ABSTRACT
The milk microbiota and mediated metabolites directly affect the health of the udder in dairy cows. Inulin, a dietary prebiotic, can modulate the profile of gastrointestinal microbiota. However, whether the inulin intake affects the milk microbial population and metabolites remains unknown. In this study, 40 subclinical mastitis (SCM) cows were randomly divided into 5 groups. Five inulin addition doses, 0, 100, 200, 300, and 400 g/day per cow, based on the same basal diet, were supplemented. The experiments lasted for 8 weeks. The results showed lower relative abundance of mastitis-causing and proinflammation microbes in milk (i.e., Escherichia-Shigella, Pseudomonas, Rhodococcus, Burkholderia-Caballeronia-Paraburkholderia, etc.) and higher abundances of probiotics and commensal bacteria, such as Lactobacillus, Bifidobacterium, etc., in the cows fed 300 g/day inulin compared to that in the control group. Meanwhile, the levels of arachidonic acid proinflammatory mediators (leukotriene E3, 20-carboxy-leukotriene B4, and 12-Oxo-c-LTB3) and phospholipid metabolites were reduced, and the levels of compounds with antibacterial and anti-inflammatory potential (prostaglandin A1, 8-iso-15-keto-prostaglandin E2 [PGE2], etc.) and participating energy metabolism (citric acid, l-carnitine, etc.) were elevated. These data suggested that inulin intake might modulate the microflora and metabolite level in extraintestinal tissue, such as mammary gland, which provided an alternative for the regulation and mitigation of SCM.
IMPORTANCE The profile of the microbial community and metabolic activity in milk are the main determinants of udder health status and milk quality. Recent studies have demonstrated that diet could directly modulate the mammary gland microbiome. Inulin is a probiotic dietary fiber which can improve the microbiota population in the gastrointestinal tract. However, whether inulin intake can further regulate the profile of the microbiota and metabolic activities in milk remains unclear. In subclinical mastitic cows, we found that inulin supplementation could reduce the abundance of Escherichia-Shigella, Pseudomonas, Rhodococcus, and Burkholderia-Caballeronia-Paraburkholderia and the levels of (±)12, 13-DiHOME, leukotriene E3 and 20-carboxy-leukotriene B4 etc., while it elevated the abundance of Lactobacillus, Bifidobacterium, and Muribaculaceae, as well as the levels of prostaglandin A1 (PGA1), 8-iso-15-keto-PGE2, benzoic acid, etc. in milk. These data suggest that inulin intake affects the profile of microorganisms and metabolites in milk, which provides an alternative for the regulation of mastitis.
KEYWORDS: inulin, subclinical mastitis, milk microbiome, metabolomics, dairy cow
INTRODUCTION
Mastitis is a major and serious disease with extremely high incidence in dairy production. Due to a shortage of visible clinical symptoms, sufficient attention, and prompt treatment, the incidence of subclinical mastitis (SCM) is much higher than that of clinical mastitis (CM), which easily causes further aggravation of inflammation and severe losses of milk yield and quality (1). In addition, an intense inflammatory reaction during intramammary infections (IMI) consumes energy, which reduces lactation performance and disease resistance (2, 3). However, it was gradually realized that the exogenous pathogenic bacterial infection in the mammary gland is not the only cause of mastitis. Actually, the gastrointestinal microflora can exert a direct role in the regulation of the occurrence and the relief of mastitis (4–6). In addition, a close microfloral relationship between the intestinal tract and mammary gland was observed (7). Similar shifts in mastitis-related microbiota between the two sites are characterized by a general increase in the abundance of Enterococcus, Streptococcus, and Staphylococcus, as well as a decrease of Lactobacillus (7). Our previous study showed that the abundance of proinflammatory bacteria (Ruminiclostridium_9, Enterorhabdus, Neisseriaceae, etc.) and metabolites (12-oxo-20-dihydroxy-leukotriene B4) in the rumen of mastitic cows was increased compared with udder-healthy cows (8). These results may be explained by a reported endogenous microbial pathway, which is the entero-mammary pathway (9–11).
Diet is a crucial factor in determining the profile of the gastrointestinal flora (12). However, recent studies have shown that diet can also directly modulate the mammary gland microbiome populations and metabolites (13), as well as influence mammary gland-related diseases, such as breast cancer (14). Shively et al. proved that the Mediterranean diet can increase the abundance of Lactobacillus and the level of bile acid metabolites and, meanwhile, reduce the abundance of Ruminococcaceae, Oscillospira, Coprococcus, and proinflammatory oxidized lipid products (13-HODE and 3-hyrdoxystearate) in the breast tissues of female monkeys (Macaca fascicularis) (13). Similarly, to probe whether the microbiome of the gut and mammary gland mediates the dietary effects on breast cancer, Soto-Pantoja et al. performed fecal transplants between mice on control and high-fat diets (HFD) and recorded mammary tumor outcomes in a chemical carcinogenesis model. The results showed proliferation of breast cancer cells and increased tumor-associated bacteria in the mammary gland after transplantation of the HFD-derived microbiome. However, fish oil supplements modulated the microbiota in tumors and normal breast tissue (14). These data suggest that dietary intervention is able to shift the microbiota diversity in the mammary gland and potentially influence the signaling pathways relevant to mammary gland-related diseases. Furthermore, oral probiotics have shown promising therapeutic results in control mastitis, which has been proposed as an effective alternative to antibiotics (15, 16). Certain ingested probiotics are still alive in the gastrointestinal tract and become members of gut microbes, which enhance stimulation of immune cells in the intestine and subsequently migrate to other organs, including the udder (15). This suggests that oral probiotics can affect the mammary gland microbiota (16).
Inulin is a prebiotic dietary fiber mainly derived from Jerusalem artichoke tubers, which can promote the proliferation of probiotics (Bifidobacterium and Lactobacillus) and inhibit the growth of potential pathogens (Clostridium, Coprococcus, Dorea, and Ruminococcus) in the gastrointestinal tract (17–19). In addition, inulin can promote the expression of interleukin-22 (IL-22) and other antimicrobial genes during inflammation and enhance immunity, owing to the proliferation effect of inulin on beneficial bacteria (20). Our previous study showed that inulin supplementation could increase the abundance of propionate and butyrate-producing bacteria (i.e., Prevotella and Butyrivibrio) and Bifidobacterium and decrease the Clostridia_UCG-014, Streptococcus, and Escherichia-Shigella in the rumen of SCM cows (21).
Milk microflora and metabolites are closely related to the incidence of mastitis (22). Our previous results revealed the significant differences in the profiles of milk microbes and metabolites between healthy, SCM, and CM cows (22). Based on that, we attempted to investigate whether it is feasible to alleviate mastitis through modulation of the profile of the milk microbiota. Until now, the effect of inulin intake on the milk microbiota and metabolites has remained unclear. However, Ravinder et al. and Shively et al. displayed the similarities in the regulation of diet on microorganisms between the gastrointestinal tract and mammary gland (12, 13). Therefore, 16S rRNA sequencing technology and liquid chromatography-mass spectrometry (LC-MS) metabolomics were utilized in the current work to investigate whether the inulin supplementation could change the microbial community and metabolic activities in milk. The results may provide new perspectives for the control of mastitis.
RESULTS
Milk compositions.
The milk yield (P = 0.031), energy-corrected milk (ECM) (P = 0.043), milk protein (P = 0.034), and lactose (P = 0.027) were increased in the I-3 group compared with the control group (the values of milk protein and lactose between the I-2 and I-3 groups were not significantly different). The fat-corrected milk (FCM) (P = 0.065) presented an increase trend with inulin supplementation. Because milk fat was shown trend down (P = 0.073) with inulin addition, the fat to protein ratio (F/P) was reduced (P = 0.044), and without significant differences among the I-2, I-3, and I-4 groups. Compared with the control group, the milk somatic cell count (SCC) was decreased in the I-3 group (P < 0.01) (Table 1).
TABLE 1.
Effect of inulin supplementation on milk compositions and performance in dairy cows with subclinical mastitisa
| Item | Data for groups (n = 8)b |
SEM | P value | ||||
|---|---|---|---|---|---|---|---|
| Con | I-1 | I-2 | I-3 | I-4 | |||
| Milk yield (kg/day) | 31.2 C | 31.7 C | 33.7 B | 34.2 A | 33.6 B | 0.54 | 0.031 |
| ECM (kg/day)c | 30.1 C | 30.6 C | 32.3 B | 33.0 A | 32.6 B | 0.52 | 0.043 |
| FCM (kg/day)d | 31.0 | 31.4 | 32.2 | 32.8 | 32.8 | 0.34 | 0.065 |
| Milk fat (%) | 3.95 | 3.93 | 3.71 | 3.73 | 3.84 | 0.044 | 0.073 |
| Milk protein (%) | 3.03 C | 3.05 C | 3.32 A | 3.38 A | 3.27 B | 0.064 | 0.034 |
| F/P | 1.30 A | 1.29 A | 1.12 B | 1.10 B | 1.17 B | 0.038 | 0.044 |
| Milk lactose (%) | 4.01 C | 4.02 C | 4.28 A | 4.31 A | 4.22 B | 0.057 | 0.027 |
| SCC (×103/mL) | 716 A | 706 A | 593 BC | 541 C | 647 B | 21.0 | <0.01 |
ECM, energy-corrected milk; FCM, fat-corrected milk; F/P, fat to protein ratio; SCC, somatic cell counts; Con, control group; I-1, inulin-1 group (the inulin addition level was 100 g/day per cow); I-2, inulin-2 group (the inulin addition level was 200 g/day per cow); I-3, inulin-1 group (the inulin addition level was 300 g/day per cow); I-4, inulin-1 group (the inulin addition level was 400 g/day per cow); SEM, standard error of mean.
Different uppercase letters in a group represent significantly different values (P < 0.05).
ECM (kg/day) = milk yield (kg/day) × (383 × fat [%] + 242 × protein [%] + 783.2)/3,140 (69).
FCM (kg/day) = 0.4 × milk yield (kg/day) + 15 × milk yield (kg/day) × fat (%) (70).
Milk fatty acids.
For saturated fatty acid (SFA), the proportion of C4 (P = 0.032), C11 (P = 0.044), C13 (P = 0.041), C15 (P = 0.022), anteiso C15:0 (P = 0.041), anteiso C17:0 (P = 0.021), and iso C17:0 (P = 0.035) in milk was increased in the I-3 group compared with the control group. Among them, there was no significant difference in the proportion of C11, C15, and anteiso C17:0 between the I-2 and I-3 groups and no significant difference in anteiso C15:0 among the I-2, I-3, and I-4 groups. Additionally, the proportion of C17 presented an increased trend with inulin addition (P = 0.057). Overall, inulin addition did not significantly affect the proportion of SFA (P = 0.115). The proportion of unsaturated fatty acids (UFA) presented a downward trend after inulin treatment (P = 0.062). Compared with the control group, the proportion of cis-9c18:1 was reduced in the I-3 group, and without significant differences among the I-2, I-3, and I-4 groups (P = 0.048). Moreover, the proportion of C20:1 (P = 0.087), C18:2n6c (P = 0.093), C18:3n3 (P = 0.081), and polyunsaturated fatty acids (PUFA) (P = 0.074) and long-chain fatty acids (LCFA) (P = 0.063) all showed a declining trend (Table 2).
TABLE 2.
