Abstract
Lanthipeptides belong to the family of ribosomally-synthesized and post-translationally peptides (RiPPs). The (methyl)lanthionine crosslinks characteristic to lanthipeptides are essential for their stability and bioactivities. In most bacteria, lanthipeptides are maturated from single precursor peptides encoded in the corresponding biosynthetic gene clusters. However, cyanobacteria engage in combinatorial biosynthesis and encode as many as 80 substrate peptides with highly diverse sequences that are modified by a single lanthionine synthetase into lanthipeptides of different lengths and ring patterns. It is puzzling how a single enzyme could exert control over the cyclization processes of such a wide range of substrates. Here, we used a library of ProcA3.3 precursor peptide variants and show that it is not the enzyme ProcM but rather its substrate sequences that determine the regioselectivity of lanthionine formation. We also demonstrate the utility of trapped ion mobility spectrometry–tandem mass spectrometry (TIMS-MS/MS) as a fast and convenient method to efficiently separate lanthipeptide constitutional isomers, particularly in cases where the isomers cannot be resolved by conventional liquid chromatography. Our data allowed identification of factors that are important for the cyclization outcome, but also showed that there are no easily identifiable predictive rules for all sequences. Our findings provide a platform for future deep learning approaches to allow such prediction of ring patterns of products of combinatorial biosynthesis.
Graphical Abstract
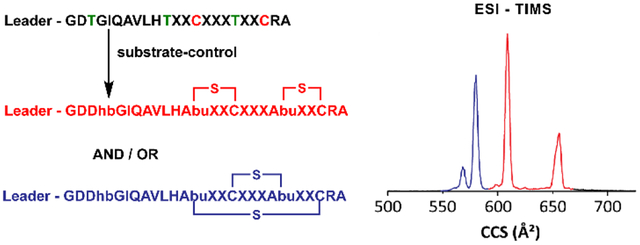
Introduction
Lanthipeptides belong to a large and diverse family of natural products called ribosomally synthesized and post-translationally modified peptides (RiPPs).1 The biosynthesis of lanthipeptides commences with ribosomally synthesized precursor peptides, termed LanAs, that contain N-terminal leader sequences followed by C-terminal core peptide sequences. The leader sequences are recognized by lanthipeptide synthetases, which catalyze dehydration of Ser/Thr residues in the core peptides to generate dehydroalanine (Dha from Ser) or (Z)-dehydrobutyrine (Dhb from Thr). Dehydration is followed by intramolecular Michael-type addition of Cys thiols to the dehydrated residues to form the class-defining lanthionine (Lan) and methyllanthionine (MeLan) linkages (Figure 1A).2 After these modifications, dedicated proteases/transporters perform leader peptide proteolysis and export the mature product. Lanthipeptide synthetases demonstrate relaxed substrate specificity as they often tolerate changes made to the core peptide sequences,3–10 a feature that has been extensively applied to bioengineer lanthipeptides.11–30, 31
Figure 1.
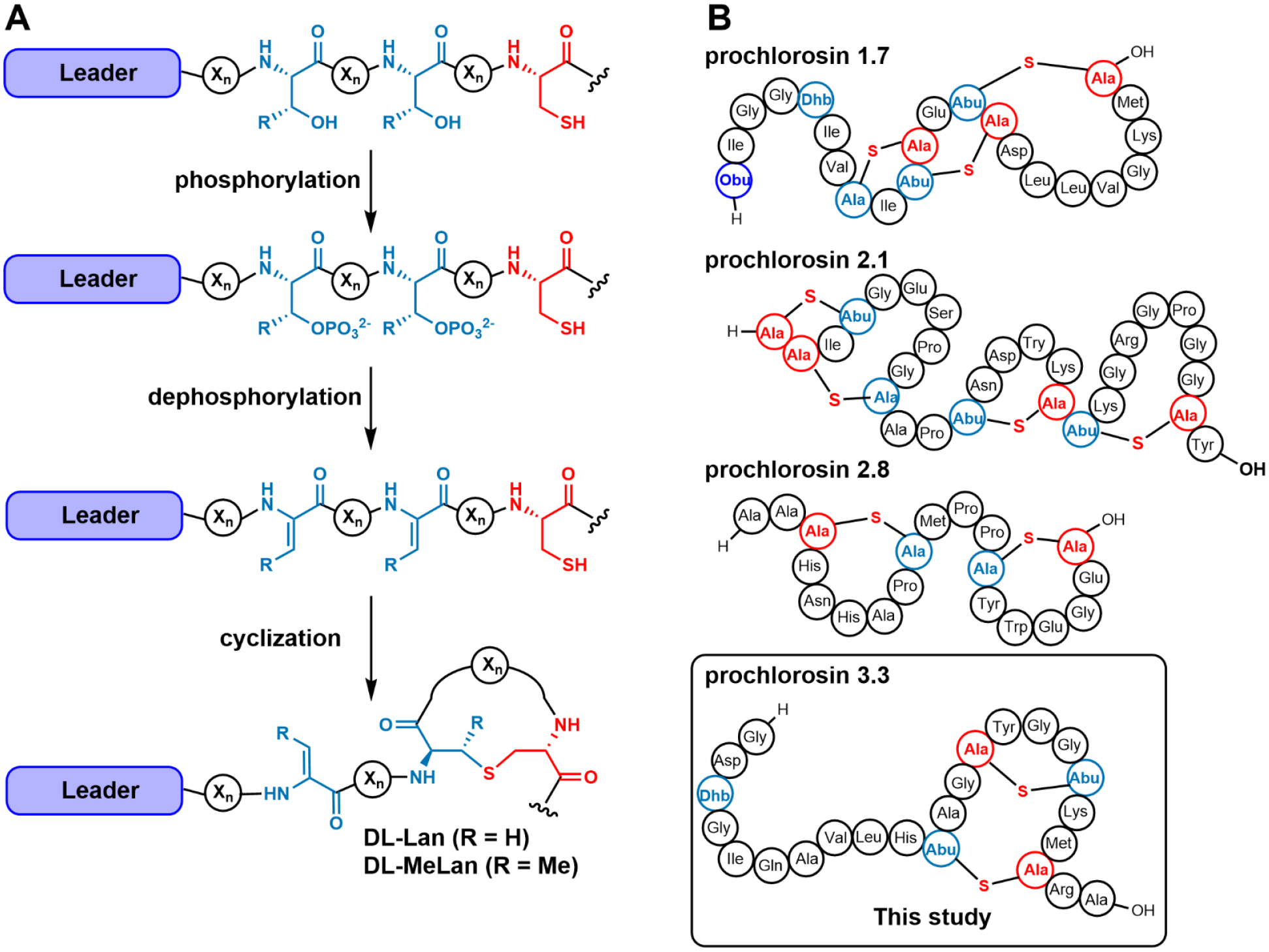
Prochlorosin biosynthesis. (A) Schematic representation of the reactions catalyzed by ProcM, a class II lanthionine synthetase that catalyzes dehydration of Ser/Thr and Michael type addition of Cys residues to the resulting dehydro amino acids. (B) Four representative prochlorosins made by ProcM, which has 30 different substrate peptides that are all converted into products with different ring patterns.
Discovered through genome-mining efforts in the cyanobacterium Prochlorococcus MIT9313, prochlorosins constitute an unusual example of substrate tolerance in lanthipeptide biosynthesis.32 The genome of this cyanobacterium encodes a single bifunctional class II lanthipeptide synthetase, termed ProcM, and 30 putative precursor peptides, termed ProcAs, scattered at different loci in the genome. ProcM catalyzes both the dehydration and cyclization steps during prochlorosin biosynthesis in two separate active sites.33 In vitro studies as well as experiments using Escherichia coli as heterologous host showed that ProcM could modify all of the 18 precursors tested thus far to form lanthipeptide products with highly diverse individual ring patterns (e.g. Figure 1B).32, 34–35 This feature is unique compared to most lanthipeptide systems where normally only a single precursor peptide (or several precursors that are closely related in sequence) is encoded in the biosynthetic gene cluster. Given that the formation of a specific ring pattern is typically critical for bioactivity of lanthipeptides,3, 36–41 the physiological relevance of the structural diversity found in prochlorosins remains to be elucidated. Since the original discovery of prochlorosins, other cyanobacteria have also been shown to encode similar combinatorial biosynthetic systems,42 with currently the largest number of unique substrate peptides (80) in Synechococcus.43 Collectively, the products of such combinatorial biosynthetic pathways in cyanobacteria have been termed cyanotins.44
From a catalyst standpoint, it is remarkable how a single enzyme, ProcM, asserts exquisite control over its many substrates to form distinct polycyclic products with high fidelity. A possible explanation that has been offered is that the conformational energy landscape of the core peptide sequences dictates the final ring patterns.44–45 In this model, the substrate peptides adopt conformations on the enzyme that favor interactions of specific Cys residue with their partner Dha/Dhb residues. By increasing the nucleophilicity of the Cys thiolates via Zn2+ coordination,46 the cyclase active site in ProcM would then accelerate the Michael-type addition to covalently lock such favorable conformations into the observed ring pattern. This model is supported by a previous study demonstrating that ProcM cyclization is irreversible and under kinetic rather than thermodynamic control so that the final outcome is dictated by the selectivity of initially formed rings.46
In this study we used ProcA3.3 as a model and show that the regioselectivity of lanthionine formation is a substrate-controlled process. We introduce trapped ion mobility spectrometry-tandem mass spectrometry (TIMS-MS/MS)47–49 as a fast and convenient method to separate lanthipeptide structural isomers, particularly for cases where the isomers cannot be resolved by conventional liquid chromatography. MD simulations demonstrate that the formation of prochlorosin 3.3 and its analogs is not under thermodynamic control, and therefore the relative kinetics of formation of the first ring determines the outcome of the cyclization process.
