Abstract
High-resolution melting (HRM) analysis was performed to detect G339D and D796Y variations in the SARS-CoV-2 Omicron variant spike protein. We employed two-step PCR consisting of the first RT–PCR and the second nested PCR to prepare the amplicon for HRM analysis. The melting temperatures (Tm) of the amplicon from the cDNA of the Omicron variant receptor binding domain (RBD) were 73.1 °C (G339D variation) and 75.1 °C (D796Y variation), respectively. These Tm values were clearly distinct from those of SARS-CoV-2 isolate Wuhan-Hu-1. HRM analysis after the two-step PCR was conducted on Omicron variant-positive specimens. The HRM curve and Tm value obtained with the Omicron variant-positive specimen were coincident with those of the amplicon from cDNA of the Omicron variant RBD. Our study demonstrates the utility of HRM analysis after two-step PCR for the detection of mutations in SARS-CoV-2 gene.
Keywords: SARS-CoV-2, Omicron, RBD, HRM analysis, Nested PCR
Abbreviations: severe acute respiratory syndrome coronavirus 2, SARS-CoV-2; high-resolution melting, HRM; melting temperature, Tm; receptor binding domain, RBD; nucleotide, nt; reverse transcriptase, RT; wild type, wt
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in December 2019 and rapidly spread globally, causing a respiratory disease named coronavirus disease 2019 (COVID-19) and leading to a pandemic [1,2]. Mutations in the wild type SARS-CoV-2 (Isolate Wuhan-Hu-1) genome have occurred frequently since the first outbreak and have increased rapidly, such as Lineage B.1.1.7 (Alpha variant) and Lineage B.1.617.2 (Delta variant). Omicron (B.1.1.529), a variant of SARS-CoV-2, has recently been reported worldwide. World Health Organization has categorized Omicron as a variant due to its higher transmissibility and infectivity. Thereafter, the basal sublineage of the Omicron (B.1.1.529) is BA.1 (B.1.1.529.1), and the B.1.1.529 lineage was subsequently subclassified into BA.1 (B.1.1.529.1), BA.2 (B.1.1.529.2), and BA.3 (B.1.1.529.3) [3].
Post-PCR high-resolution melting (HRM) analysis is a highly sensitive molecular biology method based on the melting temperature (Tm) difference between purine and pyrimidine bases that can be used for the detection of mutations in double-stranded DNA samples [4]. Several HRM analyses were employed for the detection of mutations in the SARS-CoV-2 spike protein gene, including the receptor binding domain (RBD) [[5], [6], [7], [8], [9]]. These reports show that HRM can detect the differences between the samples, whereby a single mutation can affect the melting temperature of the PCR product for the viral genome. Indeed, Gazali et al. and Aoki et al. reported the detection of D614G (nucleotide mutation: A23403G) and L452R (nucleotide mutation: T22917G) variations in the SARS-CoV-2 spike protein by post-PCR HRM analysis, respectively [5,10]. In contrast, HRM analysis has not been used to detect SARS-CoV-2 Omicron (B.1.1.529)-specific mutations.
Omicron BA.1 possesses many mutations on the Spike protein gene relating to their critical infectivity and antibody resistance, and vaccination breakthrough cases [11]. In those many mutations on the Spike protein gene, we focused on Omicron BA.1 specific G339D (Nucleotide mutation: G22578A) and D796Y (Nucleotide mutation: G23948T) variations because there is no mutation in the vicinity of those nucleotides. The combination of the first step RT-PCR and the second step post-nested PCR HRM analysis were employed for detection of those specific variations in this study.
Here, we report a sensitive method for the detection of G339D and D796Y variations of Omicron BA.1. This is the first study that detects Omicron BA.1 by the combination of the first step RT–PCR and the second step post-nested PCR HRM analysis. This combination is efficient in supporting mutation detection methods instead of relying on the time-consuming sequencing-based detection method. This report also provides a rapid experimental protocol that can be developed for the detection of different SARS-CoV-2 variants, including the ‘stealth subvariant’ BA.2.
2. Materials and methods
2.1. Ethics statement
This project was approved by the Research Ethics Committee of St. Marianna University, and it was carried out under the Infectious Diseases Control Law in Japan.