Effect of inulin supplementation on milk fatty acids in dairy cows with subclinical mastitisa
| Item (g/100 g of total FA) | FA name | Data for groups (n = 8)b |
SEM | P value | ||||
|---|---|---|---|---|---|---|---|---|
| Con | I-1 | I-2 | I-3 | I-4 | ||||
| SFA | 69.3 | 68.7 | 69.4 | 68.6 | 68.2 | 0.26 | 0.115 | |
| C4 | Butyric acid | 2.14 C | 2.17 C | 2.61 B | 2.72 A | 2.59 B | 0.035 | 0.032 |
| C6 | Caproic acid | 1.78 | 1.84 | 1.60 | 1.68 | 1.62 | 0.025 | 0.631 |
| C8 | Caprylic acid | 1.12 | 1.13 | 1.10 | 1.10 | 1.20 | 0.017 | 0.211 |
| C10 | Capric acid | 2.54 | 2.7 | 2.49 | 2.31 | 2.40 | 0.047 | 0.144 |
| C11 | Undecanoic acid | 0.11 C | 0.15 B | 0.18 A | 0.19 A | 0.16 B | 0.005 | 0.044 |
| C12 | Lauric acid | 3.17 | 3.12 | 3.09 | 3.12 | 3.07 | 0.045 | 0.256 |
| C13 | Tridecanoic acid | 0.11 C | 0.10 C | 0.17 B | 0.22 A | 0.15 B | 0.004 | 0.041 |
| C14 | Myristic acid | 10.1 | 10.4 | 10.3 | 10.1 | 10.5 | 0.10 | 0.165 |
| C15 | Pentadecanoic acid | 0.73 C | 0.79 C | 1.05 A | 1.08 A | 0.96 B | 0.017 | 0.022 |
| Anteiso C15:0 | Transisomeric pentadecanoic acid | 0.33 B | 0.35 B | 0.42 A | 0.46 A | 0.43 A | 0.011 | 0.041 |
| iso C15:0 | Isomeric pentadecanoic acid | 0.13 | 0.14 | 0.14 | 0.15 | 0.15 | 0.005 | 0.753 |
| C16 | Palmitic acid | 35.6 | 34.6 | 34.9 | 34.0 | 33.6 | 0.21 | 0.262 |
| iso C16:0 | Isomeric hexadecanoic acid | 0.18 | 0.19 | 0.16 | 0.17 | 0.16 | 0.005 | 0.130 |
| C17 | Heptadecanoic acid | 0.38 | 0.37 | 0.4 | 0.41 | 0.40 | 0.005 | 0.057 |
| anteiso C17:0 | Transisomeric heptadecanoic acid | 0.73 C | 0.79 C | 1.01 A | 1.13 A | 0.94 B | 0.03 | 0.021 |
| iso C17:0 | Isomeric heptadecanoic acid | 0.31 C | 0.34 C | 0.44 B | 0.51 A | 0.46 B | 0.016 | 0.035 |
| C18 | Stearic acid | 9.44 A | 9.19 AB | 8.94 B | 8.88 B | 9.04 AB | 0.105 | 0.024 |
| C20 | Arachidic acid | 0.21 | 0.20 | 0.16 | 0.17 | 0.18 | 0.005 | 0.094 |
| C22:0 | Behenic acid | 0.20 | 0.17 | 0.19 | 0.16 | 0.17 | 0.012 | 0.151 |
| UFA | 6.30 | 6.50 | 6.10 | 6.10 | 6.20 | 0.240 | 0.062 | |
| MUFA | 3.60 | 3.80 | 3.50 | 3.50 | 3.60 | 0.230 | 0.117 | |
| C14:1 | Myristoleic acid | 1.01 | 1.07 | 0.91 | 0.90 | 0.99 | 0.240 | 0.173 |
| C16:1 | Palmitoleic acid | 1.6 | 1.71 | 1.63 | 1.64 | 1.62 | 0.051 | 0.220 |
| Trans-9C18:1 | Elaidic acid | 0.74 | 0.76 | 0.78 | 0.77 | 0.74 | 0.022 | 0.185 |
| Cis-9C18:1 | Oleic acid | 18.7 A | 18.3 A | 16.7 B | 16.3 B | 16.8 B | 0.2 | 0.048 |
| C20:1 | Cis-11-eicosenoic acid | 0.21 | 0.23 | 0.17 | 0.18 | 0.20 | 0.004 | 0.087 |
| C22:1 | Erucic acid | 0.04 | 0.04 | 0.03 | 0.03 | 0.04 | 0.003 | 0.311 |
| PUFA | 2.70 | 2.68 | 2.61 | 2.60 | 2.58 | 0.033 | 0.074 | |
| C18:2n6c | Linolelic acid | 2.41 | 2.42 | 2.41 | 2.36 | 2.34 | 0.032 | 0.093 |
| C18:3n6 | γ-Linolenic acid | 0.03 | 0.02 | 0.02 | 0.02 | 0.03 | 0.002 | 0.149 |
| C18:3n3 | α-Linolenic acid | 0.02 | 0.04 | 0.03 | 0.05 | 0.04 | 0.002 | 0.081 |
| C20:3n6 | Cis-8,11,14-eicosatrienoic acid | 0.06 | 0.04 | 0.03 | 0.03 | 0.04 | 0.002 | 0.211 |
| C20:3n3 | Cis-11,14,17-eicosatrienoic acid | 0.18 | 0.16 | 0.12 | 0.14 | 0.13 | 0.005 | 0.305 |
| SMCFA | 22.2 | 22.8 | 22.6 | 22.5 | 22.8 | 0.088 | 0.230 | |
| LCFA | 42.1 | 41.0 | 41.0 | 39.0 | 39.7 | 0.33 | 0.063 | |
FA, fatty acids; SFA, saturated fatty acids; UFA, unsaturated fatty acids; MUFA, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids; SMCFA, short- and medium-chain fatty acids; LCFA, long-chain fatty acids; Con, control group; I-1, inulin-1 group (the inulin addition level was 100 g/day per cow); I-2, inulin-2 group (the inulin addition level was 200 g/day per cow); I-3, inulin-1 group (the inulin addition level was 300 g/day per cow); I-4, inulin-1 group (the inulin addition level was 400 g/day per cow); SEM, standard error of the mean.
Different uppercase letters in a group represent significantly different values (P < 0.05).
Overall structure, richness, diversity, and composition of milk microbiota.
A total of 1,495,060 and 1,442,675 optimized 16S rRNA gene sequences were obtained from 40 milk samples before and after inulin supplementation, respectively. According to the minimum sequence number in the samples, the reads of milk microbiota before and after inulin addition were rarefied to 44,279 and 33,766 sequencing reads. After operational taxonomic unit (OTU) clustering of nonrepeated sequences and removal of chimeras, a total of 5,651 and 4,876 OTU representative sequences with >97% similarity were obtained, respectively, before and after inulin addition. Before inulin supplementation, the alpha-diversity indexes of the milk microbiota had no significant difference among groups (see Table S3 in the supplemental material). However, inulin treatment made significant changes in the milk microbiota diversity and richness in SCM cows. The observed richness (Sobs) (P = 0.046), Shannon index (P = 0.042) abundance-based coverage estimator (ACE) (P = 0.045), and Chao (P = 0.041) results were lower in the I-3 group (the Shannon index had no significant difference in the I-2, I-3, and I-4 groups) than those in the control group. Moreover, the Good’s coverages of all samples exceeded 99%, indicating completeness of sequencing within the community (Table 3). The rarefaction curve was constructed by using the microbial alpha diversity index of each sample at different sequencing depths. As shown in Fig. S1 and S2, the curves tended to be as flat as the increase in the number of reads sampled, implying adequate sequencing data and depth.
TABLE 3.
Effect of inulin supplementation on alpha diversity of milk microorganisms in cows with subclinical mastitisa
| Item | Data for groups (n = 8)b |
SEM | P value | ||||
|---|---|---|---|---|---|---|---|
| Con | I-1 | I-2 | I-3 | I-4 | |||
| Sobs | 829 A | 725 B | 667 BC | 630 C | 699 B | 24.1 | 0.046 |
| Shannon | 4.80 A | 4.26 A | 3.38 B | 3.54 B | 3.74 B | 0.176 | 0.042 |
| Simpson | 0.10 | 0.13 | 0.10 | 0.07 | 0.10 | 0.020 | 0.299 |
| ACE | 848 A | 768 B | 771 B | 661 C | 794 B | 37.6 | 0.045 |
| Chao | 855 A | 778 B | 761 B | 667 C | 795 B | 37.6 | 0.041 |
| Coverage | 0.999 | 0.998 | 0.996 | 0.998 | 0.996 | 0.0003 | 0.603 |
Con, control group; I-1, inulin-1 group (the inulin addition level was 100 g/day per cow); I-2, inulin-2 group (the inulin addition level was 200 g/day per cow); I-3, inulin-1 group (the inulin addition level was 300 g/day per cow); I-4, inulin-1 group (the inulin addition level was 400 g/day per cow); SEM, standard error of the mean.
Different uppercase letters in a group represent significantly different values (P < 0.05).
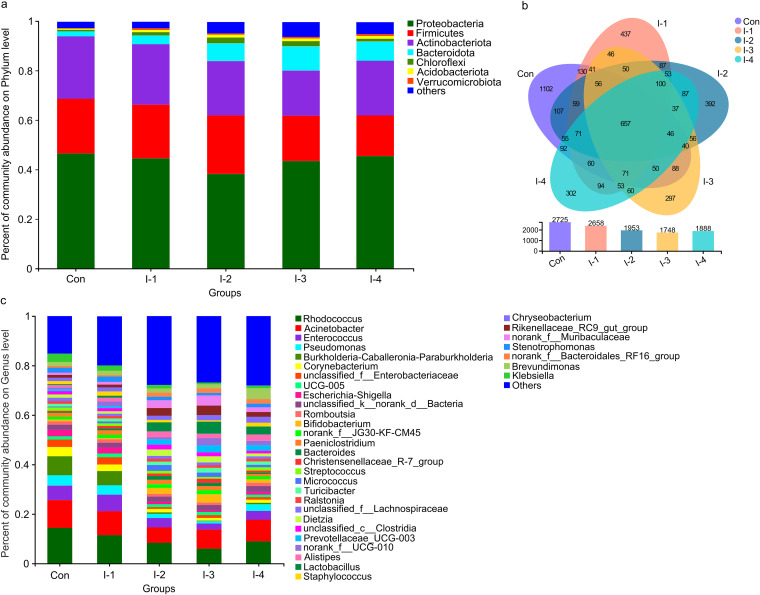
The phylum and genus compositions of milk microbiota before and after inulin addition are listed in Tables S4 to S7. At the phylum level, Proteobacteria (43.4 ± 0.25%; 41.9 ± 0.80%, respectively), Actinobacteriota (25.9 ± 0.20%; 23.9 ± 0.63%), Firmicutes (21.6 ± 0.21%; 20.4 ± 0.40%), and Bacteroidota (3.01 ± 0.09%; 5.71 ± 0.52%), were the predominant taxa (Fig. 1A and Fig. S3A). At the genus level, Rhodococcus (14.7 ± 0.45%; 10.6 ± 0.66%, respectively), Acinetobacter (11.1 ± 0.51%; 8.68 ± 0.47%), Enterococcus (5.66 ± 0.12%; 3.34 ± 0.41%), Pseudomonas (4.64 ± 0.07%; 3.29 ± 0.20%), Burkholderia-Caballeronia-Paraburkholderia (6.24 ± 0.21%; 3.03 ± 0.52%), Corynebacterium (4.64 ± 0.07%; 2.75 ± 0.26%), and unclassified_f_Enterobacteriaceae (4.25 ± 0.10%; 2.50 ± 0.33%) were the most abundant (Fig. 1C and Fig. S3B). The number of common and unique milk bacteria at the OTU level among groups after inulin addition are shown in Fig. 1B.
FIG 1.
The number of unique and common bacteria and composition of microbiol communities based on OTU level in milk samples of the control and different inulin groups. (A) Phylum level; (B) Venn diagram; (C) genus level.
Differential taxa among control and inulin-fed group milk microflora.
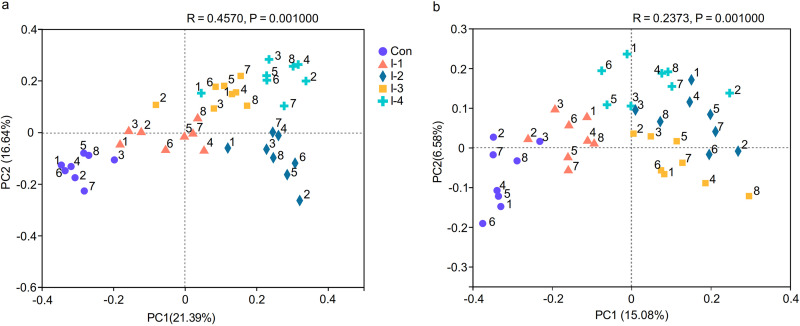
As shown in Tables S4 and S5, there was no significant differences in milk microbes among 5 groups before inulin supplementation. After inulin supplementation, the principal-coordinate analysis (PCoA) based on Bray-Curtis distance algorithm and the weighted UniFrac distance algorithm showed that the milk microbial community among different groups could be basically separated (Fig. 2A and B). To visualize the differences in community composition among different groups, hierarchical clustering analysis (HCA) of milk bacteria (top 60 of relative abundance) based on the Bray-Curtis distance algorithm on the OTU level were conducted. The milk microbiota in 40 milk samples was separated into two large clusters. Rhodococcus, Acinetobacter, Pseudomonas, Corynebacterium, Streptococcus, Enterobacter, Escherichia-Shigella, Burkholderia-Caballeronia-Paraburkholderia, and Staphylococcus were concentrated in the control and I-1 groups with high relative abundance. Lactobacillus, Bifidobacterium, norank_f_Muribaculaceae, Christensenellaceae_R-7_group, and Rikenellaceae_RC9_gut_group were clustered in the I-2, I-3, and I-4 groups, and with higher relative abundance in the I-3 group than the other two groups (Fig. S4).
FIG 2.
(A and B) Principal coordinate analysis (PCoA) based on (A) Bray-Curtis and (B) weighted UniFrac distance algorithm of microbial communities in milk samples of dairy cows with subclinical mastitis (SCM) from the control and different inulin groups. Con, control group; I-1, inulin-1 group (the inulin addition level was 100 g/day per cow); I-2, inulin-2 group (the inulin addition level was 200 g/day per cow); I-3, inulin-1 group (the inulin addition level was 300 g/day per cow); I-4,= inulin-1 group (the inulin addition level was 400 g/day per cow).