Results and Discussion
Analysis of a library of ProcA3.3 variants
A previous bioengineering study demonstrated that ProcM could modify a library of ProcA2.8 variants in which the residues within the two lanthionine rings of prochlorosin 2.8 were randomized. ProcM converted these variants into the same non-overlapping bicyclic ring patterns formed for wild type ProcA2.8 (Figure 2A).24 In the current investigation, we set out to create a library of bicyclic peptides with overlapping ring patterns using ProcA3.3 as the scaffold by randomizing all residues between the Cys and Thr residues that are involved in methyllanthionine formation (Figure 2A). The NWY codon was used (N = G,A,T,C; W = A,T; Y = T,C) for the seven randomized positions such that the library would be made up of one of eight amino acids (Asp, Phe, His, Ile, Leu, Asn, Val, Tyr) at each position. Each individual gene encoding an N-terminally hexa-histidine tagged ProcA3.3 variant was then inserted in a co-expression plasmid that already contained the gene encoding ProcM. Deep sequencing demonstrated the desired diversity of the plasmid library (Figure 2B), with a total of 1 × 106 unique peptide variants (theoretical diversity = 2.1 × 106 sequences; Figure S1). To assess the ProcM-catalyzed modification outcome, a random subset of the ProcA3.3 variants was selected to be further characterized. First, the plasmid library was used to transform E. coli cells and several clones were picked, grown in liquid media and protein expression was induced. Following purification of individual ProcA3.3 variant peptides using Ni2+ affinity chromatography, they were digested with LahT150, a protease that removes the leader peptide at the double-glycine motif.50 The prochlorosin core peptide variants thus obtained were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) to determine the dehydration state, and treated with thiol-reactive alkylating reagents such as iodoacetamide (IAA) or N-ethylmaleimide (NEM) to report on any non-cyclized or partially cyclized products.51 Finally, electrospray ionization - tandem mass spectrometry (ESI-MS/MS) analysis of the fully cyclized peptides was used to determine the ring pattern by the presence (non-overlapping rings) or absence (overlapping ring) of bi and yj fragment ions between Cys14 and dehydrated Thr18 residues.
Figure 2.
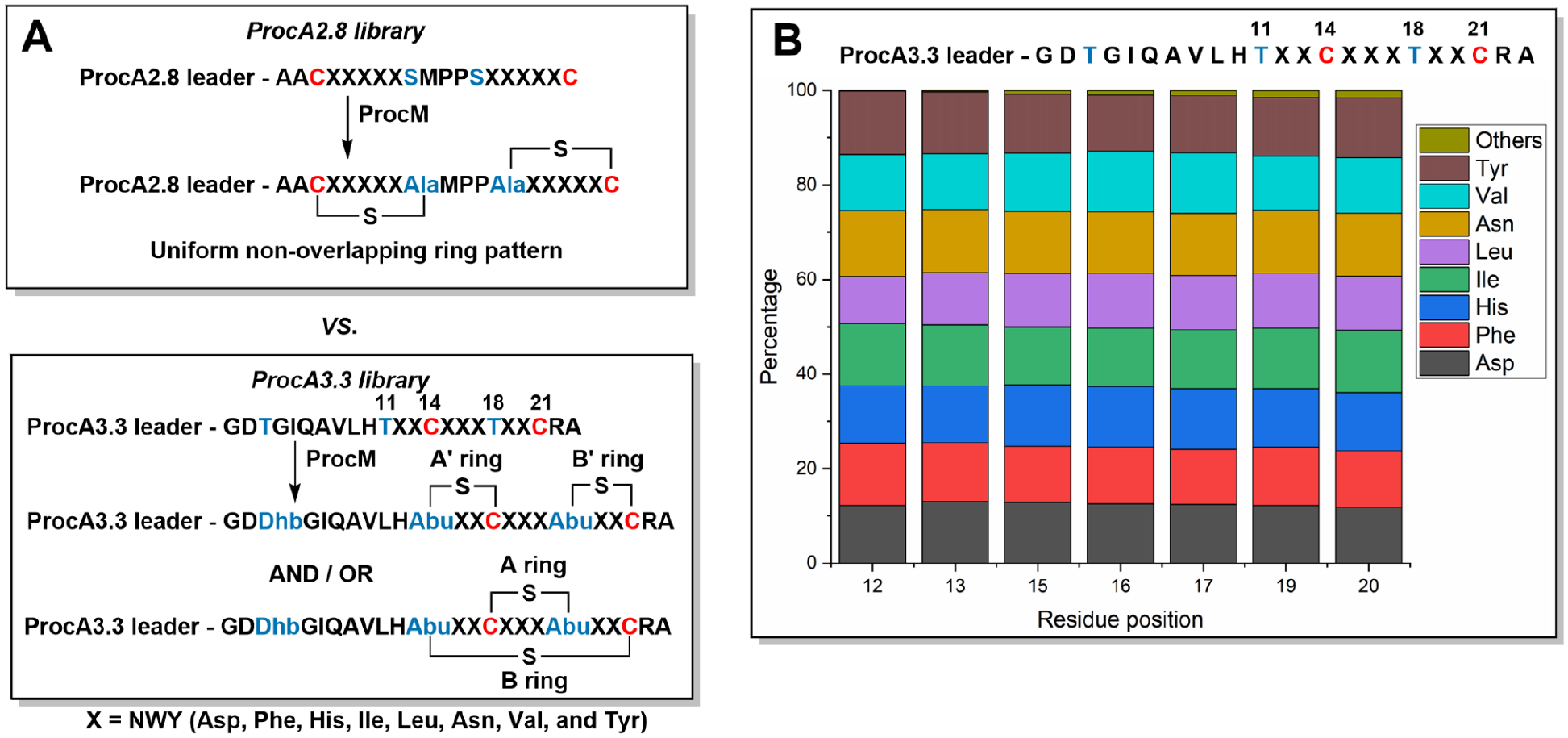
Generation of prochlorosin libraries. (A, top) A previous study showed that randomization of the residues denoted “X” in the precursor to prochlorosin 2.8 resulted in a library of variants with non-overlapping ring patterns.24 (A, bottom) In the current work randomization of the positions denoted “X” in the precursor to prochlorosin 3.3 resulted in formation of two different ring patterns depending on the substrate sequence. (B) Results of deep sequencing of the ProcA3.3 variant plasmid library.
The enzyme generally performed three dehydrations and two cyclizations of the variant peptides similar to its activity on the wild type (WT) ProcA3.3 peptide (Figure 3, Figures S2–S11). As in the case of the ProcA2.8 library, members of ProcA3.3 library (sequence and nomenclature for variants investigated in this study are shown in Table 1) were expected to possess the ring pattern observed for WT ProcA3.3, which contains two overlapping rings (Figure 1B). However, surprisingly, LC-MS data of several variants indicated the formation of two isomeric products that were both fully cyclized yet showed a different elution profile (e.g. Figures S4–S6 and S11). We first investigated whether the two peaks represented stereoisomers in which the stereochemistry of the Michael-type addition was different since previous studies have demonstrated that the anti-addition of the thiol of Cys to Dhb can occur from either the Re- or Si-face.52–54 The two products of one representative variant, ProcA3.3_6 (Figure S6), were therefore separated by HPLC and hydrolyzed in acid, and the constituent amino acids derivatized as described previously.34, 55 Analysis by gas chromatography (GC) with a chiral stationary phase and comparison with synthetic standards of known configuration demonstrated that both products of ProcA3.3_6 contained DL-methyllanthionine (Figure S12A & B) just as the product of WT ProcA3.3.34 We also investigated the stereochemistry for the product of ProcA3.3_2 by GC-MS and confirmed that this peptide also contains DL-methyllanthionine (Figure S12C).
Figure 3.
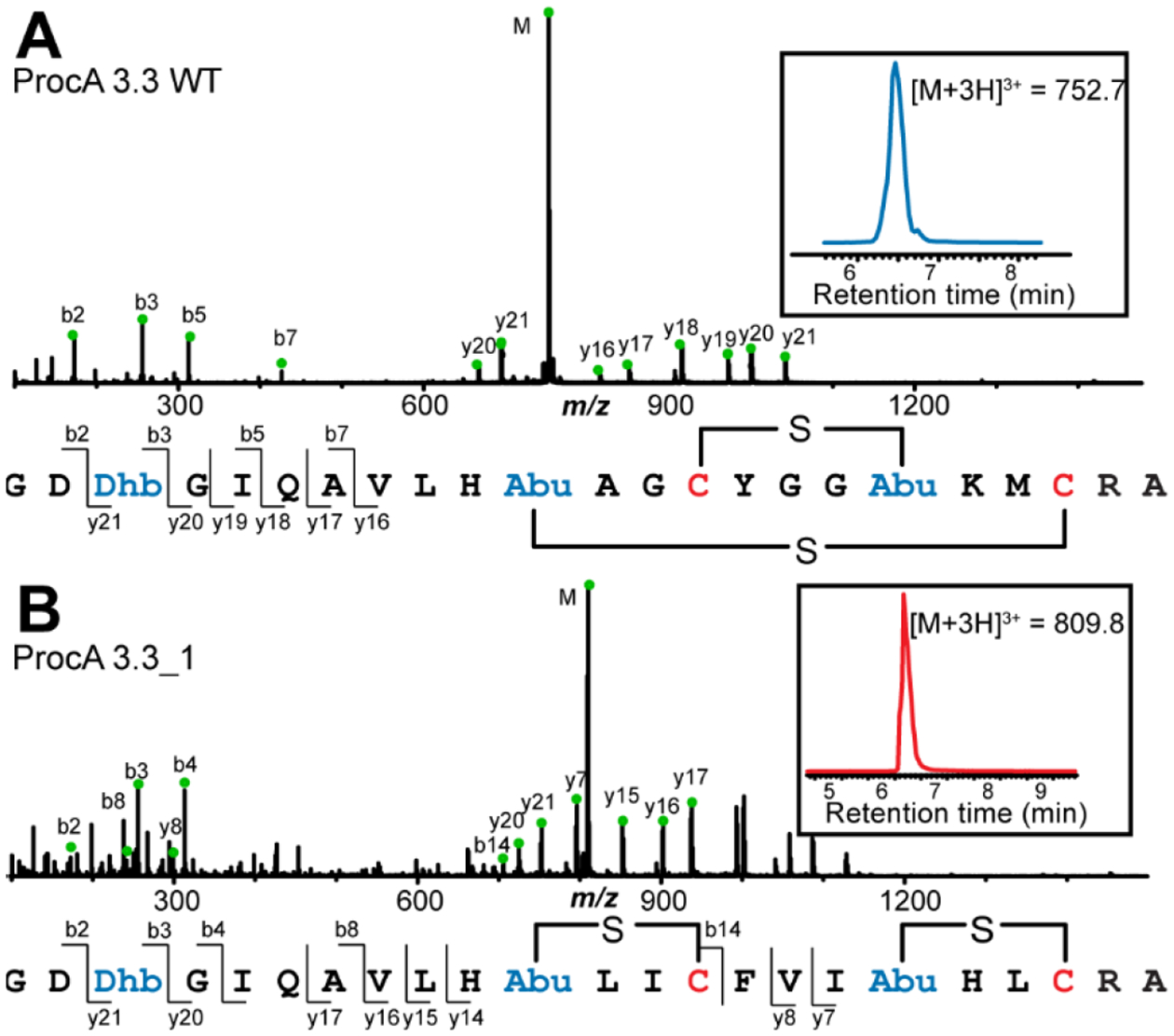
LC-MS/MS analysis of WT prochlorosin 3.3 and one of the variants (3.3_1). (A) WT ProcA3.3 is converted by ProcM into a product that is dehydrated three times and contains two overlapping rings. (B) ProcA3.3_1 is converted by ProcM into a product with non-overlapping rings.
