Abstract
Plant NADPH-dependent cytochrome P450 reductase (CPR) is a multidomain enzyme that donates electrons for hydroxylation reactions catalyzed by class II cytochrome P450 monooxygenases involved in the synthesis of many primary and secondary metabolites. These P450 enzymes include trans-cinnamate-4-hydroxylase, p-coumarate-3′-hydroxylase, and ferulate-5-hydroxylase involved in monolignol biosynthesis. Because of its role in monolignol biosynthesis, alterations in CPR activity could change the composition and overall output of lignin. Therefore, to understand the structure and function of three CPR subunits from sorghum, recombinant subunits SbCPR2a, SbCPR2b, and SbCPR2c were subjected to X-ray crystallography and kinetic assays. Steady-state kinetic analyses demonstrated that all three CPR subunits supported the oxidation reactions catalyzed by SbC4H1 (CYP73A33) and SbC3′H (CYP98A1). Furthermore, comparing the SbCPR2b structure with the well-investigated CPRs from mammals enabled us to identify critical residues of functional importance and suggested that the plant flavin mononucleotide–binding domain might be more flexible than mammalian homologs. In addition, the elucidated structure of SbCPR2b included the first observation of NADP+ in a native CPR. Overall, we conclude that the connecting domain of SbCPR2, especially its hinge region, could serve as a target to alter biomass composition in bioenergy and forage sorghums through protein engineering.
Keywords: cytochrome P450 reductase, sorghum, C4H, C3′H, F5H, monolinol, lignin, NADPH, FMN, FAD
Abbreviations: ACE, atomic contact energy; C3′H, p-coumaroyl shikimate/quinate 3′-hydroxylase; C4H, cinnamate 4-hydroxylase; cDNA, complementary DNA; CPR, cytochrome P450 reductase; DOE, Department of Energy; F5H, ferulate 5-hydroxylase; FAD, flavin adenine dinucleotide; FMNH, FMN hydroquinone; MALS, multiangle light scattering; MR, molecular replacement; PDB, Protein Data Bank; ROS, reactive oxygen species
Cytochrome P450 reductase (CPR), also known as NADPH-dependent cytochrome P450 oxidoreductase (Enzyme Commission [EC] number: 1.6.2.4), is a membrane-bound oxidoreductase embedded in the cytosolic side of the endoplasmic reticulum (1). CPR was first identified as cytochrome c reductase (2) and later found to be able to reduce cytochrome P450 and cytochrome b5 (3, 4).
CPR belongs to the diflavin oxidoreductase family, whose members are reduced by the obligatory two-electron donor NADPH. Thus, CPR has been hypothesized as a fused enzyme between FMN-containing flavodoxin and a flavin adenine dinucleotide (FAD)-containing ferredoxin–NADP+ oxidoreductase (5). Since the discovery of CPR, a series of structural and functional studies have been established mainly on rat and human CPR. Generally, the determination of their 3D structures has revealed that CPRs contain three domains: an FMN-binding domain, an FAD-binding and NADP(H)-binding domain, and a connecting (or linker) domain with a flexible hinge region that links these two domains, although other naming conventions exist including one in which the FAD-binding and NADPH-binding domains are considered separate (6).
Binding of NADPH initiates the reduction of CPR, which is a rapid and multiphase process, including (1) hydride transfer from NADPH to FAD resulting in the reduced (FADH2, FMN, and NADP+) state; (2) dissociation of the charge–transfer complex between FADH− and NADP+ and interflavin electron transfer from FADH2 to FMN with the formation of the double-semiquinone state (FADH•, FMNH•); and (3) second interflavin electron transfer with partial conversion of the double-semiquinone state to a combination of oxidized FAD with FMN hydroquinone (FAD, FMNH2), which establishes a transient equilibrium between two-electron reduced states. The electron will then be transferred from FMNH2 to cytochrome P450 or other electron acceptor proteins (7, 8). If there is an excess of NADPH, supply of another pair of electrons from NADPH results in either partial or complete reduction of the enzyme to form the reduced (FADH2, FMNH2) state.
The crystal structure shows that the FAD and FMN in oxidized rat CPR are oriented such that two isoalloxazine rings are coplanar, with 4 Å between the planes corresponding to Van der Waals contact for optimal interflavin electron transfer, and the FAD is oriented at a proper distance from the NADPH molecule for optimal hydride transfer (8, 9). However, this close proximity of FAD and FMN, the so-called locked state or closed conformation, could hinder the interaction between the internally sequestered FMN-binding domain and the target external electron acceptor, which suggests CPR should have a rapid interdomain motion in response to a different redox state. By switching between the closed and open (or extended) states, CPR allows its FMN-binding domain to perform intramolecular and intermolecular electron transfers, one at a time (10). Based on NMR and small-angle X-ray scattering, the radius of gyration of human CPR is estimated as 2.7 and 3.3 nm for the closed and the open form, respectively (11). While CPR is still reduced, release of NADP+ shifts the conformational equilibrium toward extended forms capable of electron transfer from FMN to external acceptors (12).
As in all kingdoms of life, CPRs in plants are required for the activity of endoplasmic reticulum–associated class II cytochrome P450 monooxygenases (hereafter referred to as P450s) that are essential in the synthesis of a wide variety of metabolites, such as flavonoids, terpenoids, alkaloids, and phenylpropanoids (13). In contrast to animals, yeasts, and insects that all have a single CPR gene each, vascular plants contain one to three CPR genes (14). These CPR paralogs are differentially expressed, likely to meet the reductive demand of various P450-mediated reactions across the wide range of developmental and environmental conditions observed in plants (15). Despite a second gene duplication event within monocots prior to the differentiation of grasses, most grasses still contain one to three CPR paralogs (16). Eudicots contain both CPR1 and CPR2, which are classified based on their N-terminal sequences, whereas monocots contain only CPR2. The CPR1 of Arabidopsis thaliana (ATR1) is constitutively expressed, but the expression of ATR2 appears to be induced during lignin biosynthesis and in response to wounding or light treatment (17, 18).
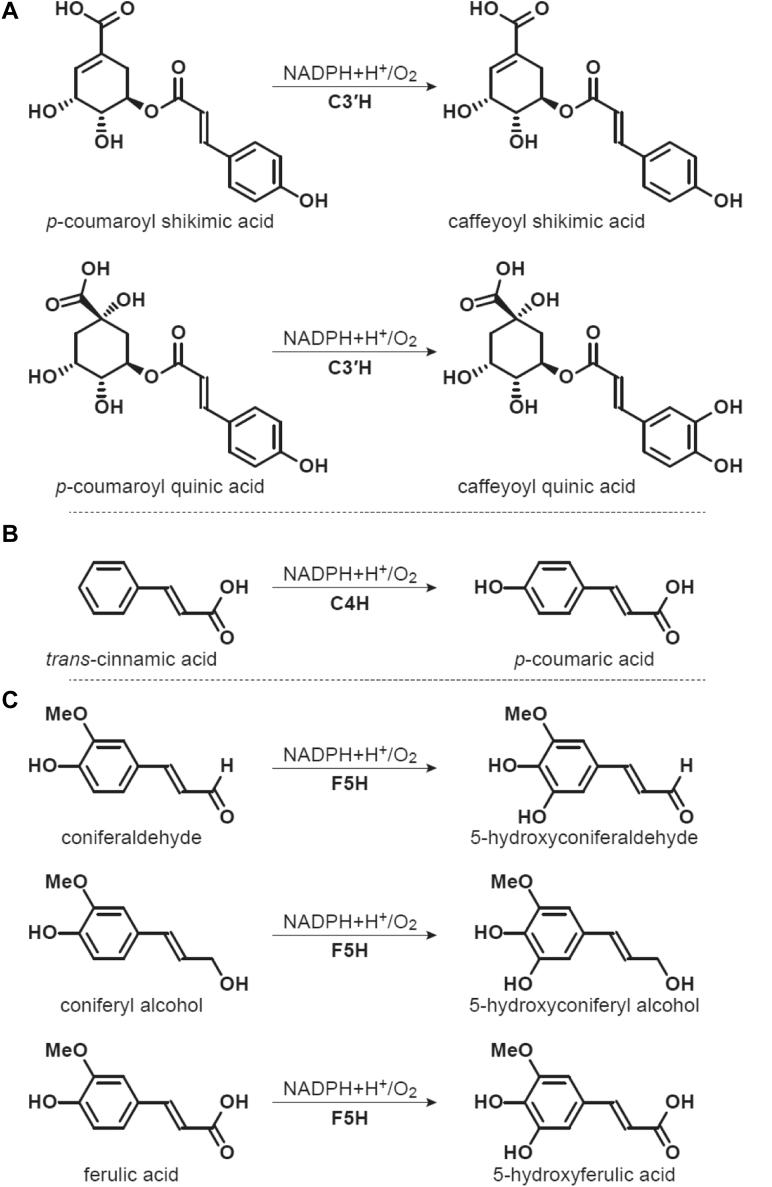
Despite their diverse and important roles in plants, the level of understanding of plant CPRs lags far behind the human and rat CPRs (19). ATR2, one of the two authentic CPRs in the model plant A. thaliana that has been physiologically and kinetically characterized, is the sole structure deposited in the Protein Data Bank (PDB) to date (PDB ID: 5GXU); it is as a complex with both FAD and FMN (20). In plant monolignol biosynthesis, CPR is required for the hydroxylation steps catalyzed by three class II P450s, cinnamate 4-hydroxylase (C4H; EC number: 1.14.13.11; (21)), p-coumaroyl shikimate/quinate 3′-hydroxylase (C3′H; EC number: 1.14.14.1; (22)), and ferulate 5-hydroxylase (F5H; EC number: 1.14.13; (23)) (Fig. 1). The hydroxylation reactions catalyzed by these three P450 enzymes give rise to p-coumaric acid, caffeoyl shikimic/quinic acid, and 5-hydroxyconiferaldehyde/5-hydroxyconiferyl alcohol, respectively (Fig. 1). These compounds are precursors for a myriad of organic compounds with agricultural, nutritional, and industrial relevance, such as stilbenes, chalcones, flavonoids, cinnamoyl anthranilates, monolignols, lignans, and lignin (21, 24, 25, 26).
Figure 1.
The reactions of cytochrome P450s in the monolignol biosynthetic pathway.A, p-Coumaroyl shikimate/quinate 3′-hydroxylase (C3′H) catalyzes the hydroxylation of p-coumaroyl shikimic acid and/or p-coumaroyl quinic acid, B, cinnamate 4-hydroxylase (C4H) catalyzes the hydroxylation of trans-cinnamic acid. C, ferulate 5-hydroxylase (F5H) catalyzes the hydroxylation of coniferaldehyde, coniferyl alcohol, and ferulic acid (97).
The first electron from CPR reduces the ferric substrate–bound heme iron of cytochrome P450 into the ferrous form, and the second electron further reduces this oxyferrous cytochrome P450 into the peroxo ferric (Fe3+OO)2− P450; then, a subsequent reaction oxidizes the substrate and yields a water molecule. The CPR is the redox partner enzyme providing electrons to most class II P450s, and thus adaptions to those individual interactions could be necessary for CPR in mammals. In plants, however, where more than one CPR isoform exists, there is a potential for specific interactions between particular P450s and one of the CPR isoforms through adaptation.
NADPH reduction of the FAD and NADP+–FADH2 charge–transfer complex formation are significantly faster in plants such as Artemisia annua CPR and A. thaliana CPR2 than in rat CPR. However, the interflavin electron transfer forming disemiquinone state is reported to be slower in plant CPRs than in rat CPRs (27).
As most P450s depend on CPR for the supply of electrons, detailed knowledge of the mechanisms for CPR–P450 interactions is important for understanding their diversification of multiple P450 and CPR genes and its impact on function. The role of CPRs in lignin deposition is especially significant because this heterogeneous aromatic polymer present in plant secondary cell walls is derived from the monolignol biosynthetic pathway that includes three P450 enzymes. The monolignol biosynthetic pathway results in the production of p-coumaryl, coniferyl, and sinapyl alcohols, which upon radical polymerization in the cell wall form p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) subunits of lignin, respectively (28, 29, 30, 31, 32). The S:G ratio in lignin has been shown to influence the efficiency of enzymatic saccharification of cell walls to produce fermentable sugars that can be converted to renewable fuels and chemicals. In dicots, a high S:G ratio appears to favor this conversion (33), whereas in grasses, a low S:G ratio appears to be more favorable (34). Thus, in-depth knowledge about the interaction among CPR, C4H, C3′H, and F5H could provide the basis for altering the carbon flux through the monolignol pathway and for modulating both lignin content and its composition. For example, it was demonstrated that a transfer DNA insertion in the AtCPR2 gene resulted in a 6% reduction in the lignin amount in the main inflorescence, a compositional shift of the lignin to a 10-fold higher fraction of H-units at the expense of S-units, and exhibited improved enzymatic saccharification, making CPR a potential target for engineering and valorizing plant cell walls for both biofuels and renewable chemicals (18). In Arabidopsis, F5H not only needs AtCPR2 but also a specific cytochrome b5 protein (AtCB5D) as the obligatory electron donor for its catalysis, and loss of functional mutations in an AtCB5D and an AtCPR2 gene impairs F5H activities to similar levels in plant extracts (35).
Both the mining of genetic variants in native populations of bioenergy crops and direct genetic manipulation of biosynthetic pathways have produced lignin feedstocks with unique properties for coproduct development (36). Sorghum is being developed as a dedicated bioenergy feedstock in the United States and abroad because it requires less water and nitrogen fertilizer than similar grasses (37, 38, 39). Given concerns over the negative impact of reducing lignin concentration on plant fitness (40), a key strategy in the development of next-generation bioenergy sorghums is the balance between agro-industrial needs and plant fitness. A more detailed mechanistic understanding of key enzymes involved in lignin biosynthesis including CPR can provide additional tools to tailor cell wall composition (41, 42, 43, 44, 45, 46, 47), by providing targets for mutational (48, 49, 50), gene editing (51, 52, 53), and transgenic approaches (54, 55, 56, 57).
Here, we characterize the three SbCPR2 isozymes of sorghum, SbCPR2a, SbCPR2b, and SbCPR2c, including crystal structures and kinetic data, to gain a mechanistic understanding of this key plant P450 reductase and to potentially enable future manipulation of monolignol biosynthesis and lignin content and composition of sorghum.