2.2. cDNA reference standard synthesis of wt SARS-CoV-2 RBD and SARS-CoV-2 RBD variants
The wild-type SARS-CoV-2 (Isolate Wuhan-Hu-1) RNA genome was purchased from VIRCELL S.L. A PrimeScript High Fidelity RT–PCR Kit (Takara Bio) was used for cDNA preparation of wt SARS-CoV-2 RBD (NCBI NC_045512.2 nt 22487–24040: 1554 base) with adequate primers according to the manufacturer's instructions. The amplified RT–PCR product (1554 bp) was purified by agarose gel electrophoresis and gel extraction using the Wizard SV Gel and PCR Clean-Up system (Promega). The sequences of the purified cDNA reference standard of wt SARS-CoV-2 RBD were confirmed by DNA sequencing.
RBD sequences of Alpha (B.1.1.7), Delta (B.1.617.2) and Omicron (B.1.1.529) variants used in this study were obtained from NCBI (https://www.ncbi.nlm.nih.gov/sars-cov-2/). Complementary DNA fragments of these RBDs were prepared by site-directed mutagenesis of the confirmed cDNA reference standard of wt SARS-CoV-2 RBD using the primer overlap extension method [12]. The purification of cDNA fragments of those RBD variants was performed in a similar manner to that of cDNA of wt SARS-CoV-2 RBD. The sequences of all constructs (1554 bp) were confirmed by DNA sequencing.
2.3. The first amplification by PCR for the cDNA reference standard of the SARS-CoV-2 RBD
The first PCR was performed using PrimeSTAR HS DNA Polymerase (Takara Bio) in accordance with the manufacturer's instructions. Each reaction mixture (25 μL) contained 0.25 nM forward and reverse primers (Fig. 1 and Supplementary Tables 1) and 1 μL of the abovementioned cDNA reference standards of RBD (1 μg/mL), and PrimeSTAR HS DNA Polymerase. PCR amplifications were performed under the following conditions: 5 s at 98 °C; 40 cycles of 10 s at 98 °C, 5 s at 57 °C, and 20 s at 72 °C; and 5 min at 72 °C. After amplification, the PCR mixture was diluted 100,000-fold with water and used as a template for the second amplification by nested PCR.
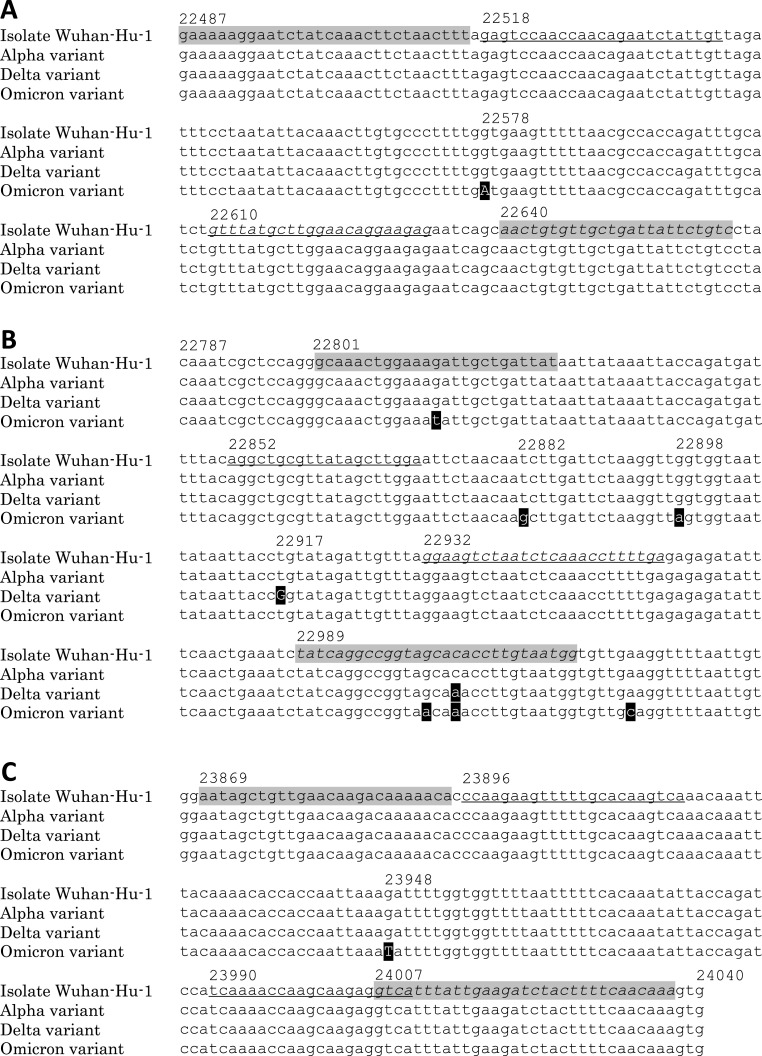
Fig. 1.
