PubMed] [Google Scholar]
- 31.Breneman CB, Tucker L. Dietary fibre consumption and insulin resistance – the role of body fat and physical activity. Br J Nutr. 2013. Jul;110(2):375–383. doi: 10.1017/S0007114512004953. [DOI] [PubMed] [Google Scholar]
- 32.Robertson MD, Wright JW, Loizon E, Debard C, Vidal H, Shojaee-Moradie F, Russell-Jones D, Umpleby AM. Insulin-Sensitizing Effects on Muscle and Adipose Tissue after Dietary Fiber Intake in Men and Women with Metabolic Syndrome. J Clin Endocrinol Metab. 2012; Sep 1;97(9):3326–3332. doi: 10.1210/jc.2012-1513. [DOI] [PubMed] [Google Scholar]
- 33.Aron-Wisnewsky J, Prifti E, Belda E, Ichou F, Kayser BD, Dao MC, Verger EO, Hedjazi L, Bouillot J-L, Chevallier J-M, et al. Major microbiota dysbiosis in severe obesity: fate after bariatric surgery. Gut. 2019. Jan 1;68(1):70–82. doi: 10.1136/gutjnl-2018-316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vieira-Silva S, Falony G, Belda E, Nielsen T, Aron-Wisnewsky J, Chakaroun R, Forslund SK, Assmann K, Valles-Colomer M, Nguyen TTD, et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature. 2020. May;581(7808):310–315. doi: 10.1038/s41586-020-2269-x. [DOI] [PubMed] [Google Scholar]
- 35.Kiewiet MBG, Elderman ME, Aidy SE, Burgerhof JGM, Visser H, Vaughan EE, Faas MM, and De Vos P. Flexibility of Gut Microbiota in Ageing Individuals during Dietary Fiber Long-Chain Inulin Intake. Mol Nutr Food Res. 2021;65(4):2000390. doi: 10.1002/mnfr.202000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deehan EC, Yang C, Perez-Muñoz ME, Nguyen NK, Cheng CC, Triador L, Zhang Z, Bakal JA, Walter J. Precision Microbiome Modulation with Discrete Dietary Fiber Structures Directs Short-Chain Fatty Acid Production. Cell Host Microbe. 2020; Mar 11;27(3):389–404.e6. doi: 10.1016/j.chom.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Feng G, Mikkelsen D, Hoedt EC, Williams BA, Flanagan BM, Morrison M, Gidley MJ. In vitro fermentation outcomes of arabinoxylan and galactoxyloglucan depend on fecal inoculum more than substrate chemistry. Food Funct. 2020; Sep 23;11(9):7892–7904. doi: 10.1039/D0FO01103G. [DOI] [PubMed] [Google Scholar]
- 38.Mewis K, Lenfant N, Lombard V, Henrissat B, Nojiri H. Dividing the Large Glycoside Hydrolase Family 43 into Subfamilies: a Motivation for Detailed Enzyme Characterization. Appl Environ Microbiol. 2016; Jan 4;82(6):1686–1692. doi: 10.1128/AEM.03453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015. Jan;517(7533):165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang J, Yang B, Ross RP, Stanton C, Zhao J, Zhang H, Chen W. Comparative Genomics of Pediococcus pentosaceus Isolated From Different Niches Reveals Genetic Diversity in Carbohydrate Metabolism and Immune System. Front Microbiol. 2020;11:253. doi: 10.3389/fmicb.2020.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Li N, Zhao S, Zhang C, Qiao N, Duan H, Xiao Y, Yan B, Zhao J, Tian F, et al. Integrated Phenotypic–Genotypic Analysis of Latilactobacillus sakei from Different Niches. Foods. 2021. Aug;10(8):1717. doi: 10.3390/foods10081717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iwai M, Kawakami T, Ikemoto T, Fujiwara D, Takenaka S, Nakazawa M, Ueda M, Sakamoto T. Molecular characterization of a Penicillium chrysogenum exo-rhamnogalacturonan lyase that is structurally distinct from other polysaccharide lyase family proteins. Appl Microbiol Biotechnol. 2015. Oct;99(20):8515–8525. doi: 10.1007/s00253-015-6600-7. [DOI] [PubMed] [Google Scholar]
- 43.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007. Jul;5(7):e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BAH, Forslund K, Hildebrand F, Prifti E, Falony G, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016. 21;535(7612):376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 45.Wang K, Liao M, Zhou N, Bao L, Ma K, Zheng Z, Wang Y, Liu C, Wang W, and Wang J. Parabacteroides distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019. Jan 2;26(1):222–235.e5. doi: 10.1016/j.celrep.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 46.Lees EK, Król E, Grant L, Shearer K, Wyse C, Moncur E, Bykowska AS, Mody N, Gettys TW, Delibegovic M, et al. Methionine restriction restores a younger metabolic phenotype in adult mice with alterations in fibroblast growth factor 21. Aging Cell. 2014. Oct;13(5):817–827. doi: 10.1111/acel.