Results
Enzyme preparation and spectral properties
The Sorghum bicolor genome contains three CPR genes, Sobic.002G295100, Sobic.007G088000, and Sobic.006G245400 that encode SbCPR2a, SbCPR2b, and SbCPR2c, respectively (14). Recombinant SbCPR2 proteins containing an N-terminal 6× His tag with and without the native N-terminal hydrophobic membrane–anchoring region (Δ1–50) were produced in Escherichia coli using expression vectors containing complementary DNAs (cDNAs) corresponding to these three genes. The membrane-anchoring region was truncated in order to produce a more soluble form of CPR for structural analyses. In this article, amino acid residue numbering is based on the full-length SbCPR2b for both clarity and consistency.
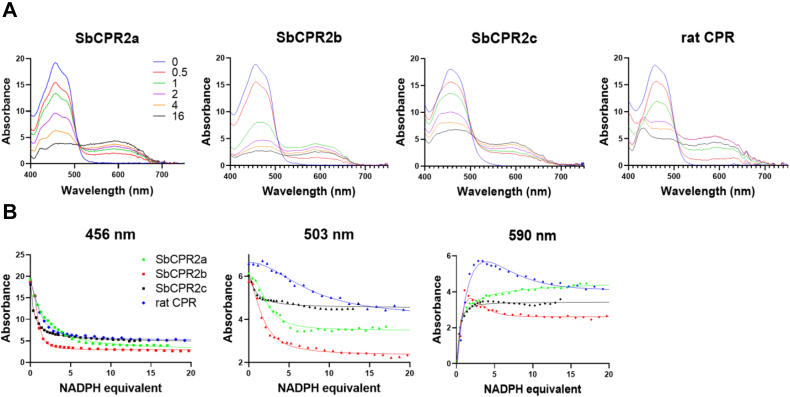
As shown in Figure 2A, where the UV–visible spectra of the oxidized SbCPR2s were shown in blue, they exhibited the main absorbance band at 456 nm with a shoulder at around 480 nm. Together with the absorbance band at 380 nm (Fig. S1), three SbCPR2s displayed a typical absorbance pattern of the flavoproteins (58). Further titration with K3Fe(CN)6 did not produce any spectral changes in the 550 to 650 nm region for SbCPR2a and SbCPR2b, confirming that those two SbCPR2s were in a fully oxidized state (59). However, SbCPR2c displayed a minor absorbance in the same region, which was absent in SbCPR2a or SbCPR2b (Fig. S1). The addition of K3Fe(CN)6 flattened the region, which indicated that purified SbCPR2c contains some flavins in a reduced state in the ambient condition that were further oxidized by this treatment (Fig. S1). This result was consistent with the greenish yellow color of the freshly purified SbCPR2c, whereas the solutions of SbCPR2a and SbCPR2b displayed a yellow color. The addition of NADPH under aerobic conditions caused the color of the SbCPR2 solutions to turn from yellow to green, indicating their flavins were being reduced.
Figure 2.
The absorbance spectra of SbCPR2s and anaerobic titration of CPR proteins with NADPH.A, exemplifies a series of spectra obtained with SbCPR2s and rat CPR. The spectra shown correspond to NADPH:CPR ratios of 0, 0.5, 1, 2, 4, and 16. B, the changes in absorbance at 456 and 595 nm. CPR concentrations were normalized to 1 mM prior to plotting. CPR, cytochrome P450 reductase.
Changes in the redox state of the enzymes observed in their anaerobic titration with NADPH
To compare the redox behavior for the three SbCPR2s, their anaerobic titrations with NADPH were performed by recording the absorbance spectra in 400 to 750 nm region at increasing NADPH:CPR molar ratio (RNADPH). A series of spectra obtained in the experiment with SbCPR2s and rat CPR are exemplified in Figure 2A. The respective changes in the absorbance at 456, 503, and 590 nm are shown in Figure 2B. The initial phase was associated with a rapid decrease in the amplitudes of the absorbance band at 456 nm, indicating the disappearance of the oxidized flavins. This decrease was associated with the appearance of the broad band centered at 590 nm, which indicated a formation of the blue (neutral) semiquinones (FADH• and/or FMNH•). In the case of SbCPR2b and rat CPR, the absorbance at 590 nm started to decrease after reaching its maximal value. This partial decrease signifies either appearance of the three- and four-electron reduced states of the flavoprotein or conversion of the neutral flavin semiquinones to the anionic states (FAD− and/or FMN−). However, in all four CPRs, the amplitude of the blue semiquinone band stabilized at RNADPH ratios of 10 to 12 and remained virtually unchanged after that point. This observation suggests that the enzymes cannot be converted entirely to their four-electron reduced state (FADH2 and FMNH2) even at high NADPH concentrations.
This finding is in good agreement with the previous findings obtained with rat CPR (58). The failure of producing a completely reduced CPR is due to the presence of increasing NADP+ concentration combined with the reversibility of the hydride transfer reaction. These circumstances establish an eventual equilibrium between the three- and four-electron reduced CPR species.
To characterize the maximal level of the four-electron reduction, the changes in the absorbance at 503 nm were analyzed. This wavelength corresponds to the isosbestic point in the absorbance spectra of the blue and red semiquinone species (60). As seen from Figure 2B, the titration curves obtained at this wavelength revealed considerable differences in both the amplitude and the shape. According to the analysis of these curves, the highest level of four-electron reduction was observed with SbCPR2b, which was followed by SbCPR2a. The amplitudes observed with SbCPR2c and rat CPR were considerably lower. Furthermore, the comparison of the shapes of the titration profiles at 503 nm revealed a significant difference between the rat CPR and its sorghum counterparts. In the case of rat CPR, the absorbance at 503 nm continued to decrease even at the highest NADPH concentrations signifying continuous reduction. The apparent RNADPH ratio needed to reach 50% of the maximal amplitude of this decrease (R50%) was as high as 7.5. In contrast, the maximal level of the apparent formation of the fully reduced state in SbCPR2s was reached at much lower NADPH concentrations. The R50% ratios estimated for SbCPR2a, SbCPR2b, and SbCPR2c were 2.3, 2.0, and 1.2, respectively.
This notable distinction may indicate a substantial difference between the rat CPR and SbCPR2s in the thermodynamic parameters of the interdomain electron transfer, which could be translated into a fundamental difference between mammalian and plant CPRs in the kinetics of their reduction (27).
Steady-state kinetics of three SbCPR isozymes
To compare the reductase activity of the three isoforms, SbCPR2a, SbCPR2b, and SbCPR2c were assayed for NADPH-dependent cytochrome c and cytochrome P450 reduction activities. Because of the insoluble nature of F5H in the E. coli expression system, the kinetic assay was carried out for SbC3′H and SbC4H1.
Cytochrome c
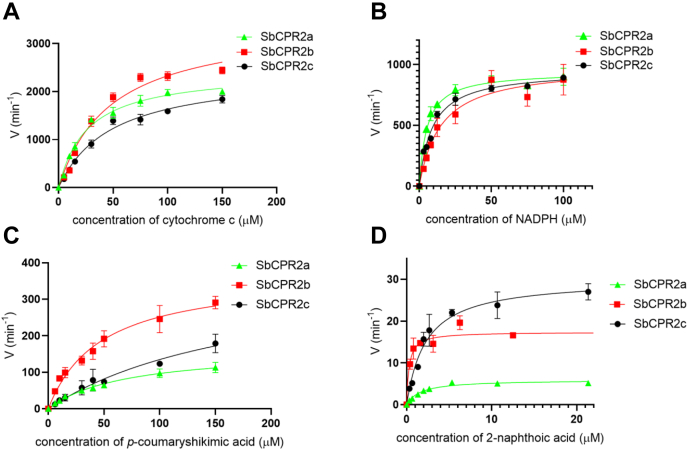
The activities of full-length SbCPR2s were assayed for their concentration dependence on NADPH (0–100 μM) with a fixed amount of cytochrome c (40 μM). In addition, the truncated (Δ1–50) SbCPR2s were assayed for their concentration dependence on cytochrome c (0–150 μM) with a fixed amount of NADPH (100 μM). The reduction of cytochrome c3+ to cytochrome c2+ was monitored at 550 nm (ε = 21 mM−1 cm−1). The results showed that all three SbCPR2s in either full-length or truncated form efficiently served as the electron donor to reduce cytochrome c, displaying typical Michaelis–Menten type dependency for both NADPH and cytochrome c (Fig. 3, A and B). For NADPH and cytochrome c, the corresponding Vmax and KM for SbCPR2a, SbCPR2b, and SbCPR2c are provided in Table 1. Overall, SbCPR2a was the most efficient in cytochrome c reduction according to the Vmax/KM values exhibited by the three enzymes. All three SbCPR2s displayed close to the value of 6.3 μM reported for the rat CPR (61). However, the values of of exhibited by SbCPR2s are noticeably higher than that observed with the rat CPR (16 μM, (61)).
Figure 3.
Kinetic assays of SbCPR2s with different electron acceptors.A, kinetic assay in dependence on cytochrome c concentration with constant NADPH concentration, B, kinetic assay for dependence on NADPH concentration with constant cytochrome c concentration. C, kinetic assay with C3′H in dependence on p-coumaroyl shikimic acid. D, kinetic assay with C4H in dependence on 2-naphthoic acid as an alternative substrate. Velocities are normalized by enzyme concentrations. C3′H, p-coumaroyl shikimate/quinate 3′-hydroxylase; C4H, cinnamate 4-hydroxylase.
Table 1.
Enzymatic parameters showing the comparison of SbCPR2s
| CPR type | Cytochrome c reduction |
C3′H reduction |
C4H reduction |
|||||
|---|---|---|---|---|---|---|---|---|
| Substrate | ||||||||
| Cytochrome c |
NADPH |
p-Coumaroyl shikimic acid |
2-Naphthoic acid |
|||||
| KM (μM) | Vmax (min−1)a | KM (μM) | Vmax (min−1)a | KM (μM) | Vmax (min−1)a | KM (μM) | Vmax (min−1)a | |
| SbCPR2a | 26.6 ± 3.3 | 2438.2 ± 99.7 | 5.3 ± 0.5 | 940.9 ± 22.8 | 69.6 ± 7.0 | 163.9 ± 69.6 | 1.6 ± 0.3 | 6.0 ± 0.3 |
| SbCPR2b | 48.9 ± 7.8 | 2432.8 ± 159.7 | 8.9 ± 1.0 | 950.7 ± 29.5 | 47.1 ± 6.6 | 369.8 ± 22.1 | 2.3 ± 0.4 | 30.1 ± 1.7 |
| SbCPR2c | 48.6 ± 11.5 | 3460.7 ± 337.4 | 14.9 ± 3.1 | 992.0 ± 14.9 | 186.8 ± 48.8 | 387.0 ± 66.5 | 0.3 ± 0.1 | 17.4 ± 0.9 |
Velocity was normalized by enzyme concentration.
C3′H (CYP98A1)
C3′H converts p-coumaroyl shikimic acid and/or p-coumaroyl quinic acid into caffeoyl shikimic acid and/or caffeoyl quinic acid. To compare the enzymatic activity of the three SbCPR2 isozymes to reduce the SbC3′H, a reconstituted membranous system in which SbC3′H interacts with preformed proteoliposomes containing one of each full-length SbCPR2 enzyme was used. We maintained the SbCPR concentration at two times higher than the concentration of SbC3′H to achieve the optimal activity of the system. For the steady-state kinetics experiments, the natural substrate of C3′H, p-coumaroyl shikimic acid, was used, and product formation of caffeoyl shikimic acid was monitored with HPLC (320 nm) (Fig. 3C). The Vmax and KM for SbCPR2a, SbCPR2b, and SbCPR2c are provided in Table 1. The corresponding Vmax/KM values are 4.0 × 104, 1.3 × 105, and 3.5 × 104 M−1 s−1, respectively, demonstrating that SbCPR2b is the most efficient electron donor for SbC3′H. SbCPR2c has the lowest catalytic efficiency, which is one fourth of SbCPR2bs. However, all three SbCPR isozymes displayed similar efficiencies and substrate-binding affinities, suggesting that C3′H could obtain similar reduction from any of them as long as they are in close proximity.
C4H1 (CYP73A33)
C4H catalyzes the 4-hydroxylation of trans-cinnamic acid using two electrons from reduced CPR. Similarly to the assay with C3′H, three full-length SbCPR2 isozymes were incorporated into a liposome and incubated overnight with SbC4H1 in a 2:1 ratio. Kinetic parameters for SbCPR2b pairing with SbC4H1 have been reported previously using either the natural substrate trans-cinnamic acid or a fluorometric substrate, 2-naphthoic acid (47). The use of the fluorometric substrate enables measurement of the product formation by monitoring its emission at 450 nm. Kinetic parameters obtained with both substrates were quite similar in terms of the Vmax and KM values determined (47). Therefore, the fluorometric substrate was used to compare the activity of SbCPR2a, SbCPR2b, and SbCPR2c with SbC4H1 (Fig. 3D). The Vmax and KM values for SbCPR2a, SbCPR2b, and SbCPR2c are provided in Table 1. The corresponding Vmax/KM values were 6.0 × 104, 2.2 × 105, and 1.0 × 106 M−1 s−1, respectively, indicating that all three SbCPR2s supported SbC4H1 activity. These results were comparable to SbC3′H results obtained previously, in which all three isozymes displayed similar values of Vmax and KM. SbCPR2c provided the best catalytic efficiency, and SbCPR2a displayed lowest catalytic efficiency that is 17% of the SbCPR2cs.
