ABSTRACT
This study aims to probe the mechanism by which circPVT1 in lung cancer (LC)-derived exosomes (Exos) promotes proliferation, invasion and migration of LC cells by regulating macrophage polarization through miR-124-3p/EZH2 axis. The expressions of circPVT1 in blood-derived Exos extracted from lung adenocarcinoma (LA) patients were measured. Loss or gain-of-function experiments of circPVT1 and miR-124-3p were carried out to evaluate the effects of circPVT1 in LC-derived Exos and miR-124-3p on macrophage polarization toward M2 phenotype, whichi found that incubation of Exo-A with macrophages induced macrophage polarization to M2 type and M2-polarized macrophages co-incubated with A549 cells enhanced the biological function of LC cells. Co-incubation with M+ Exo-A-oecircPVT1 increased the proliferation, migration and invasion abilities of LC cells, while coculture with M+ Exo-A-si-circPVT1 reversed these abilities. The verification among circPVT1, miR-124-3p and EZH2 showed that miR-124-3p was negatively related to circPVT1 and EZH2, and EZH2 was positively related to circPVT1. CircPVT1 in LC-derived Exos increased EZH2 expression through inhibiting miR-124-3p expression level in macrophage. Taken togther, exosomal circPVT1 derived from LC mediates macrophage polarization via the miR-124-3p/EZH2 axis to potentiate LC cells’ proliferation, invasion and migration.
KEYWORDS: Lung cancer, circPVT1, hsa-miR-124-3p, EZH2, proliferation, migration, invasion
Introduction
Lung cancer (LC), a heterogeneous disease, is the leading cause of cancer-related death in both men and women worldwide [1,2]. Commonly, LC is histologically classified into two main types with distinct clinical features, namely, non-small cell lung cancer, accounting for around 85% of all cases, and small cell lung cancer, approximately in the proportion of 15% [3]. The epidemiologic risk factors for lung cancer include cigarette smoking, family history, prior respiratory diseases, environmental and occupational factors, physical activity, diet, and nutrition [4]. The 5-y survival in populations with LC varies from 4% to 17% depending on the stage and regional differences [5]. Depending on the type of malignancy and stage at the time of diagnosis, LC treatment often involves a combination of surgery, chemotherapy and radiation therapy as well as a systemic chemotherapy treatment [6]. However, the therapeutic effect remains largely unsatisfactory. Hence, it is of vital importance for the development of effective drugs to explore the mechanism underlying the carcinogenesis of LC.
Exosomes (Exos), usually 30 –150 nm in diameter, are nanoscale extracellular vesicles of endocytic origin and are secreted by the majority of cell types [7]. Exos act as message transmitters in intercellular communication by delivering lipids, circular RNAs (circRNAs) and microRNAs (miRNA) [8]. In colorectal cancer, exosomal circRNA may prompt glycolysis to contribute to chemoresistance through the miR-122-PKM2 axis [9]. CircRNA is a novel subtype of endogenous noncoding RNAs featuring stable structure and high tissue-specific expression, and arising from a particular alternative splicing mechanism of precursor mRNAs [10,11]. CircPVT1, an oncogenic non-coding RNA, has reported to serve as a promising therapeutic target for non-small cell lung cancer patients [12]. M2-polarized macrophages are commonly called tumor-associated macrophages, and these promote cancer cell growth, invasion and metastasis [13]. Thus, M2-polarized macrophages are considered to be a potential target for adjuvant anticancer therapies. Furthermore, in tumor microenvironment, tumor-derived exosomes can be ingested by macrophages and ultimately promote tumor progression and metastasis [14]. In cigarette smoke-related lung diseases, Exos and exosomal miRNAs is responsible for the cross-talk between alveolar macrophages and lung epithelial cells so as to maintain lung homeostasis [15]. Nevertheless, how exosomal circPVT1 can regulate macrophage polarization LC remains largely unexplored.
One of the main ways that circRNAs perform their regulation of gene expression is by interacting with miRNAs and serving as an efficient miRNA sponge [16]. For example, circPVT1 facilitates the resistance of gastric cancer cells to paclitaxel by miR-124-3p-mediated ZEB1 [17]. Recently, miR-124-3p, a form of mature miR-124, has been introduced to be dysregulated in LC [18]. The miR-124-3p/EZH2 axis plays a pivotal role in various types of tumors [19,20]. However, whether the biological function of circPVT1 involves in LC through the mediation of the miR-124-3p/EZH2 axis remains not fully understood.
Herein, our results exhibited high circPVT1 expression in blood-derived Exos extracted from lung adenocarcinoma (LA) patients and low miR-124-3p expression in LA tissues. Importantly, circPVT1 can bind to miR-124-3p as a miRNA sponge to upregulate EZH2 expression, thereby expediting the macrophage polarization toward M2 and boosting the proliferation, migration and invasion of LC cells. Overall, our findings emphasized the importance of the circPVT1/miR-124-3p/EZH2 axis in LC progression.
Materials and Methods
Ethical statement
The study was conducted under the approval of the Ethics Committee of the Second Hospital of Shanxi Medical University. All participating patients signed informed consent documentation. All experiments were conducted in strict accordance with the Declaration of Helsinki.
Subjects and sample collection
From 2018 April to 2020 September, a total of 60 pathologically diagnosed lung adenocarcinoma (LA) patients were recruited consecutively from the Second Hospital of Shanxi Medical University and People’s Hospital of Shanxi Province. The blood samples from 60 LA patients and 60 healthy volunteers were collected. The LA tissues and adjacent tumor tissues were collected from 60 patients undergoing surgery in the Second Hospital of Shanxi Medical University and People’s Hospital of Shanxi Province. The collected tissues were immediately frozen in liquid nitrogen until further extraction of RNA or protein.
Cell culture
The human LA cell line A549, human bronchial epithelial (HBE) cells and human monocytic cell line THP-1 were obtained from Shanghai Institutes for Biological Science, CAS. Cells were given Dulbecco’s Modified Eagle Medium (Gibco, Grand Island, NY, USA) covering 10% fetal bovine serum (FBS), 1% penicillin and 1% streptomycin in an 37°C incubator with 5% CO2. THP-1 was subjected to 48 h of treatment with 100 ng/mL phorbol-12-myristate-13-acetate (PMA, Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) for differentiation into macrophages.
Extraction of Exo by differential centrifugation
The culture medium was replaced with Exo-depleted FBS for 48 h of incubation at 37°C with 5% CO2 when the confluence of HBE or A549 cells reached 80–90%. Exos were harvested by differential centrifugation: the supernatant of culture medium was obtained followed by low-speed centrifugation (300 g × 10 min, 2,000 g × 10 min) and ultracentrifugation (10,000 g × 30 min, 100,000 g × 70 min) at 4°C. Cell precipitates were resuspended in phosphate-buffered saline (PBS) and then subjected to ultracentrifugation (100,000 g × 70 min). Then, cell precipitates were resuspended in 200 µL PBS prior to protein concentration detection by using the BCA method or stored at −80°C for use.
Transmission electron microscopy (TEM) observation
Ten microliters of Exo suspension was diluted by PBS and added to the 2-mm formvar-coated copper grid for 1 min of standing at room temperature. After the removal of excess liquid with filter paper, Exos were negatively stained with 3% (w/v) sodium phosphotungstate solution (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) for 5 min. Exos were washed in PBS and dried naturally in air prior to observation under a TEM (H-8100, Hitachi, Tokyo, Japan) (Scare bar = 0.2 μm) at 80–120 kV.
Measurement of Exo particle size
Exos were diluted in the proportion of 1:100, and 1 mL of Exo solution was added in a quartz colorimetric utensil. Then, the distribution of particle size was analyzed by a dynamic light-scattering instrument (NanoS90, Marvin, UK).
Fluorescence labeling of Exos for detecting the uptake of Exos
Exos were stained with green fluorescent linker dye PKH67 (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) according to the manufacturer’s instructions. The PKH67-labeled Exos were resuspended and incubated with unstained macrophages for 4 h to analyze the uptake of Exos. Pictures were obtained by using a fluorescence microscope.