Table 1.
Summary of LC-ESI-MS/MS ring pattern results of tested ProcA3.3 mutants. All mutants are mostly dehydrated at all three available Thr residues.
| Core peptide sequence | Ring pattern | Ratio LC-MSa | Ratio TIMS | |
|---|---|---|---|---|
| WT | GDTGIQAVLHTAGCYGGTKMCRA | overlapping | >100:1b | >100:1b |
| ProcA3.3_1 | GDTGIQAVLHTLICFVITHLCRA | both | <1:100 | 5:95 |
| ProcA3.3_2 | GDTGIQAVLHTIDCVVITFYCRA | non-overlapping | <1:100 | <1:100 |
| ProcA3.3_3 | GDTGIQAVLHTDNCLVFTFVCRA | non-overlapping | <1:100 | <1:100 |
| ProcA3.3_4 | GDTGIQAVLHTIYCNNLTVICRA | both | 20:80 | 25:75 |
| ProcA3.3_5 | GDTGIQAVLHTHNCVINTFVCRA | both | 40:60 | 30:70 |
| ProcA3.3_6 | GDTGIQAVLHTIYCILDTHLCRA | both | 45:55 | 52:48 |
| ProcA3.3_7 | GDTGIQAVLHTHFCNYLTYICRA | non-overlapping | <1:100 | <1:100 |
| ProcA3.3_8 | GDTGIQAVLHTYNCYYYTYICRA | non-overlapping | <1:100 | <1:100 |
| ProcA3.3_9 | GDTGIQAVLHTILCLIFTVDCRA | both | N.D. | 42:58 |
| ProcA3.3_10 | GDTGIQAVLHTDFCLLVTHDCRA | both | N.D. | 30:70 |
| ProcA3.3_11 | GDTGIQAVLHTHFCINLTYVCRA | both | 75:25 | 75:25 |
| ProcA3.3_12 | GDTGIQAVLHTNYCNLVTHLCRA | non-overlapping | N.D. | <1:100c |
| ProcA3.3_13 | GDTGIQAVLHTLFCYGGTFDCRA | both | 40:60 | d |
The integration ratio is estimated for overlapping/non-overlapping ring patterns. N.D. ratio could not be determined because of co-elution.
Estimated detection limit.
Isomers were separated by LC prior to analysis by TIMS. See Figure S30.
We next investigated whether the two products observed for a subset of ProcA3.3 variants could be isomers with different ring patterns. Analysis by ESI-MS/MS confirmed that the two compounds giving rise to the peaks are indeed constitutional isomers (Figures S4–S6 and S11). In addition to the ring pattern observed for WT ProcA3.3, which consists of two overlapping rings (Figure 1B), fragmentation patterns indicative of two non-overlapping rings were observed for several variants (Figure 2A; Figures S2–S11). In some cases, the two constitutional isomers could be separated by LC and their ratio determined, whereas for other variants only a single LC peak was seen that could either indicate co-elution of two isomers or generation of a single isomer. When two products were formed that were chromatographically separable, the elution order proved not to be predictive of the ring pattern. For instance, for ProcA3.3_4 the isomer with the non-overlapping ring pattern eluted after the isomer with the overlapping ring pattern whereas for ProcA3.3_5 the elution order was reversed (Figures S4 and S5, respectively). Altogether, these results demonstrated that ProcA3.3 substrate sequences can heavily influence the ring pattern outcome of ProcM modification.
The cyanotins constitute a unique group of lanthipeptides
If the outcome of cyclization by ProcM is indeed dictated by its substrate sequences, then deep learning approaches may potentially be used to predict the ring patterns produced from the thousands of ProcA homologs that are encoded in the marine metagenomes.43 Such a deep learning approach has previously been attempted to predict chemical cross-linking patterns in several RiPP classes, including lanthipeptides, using a database of structurally characterized RiPPs as training set.56–57 However, the reliability and applicability of such approaches are currently still mostly limited to predicting the leader peptide cleavage site rather than predicting the ring pattern. This limitation likely stems from multiple factors. First, the training set of structurally characterized lanthipeptides is comprised of fairly conserved substrate sequences that are biased towards a relatively small subset of ring patterns with bioactivities. Second, a recent analysis of all then-known lanthipeptide precursor peptides and categorization of their sequences by sequence similarity networks demonstrated that a very large number of sequence families have currently no structures associated with even a single member.58 Thus, the sequence-structure space of lanthipeptides is still vastly under sampled.
Within the structural landscape of lanthipeptides, the cyanotins including the prochlorosins are unique. For nearly all structurally characterized lanthipeptides, the cognate enzyme acts on just one precursor peptide and morphs it into a well-defined structure that is the result of co-evolution of enzyme and substrate to efficiently make a product with some biological function.38 These enzymes do not appear to generate ring patterns that are determined by their substrates. In contrast, thousands of ProcA peptides containing very diverse core peptides are modified by a small set of ProcM enzymes that are highly conserved in sequence.42–43 For these systems, the substrate does appear to determine the outcome of the cyclization process, but at present less than ten prochlorosins have been structurally characterized.34–35 Increasing the size of the training set is non-trivial as structural characterization for products with overlapping ring patterns requires relatively large-scale purification for extensive nuclear magnetic resonance (NMR) analysis of each individual peptide.34–35 The discovery that ProcM generates two different ring patterns from ProcA3.3 substrate variants may provide a unique opportunity to analyze the ring patterns of the products for a large number of analogs from the library of substrates, and then use the data for deep learning approaches. This strategy would require a method to determine the ring pattern that is faster than LC-MS and ideally a method that would not rely on chromatographic separation of isomers, which is highly case-dependent.
ESI-TIMS-MS/MS can characterize isomers with different ring patterns
We sought to implement nESI-TIMS-q-TOF MS/MS as a method to separate constitutional isomers and obtain information regarding the ring patterns in a high-throughput manner. TIMS provides fast gas-phase separation of ions with differences in their mobility. ProcA3.3 mutants that form overlapping or non-overlapping ring patterns would be expected to induce structural changes allowing for their mobility separation (owing to differences in their collision cross sections, CCS). As a proof-of-concept, we analyzed 12 different products of ProcM-catalyzed post-translational modification of ProcA3.3 variants. We first validated the method with two peptides, ProcA3.3_4 and ProcA3.3_6 that we could separate by LC. As shown in Figure 4A and 4B, the ratios of products determined from the TIMS mobility profiles are similar to those determined using LC (Figures S4, S6; Table 1). We also note that the TIMS mobility profiles are more complicated than those obtained by LC since TIMS not only resolves constitutional and configurational isomers but also conformational isomers as has been shown for several RiPPs.59–62 For example, ProcA3.3_4 and ProcA3.3_6 exhibited a CCS distribution consisting of 3 and 5 main IMS bands, respectively. In addition, the nESI-TIMS-q-TOF MS/MS platform provides mass-selected ion alignment with the mobility scan steps allowing for precursor-fragment mobility alignment. Then, TIMS-MS/MS analysis for each IMS band can be obtained and correlated to a lanthipeptide ring pattern. The TIMS-MS/MS analysis of ProcA3.3_4 revealed the presence of an overlapping ring pattern for the IMS band 1 and non-overlapping-ring pattern for IMS bands 2/3, whereas ProcA3.3_6 displayed an overlapping ring pattern for IMS bands 1–3 and non-overlapping ring pattern for IMS bands 4/5 (Figure S13–S14).
Figure 4.