Comparison between full-length and truncated SbCPR2b
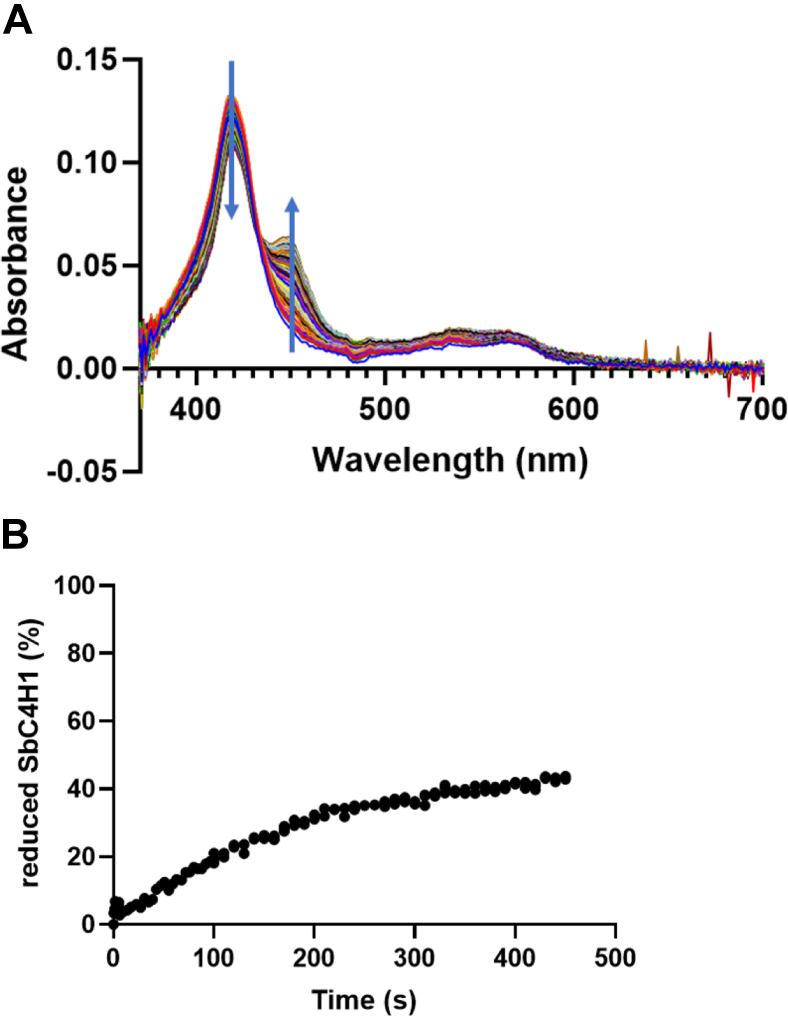
Our enzymatic experiments for SbCPR2s and SbC4H1 were performed in a membranous system containing SbCPR2s incorporated in proteoliposomes. The ability of truncated CPR to supply the electrons to C4H in solution has not been investigated to date. To probe this ability, changes in the absorbance of the system were monitored after adding NADPH to the mixture of SbC4H1 with the soluble form of SbCPR2b (Δ1–50) in CO-saturated solution. As shown in Figure 4, upon addition of NADPH to the system under anaerobic conditions, the absorbance band of the reduced carboxyl complex of P450 at 450 nm increased until it reached a plateau, equating to only 40% of the SbC4H1 in the membranous system. This result may suggest that without being membrane anchored, only a fraction of the SbC4H1 was available for the interaction with SbCPR2b, possibly because of aggregation of SbC4H1 in solution.
Figure 4.
Interaction between SbC4H1 and truncated SbCPR2b.A, continuous absorbance spectrum for SbC4H1 when reduced by CPR. Arrows showed the direction of spectrum change through time. The experiment was conducted with both enzymes’ membrane-anchoring peptide being truncated. Low ionic strength was provided with CO-saturated 20 mM Hepes buffer. B, the percentage of reduced SbC4H1 is plotted versus time, and the total amount of SbC4H1 was 1 μM. CPR, cytochrome P450 reductase.
Stoichiometry of substrate oxidation and futile cycling of the SbCPR2b pair with SbC4H1
Oxidation of a substrate molecule by cytochrome P450 requires a supply of two electrons from CPR. However, utilization of supplied electrons through the intrinsic oxidase and peroxidase shunts can also result in a futile cycle producing water and reactive oxygen species (ROS; H2O2 and superoxide), respectively. Both substrate monooxygenation and ROS production through the peroxidase shunt involve consuming 1 mol of oxygen per 1 mol of NADPH oxidized. In contrast, water production through the oxidase shunt consumes 2 mol of NADPH per 1 mol of oxygen (62). Thus, comparing the rate of NADPH oxidation with oxygen consumption and product formation, one can deduce the degree of coupling of the CPR–P450 system and assess the partitioning between the water-producing oxidase pathway and ROS-producing peroxidase shunt.
As we reported earlier (47), the turnover rate of SbC4H1 in substrate oxidation was much slower when the rat CPR was used as a surrogate of the native redox partner, SbCPR2b. To further probe the differences in catalytic efficiency and coupling between the homologous and heterologous redox pairs, the oxygen and NADPH consumption rates were measured during futile cycling of the systems (no substrate added) and in NADPH-dependent oxidation of 2-naphthoic acid to the product 6-hydroxy-2-naphthoic acid.
As presented in Table 2, in the absence of substrate, SbC4H1 was prone to futile cycling when paired with either of the two CPRs. The utilization of NADPH in the absence of substrate differed only slightly. In both systems, production of ROS amounted to ∼100% of the NADPH consumption. In the case of the heterologous pair of SbC4H1 with rat CPR, the addition of substrate did not result in any considerable increase in the NADPH consumption (Table 2), and the degree of its coupling to substrate oxidation was very low (∼6%). In contrast, adding the substrate to the SbCPR2b–SbC4H1 pair resulted in an approximate fourfold increase in NADPH and oxygen consumption rates, and the coupling degree was ∼25%. In both systems, the water production was about 20% with the presence of substrate. However, because of lower degree of coupling of NADPH consumption to product formation, the partitioning of the electron flow through the peroxidase shunt, and consequently, production of H2O2 and/or superoxide in the pair of SbC4H1 with rat CPR, was considerably higher than in the case of its pairing with SbCPR2b.
Table 2.
Parameters of CPR-dependent reduction of SbC4H in proteoliposomes
| System | Substrate | NADPH consumption, min−−1 | Oxygen consumption, min−1 | Product formation, min−1 | Coupling, % | H2O production, % |
|---|---|---|---|---|---|---|
| SbCPR–SbC4H1 | None | 28.5 ± 4.2 | 27.0 ± 0.3 | N/A | N/A | 5.5 ± 15.1 |
| 2-Naphthoic acid | 108.2 ± 7.0 | 88.1 ± 8.2 | 26.3 ± 4.4 | 25.6 ± 1.5 | 23.5 ± 9.0 | |
| Rat CPR–SbC4H1 | None | 12.5 ± 0.2 | 11.6 ± 0.3 | N/A | N/A | 8.0 ± 1.5 |
| 2-Naphthoic acid | 12.3 ± 1.3 | 10.2 ± 0.2 | 0.8 ± 0.2 | 6.2 ± 0.7 | 20.6 ± 15.0 |
Abbreviation: NA, not applicable.
Oligomeric state and overall structure of SbCPR2
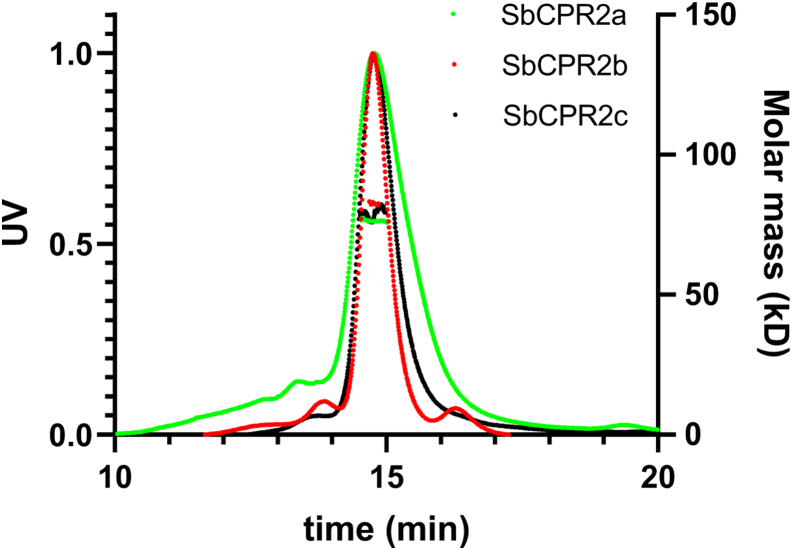
Truncated recombinant SbCPR2b and SbCPR2c (Δ1–50), which lacked the N-terminal 50 amino acids spanning both signal peptide and hydrophobic transmembrane-anchoring regions, were purified and crystallized. Three different forms of SbCPR2b were obtained by cocrystallization—no NADP+, NADP+, and NADP+—with potassium ferricyanide pretreatment to maintain its fully oxidized state. All those were crystallized in a same orthorhombic space group P212121 and diffracted up to 2.50, 2.36, and 2.70 Å resolution, respectively. Quality of the diffraction data and refinement statistics are listed in Table 3. The lattice packing of SbCPR2b crystal was through one molecule in the asymmetric unit without any extensive intermolecular interactions. Confirming this monomeric nature of SbCPR2b in crystal lattice, the multiangle light scattering (MALS) experiment also indicated that the solutional molar mass was ∼77 kDa (Fig. 5), matching the expected mass calculated from the amino acid sequence of SbCPR2b. SbCPR2a, and SbCPR2c that had similar molar masses, which indicated all SbCPR2s exist as a monomer in solution.
Table 3.
X-ray diffraction data and refinement statistics for SbCPR2
| Crystal and statistics | SbCPR2b apo |
SbCPR2b (ambient NADP+ complex) |
SbCPR2b (oxidized NADP+ complex) |
|---|---|---|---|
| PDB ID: 7SUZ | PDB ID: 7SUX | PDB ID: 7SV0 | |
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 47.143, 68.894, 154.265 | 47.127, 69.653, 153.271 | 46.947, 69.158, 150.949 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 38.91–2.501 (2.591–2.501) | 34.78–2.36 (2.444–2.36) | 44.83–2.7 (2.797–2.7) |
| Rmerge | 0.09279 (1.029) | 0.1129 (1.768) | 0.123 (1.537) |
| Wavelength (Å) | 1.000 | 1.000 | 1.000 |
| Unique reflections | 18,037 (1754) | 21,324 (1939) | 14,106 (1372) |
| Completeness (%) | 99.78 (98.76) | 99.08 (92.07) | 99.82 (99.85) |
| <I>/σI | 13.17 (2.21) | 13.58 (0.88) | 10.18 (0.96) |
| CC1/2 | 0.997 (0.729) | 0.997 (0.409) | 0.994 (0.841) |
| Redundancy | 7.1 (7.1) | 6.9 (5.2) | 5.5 (5.6) |
| Refinement | |||
| Rwork/Rfree | 0.2098/0.2499 (0.4020/0.4639) | 0.2321/0.2651 (0.3552/0.3573) | 0.2232/0.2659 (0.3084/0.3480) |
| Number of atoms | |||
| Protein and ligand | 3026 | 3019 | 3026 |
| Water | 84 | 40 | 17 |
| B-factors (Å2) | |||
| All atoms | 63.86 | 75.64 | 86.31 |
| Solvent | 64.69 | 76.08 | 86.53 |
| RMSD | |||
| Bonds (Å) | 0.012 | 0.012 | 0.013 |
| Angles (º) | 1.18 | 1.38 | 1.30 |
| Ramachandran plot | |||
| % Favored | 93.25 | 95.31 | 94.29 |
| % Outliers | 0.52 | 0 | 0 |
| Clashscore | 15.67 | 19.28 | 28.7 |
| TLS groups | 3 | 3 | 3 |
Abbreviation: TLS, translation/libration/screw.
Figure 5.
Oligomeric state of SbCPR2a, SbCPR2b, and SbCPR2c in solution. The elution profile for SbCPR2s was monitored with multiangle laser light scattering and is shown as absorbance (left y-axis) and molecular weight (right y-axis) versus elution time (min). The flow rate was 0.5 ml min−1. The solid line represents changes in absorption at 280 nm (green for SbCPR2a, red for SbCPR2b, and black for SbCPR2c). The thick cluster in the middle of the peak indicated the calculated molecular mass (∼77 kD) from light scattering, illustrating the monomeric nature of SbCPR2s. CPR, cytochrome P450 reductase.
From the early stage of crystallographic refinement, the electron density corresponding to the N-terminal FMN-binding domain and the connecting (or linker) domain were not visible in any of the three datasets, revealing the electron density corresponding only to NADP(H)-binding and FAD-binding domains. The resulting crystal structures of NADP(H)-binding and FAD-binding domains of SbCPR2b contained 15 α-helices and 13 β-strands and was superimposable with those of rat CPR, human CPR, yeast CPR, and Arabidopsis CPR2 with RMSD values of 0.89, 0.93, 0.93, and 0.68 Å, respectively. Despite numerous purification and recrystallization attempts with various protease inhibitors, the resulting crystals of SbCPR2b were consistently without FMN-binding and connecting domains. Purified SbCPR2b started to show a significant amount (∼50%) of cleaved proteins after 2 weeks. Small crystals were visible within 2 to 4 days, which indicated the truncated forms selectively crystallized from the mixture with full-length protein. This explanation was confirmed using SDS-PAGE analysis of the freshly formed crystal.
To identify the cleavage site, the crystals of SbCPR2b were collected and separated by SDS-PAGE. The major band of ∼45 kDa was cut, extracted, and then digested by trypsin and analyzed by MALDI–MS/MS. According to the data, the corresponding band uniquely contains the peptide 295NGHAVHDAQHPCQANVAVR313 of 2082 Da along with many other fragments from the tryptic digest, and thus, the cutting site was deduced as the peptide bond between Ala294 and Asn295. This was consistent with the presence of a ∼45 kDa fragment in our SDS-polyacrylamide gel and implied the cleavage site was located in the middle of a connecting domain. Confirming this, the tryptic digest of intact full-length SbCPR2b contains a 2614 Da peptide 290NFSLANGHAVHDAQHPCQANVAVR313, including all the other same tryptic peptides as dissolved crystals. Similar to SbCPR2b, a truncated form of SbCPR2a became predominant after 1 week of storage. LC–MS/MS results of SbCPR2a showed that its N-terminal ends either at 294SNGHAVYDIQHPCR307 or 292TLSNGHAVYDIQHPCR307, which is near the cleavage site in SbCPR2b. In short, the relatively rapid degradation was observed in all three SbCPR isoforms but was not observed in rat CPR (Fig. S2). Because of a fast cleavage event of the FMN-binding domain in SbCPR2a, its crystallization was not attempted.