Sequence comparisons between SARS-CoV-2 isolate Wuhan-Hu-1, alpha variant, delta variant and Omicron variant.
(A) Sequence position at SARS-CoV-2 Isolate Wuhan-Hu-1: nt22487 – nt22666. (B) Sequence position at SARS-CoV-2 Isolate Wuhan-Hu-1: nt22787 – nt23026. (C) Sequence position at SARS-CoV-2 Isolate Wuhan-Hu-1: nt23867 – nt24040.
Shading indicates the sequence of the primer for the first amplification by PCR and RT–PCR, and plain and italic letters indicate the sequences of the forward and reverse primers, respectively. The underline shows the sequence of the primer for the second step nested PCR, and plain and italic letters indicate the sequences of the forward and reverse primers, respectively. Mutations of the detection target of HRM analysis are shown in capital letters. Numbers are positioned at SARS-CoV-2 isolate Wuhan-Hu-1 (NC_045512.2). All amplicons from the second amplification by nested PCR are obtained from the product of the first amplification by PCR against cDNA reference standard (nt 22487–24040) of wt SARS-CoV-2 RBD and SARS-CoV-2 RBD variants. Amplicons from the second amplification by nested PCR are 117 bp (nt22518 - nt22632), 105 bp (nt22852 - nt22956) and 115 bp (nt23896 - nt24010) respectively.
2.4. Viral RNA extraction and the first amplification by RT–PCR
After study approval by the institutional ethics and biosafety committees, saliva samples were obtained from two patients showing Delta variant-positive D1 and Omicron variant-positive O1. Viral RNA was extracted into a 50 μL aliquot from saliva using the QIAamp Viral RNA Mini Kit (Qiagen GmbH) according to the manufacturer's instructions.
Complementary DNA preparation from extracted viral RNA was performed by using the PrimeScript High Fidelity RT–PCR Kit according to the manufacturer's instructions. Each reaction mixture (25 μL) contained 0.25 nM forward and reverse primers (Fig. 1 and Supplementary Tables 1) and 5 μL of the extracted viral RNA, PrimeScript RTase, RNase Inhibitor, and PrimeSTAR Max Premix. RT–PCR was performed under the following conditions: 30 min at 50 °C; 2 min at 94 °C; 40 cycles of 10 s at 98 °C, 5 s at 55 °C, and 15 s at 72 °C; and 5 min at 72 °C. After amplification, the PCR mixture was diluted 10,000-fold with water and used as a template for the second amplification by nested PCR.
2.5. The second step post-nested PCR HRM analysis
The second step post-nested PCR HRM analysis was performed using a MeltDoctor HRM Master Mix (Thermo Fisher Scientific) according to the manufacturer's instructions. Primer pairs for the second step nested PCR are shown in Fig. 1 and Supplementary Table 1. The primer pairs were designed to detect Delta (B.1.617.2)-specific L452R (T22917G mutation) and Omicron BA.1-specific G339D (G22578A mutation) and D796Y (G23948T mutation) variations. Each reaction mixture (20 μL) contained 1 μL of the first step PCR or RT–PCR 10,000-fold dilution mixture, 300 nmol/L of each primer, and 1 × MeltDoctor HRM Master Mix. All reactions were performed on a QuantStudio 5 real-time PCR system (Thermo Fisher Scientific). PCR amplification was carried out under the following conditions: 10 min at 95 °C; 40 cycles of 10 s at 95 °C and 30 s at 60 °C. After amplification, HRM was performed with denaturation at 95 °C for 10 s, cooling at 60 °C for 60 s and melting curve generation from 60 °C to 95 °C in 0.025 °C/s increments. High Resolution Melt Software v.3.2 (Applied Biosystem) was used to discriminate between the high-resolution melting curves and obtain melting temperature (Tm) values from the derivative HRM curves. After HRM analysis, the sequences of all amplicons were confirmed by DNA sequencing.