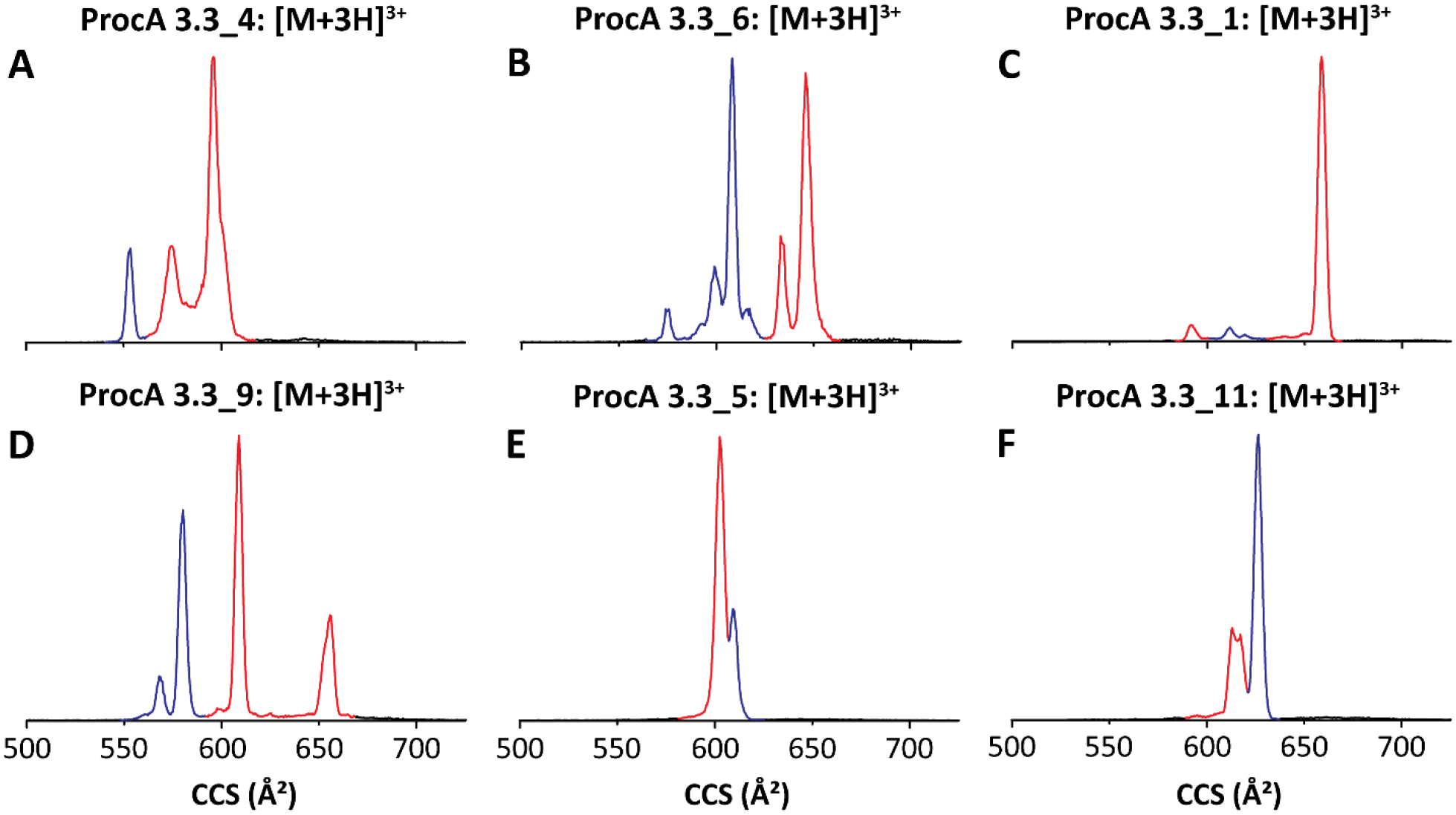
TIMS spectra of (A) ProcM-modified ProcA3.3_4; (B) ProcM-modified ProcA3.3_6; (C) ProcM-modified ProcA3.3_1, (D) ProcM-modified ProcA3.3_9, (E) ProcM-modified ProcA3.3_5 and (F) ProcM-modified ProcA3.3_11. Peaks in red represent product with non-overlapping ring pattern and peaks in blue correspond to overlapping ring patterns. For TIMS-MS/MS spectra used to assign the ring patterns, see Figures S13–S18.
We next moved to three of the peptides that showed only a single LC peak (ProcA3.3_1, ProcA3.3_9 and ProcA3.3_10). The TIMS-MS/MS analysis of these peptides exhibited multiple features in the mobility domain (Figure 4C, D and S17), for which IMS bands 2 (Figure S15), IMS bands 1/2 (Figure S16) and IMS band 1 (Figure S17) were assigned to overlapping ring patterns, while IMS bands 1/3, IMS bands 3/4 and IMS band 2 were assigned to non-overlapping ring patterns, respectively. Thus, the single peaks observed by LC (Figures S9 and S10) were the result of co-elution of the two constitutional isomers. The superior analytical power of TIMS-MS/MS as compared to traditional LC-MS/MS and the much shorter timeframe of acquiring the data because of the absence of LC makes the present workflow highly suitable for the rapid identification of and discrimination between lanthipeptide constitutional isomers. Generally, products with overlapping rings would be expected to be more compact than the corresponding isomer with non-overlapping rings, and indeed the relative CCS was generally smaller for the product with overlapping rings. However, for some peptides this expectation was not observed (e.g. ProcA3.3_5 and ProcA3.3_11, Figures 4E and F; Figures S18–S19).
Factors that may control ring pattern
The factors that determine the final ring pattern depend on whether product formation is under thermodynamic or kinetic control. Our previous mechanistic study did not provide any evidence for reversibility of the cyclization events for ProcA3.3,46 but those studies investigated only the WT sequence. Although we anticipate the same will be true for the variants, we cannot rule out thermodynamic control. In an alternative approach, in this work we performed Molecular Dynamics (MD) calculations to compare the energies of the final products. We first chose four prochlorosins that all contain two rings and for which the structures have been determined, prochlorosins 1.1, 2.8, 3.3 and 4.3 (Figures 1B and S20). Calculations were performed for products with either overlapping or non-overlapping ring patterns for each peptide (Table 2, Figure S21). For prochlorosins 1.1, 2.8 and 4.3, the structures experimentally generated by ProcM with non-overlapping ring patterns were also favored in terms of thermodynamic stability. However, for prochlorosin 3.3, the thermodynamically most favored ring pattern is again the non-overlapping structure and not the experimentally determined overlapping ring pattern. Thus, in support of previous studies, these calculations demonstrate that for prochlorosin 3.3, the ring pattern observed indicates that the system is not under thermodynamic control.
Table 2.
Relative energies of isomers of prochlorosins (Pcn) 1.1, 2.8, 3.3, 3.3_1, and 4.3 by molecular dynamics simulations. For structures see Figures 1B and S20. See also Figure S21.
| Pcn | Pre-cyclization sequence | Non-overlapping rings product (kcal/mol) | Overlapping rings product (kcal/mol) |
|---|---|---|---|
| 1.1 | FFCysVQGDhbANRFDhbINVCys | 0.0 | +3.7 |
| 2.8 | AACysHNHAPDhaMPPDhaYWEGECys | 0.0 | +4.2 |
| 3.3 | GDDhbGIQAVLHDhbAGCysYGGDhbKMCysRA | 0.0 | +8.7 |
| 3.3_1 | GDDhbGIQAVLHDhbLICysFVIDhbHLCysRA | 0.0 | +10.4 |
| 4.3 | GVAGGDhbADhaGGCysDDhbSMFCysYa | 0.0 | +7.2 |
The sequence in italics for Pcn4.3 derives from the leader peptide. If this sequence is removed, the N-terminal Dhb hydrolyzes to a ketone and would not be representative of the peptide encountered by the cyclization domain of ProcM. For the other peptides the leader peptide was not included to limit the influence of the leader peptide on the energies calculated. Dhb residues that do not engage in ring formation are in green font. For all peptides except Pcn3.3 (red entries), the calculations are consistent with the observed preference of ProcM to generate products with non-overlapping rings.
The consistent preference for non-overlapping patterns across different prochlorosins (Pcns) was analyzed in terms of the geometries modelled for Pcn 3.3 WT and Pcn 3.3_1 through MD simulations (Figures 5 and 6). For both peptides, the local structures of rings A’ and B’ in non-overlapping isomers are quite conserved and the flexibility of these motifs is relatively small. However, a larger difference is observed for rings A and B in the overlapping isomers: while for Pcn 3.3_1 both motifs are quite rigid and ring A is twisted, for Pcn 3.3 WT, the rings are more flexible and interlaced with no twisting. The different geometric constraints imposed by the differently sized methyllanthionine rings are apparent when inspecting the Ramachandran plots, i.e. the φ/ψ backbone dihedral angles distributions, of residues 11–21 (Figures S22–S25). As expected, the non-overlapping ring pattern allows a much more relaxed geometry of the peptide triad involved in formation of ring A (Tyr-Gly-Gly in Pcn 3.3 and Phe-Val-Ile in Pcn 3.3_1). On the contrary, this motif is highly constrained to very unusual (i.e. high-energy) conformations in the overlapping isomers, particularly in Pcn 3.3_1; the comparatively more relaxed Tyr-Gly-Gly motif due to the presence of two consecutive glycine residues, might be related to the preference towards the non-overlapping product in wild-type Pcn 3.3 (see below). Of note, such ability of the Tyr-Gly-Gly to better accommodate ring A is transmitted to the rest of the amino acids comprising the exterior ring B, which are also more flexible in Pcn 3.3. Overall, our MD simulations suggest that the overlapping ring pattern imposes a high conformational penalty in prochlorosins so that the non-overlapping products are generally more stable.
Figure 5.