FAD-binding and NADP(H)-binding site of SbCPR2b
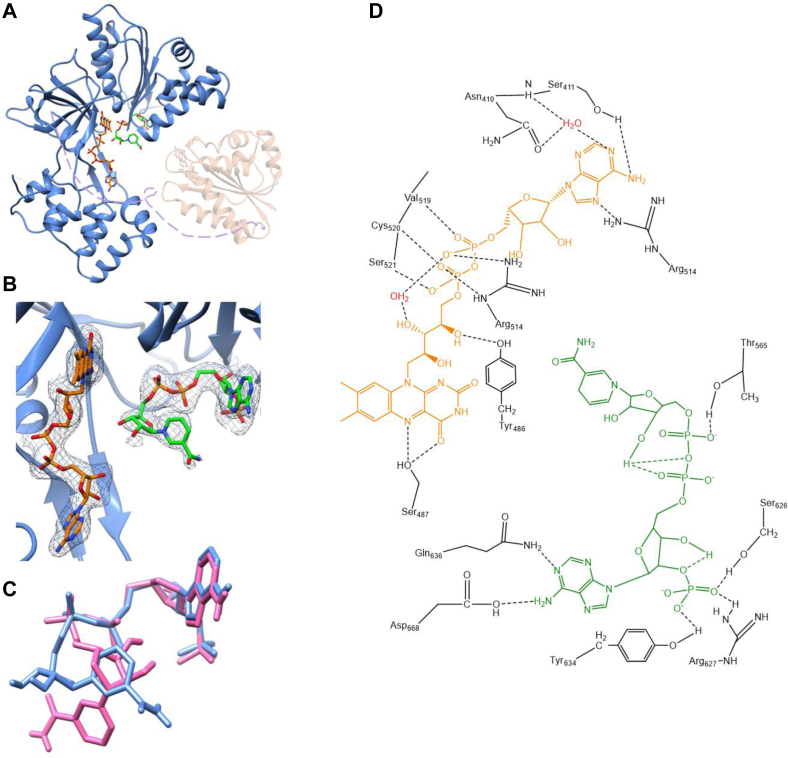
Although the crystallization buffer of SbCPR2b did not contain FAD, the electron density corresponding to the entire FAD molecule was well defined from the early stages of refinement of the diffraction data from all three crystallization conditions (Fig. 6A).
Figure 6.
The crystal structure of SbCPR2b.A, ribbon diagram representing the crystal structure of SbCPR2b (blue) with the FMN-binding domain (orange) that was built by the homology model (Phyre2 (68)), and the connecting domain is shown in violet dashed line. The position and orientation of FMN-binding domain is arbitrary; B, FAD and NADP+ with 2mFo–DFc (0.97) map in the naturally oxidized SbCPR2b. C, the NADP+ molecules in the CPR structures: NADP+ in naturally oxidized SbCPR2b (blue) and fully oxidized (pink). D, hydrogen-bonding network that anchors FAD and NADPH in SbCPR2b. Side chain and backbone of amino acids are shown in black. FAD is shown in orange, and NADP+ is shown in green. Water molecules are shown in red. CPR, cytochrome P450 reductase; FAD, flavin adenine dinucleotide.
The FAD cofactor was bound through a network of hydrogen bonds, which was similar to the observations from other CPR structures. The pyrophosphate group of the FAD was within hydrogen-bonding distance to the backbone amide nitrogen of Ser521 and Cys520 (Fig. 6D), and this group was in an N-capping position compensating for the macrodipole of the nearby α-helix. The isoalloxazine ring of FAD established hydrogen bonds with the side chain of Ser487 (Fig. 6D), backbone carbonyl oxygen of Thr502, and backbone nitrogen atoms of Ser487 and Ala504. In addition, the re-side of the same isoalloxazine ring was in a parallel p–p stacking interaction with the indole side chain of Trp706. On the other hand, the side chain of Tyr486 established an almost perpendicular p–p interaction with the si-side of isoalloxazine ring of FAD with an angle of ∼70°. There were three bound water molecules near the isoalloxazine ring, and one of them formed a bridge between O4 of isoalloxazine and the backbone of Thr502 and Ser487. The other water molecule was within a hydrogen-bond distance from the ribitol hydroxyl group and O2 of isoalloxazine and the backbone amide of Ala504. The third water was connecting O2 of isoalloxazine and the indole ring of Trp706. The ribitol hydroxyl groups also formed hydrogen bonds with the side chain of Tyr486 and the backbone of Tyr485. Both electrostatic interaction with the guanidium side chain of Arg484 and polar interaction with imidazole side chain of His516 stabilized the pyrophosphate group of FAD. The same pyrophosphate group also established polar interactions with the side chain of Ser521 and the backbone amide groups of Val519 and Cys520 (Fig. 6D). In addition, there were four water molecules within a hydrogen-bond distance from the same pyrophosphate group. The adenosine group of FAD was hydrogen bonded to the side chain of Ser411 and Arg514 (Fig. 6D). In contrast to animal CPRs, where the tyrosine side chain undergoes a π–π interaction with the adenine ring, this interaction was not observed in the structure of SbCPR2b. Based on the sequence alignment, the corresponding residue in SbCPR2b is glutamic acid.
Similarly to the failed attempt to cocrystallize AtCPR2 with NADPH (20), our attempt of crystallizing SbCPR2b with NADPH was also unsuccessful, and thus efforts were focused on obtaining the oxidized form of SbCPR2b through addition of NADP+ instead of NADPH. When SbCPR2b was crystallized with NADP+ under atmospheric conditions, the electron density of the entire NADP+ molecule was well defined, in contrast to the rat CPR (63) and human CPR (64), where the ribosyl-nicotinamide moiety of the bound NADP+ was significantly disordered. The diffraction data obtained from crystals of SbCPR2b that had been in a solution containing NADP+ with potassium ferricyanide pretreatment to maintain its fully oxidized state showed the electron density for the entire NADP+. This is the first observation of ribosyl-nicotinamide in any native CPR. In Figure 6A, several residues established noncovalent interactions with the 2′,5′-ADP portion of NADP+, which included Gly564, Thr565, Ala504, Gly595, Arg597, Ser626, Arg627, Tyr634, Gln636, Gly664, Met665, and Asp668 (Fig. 6D). A significant finding was that the position of the indole ring from Trp706 (Fig. 6B) blocked a potential interaction between the nicotinamide ring of NADP+ and the flavin ring of FAD. The nicotinamide moiety of NADP+ adapts different conformations in the atmospheric versus fully oxidized form of SbCPR2b. In the fully oxidized form, the nicotinamide ring that was in the syn-conformation relative to the ribosyl ring was extended to a position between the side chain of Glu508 and Arg323, being away from isoalloxazine ring of FAD. In the atmospheric form, the nicotinamide ring was in anti-conformation and near the side chain of Asp661 (Fig. 6C), which was located exactly between the position of the reduced NADP+ in rat CPR (PDB ID: 5URE) (65) and the position where indole ring and isoalloxazine ring stacked in our structure. The comparison between these two structures illustrates different conformations of NADP+ in different redox states in sorghum CPR.
Structure of SbCPR2c
The soluble form of SbCPR2c without its membrane-anchoring region (Δ1–50), which had the slowest cleavage rate, was cocrystallized with NADP+ within a couple of days. The resulting crystal lattice in the P43212 space group with the unit cell parameter of a = 116.82, b = 116.82, and c = 431.05 Å diffracted only up to ∼4.5 to 6 Å resolution. All attempts including transferring crystals to the reservoir with higher concentration of precipitant up to 50% PEG-3350 for dehydration and crosslinking crystals with glutaraldehyde (66) were applied to improve resolution, but the best diffracted crystals yielded only a 4.5 Å resolution at best. The initial trials of molecular replacement (MR) for finding the position and orientation for the whole SbCPR2c molecule did not yield any solutions, but a search for NADPH-binding or FAD-binding domain offered a solution and produced a good quality electron density containing the FAD and 2′,5′-ADP moiety of NADP+. However, despite our numerous attempts, the MR did not yield any solution on the FMN-binding domain. The SDS-PAGE of the dissolved SbCPR2c crystals showed a single band for the SbCPR2c at ∼70 kDa, which indicated the protein was fully intact, unlike what had been observed for SbCPR2b. It is likely that the FMN-binding domain of SbCPR2c was not establishing any specific interface with the FAD-binding domain existing in an ensemble of structures. The crystal lattice was mainly built through interaction amongst FAD-binding domains (Fig. S3), which may explain the low-resolution diffraction. The RMSD between SbCPR2c and SbCPR2b was 1.14 Å, and the conformation of the 2′,5′-ADP-moiety and FAD in these two structures was readily superimposed.
FMN-binding domain model and docking
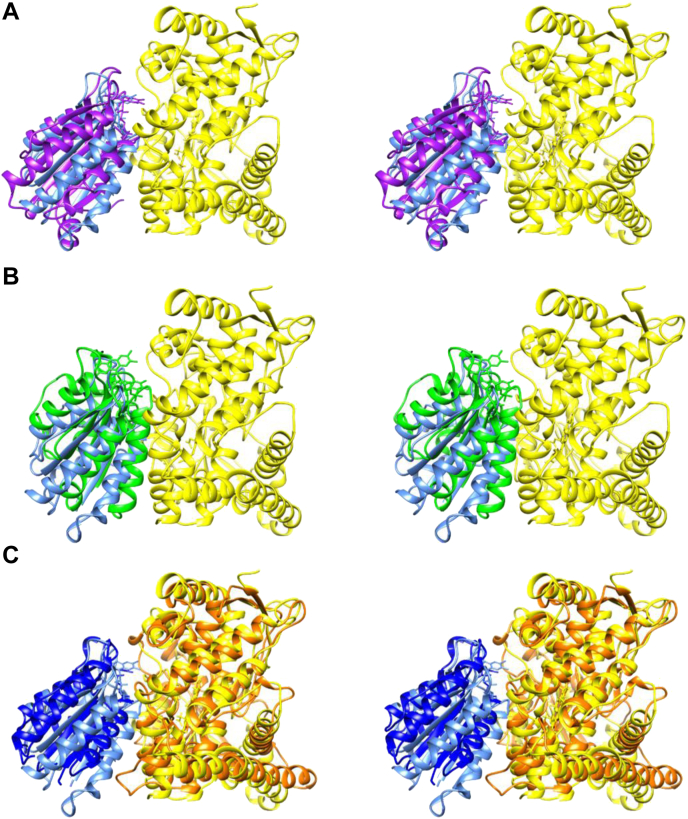
Molecular docking experiments were performed using the ‘PatchDock’ algorithm (http://bioinfo3d.cs.tau.ac.il/PatchDock/patchdock.html) (67) between the crystal structure of SbC4H1 (PDB ID: 6VBY) and the FMN-binding domain model of SbCPR2b, which was built with Phyre2 server (Structural Bioinformatics Group) (68). The FMN-binding domain of rat CPR (PDB ID: 1AMO) and the FMN-binding domain of ATR2 (PDB ID: 5GXU) were used as positive controls for this docking experiment (Fig. 7, A and B). Among the top 10 solutions of both FMN-binding domains, four of each located an FMN-binding domain at the proper position at the heme-proximal side of SbC4H1, which would allow electron transfer from FMNH2 to heme iron. Those docking solutions for SbCPR2b had an average score of 11,609.5, a binding area of 1591.05 Å2, and atomic contact energy (ACE) of 360.9 cal mol−1. This solution for rat CPR at the similar orientation and position displayed an average score of 12,536, a binding area of 1736.7 Å2,and ACE of 294.35 cal mol−1 (Fig. 7A). The solution for ATR2 at the similar orientation and position displayed an average score of 12,066, a binding area of 1763.0 Å2, and ACE of 449.98 cal mol−1 (Fig. 7B). Furthermore, the location of the interface between FMN-binding domain of SbCPR2b and SbC4H1 is very similar to that between the FMN-binding domain and the heme-binding domain in the structure of Bacillus megaterium P450BM3 (PDB ID: 1BVY) (69) (Fig. 7C). In addition, the key secondary structural elements of SbC4H1 and P450BM3 align well. Consequently, the distance and angle between FMN and heme are very similar in those complexes.
Figure 7.
Molecular docking of FMN-binding domain with SbC4H1.A, stereo view of SbCPR2b FMN-binding domain (light blue) docked to SbC4H1 (yellow, PDB ID: 6VBY). The rat CPR FMN-binding domain (purple, PDB ID: 1AMO) docked to SbC4H1 at the same position was superimposed. B, stereo view of SbCPR2b FMN-binding domain (light blue) docked to SbC4H1 (yellow, PDB ID: 6VBY). The ATR2 FMN-binding domain (green, PDB ID: 5GXU) docked to SbC4H1 at the same position is superimposed. C, stereo view of SbCPR2b FMN-binding domain (light blue) docked to SbC4H1 (yellow, PDB ID: 6VBY) is superimposed with the heme (orange) domain and FMN-binding domain (dark blue) of the cytochrome P450 (BM-3) (PDB ID: 1BVY). The molecular docking was performed using the “PatchDock” algorithm (67), and the FMN-binding domain model of SbCPR2b was built with Phyre2 server (68). CPR, cytochrome P450 reductase.
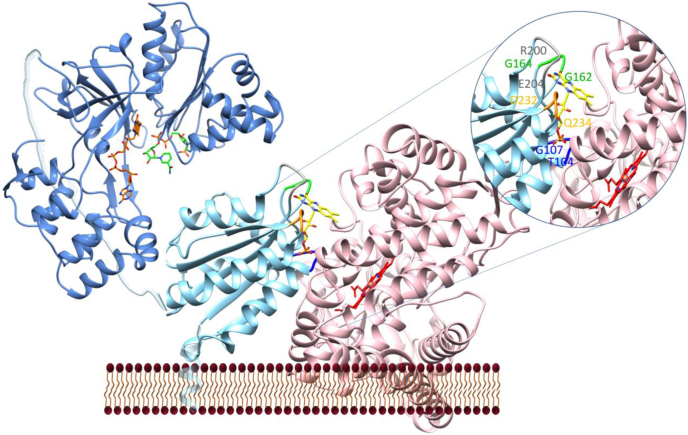
Two particular sequences in SbCPR that form the interface with the P450 enzyme, 160TYGDGEPTD168 and 229LGDDDQCIEDD239, are highly conserved among the FMN-binding domain of CPR from various species (Fig. S4). Our docked model of the FMN-binding domain of SbCPR to SbC4H1 adopted both peptide motifs, especially the tip of the loop in the two sequence motifs 232DDQ234 and 162GDG164 were interacting with the surface of SbC4H1 (Fig. 8). In addition, two other loops, 200RQYE203 and 104TQTG107, constituted the interface with SbC4H1. Significantly, those four loops contain the residues that establish the FMN-binding pocket. In addition, the four sequence motifs of rat CPR exactly establish the interface with rat heme oxygenase (PDB ID: 3WKT) (70), thus strongly supporting our docked interface between SbC4H1 and the FMN-binding domain of SbCPR2b. The backbone amide nitrogen and carbonyl oxygen of Gly141 in the first peptide mentioned previously, 139TYGEGDPTD147 in rat and human CPRs, form hydrogen bonds to either oxidized or reduced N5 in the isoalloxazine ring of bound FMN and FMNH2, respectively, which is referred to as either the O-down or O-up conformation. Deletion of this completely conserved Gly141, which is equivalent to Gly162 in SbCPR, abolishes the catalytic activity of CPR because of the destabilization of the FMN semiquinone state (71). It was also proposed that the consequential reorientation of Gly141 upon altering the oxidation state disrupts the network of interactions with neighboring residues and interdomain interactions, leading to the open conformation of CPR (11, 72).
Figure 8.