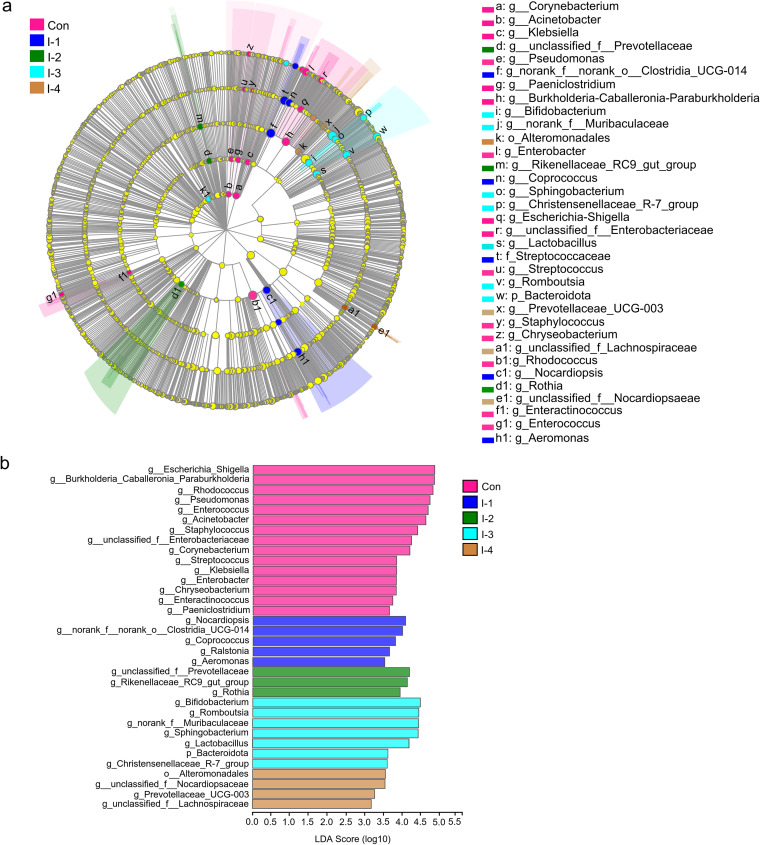
To further obtain the significant difference of milk microbiota among groups, linear discriminant analysis effect size (LEfSe) and Kruskal-Wallis H test analysis were used to perform significant difference tests among multiple groups. The significantly differential bacteria selected are shown in Fig. 3 and Table 4, with a linear discriminant analysis (LDA) score of >3.0 and a false-discovery rate (FDR)-adjusted P value of <0.05. At the phylum level, compared with the control group, the relative abundance of Bacteroidota (FDR-adjusted P value = 0.032) was increased in the I-3 group, without a significant difference between the I-2, I-3, and I-4 groups. However, the relative abundance of Proteobacteria (FDR-adjusted P value = 0.085) and Actinobacteriota (FDR-adjusted P value = 0.081) tended to be decreased in the I-3 group.
FIG 3.
The linear discriminant analysis effect size (LEfSe) analysis of milk microbial taxa among the control and different inulin groups. (A) The cladogram shows the taxa with significant differences in abundance (from phylum to genus level). (B) The LDA bar chart shows the influence of different bacteria on the differences among groups. The LDA score is >3.0. Con, control group; I-1, inulin-1 group (the inulin addition level was 100 g/day per cow); I-2, inulin-2 group (the inulin addition level was 200 g/day per cow); I-3, inulin-1 group (the inulin addition level was 300 g/day per cow); I-4, inulin-1 group (the inulin addition level was 400 g/day per cow).
TABLE 4.
Significantly differential milk microbiota among control and inulin groupsa
| Item | Data for groups (n = 8)b |
SEM | P value | FDR | ||||
|---|---|---|---|---|---|---|---|---|
| Con | I-1 | I-2 | I-3 | I-4 | ||||
| Proteobacteria | 45.15 | 42.21 | 35.22 | 42.03 | 44.94 | 0.803 | 0.017 | 0.085 |
| Acinetobacter | 11.70 A | 10.51 A | 6.16 B | 7.11 B | 7.92 B | 0.468 | 2.35 × 10−4 | 0.035 |
| Burkholderia-Caballeronia-Paraburkholderia | 6.33 A | 5.40 A | 1.04 B | 0.93 B | 1.47 B | 0.523 | 4.01 × 10−4 | 0.037 |
| Pseudomonas | 4.78 A | 4.57 A | 2.11 B | 1.65 B | 3.34 AB | 0.198 | 4.61 × 10−4 | 0.038 |
| Escherichia -Shigella | 3.07 A | 3.18 A | 1.23 B | 1.04 B | 1.47 B | 0.200 | 1.54 × 10−3 | 0.045 |
| unclassified_f_Enterobacteriaceae | 4.38 A | 4.12 A | 0.85 B | 1.61 B | 1.53 B | 0.326 | 6.11 × 10−4 | 0.040 |
| Klebsiella | 0.44 A | 0.48 A | 0.22 B | 0.11 C | 0.13 C | 0.033 | 1.31 × 10−3 | 0.046 |
| Brucella | 0.08 | 0.08 | 0.05 | 0.14 | 0.10 | 0.007 | 0.011 | 0.069 |
| Succinivibrio | 0.02 | 0.02 | 0.04 | 0.10 | 0.04 | 0.006 | 0.014 | 0.080 |
| Firmicutes | 20.72 | 20.00 | 23.63 | 19.42 | 18.42 | 0.395 | 0.086 | 0.107 |
| Lactobacillus | 0.09 C | 0.13 C | 0.50 A | 0.62 A | 0.34 B | 0.050 | 9.47 × 10−4 | 0.041 |
| Staphylococcus | 0.56 A | 0.54 A | 0.16 B | 0.11 B | 0.50 A | 0.044 | 1.19 × 10−3 | 0.040 |
| Enterococcus | 5.37 A | 5.90 A | 2.07 B | 1.67 B | 2.12 B | 0.407 | 7.04 × 10−4 | 0.042 |
| Streptococcus | 0.80 A | 0.70 A | 0.48 B | 0.24 C | 0.46 B | 0.044 | 1.26 × 10−3 | 0.045 |
| Romboutsia | 0.68 | 0.81 | 1.24 | 1.77 | 0.96 | 0.086 | 0.014 | 0.084 |
| Paeniclostridium | 0.97 | 0.61 | 0.53 | 0.58 | 0.39 | 0.043 | 0.004 | 0.070 |
| Christensenellaceae_R-7_group | 0.27 | 0.50 | 0.62 | 0.86 | 0.61 | 0.043 | 0.005 | 0.068 |
| norank_f_UCG-010 | 0.12 | 0.87 | 0.11 | 0.52 | 0.32 | 0.064 | 0.010 | 0.062 |
| Coprococcus | 0.13 | 0.10 | 0.07 | 0.04 | 0.04 | 0.008 | 0.024 | 0.094 |
| Bacteroidota | 2.71 B | 3.33 B | 7.00 A | 8.86 A | 6.65 A | 0.521 | 0.001 | 0.032 |
| Prevotellaceae_UCG-003 | 0.14 C | 0.17 C | 0.48 B | 0.65 A | 0.66 A | 0.051 | 1.32 × 10−3 | 0.043 |
| norank_f_Muribaculaceae | 0.10 C | 0.16 C | 0.43 B | 0.67 A | 0.35 B | 0.046 | 2.16 × 10−3 | 0.048 |
| Rikenellaceae_RC9_gut_group | 0.15 | 0.14 | 0.45 | 0.69 | 0.32 | 0.046 | 0.005 | 0.068 |
| Sphingobacterium | 0.07 | 0.14 | 0.23 | 0.33 | 0.17 | 0.019 | 0.006 | 0.071 |
| Actinobacteriota | 27.29 | 26.31 | 24.03 | 19.56 | 22.06 | 0.629 | 0.012 | 0.081 |
| Rhodococcus | 15.53 A | 11.66 AB | 8.76 B | 6.76 B | 10.46 AB | 0.660 | 1.04 × 10−4 | 0.031 |
| Corynebacterium | 4.61 A | 3.86 A | 1.97 B | 1.56 B | 1.76 B | 0.257 | 1.02 × 10−4 | 0.043 |
| Bifidobacterium | 0.35 C | 0.33 C | 0.90 A | 1.18 A | 0.74 B | 0.073 | 5.08 × 10−4 | 0.039 |
| Micrococcus | 0.18 | 0.25 | 0.92 | 0.72 | 0.58 | 0.062 | 0.008 | 0.057 |
Con, control group; I-1, inulin-1 group (the inulin addition level was 100 g/day per cow); I-2, inulin-2 group (the inulin addition level was 200 g/day per cow); I-3, inulin-1 group (the inulin addition level was 300 g/day per cow); I-4, inulin-1 group (the inulin addition level was 400 g/day per cow); SEM, standard error of the mean; FDR, false-discovery rate.
Different uppercase letters in a group represent significantly different values (FDR < 0.05).
At the genus level, 11 taxa were significantly decreased with inulin supplementation, including Acinetobacter (FDR-adjusted P value = 0.035), Burkholderia-Caballeronia-Paraburkholderia (FDR-adjusted P value = 0.037), Pseudomonas (FDR-adjusted P value = 0.038), Escherichia-Shigella (FDR-adjusted P value = 0.045), unclassified_f__Enterobacteriaceae (FDR-adjusted P value = 0.040), Klebsiella (FDR-adjusted P value = 0.046), Staphylococcus (FDR-adjusted P value = 0.040), Enterococcus (FDR-adjusted P value = 0.042), Streptococcus (FDR-adjusted P value = 0.045), Rhodococcus (FDR-adjusted P value = 0.031), and Corynebacterium (FDR-adjusted P value = 0.043).
Among them, the relative abundance of Staphylococcus, Enterococcus, and Rhodococcus had no significant difference between the I-2 and I-3 groups. The relative abundance of Acinetobacter, Burkholderia-Caballeronia-Paraburkholderia, Escherichia-Shigella, unclassified_f__Enterobacteriaceae, Enterococcus, and Corynebacterium had no significant difference among the I-2, I-3, and I-4 groups.
On the other hand, 4 taxa were significantly enriched after inulin treatment, Lactobacillus (FDR-adjusted P value = 0.044), Bifidobacterium (FDR-adjusted P value = 0.026), norank_f_Muribaculaceae (FDR-adjusted P value = 0.030), and Prevotellaceae_UCG-003 (FDR-adjusted P value = 0.046). The highest abundance of Lactobacillus and Bifidobacterium had no significant difference among the I-2 and I-3 groups. The highest abundance of Prevotellaceae_UCG-003 had no significant difference between the I-3 and I-4 groups. The highest value of norank_f_Muribaculaceae was in the I-3 group (Table 4).
LC-MS metabolite profiling of milk samples.
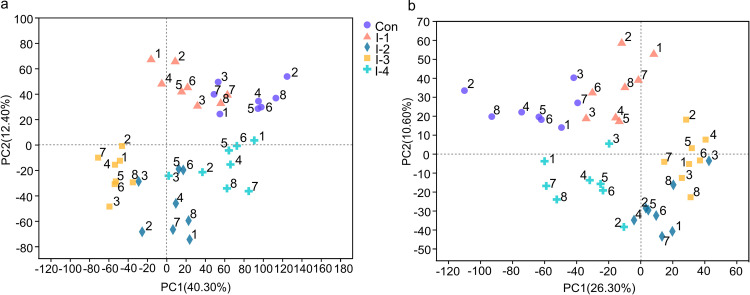
To characterize the milk metabolic profile, LC-MS metabolomic profiling was carried out in nontargeted mode. Total ion chromatograms (TIC) of quality control (QC) samples in positive and negative ionization mode are shown in Fig. S4. The total ion intensity and retention time of different QC samples were basically consistent, implying the reliability of the experimental data. To provide a global difference among groups and the degree of variation within groups, an unsupervised multivariate statistical analysis method, principal-component analysis (PCA), was conducted in positive and negative ion mode (Fig. 4A and B). Orthogonal partial least-squares discriminant analysis was performed to further distinguish the difference between groups in positive and negative ion mode. Meanwhile, response permutation testing (RPT) was conducted to assess the accuracy of the orthogonal least partial square discriminant analysis (OPLS-DA) model. In OPLS-DA, R2X(cum) and R2Y(cum) indicate the cumulative interpretation rate of the X and Y matrix of the model, respectively. The closer relationship between these two indicators was to 1, the more the model was stable and reliable; Q2(cum) denoted the predictive ability of the model, which was above 0.5 and represented a credible predictive ability of the model (23). In RPT, the intercept value of R2 and Q2 were >0.70 and <0, respectively, implying that the OPLS-DA model was not been overfitted (Fig. S5 and S6).
FIG 4.
(A and B) Principal-component analysis (PCA) plots of the milk metabolome of the control and different inulin groups in (A) positive and (B) negative ion modes. Con, control group; I-1, inulin-1 group (the inulin addition level was 100 g/day per cow); I-2, inulin-2 group (the inulin addition level was 200 g/day per cow); I-3, inulin-1 group (the inulin addition level was 300 g/day per cow); I-4, inulin-1 group (the inulin addition level was 400 g/days per cow).
Metabolite identification and difference analysis among groups.
After filtering and normalization of the raw data and QC verification, the metabolites in 40 milk samples added up to 891, with 522 and 369 metabolites by positive and negative ion modes, respectively. At the superclass level, 38.85% were classified as lipids and lipid-like molecules, 17.15% as organic acids and derivatives, 15.32% as organoheterocyclic compounds, and 9.71% as organic oxygen compounds (Fig. S7A). At the class level, the metabolites belonging to carboxylic acids and derivatives (14.50%), glycerophospholipids (13.41%), fatty acyls (11.08%), and organooxygen compounds (9.71%) were the most abundant (Fig. S7B). At the subclass level, the top four classifications of metabolites in abundance were amino acids (AAs), peptides, and analogues (14.09%), not available (10.40%), carbohydrates and carbohydrate conjugates (6.43%), and glycerophosphocholines (4.65%) (Fig. S7C).