Cell transfection and grouping
The si-circPVT1 (2 μg), oecircPVT1 (2 μg), miR-124-3p mimic (30 nM), miR-124-3p inhibitor (30 nM), si-NC, vector, mimic NC and inhibitor NC were synthesized and constructed by Shanghai GenePharma Co., Ltd. (Shanghai, China). Transfection was conducted utilizing the LipoFiterTM transfection kit (Hanbio Biotech, Shanghai, China).
After cell transfection, the A549 cells were grouped into the Blank group, si-circPVT1 group, si-NC group and vector group. The extracted Exos after 24 h of transfection were classified into Exo-A, Exo-A-si-circPVT1, Exo-A-si-NC, Exo-A-oecircPVT1 and Exo-A-vector groups.
Following transfection or cotransfection, the macrophages were assigned into M/Blank, M/miR-124-3p mimic, M/mimic NC, M/si-circPVT1, M/si-NC, M/oecircPVT1, M/vector, M/inhibitor, M/inhibitor NC and M/inhibitor-miR-124-3p+si-circPVT1 groups. Each experiment was independently repeated three times. After 24 h of transfection, the subsequent experiments were conducted.
qRT-PCR
Total RNA from A549 cells or macrophages was extracted according to instructions of the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The concentration and quality of total RNA were determined and adjusted to the appropriate concentration for reverse transcription. Random primers were applied as reverse transcription primers, and the operation was executed according to the instructions of the reverse transcription kit (TaKaRa, Tokyo, Japan) and random primers. Gene expression was detected by using a LightCycler 480 real-time polymerase chain reaction instrument (Roche, Indianapolis, IN, USA). The reaction conditions were consistent with the instructions of the fluorescent quantitative polymerase chain reaction kit (SYBR Green Mix, Roche Diagnostics, Indianapolis, IN). The thermal cycle parameters were 95°C for 5 min, followed by 40 cycles of 95°C for 10 s, 60°C for 10 s and 72°C for 20 s. Three replicates were run in parallel. The quantification of mRNA and circRNA was normalized to GAPDH, while that of miRNAs was normalized to U6. All primers are listed in Table 1. The fold changes were calculated by the 2−ΔΔCt method.
Table 1.
Primer sequence information
| Name of primer | Sequences |
|---|---|
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | ACGCTTCACGAATTTGCGT |
| Hsa-miR-124-3p-F | TGTGATGAAAGACGGCACAC |
| Hsa-miR-124-3p-R | CTTCCTTTGGGTATTGTTTGG |
| GAPDH-F | GCAAGGATGCTGGCGTAATG |
| GAPDH-R | TACGCGTAGGGGTTTGACAC |
| CircPVT1-F | GGTTCCACCAGCGTTATTC |
| CircPVT1-R | CAACTTCCTTTGGGTCTCC |
| EZH2-F | ATGGCCAAGACACACCTTCC |
| EZH2-R | GGAGTAGGGGCGTATCTGGT |
| CD68-F CD68-R |
CTTCTCTCATTCCCCTATGGACA GAAGGACACATTGTACTCCACC |
| CD206-F | GGGTTGCTATCACTCTCTATGC |
| CD206-R | TTTCTTGTCTGTTGCCGTAGTT |
| CD163-F | TTTGTCAACTTGAGTCCCTTCAC |
| CD163-R | TCCCGCTACACTTGTTTTCAC |
| Arginase-1-F | GGTTTTTGTTGTTGCGGTGTTC |
| Arginase-1-R | CTGGGATACTGATGGTGGGATGT |
| iNOS-F | AGGGACAAGCCTACCCCTC |
| iNOS-R | CTCATCTCCCGTCAGTTGGT |
| IL1β-F | ATGATGGCTTATTACAGTGGCAA |
| IL1β-R | GTCGGAGATTCGTAGCTGGA |
Western blot
Cells or Exos were lysed with RIPA lysis buffer (Beyotime Biotech, Shanghai, China) and centrifuged to obtain protein samples. The protein concentration was evaluated by BCA kit (Beyotime), and the corresponding volume of protein was mixed with loading buffer (Beyotime) followed by 3 min of denaturation in a boiling-water bath. The 10% SDS-PAGE was configured in complied with the direction of the SDS-PAGE preparation kit (Beyotime) and then was applied to separate proteins. Electrophoresis was conducted at 80 V, and then for 1 –2 h at 120 V once bromphenol blue reached the separation gel. Then, proteins were transferred to membranes at 300 mA for 60 min in an ice-bath. The membranes were rinsed 1 –2 min with washing solution and sealed in the blocking solution at room temperature for 60 min, or sealed overnight at 4°C. The membranes were incubated with the primary antibodies against rabbit anti-human GAPDH (5174S, 1:1000, Cell Signaling, Boston, USA), CD9 (sc-13,118, 1:500, Santa Cruz, Texas, USA), CD81 (sc-166,029, 1:500, Santa Cruz, Texas, USA), TSG101 (sc-7964, 1:500, Santa Cruz, Texas, USA), CD206 (sc-376,108, 1:500, Santa Cruz, Texas, USA), arginase-1 (sc-166,920, 1:500, Santa Cruz, Texas, USA), CD63 (ab134045, 1:1000, Abcam, MA, USA), CD163 (ab182422, 1:1000, Abcam, MA, USA) and EZH2 (ab186006, 1:2000, Abcam, MA, USA) at room temperature in a shaking table for 1 h. Following primary incubation, the membranes were washed with the washing solution for 3 × 10 min and then incubated with the secondary antibody against horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5000, Beijing ComWin Biotech Co., Ltd., Beijing, China) for 1 h at room temperature. Then, the membranes were washed 3 × 10 min with washing solution followed by X-ray film exposure, color development and color fixation. GAPDH served as an internal reference.
Dual-luciferase reporter assay
TargetScan (http://starbase.sysu.edu.cn/agoClipRNA.php?source=circRNA) was employed to predict the binding site of miR-124-3p and circPVT1, and the online software PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_data.html) was applied to predict the binding region of miR-124-3p and EZH2. The mutated type and wild-type sequences (mut-circPVT1, wt-circPVT1, mut-EZH2 and wt-EZH2) were designed and synthesized in accordance with the predicted results, and cloned into luciferase reporter vector (pGL3-Basic, Promega, Madison, WI, USA). The vectors were cotransfected with miR-124-3p mimic or mimic-NC (30 nM, Gene Pharma), respectively, into HEK293T cells. After transfection, the dual-luciferase reporter assay kit (Promega, Madison, WI, USA) was utilized to measure the fluorescence intensity of cells in each group and determine the binding of miR-124-3p to circPVT1 and miR-124-3p to EZH2. Cells were grouped into mimic+mut-circPVT1 group, mimic+wt-circPVT1 group, mimic NC+mut-circPVT1 group, mimic NC+wt-circPVT1 group, mimic+mut-EZH2 group, mimic+wt-EZH2 group, mimic NC+mut-EZH2 group and mimic NC+wt-EZH2 group. Three replicates were used for this test.
Transwell assay
Transwell chamber: the THP-1 cells were pipetted into the apical chamber and exposed to PMA to differentiate into macrophages. Then, 100 μg/mL Exos and Exo-free complete medium were aspirated into the apical chamber for 24 h of incubation. The A549 cells were placed on the basolateral chamber, and the apical chamber was washed with PBS when THP-1 cells were induced into macrophages. The two chambers were cocultured for 48 h with Exo-free serum complete medium. The A549 cells in the basolateral chamber were digested with trypsin and resuspended in serum-free culture medium for subsequent experiments.
Matrigel matrix-precoated Transwell chamber: Transwell chamber (Corning, New York, USA) that stored at −20°C and coated with Matrigel was melted at room temperature, and a Transwell or a 24-well plate was added with 0.5 mL serum-free medium and maintained at 37°C in 5% CO2 for 2 h. Then, 1 × 105 A549 cells and 500 μL culture medium covering 1% FBS were aspirated into pretreated Transwell chamber, and 0.75 mL culture medium containing 15% FBS was pipetted into the 24-well plate for 20 h of incubation at 37°C with 5% CO2 maintained constant temperature and humidity. The Transwell chamber was washed with calcium-free PBS twice, followed by 10 min of fixation in 4% paraformaldehyde, 2 × 2 min of PBS washing and 10 min of crystal violet staining. The non-invaded cells and Matrigel matrix were removed using a cotton swab and then washed thrice with PBS. Five fields were selected randomly under a microscope (× 20) to count invaded cells and capture pictures.