Plausible interaction between SbCPR2b and SbC4H1. The FMN-binding domain of SbCPR2b is shown in light blue and FMN molecule as yellow. The FAD-binding domain is shown in dark blue with FAD in orange and NADP+ in green. The relative orientation between the FAD-binding domain and the FMN-binding domain is arbitrary. SbC4H1 is shown in pink with heme as red. Inset shows the interface between the FMN-binding domain and SbC4H1 with the four interacting loops highlighted. FAD, flavin adenine dinucleotide.
Discussion
Analysis of sequence and expression data from various plant species has indicated the existence of divergent CPR genes in the plant kingdom that are either constitutively expressed or regulated in response to the various environmental and developmental cues. The occurrence of multiple CPR isoenzymes in plants may also reflect the diversity of P450s required for synthesis of the many metabolites that are essential for plant growth, development, and protection from environmental challenges.
Three CPR isozymes, SbCPR2a, SbCPR2b, and SbCPR2c, have been identified in sorghum (14), which have on average 72% amino acid sequence identity and 87% sequence similarity. Those identity and similarity values increase to 80% and 89%, respectively, when the N-terminal 70 amino acids containing the leader and transmembrane-anchoring sequences are excluded from the analysis. Significantly, the similar NADPH-dependent reduction behaviors of SbCPR2a and SbCPR2c (Fig. 2) are consistent with the observed phylogenetic distance among three SbCPR2s (Fig. S5). As indicated in the phylogenetic tree, the three subclasses of CPR2 from the monocots for which the sequences were analyzed clustered together. A BLAST (73) search to identify proteins with similar amino acid sequences in the nonredundant database showed over 90% identity with CPRs from several other grasses, including Miscanthus lutarioriparius, Zea mays, Setaria italica, Digitaria exilis, Eragrostis curvula, and Oryza sativa. As expected, three SbCPR2s are more closely related to ATR2 than ATR1, in terms of both amino acid identity and similarity values. The phylogenetic distance to the animal CPRs is even greater than to both Arabidopsis CPR1 and CPR2. One notable difference between the sequences of ATR1 and ATR2 is the Ser/Thr-rich sequence 2SSSSSSSTS10 at its N terminus, for which the similarity to the chloroplast target sequence has been reported (74). The corresponding sequence does not exist in the N terminus of SbCPR2s.
A Dali (http://ekhidna2.biocenter.helsinki.fi/dali/) (75) search also indicated that CPR2 from Arabidopsis (PDB ID: 5GXU) showed the highest Z-score of 52.8, followed by human CPR (PDB ID: 3QFS) with an Z-score of 45.2, rat CPR (PDB ID: 4Y7C) with an Z-score of 44.9, and the FAD-binding domain of BM3 from Priestia megaterium (PDB ID: 4DQK) with an Z-score of 44.6. Among those structural homologs, the conserved residues were distributed throughout all secondary structural elements. In general, the CPRs with the highest similarity scores have the same topological order for the secondary structural elements as observed in SbCPR2b. However, detailed visual inspection revealed a few structural heterogeneities in terms of the number and size of helices and sheets.
Structural and mechanistical comparison with rat and human CPRs
The observed high sequence similarity among the three SbCPR2 isozymes is consistent with the similar activities they displayed in conjunction with cytochrome c and cytochrome P450s (SbC4H1 and SbC3′H). Although our crystal structures of SbCPR2b and SbCPR2c did not provide structural data pertaining to the FMN-binding domain, the structure of the FMN-binding domain is expected to be very similar to other reported structures considering the high level of sequence identity (∼80%).
FAD-binding and/or NADPH-binding domain
Similar to the highly conserved sequence of the FMN-binding domain among CPRs, both structure and sequence of the FAD-binding domain among well-investigated CPRs are highly conserved. Among the 25 residues, which constitute the FAD-binding pocket, 14 of them are invariant in all the CPRs we compared and seven of them are conserved in only plant CPRs (Fig. S4). Among those conserved residues, Ser411, Arg484, Tyr485, Ser487, Arg514, Val519, and Ser521 form hydrogen bonds with a bound FAD molecule in SbCPR2b (Fig. 6D). The position and conformation of the FAD molecule in the three different forms of SbCPR2b were not noticeably different, indicating that the association of NADP+ did not affect the conformation of FAD. In all three forms of SbCPR2b, the indole side chain of Trp706 was stacked parallel to the isoalloxazine ring of FAD shielding its re-face and thus preventing the nicotinamide ring from occupying the same position. The corresponding tryptophan residue has been proposed to regulate the binding and release of NADPH and NADP+, respectively.
Fourteen residues were within a distance of 3.5 Å from the 2′AMP-PPi moiety of NADP+ in our crystal structure of SbCPR2b. Thirteen of them are completely conserved among all CPRs, and the remaining Gly664 is conserved among plant CPRs (Fig. S4). Among those completely conserved residues, Arg323, Thr565, Arg627, Ser626, Asp668, Tyr634, and Gln636 established hydrogen bonds with the 2′AMP-PPi moiety of bound NADP+, which indicates that the binding mode for 2′AMP-PPi moiety is conserved among plant and animal CPRs. The conformation of the 2′AMP-PPi moiety of NADP+ in the open-air and fully oxidized SbCPR2b structures was quite similar. Its pyrophosphate and 2′ phosphate moieties were stabilized by guanidium side chains of the Arg323, Arg597, and Arg627, which are conserved among compared CPRs (Fig. S4). The guanidium side chain of Arg323 interacted with the pyrophosphate group of NADP+, and Arg597 interacted with the 2′ monophosphate, whereas Arg627 interacted with both. The whole NADP+ molecule bound in the open-air SbCPR2b was bent at its pyrophosphate group, and its positively charged nicotinamide ring was extended to the terminal carboxyl group of Trp706. The ribosyl-nicotinamide of NADP+ was proximal to the re-face of the isoalloxazine ring similar to the position observed in the structure of the S457A/C630A/D675N triple mutant of rat CPR ((63), PDB ID: 1JA1), although the ring orientation was different. Close inspection of the SbCPR2b structures with/without NADP+ revealed that the position of the phenolic side chain of Tyr634 was adjusted to establish a parallel orientation with the adenine ring of the bound NADP+ molecule.
Significantly, the positively charged nicotinamide ring in SbCPR2b was located near the terminal carboxyl end of Trp706 and the carboxyl side chains of Glu508 and Asp661. Although Glu508 is not conserved, both Trp706 and Asp661 are completely conserved among the CPRs we compared. Trp706 is the C-terminal residue (703RDVW706) conserved in plant CPRs, but in rat and human CPRs, it precedes a serine residue (654LDVWS678). In addition, the amide and carboxyl groups of Trp706 in all three structures of SbCPR2b were hydrogen bonded to the carbonyl oxygen of Gly660 and the backbone amide nitrogen of Ala662. Asp661 was located at the center of the loop at the N terminus of a helix, which is conserved in plants (660GDAKGM665) and animals (631GDARNM636). Movement of this so-called “Asp-loop” has been proposed to modulate catalytic activity through controlling the binding of NADPH and release of NADP+ in both rat and human CPRs (63, 76). Upon NADPH binding, charge and steric repulsion force the side chain of this aspartic acid away from NADPH, and it forms three hydrogen bonds with the backbone amide groups of Arg634, Asn635, and Met636 of 631GDARNM636 in rat/human CPR, which results in a tight and retracted loop conformation (65). Similarly, the side chain of Asp661 in SbCPR2b structure established hydrogen bonds with equivalent backbone residues, Lys663, Gly664, and Met665 in 660GDAKGM665. However, comparison of our NADP+-free SbCPR2b structure with two NADP+-bound structures did not reveal any significant shift in position of this loop as previously observed in the mammalian CPRs. Likewise, the tight and contracted conformation of the Asp-loop was observed in the crystal structure of ATR2 without bound NADP+ ((20), PDB ID: 5GXU), which suggests that this NADP+-dependent conformational change does not occur in plant CPRs. The observed hydrogen-bond interaction between the backbone of the C-terminal Trp706 and the Asp-loop in SbCPR2b supports the hypothesis that the binding and release of NADP(H) and NADP+ closely coordinate a concerted motion between the Asp-loop and the rotation/retraction of the indole ring away from the flavin (65), although retraction of the Asp-loop was not visible in SbCPR2b. In addition, the observed potential electrostatic interaction between the nicotinamide ring and the C terminus of Trp706 could differentially push and pull the indole ring of Trp706 depending on the association of NADP+ or NADPH, which could be a plant-specific shift. Thus, a concerted movement of Trp706 and the 660GDAKGM665 loop (Asp-loop) upon NADP(H) binding will likely affect the domain interface in plant CPRs and alter its equilibrium between the compact and extended states proposed in animal CPR (77).
Considering our crystal structures of three different states, NADPH binding could be initiated by electrostatic attractions between the negatively charged phosphate groups in 2′-AMP-5′PPi of NADPH and the positively charged amino acids (Arg323, Arg597, and Arg627). Next, the conserved negatively charged carboxyl group of Trp706 likely anchors the positively charged nicotinamide ring of NADPH within the binding pocket and then swings the nicotinamide ring into the re-face of the isoalloxazine ring of FAD, simultaneously displacing its side-chain indole. The NADP+ molecule is proposed to be displaced from CPR prior to electron transfer from FADH2 to the FMN (63). Weakened p–p orbital interactions between the oxidized nicotinamide and isoalloxazine allow the indole side chain of tryptophan to displace the nicotinamide ring from its binding site. In the SbCPR2b structure, the N3 and O2 atoms of FAD isoalloxazine formed hydrogen bonds with the backbone of Thr502 and Ala504. In addition, its N5 atom was located within a hydrogen-bond distance to the hydroxyl side chain of highly conserved Ser487. Furthermore, the backbone and side chains of the completely conserved surrounding residues His344 and Arg544 established a hydrogen-bond network with Asp704. Protonation at the N5 atom of the isoalloxazine ring upon hydride transfer from C4 of the nicotinamide ring was proposed to substantially alter those hydrogen-bonding interactions (27). The oxidized nicotinamide in NADP+ re-establish the hydrogen bond between Ser487 and Asp704, which stabilizes the ensuing FAD semiquinone intermediate. Given that realignment of the C-terminal tryptophan precedes interflavin electron transfer, it may be these conformational changes rather than the shuttling of electrons between the redox centers that limits the overall flux of reducing equivalents in the plant enzymes (27).
Based upon the SbCPR2b structures, most mechanistic hypotheses are similar to the ones proposed for mammalian CPRs. The observed difference in activity between animal and plant CPRs is therefore likely because of the heterogeneity of amino acid sequences within the connecting domain that joins the FMN-binding and FAD-binding domains that also guides the relative orientation of both domains.
Connecting domain
The interaction between CPR and the partner P450 and the subsequent electron transfer require a conformational transition of the CPR from the closed to the open state in a redox-dependent manner. The open conformation of CPR is required for delivery of electrons from the reduced FMN cofactor to the P450. Directly following the first interflavin electron transfer, CPR adopts an “open” conformation to transfer an electron to the receptor enzyme (78). So far, however, all crystal structures of wildtype rat, yeast, and human CPRs are in their closed conformation with the isoalloxazine rings of FAD and FMN in close proximity to each other, which is required for electron transfer between two rings (77). The same conformational transition between the open and closed conformations may not only regulate a hydride transfer but also control interflavin electron transfer. However, the conformational transition of the CPR from a closed state, which favors intramolecular electron transport, to an open state that is capable of transferring electrons to its redox partners, remains insufficiently well understood (72, 79, 80).
The asymmetric unit of the SbCPR2b crystal lattice was one molecule, and all three SbCPR2 isozymes exist in monomeric form in solution (Fig. 5). However, the FMN-binding domain is missing in our crystal structures of SbCPR2b, and the experimentally determined cleavage site is present in SbCPR2 isozymes within the connecting domain. A similar behavior was with the crystal structure of Arabidopsis CPR2 (AtCPR2, PDB ID: 5GXU). The asymmetric unit of AtCPR2 consists of two molecules; one molecule has neither the connecting domain nor the FMN-binding domain like our SbCPR2b structure (20). Even though the other molecule contained the FMN-binding domain and the connecting domain, the 43 amino acids of the connecting domain were completely disordered. In contrast, the corresponding disordered area of the connecting domain is less than 10 amino acids in crystal structures of mammalian CPRs. These findings from the Arabidopsis and sorghum CPRs may indicate higher flexibility of the FMN-binding domain in plant CPRs than in the animal CPRs, which exposes their connecting domain to proteolysis.
The conformational change between the open and closed states is an essential process for electron transfer between CPR and redox partners. The solutional small-angle X-ray scattering data on human CPR indicate its conformation transitions between a closed state and an open state (6, 11, 12, 81). The corresponding exchange frequency between the two states has been estimated to be at a minimum 600 s−1 (12). In the neutron reflectometry study on SbCPR2b, the coexistence of the compact and extended forms with a thickness of 44 and 79 Å, respectively, was found, and this conformational equilibrium shifts toward the compact form upon complete reduction by NADPH (82). In addition, several closed conformations might coexist, and not all of them are able to perform the interflavin electron transfer (83).
So far, the structural determinants that guide this conformational transition remain unidentified (81). The middle of the connecting domain contains a flexible hinge region that allows the FMN-binding domain to move on a large scale in order to interact with both the FAD-binding domain and its partner protein for electron transfer. The flexibility of this hinge region is an important factor determining the conformational equilibrium between open and closed states, and therefore, the reaction rate of CPR (81). The crystal structure of the deletion mutant ΔTGEE of rat CPR at the center of the hinge region, 235GVEATGEESSIR248, displayed a reoriented FMN-binding domain in open conformation relative to the FAD-binding domain (10). The chimeric yeast–human CPR (84), in which the connecting domain of human CPR was replaced with the yeast one, yielded an open conformation where the FMN and FAD cofactors were separated by 86 Å, which is compatible with FMN-to-P450 electron transfer. As shown in Fig. S4, the corresponding peptide in the aligned connection domain of SbCPR2s is TP(T)YT, which would produce quite different conformational dynamics.
In general, the amino acid sequence of the connecting domain among plants is quite different from those sequences in animals (Fig. S4). The entire region in the crystal structure of AtCPR2, SbCPR2b, and SbCPR2c was severely disordered, and the rapid cleavage within the region that was observed in all three SbCPR2 isozymes was located at the connecting domain.
Several pairs such as G240/S243 (84), I245/R246 (79), and S243/R246 (12) in the connecting domain of rat and human CPRs have been proposed to be important for the conformational changes between the open and closed states. Among those residues, only I245 is conserved in SbCPR2s, thus dynamic behavior of the FMN-binding domain of SbCPR2s is expected to be quite different. The f/y angles around R246 have also been proposed to enable this conformational exchange in mammalian versions, but the proline at this position in SbCPR2s would be expected to limit motion significantly.