3. Results
3.1. HRM analysis of the amplicon of the second-step nested PCR wt SARS-CoV-2 RBD and SARS-CoV-2 RBD variants
Post-nested PCR HRM analyses of cDNA reference standards of wt SARS-CoV-2 RBD and SARS-CoV-2 RBD variants were carried out to confirm the efficacy of the combination of the first-step PCR and the second-step post-nested PCR HRM analysis. Normalized and derivative HRM curves for amplicons of the second step nested PCR are shown in Fig. 2 and Supplementary Fig. 1. These HRM analyses were performed in triplicate.
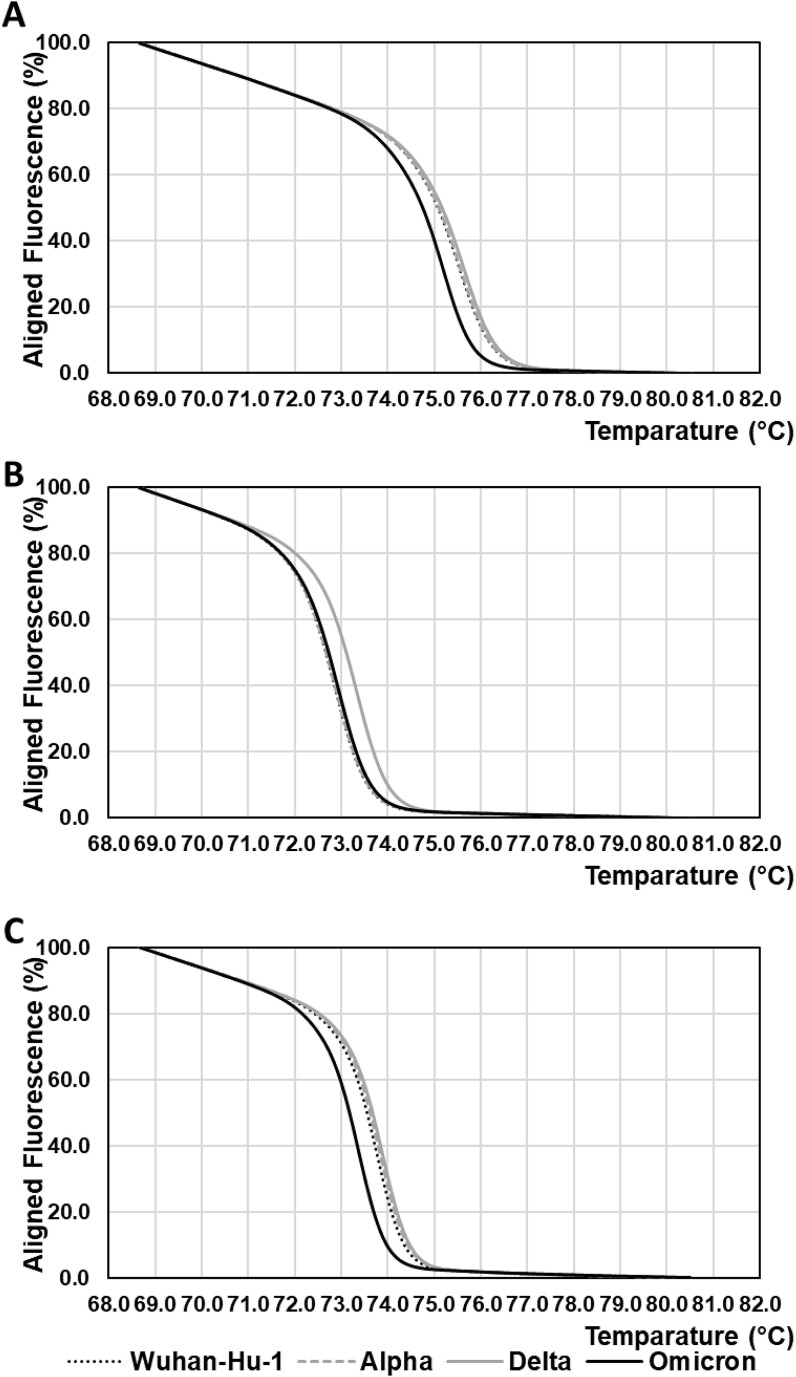
Fig. 2.
Normalized HRM curves for amplicons from cDNA reference standards of SARS-CoV-2 isolate Wuhan-Hu-1 RBD and SARS-CoV-2 RBD variants.