Cell scratch assay
The A549 cells were cocultured with macrophages polarized to M2 phenotype, and then A549 cells were made into cell suspension, which was uniformly inoculated on a six-well plate. After 24 h, cells of each group were treated according to the corresponding grouping requirements and incubated at 37°C with 5% CO2. Following incubation, cells were scratched using a 100-µL sterile pipette tip, which was perpendicular to the cells as possible to ensure that the scratch width of each group was basically the same. The cells were washed twice with PBS and then grown in serum-free medium for 24 h of incubation at 37°C gassed with 5% CO2. Photographs were captured at time 0 h and 24 h post-cell scratch, and the cell migration rate was calculated based on changes in scratch distance.
CCK-8 assay
After coculture of A549 cells with macrophages polarized to M2 phenotype, A549 cells in 100 μL suspension were seeded into a ninety-six-well plate. Each sample had three duplicates. Ten microliters of CCK-8 solution (Tokyo, Dojindo, Japan) was added to each well for 1–4 h of incubation after 12, 24, 36 or 48 h of cell culture in an incubator. The absorbance (optical density, OD) was assessed at 450 nm wavelength.
Statistical analysis
Statistical analysis was conducted utilizing GraphPad Prism 7 software, and data are displayed as the mean ± standard deviation (SD). The overall survival rate was displayed by the Kaplan–Meier method, and the difference between the curves was analyzed by the log-rank test. The T-test was employed for comparisons between two groups. The one-way analysis of variance (ANOVA) was adopted followed by Tukey’s multiple comparison tests for comparisons among multiple groups. P values of significance were those less than 0.05.
Results
Isolation and identification of LC-derived Exos
The A549 cell-derived Exos (Exo-A) and HBE cell-derived Exos (Exo-B) were extracted by differential centrifugation. The TEM result observed that there were small vesicles with uniform size and shape in round or oval (Figure 1a). The diameter of particles ranged from 50 to 200 nm, and the particles had the highest proportion at 100 nm (Figure 1b). Western blot displayed high expression of Exo surface protein markers (CD9, CD63, CD81 and TSG101) (Figure 1c). These data indicated the successful extraction of Exos.
Figure 1.
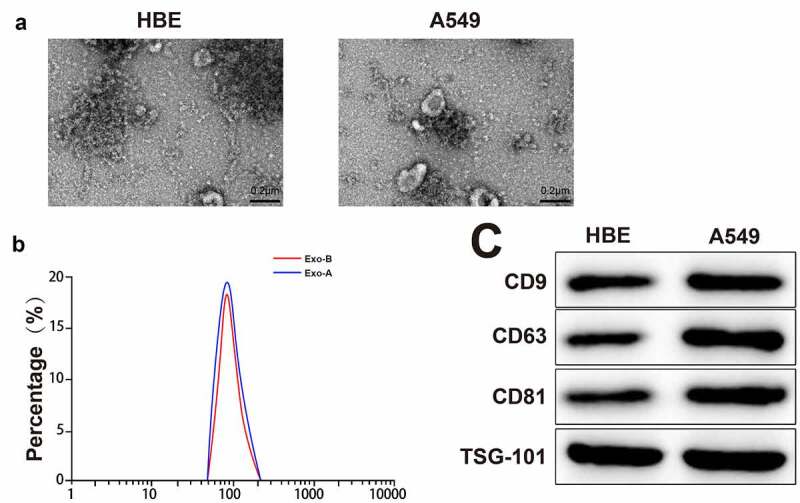
Extraction and identification of LC-derived Exos.
Notes: The morphological characteristic of Exos was observed by TEM (A) (n = 3). The particle size of Exos was analyzed by NanoS90 (B) (n = 3). The expressions of marker proteins of Exos were measured by Western blot (C) (n = 3); ***P < 0.001; LC, lung cancer; Exos, exosomes; TEM, transmission electron microscope.
LC-derived Exos induce M2 phenotype macrophage polarization
After THP-1 cells were stimulated by PMA, we found that THP-1 changed from a suspended state to an adherent state (Figure 2a). The specific marker of macrophage CD68 was determined by Western blot, and the results showed that adherent macrophages had high expression of CD68 (Figure 2b, ***P < 0.001). These findings implied that THP-1 was successfully induced to differentiate into macrophages.
Figure 2.
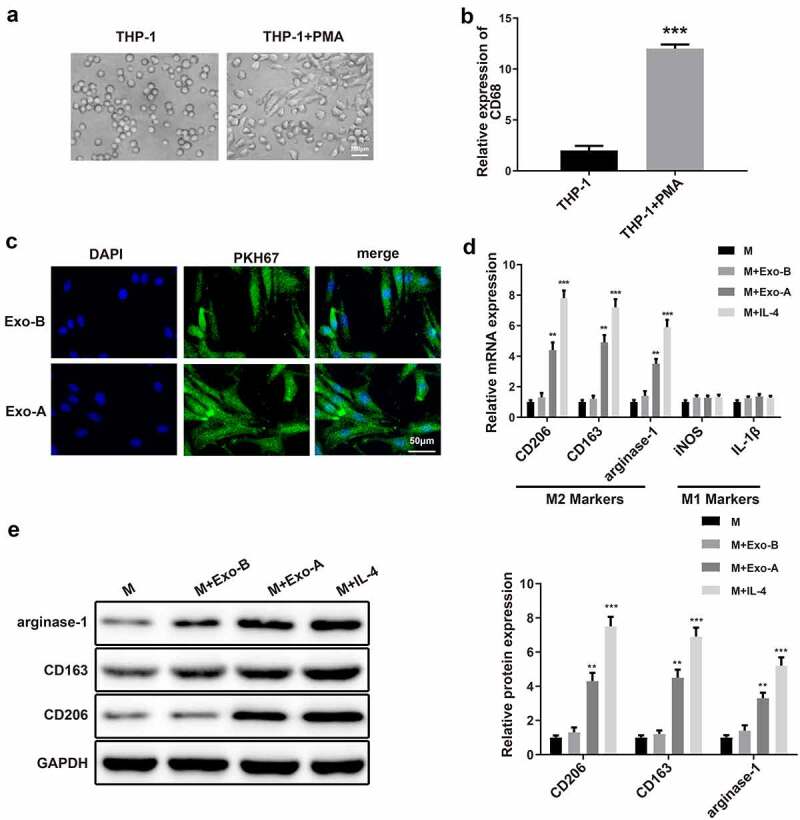
LC-derived Exos induce macrophage polarization toward to M2 phenotype.
Notes: After PMA induction, the morphology of THP-1 cells was observed under a microscope (A) (n = 3). qRT-PCR was applied to detect the expression of macrophage surface marker CD68 (B) and surface markers of M1 (iNOS and IL-1β) and M2 (CD206, CD163 and arginase-1) (D) (n = 3). The uptake of Exos in macrophages was inspected by fluorescence labeling of Exos (C) (n = 3). The protein levels of M2 markers CD206, CD163 and arginase-1 in macrophages were assessed by Western blot (E) (n = 3); **P < 0.01, ***P < 0.001, compared to the M group; LC, lung cancer; Exos, exosomes; PMA, phorbol-12-myristate-13-acetate; M, macrophage.
To explore whether LC-derived Exos affect macrophage polarization, macrophages were incubated with PKH67-labeled Exo-A or PKH67-labeled Exo-B for 4 h. A fluorescence microscope observed that there was a large amount of fluorescence enrichment around the nucleus of macrophages, suggesting that a large number of Exos entered macrophages (Figure 2c). Then, 100 μg/mL Exos or IL-4 (positive control) were exposed to macrophages for 48 h of incubation, and the polarization of macrophages was inspected by measuring the expressions of the surface markers of M1 (iNOS and IL-1β) and M2 (CD206, CD163 and arginase-1). Analyses of qRT-PCR and Western blot manifested that compared with macrophages cultured alone, incubation with Exo-A or IL-4 enhanced expressions of CD206, CD163 and arginase-1 in macrophages (Figure 2d-e, **P < 0.01) and incubation with Exo-B did not change levels of M2 markers. Additionally, expressions of iNOS and IL-1β remained unchanged when macrophages were incubated with IL-4, Exo-A or Exo-B. The above results suggested that LC-derived Exos induce macrophage polarization toward to M2 phenotype.