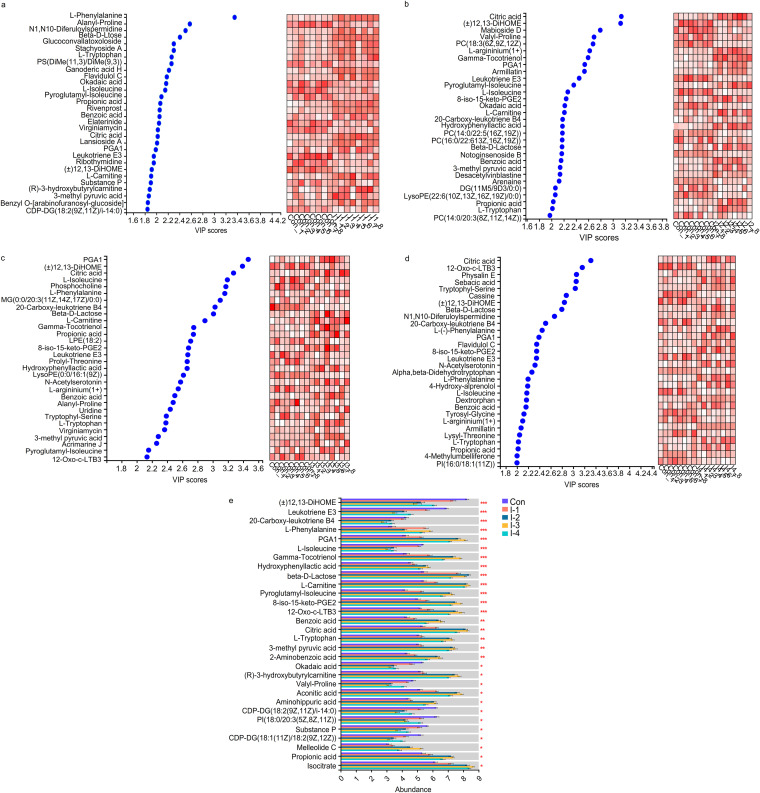
Differential metabolites were identified by variable importance in projection (VIP) analysis and multigroup comparison. Variable important in projection based on OPLS-DA was performed to select the differential metabolites between control and I-1, control and I-2, control and I-3, control and I-4. The differential metabolites with the top 50 VIP scores are listed in Fig. 5A to D and Tables S8 to S11. Further, the Kruskal-Wallis H test was used to conduct multigroup analysis of variance. The differential metabolites with an FDR-adjusted P value of <0.05 among 5 groups are shown in Fig. 5E and Table S12. Among them, compared with the control group, the abundance of (±)12,13-DiHOME, leukotriene E3, valyl-proline, phosphatidylinositol (PI) [18:0/20:3(5Z,8Z,11Z)], and cytidine diphosphate diacylglycerol (CDP-DG) [18:1(11Z)/18:2(9Z,12Z)] were significantly decreased in the I-2 and I-3 groups, and without significant difference between the two groups. The lowest value of 20-carboxy-leukotriene B4, 12-Oxo-c-LTB3, CDP-DG [18:2(9Z,11Z)/i-14:0], and substance P were in the I-3 group. Moreover, the lowest value of okadaic acid had no significant difference among the I-2, I-3, and I-4 groups.
FIG 5.
Significance test of differences in milk metabolites based on pairwise comparison and multigroup tests. (A to D) Variable importance in projection (VIP) plots showing the significant differential metabolites (top 30 VIP scores) in milk samples between the control and I-1 groups, the control and I-2 groups, the control and I-3 groups, and the control and I-4 groups, respectively. Cells on the right are colored based on the relative abundance of metabolites in milk; red represents a high level, while white represents a low level. (E) Multigroup comparison bar chart showing the significantly differing metabolites (top 30 FDR-adjusted P values) in milk samples among the 5 groups. Con, control group; I-1, inulin-1 group (the inulin addition level was 100 g/day per cow); I-2, inulin-2 group (the inulin addition level was 200 g/day per cow); I-3, inulin-1 group (the inulin addition level was 300 g/day per cow); I-4, inulin-1 group (the inulin addition level was 400 g/day per cow). *, 0.01 < FDR-adjusted P value ≤ 0.05; **, 0.001 < FDR-adjusted P value ≤ 0.01; ***, FDR-adjusted P value ≤ 0.001.
On the other hand, the abundance of l-phenylalanine, gamma-tocotrienol, hydroxyphenyllactic acid, beta-d-lactose, l-carnitine, pyroglutamyl-isoleucine, 8-iso-15-keto-prostaglandin E2 (PGE2), citric acid, benzoic acid, l-tryptophan, 3-methyl pyruvic acid, 2-aminobenzoic acid, (R)-3-hydroxybutyrylcarnitine, aconitic acid, aminohippuric acid, propionic acid, and isocitrate were significantly increased in the I-2 and I-3 groups compared with the control group, and without significant difference between the two groups. The highest value of prostaglandin A1 (PGA1) and melleolide C were in the I-3 group.
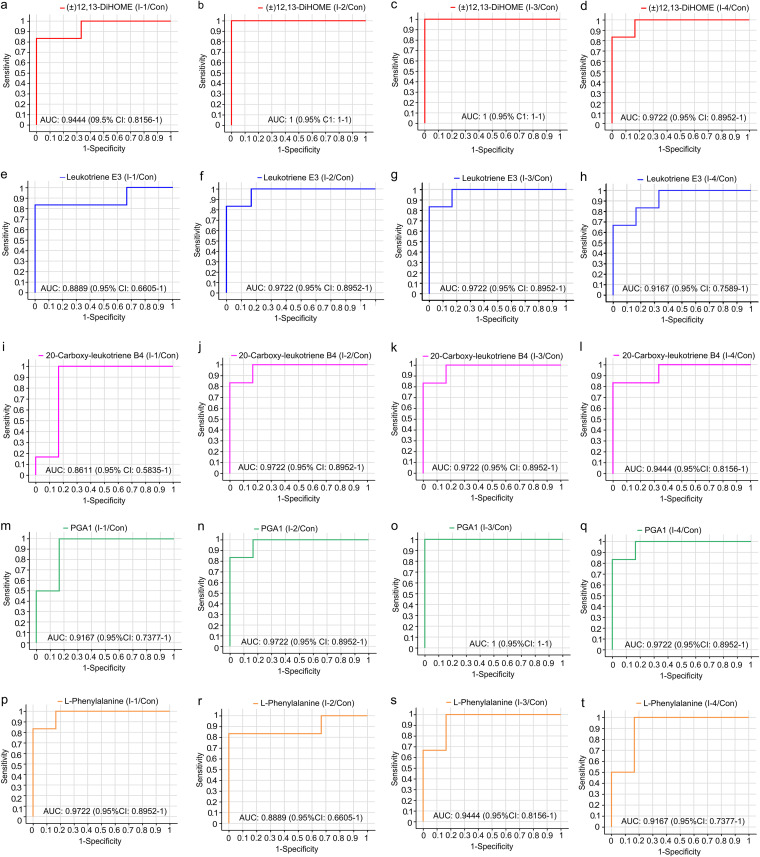
The top 5 differential metabolites with a lower FDR-adjusted P value were used for receiver operator characteristic curve (ROC) analysis. As shown in Fig. 6, the area under the curve (AUC) of (±)12, 13-DiHOME (1 and 1), leukotriene E3 (0.972 and 0.972), 20-carboxy-leukotriene B4 (0.972 and 0.972) and PGA1 (0.972 and 1) from the control versus the I-2 group and the control versus the I-3 group, respectively, were higher than those of the control versus the I-1 group (0.944, 0.889, 0.861, and 0.917, respectively) and the control versus the I-4 group (0.972, 0.917, 0.944 and 0.972, respectively), which meant that the above-mentioned 4 biomarkers in the I-2 and I-3 groups had a reliable assessment ability to the effect of inulin on SCM. However, the AUC of l-phenylalanine in the control versus the I-1 group and the control versus the I-3 group (0.972 and 0.944, respectively) were higher than those in the control versus the I-2 group (0.889) and the control versus the I-4 group (0.917).
FIG 6.
Receiver operator characteristic (ROC) curve of significantly differential metabolites (top 5 FDR P value). (A to D) (±)12, 13-DiHOME between Con and I-1, Con and I-2, Con and I-3, and Con and I-4, respectively. (E to H) Leukotriene E3 between Con and I-1, Con and I-2, Con and I-3, and Con and I-4, respectively. (I to L) 20-Carboxy-leukotriene B4 between Con and I-1, Con and I-2, Con and I-3, and Con and I-4, respectively. (M to P) PGA1 between Con and I-1, Con and I-2, Con and I-3, and Con and I-4, respectively. (Q to T) l-Phenylalanine between Con and I-1, Con and I-2, Con and I-3, and Con and I-4, respectively. Con, control group; I-1 = inulin-1 group (the inulin addition level was 100 g/day per cow); I-2, inulin-2 group (the inulin addition level was 200 g/day per cow); I-3, inulin-1 group (the inulin addition level was 300 g/day per cow); I-4, inulin-1 group (the inulin addition level was 400 g/day per cow).
Metabolic pathway enrichment analysis.
In the present study, the downregulated metabolic pathway included linoleic acid metabolism, arachidonic acid metabolism, and phospholipid metabolism (FDR-adjusted P value < 0.05). On the contrary, several AAs’ biosynthesis and degradation, aminoacyl-tRNA biosynthesis, citrate cycle, fatty acid degradation, galactose metabolism, ubiquinone, and other terpenoid-quinone biosynthesis and eicosanoid metabolism were upregulated (FDR-adjusted P value < 0.05) (Table 5).
TABLE 5.
Metabolic pathway enrichment analysis of significant differential milk metabolites among control and inulin groupsa
| Metabolites | KEGG pathway | P value | FDR |
|---|---|---|---|
| Downregulated | |||
| (±)12,13-DiHOME | Linoleic acid metabolism | 0.003 | 0.034 |
| Leukotriene E3 | Arachidonic acid metabolism | 0.001 | 0.009 |
| 20-Carboxy-leukotriene B4 | Arachidonic acid metabolism | 1 × 10−4 | 0.003 |
| 12-Oxo-c-LTB3 | Arachidonic acid metabolism | 0.001 | 0.009 |
| CDP-DG [18:2(9Z,11Z)/i-14:0] | Phospholipid metabolism | 0.002 | 0.024 |
| PI [18:0/20:3(5Z,8Z,11Z)] | Phospholipid metabolism | 0.002 | 0.025 |
| CDP-DG [18:1(11Z)/18:2(9Z,12Z)] | Phospholipid metabolism | 0.004 | 0.042 |
| Okadaic acid | – | – | – |
| Valyl-proline | Protein degradation | 0.002 | 0.027 |
| Substance P | Neuroactive ligand-receptor interaction | 0.004 | 0.046 |
| Upregulated | |||
| l-Phenylalanine | Phenylalanine metabolism | 0.001 | 0.018 |
| ABC transporters | 0.003 | 0.037 | |
| Benzoic acid | Phenylalanine, tyrosine, and tryptophan biosynthesis | 0.005 | 0.049 |
| Phenylalanine metabolism | 1.00 × 10−4 | 0.003 | |
| Tryptophan metabolism | infinitesimal | infinitesimal | |
| 2-Aminobenzoic acid | Phenylalanine, tyrosine, and tryptophan biosynthesis | 0.005 | 0.049 |
| l-Tryptophan | Phenylalanine, tyrosine, and tryptophan biosynthesis | 0.005 | 0.049 |
| Tryptophan metabolism | infinitesimal | infinitesimal | |
| 3-Methyl pyruvic acid | Glycine, serine, and threonine metabolism | 0.003 | 0.036 |
| Cysteine and methionine metabolism | 0.001 | 0.031 | |
| Aminohippuric acid | Phenylalanine metabolism | 1.00 × 10−4 | 0.003 |
| Propionic acid | Carbohydrate digestion and absorption | 0.004 | 0.041 |
| Propanoate metabolism | 0.001 | 0.023 | |
| Phenylalanine metabolism | 1.00 × 10−4 | 0.003 | |
| Hydroxyphenyllactic acid | Ubiquinone and other terpenoid-quinone biosynthesis | 0.002 | 0.039 |
| Tyrosine metabolism | 0.007 | 0.061 | |
| Pyroglutamyl-isoleucine | Peptide and amino acid metabolism | 0.006 | 0.054 |
| Citric acid | Citrate cycle (TCA cycle) | 0.001 | 0.038 |
| Alanine, aspartate, and glutamate metabolism | 0.006 | 0.057 | |
| Aconitic acid | Citrate cycle (TCA cycle) | 0.004 | 0.038 |
| Glyoxylate and dicarboxylate metabolism | 0.006 | 0.054 | |
| Isocitrate | Citrate cycle (TCA cycle) | 0.002 | 0.038 |
| l-Carnitine | Fatty acid degradation | 0.005 | 0.047 |
| (R)-3-hydroxybutyrylcarnitine | Fatty acid degradation | 0.005 | 0.049 |
| Beta-d-lactose | Galactose metabolism | 0.002 | 0.037 |
| Gamma-tocotrienol | Ubiquinone and other terpenoid-quinone biosynthesis | 0.002 | 0.031 |
| PGA1 | Eicosanoid metabolism | 0.002 | 0.033 |
| 8-Iso-15-keto-PGE2 | Eicosanoid metabolism | 0.006 | 0.052 |
| Melleolide C | – | – | – |
PGA1, prostaglandin A1; ABC transporters, ATP binding cassette transporter; TCA cycle, tricarboxylic acid cycle; –, the KEGG pathway was unknown; FDR, false discovery rate.
Analysis of correlation between differential milk microbiota and milk compositions, differential metabolites and milk compositions, and differential microbiomes and metabolites in milk.