(A) Normalized HRM curves of amplicons from the second amplification by nested PCR with the primer pair “Second G339D forward” and “Second G339D reverse” to detect the G22578A mutation.
(B) Normalized HRM curves of amplicons from the second amplification by nested PCR with the primer pair “Second L452R forward” and “Second L452R reverse” to detect the T22917G mutation.
(C) Normalized HRM curves of amplicons from the second amplification by nested PCR with the primer pair “Second D796Y forward” and “Second D796Y reverse” to detect the G23948T mutation.
Black dotted lines indicate HRM curves for amplicons from the cDNA reference standard of SARS-CoV-2 isolate Wuhan-Hu-1 RBD. Grey broken lines indicate HRM curves for amplicons from the cDNA reference standard of SARS-CoV-2 Alfa variant RBD. Grey solid lines indicate HRM curves f for amplicons from the cDNA reference standard of SARS-CoV-2 Delta variant RBD. Black solid lines indicate HRM curves for amplicons from the cDNA reference standard of SARS-CoV-2 Omicron variant RBD. HRM analyses were performed three times to obtain the mean value of Tm. A representative curve is shown in this figure.
Normalized HRM curves for cDNA fragments of the Omicron variant RBD showed significant differences from the other curves for the detection of mutations G22578A and G23948T (Fig. 2A and C). For the detection of the mutation T22917G, there were no significant differences in the HRM curve between the Omicron variant RBD and the other variants, except for the Delta variant RBD (Fig. 2B).
3.2. Tm values of amplicon of the second step nested PCR wt SARS-CoV-2 RBD and SARS-CoV-2 RBD variants
Tm values were obtained from the derivative HRM curves (Supplementary Fig. 1A, B and C), as shown in Table 1 . The Tm values of the amplicon from the Omicron variant RBD were 75.1 and 73.3 °C in HRM analyses designed to target G339D and D796Y variations (G22578A and G23948T mutations), as shown in Table 1. These Tm values were distinct from the others (75.5–75.6 and 73.7–73.8 °C). Additionally, the Tm value (73.3 °C) of the RBD Delta variant to detect the L452R variation (T22917G mutation) was distinct from the others (72.8–72.9 °C).
Table 1.
Tm values of amplicons from the second amplification by nested PCR for cDNA reference standards of SARS-CoV-2 RBD Isolate Wuhan-Hu-1 and SARS-CoV-2 RBD variants.
| Target variation (mutation) | Mean value of Tm of isolate Wuhan-Hu-1 RBD (°C) | Mean value of Tm of Alfa variant RBD (°C) | Mean value of Tm of Delta variant RBD (°C) | Mean value of Tm of Omicron variant RBD (°C) |
|---|---|---|---|---|
| G339D (mutation G22578A) | 75.5 | 75.5 | 75.6 | 75.1 |
| L452R (mutation T22917G) | 72.8 | 72.8 | 73.3 | 72.9 |
| D796Y (mutation G23948T) | 73.7 | 73.7 | 73.8 | 73.3 |
Tm values were calculated from the derivative HRM curves (Supplementary Fig. 1A, B and C). HRM analyses were performed three times to obtain the mean value of Tm.
3.3. HRM analysis of the amplicon of the second step nested PCR of viral RNA
Viral RNA was analysed through the use of a combination of the first step RT–PCR and the second step post-nested PCR HRM analysis. Normalized and derivative HRM curves for amplicons of the second step nested PCR for viral RNA were compared with those of cDNA of reference standards of wt SARS-CoV-2 RBD and SARS-CoV-2 RBD variants, as shown in Fig. 3 and Supplementary Fig. 2.