M2-polarized macrophages enhance the proliferative, migratory and invasive properties of LC cells
To investigate the effect of M2 phenotype macrophage polarization induced by LC-derived Exos on the biological function of LC cells, macrophages polarized to M2 phenotype were cocultured with LC cells. In brief, Exo-A was isolated from A549 cells and then incubated with macrophages to obtain M2-polarized macrophages. Then, M2-polarized macrophages were co-cultured with A549 cells, and the abilities of LC cell proliferation, migration and invasion were measured. CCK-8 assay, Transwell and cell scratch assay results addressed that the elevations in cell proliferation (Figure 3a, **P < 0.01), invasion and migration abilities (Figure 3b–c) were noticed in the A549 + M-Exo-A group rather than the A549 + M group. No obvious differences were observed between the A549 + M-Exo-B and A549 + M group. These findings illustrated that M2 macrophages induced by LC-derived Exos promote the propagation and aggressiveness of LC cells.
Figure 3.
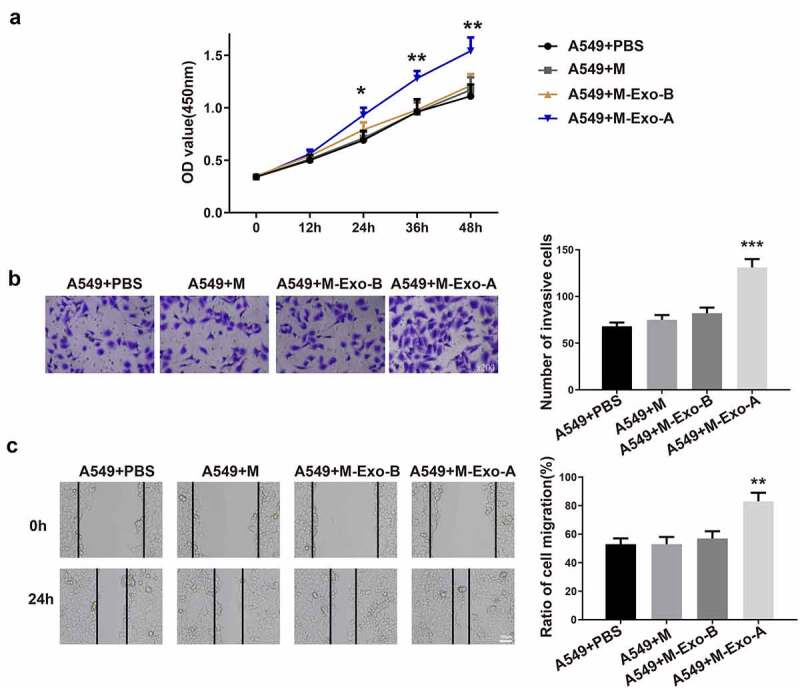
Macrophages polarized to M2 phenotype accelerate the biological process of LC cells.
Notes: Exo was extracted from A549 cells or HBE cells and then incubated with macrophages. Then, M-Exo-A or M-Exo-B was cocultured with A549 cells. The OD value of A549 cells was evaluated by CCK-8 assay (A) (n = 3). The invasion ability of A549 cells was assessed by Transwell (B) (n = 3), and the migration ability of A549 cells was measured by cell scratch assay (C) (n = 3); **P < 0.01, ***P < 0.001, compared to the A549 + M group; LC, lung cancer; Exos, exosomes; M, macrophage; OD, optical density.
High circPVT1 expression in LC-derived Exos
qRT-PCR was utilized to analyze circPVT1 expression in Exo-A and Exo-B. We found that Exo-A had a higher level of circPVT1 than Exo-B (Figure 4a, **P < 0.01). Subsequently, the expression of circPVT1 in blood-derived Exos extracted from 60 healthy volunteers and 60 LA patients was analyzed. The results showed that the circPVT1 level in blood-derived exosomes of LA patients was strikingly higher than that of healthy volunteers (Figure 4b, **P < 0.01). Sixty LA patients were assigned into two groups: the circPVT1 low-expression group (n = 30) and the circPVT1 high-expression group (n = 30) using the median of circPVT1 expression in LA patients (accurate value: 2.79) as the critical value, according to the expression of circPVT1 in blood-derived Exos. Kaplan–Meier survival curve analysis displayed that the overall survival of the circPVT1 high-expression group was shorter than that of the circPVT1 low-expression group (Figure 4c, p = 0.021).
Figure 4.
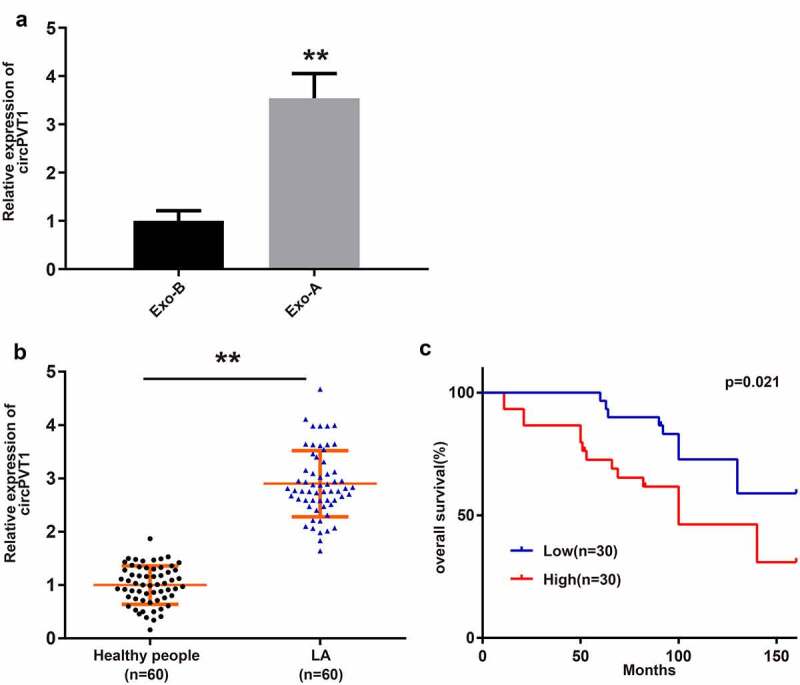
LC-derived Exos have high expression of circPVT1.
Notes: The expression of circPVT1 in HBE cell-derived Exos and A549 cell-derived Exos was performed by qRT-PCR (A) (n = 3). qRT-PCR was used to detect the circPVT1 level in blood-derived Exos from 60 healthy volunteers and 60 LA patients (B) (n = 60). Kaplan–Meier survival curve was employed to analyze the overall survival (C); **P < 0.01, compared to the Exo-B group and Healthy people group; *P < 0.05, compared to the Low group; LC, lung cancer; Exos, exosomes; LA, lung adenocarcinoma.
Exosomal circPVT1 derived from LC induces macrophage polarization toward M2 phenotype and thus enhancing the malignancy of LC cells
To probe the role of circPVT1 in the polarization of macrophages toward M2 type, the A549 cells were transfected with oecircPVT1 or si-circPVT1. qRT-PCR results exhibited that transfection with oecircPVT1 elevated circPVT1 expression in A549 cells and Exo-A, whereas transfection with si-circPVT1 suppressed circPVT1 level (Figure 5a-b, ***P < 0.001). Then, Exos were extracted from the transfected A549 cells (Exo-A-oecircPVT1 or Exo-A-si-circPVT1), followed by coincubation with macrophages (M+ Exo-A-oecircPVT1 or M+ Exo-A-si-circPVT1). The expressions of M2 markers in macrophages incubated with Exo-A-oecircPVT1 or Exo-A-si-circPVT1 were examined by qRT-PCR and Western blot. We discovered that macrophages in the M+ Exo-A-oecircPVT1 group had increased expressions of CD206, CD163 and arginase-1 in comparison to cells in the M+ Exo-A-vector group (Figure 5c-d, **P < 0.01), while macrophages in the M+ Exo-A-si-circPVT1 group possessed decreased levels of M2 makers when compared with cells in the M+ Exo-A-si-NC group (Figure 5c-d, *P < 0.05). CCK-8 assay, Transwell and cell scratch assay displayed that coincubation with M+ Exo-A-oecircPVT1 enhanced the proliferative, invasive and migratory properties of A549 cells, while coincubation with the M+ Exo-A-si-circPVT1 restrained these abilities of A549 cells (Figure 5e-g, *P < 0.05).