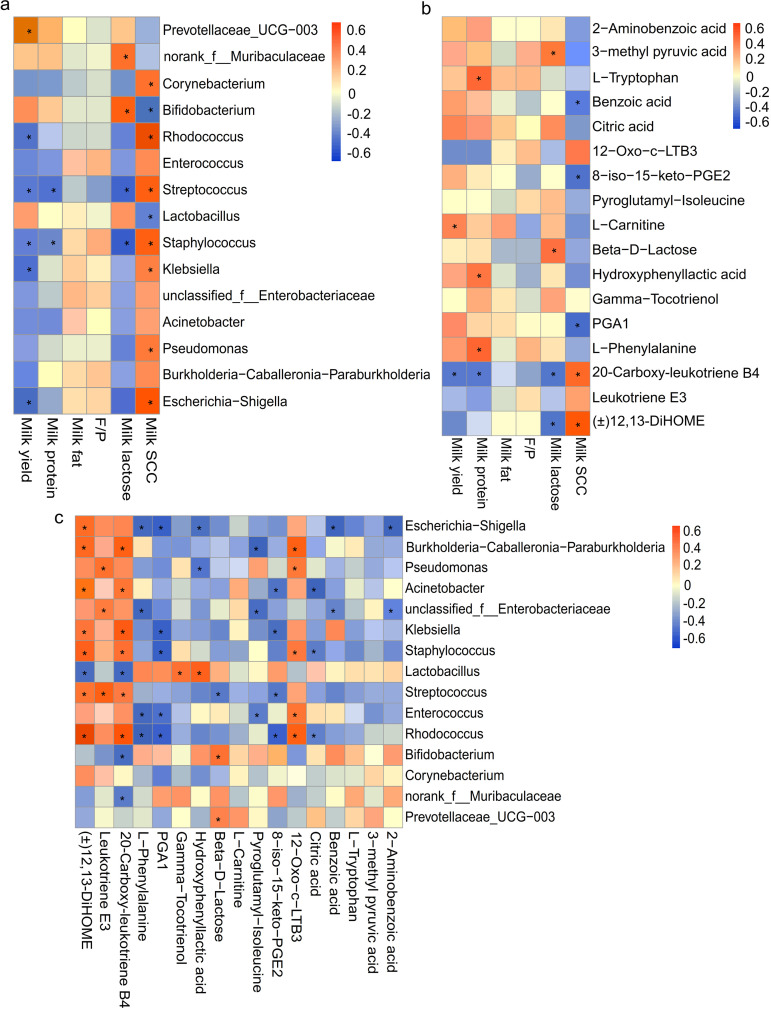
Spearman correlation analysis was used to determine the correlation. The r of >0.40 and FDR-adjusted P value of <0.05 are listed in Tables S13 to S15. As shown in Fig. 7A and Table S13, the milk yield was positively associated with Prevotellaceae_UCG-003 while negatively associated with Streptococcus, Staphylococcus, Rhodococcus, Klebsiella, and Escherichia-Shigella. The milk proteins were negatively associated with Staphylococcus and Streptococcus. The milk lactose was positively associated with Bifidobacterium and norank_f_Muribaculaceae but negatively associated with Staphylococcus and Streptococcus. The milk SCC was positively correlated with Rhodococcus, Escherichia-Shigella, Streptococcus, Staphylococcus, Pseudomonas, Klebsiella, and Corynebacterium while negatively associated with Bifidobacterium, Lactobacillus, and Acinetobacter.
FIG 7.
(A to C) Analysis of Spearman correlation between (A) significantly abundant milk microorganisms and milk compositions, (B) significantly differing milk metabolites and milk compositions, and (C) significantly differential bacteria and metabolites in milk samples among the control and different inulin treatment groups. The red color represents a positive correlation, while the blue color represents a negative correlation. *, significant correlation between the factors (FDR-adjusted P value < 0.05).
In addition, the milk yield was positively correlated with l-carnitine and negatively correlated with 20-carboxy-leukotriene B4. The milk proteins were positively correlated with l-phenylalanine, l-tryptophan, and hydroxyphenyllactic acid but negatively correlated with 20-carboxy-leukotriene B4. The milk lactose was positively correlated with beta-d-lactose and 3-methyl pyruvic acid, whereas it was negatively correlated with 20-carboxy-leukotriene B4 and (±)12,13-DiHOME. The milk SCC was positively correlated with (±)12,13-DiHOME and 20-carboxy-leukotriene B4 while negatively correlated with benzoic acid, 8-iso-15-keto-PGE2, and PGA1 (Fig. 7B and Table S14).
As shown in Fig. 7C and Table S15, (±)12,13-DiHOME was positively correlated with Rhodococcus, Burkholderia-Caballeronia-Paraburkholderia, Staphylococcus, Escherichia-Shigella, Klebsiella, Streptococcus, and Acinetobacter but negatively correlated with Lactobacillus. Leukotriene E3 was positively correlated with Streptococcus, Pseudomonas, and unclassified_f_Enterobacteriaceae. 20-carboxy-leukotriene B4 was positively correlated with Rhodococcus, Klebsiella, Burkholderia-Caballeronia-Paraburkholderia, and Staphylococcus while negatively correlated with norank_f_Muribaculaceae, Lactobacillus, and Bifidobacterium. l-phenylalanine was negatively correlated with Escherichia-Shigella, Enterococcus, unclassified_f_Enterobacteriaceae, and Rhodococcus. PGA1 was negatively correlated with Enterococcus, Klebsiella, Staphylococcus, and Rhodococcus. Gamma-tocotrienol was positively associated with Lactobacillus. Hydroxyphenyllactic acid was negatively associated with Pseudomonas and Escherichia-Shigella but positively associated with Lactobacillus. Beta-d-lactose was positively correlated with Prevotellaceae_UCG-003 while negatively correlated with Streptococcus. Pyroglutamyl-isoleucine was negatively correlated with Burkholderia-Caballeronia-Paraburkholderia and unclassified_f_Enterobacteriaceae. 8-iso-15-keto-PGE2 was negatively correlated with Rhodococcus, Streptococcus, and Klebsiella. 12-Oxo-c-LTB3 was positively correlated with Rhodococcus, Burkholderia-Caballeronia-Paraburkholderia, Enterococcus, and Pseudomonas. Citric acid was negatively correlated with Rhodococcus, Acinetobacter, and Staphylococcus. Benzoic acid and 2-aminobenzoic acid were all negatively correlated with unclassified_f_Enterobacteriaceae and Escherichia-Shigella.
DISCUSSION
To our knowledge, the effects of dietary inulin supplementation on milk yield and composition in dairy cows have not been reported. However, one of the reasons might be attributed to the shifts in the rumen internal environment. Consistent with Zhao et al. (24) and Tian et al. (25), our previous study also showed that the addition of inulin could increase the concentration of propionate, butyrate, and total volatile fatty acid (VFAs) and decrease the NH3-N concentrations in the rumen of SCM cows (21). As a precursor to lactose synthesis, the increase in propionate promotes milk lactose synthesis. Moreover, the decreased NH3-N concentration resulted in the increase of microbial protein synthesis in the rumen, which might contribute to the mammary gland ingesting protein to synthesize milk protein (26). Meanwhile, additional VFAs could provide energy for lactation. However, our previous study found no significant increase in acetate concentration after inulin supplementation compared to the control group (21). Also, several studies have reported a decrease in acetate and increased butyrate in the rumen after inulin supplementation (24, 27). As a precursor of milk fat synthesis (28), the decrease in ruminal acetate might explain the downward trend in milk fat.
Further, we investigated the effects of inulin on milk fatty acids (FAs), which might be closely related to ruminal microbial community structure and VFA concentration. In the current study, the increase in percentage of C4 might be attribute to the increase of butyrate concentration in the rumen (24). In addition, propionyl coenzyme A is the precursor of linear odd-number-chain FA synthesis in the mammary gland tissue (29). Thus, the increased percentage of C13, C15, and C17 might reflect the increased propionate production in the rumen. The odd- and branched-chain FAs in milk mostly come from the rumen bacteria (30). Our previous study showed increased Bacteroidetes and decreased Firmicutes in the rumen of SCM cows in the I-3 group compared to the control group (21). Jami et al. reported a positive correlation between the milk fat and the ratio of Firmicutes to Bacteroidetes (31). The increase of Bacteroidetes in the gut indicated a decreased fat concentration in blood and tissues (32). Meanwhile, the decrease in PUFA was also reported to be related to the increase in Bacteroidetes and decrease in Firmicutes (33). Increased anteiso C15:0 might be related to Prevotella, which is reported to be rich in anteiso C15:0 (30). The iso C17 was reported to be strongly correlated with the ratio of adenine:N of rumen bacteria (34). In addition, Cabrita et al. suggested that the concentration of anteiso C17:0 in milk was significantly negatively related with the rumen NH3-N concentration (35). Therefore, the content of anteiso C17:0 and iso C17 might imply the synthesis of microbial proteins in rumen. Moreover, cows with mastitis are often accompanied by negative energy balance (36). During mastitis, metabolites involved in energy metabolism were reported to be decreased (23). The LCFA, stearic acid, and in particular, oleic acid were reported to be compensatorily increased during the low body energy level (37, 38). In the present study, the decrease in the percentage of cis-9c18:1 and the downward trend of LCFA after the addition of inulin might reflect the elevated energy level in SCM cows.
The function of diet modulation on mammary gland microflora has been demonstrated (13–15). In the current study, inulin supplementation reduced the richness and diversity of mastitic milk microorganisms, which were mainly from the phyla Proteobacteria and Firmicutes, which were the main bacteria phyla enriched in etiologic agents from mastitic milk, including Escherichia-Shigella, Pseudomonas, Acinetobacter, Klebsiella, Staphylococcus, Streptococcus, and Corynebacterium (39, 40). The positive correlation between these taxa and milk SCC has also been shown (41). Interestingly, our previous study found that Streptococcus, Staphylococcus, and Escherichia-Shigella were also reduced in the rumen of SCM cows that consumed inulin (21), which might indicate that several microbiomes were coregulated between the rumen and mammary gland. Rhodococcus has previously been misidentified as Corynebacterium bovis in mastitic milk (42). Angeliki et al. found that the abundance of Rhodococcus was the highest among 18 of 50 clinical mastitic milk samples (40).
In addition, several potentially proinflammatory bacteria rarely reported in mastitis were identified in the present study. Burkholderia-Caballeronia-Paraburkholderia, a kind of Burkholderia complex bacteria, was an opportunistic pathogen (43). It has been reported that the ability of Burkholderia complex bacteria to induce proinflammatory cytokines was 10 times greater than that of P. aeruginosa (44). Thus, the current study speculated that Burkholderia-Caballeronia-Paraburkholderia might be responsible for the increase of milk SCC during SCM.
However, inulin supplementation increased the relative abundance of the phylum Bacteroidota in SCM milk, which was analogous with the variation tendency of ruminal bacteria (21). The phylum Bacteroidota, including norank_f_Muribaculaceae and Rikenellaceae_RC9_gut_group, is known to produce propionate (45). In addition, Prevotellaceae_UCG-003 could produce succinate, which could further produce propionate (46). This might explain the positive association between the above-mentioned bacteria and milk lactose. The accumulating evidence showed that inulin intake could stimulate the proliferation of Lactobacillus and Bifidobacterium in the intestinal tract (18, 19). Our previous study also found the increased relative abundance of these two bacteria in the rumen of SCM cows after being fed inulin (21). In the current study, we further observed the increase of these two bacteria in the sampled milk. These observations might support the existence of the endogenous milk microbiota pathway (9–11). In fact, during lactation, several commensal microbiotas in the gastrointestinal tract can be carried out by intestinal immune cells (such as dendritic cells) and transferred to the udder via lymphoid tissue and peripheral blood circulation (11, 47). Lactobacillus in the udder could exert antimicrobial activity by producing antibacterial substances, such as bacteriocins, organic acids, etc. (48). However, during IMI, some pathogens, such as Streptococcus and Staphylococcus, could produce anti-Lactobacillus substances (i.e., lantibiotics) to resist Lactobacillus (41). In addition, Ma et al. reported that mastitis might be correlated with the absence of Lactobacillus, which would increase the susceptibility to mastitis (7). The negative correlation between Lactobacillus and SCC was also verified (7, 41). Similarly, Bifidobacterium in milk and the gastrointestinal tract plays an important role in the host’s symbiotic relationship of maintaining microbiota homeostasis (49). Nagahata et al. reported that injection of Bifidobacterium into the udder could trigger immune cell infiltration and enhance the immune response of the mammary gland, which further eliminates secondary pathogens of SCM (50). However, the correlation between Bifidobacterium and SCC needs to be further verified.
Similarities exist in the effects of diet on the microbiome population between the gastrointestinal tract and mammary glands (12, 13). Combined with our previous studies, the current study shows that inulin intake could regulate the milk microbiota of SCM cows. The effect of inulin on the milk microbiota compared with the ruminal microbiome displayed similarities (21).
The enrichment of proinflammatory mediators, including leukotriene E3, 20-carboxy-leukotriene B4, and 12-Oxo-c-LTB3, in milk during SCM upregulated arachidonic acid metabolism. Among them, the 20-carboxyleukotriene B4 is an omega-oxidized metabolite of leukotriene B4, which was reported to be significantly increased in Klebsiella-induced bovine mastitis (51). Meanwhile, another leukotoxin produced by neutrophils, (±)12, 13-DIHOME, could result in cellular oxidative stress and specifically inhibit mitochondrial function (52). The above inflammation-related metabolites were closely related to the chemotaxis and production of neutrophils, which might explain their positive correlation with milk SCC. In addition, a neuropeptide, substance P, with the potential for proinflammation in intestinal inflammation was identified in SCM cow milk (53). It might imply the correlation between the occurrence of cow mastitis and intestinal health status. Moreover, the increase of several phospholipid metabolites, such as CDP-DG [18:2(9Z,11Z)/i-14:0] and PI [18:0/20:3(5Z,8Z,11Z)] confirmed the increased lipolysis during SCM, which resulted in an increase in free fatty acids (54). This also reflected a decrease in energy level of mastitic cows (38). Thus, our data indicated that the proinflammatory metabolites and energy consumption in cows were raised during SCM.