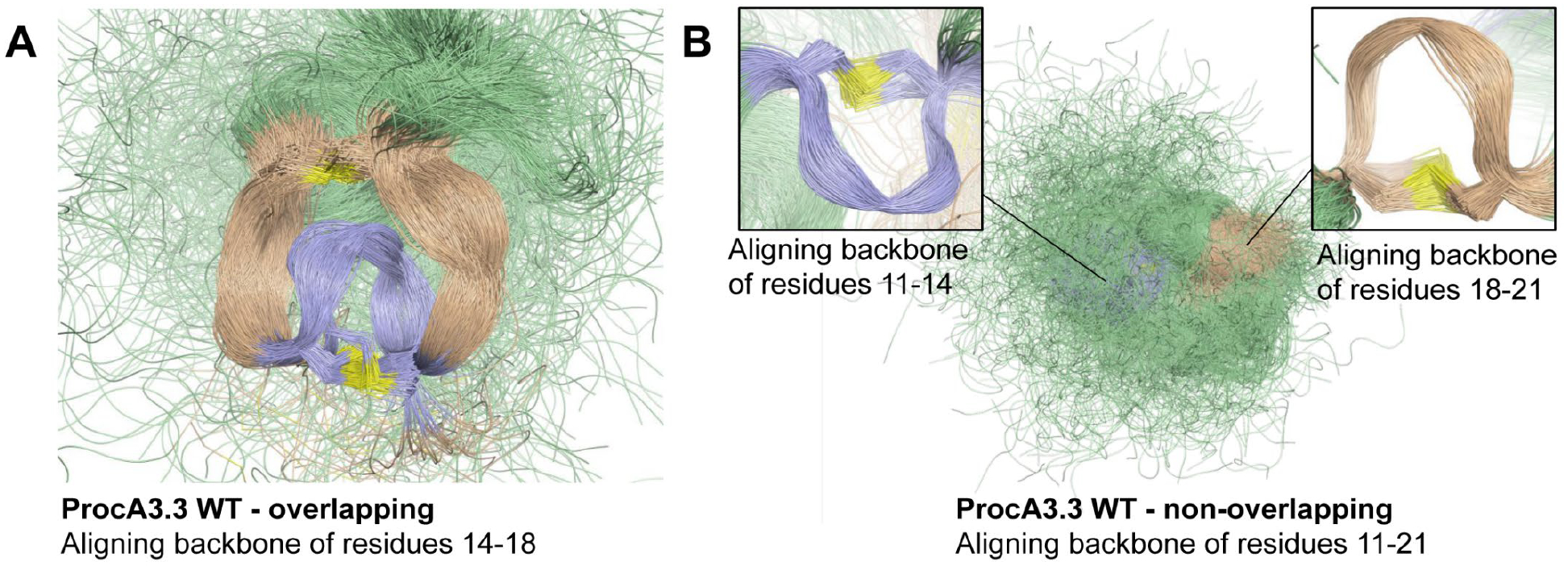
Overlay of 1,000 snapshots extracted from 1 μs simulations of Pcn 3.3 WT isomers with overlapping (A) and non-overlapping (B) ring patterns, in ribbon representation. The A- and A’-rings are shown in blue, the B- and B’-rings are shown in wheat, and the remainder of the peptide is shown in green; sulfur atoms are shown in yellow. Different alignments of the snapshots are shown for a better representation of the local structures of rings A, A’, B and B’.
Figure 6.
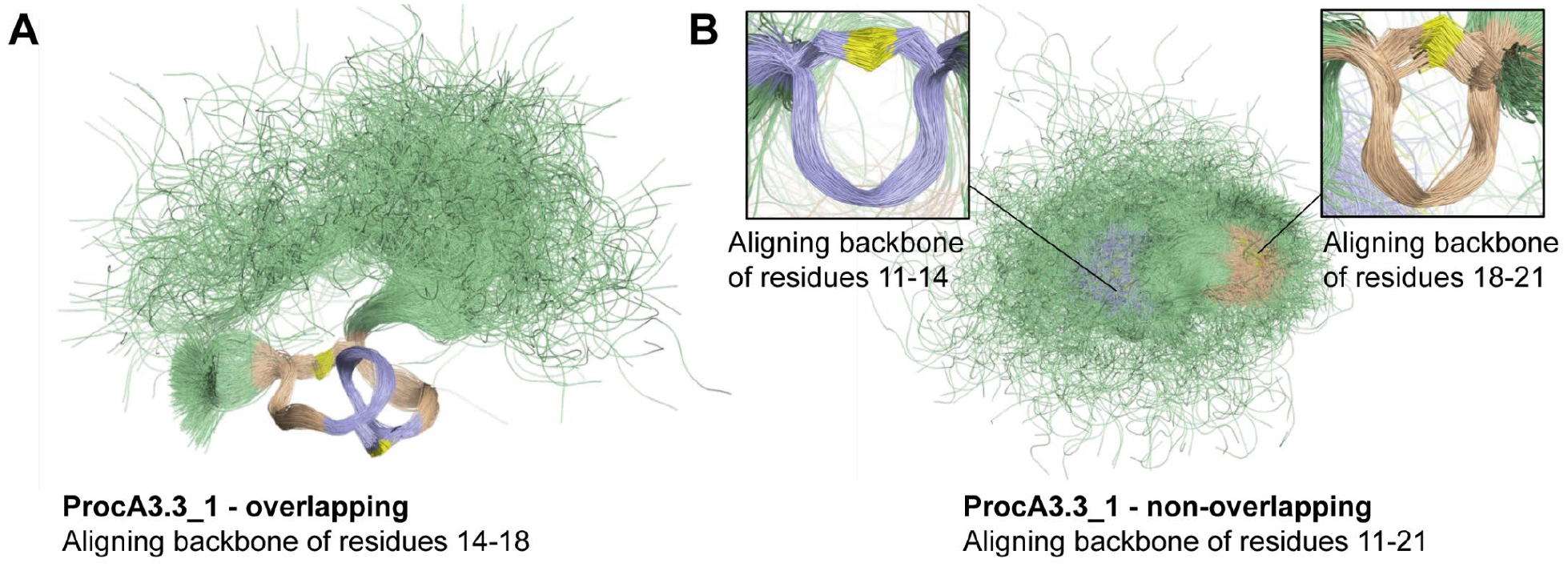
Overlay of 1,000 snapshots extracted from 1 μs simulations of Pcn 3.3_1 isomers with overlapping (A) and non-overlapping (B) ring patterns, in ribbon representation. The A- and A’-rings are shown in blue, the B- and B’-rings are shown in wheat, and the remainder of the peptide is shown in green; sulfur atoms are shown in yellow. Different alignments of the snapshots are shown for a better representation of the local structures of rings A, A’, B and B’.
Previous studies have shown that ProcM first completes all dehydrations and then in a slower process catalyzes the cyclization reactions.46, 63–64 Furthermore, for WT ProcA3.3, ProcM first installs the inner A-ring before cyclization of the larger B-ring, and intermediates with a single ring formed are committed to their final structures and do not return to a non-cyclized state.46 Thus, the pattern-determining step is the first cyclization event, which is a competition between formation of the A-ring (which sets the peptide en route to an overlapping ring product) or formation of either the A’ or B’ rings (which will result in a non-overlapping product, Figure 2A). Comparing WT ProcA3.3, which has the sequence Tyr-Gly-Gly in the A-ring, with ProcA3.3_1 for which the corresponding sequence is Phe-Val-Ile, we wondered whether the bulky amino acids in this sequence could have slowed down A-ring formation, making either A’ or B’ ring formation the predominant first cyclization event. In order to test the contribution of the Phe-Val-Ile motif for the outcome of the cyclization process, we swapped these residues in 3.3_1 and the corresponding Tyr-Gly-Gly in the WT sequence to create ProcA3.3_1-F15Y/V16G/I17G and ProcA WT-Y15F/G16V/G17I, respectively. Modification of these mutants by ProcM resulted in almost complete reversion of ring pattern outcome, with ProcA3.3_1-F15Y/V16G/I17G converted mostly into a product with overlapping ring pattern (Figure 7) and ProcA WT-Y15F/G16V/G17I transformed into a product with non-overlapping rings as shown by LC-MS/MS and TIMS-MS/MS (Figure S26). The reversal of regioselectivity upon exchange of the residues at positions 15–17 supports the importance of these residues in governing the ring pattern in these two peptides.
Figure 7.
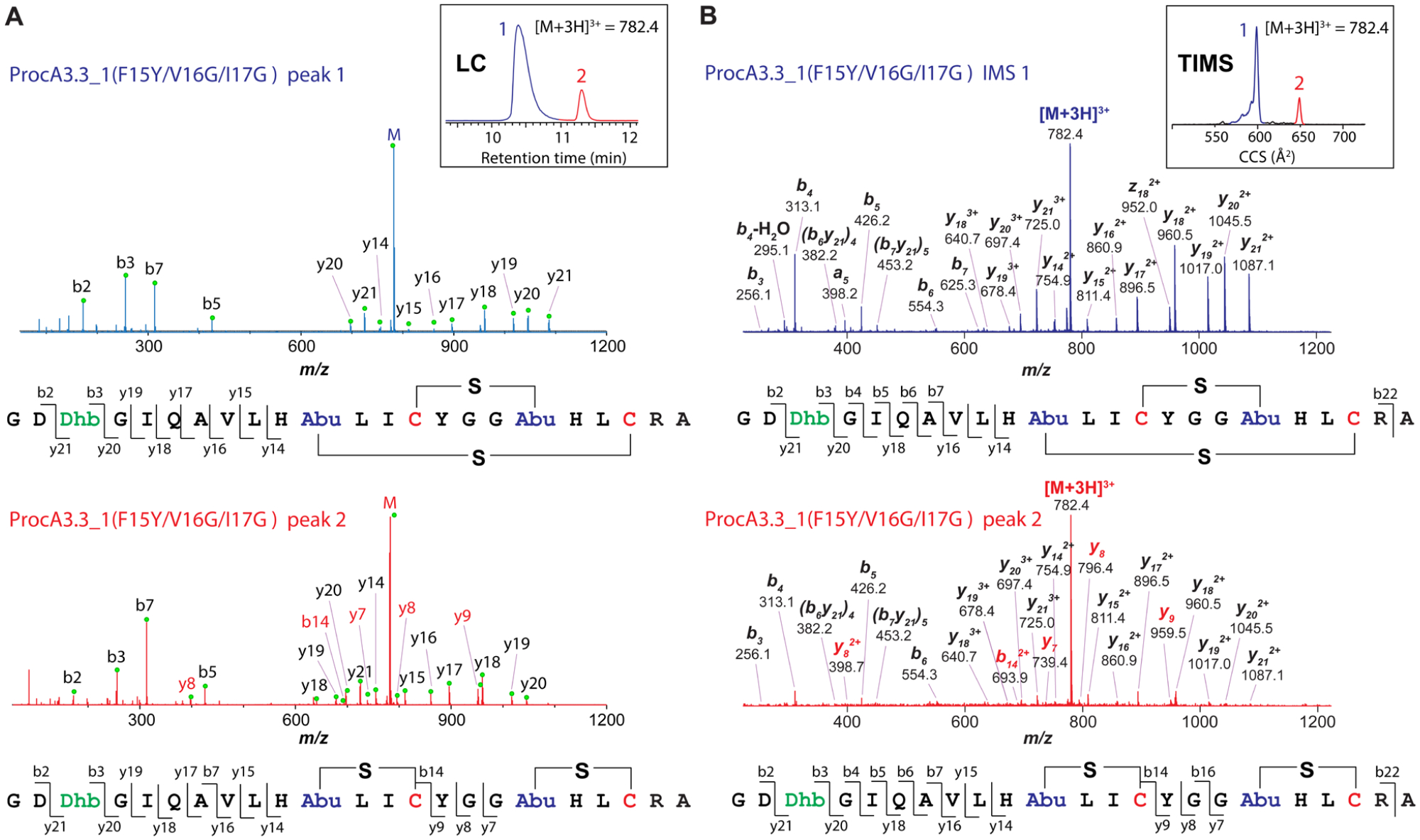
Analysis of ProcA3.3_1-F15Y/V16G/I17G by (A) LC-MS/MS, and (B) TIMS-MS/MS.