Figure 5.
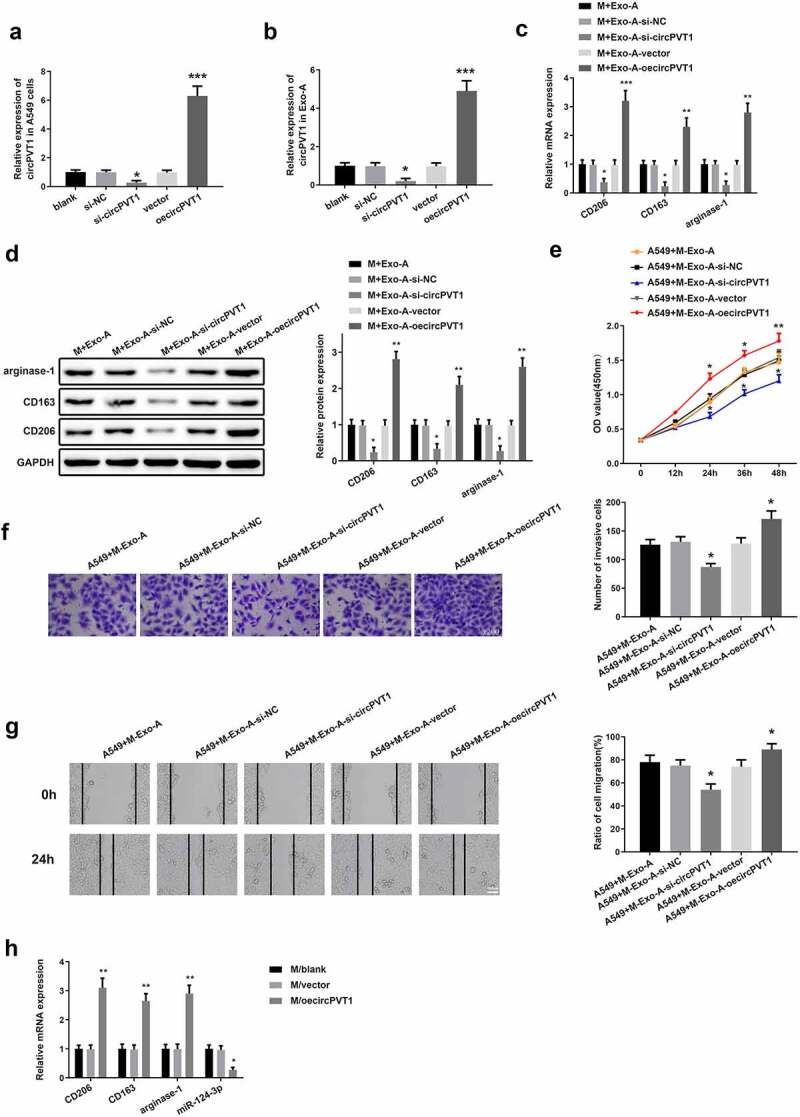
Exosomal circPVT1 stimulates macrophage polarization to M2 type and enhances the biological function of LC cells.
Notes: The A549 cells were transfected with oecircPVT1 or si-circPVT1. Exos were extracted from the transfected A549 cells (Exo-A-oecircPVT1 or Exo-A-si-circPVT1) and coincubated with macrophages (M+ Exo-A-oecircPVT1 or M+ Exo-A-si-circPVT1). Then, qRT-PCR was utilized to measure the mRNA level of circPVT1 in A549 cells (A) and Exo-A (B) as well as the expressions of CD206, CD163 and arginase-1 in M+ Exo-A-oecircPVT1 or M+ Exo-A-si-circPVT1 (C) (n = 3). The protein expressions of M2 markers in M+ Exo-A-oecircPVT1 or M+ Exo-A-si-circPVT1 were detected by Western blot (D) (n = 3). Subsequently, the proliferation, invasion and migration abilities of A549 cells that coincubated with M+ Exo-A-oecircPVT1 or M+ Exo-A-si-circPVT1 were determined by CCK-8 assay (E), Transwell (F) and cell scratch assay (G), respectively (n = 3). The mRNA expressions of M2 markers and miR-124-3p in the transfected macrophages were inspected by qRT-PCR (H) (n = 3); *P < 0.05, ***P < 0.001, compared to the Blank group; *P < 0.05, **P < 0.01, ***P < 0.001, compared to the M+ Exo-A-si-NC or M+ Exo-A-vector group; *P < 0.05, **P < 0.01, compared to the M/Blank group; M, macrophage; LC, lung cancer; Exos, exosomes.
Analysis of qRT-PCR showed that macrophages transfected with oecircPVT1 heightened expressions of M2 markers (Figure 5h, **P < 0.01) and repressed miR-124-3p level in macrophages (Figure 5h, *P < 0.05), suggesting that miR-124-3p may be implicated in the progression of macrophage polarization toward M2 type.
The above data manifested that exosomal circPVT1 may induce macrophage polarizing to M2 phenotype and then facilitate LC cell proliferation, migration and invasion.
CircPVT1 targets miR-124-3p
qRT-PCR was used to detect miR-124-3p expression in LA tissues and adjacent tumor tissues from 60 LA patients. We found that the mRNA level of miR-124-3p in LA tissues was obviously lower than that in adjacent tumor tissues (Figure 6a, **P < 0.01). Additionally, transfection with miR-124-3p inhibitor decreased the level of miR-124-3p in macrophages (Figure 6b, *P < 0.05). Transfection with si-circPVT1 increased miR-124-3p expression in macrophages, while the coeffect of miR-124-3p inhibitor and si-circPVT1 in macrophages reduced miR-124-3p level (Figure 6b, ***P < 0.001). The mutated type sequences and wild type sequences containing the 3ʹUTR region of circPVT1 were constructed (Figure 6c). The binding of miR-124-3p and circPVT1 presented that cotransfection with wt-circPVT1 and miR-124-3p mimic had lower relative luciferase activity than cells cotransfected with wt-circPVT1 and mimic NC (Figure 6d, **P < 0.01). However, the relative luciferase activity in cells cotransfected with mut-circPVT1 and miR-124-3p mimic was not statistically different from that in cells cotransfected with mimic NC and mut-circPVT1 or cells cotransfected with wt-circPVT1 and mimic NC (Figure 6d, **P > 0.01). These findings indicated that circPVT1 may negatively mediate miR-124-3p.
Figure 6.
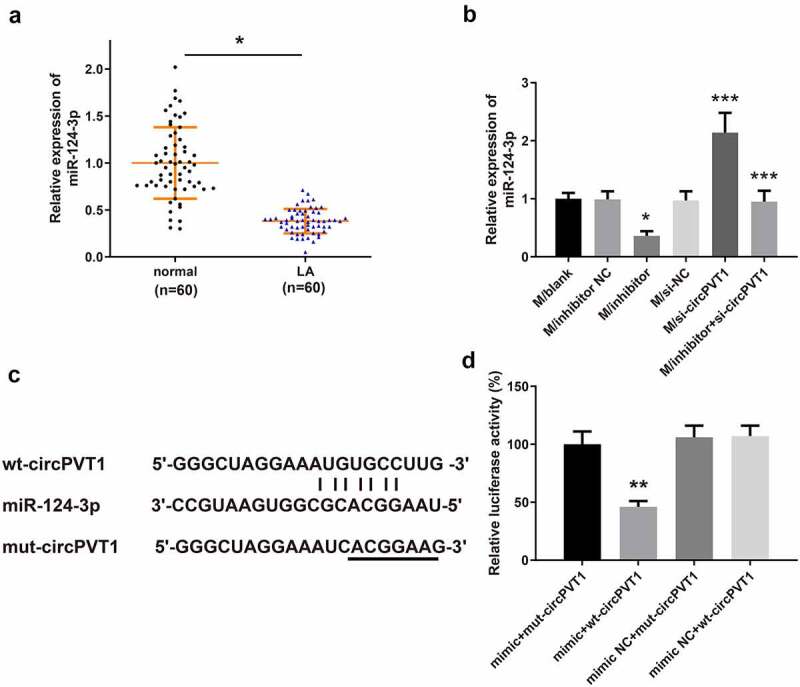
CircPVT1 negatively targets miR-124-3p.