However, inulin supplementation in the current study seem to relieve the above-described symptoms to a certain extent. Elevated l-phenylalanine was the precursor of tyrosine and has been reported to be concentrated in foods with high protein (55). This finding might mirror the increase in milk protein after feeding inulin. Further, two phenylalanine metabolites, benzoic acid and aminohippuric acid, were also identified. Benzoic acid could diminish the viability of Escherichia coli (56). Hippurate was reported to be associated with increased consumption of dietary fiber and negatively associated with the risk of metabolic syndrome (57). Pyroglutamyl-isoleucine could exert the anti-inflammatory activity by inhibiting the production of nitric oxide (58). Hydroxyphenyll-actic acid is a tyrosine analogue, which is produced by Lactobacillus (59). Besides, elevated PGA1 could cause anti-inflammation properties by inhibiting phosphorylation and preventing degradation of IkB-α, which is the NF-κB inhibitor (60). 8-iso-15-keto-PGE2 and gamma-tocotrienol were reported to have potentially antiproliferative effects on neoplastic mammary epithelial cells (61, 62). The increased metabolites suggested the roles of inulin supplementation to prevent pathogen proliferation and inflammatory response.
On the other hand, the increased abundance of 3-methyl pyruvic acid, citric acid, aconitic acid, and isocitrate upregulated the tricarboxylic acid (TCA) cycle pathway. l-carnitine was involved in FA β-oxidation in mitochondria, and finally, in upgrading energy metabolism (63). Therefore, the increase of these metabolites suggested that inulin supplementation could enhance the energy level in SCM cows, which also explains their positive correlation with milk yield. The increase of propionic acid and beta-d-lactose showed that inulin supplementation enhanced propanoate and galactose metabolism, respectively. Our previous study reported the increased propionate in the rumen of cows fed inulin (21), which might promote milk lactose synthesis. Ma et al. showed that probiotic intake could improve galactose, pyruvate metabolism, and glycolysis/gluconeogenesis (5). Thus, the elevated energy metabolism level might be due to the increased abundance of probiotics after inulin supplementation.
Collectively, our data highlight the impact of inulin supplementation on the profile of milk microflora and metabolites in SCM cows, which support that the diet could affect the mammary gland microbiota. In combination with the findings from our previous study, the alteration of milk microflora in SCM dairy cows after inulin supplementation was partially similar to that in the ruminal bacterial community, suggesting the existence of a coregulated microbial community in rumen and milk. Moreover, the effect of the addition of inulin on milk compositions might be associated with the ruminal bacterial population and its mediated metabolites, such as propionate. In the present study, 300 g/day per cow inulin showed a better response to most detected indicators, which might be attributed to the degradation of inulin in the rumen (Yue Wang, unpublished data), the changes in the internal environment of rumen (21), and the cow’s own metabolic adaptation after inulin supplementation. Our previous study investigated the degradation rate of inulin in the rumen through a rumen simulation technique. The results showed that after adding more than 1.2% dry matter (DM) of inulin to the rumen fermentation liquor containing 0.5 g dried total mixed rations (TMR), the degradation rate of inulin in the rumen significantly decreased and leveled off (unpublished data). The inulin addition level in the current study was calculated based on the in vitro study and average dry matter intake (25 kg/day per cow) of cows. Moreover, elevated numbers of propionate- and butyrate-producing bacteria, and the metabolites related to energy and amino acid metabolism, and suppressed proinflammatory bacteria accompanied by proinflammatory metabolites were observed in the rumen of cows fed 300 g/day inulin (21). However, due to the variations in milk composition at different time points, further study will be necessary to increase the sampling time to verify the effectiveness of inulin.
Our data suggest that the dietary supplementation of inulin (300 g/day per cow) in cows with SCM could increase the relative abundance of commensal microflora, including Lactobacillus, Bifidobacterium, norank_f_Muribaculaceae, Rikenellaceae_RC9_gut_group, etc. and reduce mastitis-causing pathogens (Escherichia-Shigella, Klebsiella, Staphylococcus, Rhodococcus, etc.) as well as potentially proinflammation bacteria (Burkholderia-Caballeronia-Paraburkholderia and Ralstonia) in milk. Meanwhile, the expression of metabolites related to anti-inflammatory (PGA1, gamma-tocotrienol, 8-iso-15-keto-PGE2, etc.) and energy metabolism (citric acid, aconitic acid, l-carnitine, beta-d-lactose, etc.) were upregulated, while some proinflammatory mediators [(±)12,13-DiHOME, leukotriene E3, 20-carboxy-leukotriene B4, etc.] and phospholipid metabolites (CDP-DG [18:2(9Z,11Z)/i-14:0] and PI [18:0/20:3(5Z,8Z,11Z)]) were downregulated. The improvement of the milk microbiome and metabolite profiling might be conducive to restoring the loss of milk production and quality during SCM.
MATERIALS AND METHODS
Ethics statement.
The experimental animal welfare and ethics protocol of the current trial was inspected and approved by the Animal Ethics Committee of the Chinese Academy of Agricultural Sciences (Beijing, China; approval number IAS-2020-92). The animal management and experimental operation were in accordance with the academy’s guidelines for animal research.
Animals, diets, experimental design, and feeding regimen.
The trial was implemented in a dairy farm in the suburbs of Beijing for 56 days. A total of 40 dairy cows suffering from SCM (days in milk = 133 ± 2.4 days; parity = 3.38 ± 0.32; milk yield = 29.7 ± 1.42 kg/day; milk somatic cell counts [SCC] = 716,000 ± 17,500 cells/mL; the results of California mastitis test [CMT] were weakly positive and positive; no clinical symptoms in udders) were screened and randomly divided into 5 groups (n = 8 per group). The diagnostic criterion of CMT results was referred to Wang et al. (22). The information of initial body condition of SCM cows in 5 groups is given in Table S1. The cows were driven to the milking parlor and milked 3 times per day at 6:30, 12:30, and 20:30 with automatic milking equipment (Afimilk, Israel). The cows in the 5 groups were offered the same TMR with a concentrate to roughage ratio of 40:60 as the basal diet. The ingredients and chemical composition of the TMR are shown in Table S2. The TMR was provided three times a day, at 7:00, 13:00, and 19:00. Cows were housed in individual tie stalls and given ad libitum access to feed and water.
Inulin used in the present study was derived from the tuber of Jerusalem artichoke, which was provided by the Langfang Academy of Agriculture and Forestry Sciences (Hebei, China). According to the additive doses in the different groups, inulin was accurately weighed and pilled in the same size. A 1-m stainless steel pill propellant gun (Boehringer-Ingelheim, Ingelheim, Germany) was used to supplement inulin through the oral cavity to ensure accurate inulin intake of each cow in accordance with the experiment’s requirements. Inulin was given 3 times a day at the time of feeding TMR.
Milk sample collection and analysis.
Milk samples were collected on day 1 before inulin supplementation and day 56 after inulin supplementation. In the last week of the experiment (week 8), the daily milk production was recorded for 7 consecutive days using the Afimilk computerized dairy farm management system (Afimilk, Israel). The average milk yield of each group from the 7 consecutive days represented the milk yield of that group. Milk samples were collected 3 times at the time of milking on the last day of the trial (day 56). Two 50-mL milk samples were taken from each cow at a time. The milk samples of each cow collected in the morning, noon, and afternoon were mixed in a 4:3:3 ratio (64, 65). A piece of potassium dichromate with 0.6 mg/mL as an antiputrefactive was added in one of the composited milk samples of each cow, which was stored at 4°C for measurement of milk composition, including milk protein, milk fat, milk lactose, and SCC. Another composited milk sample was divided into three 10-mL sterile centrifugal tubes. One of them was stored at −20°C for analysis of milk FAs. The other two were store at −80°C for analysis of milk microbiota and metabolites. The protein, fat, lactose, and SCC in the milk samples were detected by midinfrared spectroscopy using MilkoScan Minor device (FOSS, Denmark).
Milk fatty acid assay.
Milk FAs were detected by using a gas chromatograph (Agilent 7890A; Agilent, CA, USA) with a flame ionization detector and external standard method for quantitative analysis. A 2-mL milk sample collected in a 25-mL centrifuge tube with high temperature resistance, which contained 2.5 mL methanol and 1.25 mL chloroform, and mixed well. After 1 h, the solution was added to 1.25 mL chloroform, 1.15 mL water, and 0.1 mL 3 mol/L HCl, which was centrifuged at 1,200 × g for 3 min. The bottom solution was dried by anhydrous sodium sulfate and filtered. The filtrate was collected in a centrifuge tube and blow-dried by nitrogen gas, which was then added to 1 mL toluene and mixed well. A 200-μL sodium hydroxide alcohol solution (0.2 mg NaOH dissolved in methanol) was added into the above-described solution to methylate the fat. After 25 min, 11 mL methanolized sulfuric acid (2.8 mL 96% sulfuric acid added to 100 mL methanol) was added and mixed thoroughly. After a water bath for 15 min at 50°C, the solution was preserved at −20°C for 3 min and then added to 1 mL water and 1 mL n-hexane. After being mixed well, it was centrifuged at 1,200 × g at room temperature for 3 min. The supernatant (2 μL) was filtered by a 0.22-μm filter membrane for analysis. The gas chromatographic conditions were as follows: the inlet temperature was 240°C, and the pressure was 266.9 kPa. The initial temperature of the column was 170°C and was kept for 30 min and then increased to 200°C at 1.5/min for 20 min and finally increased to 230°C at 5°C/min for 5 min. Each peak was identified using known FA methyl ester standards, which were purchased from Sigma-Aldrich (Saint Louis, MO).
Total DNA extraction and PCR amplification.
Milk microbiota in SCM cows before and after inulin supplementation were all analyzed in the current study. The total milk microbial DNA was extracted using a milk microbial DNA extraction kit (Norgen Biotek, Canada) following the manufacturer’s instructions. A spectrophotometer (Orion AquaMate 8000; Thermo Fisher Scientific, Waltham, USA) and 1% agarose gel electrophoresis (Beijing Liuyi Biological Technology Co., Ltd., Beijing, China) were used to detect the purity and integrity of DNA, respectively. The primer regions of PCR amplification were the V3 and V4 regions of the microbial 16S rRNA gene, with forward and reverse primers being 338F ACTCCTACGGGAGGCAGCAG and 806R GGACTACHVGGGTWTCTAAT. The PCR system was as follows: 4 μL of 5 × FastPfu buffer, 2 μL of 2.5 mM dNTPs (deoxynucleoside triphosphates), 0.8 μL of 5 μM forward primer, 0.8 μL of 5 μM reverse primer, 0.4 μL of FastPfu polymerase, 0.2 μL of bovine serum albumin (BSA), and 10 ng of template. Double-distilled water was added to make the total volume 20 μL. The PCR was through the following process: 95°C for 3 min, 30 cycles of 95°C for 30 s, 50°C for 30 s and 72°C for 45 s and 72°C for 10 min, followed by 10°C until halted by user. Then, 2% agarose gel electrophoresis (Beijing Liuyi Biological Technology Co., Ltd.), GenElute PCR extraction kit (Sigma-Aldrich, Saint Louis, MO, USA), and a Quantus fluorometer were used to perform identify, purify, and quantify the PCR products, respectively.
Amplicon sequencing and bioinformatics analysis for sequencing data.
A TargetSeq next-generation sequencing (NGS) library prep kit (iGeneTech Biotechnology Co., Ltd., Beijing, China) was used to construct sequencing libraries. Amplicon sequencing was conducted on a Miseq PE300 platform (Illumina). The raw sequence was quality-controlled using Trimmomatic software (version 0.36). Splicing of paired-end reads was implemented by FLASh software (https://ccb.jhu.edu/software/FLASH/index.shtml). Uparse software (version 7.1) was used to conducted operational taxonomic unit (OTU) clustering. The nonrepetitive sequence was extracted from the optimized sequence (http://drive5.com/usearch/manual/dereplication.html). To compare each sample at the same OTU sequence number level, the sequences of all milk samples were randomly sampled and unified to the minimum sequence number. After removal of nonrepetitive singletons, the OTU with >97% similarity was clustered. In the process of OTU clustering, the chimeras were removed to obtain representative sequences of OTU (63). To obtain the taxonomic information of each OTU, the Ribosomal Database Project (RDP) classifier Bayesian algorithm was carried out to perform taxonomic analysis on OTU representative sequences with >97% similarity (66). Silva (release 132; http://www.arb-silva.de) was used to provide databases of aligned 16S rRNA sequences. A rarefaction curve and a Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) were drawn with the package R. Alpha diversity was analyzed with mothur software (version 1.30.1; https://www.mothur.org/wiki/Download_mothur) to show the richness and diversity of microorganisms. To visualize the degree of similarity and difference of community composition in different samples, principal-coordinate analysis (PCoA) based on Bray-Curtis analysis and a weighted UniFrac distance algorithm was conducted using Qiime (version 1.9.1 http://qiime.org/install/index.html) (67). SciPy in Python (version 1.0.0) was used to perform the hierarchical clustering analysis (HCA) on abundances of bacteria with the top 60 relative abundances in milk samples among 5 groups. The Kruskal-Wallis H test was used to analyze the significant differential microbiota among multiple groups. The false-discovery rate (FDR) been applied in multiple test correction. Further, linear discriminant analysis effect size (LEfSe) analysis (https://huttenhower.sph.harvard.edu/galaxy/) was conducted to evaluate the influence of these differential bacteria on the differences among groups.