To investigate the generality of the importance of the three-residue sequence, we also swapped these respective linker residues between WT ProcA3.3 and ProcA3.3_10, a variant which results in approximately 30:70 ring pattern ratio (overlapping:non-overlapping) as judged by TIMS-MS/MS (Figure S17). After ProcM modification, ProcA3.3_10-L15Y/L16G/V17G did not show a significant change in the ring pattern ratio (Figure S27), whereas ProcA WT-Y15L/G16L/G17V displayed only a modestly increased amount of the non-overlapping ring pattern (Figure S28). These results illustrate that predicting the cyclization pattern is difficult because the outcome is dependent not only on the rate of the formation of the A-ring but also on the relative rates of cyclization of two potentially competing other rings for ProcA3.3 variants (A’, and B’).
These data also show that it will be challenging to assemble a set of guidelines to predict the cyclization pattern for the thousands of prochlorosins encoded in the genomes of cyanobacteria.43 Experimental determination of these rates will be an arduous task given the numbers of variants involved. For instance, to determine all cyclization rates for a lanthionine ring containing four-amino acids (i.e. with two variable residues between Dha and Cys such as the A’ and B’ rings of the non-overlapping products for ProcA3.3) would require 484 individual kinetic experiments for 22 possible amino acids in the variable positions (the 20 proteinogenic amino acids and Dha and Dhb). To include both Lan and MeLan rings this number would be 968. Considering that rings of five amino acids are also common in lanthipeptides (e.g., the A-ring of prochlorosin 3.3), the number of kinetic experiments to determine rates of all possible Lan/MeLan formation of this ring size would be 21,296. These numbers assume that the rates are only dependent on amino acids in the rings and not on any other sequences in the peptide.
With these considerations that demonstrate the impracticality of acquiring kinetic data to predict ring patterns, we investigated the possibility to obtain relative rates in a high throughput fashion by obtaining the ratios of overlapping and non-overlapping ring products by TIMS-MS/MS. Instead of analyzing single peptides, a mixture of twelve ProcA3.3 variants was analyzed by TIMS-MS and mobility spectra were extracted for each variant based on their m/z (Figure 8). The results were consistent with the ratios of peptides obtained using single peptide TIMS-MS analysis (Figure S29). Thus, TIMS-MS/MS provides a technology for fast and direct (no purification) identification of the lanthipeptide ring patterns of the ProcA3.3 variants. The findings in this study demonstrate an opportunity to scale TIMS-MS analysis of many ProcA3.3 variants and use the data for deep learning strategies to determine the preferences of sequences that can form four- and five-amino acid containing (Me)Lan rings (A’, B’, and A). These ring sizes together account for nearly half of all rings in naturally occurring class II lanthipeptides of known structure (16% four-amino acid rings and 27% five amino acid rings, Table S3).58 In turn, the data can then be utilized for future prediction of lanthipeptide structure for sequences that do not have structural information.
Figure 8.
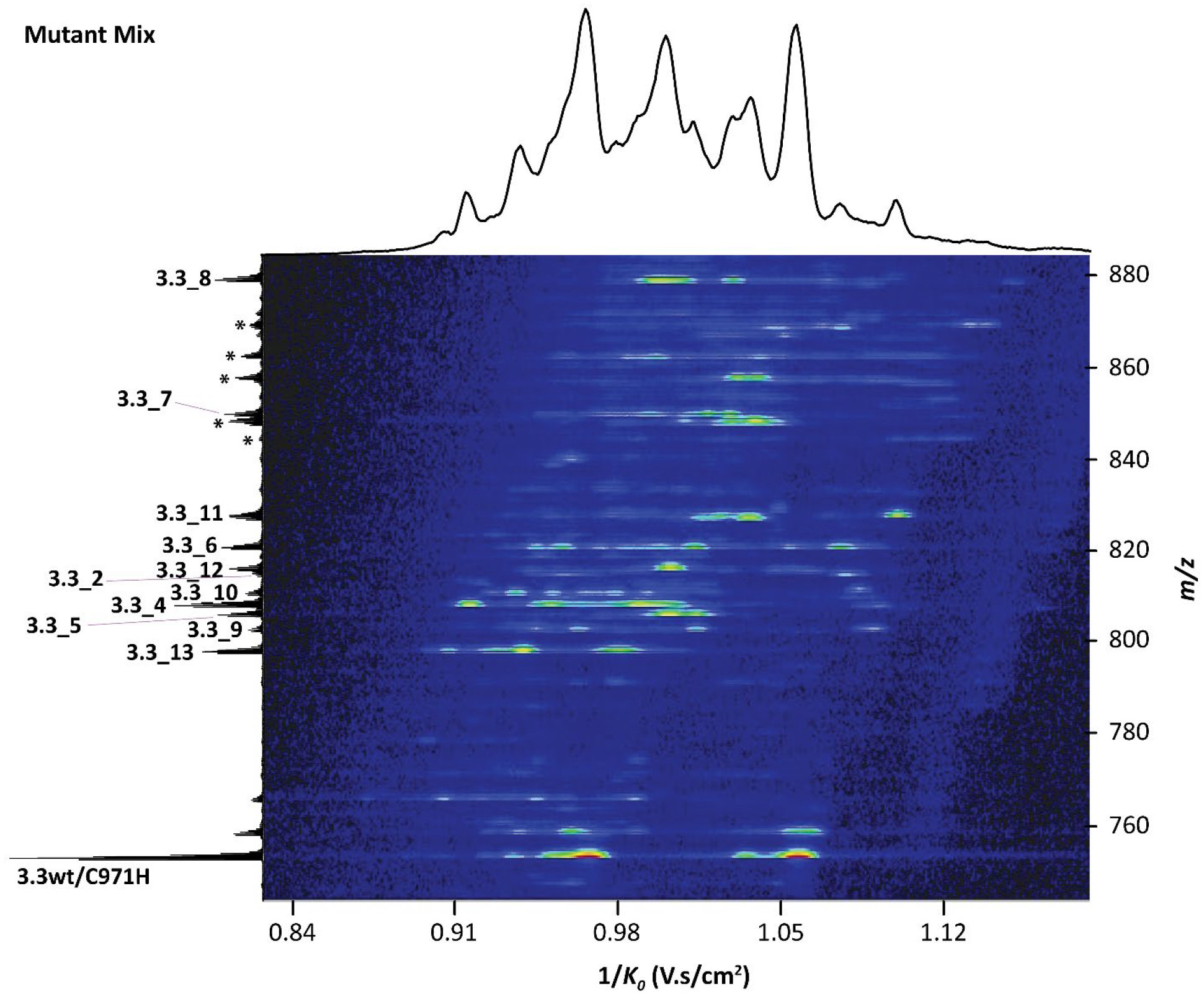
TIMS spectra of a mixture of twelve different ProcA3.3 variants after modification by ProcM and removal of the leader peptide. For comparison of the extracted data with data on individual purified peptides, see Figure S29. Peaks annotated with * correspond to partially cyclized lanthipeptide variants that contain a free Cys and that were alkylated with NEM in the work-flow (see experimental details). ProcA3.3wt/C971H represents a mixture of peptides with overlapping and non-overlapping rings reported previously46 that was used as standard.
Conclusions
Using ProcA3.3 as a model, we show that the regioselectivity of the Michael addition reactions to form methyllanthionine catalyzed by the substrate tolerant ProcM is controlled by substrate sequence. This study provides a clear demonstration that the kinetics of formation of competing rings can lead to alternative ring patterns with ProcM. Additional evidence was obtained against thermodynamic control via MD simulations that demonstrated that the experimentally observed preferences with ProcA3.3 cannot be explained by the relative energies of the products. This study also illustrates that predicting ring patterns from sequence is challenging owing to the complex interplay of the kinetics of competing reactions and requires much more data than currently available. We demonstrate that TIMS can greatly facilitate structural characterization of lanthipeptide variants, especially in analyzing constitutional isomers with identical sequence but different ring patterns. The method is much faster than LC and can be used with mixtures of peptides. We demonstrated previously that TIMS-MS/MS can also be used for analysis of complex biological samples in combination with LC separation,65–67 suggesting that even cell extracts of RiPP-producing bacteria may be analyzed in future work. The throughput of TIMS technology will allow large-scale study of the relationship between lanthipeptide sequence and the resulting ring pattern, which ultimately may facilitate prediction of the structure of the products of combinatorial biosynthesis in marine environments.
Experimental
Expression and purification of modified core ProcA3.3 variants
Plasmids extracted from pRSF_procA3.3NWY_procM stock (see Supporting Information) were used to transform E. coli BL21(DE3)-T1R. Random colonies were picked and used to inoculate 2 mL of TB/kanamycin. Then 1 mL of each overnight culture was miniprepped for sequencing. The remaining culture was stored at 4 °C for 1 day. After sequencing, the stored culture aliquot was used to inoculate (1:100) 15 mL of TB/kanamycin and the cells were cultured at 200 rpm, 37 °C. At OD 0.6–0.8, the culture was chilled on ice, followed by IPTG induction (0.25 mM) and shaking at 200 rpm, 18 °C for 18–20 h.