Notes: qRT-PCR detected the expression of miR-124-3p in LA tissues and adjacent tumor tissues from 60 LA patients (A) and in macrophages after transfection (B) (n = 3). The binding of miR-124-3p and circPVT1 in the 3ʹUTR region was predicted by TargetScan (C). The dual-luciferase reporter assay was used to verify the binding relationship between miR-124-3p and circPVT1 (D) (n = 3); **P < 0.01, compared to the Normal group or the mimic+mut-circPVT1 group; *P < 0.05, ***P < 0.001, compared to the si-NC group, inhibitor NC group or si-circPVT1 group; LA, lung adenocarcinoma.
Effect of miR-124-3p on macrophage polarization toward M2 type
The macrophages transfected with miR-124-3p mimic had increased miR-124-3p level (Figure 7a, **P < 0.01). Then, qRT-PCR and Western blot were utilized to examine the expressions of M2 markers. We discovered that overexpression of miR-124-3p downregulated the mRNA and protein levels of CD206, CD163 and arginase-1 in macrophages (Figure 7b-c, **P < 0.01). These results suggested that miR-124-3p upregulation may inhibit macrophage polarization toward M2 phenotype.
Figure 7.
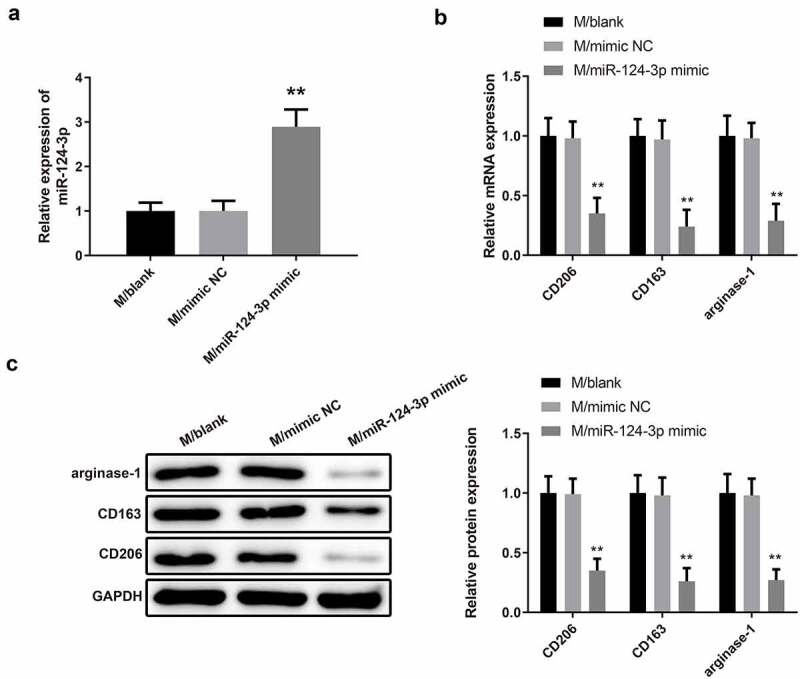
MiR-124-3p upregulation impedes M2 type macrophage polarization.
Note: After macrophages were transfected with miR-124-3p mimic, the expression of miR-124-3p in macrophages was measured by qRT-PCR (A), and the expressions of M2 markers were evaluated by qRT-PCR (B) and Western blot (C) (n = 3); **P < 0.01, compared to the M/mimic NC group; M, macrophage.
The negative relationship between EZH2 and miR-124-3p as well as a positive relationship between EZH2 and circPVT1
The mRNA level of EZH2 in LA tissues and adjacent tumor tissues from 60 LA patients was examined by qRT-PCR, which exhibited that the EZH2 level in LA tissue was prominently higher than that in adjacent tumor tissues (Figure 8a, **P < 0.01). Furthermore, silencing of miR-124-3p elevated EZH2 expression in macrophages (Figure 8b-c, ***P < 0.001). Knockdown of circPVT1 decreased the mRNA and protein expressions of EZH2 in macrophages, while cotransfection of miR-124-3p inhibitor and si-circPVT1 reversed EZH2 expression (Figure 8b-c, *P < 0.05). The mutated type and wild-type sequences containing the 3ʹUTR region of EZH2 were constructed (Figure 8d). The dual-luciferase reporter assay result showed that the mimic+wt-EZH2 group possessed diminished luciferase activity in comparison to the mimic+mut-EZH2 group or the mimic NC+wt-EZH2 group (Figure 8e, **P < 0.01). No obvious change in relative luciferase activity was observed among the mimic+mut-EZH2, mimic NC+mut-EZH2 and mimic NC+wt-EZH2 groups (Figure 8e, **P > 0.01). The above results implied that circPVT1 may upregulate EZH2 expression by negatively regulating the expression of miR-124-3p, thereby promoting the polarization of macrophages toward M2 type.
Figure 8.
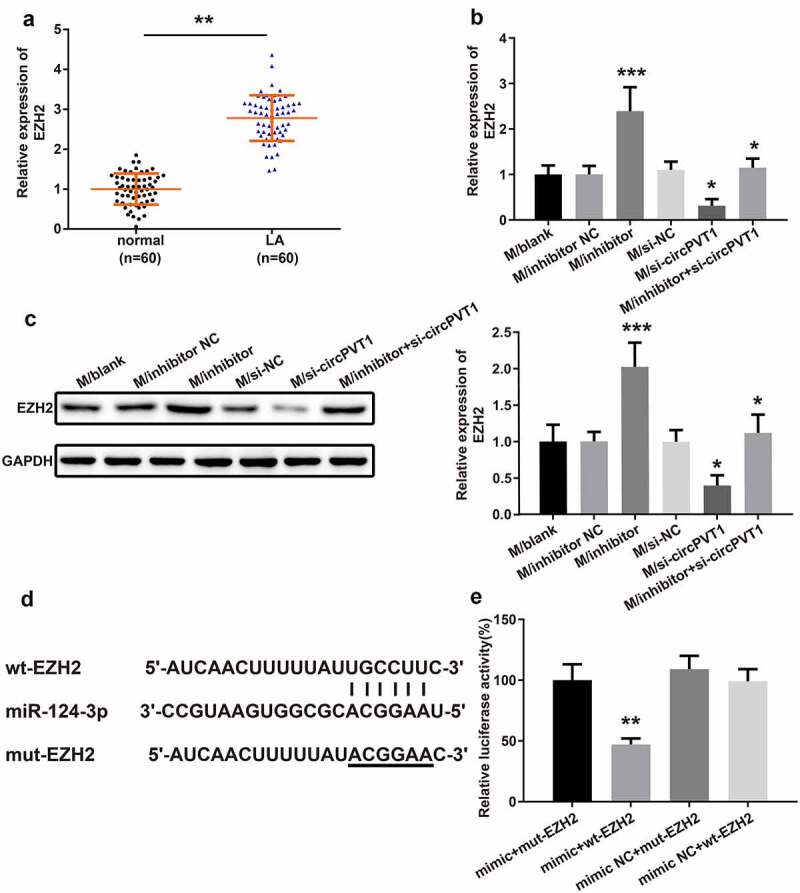
The relationship between circPVT1, miR-124-3p and EZH2.