Milk sample preparation for LC-MS analysis.
After thawing and mixing well, an aliquot of 100 μL of each milk sample was pipetted into a 1.5-mL centrifuge tube with 200 μL of chromatographic-grade acetonitrile and 200 μL of chromatographic-grade methanol (Thermo Fisher Scientific, Waltham, USA), which were thoroughly vortex-mixed for 30 s and extracted by ultrasonography (SBL-10TD ultrasonic cleaner; Xinzhi Biotechnology Co., Ltd., Ningbo, China) at 5°C and 40 KHz for 30 min. Further, the samples were centrifuged for 15 min. After the supernatant was removed, the sediment at the bottom was blow-dried with N2 (JXDC-20 nitrogen purge instrument; Jingxin Industrial Development Co., Ltd., Shanghai, China); 60 μL chromatographic-grade acetonitrile and 60 μL purified water (Thermo Fisher Scientific) were added to redissolve the mixture, and ultrasonic extraction (SBL-10TD ultrasonic cleaner, Xinzhi Biotechnology Co., Ltd.) was performed at 5°C and 40 KHz for 5 min. Next, the samples were centrifuged at 13,000 × g for 5 min at 4°C, and the supernatant was transferred to an injection vial for LC-MS analysis. Meanwhile, to assess the repeatability of metabolomics data, the equal-volume samples were pipetted from any 10 samples and were mixed to make a quality control (QC) sample. A total of 4 QC samples were set in positive and negative ion modes.
LC-MS metabolomic profile processing.
The instrument platform for LC-MS analysis was the ultraperformance liquid chromatography tandem time of flight system mass spectrometry (UPLC-triple TOF MS; AB SCIEX). A 10-μL amount of each milk sample was separated through a high-strength silica (HSS) T3 column (100 mm by 2.1 mm inside diameter [i.d.], 1.8 μm; Waters, MA, USA) with a flow rate of 0.40 mL/min and column temperature of 40°C. The chromatographic conditions were as follows: mobile phase A, chromatographic-grade ultrapure water containing 0.1% chromatographically pure formic acid; mobile phase B, chromatographically pure acetonitrile and isopropanol (1:1) containing 0.1% formic acid. The separation gradient was as follows: from 0 to3 min, 95 to 80% A, and 5 to 20% B; from 3 to 9 min, 80 to 5% A and 20 to 95% B; from 9 to13 min, 5% A and 95% B was maintained; 5 to 95% A and 95 to 5% B over 13 to 16 min. The MS conditions were as follows: the mass signal was collected through positive (+) and negative (–) ion scan patterns with a mass scan range of 50 to 1,000 m/z; the ion spray voltage was 5 kV (+) and 4 kV (–); the declustering potential was 80 V; the heating temperature of the ion source was 500°C, and the cyclic collision energy was set from 20 to 60 V.
Metabolome data analyses.
LC-MS raw data were processed using ProGenesis Qi software (Waters, MA, USA) for baseline filtering, peak identification, integral pretreatment, retention time correction, and peak alignment. Finally, a data matrix with retention time, mass to charge ratio (m/z), and peak strength was obtained. To reduce trial error, the sum normalization method was used to normalize the response intensity of the sample mass spectrum peak. Meanwhile, the variate with relative standard deviation of >30% of the QC sample was deleted to get the final data matrix for subsequent analysis.
The MS information was matched with the Human Metabolome Database (HMDB; version 4.0; http://www.hmdb.ca/) to obtain metabolite information. The ropls package in R (version 1.6.2) was used for principal-component analysis (PCA) and orthogonal least partial square discriminant analysis (OPLS-DA). The stability of the model was evaluated by response permutation testing (RPT). The identification of significantly differential metabolites through pairwise comparison was determined based on the variable importance in projection (VIP) obtained from the OPLS-DA model and the P value of Student’s t test, which was corrected through FDR. A metabolite with a VIP of >1.5-fold change (FC) >1 or <0.67 and an FDR-adjusted P value of <0.05 was recognized as a significantly differential metabolite. The multigroup difference significance test used was the Kruskal-Wallis H test. The P value was corrected through FDR. The metabolic pathways of different metabolites involved were annotated through KEGG database (https://www.kegg.jp/kegg/pathway.html). SciPy in Python (version 1.0.0) was used to conduct the pathway enrichment analysis. To depict the ability of different metabolites to differentiate among groups, a receiver operator characteristic curve (ROC) was plotted with the true-positive rate (sensitivity) as the ordinate and the false-positive rate (specificity) as the abscissa. The area under the curve (AUC) was used to measure classification effectiveness. The threshold value of the AUC was from 0 to 1. AUC values close to 1 indicate a satisfied classifier (68). The ROC analysis was implemented through SPSS Statistics (version 22.0; IBM). Spearman’s correlation analysis was conducted using the SciPy in Python (version 1.0.0). The threshold value of correlation coefficient (r) was from −1 to 1. The r < 0 represented a negative correlation, while r > 0 represented a positive correlation. The absolute value range of r and the degree of correlation were as follows: 0 to 2 indicated very weak or no correlation, 0.2 to 0.4 indicated weak correlation, 0.4 to 0.6 indicated moderate correlation, 0.6 to 0.8 indicated strong correlation, and 0.8 to 1.0 indicated extremely strong correlation (8). The significance of correlation was corrected by FDR, and an FDR-adjusted P value of <0.05 was considered a significant correlation.
Statistical analysis.
Data of the initial body condition of SCM cows, milk yield, milk compositions, milk FAs, and the alpha diversity of milk microbiota were analyzed through one-way analysis of variance (ANOVA) with SPSS Statistics (version 22.0; IBM). Statistical significance was accepted at a P value of <0.05; a tendency was accepted at 0.05 < P value < 0.1.
Data availability.
The raw sequencing data have been deposited in the Sequence Read Archive at the National Center for Biotechnology Information database (accession no. PRJNA782675).
ACKNOWLEDGMENTS
We thank the Langfang Academy of Agriculture and Forestry Sciences, Hebei, China, for providing the experimental equipment.
This study was funded by the National Key R&D Program of China (grant no. 2019YFE0125600) and Beijing Dairy Industry Innovation Team (bjcystx-ny-1).
Footnotes
Supplemental material is available online only.
Contributor Information
Junhu Yao, Email: yaojunhu2004@sohu.com.
Liang Yang, Email: yangliang@caas.cn.
Benhai Xiong, Email: xiongbenhai@caas.cn.
Johanna Björkroth, University of Helsinki.
REFERENCES
- 1.Song XB, Huang XP, Xu HY, Zhang C, Chen S, Liu F, Guan S, Zhang S, Zhu K, Wu CM. 2020. The prevalence of pathogens causing bovine mastitis and their associated risk factors in 15 large dairy farms in China: an observational study. Vet Microbiol 247:108757. 10.1016/j.vetmic.2020.108757. [DOI] [PubMed] [Google Scholar]
- 2.Yeiser EE, Leslie KE, Mcgilliard ML, Petersson-Wolfe CS. 2012. The effects of experimentally induced Escherichia coli mastitis and flunixin meglumine administration on activity measures, feed intake, and milk parameters. J Dairy Sci 95:4939–4949. 10.3168/jds.2011-5064. [DOI] [PubMed] [Google Scholar]
- 3.Moyes KM, Larsen T, Srensen P, Ingvartsen KL. 2015. Changes in various metabolic parameters in blood and milk during experimental Escherichia coli mastitis for primiparous Holstein dairy cows during early lactation. J Anim Sci Biotechnol 5:47–57. 10.1186/2049-1891-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang JQ, Tan QW, Fu QY, Zhou YJ, Hu YY, Tang SL, Zhou YT, Zhang JH, Q JJ, Lv Q. 2017. Gastrointestinal microbiome and breast cancer: correlations, mechanisms and potential clinical implications. Breast Cancer 24:220–224. 10.1007/s12282-016-0734-z. [DOI] [PubMed] [Google Scholar]
- 5.Ma C, Sun Z, Zeng B, Huang S, Zhao J, Zhang Y, Sun XQ, Xu J, Wei H, Zhang HP. 2018. Cow-to-mouse fecal transplantations suggest intestinal microbiome as one cause of mastitis. Microbiome 6:200–217. 10.1186/s40168-018-0578-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu XY, Guo J, Zhao CJ, Jiang P, Maimai T, Li YY, Cao YG, Fu YH, Zhang NS. 2020. The gut microbiota contributes to the development of staphylococcus aureus-induced mastitis in mice. ISME J 14:1897–1910. 10.1038/s41396-020-0651-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma C, Zhao J, Xi X, Ding J, Wang H, Zhang H, Kwok LY. 2016. Bovine mastitis may be associated with the deprivation of gut Lactobacillus. Benef Microbes 7:95–102. 10.3920/BM2015.0048. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Nan XM, Zhao YG, Jiang LS, Wang ML, Wang H, Zhang F, Xue FG, Hua DK, Liu J, Yao JH, Xiong BH. 2021. Rumen microbiome structure and metabolites activity in dairy cows with clinical and subclinical mastitis. J Anim Sci Biotechnol 12:36–57. 10.1186/s40104-020-00543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodríguez JM. 2014. The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv Nutr 5:779–784. 10.3945/an.114.007229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young W, Hine BC, Wallace OAM, Callaghan M, Bibiloni R. 2015. Transfer of intestinal bacterial components to mammary secretions in the cow. PeerJ 3:e888. 10.7717/peerj.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Addis MF, Tanca A, Uzzau S, Oikonomou G, Bicalho RC, Moroni P. 2016. The bovine milk microbiota: insights and perspectives from -omics studies. Mol Biosyst 12:2359–2372. 10.1039/C6MB00217J. [DOI] [PubMed] [Google Scholar]
- 12.Nagpal R, Shively CA, Appt SA, Register TC, Michalson KT, Vitolins MZ, Yadav H. 2018. Gut microbiome composition in non-human primates consuming a Western or Mediterranean diet. Front Nutr 5:28–37. 10.3389/fnut.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shively CA, Register TC, Appt SE, Clarkson TB, Uberseder B, Clear KYJ, Wilson AS, Chiba A, Tooze JA, Cook KL. 2018. Consumption of Mediterranean versus western diet leads to distinct mammary gland microbiome populations. Cell Rep 25:47–56. 10.1016/j.celrep.2018.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soto-Pantoja DR, Gaber M, Arnone AA, Bronson SM, Cruz-Diaz N, Wilson AS, Clear KYJ, Ramirez MU, Kucera GL, Levine EA, Lelièvre SA, Chaboub L, Chiba A, Yadav H, Vidi PA, Cook KL. 2021. Diet alters entero-mammary signaling to regulate the breast microbiome and tumorigenesis. Cancer Res 81:3890–3904. 10.1158/0008-5472.CAN-20-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leblanc ADMD, Matar C, Thériault C, Perdigón G. 2005. Effects of milk fermented by Lactobacillus helveticus R389 on immune cells associated to mammary glands in normal and a breast cancer model. Immunobiology 210:349–358. 10.1016/j.imbio.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 16.Jimenez E, Fernandez L, Maldonado A, Martin R, Olivares M, Xaus J, Rodriguez JM. 2008. Oral administration of Lactobacillus strains isolated from breast milk as an alternative for the treatment of infectious mastitis during lactation. Appl Environ Microbiol 74:4650–4655. 10.1128/AEM.02599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A, Zapata RC, Adel P, Reidelberger RD, Chelikani PK. 2018. Inulin fiber dose-dependently modulates energy balance, glucose tolerance, gut microbiota, hormones and diet preference in high-fat fed male rats. J Nutr Biochem 59:142–152. 10.1016/j.jnutbio.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 18.Healey G, Murphy R, Butt C, Brough L, Whelan K, Coad J. 2018. Habitual dietary fibre intake influences gut microbiota response to an inulin-type fructan prebiotic: a randomised, double-blind, placebo-controlled, cross-over, human intervention study. Br J Nutr 12:176–189. 10.1017/S0007114517003440. [DOI] [PubMed] [Google Scholar]
- 19.Guo L, Xiao P, Zhang X, Yang Y, Yang M, Wang T, Lu H, Tian H, Wang H, Liu J. 2021. Inulin ameliorates schizophrenia via modulation of the gut microbiota and anti-inflammation in mice. Food Funct 12:1156–1175. 10.1039/D0FO02778B. [DOI] [PubMed] [Google Scholar]