The following day, the culture was harvested at 5000×g for 15 min. The cell pellet was resuspended in 1 mL of LanB Start buffer (100 mM NaH2PO4, 10 mM Tris, 6 M guanidine, 10 mM imidazole, pH 8.0), and lysed by 5 rounds of freeze-thaw with liquid nitrogen-ultrasound water bath. The lysate was clarified by centrifugation for 10 min at 16,000×g. The supernatant was mixed with 100 μL Ni resin (Takara) pre-equilibrated with LanB Start buffer, shaken for 20 min at 20 °C, followed by centrifugation at 2000×g for 2 min. The resin was washed with 2–3 mL of LanB Wash buffer (100 mM NaH2PO4, 10 mM Tris, 6 M guanidine, 30 mM imidazole, pH 8.0). Finally, His-tagged peptide was eluted twice with 2×250 μL of LanB Elution buffer (20 mM Tris, 500 mM NaCl, 500 mM imidazole, pH 8.0). The elution fraction was concentrated with an Amicon Ultra-0.5 spin column (Millipore) for 20 min at 13,000×g (3 kDa MWCO) until ~20 μL solution remained. This fraction was recovered with 150 μL of LahT buffer (25 mM HEPES, 50 mM NaCl, 1 mM TCEP, pH 7.5, containing His6-LahT150 (200 μg/mL)). The digestion reaction was carried out at 37 °C for 4–5 h and monitored by MALDI-TOF MS.
Alkylation assay
N-ethylmaleimide (NEM) was dissolved in ddH2O to 100 mM. This stock solution was added 1:10 (v/v) to LahT150 digestion reactions and the mixture was incubated at 37 °C for 30 min. NEM alkylates any Cys residues that have not been cyclized such that they are no longer isobaric with cyclized peptides and do not appear in extracted ion chromatograms (they do appear in TIMS, e.g. Figure S22). The alkylation reaction was monitored by MALDI-TOF MS and alkylated peptides constituted minor products indicating that the variants were mostly cyclized by ProcM. These samples were then used for LC-MS and TIMS/MS.
Supplementary Material
ACKNOWLEDGEMENTS
We dedicate this study to the memory of Prof. Dan Tawfik (Weizmann Institute of Science) and fondly remember the stimulating discussions with him regarding the prochlorosins. We thank Dr. Alex V. Ulanov (Carver Biotechnology Center at UIUC) for help with the GC-MS experiments. This work was funded by the National Institutes of Health (R37 GM058822 to W.A.V.), the National Science Foundation Division of Chemistry (under CAREER award CHE-1654274 to F.F.L.) with co-funding from the Division of Molecular and Cellular Biosciences, the Agencia Estatal Investigación of Spain (AEI; Grant RTI2018-099592-B-C22 to G.J.O.), and Severo Ochoa Excellence Accreditation (SEV-2016-0644 to CIC bioGUNE).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Experimental procedures, supporting figures and tables.
References
- (1).Montalbán-López M; Scott TA; Ramesh S; Rahman IR; van Heel AJ; Viel JH; Bandarian V; Dittmann E; Genilloud O; Goto Y; Grande Burgos MJ; Hill C; Kim S; Koehnke J; Latham JA; Link AJ; Martínez B; Nair SK; Nicolet Y; Rebuffat S; Sahl H-G; Sareen D; Schmidt EW; Schmitt L; Severinov K; Süssmuth RD; Truman AW; Wang H; Weng J-K; van Wezel GP; Zhang Q; Zhong J; Piel J; Mitchell DA; Kuipers OP; van der Donk WA, New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep 2021, 138 (1), 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Repka LM; Chekan JR; Nair SK; van der Donk WA, Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem. Rev 2017, 117, 5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Cotter PD; Deegan LH; Lawton EM; Draper LA; O’Connor PM; Hill C; Ross RP, Complete alanine scanning of the two-component lantibiotic lacticin 3147: generating a blueprint for rational drug design. Mol. Microbiol 2006, 62 (3), 735. [DOI] [PubMed] [Google Scholar]
- (4).Field D; Collins B; Cotter PD; Hill C; Ross RP, A system for the random mutagenesis of the two-peptide lantibiotic lacticin 3147: analysis of mutants producing reduced antibacterial activities. J. Mol. Microbiol. Biotechnol 2007, 13 (4), 226. [DOI] [PubMed] [Google Scholar]
- (5).Appleyard AN; Choi S; Read DM; Lightfoot A; Boakes S; Hoffmann A; Chopra I; Bierbaum G; Rudd BA; Dawson MJ; Cortés J, Dissecting structural and functional diversity of the lantibiotic mersacidin. Chem. Biol 2009, 16 (5), 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Islam MR; Shioya K; Nagao J; Nishie M; Jikuya H; Zendo T; Nakayama J; Sonomoto K, Evaluation of essential and variable residues of nukacin ISK-1 by NNK scanning. Mol. Microbiol 2009, 72 (6), 1438. [DOI] [PubMed] [Google Scholar]
- (7).Caetano T; Krawczyk JM; Mosker E; Süssmuth RD; Mendo S, Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli. Chem. Biol 2011, 18 (1), 90. [DOI] [PubMed] [Google Scholar]
- (8).Field D; Molloy EM; Iancu C; Draper LA; PM OC; Cotter PD; Hill C; Ross RP, Saturation mutagenesis of selected residues of the alpha-peptide of the lantibiotic lacticin 3147 yields a derivative with enhanced antimicrobial activity. Microb. Biotechnol 2013, 6 (5), 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Krawczyk JM; Völler GH; Krawczyk B; Kretz J; Brönstrup M; Süssmuth RD, Heterologous expression and engineering studies of labyrinthopeptins, class III lantibiotics from Actinomadura namibiensis. Chem. Biol 2013, 20 (1), 111. [DOI] [PubMed] [Google Scholar]
- (10).Chen S; Wilson-Stanford S; Cromwell W; Hillman JD; Guerrero A; Allen CA; Sorg JA; Smith L, Site-directed mutations in the lanthipeptide mutacin 1140. Appl. Environ. Microbiol 2013, 79 (13), 4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Bierbaum G; Szekat C; Josten M; Heidrich C; Kempter C; Jung G; Sahl HG, Engineering of a novel thioether bridge and role of modified residues in the lantibiotic Pep5. Appl. Environ. Microbiol 1996, 62 (2), 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kuipers OP; Bierbaum G; Ottenwälder B; Dodd HM; Horn N; Metzger J; Kupke T; Gnau V; Bongers R; van den Bogaard P; Kosters H; Rollema HS; de Vos WM; Siezen RJ; Jung G; Götz F; Sahl HG; Gasson MJ, Protein engineering of lantibiotics. Antonie van Leeuwenhoek 1996, 69 (2), 161. [DOI] [PubMed] [Google Scholar]
- (13).Shi Y; Yang X; Garg N; van der Donk WA, Production of lantipeptides in Escherichia coli. J. Am. Chem. Soc 2011, 133 (8), 2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Bosma T; Kuipers A; Bulten E; de Vries L; Rink R; Moll GN, Bacterial display and screening of posttranslationally thioether-stabilized peptides. Appl. Environ. Microbiol 2011, 77 (19), 6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Oldach F; Al Toma R; Kuthning A; Caetano T; Mendo S; Budisa N; Süssmuth RD, Congeneric lantibiotics from ribosomal in vivo peptide synthesis with noncanonical amino acids. Angew. Chem. Int. Ed 2012, 51, 415. [DOI] [PubMed] [Google Scholar]
- (16).Field D; Cotter PD; Hill C; Ross RP, Bioengineering lantibiotics for therapeutic success. Front Microbiol 2015, 6, 1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zhou L; Shao J; Li Q; van Heel AJ; de Vries MP; Broos J; Kuipers OP, Incorporation of tryptophan analogues into the lantibiotic nisin. Amino Acids 2016, 48 (5), 1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Montalbán-López M; van Heel AJ; Kuipers OP, Employing the promiscuity of lantibiotic biosynthetic machineries to produce novel antimicrobials. FEMS Microbiol. Rev 2016, 41 (1), 5. [DOI] [PubMed] [Google Scholar]
- (19).Kuthning A; Durkin P; Oehm S; Hoesl MG; Budisa N; Süssmuth RD, Towards biocontained cell factories: an evolutionarily adapted Escherichia coli strain produces a new-to-nature bioactive lantibiotic containing thienopyrrole-alanine. Sci. Rep 2016, 6, 33447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Burkhart BJ; Kakkar N; Hudson GA; van der Donk WA; Mitchell DA, Chimeric leader peptides for the generation of non-natural hybrid RiPP products. ACS Cent. Sci 2017, 3 (6), 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Urban JH; Moosmeier MA; Aumüller T; Thein M; Bosma T; Rink R; Groth K; Zulley M; Siegers K; Tissot K; Moll GN; Prassler J, Phage display and selection of lanthipeptides on the carboxy-terminus of the gene-3 minor coat protein. Nat. Commun 2017, 8, 1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Hetrick KJ; Walker MC; van der Donk WA, Development and application of yeast and phage display of diverse lanthipeptides. ACS Cent. Sci 2018, 4, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Si T; Tian Q; Min Y; Zhang L; Sweedler JV; van der Donk WA; Zhao H, Rapid screening of lanthipeptide analogs via in-colony removal of leader peptides in Escherichia coli. J. Am. Chem. Soc 2018, 140 (38), 11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yang X; Lennard KR; He C; Walker MC; Ball AT; Doigneaux C; Tavassoli A; van der Donk WA, A lanthipeptide library used to identify a protein-protein interaction inhibitor. Nat. Chem. Biol 2018, 14 (4), 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kakkar N; Perez JG; Liu WR; Jewett MC; van der Donk WA, Incorporation of nonproteinogenic amino acids in class I and II lantibiotics. ACS Chem. Biol 2018, 13, 951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Geng M; Ravichandran A; Escano J; Smith L, Efficacious analogs of the lantibiotic mutacin 1140 against a systemic methicillin-resistant Staphylococcus aureus infection. Antimicrob. Agents Chemother 2018, 62 (12), e01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Geng M; Smith L, Modifying the lantibiotic mutacin 1140 for increased yield, activity, and stability. Appl. Environ. Microbiol 2018, 84 (15), e00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Li Q; Montalban-Lopez M; Kuipers OP, Increasing the antimicrobial activity of nisin-based lantibiotics against gram-negative pathogens. Appl. Environ. Microbiol 2018, 84 (12), e00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Schmitt S; Montalban-Lopez M; Peterhoff D; Deng J; Wagner R; Held M; Kuipers OP; Panke S, Analysis of modular bioengineered antimicrobial lanthipeptides at nanoliter scale. Nat. Chem. Biol 2019, 15 (5), 437. [DOI] [PubMed] [Google Scholar]