Notes: The level of EZH2 in LA tissues and adjacent tumor tissues from 60 LA patients was determined by qRT-PCR (A) (n = 60). After transfection, qRT-PCR (B) and Western blot (C) were employed to measure the EZH2 expression in macrophages. The binding of miR-124-3p and EZH2 in the 3ʹUTR region of EZH2 was predicted by TargetScan (D) (n = 3). The binding relationship between miR-124-3p and EZH2 was verified by dual-luciferase reporter assay (E) (n = 3); *P < 0.01, compared to the Normal group; *P < 0.05, ***P < 0.001, compared to the si-NC group, inhibitor NC group or si-circPVT1 group; **P < 0.01, compared to the mimic+mut-EZH2 group; LA, lung adenocarcinoma.
CircPVT1 regulates the expressions of miR-124-3p and EZH2
The A549 cells were subjected to Exo inhibitor GW4869, and TEM was adopted to observe the secretion of Exos. We found that treatment with GW4869 markedly inhibited Exo secretion (Figure 9a). Then, secreted Exos were labeled with PKH67 followed by 4 h of incubation with macrophages. Compared with the NC group, the GW4869 group had diminished fluorescence intensity (Figure 9b, *P < 0.05), verifying that GW4869 may suppress the secretion of Exos. The measurement showed that A549 cells were firstly transfected with si-circPVT1 and then co-cultured with macrophages, in which elevated miR-124-3p expression was found compared with those in the si-NC group (Figure 9c, *P < 0.05), while transfection with oecircPVT1 resulted in decreased expression of miR-124-3p in macrophages (Figure 9c, *P < 0.05). Then, the A549 cells were treated by exosomal inhibitor GW4869 or transfected with si-circPVT1 or oecircPVT1 before co-culture with macrophages. The detection on EZH2 expression showed that co-culture with A549 cells lead to elevated expression of EZH2 in macrophages (Figure 9d-e, *P < 0.05) and pretreatment by GW4869 in A549 cells showed no significant difference on EZH2 expression in macrophages, while transfection of si-circPVT1 or oecircPVT1 in A549 cells respectively lead to decreased expression and elevated expression of EZH2 in macrophages (Figure 9d-e, *P < 0.05). The above data suggested that exosomal circPVT1 derived from LC may inhibit the expression of miR-124-3p and enhance the expression of EZH2 in macrophages.
Figure 9.
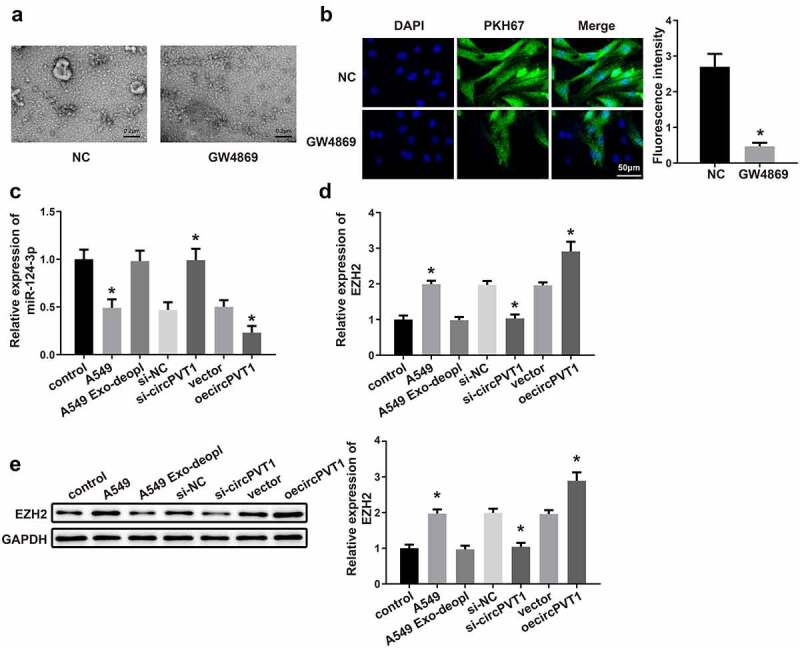
CircPVT1 in LC-derived Exos mediates the expressions of miR-124-3p and EZH2 in macrophages.
Notes: The secretion of Exos was observed by TEM (A) (n = 3). The A549 cells were subjected to GW4869, and the secreted Exos were labeled with PKH67 and incubated with macrophages. Then, the fluorescence intensity was observed under a fluorescence microscope (B) (n = 3). qRT-PCR was employed to determine the mRNA expressions of miR-124-3p (C) and EZH2 (D) in macrophages (n = 3). Western blot was utilized to measure the protein level of EZH2 (E) in macrophages (n = 3); *P < 0.05, compared to the NC group, control group, si-NC group or vector group; LC, lung cancer; Exos, exosomes; TEM, transmission electron microscope.
Discussion
CircRNAs are covalently closed circular RNAs that have been originally identified in plant viroid, virus, archaea, and metazoans [21]. Currently, with deep RNA sequencing technologies and bioinformatic approaches developed, an increasing body of circRNAs has been gradually identified in various diseases, including cancers [7]. Synchronously, the presence of several circRNAs with potential biological function in Exos has been revealed to interfere with cancer growth, such as circ-CCDC66, circ-0051443 and circPDE8A [22–24]. In this study, Exos were extracted from LA cell line A549, and THP-1 cells were induced into macrophages by exposure to PMA. Then, exosomal circPVT1 downregulated or upregulated in A549 cells was incubated with macrophages followed by coculture with A549 cells to observe the performance of circPVT1 on the polarization of macrophages and the proliferation, invasion and migration of LC cells. We found that Exos derived from LC induce M2 phenotype macrophage polarization, and M2-polarized macrophages enhance the proliferative, invasive and migratory properties of LC cells. Besides, this effect may exert by exosomal circPVT1 derived from LC.
In recent years, Exos have been widely studied as an important mediator for the intercellular interaction of cells [25]. Here, LC-derived Exos were found to induce macrophages to polarize to M2 phenotype as evidenced by potentiated expressions of M2 surface markers CD206, CD163 and arginase-1 in macrophages cultured with Exo-A, and unchanged levels of M2 markers in macrophages incubated with Exo-B. Additionally, we further discovered that the M2-polarized macrophages coincubated with A549 cells contributed to the increase in the proliferation, invasion and migration of LC cells regarding proliferation, invasion and migration. Our findings were supported by the recent work of Wang et al., who reported that hypoxic tumor-derived exosomal miR-301a is responsible for the metastasis of pancreatic cancer by regulating M2 macrophage polarization through the PTEN/PI3Kg pathway [26]. Interestingly, Exos derived from tumor cells can package circRNAs and be released into the circulation system [27]. Two circular RNAs, circ-MEMO1 and hsa_circRNA_0056616 have been found to be overexpressed in LSCLC and in lung adenocarcinoma [28,29], and evidence supported that high expression of serum exosomal circ-MEMO1 and exo-hsa_circRNA_0056616 might be valuable diagnostic markers for LC. Our study highlighted the behavior of exosomal circPVT1 in the course of LC. Consistent with previous research, we found elevated expression of circPVT1 in blood-derived Exos extracted from LA patients and Exo-A isolated from LA cell line. Kaplan–Meier survival curve analysis confirmed that the circPVT1 high-expression group had significantly shorter survival time than the circPVT1 low-expression group. These results prompted us to investigate whether LC-derived Exos affect the polarization of macrophages and whether the biological behavior of LC cells is elicited by exosomal circPVT1. Toward this end, circPVT1 was overexpressed or silenced in the A549 cells. We found that the extracted Exo-A after circPVT1 upregulation conferred a supportive role in the macrophage polarization to M2 type. As expected, consistent evidence was noticed in macrophages transfected with oecircPVT1 that overexpression of circPVT1 facilitated macrophages to polarize M2 type. Additionally, results of CCK-8 assay, Transwell and cell scratch assay further proposed that A549 cells cocultured with exosomal circPVT1-activated macrophages had enhanced proliferative, invasive and migratory properties. These data confirmed that exosomal circPVT1 derived from LC may interfere with the polarization of macrophages and thus influencing LC development.