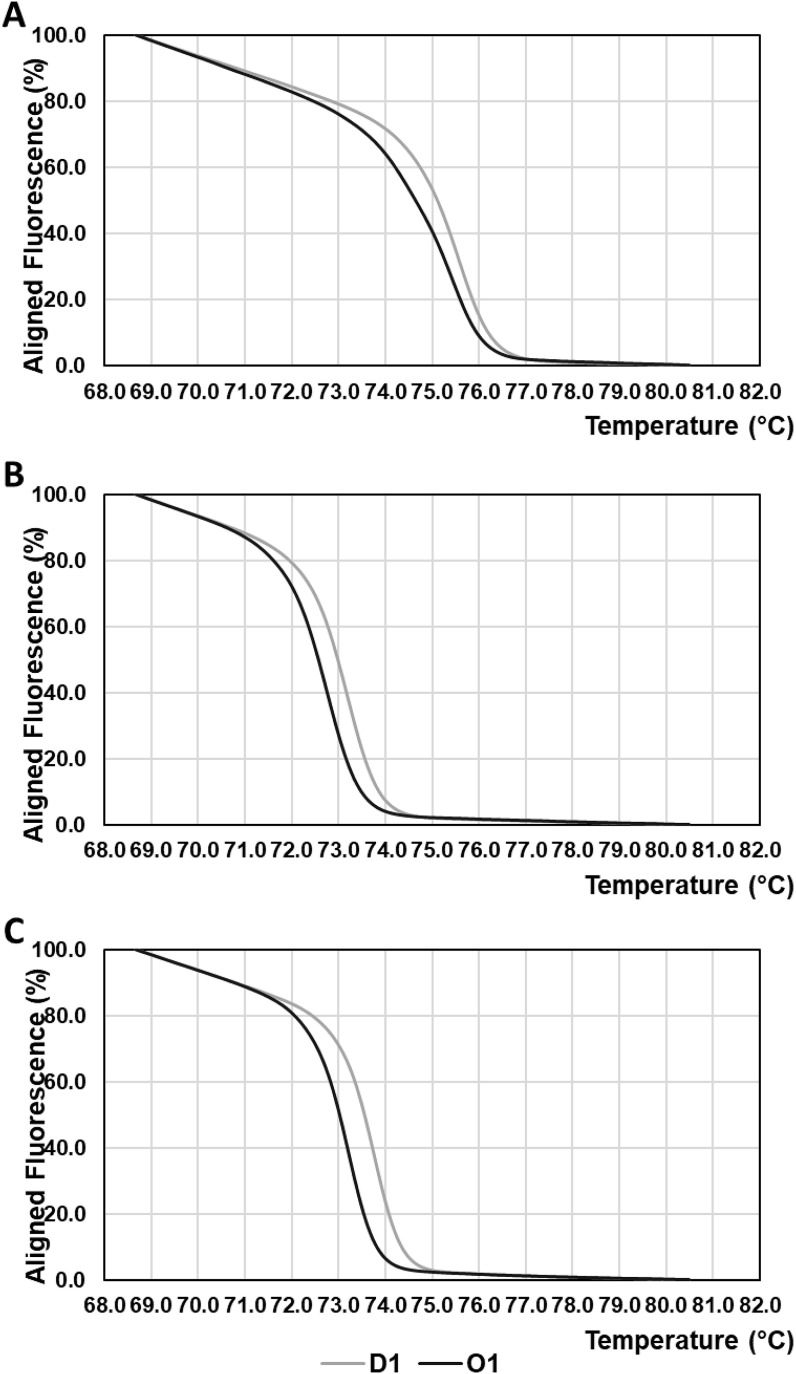
Fig. 3.
Normalized HRM curves of amplicons of extracted viral RNA from Delta variant-positive D1 specimens and Omicron variant-positive O1 specimens.
(A) Normalized HRM curves of amplicons from the second amplification by nested PCR with the primer pair “Second G339D forward” and “Second G339D reverse” to detect the G22578A mutation.
(B) Normalized HRM curves of amplicons from the second amplification by nested PCR with the primer pair “Second L452R forward” and “Second L452R reverse” to detect the T22917G mutation.
(C) Normalized HRM curves of amplicons from the second amplification by nested PCR with the primer pair “Second D796Y forward” and “Second D796Y reverse” to detect the G23948T mutation
Grey solid lines indicate HRM curves of amplicons of extracted viral RNA from Delta variant-positive D1 specimens. Black solid lines indicate HRM curves of amplicons of extracted viral RNA from Omicron variant-positive O1 specimens.
In the detection of mutations G22578A and G23948T (Fig. 3A and C), the normalized HRM curves of the Omicron variant-positive O1 specimen showed similar profiles to those of the Omicron variant RBD in Fig. 2A and C. In contrast, HRM curves (Fig. 3A and C) of Delta variant-positive D1 specimens showed analogous profiles to those of wt Isolate Wuhan-Hu-1 RBD, Alfa variant RBD and Delta variant RBD in Fig. 2A and C. For the detection of mutation T22917G, HRM curves derived from O1 specimens were similar to HRM curves of wt Isolate Wuhan-Hu-1 RBD and Omicron variant RBD. In addition, the HRM curve from the D1 specimen exhibited a profile similar to that of the Delta variant RBD (Fig. 3B).