- (30).Kuipers A; Moll GN; Wagner E; Franklin R, Efficacy of lanthionine-stabilized angiotensin-(1–7) in type I and type II diabetes mouse models. Peptides 2019, 112, 78. [DOI] [PubMed] [Google Scholar]
- (31).Moll GN; Kuipers A; Rink R; Bosma T; de Vries L; Namsolleck P, Biosynthesis of lanthionine-constrained agonists of G protein-coupled receptors. Biochem. Soc. Trans 2020, 48 (5), 2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Li B; Sher D; Kelly L; Shi Y; Huang K; Knerr PJ; Joewono I; Rusch D; Chisholm SW; van der Donk WA, Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A 2010, 107 (23), 10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Dong SH; Tang W; Lukk T; Yu Y; Nair SK; van der Donk WA, The enterococcal cytolysin synthetase has an unanticipated lipid kinase fold. eLife 2015, 4, e07607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tang W; van der Donk WA, Structural characterization of four prochlorosins: a novel class of lantipeptides produced by planktonic marine cyanobacteria. Biochemistry 2012, 51 (21), 4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Bobeica SC; Zhu L; Acedo JZ; Tang W; van der Donk WA, Structural determinants of macrocyclization in substrate-controlled lanthipeptide biosynthetic pathways. Chem. Sci 2020, 11, 12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Lubelski J; Rink R; Khusainov R; Moll GN; Kuipers OP, Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci 2008, 65 (3), 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Ross AC; Vederas JC, Fundamental functionality: recent developments in understanding the structure-activity relationships of lantibiotic peptides. J. Antibiot 2011, 64 (1), 27. [DOI] [PubMed] [Google Scholar]
- (38).Zhang Q; Yu Y; Velásquez JE; van der Donk WA, Evolution of lanthipeptide synthetases. Proc. Natl. Acad. Sci. U. S. A 2012, 109 (45), 18361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Knerr PJ; van der Donk WA, Chemical synthesis and biological activity of analogues of the lantibiotic epilancin 15X. J. Am. Chem. Soc 2012, 134 (18), 7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Oman TJ; van der Donk WA, Insights into the mode of action of the two-peptide lantibiotic haloduracin. ACS Chem. Biol 2009, 4, 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Teng K; Zhang J; Zhang X; Ge X; Gao Y; Wang J; Lin Y; Zhong J, Identification of ligand specificity determinants in lantibiotic bovicin HJ50 and the receptor BovK, a multitransmembrane histidine kinase. J. Biol. Chem 2014, 289 (14), 9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Zhang Q; Yang X; Wang H; van der Donk WA, High divergence of the precursor peptides in combinatorial lanthipeptide biosynthesis. ACS Chem. Biol 2014, 9 (11), 2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Cubillos-Ruiz A; Berta-Thompson JW; Becker JW; van der Donk WA; Chisholm SW, Evolutionary radiation of lanthipeptides in marine cyanobacteria. Proc. Natl. Acad. Sci. USA 2017, 114 (27), E5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Le T; van der Donk WA, Mechanisms and evolution of diversity-generating RiPP biosynthesis. Trends Chem 2021, 3 (4), 266. [Google Scholar]
- (45).Yu Y; Zhang Q; van der Donk WA, Insights into the evolution of lanthipeptide biosynthesis. Protein Sci 2013, 22 (11), 1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Yu Y; Mukherjee S; van der Donk WA, Product formation by the promiscuous lanthipeptide synthetase ProcM is under kinetic control. J. Am. Chem. Soc 2015, 137 (15), 5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ridgeway ME; Lubeck M; Jordens J; Mann M; Park MA, Trapped ion mobility spectrometry: A short review. Int. J. Mass Spectrom 2018, 425, 22. [Google Scholar]
- (48).Jeanne Dit Fouque K; Fernandez-Lima F, Recent advances in biological separations using trapped ion mobility spectrometry – mass spectrometry. TrAC Trends Anal. Chem 2019, 116, 308. [Google Scholar]
- (49).Ridgeway ME; Bleiholder C; Mann M; Park MA, Trends in trapped ion mobility – Mass spectrometry instrumentation. TrAC Trends Anal. Chem 2019, 116, 324. [Google Scholar]
- (50).Bobeica SC; Dong SH; Huo L; Mazo N; McLaughlin MI; Jimenéz-Osés G; Nair SK; van der Donk WA, Insights into AMS/PCAT transporters from biochemical and structural characterization of a double glycine motif protease. eLife 2019, 8, e42305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Bobeica SC; van der Donk WA, The enzymology of prochlorosin biosynthesis. Methods Enzymol 2018, 604, 165. [DOI] [PubMed] [Google Scholar]
- (52).Tang W; van der Donk WA, The sequence of the enterococcal cytolysin imparts unusual lanthionine stereochemistry. Nat. Chem. Biol 2013, 9 (3), 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Acedo JZ; Bothwell IR; An L; Trouth A; Frazier C; van der Donk WA, O-methyltransferase-mediated incorporation of a β-amino acid in lanthipeptides. J. Am. Chem. Soc 2019, 141 (42), 16790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Lohans CT; Li JL; Vederas JC, Structure and biosynthesis of carnolysin, a homologue of enterococcal cytolysin with D-amino acids. J. Am. Chem. Soc 2014, 136, 13150. [DOI] [PubMed] [Google Scholar]
- (55).Liu W; Chan ASH; Liu H; Cochrane SA; Vederas JC, Solid supported chemical syntheses of both components of the lantibiotic lacticin 3147. J. Am. Chem. Soc 2011, 133 (36), 14216. [DOI] [PubMed] [Google Scholar]
- (56).Agrawal P; Khater S; Gupta M; Sain N; Mohanty D, RiPPMiner: a bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and crosslinks. Nucleic Acids Res 2017, 45 (W1), W80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Merwin NJ; Mousa WK; Dejong CA; Skinnider MA; Cannon MJ; Li H; Dial K; Gunabalasingam M; Johnston C; Magarvey NA, DeepRiPP integrates multiomics data to automate discovery of novel ribosomally synthesized natural products. Proc. Natl. Acad. Sci. USA 2020, 117 (1), 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Walker MC; Eslami SM; Hetrick KJ; Ackenhusen SE; Mitchell DA; van der Donk WA, Precursor peptide-targeted mining of more than one hundred thousand genomes expands the lanthipeptide natural product family. BMC Genomics 2020, 21 (1), 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Dit Fouque KJ; Moreno J; Hegemann JD; Zirah S; Rebuffat S; Fernandez-Lima F, Identification of lasso peptide topologies using native nanoelectrospray ionization-trapped ion mobility spectrometry-mass spectrometry. Anal. Chem 2018, 90 (8), 5139. [DOI] [PubMed] [Google Scholar]
- (60).Jeanne Dit Fouque K; Bisram V; Hegemann JD; Zirah S; Rebuffat S; Fernandez-Lima F, Structural signatures of the class III lasso peptide BI-32169 and the branched-cyclic topoisomers using trapped ion mobility spectrometry-mass spectrometry and tandem mass spectrometry. Anal. Bioanal. Chem 2019, 411 (24), 6287. [DOI] [PubMed] [Google Scholar]
- (61).Dit Fouque KJ; Scutelnic V; Hegemann JD; Rebuffat S; Maître P; Rizzo TR; Fernandez-Lima F, Structural insights from tandem mass spectrometry, ion mobility-mass spectrometry, and infrared/ultraviolet spectroscopy on sphingonodin i: lasso vs branched-cyclic topoisomers. J. Am. Soc. Mass Spectrom 2021, 32 (4), 1096. [DOI] [PubMed] [Google Scholar]
- (62).Jeanne Dit Fouque K; Hegemann JD; Santos-Fernandez M; Le TT; Gomez-Hernandez M; van der Donk WA; Fernandez-Lima F, Exploring structural signatures of the lanthipeptide prochlorosin 2.8 using tandem mass spectrometry and trapped ion mobility-mass spectrometry. Anal. Bioanal. Chem 2021, 413 (19), 4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Mukherjee S; van der Donk WA, Mechanistic studies on the substrate-tolerant lanthipeptide synthetase ProcM. J. Am. Chem. Soc 2014, 136 (29), 10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Thibodeaux CJ; Ha T; van der Donk WA, A price to pay for relaxed substrate specificity: a comparative kinetic analysis of the class II lanthipeptide synthetases ProcM and HalM2. J. Am. Chem. Soc 2014, 136 (50), 17513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Jeanne Dit Fouque K; Ramirez CE; Lewis RL; Koelmel JP; Garrett TJ; Yost RA; Fernandez-Lima F, Effective Liquid Chromatography-Trapped Ion Mobility Spectrometry-Mass Spectrometry Separation of Isomeric Lipid Species. Anal Chem 2019, 91 (8), 5021. [DOI] [PubMed] [Google Scholar]
- (66).Adams KJ; Ramirez CE; Smith NF; Munoz-Munoz AC; Andrade L; Fernandez-Lima F, Analysis of isomeric opioids in urine using LC-TIMS-TOF MS. Talanta 2018, 183, 177. [DOI] [PubMed] [Google Scholar]
- (67).Adams KJ; Smith NF; Ramirez CE; Fernandez-Lima F, Discovery and targeted monitoring of polychlorinated biphenyl metabolites in blood plasma using LC-TIMS-TOF MS. Int J Mass Spectrom 2018, 427, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