Although all residues that constitute the FAD-binding and NADP-binding pockets are highly conserved, the rate-limiting step in rat and human CPR has been found to be the hydride transfer from NADPH to FAD and reduction of the FAD is ∼50-fold faster in Arabidopsis ATR2 and A. annua CPR (27). NADP+–FADH2 charge–transfer complex formation is also significantly faster in the plant CPRs. In contrast, interflavin electron transfer is slower and represents the rate-limiting step in plant CPRs (27, 85). It is tempting to speculate that the apparent kinetic difference between plant and animal CPR is mainly because of the amino acid sequences of their connecting domains.
Conclusion
Although lignin is one of the earth's most abundant biopolymers, converting it to fuels or commodity chemicals is challenging because of its heterogeneity and recalcitrant nature (86, 87). Engineering plant feedstocks with modified lignin structure and tailored functionality has shown promise toward facilitating recovery and chemical transformation of lignin (36, 88, 89, 90). In addition, flavonoids are of growing interest because of their health benefits (91). CPR is a strategic target to manipulate both the monolignol and flavonoid biosynthetic pathways, given its pivotal role in providing activity to the P450 enzymes, C4H, C3′H, F5H, F3′H, and F3′5′H, which generate precursors for lignin and various flavonoids. In Arabidopsis, CPR2-deficient plants have 6% less lignin with a 10-fold to 15-fold increase in H-units without any morphological defects, establishing CPR as a good candidate for the genetic engineering of lignocellulosic crops (18). SbCPR2a, SbCPR2b, and SbCPR2c are the only bona fide CPR genes in sorghum; only their gene products have the ability to transfer electrons from NADPH to P450s. Through crystal structure determination, kinetic analyses, and phylogenetic analyses, we have characterized the three sorghum CPR isozymes and examined the differences between animal and plant CPRs. Changing key features of these enzymes could subtly alter their enzyme kinetics. This study therefore reveals potential targets, especially the linker peptides, for gene-editing approaches aimed at tailoring lignin levels and lignin composition in plants to improve the conversion of biomass into biofuels or the forage utilization by livestock without negatively affecting plant growth and responses to biotic and abiotic stresses.
Experimental procedures
Chemicals and general
Analytical-grade chemicals were obtained from Sigma–Aldrich, Thermo Fisher Scientific, and Alfa Aesar. Screening solutions for crystallization were obtained from Hampton Research. p-Coumaroyl-shikimic acid was synthesized (92). Molecular graphics images were produced using the Chimera package (University of California San Francisco, NIH P41 RR-01081). The plotted figures were generated by GraphPad Prism (GraphPad Software, Inc).
Recombinant enzyme expression and purification
The wildtype SbCPR cDNA sequences were cloned and expressed in E. coli, and the recombinant proteins were purified as previously described (47). Briefly, the SbCPR cDNA sequences excluding the sequences encoding the transmembrane domain were cloned into vector pET-30a(+) (MilliporeSigma) for overexpression. The vector was introduced into E. coli Rosetta 2(DE3) cells via transformation. Three liter LB medium complemented with 25 μg ml−1 chloramphenicol and 50 μg ml−1 kanamycin was inoculated with 20 ml from an overnight culture. The cells were grown at 37 °C until the culture reached absorbance of 0.6 to 0.8 at 600 nm, and then the temperature was reduced to 25 °C before adding IPTG to a final concentration of 0.5 mM. After being induced for 16 h, the cells were harvested by centrifugation at 5000 rpm for 20 min at 4 °C and resuspended in buffer A (50 mM Tris–HCl, 300 mM NaCl, pH 8.0) with 20 mM imidazole. After sonicating on ice for 30 min (model 450 sonifier; Branson Ultrasonics), the cell debris was removed by ultracentrifuge at 17,000 rpm for 1 h. The clear lysate was loaded on a column containing nickel–nitrilotriacetic acid resin (Qiagen) and washed with the same buffer. Modified SbCPR protein was eluted with buffer A containing 250 mM imidazole, pH 8.0. The protein was concentrated, and the buffer was exchanged against 5 mM potassium phosphate buffer and loaded onto hydroxyapatite column. The fraction containing SbCPR2s was eluted by a linear gradient of potassium phosphate and then concentrated to 3 ml. Final purity was analyzed by SDS-PAGE, and the concentration was determined by the Bradford assay (Bio-Rad).
Absorbance spectroscopy and redox titration studies
The NADPH-titration experiments were performed with 15 to 25 μM solutions of the CPRs in 20 mM Hepes buffer, pH 7.5 placed into semimicro optical cell with a path length of 1 cm. The anaerobic environment during the assay was maintained by adding glucose (60 mM), glucose oxidase (30 U ml−1), and catalase (2000 U ml−1). The absorbance spectra were acquired with a rapid scanning MC2000-2 CCD-spectrometer (Ocean Optics, Inc) equipped with DT-Mini-2 deuterium/tungsten light source and a custom-made thermostated cell holder with magnetic stirrer. Titrations were performed at continuous stirring at 25 °C by consequential additions of the aliquots of 20 mM NADPH solution. In each titration, a series of 20 to 25 spectra corresponding to the NADPH-to-CPR molar ratios (RNADPH) varying from 0 to 15 to 20 were recorded. After each addition of NADPH, the sample was incubated for 1 min prior to recording the spectrum.
Crystallization and structure determination
Prior to crystallization, the buffer containing the apo SbCPR2b protein was exchanged against 20 mM Tris, pH 7.5, 50 mM NaCl, and concentrated to 15 mg ml−1 by using an Amicon 8050 ultrafiltration cell with a 30 kDa cutoff membrane (EMDMillipore). For the complex formation, the SbCPR2b solution was pretreated with the NADP+ in 3× molar excess. For the NADP+ complex structure in oxidized SbCPR2b, 1 mM potassium ferricyanide was added prior to addition of NADP+, and the potassium ferricyanide was removed by Bio-gel P6 gel (Bio-Rad). A commercial crystallization kit, MCSG-1 (Anatrace), was used for crystal screening through the sitting-drop vapor-diffusion method by Crystal Phoenix (Art Robbins Instruments). The initial crystal appeared in the solution of 0.2 M sodium thiocyanate, pH 6.9, and 20% (w/v) PEG-3350 at room temperature. Then the larger crystals were reproduced by the hanging-drop vapor-diffusion method. Small crystals appeared in 2 days and grew to 0.1 mm × 0.2 mm in 4 days. The crystallization for SbCPR2a was not attempted because of the fast cleavage of the FMN-binding domain.
The buffer containing SbCPR2c was exchanged against 50 mM potassium phosphate buffer, pH 6.8, and concentrated to 30 mg ml−1. Five microliters of SbCPR2c solution were mixed with 4 μl 0.1 M Hepes, pH 7.5, 0.2 M NaCl, 25% (w/v) PEG-3350, and 1 μl 1,5-diaminopentane dihydrochloride (Hampton Research). After the formation of small crystals, a single crystal was transferred to the identical crystallization solution as seed. The large crystals formed in 1 week and were transferred to a reservoir solution with a 50% (w/v) PEG-3350 concentration for further dehydration.
The crystals were transferred to cryoprotectant and flash frozen in liquid nitrogen. The diffraction data were collected in Advanced Light Source Beamline 5.0.2 and 5.0.3 at 100 K. Initial phasing of diffraction data was performed by MR with the Phaser in the PHENIX package (https://phenix-online.org/) (93) using the coordinate of Arabidopsis CPR (PDB ID: 5GXU) as a search model. The conformation and position of the initial solutions were refined further using PHENIX and manually adjusted with Coot (https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/) software (94). The final Rwork was 20.75%, 23.17%, and 22.32%, respectively; Rfree was 24.67%, 26.52%, and 26.59% for apo SbCPR2b, naturally oxidized SbCPR2b with NADP+, and fully oxidized SbCPR2b with NADP+, respectively. The statistics for the diffraction data are listed in Table 3.
Modeling and molecular docking
The FMN-binding domain model of SbCPR2b was built with Phyre2 server (68) by submitting the amino acid sequence. Molecular docking experiments were performed using the “PatchDock” algorithm (67) between the crystal structure of SbC4H1 (PDB ID: 6VBY) and SbCPR2b FMN-binding domain model, the FMN-binding domain of ATR2 (PDB ID: 5GXU), or the FMN-binding domain of rat CPR (PDB ID: 1AMO). The top 10 docking results were analyzed.
MALS experiment
The DAWN 8+ MALS detector (Wyatt Technology Corporation) was connected to 1260 Infinity II LC System (Agilent) with Yarra 3 μM SEC-2000 LC column (Phenomenex). The column was pre-equilibrated with 20 mM sodium phosphate buffer, pH 7. The SbCPR2b sample had a concentration of 2 mg ml−1 and was filtered through a syringe filter of 0.22 μm (Fisher Scientific) before injection into the system. The injector flow rate was 0.5 ml/min. The molar mass of the defined main peak was analyzed by ASTRA software (Wyatt Technology Corporation).
Steady-state kinetics of three SbCPR isozymes
Cytochrome c
To compare the enzyme activity of the three SbCPR2s, NADPH-dependent cytochrome c reductase activity was measured at 25 °C. The corresponding kinetic assay was conducted in 1 ml assay buffer (0.125 M potassium phosphate solution, pH 7.4, 100 μM NADPH, and 0.1 mM EDTA). The cytochrome c concentration was varied from 5 μM to 150 μM. A volume of 2.5 to 5 μl of truncated SbCPR2s was added to initiate the reaction; the final concentration of SbCPR2s was 10 to 20 nM. To determine the KM of NADPH, the assay buffer contained 40 μM cytochrome c with no NADPH. Then the NADPH concentration was varied from 3 to 100 μM. The time-dependent change in the absorbance at 550 nm was monitored up to 5 min by a GENESYS 10S UV–Vis Spectrophotometer (Thermo Fisher Scientific). The molar extinction coefficient is 21 mM−1 cm−1 for reduced cytochrome c. Kinetic parameters were calculated with GraphPad Prism.
SbC4H1 and SbC3′H
The fluorometric assay to measure the activity of SbC4H1 was previously reported (47). The final concentrations of SbCPR2a, SbCPR2b, and SbCPR2c used were 7.9, 10, and 10 nM, respectively. The kinetic assay with SbC3′H was performed with the same protocol; the full-length CPR incorporated in liposome was incubated overnight with truncated SbC3′H in a 2:1 ratio. The kinetic assay was performed with the substrate p-coumaroyl shikimic acid. The reaction was conducted in 200 μl 125 mM potassium phosphate buffer (pH 7.4). SbC3′H reconstituted with SbCPR2s in a proteoliposomal system was added to the assay mixture to a final concentration of 20 to 40 nM. The reaction was initiated by addition of 20 mM stock solution of NADPH to a final concentration of 100 μM. The reaction system was maintained at 30 °C for 5 min, and glacial acetic acid was added to a final concentration of 30% (v/v) to quench the reaction. The reaction mixture was injected onto a Luna 5 μm C18 column, and the product was monitored by HPLC (Hitachi Elite LaChrom L-2100; Hitachi High-Tech) operating at a flow rate of 1 ml min−1, with a gradient of solvent A (0.1% trifluoroacetic acid in deionized water) and solvent B (100% acetonitrile) varying from 95% A and 5% B to 0% A and 100% B over a period of 30 min. Products were quantified by detection at 320 nm using the Hitachi Elite LaChrom L-2400 detector. All experiments were performed in triplicates. The kinetic data were analyzed using Michaelis–Menten equations.
Comparison between full-length and truncated SbCPR2b
SbC4H1 was expressed and purified as described previously (47). The anaerobic environment during the assay was maintained with the concentrations of glucose, glucose oxidase, and catalase at 60 mM, 30 U ml−1, and 2000 U ml−1, respectively, in CO-saturated 20 mM Hepes buffer (pH 7.4). Taking the solution containing 2 μM truncated SbCPR2b and 500 μM NADPH as baseline, the absorbance spectrum at 450 nm was recorded with 1 μM SbC4H1 in the aforementioned buffer that was presaturated with CO. Continuous absorbance spectra were recorded every 4 s with an S2000 rapid-scanning CCD spectrometer (Ocean Optics). The absorbance at 450 nm was plotted against time to visualize the kinetics.
Stoichiometry study of NADPH-dependent reduction
The preparation of the SbCPR–SbC4H1 and rCPR–SbC4H1 complexes and fluorometric product measurement were described previously (47). Monitoring the consumption of NADPH and oxygen were performed with a measurement setup combining an MC2000-2 two-channel CCD spectrometer (Ocean Optics, Inc) with an OXROB10 robust optical oxygen probe (PyroScience). We used a PX-2 pulsed xenon light source (Ocean Optics) and a 617 nm M617L3 LED (Thorlabs) as the light sources for the absorbance and luminescence channels, respectively.
The experiments were performed at 30 °C in 125 mM potassium phosphate buffer at the initial NADPH concentration of 100 μM. The final concentration of CPR and SbC4H1 was equal to 75 and 150 nM, respectively. The sample with a volume of 1 ml was placed into a cylindrical optical cell (path length of 1 cm) inserted into a custom-made thermostated cell holder equipped with a motorless magnetic stirrer (Variomag-USA). The absorbance-beam optical windows of the holder were connected via optical fiber to a PX-2 light source and a master channel of the MC2000-2 spectrometer for absorbance measurements. The cell was closed with a Teflon stopper with a 1.5 mm bore for insertion of the OXR430 sensor. The stopper was designed in such a way as to ensure direct contact of its surface with the sample and eliminate air bubbles. After inserting the sensor connected via a bifurcated fiber bundle to the slave channel of MC2000-2 and an M617L3 LED light source, the reaction was initiated by adding 25 μl of P450–CPR mixture with a gas-tight syringe through a bore in the stopper. Absorbance (320–500 nm) and luminescence (700–900 nm) spectra were collected simultaneously in time intervals of 1 to 2 s for 5 to 10 min with a custom-designed software. For the experiment with substrate, 3 μM 2-napthoic acid was added 5 min after the addition of the microsomal suspension. The rates of utilization of NADPH and consumption of oxygen were determined from the changes in absorbance at 340 nm and the integral intensity of luminescence of the oxygen probe, respectively.
The ratio of the rates of NADPH and oxygen consumption (R) determined in these experiments was used to estimate the fraction of reducing equivalents used through the oxidase shunt (uncoupled production of water) that requires two NADPH molecules per one oxygen molecule as opposed to the fraction used for substrate monooxygenation and generation of superoxide and H2O2 that require one molecule of NADPH per one molecule of oxygen. The percent of reducing equivalents utilized through the oxidase shunt was calculated as FH2O = 100 × (R − 1).