3.4. Tm values of amplicon of the second step nested PCR of viral RNA
The Tm values of the O1 and D1 specimens were obtained from the derivative HRM curves (Supplementary Fig. 2A, B and C), as shown in Supplementary Table 2. The Tm values of the O1 specimen obtained from the derivative HRM curves (Supplementary Fig. 2A, B and C) were 75.3 °C (target: G22578A mutation), 72.7 °C (target: T22917G mutation) and 73.2 °C (target: G23948T mutation). In the case of the D1 specimen, the Tm values obtained from the derivative HRM curves (Supplementary Fig. 2A, B and C) were 75.5 °C (target: G22578A mutation), 73.2 °C (target: T22917G mutation) and 73.7 °C (target: G23948T mutation).
4. Discussion
We employed the same primer pair (“Second L452R forward” and “Second L452R reverse” in Supplementary Table 1) as reported by Aoki et al. for the detection of mutation T22917G to confirm the efficacy of the post-nested PCR HRM analysis. In Fig. 2B and Supplementary Fig. 1B, for the detection of the mutation T22917G, HRM curves of the amplicon from the cDNA reference standard of the RBD Delta variant showed a significant difference from the others. This significant difference was consistent with the results previously reported by Aoki et al. [10]. Thus, these results indicate the efficacy of the post-nested PCR HRM analysis in this study.
Despite the fact that there are T22882G and G22898A mutations in the amplicon of the cDNA reference standard of the Omicron variant RBD (Fig. 1B), the HRM curve of the cDNA reference standard of the Omicron variant RBD did not show a significant difference in the others, except for the cDNA reference standard of the Delta variant RBD (Fig. 2B). This means that the positive effect of the T22882G mutation on the Tm of the amplicon counteracts the negative effect of the G22898A mutation on the Tm of the amplicon. Therefore, a pair of “Second L452R forward” and “Second L452R reverse” primers was not suitable to detect mutations in RBD nt 22852–22956.
In HRM analyses designed to target G339D and D796Y variations (nt G22578A and G23948T), Tm values of amplicon from cDNA reference standard of Omicron variant RBD were 0.4–0.5 °C higher than the others (Table 1). In the case of detection for the D614G variation (mutation A23403G) reported by Gazali et al., the difference in the Tm value was 0.23 °C [5]. Gazali et al. utilized 172 bp amplicon for HRM analysis, as opposed to 115 bp amplicons in this study, so the difference of Tm value was smaller than our results (0.4–0.5 °C). Evidently, the larger the value of the Tm difference is, the easier the detection of mutation. In conclusion, our results clearly indicate that the combination of the first PCR and the second nested PCR is useful for HRM analysis and that the primer pairs for the first PCR and the second nested PCR are valuable for the detection of G339D and D796Y variations.
In HRM analysis after the second amplification by RT–PCR of extracted viral RNA, the HRM curve profiles and the calculated Tm values of the O1 specimen corresponded to those of the amplicon from the cDNA reference standard of the Omicron variant RBD (Fig. 2 and Supplementary Fig. 1). Similarly, the HRM curve profiles and the calculated Tm values of the D1 specimen coincided with those of the amplicon from the cDNA reference standard of the RBD Delta variant (Fig. 2 and Supplementary Fig. 1). Additionally, we attempted HRM analysis of viral RNA without nested PCR, which is the direct HRM analysis of amplicons from RT–PCR by using a primer pair for nested PCR (i.e., “Second G339D forward” and “Second G339D reverse” in Supplementary Table 1). However, we could not obtain valuable HRM curve profiles from HRM analysis of viral RNA without nested PCR because of byproduct formation of RT–PCR amplification (data not shown). Taken together, these results indicate that HRM analysis after the second amplification by nested PCR can be applied to detect specific G339D (G22578A mutation) and D796Y (G23948T mutation) variations in SARS-CoV-2 Omicron variant-positive specimens.
