Enterococcus faecium has emerged as a major cause of nosocomial infections. Correspondingly, this species has become increasingly resistant to a broad range of antimicrobial agents, including aminoglycosides, penicillins, and glycopeptides (4). Quinupristin-dalfopristin, an injectable streptogramin, is proposed as an alternative drug for severe infections caused by multiply resistant E. faecium and in many cases could represent the only therapy available (3).
We have isolated from urine a strain of E. faecium, HM1032, which was resistant to both vancomycin and quinupristin-dalfopristin, and we have studied the localization of the genetic determinants for these resistances. E. faecium HM1032 was resistant to erythromycin (MIC > 128 μg/ml), quinupristin-dalfopristin (MIC = 16 μg/ml), vancomycin (MIC >128 μg/ml), and tetracycline. We found that resistance to macrolides and vancomycin was related to the presence of an ermAM (ermB)-like gene and a vanA gene, respectively, as shown by PCR experiments (2, 6). Resistance to quinupristin-dalfopristin was due to inactivation of quinupristin and dalfopristin (MICs = 64 μg/ml), as suggested by a microbiological screen test, Gots’ test, and confirmed in PCR experiments by amplification of vgb- and satA-like genes responsible for acetylation and hydrolysis of quinupristin and dalfopristin, respectively. The nucleotide sequences of amplicons were nearly identical to those of the prototype satA and vgb genes (1, 5).
The resistances could not be transferred by mating on filters to E. faecium HM1070, a plasmid-free recipient strain, except for tetracycline resistance, which transferred with a frequency of 10−6 per donor colony. Transformation of E. faecium HM1070 with plasmid extracts of E. faecium HM1032 yielded clones coresistant to vancomycin, quinupristin-dalfopristin (MIC = 8 μg/ml), and erythromycin. However, the transformants inactivated dalfopristin but not quinupristin. One of the transformants, E. faecium HM1070SR, was studied further, and PCR experiments indicated that the strain contained the vanA, ermAM, and satA genes but not the vgb gene.
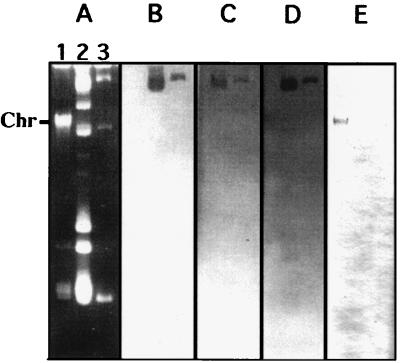
Total DNA, plasmid DNA of E. faecium HM1032, and plasmid DNA of E. faecium HM1070SR were analyzed by agarose gel electrophoresis and hybridization with vanA, ermAM, vgb, and satA probes (Fig. 1). E. faecium HM1032 contained at least six plasmids, three of which were present in E. faecium HM1070SR. The ermAM, satA, and vanA genes were localized on the same large plasmid (>60 kb), while the vgb gene was apparently chromosomal in the wild-type strain (lack of hybridization to plasmid bands with hybridization to the region with linearized DNA, including fragments of chromosomal DNA) and absent from the transformant. After plasmid digestion with EcoRI, vanA, satA, and ermAM probes hybridized to EcoRI fragments, of 5.5, 10, and 10 kb, respectively (data not shown).
FIG. 1.
Analysis of non-digested genomic DNA (lane 1) and plasmid DNA (lane 2) from E. faecium HM1032 and of nondigested plasmid DNA from the transformant E. faecium HM1070SR (lane 3) by agarose gel electrophoresis and hybridization. (A) Agarose gel electrophoresis of DNA; (B to E) hybridization to ermAM, satA, vanA, and vgb probes, respectively. Linear (chromosomal) DNA is indicated on the left of the gel (Chr). ermAM, satA, and vanA probes hybridized to the same large plasmid; the vgb probe hybridized to the chromosomal DNA of E. faecium HM1032.
Although uncommon, quinupristin-dalfopristin resistance in glycopeptide-resistant E. faecium has already been reported for human and animal isolates from Germany, the United Kingdom, and the United States (3, 7, 8). The presence of genes responsible for glycopeptide and streptogramin resistance linked on the same plasmid is an additional cause of concern if this plasmid happened to disseminate. Although we were unable to transfer by conjugation this resistance plasmid to a recipient strain, transformation experiments showed that the combination of genes for resistance to vancomycin and streptogramins borne by this plasmid was sufficient to confer resistance to both antibiotics.
REFERENCES
- 1.Allignet J, Loncle V, Mazodier P, El Solh N. Nucleotide sequence of a staphylococcal plasmid gene, vgb, encoding a hydrolase inactivating the B components of virginiamycin-like antibiotics. Plasmid. 1988;20:271–275. doi: 10.1016/0147-619x(88)90034-0. [DOI] [PubMed] [Google Scholar]
- 2.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eliopoulos G M, Wennersten C B, Gold H S, Schulin T, Souli M, Farris M G, Cerwinka S, Nadler H L, Dowzicky M, Talbot G H, Moellering R C., Jr Characterization of vancomycin-resistant Enterococcus faecium isolates from the United States and their susceptibility in vitro to dalfopristin-quinupristin. Antimicrob Agents Chemother. 1998;42:1088–1092. doi: 10.1128/aac.42.5.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leclercq R, Courvalin P. Resistance to glycopeptides in enterococci. Clin Infect Dis. 1997;24:545–554. doi: 10.1093/clind/24.4.545. [DOI] [PubMed] [Google Scholar]
- 5.Rende-Fournier R, Leclercq R, Galimand M, Duval J, Courvalin P. Identification of the satA gene encoding a streptogramin A acetyltransferase in Enterococcus faecium BM4145. Antimicrob Agents Chemother. 1993;37:2119–2225. doi: 10.1128/aac.37.10.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werner G, Klare I, Witte W. Association between quinupristin/dalfopristin resistance in glycopeptide-resistant Enterococcus faecium and the use of additives in animal feed. Eur J Clin Microbiol Infect Dis. 1998;17:401–402. doi: 10.1007/BF01691571. [DOI] [PubMed] [Google Scholar]
- 8.Woodford N, Palepou M F, Johnson A P, Chadwick P R, Bates J. Methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci. Lancet. 1997;350:738. doi: 10.1016/s0140-6736(05)63544-1. [DOI] [PubMed] [Google Scholar]