Calibration of the oxygen sensor used in these measurements was performed following the kinetics of oxygen scavenging by protocatechuic acid dioxygenase. In these calibration assays, we followed the changes in the intensity in luminescence of the oxygen probe in parallel with monitoring the changes in absorbance at 290 nm. The decrease in absorbance at this wavelength during the conversion of protocatechuic acid to b-ketoadipic acid by protocatechuic acid dioxygenase was used to determine the changes in oxygen concentration. In these calculations, we used the differential extinction coefficient of 2.3 mM−1 cm−1 (95). Resulting nonlinear calibration curves were approximated with the third-order polynomial, which was further used to calculate the molar concentration of oxygen in the samples based on the intensity of luminescence of the probe.
Cutting-site determination by mass spectroscopy
The gel was sliced and digested with trypsin following the established method (96). The corresponding fragments were identified with MALDI–MS/MS and LC–MS/MS. For MALDI–MS/MS, the peptide mass spectra were obtained using procedure and collection programs supplied by the manufacturer (Applied Biosystems). The matrix, a-cyano-4-hydroxycinnamic acid (Sigma–Aldrich), was prepared as a solution of 10 mg ml−1 in 50% water/acetonitrile with 0.1% TFA. The matrix solution was mixed in ratio of 1:1 with the trypsin digest, applied to the sample plate, and dried. Spectra were collected using a 4800 MALDI TOF/TOF Analyzer (Applied Biosystems), using the data collection programs in positive mode for both MS and MS/MS spectra. The same trypsin digestion treatment was applied to this SDS-PAGE band of CPR2a. MALDI MS/MS was not able to give conclusive result on N-terminal peptide, so we applied the LC–MS/MS aiming the N-terminal peptide and the total mass of the band.
For LC–MS/MS, samples were analyzed on an RSLCnano system (Thermo Fisher Scientific) coupled to a Q-Exactive HF mass spectrometer (Thermo Fisher Scientific). The samples were first injected onto a trap column (Acclaim PepMap 100, 75 μm × 2 cm; Thermo Fisher Scientific) for 3.0 min at a flow rate of 5 μl/min, 1.5% acetonitrile, and 0.2% formic acid before switching in line with the main column. Separation was performed on a C18 nano column (Acquity UPLC M-class, Peptide CSH 130A, 1.7 μm 75 μm × 250 mm; Waters Corp) at 260 nl/min with a linear gradient from 5 to 35% over 96 min. The LC aqueous mobile phase contained 0.1% (v/v) formic acid in water, and the organic mobile phase contained 0.1% (v/v) formic acid in 80% (v/v) acetonitrile. Mass spectra for the eluted peptides were acquired on a Q Exactive HF mass spectrometer in data-dependent mode using a mass range of m/z 375 to 1500, resolution 120,000, automated gain control target of 3 × 106, and maximum injection time of 60 ms for the MS1 peptide measurements. Data-dependent MS2 spectra were acquired by higher-energy collisional dissociation as a Top20 experiment with a normalized collision energy set at 33%, automated gain control target set to 1 × 105, 45,000 resolution, intensity threshold 1 × 105, and a maximum injection time of 86 ms. Dynamic exclusion was set at 45 s, and the isolation window was set to 1.2 m/z to reduce coisolation. MS spectra are shown in Fig. S6.
Data availability
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers XP_002462826.1, XP_002444097.1, and XP_002447240.1. The structure discussed in this article can be found at www.rcsb.org deposited under the PDB IDs: 7SUX, 7SUZ, and 7SV0.
Supporting information
This article contains supporting information (98).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Ms Tammy Gries for technical assistance with the expression of recombinant proteins in E. coli; Olatunji Feyisola and Emily Savoy for technical assistance with HPLC for kinetic assay. Drs Sophie Alvarez and Michael Naldret from Proteomics and Metabolomics Facility at the University of Nebraska-Lincoln. This research used resources of the Advanced Light Source (beamlines 5.0.2 and 5.0.3), which is a Department of Energy Office of Science User Facility under contract no. DE-AC02-05CH11231.
Author contributions
B. Z., S. S., and C. K. conceptualization; B. Z. and D. D. methodology; B. Z. validation; B. Z., G. M., D. D., and S. S. formal analysis; V. T. and S. S. resources; B. Z. writing–original draft; J. R., D. D., W. V., S. S., and C. K. writing–review & editing; C. K. project administration; C. K. funding acquisition.
Funding and additional information
Original research was supported by grant from the National Science Foundation (grant numbers: CHE-1804699 and MCB-2043248) and Murdock Charitable Trust (to C. H. K.). W. V. acknowledges financial support from the US Department of Energy’s (DOE) Office of Energy Efficiency and Renewable Energy, Bioenergy Technologies Office and sponsored by the US DOE’s International Affairs (grant no.: DE-PI0000031) and the US Department of Agriculture-National Institute of Food and Agriculture Biomass Research and Development Initiative (grant no.: 2011-1006-30358). S. E. S. acknowledges support from US Department of Agriculture-Agriculture Research Service project (grant no.: 3042-21220-033-000-D). V. I. T. and J. R. were funded by the DOE Great Lakes Bioenergy Research Center, DOE Biological and Environmental Research Office of Science (grant no.: DE-SC0018409). And we thank Sebastien Santini—CNRS/Aix-Marseille University IGS UMR7256 for releasing the Phylogeny.fr for phylogenetic tree building.
Edited by Joseph Jez
Supporting information
References
- 1.Phillips A.H., Langdon R.G. Hepatic triphosphopyridine nucleotide-cytochrome c reductase: Isolation, characterization, and kinetic studies. J. Biol. Chem. 1962;237:2652–2660. [PubMed] [Google Scholar]
- 2.Horecker B., Heppel L.A. The reduction of cytochrome c by xanthine oxidase. J. Biol. Chem. 1949;178:683–690. [PubMed] [Google Scholar]
- 3.Omura T., Sato R. A new cytochrome in liver microsomes. J. Biol. Chem. 1962;237:PC1375–PC1376. [PubMed] [Google Scholar]
- 4.Raw I., Mahler H. Studies of electron transport enzymes III. Cytochrome b5 of pig liver mitochondria. J. Biol. Chem. 1959;234:1867–1873. [PubMed] [Google Scholar]
- 5.Porter T.D., Kasper C.B. NADPH-cytochrome P-450 oxidoreductase: Flavin mononucleotide and flavin adenine dinucleotide domains evolved from different flavoproteins. Biochemistry. 1986;25:1682–1687. doi: 10.1021/bi00355a036. [DOI] [PubMed] [Google Scholar]
- 6.Ellis J., Gutierrez A., Barsukov I.L., Huang W.-C., Grossmann J.G., Roberts G.C. Domain motion in cytochrome P450 reductase conformational equilibria revealed by NMR and small-angle X-ray scattering. J. Biol. Chem. 2009;284:36628–36637. doi: 10.1074/jbc.M109.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyanagi T., Mason H. Properties of hepatic reduced nicotinamide adenine dinucleotide phosphate-cytochrome c reductase. Biochemistry. 1973;12:2297–2308. doi: 10.1021/bi00736a018. [DOI] [PubMed] [Google Scholar]
- 8.Munro A.W., Noble M.A., Robledo L., Daff S.N., Chapman S.K. Determination of the redox properties of human NADPH-cytochrome P450 reductase. Biochemistry. 2001;40:1956–1963. doi: 10.1021/bi001718u. [DOI] [PubMed] [Google Scholar]
- 9.Wang M., Roberts D.L., Paschke R., Shea T.M., Masters B.S.S., Kim J.-J.P. Three-dimensional structure of NADPH–cytochrome P450 reductase: Prototype for FMN-and FAD-containing enzymes. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamdane D., Xia C., Im S.-C., Zhang H., Kim J.-J.P., Waskell L. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J. Biol. Chem. 2009;284:11374–11384. doi: 10.1074/jbc.M807868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang W.-C., Ellis J., Moody P.C., Raven E.L., Roberts G.C. Redox-linked domain movements in the catalytic cycle of cytochrome p450 reductase. Structure. 2013;21:1581–1589. doi: 10.1016/j.str.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frances O., Fatemi F., Pompon D., Guittet E., Sizun C., Pérez J., Lescop E., Truan G. A well-balanced preexisting equilibrium governs electron flux efficiency of a multidomain diflavin reductase. Biophys. J. 2015;108:1527–1536. doi: 10.1016/j.bpj.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizutani M., Ohta D. Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 2010;61:291–315. doi: 10.1146/annurev-arplant-042809-112305. [DOI] [PubMed] [Google Scholar]
- 14.Jensen K., Møller B.L. Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry. 2010;71:132–141. doi: 10.1016/j.phytochem.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 15.Ro D.K., Ehlting J., Douglas C.J. Cloning, functional expression, and subcellular localization of multiple NADPH-cytochrome P450 reductases from hybrid poplar. Plant Physiol. 2002;130:1837–1851. doi: 10.1104/pp.008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paquette S.M., Jensen K., Bak S. A web-based resource for the Arabidopsis P450, cytochromes b5, NADPH-cytochrome P450 reductases, and family 1 glycosyltransferases (http://www. P450. kvl. dk) Phytochemistry. 2009;70:1940–1947. doi: 10.1016/j.phytochem.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 17.Mizutani M., Ohta D. Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol. 1998;116:357–367. doi: 10.1104/pp.116.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sundin L., Vanholme R., Geerinck J., Goeminne G., Höfer R., Kim H., Ralph J., Boerjan W. Mutation of the inducible Arabidopsis thaliana cytochrome P450 reductase2 alters lignin composition and improves saccharification. Plant Physiol. 2014;166:1956–1971. doi: 10.1104/pp.114.245548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werck-Reichhart D., Feyereisen R. Cytochromes P450: A success story. Genome Biol. 2000;1:3003. doi: 10.1186/gb-2000-1-6-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niu G., Zhao S., Wang L., Dong W., Liu L., He Y. Structure of the Arabidopsis thaliana NADPH-cytochrome P450 reductase 2 (ATR2) provides insight into its function. FEBS J. 2017;284:754–765. doi: 10.1111/febs.14017. [DOI] [PubMed] [Google Scholar]
- 21.Russell D.W. The metabolism of aromatic compounds in higher plants X. Properties of the cinnamic acid 4-hydroxylase of pea seedlings and some aspects of its metabolic and developmental control. J. Biol. Chem. 1971;246:3870–3878. [PubMed] [Google Scholar]
- 22.Schoch G., Goepfert S., Morant M., Hehn A., Meyer D., Ullmann P., Werck-Reichhart D. CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 2001;276:36566–36574. doi: 10.1074/jbc.M104047200. [DOI] [PubMed] [Google Scholar]
- 23.Meyer K., Shirley A.M., Cusumano J.C., Bell-Lelong D.A., Chapple C. Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6619–6623. doi: 10.1073/pnas.95.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheynier V., Comte G., Davies K.M., Lattanzio V., Martens S. Plant phenolics: Recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol. Biochem. 2013;72:1–20. doi: 10.1016/j.plaphy.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Schreiner M., Mewis I., Huyskens-Keil S., Jansen M., Zrenner R., Winkler J., O’Brien N., Krumbein A. UV-B-induced secondary plant metabolites-potential benefits for plant and human health. Crit. Rev. Plant Sci. 2012;31:229–240. [Google Scholar]
- 26.Shahidi F., Ambigaipalan P. Phenolics and polyphenolics in foods, beverages and spices: Antioxidant activity and health effects–a review. J. Funct. Foods. 2015;18:820–897. [Google Scholar]
- 27.Whitelaw D.A., Tonkin R., Meints C.E., Wolthers K.R. Kinetic analysis of electron flux in cytochrome P450 reductases reveals differences in rate-determining steps in plant and mammalian enzymes. Arch. Biochem. Biophys. 2015;584:107–115. doi: 10.1016/j.abb.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Boerjan W., Ralph J., Baucher M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 29.Dixon R.A., Chen F., Guo D., Parvathi K. The biosynthesis of monolignols: A “metabolic grid”, or independent pathways to guaiacyl and syringyl units? Phytochemistry. 2001;57:1069–1084. doi: 10.1016/s0031-9422(01)00092-9. [DOI] [PubMed] [Google Scholar]
- 30.Guillaumie S., Pichon M., Martinant J.-P., Bosio M., Goffner D., Barrière Y. Differential expression of phenylpropanoid and related genes in brown-midrib bm1, bm2, bm3, and bm4 young near-isogenic maize plants. Planta. 2007;226:235–250. doi: 10.1007/s00425-006-0468-9. [DOI] [PubMed] [Google Scholar]
- 31.Rastogi S., Dwivedi U.N. Manipulation of lignin in plants with special reference to O-methyltransferase. Plant Sci. 2008;174:264–277. [Google Scholar]
- 32.Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. Lignin biosynthesis and structure. Plant Physiol. 2010;153:895–905. doi: 10.1104/pp.110.155119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansfield S.D., Kang K.Y., Chapple C. Designed for deconstruction –poplar trees altered in cell wall lignification improve the efficacy of bioethanol production. New Phytol. 2012;194:91–101. doi: 10.1111/j.1469-8137.2011.04031.x. [DOI] [PubMed] [Google Scholar]
- 34.Sattler S.E., Palmer N.A., Saballos A., Greene A.M., Xin Z., Sarath G., Vermerris W., Pedersen J.F. Identification and characterization of four missense mutations in brown midrib 12 (Bmr12), the caffeic O-methyltranferase (COMT) of sorghum. Bioenergy Res. 2012;5:855–865. [Google Scholar]
- 35.Gou M., Yang X., Zhao Y., Ran X., Song Y., Liu C.-J. Cytochrome b5 is an obligate electron Shuttle protein for syringyl lignin biosynthesis in Arabidopsis. Plant Cell. 2019;31:1344–1366. doi: 10.1105/tpc.18.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ragauskas A.J., Beckham G.T., Biddy M.J., Chandra R., Chen F., Davis M.F., Davison B.H., Dixon R.A., Gilna P., Keller M. Lignin valorization: Improving lignin processing in the biorefinery. Science. 2014;344:1246843. doi: 10.1126/science.1246843. [DOI] [PubMed] [Google Scholar]