We then investigated whether other RNA extraction methods from viruses are available for the first amplification by RT–PCR. Phenol–chloroform extraction [13] from viral RNA was employed for the first amplification by RT–PCR. Briefly, Omicron-positive saliva samples (O2) were mixed with equal volumes of TE-saturated phenol:chloroform (1:1) mixture. After centrifugation, the upper aqueous phase was pipetted off. This procedure was performed three times to increase the purity of SARS-CoV-2 RNA. The phenol–chloroform extracted viral RNA was compared with the extracted RNA by a QIAamp Viral RNA Mini Kit analysed by the abovementioned HRM. The measured normalized and derivative HRM curves of the detection of the D796Y variation (mutation: G23948T) are shown in Supplementary Fig. 3. The Tm values obtained from the derivative HRM curves were 73.3 °C, which was close to 73.2 °C in the case of using the QIAamp Viral RNA Mini Kit (Supplementary Fig. 3B and Supplementary Table 2). These results clearly show that several procedures of viral RNA extraction can be applied to the first amplification by RT–PCR for HRM analysis.
Here, we report the detection of G339D and D796Y variations in the SARS-CoV-2 spike protein by HRM analysis of nested PCR amplicons. This procedure could be applied to the detection of various mutations by designing an adequate oligonucleotide. Thus, this strategy could be adapted to screen a considerable number of SARS-CoV-2 variants in the future.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2022.03.083.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. 2021. https://www.who.int/news/item/26-11-2021-classification-of-Omicron-(B.1.1.529)-sars-cov-2-variant-of-concern
- 4.Reed G.H., Kent J.O., Wittwer C.T. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;8:597–608. doi: 10.2217/14622416.8.6.597. [DOI] [PubMed] [Google Scholar]
- 5.Gazali F.M., Nuhamunada M., Nabilla R., Supriyati E., Hakim M.S., Arguni E., Daniwijaya E.W., Nuryastuti T., Haryana S.M., Wibawa T., Wijayanti N. Detection of SARS-CoV-2 spike protein D614G mutation by qPCR-HRM analysis. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira BldS., da Silva-Gomes N.L., Coelho WldCNP., da Costa V.D., Carneiro VcdS., Kader R.L., Amaro M.P., Villar L.M., Miyajima F., Alves-Leon S.V., de Paula V.S., Leon L.A.A., Moreira O.C. Validation of a novel molecular assay to the diagnostic of COVID-19 based on real time PCR with high resolution melting. PLoS One. 2021;16 doi: 10.1371/journal.pone.0260087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz-Garcia H., Guzmán-Ortiz A.L., Angeles-Floriano T., Parra-Ortega I., López-Martínez B., Martínez-Saucedo M., Aquino-Jarquin G., Sánchez-Urbina R., Quezada H., Granados-Riveron J.T. Genotyping of the major SARS-CoV-2 clade by short-amplicon high-resolution melting (SA-HRM) analysis. Genes. 2021;12:531. doi: 10.3390/genes12040531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalita M.J., Dutta K., Hazarika G., Dutta R., Kalita S., Das P.P., Sarma M.P., Banu S., Idris M.G., Talukdar A.J., Dutta S., Sharma A., Medhi S. In-house reverse transcriptase polymerase chain reaction for detection of SARS-CoV-2 with increased sensitivity. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-97502-1. 17878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoki A., Mori Y., Okamoto Y., Jinno H. Development of a genotyping platform for SARS-CoV-2 variants using high-resolution melting analysis. J. Infect. Chemother. 2021;27:1336–1341. doi: 10.1016/j.jiac.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aoki A., Adachi H., Mori Y., Ito M., Sato K., Okuda K., Sakakibara T., Okamoto Y., Jinno H. A rapid screening assay for L452R and T478K spike mutations in SARS-CoV-2 Delta variant using high-resolution melting analysis. J. Toxicol. Sci. 2021;46:471–476. doi: 10.2131/jts.46.471. [DOI] [PubMed] [Google Scholar]
- 11.Saxena S.K., Kumar S., Ansari S., Paweska J.T., Maurya V.K., Tripathi A.K., Abdel-Moneim A.S. Transmission dynamics and mutational prevalence of the novel SARS-CoV-2 Omicron Variant of Concern. J. Med. Virol. 2022:1–7. doi: 10.1002/jmv.27611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton R.M., Hunt H.D., Ho S.N., Pullen J.K., Pease L.R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 13.Chan P.K., Chan D.P., To K.F., Yu M.Y., Cheung J.L., Cheng A.F. Evaluation of extraction methods from paraffin wax embedded tissues for PCR amplification of human and viral DNA. J. Clin. Pathol. 2001;54:401–403. doi: 10.1136/jcp.54.5.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.