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orgeron ML, Stone KP, Wanders D, Cortez CC, Van NT, Gettys TW. The Impact of Dietary Methionine Restriction on Biomarkers of Metabolic Health. Prog Mol Biol Transl Sci. 2014;121:351–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang N, Yao Z, Miao L, Liu J, Gao X, Fan H, Hu Y, Zhang H, Xu Y, Qu A, et al. Novel Clinical Evidence of an Association between Homocysteine and Insulin Resistance in Patients with Hypothyroidism or Subclinical Hypothyroidism. PloS One. 2015;10(5):e0125922. doi: 10.1371/journal.pone.0125922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ying Z, Zhang H, Su W, Zhou L, Wang F, Li Y, Zhang L, and Wang T. Dietary Methionine Restriction Alleviates Hyperglycemia in Pigs with Intrauterine Growth Restriction by Enhancing Hepatic Protein Kinase B Signaling and Glycogen Synthesis. J Nutr. 2017Oct, 147(10), 1892–1899. [DOI] [PubMed] [Google Scholar]
- 50.Ebrahimpour A, Vaghari-Tabari M, Qujeq D, Moein S, Moazezi Z. Direct correlation between serum homocysteine level and insulin resistance index in patients with subclinical hypothyroidism: does subclinical hypothyroidism increase the risk of diabetes and cardio vascular disease together? Diabetes Metab Syndr. 2018. Nov;12(6):863–867. doi: 10.1016/j.dsx.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Jiang X-C, Liang Z-D, Chen D-L, Jia J-P, Hu J-R HL. Correlation of Homocysteine, AHSG, CRP with Insulin Resistance, 25-(OH)2-VitD, Blood Lipids in Gestational Diabetes Patients. Clin Lab. 2021. Feb 1;67:2. doi: 10.7754/Clin.Lab.2020.200609. [DOI] [PubMed] [Google Scholar]
- 52.Graf D, Di Cagno R, Fåk F, Flint HJ, Nyman M, Saarela M, and Watzl B. Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis. Internet]. 2015 Feb 4cited 2019. Apr 26];26. Available from. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4318938/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen C, Zheng Z, Shao T, Liu L, Xie Z, Le Chatelier E, He Z, Zhong W, Fan Y, Zhang L, et al. Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis. Genome Biol. 2017. Jul 27;18(1):142. doi: 10.1186/s13059-017-1271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plaza Onate F, Pons N, Gauthier F, Almeida M, Ehrlich SD, Le Chatelier E. Updated Metagenomic Species Pan-genomes (MSPs) of the human gastrointestinal microbiota [Internet]. Portail Data INRAE; 2021. [cited 2021 Jun 22]. Available from: https://data.inrae.fr/dataset.xhtml?persistentId=doi:10.15454/FLANUP
- 55.B L SS, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012; Mar 4;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018. Nov;36(10):996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 57.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015. Jan;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 58.Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; Jan 1;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vieira-Silva S, Falony G, Darzi Y, Lima-Mendez G, Garcia Yunta R, Okuda S, Vandeputte D, Valles-Colomer M, Hildebrand F, and Chaffron S. Species-function relationships shape ecological properties of the human gut microbiome. Nat Microbiol. 2016. Jun 13;1(8):16088. doi: 10.1038/nmicrobiol.2016.88. [DOI] [PubMed] [Google Scholar]
- 60.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014. Jan;42(Database issue):D490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R: a language and environment for statistical computing [Internet]. Accessed 26 March 2021. https://www.gbif.org/fr/tool/81287/r-a-language-and-environment-for-statistical-computing
- 62.Noguchi K, Gel YR, Brunner E, Konietschke F. nparLD: an R Software Package for the Nonparametric Analysis of Longitudinal Data in Factorial Experiments. J Stat Softw. 2012; Sep 18;50(1):1–23. doi: 10.18637/jss.v050.i12.25317082 [DOI] [Google Scholar]
- 63.Torchiano M Effsize: Efficient Effect Size Computation. 2014.
- 64.Appropriate statistics for ordinal level data: Should we really be using t-test and Cohen’sd for evaluating group differences on the NSSE and other surveys? BibSonomy [Internet]. Accessed 22 June 2021. https://www.bibsonomy.org/bibtex/216a5c27e770147e5796719fc6b68547d/kweiand
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw sequencing data are deposited into the European Nucleotide Archive (ENA) of EBI (https://www.ebi.ac.uk/ena/browser/home) under PRJEB48663. The study protocol and the datasets generated during and/or analyzed during the current study, including deidentified participant data will be available from the corresponding author on reasonable request.