- 37.Feltus F.A., Vandenbrink J.P. Bioenergy grass feedstock: Current options and prospects for trait improvement using emerging genetic, genomic, and systems biology toolkits. Biotechnol. Biofuels. 2012;5:80. doi: 10.1186/1754-6834-5-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarath G., Mitchell R.B., Sattler S.E., Funnell D., Pedersen J.F., Graybosch R.A., Vogel K.P. Opportunities and roadblocks in utilizing forages and small grains for liquid fuels. J. Ind. Microbiol. Biotechnol. 2008;35:343–354. doi: 10.1007/s10295-007-0296-3. [DOI] [PubMed] [Google Scholar]
- 39.Weijde T.v. d., Alvim Kamei C.L., Torres A.F., Vermerris W., Dolstra O., Visser R.G.F., Trindade L.M. The potential of C4 grasses for cellulosic biofuel production. Front. Plant Sci. 2013;4:107. doi: 10.3389/fpls.2013.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casler M., Buxton D., Vogel K.P. Genetic modification of lignin concentration affects fitness of perennial herbaceous plants. Theor. Appl. Genet. 2002;104:127–131. doi: 10.1007/s001220200015. [DOI] [PubMed] [Google Scholar]
- 41.Walker A.M., Hayes R.P., Youn B., Vermerris W., Sattler S.E., Kang C. Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme A-dependent transferases and synthases. Plant Physiol. 2013;162:640–651. doi: 10.1104/pp.113.217836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker A.M., Sattler S.A., Regner M., Jones J.P., Ralph J., Vermerris W., Sattler S.E., Kang C. The structure and catalytic mechanism of Sorghum bicolor caffeoyl-CoA O-methyltransferase. Plant Physiol. 2016;172:78–92. doi: 10.1104/pp.16.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green A.R., Lewis K.M., Barr J.T., Jones J.P., Lu F., Ralph J., Vermerris W., Sattler S.E., Kang C. Determination of the structure and catalytic mechanism of Sorghum bicolor caffeic acid O-methyltransferase and the structural impact of three brown midrib12 mutations. Plant Physiol. 2014;165:1440–1456. doi: 10.1104/pp.114.241729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sattler S.A., Walker A.M., Vermerris W., Sattler S.E., Kang C. Structural and biochemical characterization of cinnamoyl-CoA reductases. Plant Physiol. 2017;173:1031–1044. doi: 10.1104/pp.16.01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jun S.-Y., Walker A.M., Kim H., Ralph J., Vermerris W., Sattler S.E., Kang C. The enzyme activity and substrate specificity of two major cinnamyl alcohol dehydrogenases in sorghum (Sorghum bicolor), SbCAD2 and SbCAD4. Plant Physiol. 2017;174:2128–2145. doi: 10.1104/pp.17.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moural T.W., Lewis K.M., Barnaba C., Zhu F., Palmer N.A., Sarath G., Scully E.D., Jones J.P., Sattler S.E., Kang C. Characterization of class III peroxidases from switchgrass. Plant Physiol. 2017;173:417–433. doi: 10.1104/pp.16.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang B., Lewis K.M., Abril A., Davydov D.R., Vermerris W., Sattler S.E., Kang C. Structure and function of the cytochrome P450 monooxygenase cinnamate 4-hydroxylase from Sorghum bicolor. Plant Physiol. 2020;183:957–973. doi: 10.1104/pp.20.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xin Z., Wang M.L., Barkley N.A., Burow G., Franks C., Pederson G., Burke J. Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol. 2008;8:103. doi: 10.1186/1471-2229-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiao Y., Burke J., Chopra R., Burow G., Chen J., Wang B., Hayes C., Emendack Y., Ware D., Xin Z. A sorghum mutant resource as an efficient platform for gene discovery in grasses. Plant Cell. 2016;28:1551–1562. doi: 10.1105/tpc.16.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sattler S.E., Saballos A., Xin Z., Funnell-Harris D.L., Vermerris W., Pedersen J.F. Characterization of novel sorghum brown midrib mutants from an EMS-mutagenized population. G3 (Bethesda) 2014;4:2115–2124. doi: 10.1534/g3.114.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang W., Zhou H., Bi H., Fromm M., Yang B., Weeks D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scully E.D., Gries T., Sarath G., Palmer N.A., Baird L., Serapiglia M.J., Dien B.S., Boateng A.A., Ge Z., Funnell-Harris D.L. Overexpression of SbMyb60 impacts phenylpropanoid biosynthesis and alters secondary cell wall composition in Sorghum bicolor. Plant J. 2016;85:378–395. doi: 10.1111/tpj.13112. [DOI] [PubMed] [Google Scholar]
- 55.Scully E.D., Gries T., Palmer N.A., Sarath G., Funnell-Harris D.L., Baird L., Twigg P., Seravalli J., Clemente T.E., Sattler S.E. Overexpression of SbMyb60 in Sorghum bicolor impacts both primary and secondary metabolism. New Phytol. 2018;217:82–104. doi: 10.1111/nph.14815. [DOI] [PubMed] [Google Scholar]
- 56.Tetreault H.M., Gries T., Palmer N., Funnell-Harris D.L., Sato S., Ge Z., Sarath G., Sattler S. Overexpression of ferulate 5-hydroxylase increases syringyl units in Sorghum bicolor. Plant Mol. Biol. 2020;103:269–285. doi: 10.1007/s11103-020-00991-3. [DOI] [PubMed] [Google Scholar]
- 57.Tetreault H.M., Scully E.D., Gries T., Palmer N.A., Funnell-Harris D.L., Baird L., Seravalli J., Dien B.S., Sarath G., Clemente T.E. Overexpression of the Sorghum bicolor SbCCoAOMT alters cell wall associated hydroxycinnamoyl groups. PLoS One. 2018;13 doi: 10.1371/journal.pone.0204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vermilion J.L., Coon M.J. Purified liver microsomal NADPH-cytochrome P-450 reductase. Spectral characterization of oxidation-reduction states. J. Biol. Chem. 1978;253:2694–2704. [PubMed] [Google Scholar]
- 59.Benveniste I., Gabriac B., Durst F. Purification and characterization of the NADPH-cytochrome P-450 (cytochrome c) reductase from higher-plant microsomal fraction. Biochem. J. 1986;235:365–373. doi: 10.1042/bj2350365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sevrioukova I., Shaffer C., Ballou D.P., Peterson J.A. Equilibrium and transient state spectrophotometric studies of the mechanism of reduction of the flavoprotein domain of P450BM-3. Biochemistry. 1996;35:7058–7068. doi: 10.1021/bi960060a. [DOI] [PubMed] [Google Scholar]
- 61.Shen A.L., Kasper C.B. Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding and catalysis. J. Biol. Chem. 2000;275:41087–41091. doi: 10.1074/jbc.M008380200. [DOI] [PubMed] [Google Scholar]
- 62.Grinkova Y.V., Denisov I.G., McLean M.A., Sligar S.G. Oxidase uncoupling in heme monooxygenases: Human cytochrome P450 CYP3A4 in nanodiscs. Biochem. Biophys. Res. Commun. 2013;430:1223–1227. doi: 10.1016/j.bbrc.2012.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hubbard P.A., Shen A.L., Paschke R., Kasper C.B., Kim J.-J.P. NADPH-cytochrome P450 oxidoreductase structural basis for hydride and electron transfer. J. Biol. Chem. 2001;276:29163–29170. doi: 10.1074/jbc.M101731200. [DOI] [PubMed] [Google Scholar]
- 64.Xia C., Panda S.P., Marohnic C.C., Martásek P., Masters B.S., Kim J.-J.P. Structural basis for human NADPH-cytochrome P450 oxidoreductase deficiency. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13486–13491. doi: 10.1073/pnas.1106632108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia C., Rwere F., Im S., Shen A.L., Waskell L., Kim J.-J.P. Structural and kinetic studies of Asp632 mutants and fully reduced NADPH-cytochrome P450 oxidoreductase define the role of Asp632 loop dynamics in the control of NADPH binding and hydride transfer. Biochemistry. 2018;57:945–962. doi: 10.1021/acs.biochem.7b01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lusty C.J. A gentle vapor-diffusion technique for cross-linking of protein crystals for cryocrystallography. J. Appl. Crystallogr. 1999;32:106–112. [Google Scholar]
- 67.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sevrioukova I.F., Li H., Zhang H., Peterson J.A., Poulos T.L. Structure of a cytochrome P450–redox partner electron-transfer complex. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1863–1868. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sugishima M., Sato H., Higashimoto Y., Harada J., Wada K., Fukuyama K., Noguchi M. Structural basis for the electron transfer from an open form of NADPH-cytochrome P450 oxidoreductase to heme oxygenase. Proc. Natl. Acad. Sci. U. S. A. 2014;111:2524–2529. doi: 10.1073/pnas.1322034111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rwere F., Xia C., Im S., Haque M.M., Stuehr D.J., Waskell L., Kim J.-J.P. Mutants of cytochrome P450 reductase lacking either Gly-141 or Gly-143 destabilize its FMN semiquinone. J. Biol. Chem. 2016;291:14639–14661. doi: 10.1074/jbc.M116.724625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sellner M., Fischer A., Don C.G., Smieško M. Conformational landscape of cytochrome P450 reductase interactions. Int. J. Mol. Sci. 2021;22:1023. doi: 10.3390/ijms22031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watson C.J., Froehlich J.E., Josefsson C.A., Chapple C., Durst F., Benveniste I., Coolbaugh R.C. Localization of CYP86B1 in the outer envelope of chloroplasts. Plant Cell Physiol. 2001;42:873–878. doi: 10.1093/pcp/pce110. [DOI] [PubMed] [Google Scholar]
- 75.Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 76.Mothersole R.G., Meints C.E., Louder A., Wolthers K.R. Role of active site loop in coenzyme binding and flavin reduction in cytochrome P450 reductase. Arch. Biochem. Biophys. 2016;606:111–119. doi: 10.1016/j.abb.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 77.Freeman S.L., Martel A., Raven E.L., Roberts G.C.K. Orchestrated domain movement in catalysis by cytochrome P450 reductase. Sci. Rep. 2017;7:11. doi: 10.1038/s41598-017-09840-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Halaka F.G., Babcock G.T., Dye J.L. The use of principal component analysis to resolve the spectra and kinetics of cytochrome c oxidase reduction by 5, 10-dihydro-5-methyl phenazine. Biophys. J. 1985;48:209–219. doi: 10.1016/S0006-3495(85)83774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sündermann A., Oostenbrink C. Molecular dynamics simulations give insight into the conformational change, complex formation, and electron transfer pathway for cytochrome P450 reductase. Protein Sci. 2013;22:1183–1195. doi: 10.1002/pro.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ebrecht A.C., van der Bergh N., Harrison S.T., Smit M.S., Sewell B.T., Opperman D.J. Biochemical and structural insights into the cytochrome P450 reductase from Candida tropicalis. Sci. Rep. 2019;9:20088. doi: 10.1038/s41598-019-56516-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Campelo D., Lautier T., Urban P., Esteves F., Bozonnet S., Truan G., Kranendonk M. The hinge segment of human NADPH-cytochrome P450 reductase in conformational switching: The critical role of ionic strength. Front. Pharmacol. 2017;8:755. doi: 10.3389/fphar.2017.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wadsater M., Laursen T., Singha A., Hatzakis N.S., Stamou D., Barker R., Mortensen K., Feidenhans'l R., Moller B.L., Cardenas M. Monitoring shifts in the conformation equilibrium of the membrane protein cytochrome P450 reductase (POR) in nanodiscs. J. Biol. Chem. 2012;287:34596–34603. doi: 10.1074/jbc.M112.400085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hay S., Brenner S., Khara B., Quinn A.M., Rigby S.E., Scrutton N.S. Nature of the energy landscape for gated electron transfer in a dynamic redox protein. J. Am. Chem. Soc. 2010;132:9738–9745. doi: 10.1021/ja1016206. [DOI] [PubMed] [Google Scholar]
- 84.Aigrain L., Pompon D., Morera S., Truan G. Structure of the open conformation of a functional chimeric NADPH cytochrome P450 reductase. EMBO Rep. 2009;10:742–747. doi: 10.1038/embor.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simtchouk S., Eng J.L., Meints C.E., Makins C., Wolthers K.R. Kinetic analysis of cytochrome P450 reductase from Artemisia annua reveals accelerated rates of NADPH-dependent flavin reduction. FEBS J. 2013;280:6627–6642. doi: 10.1111/febs.12567. [DOI] [PubMed] [Google Scholar]
- 86.Terrett O.M., Dupree P. Covalent interactions between lignin and hemicelluloses in plant secondary cell walls. Curr. Opin. Biotechnol. 2019;56:97–104. doi: 10.1016/j.copbio.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 87.Linger J.G., Vardon D.R., Guarnieri M.T., Karp E.M., Hunsinger G.B., Franden M.A., Johnson C.W., Chupka G., Strathmann T.J., Pienkos P.T. Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12013–12018. doi: 10.1073/pnas.1410657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stone M.L., Anderson E.M., Meek K.M., Reed M., Katahira R., Chen F., Dixon R.A., Beckham G.T., Román-Leshkov Y. Reductive catalytic fractionation of C-lignin. ACS Sustain. Chem. Eng. 2018;6:11211–11218. [Google Scholar]
- 89.Shuai L., Amiri M.T., Questell-Santiago Y.M., Héroguel F., Li Y., Kim H., Meilan R., Chapple C., Ralph J., Luterbacher J.S. Formaldehyde stabilization facilitates lignin monomer production during biomass depolymerization. Science. 2016;354:329–333. doi: 10.1126/science.aaf7810. [DOI] [PubMed] [Google Scholar]
- 90.Li Y., Shuai L., Kim H., Motagamwala A.H., Mobley J.K., Yue F., Tobimatsu Y., Havkin-Frenkel D., Chen F., Dixon R.A. An “ideal lignin” facilitates full biomass utilization. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aau2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ballard C.R., Maróstica M.R. In: Bioactive Compounds. Campos M.R.S., editor. Woodhead Publishing; Sawston, Cambridge: 2019. Chapter 10 - health benefits of flavonoids; pp. 185–201. [Google Scholar]
- 92.Padmakshan D., Timokhin V., Lu F., Schatz P., Vanholme R., Boerjan W., Ralph J. Synthesis of hydroxycinnamoyl shikimates and their role in monolignol biosynthesis. Holzforschung. 2022;76:133–144. [Google Scholar]
- 93.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.-W., Jain S., McCoy A.J. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stanier R.Y., Ingraham J.L. Protocatechuic acid oxidase. J. Biol. Chem. 1954;210:799–808. [PubMed] [Google Scholar]
- 96.Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 97.Humphreys J.M., Hemm M.R., Chapple C. New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10045–10050. doi: 10.1073/pnas.96.18.10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M. Phylogeny.Fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers XP_002462826.1, XP_002444097.1, and XP_002447240.1. The structure discussed in this article can be found at www.rcsb.org deposited under the PDB IDs: 7SUX, 7SUZ, and 7SV0.