- 20.Ray K. 2018. Gut microbiota: filling up on fibre for a healthy gut. Nat Rev Gastroenterol Hepatol 15:67. 10.1038/nrgastro.2018.2. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Nan XM, Zhao YG, Jiang LS, Wang H, Zhang F, Hua DK, Liu J, Yao JH, Yang L, Luo QY, Xiong BH. 2021. Dietary supplementation of inulin ameliorates subclinical mastitis via regulation of rumen microbial community and metabolites in dairy cows. Microbiol Spectr 8:e00105-21. 10.1128/Spectrum.00105-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Nan XM, Zhao YG, Wang H, Wang ML, Jiang LS, Zhang F, Xue FG, Hua DK, Li KM, Liu J, Yao JH, Xiong BH. 2020. Coupling 16S rDNA sequencing and untargeted mass spectrometry for milk microbial composition and metabolites from dairy cows with clinical and subclinical mastitis. J Agric Food Chem 68:8496–8508. 10.1021/acs.jafc.0c03738. [DOI] [PubMed] [Google Scholar]
- 23.Xi XM, Kwok LY, Wang YN, Chen M, Mi ZH, Zhang HP. 2017. Ultra-performance liquid chromatography-quadrupole-time of flight mass spectrometry MSE-based untargeted milk metabolomics in dairy cows with subclinical or clinical mastitis. J Dairy Sci 100:4884–4896. 10.3168/jds.2016-11939. [DOI] [PubMed] [Google Scholar]
- 24.Zhao XH, Gong JM, Zhou S, Liu CJ, Qu MR. 2014. The effect of starch, inulin, and degradable protein on ruminal fermentation and microbial growth in rumen simulation technique. Ital J Anim Sci 13:3121–3127. 10.4081/ijas.2014.3121. [DOI] [Google Scholar]
- 25.Tian K, Liu JH, Sun YW, Wu YJ, Chen JC, Zhang RM, He TL, Dong GZ. 2019. Effects of dietary supplementation of inulin on rumen fermentation and bacterial microbiota, inflammatory response and growth performance in finishing beef steers fed high or low-concentrate diet. Anim Feed Sci Tech 258:114299–114311. 10.1016/j.anifeedsci.2019.114299. [DOI] [Google Scholar]
- 26.Nousiainen J, Shingfield KJ, Huhtanen P. 2004. Evaluation of milk urea nitrogen as a diagnostic of protein feeding. J Dairy Sci 87:386–398. 10.3168/jds.S0022-0302(04)73178-1. [DOI] [PubMed] [Google Scholar]
- 27.Umucalilar HD, Gulsen N, Hayirli A, Alata MS. 2010. Potential role of inulin in rumen fermentation. Rev Méd Vét 161:3–9. [Google Scholar]
- 28.Smith GH, McCarthy S, Rook JA. 1974. Synthesis of milk fat from β-hydroxybutyrate and acetate in lactating goats. J Dairy Res 41:175–191. 10.1017/s0022029900019609. [DOI] [PubMed] [Google Scholar]
- 29.Emmanuel B, Kennelly JJ. 1985. Measures of de novo synthesis of milk components from propionate in lactating goats. J Dairy Sci 68:312–319. 10.3168/jds.S0022-0302(85)80827-4. [DOI] [PubMed] [Google Scholar]
- 30.Vlaeminck B, Fievez V, Cabrita ARJ, Fonseca AJM, Dewhurst RJ. 2006. Factors affecting odd- and branched-chain fatty acids in milk: a review. Anim Feed Sci Tech 131:389–417. 10.1016/j.anifeedsci.2006.06.017. [DOI] [Google Scholar]
- 31.Jami E, White BA, Itzhak M, Heimesaat MM. 2014. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS One 9:e85423. 10.1371/journal.pone.0085423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031. 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 33.Pitta DW, Indugu N, Vecchiarelli B, Rico DE, Harvatine KJ. 2018. Alterations in ruminal bacterial populations at induction and recovery from diet-induced milk fat depression in dairy cows. J Dairy Sci 101:295–309. 10.3168/jds.2016-12514. [DOI] [PubMed] [Google Scholar]
- 34.Vlaeminck B, Fievez V, Demeyer D, Dewhurst RJ. 2006. Effect of forage: concentrate ratio on fatty acid composition of rumen bacteria isolated from ruminal and duodenal digesta. J Dairy Sci 89:2668–2678. 10.3168/jds.S0022-0302(06)72343-8. [DOI] [PubMed] [Google Scholar]
- 35.Cabrita ARJ, Fonseca AJM, Dewhurst RJ, Gomes E. 2003. Nitrogen supplementation of corn silages. 2. Assessing rumen function using fatty acid profiles of bovine milk. J Dairy Sci 86:4020–4032. 10.3168/jds.S0022-0302(03)74013-2. [DOI] [PubMed] [Google Scholar]
- 36.Jánosi S, Kulcsár M, Kóródi P, Katái L, Reiczigel J, Dieleman SJ, Nikolic JA, Sályi G, Ribiczey-Szabó P, Huszenicza G. 2003. Energy imbalance related predisposition to mastitis in group-fed high-producing postpartum dairy cows. Acta Vet Hung 51:409–424. 10.1556/AVet.51.2003.3.14. [DOI] [PubMed] [Google Scholar]
- 37.Luick JR, Smith LM. 1963. Fatty acid synthesis during fasting and bovine ketosis1. J Dairy Sci 46:1251–1255. 10.3168/jds.S0022-0302(63)89254-1. [DOI] [Google Scholar]
- 38.Gross J, Dorland H, Bruckmaier RM, Schwarz FJ. 2011. Milk fatty acid profile related to energy balance in dairy cows. J Dairy Res 78:479–488. 10.1017/S0022029911000550. [DOI] [PubMed] [Google Scholar]
- 39.Patel SH, Vaidya YH, Patel RJ, Pandit RJ, Joshi CG, Kunjadiya AP. 2017. Culture independent assessment of human milk microbial community in lactational mastitis. Sci Rep 7:7804–7815. 10.1038/s41598-017-08451-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Angelopoulou A, Holohan R, Rea MC, Warda AK, Hill C, Ross RP. 2019. Bovine mastitis is a polymicrobial disease requiring a polydiagnostic approach. Int Dairy J 99:104539. 10.1016/j.idairyj.2019.104539. [DOI] [Google Scholar]
- 41.Qiao J, Kwok L, Zhang J, Gao P, Zheng Y, Guo Z, Hou Q, Hou D, Huang W, Zhang H. 2015. Reduction of Lactobacillus in the milks of cows with subclinical mastitis. Benef Microbes 6:485–490. 10.3920/BM2014.0077. [DOI] [PubMed] [Google Scholar]
- 42.Watts JL, Lowery DE, Teel JF, Rossbach S. 2000. Identification of Corynebacterium bovis and other Coryneforms isolated from bovine mammary glands. J Dairy Sci 83:2373–2379. 10.3168/jds.S0022-0302(00)75126-5. [DOI] [PubMed] [Google Scholar]
- 43.Mahenthiralingam E, Baldwin A, Dowson CG. 2008. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J Appl Microbiol 104:1539–1551. 10.1111/j.1365-2672.2007.03706.x. [DOI] [PubMed] [Google Scholar]
- 44.Zughaier SM, Ryley HC, Jackson SK. 1999. Lipopolysaccharide (LPS) from Burkholderia cepacia is more active than LPS from Pseudomonas aeruginosa and Stenotrophomonas maltophilia in stimulating tumor necrosis factor alpha from human monocytes. Infect Immun 67:1505–1507. 10.1128/IAI.67.3.1505-1507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macy JM, Ljungdahl LG, Gottschalk G. 1978. Pathway of succinate and propionate formation in Bacteroides fragilis. J Bacteriol 134:84–91. 10.1128/jb.134.1.84-91.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Björck I, Bäckhed F. 2015. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab 22:971–982. 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Macpherson AJ, Uhr T. 2004. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303:1662–1665. 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 48.Espeche MC, Pellegrino M, Frola I, Larriestra A, Bogni C, Nader-Macías MEF. 2012. Lactic acid bacteria from raw milk as potentially beneficial strains to prevent bovine mastitis. Anaerobe 18:103–109. 10.1016/j.anaerobe.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Mayo B, Sinderen DV. 2010. Bifidobacteria genomics and molecular aspects. Caister Academic, Norfolk, UK. [Google Scholar]
- 50.Nagahata H, Mukai T, Natsume Y, Okuda M, Ando T, Hisaeda K, Gondaira S, Higuchi H. 2020. Effects of intramammary infusion of Bifidobacterium breve on mastitis pathogens and somatic cell response in quarters from dairy cows with chronic subclinical mastitis. Anim Sci J 91:e13406. 10.1111/asj.13406. [DOI] [PubMed] [Google Scholar]
- 51.Rose DM, Giri SN, Wood SJ, Cullor JS. 1989. Role of leukotriene B4 in the pathogenesis of Klebsiella pneumoniae-induced bovine mastitis. Am J Vet Res 50:915–918. [PubMed] [Google Scholar]
- 52.Thompson DA, Hammock BD. 2007. Dihydroxyoctadecamonoenoate esters inhibit the neutrophil respiratory burst. J Biosci 32:279–291. 10.1007/s12038-007-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koon HW, Pothoulakis C. 2006. Immunomodulatory properties of substance p: the gastrointestinal system as a model. Ann N Y Acad Sci 1088:23–40. 10.1196/annals.1366.024. [DOI] [PubMed] [Google Scholar]
- 54.Gudding R. 1982. Increased free fatty acid concentrations in mastitic milk. J Food Prot 45:1143–1144. 10.4315/0362-028X-45.12.1143. [DOI] [PubMed] [Google Scholar]
- 55.Jorgensen GN, Larson BL. 1968. Conversion of phenylalanine to tyrosine in the bovine mammary secretory cell. Biochim Biophys Acta 165:121–126. 10.1016/0304-4165(68)90196-7. [DOI] [PubMed] [Google Scholar]
- 56.Knarreborg A, Miquel N, Granli T, Jensen BB. 2002. Establishment and application of an in vitro methodology to study the effects of organic acids on coliform and lactic acid bacteria in the proximal part of the gastrointestinal tract of piglets. Anim Feed Sci Tech 99:131–140. 10.1016/S0377-8401(02)00069-X. [DOI] [Google Scholar]
- 57.Pallister T, Jackson MA, Martin TC, Zierer J, Jennings A, Mohney RP, MacGregor A, Steves CJ, Cassidy A, Spector TD, Menni C. 2017. Hippurate as a metabolomic marker of gut microbiome diversity: modulation by diet and relationship to metabolic syndrome. Sci Rep 7:13670–13679. 10.1038/s41598-017-13722-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wada S, Kiyono T, Fukunaga S, Aoi W, Higashi A. 2018. Japanese rice wine (sake)-derived pyroglutamyl peptides have anti-inflammatory effect on dextran sulfate sodium (DSS)-induced acute colitis in mice. Free Rad Bio Med 120:S141. 10.1016/j.freeradbiomed.2018.04.465. [DOI] [Google Scholar]
- 59.Mu W, Yu Y, Jia J, Tao Z, Bo J. 2010. Production of 4-hydroxyphenyllactic acid by Lactobacillus sp. SK007 fermentation. J Biosci Bioeng 109:369–371. 10.1016/j.jbiosc.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 60.Rossi A, Elia G, Santoro MG. 1997. Inhibition of nuclear factor κB by prostaglandin A1: an effect associated with heat shock transcription factor activation. Proc Natl Acad Sci USA 94:746–750. 10.1073/pnas.94.2.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee EJ, Hahn YI, Kim SJ, Surh YJ, Na HK. 2017. Abstract 2231: 15-keto prostaglandin E2 inhibits STAT3 signaling in H- Ras transformed mammary epithelial cells. Cancer Res 77:2231–2231. 10.1158/1538-7445.AM2017-2231.28235762 [DOI] [Google Scholar]
- 62.Samant GV, Sylvester PW. 2006. Gamma-tocotrienol inhibits ErbB3-dependent PI3K/AKt mitogenic signalling in neoplastic mammary epithelial cells. Cell Prolif 39:563–574. 10.1111/j.1365-2184.2006.00412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yano H, Oyanagi E, Kato Y, Samejima Y, Sasaki J, Utsumi K. 2010. L-carnitine is essential to β-oxidation of quarried fatty acid from mitochondrial membrane by PLA2. Mol Cell Biochem 342:95–100. 10.1007/s11010-010-0472-z. [DOI] [PubMed] [Google Scholar]
- 64.Pu J, Vinitchaikul P, Gu Z, Mao H, Zhang F. 2021. The use of metabolomics to reveal differences in functional substances of milk whey of dairy buffaloes raised at different altitudes. Food Funct 12:5440–5450. 10.1039/d0fo03231j. [DOI] [PubMed] [Google Scholar]
- 65.Wang B, Tu Y, Zhao SP, Hao YH, Liu JX, Liu FH, Xiong BH, Jiang LS. 2017. Effect of tea saponins on milk performance, milk fatty acids, and immune function in dairy cow. J Dairy Sci 100:8043–8052. 10.3168/jds.2016-12425. [DOI] [PubMed] [Google Scholar]
- 66.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 67.Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, Collman RG, Bushman FD, Li HZ. 2012. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 28:2106–2113. 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qamar K, Khan MA, Saleem S. 2018. Diagnostic accuracy study of internal assessment using receiver operator characteristic curve analysis. J Pak Med Assoc 68:721–724. [PubMed] [Google Scholar]
- 69.Sjaunja LO, Baevre L, Junkkarinen L, Pedersen JA. 1990. Nordic proposal for an energy corrected milk (ECM) formula. 27th Session International Committee for Recording and Productivity of Milk Animals, 2–6 July, Paris, France. [Google Scholar]
- 70.Gaines WL. 1928. The energy basis of measuring milk yield in dairy cows. University of Illinois Agricultural Experiment Station, Urbana, IL. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S15 and Fig. S1 to S8. Download aem.02059-21-s0001.pdf, PDF file, 4.3 MB (4.3MB, pdf)
Data Availability Statement
The raw sequencing data have been deposited in the Sequence Read Archive at the National Center for Biotechnology Information database (accession no. PRJNA782675).