CircRNAs perform regulatory roles, including modulation of gene transcription, interaction with RNA-binding proteins and acting as miRNA sponges, mainly at the transcriptional and post-transcriptional levels [30]. As a frequently researched miRNA, miR-124-3p plays an inhibitory role in the progression of cancers, such as bladder cancer and breast cancer [31,32]. Under this setting, the lowly expressed miR-124-3p was detected in LA tissues rather than in adjacent tumor tissues. Furthermore, miR-124-3p was identified as a target gene of circPVT1 in our study by TargetScan and dual-luciferase reporter gene assay. The loss-of-function experimental results showed that circPVT1 negatively regulated miR-124-3p expression. Analyses of qRT-PCR and Western blot manifested that overexpression of miR-124-3p impeded macrophage polarization toward M2 phenotype. Subsequently, EZH2 was proven to be a target gene of miR-124-3p, and the strikingly high expression of EZH2 was discovered in LA tissues. Based on this finding, we boldly speculated that the miR-124-3p/EZH2 axis may confer an indispensable factor for the oncogenic effect of exosomal circPVT1 on LC. Then, the prediction was verified by our results that exosomal circPVT1 restrained the expression of miR-124-3p and enhanced the expression of EZH2 in macrophages. Our findings were similar to those reported by He et al. in 2019, which showed that the overexpressed circPVT1 in oral squamous cell carcinoma encourages cancer cell proliferation through functioning as a sponge of miR‑125b [33].
In summary, this study sheds light on the central role of exosomal circPVT1 in macrophage polarization toward M2 type and the proliferative, migratory and invasive properties of LC cells. Furthermore, the circPVT1/miR-124-3p/EZH2 axis may be a promising predictive biomarker and therapeutic target for LC patients. However, these studies are generally found with small samples and poor repeatability, so large multicenter studies are necessary to develop preciseness. Additionally, due to time and budget limitations, we failed to set up an in vivo model to verify our speculation. But this indicates a promising future direction for our future studies.
Acknowledgments
We would like to acknowledge Xiaoyu He for his support for experimental design, biomarker selection and the collection on blood sample. Additionally, we also want to acknowledge Guohua Cheng for his valuable suggestion on experimental design and sample detection.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Romaszko AM, Doboszynska A.. Multiple primary lung cancer: a literature review. Adv Clin Exp Med. 2018;27(5):725–730. [DOI] [PubMed] [Google Scholar]
- [2].Villalobos P, Wistuba II, Wistuba II.. Lung cancer biomarkers. Hematol Oncol Clin North Am. 2017;31(1):13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Xu N, Chen S, Liu Y, et al. Profiles and bioinformatics analysis of differentially expressed circrnas in taxol-resistant non-small cell lung cancer cells. Cell Physiol Biochem. 2018;48(5):2046–2060. [DOI] [PubMed] [Google Scholar]
- [4].Hu W, Bi ZY, Chen ZL, et al. Emerging landscape of circular RNAs in lung cancer. Cancer Lett. 2018;427:18–27. [DOI] [PubMed] [Google Scholar]
- [5].Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. [DOI] [PubMed] [Google Scholar]
- [6].Hussain S. Nanomedicine for treatment of lung cancer. Adv Exp Med Biol. 2016;890:137–147. [DOI] [PubMed] [Google Scholar]
- [7].Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang H, Deng T, Ge S, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38(15):2844–2859. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [9].Wang X, Zhang H, Yang H, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Huang X, Zhang W, Shao Z. Prognostic and diagnostic significance of circRNAs expression in lung cancer. J Cell Physiol. 2019;234:18459–18465. [DOI] [PubMed] [Google Scholar]
- [11].Zong L, Sun Q, Zhang H, et al. Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed Pharmacother. 2018;102:639–644. [DOI] [PubMed] [Google Scholar]
- [12].Li X, Zhang Z, Jiang H, et al. Circular RNA circPVT1 promotes proliferation and invasion through sponging miR-125b and activating E2F2 signaling in non-small cell lung cancer. Cell Physiol Biochem. 2018;51(5):2324–2340. [DOI] [PubMed] [Google Scholar]
- [13].Xu F, Cui WQ, Wei Y, et al. Astragaloside IV inhibits lung cancer progression and metastasis by modulating macrophage polarization through AMPK signaling. J Exp Clin Cancer Res. 2018;37(1):207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shen M, Shen Y, Fan X, et al. Roles of macrophages and exosomes in liver diseases. Front Med (Lausanne). 2020;7:583691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chen Z, Wu H, Shi R, et al. miRNAomics analysis reveals the promoting effects of cigarette smoke extract-treated Beas-2B-derived exosomes on macrophage polarization. Biochem Biophys Res Commun. 2021;572:157–163. [DOI] [PubMed] [Google Scholar]
- [16].Zhang H, Zhu L, Bai M, et al. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int J Cancer. 2019;144(10):2501–2515. [DOI] [PubMed] [Google Scholar]
- [17].Liu YY, Zhang LY, Du WZ. Circular RNA circ-PVT1 contributes to paclitaxel resistance of gastric cancer cells through the regulation of ZEB1 expression by sponging miR-124-3p. Biosci Rep. 2019;39:BSR20193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tang LX, Chen GH, Li H, et al. Long non-coding RNA OGFRP1 regulates LYPD3 expression by sponging miR-124-3p and promotes non-small cell lung cancer progression. Biochem Biophys Res Commun. 2018;505(2):578–585. [DOI] [PubMed] [Google Scholar]
- [19].Huang HG, Tang XL, Huang XS, et al. Long noncoding RNA LINC00511 promoted cell proliferation and invasion via regulating miR-124-3p/EZH2 pathway in gastric cancer. Eur Rev Med Pharmacol Sci. 2020;24(8):4232–4245. [DOI] [PubMed] [Google Scholar]
- [20].Xiao D, Cui X, Wang X. Long noncoding RNA XIST increases the aggressiveness of laryngeal squamous cell carcinoma by regulating miR-124-3p/EZH2. Exp Cell Res. 2019;381(2):172–178. [DOI] [PubMed] [Google Scholar]
- [21].Zhao J, Li L, Wang Q, et al. CircRNA expression profile in early-stage lung adenocarcinoma patients. Cell Physiol Biochem. 2017;44(6):2138–2146. [DOI] [PubMed] [Google Scholar]
- [22].Chen W, Quan Y, Fan S, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128. [DOI] [PubMed] [Google Scholar]
- [23].Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77(9):2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li Z, Yanfang W, Li J, et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–250. [DOI] [PubMed] [Google Scholar]
- [25].Masaoutis C, Mihailidou C, Tsourouflis G, et al. Exosomes in lung cancer diagnosis and treatment. From the translating research into future clinical practice. Biochimie. 2018;151:27–36. [DOI] [PubMed] [Google Scholar]
- [26].Wang X, Luo G, Zhang K, et al. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kgamma to promote pancreatic cancer metastasis. Cancer Res. 2018;78(16):4586–4598. [DOI] [PubMed] [Google Scholar]
- [27].Pan B, Qin J, Liu X, et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ding C, Xi G, Wang G, et al. Exosomal Circ-MEMO1 promotes the progression and aerobic glycolysis of non-small cell lung cancer through targeting MiR-101-3p/KRAS axis. Front Genet. 2020;11:962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].He F, Zhong X, Lin Z, et al. Plasma exo-hsa_circRNA_0056616: a potential biomarker for lymph node metastasis in lung adenocarcinoma. J Cancer. 2020;11(14):4037–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li J, Li Z, Jiang P, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fu W, Wu X, Yang Z, et al. The effect of miR-124-3p on cell proliferation and apoptosis in bladder cancer by targeting EDNRB. Arch Med Sci. 2019;15(5):1154–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yan G, Li Y, Zhan L, et al. Decreased miR-124-3p promoted breast cancer proliferation and metastasis by targeting MGAT5. Am J Cancer Res. 2019;9(3):585–596. [PMC free article] [PubMed] [Google Scholar]
- [33].He T, Li X, Xie D, et al. Overexpressed circPVT1 in oral squamous cell carcinoma promotes proliferation by serving as a miRNA sponge. Mol Med Rep. 2019;20(4):3509–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]


