Due to the multiplicity of antibiotics, infections caused by drug-resistant bacteria did not represent a medical problem until the early 1980s. However, evolution of bacteria towards resistance has been considerably accelerated by the selective pressure exerted by overprescription of drugs in clinical settings and their heavy use as growth promoters for farm animals. Since bacteria have the remarkable ability to develop resistance to every antibiotic, we can anticipate that even bacterial species such as Listeria, which are still considered to be susceptible to almost all antibiotics, will evolve toward multiresistance.
Listeria spp. are ubiquitous bacteria widely distributed in the environment (20, 41). Among the seven species of Listeria, only Listeria monocytogenes is commonly pathogenic for humans. It can cause serious infections such as meningitis or septicemia in newborns, immunocompromised patients, and the elderly or lead to abortion (16, 20, 27, 41, 43). Although human listeriosis occurs only sporadically (16, 41, 43) several outbreaks have been observed during the last two decades (20, 25, 43). It is now established that food-borne transmission constitutes the main route of acquisition of listeriosis (16, 34, 41, 43). Despite efficient antibiotic therapy, listeriosis represents a public health problem since it is fatal in up to 30% of cases (16, 26). In general, isolates of L. monocytogenes, as well as strains of other Listeria spp., are susceptible to a wide range of antibiotics except cephalosporins and fosfomycin (23, 24). The treatment of choice for listeriosis remains the administration of ampicillin or penicillin G combined with an aminoglycoside, classically gentamicin (4, 12, 18, 20, 26, 27, 29, 31, 32). The association of trimethoprim with a sulfonamide, such as sulfamethoxazole in co-trimoxazole, is considered to be a second-choice therapy. The most active agent in the combination seems to be trimethoprim, which is synergized by sulfamethoxazole (4, 26, 27, 29, 32, 43). Most isolates from clinical as well as food-borne and environmental sources are susceptible to the antibiotics active against gram-positive bacteria. The first L. monocytogenes strains resistant to antibiotics were reported in 1988 (35). The strains were resistant to >10 μg of tetracycline per ml. The first multiresistant strain of L. monocytogenes was isolated in France in 1988 (35). Since then, other strains of Listeria spp. isolated from food or the environment or in sporadic cases of human listeriosis resistant to one or several antibiotics have been described (2, 9, 14, 15, 18, 21, 30, 36, 38, 44). This review will focus on the recent emergence of antibiotic resistance in Listeria spp. by acquisition of three types of mobile genetic elements: self-transferable and mobilizable plasmids and conjugative transposons (2, 8, 9, 21, 35, 36).
IN VITRO AND IN VIVO CONJUGATIVE TRANSFER OF RESISTANCE IN LISTERIA SPP.
Several studies have described the transfer by conjugation of enterococcal and streptococcal plasmids and transposons carrying antibiotic resistance genes from Enterococcus-Streptococcus to Listeria and between species of Listeria (17, 33, 47). Plasmid pIP501, initially detected in Streptococcus agalactiae, has a broad host range and confers resistance to chloramphenicol, macrolides, lincosamides, and streptogramins (13). Transfer by conjugation of pIP501 from S. agalactiae to L. monocytogenes, L. murrayi, and L. grayi was obtained at frequencies of approximately 10−6. Plasmid pIP501 was then able to replicate in Listeria and to promote its own transfer between strains of Listeria and from Listeria back to Streptococcus (33, 47). The broad-host-range plasmid pAMβ1 of Enterococcus faecalis, conferring resistance to erythromycin, was transferred successfully by conjugation from E. faecalis to L. monocytogenes at frequencies ranging from 10−4 to 10−8 (17). This plasmid could replicate in the new host and be transferred by conjugation between strains of L. monocytogenes and from L. monocytogenes back to E. faecalis. Plasmid pAMβ1 was also compatible with two cryptic plasmids of L. monocytogenes (17). Plasmid pRYC16 is a conjugative cryptic replicon which is widespread in Listeria (33). This plasmid could mobilize pDB1, a nonconjugative plasmid of Bacillus subtilis, from L. grayi to L. monocytogenes (47). Another study reported the transfer of a plasmid carrying the vanA gene cluster, conferring glycopeptide resistance, from E. faecium to L. monocytogenes, L. ivanovii, and L. welshimeri (3). In retransfer experiments using Listeria transconjugants as donors, vancomycin resistance was transferred to L. monocytogenes, L. ivanovii, L. welshimeri, L. innocua, and L. seeligeri with a frequency higher than that in the primary matings (3).
Tn916, initially detected in E. faecalis, is a broad-host-range conjugative transposon carrying the tet(M) gene conferring resistance to tetracycline-minocycline (47). Transfer by conjugation of Tn916 was obtained at a frequency of 10−6 from E. faecalis to L. innocua. Then, Tn916 mediated its own transfer from L. innocua to L. murrayi and L. grayi and from these three species of Listeria to E. faecalis, S. agalactiae, Streptococcus pneumoniae, and Erysipelothrix rhusiopathiae (47). Conjugative transfer of the Tn916-related transposon Tn1545, initially found in S. pneumoniae, was obtained from E. faecalis to L. monocytogenes in vitro and in vivo (11, 36). The in vitro transfer frequency from E. faecalis to L. monocytogenes was increased when the experiments were performed in the presence of subinhibitory concentrations of tetracycline. In addition, low doses of tetracycline increased transfer in vivo in the intestinal tract of gnotobiotic mice (11). L. monocytogenes is frequently found in the digestive tract in humans and animals where diverse species of Enterococcus and Streptococcus harboring conjugative plasmids and transposons are also present at very high numbers. This observation reinforces the notion that the intestinal tract represents an ecosystem most favorable to direct exchange of genetic information between these two bacterial genera.
ACQUIRED MULTIPLE RESISTANCE IN L. MONOCYTOGENES MEDIATED BY SELF-TRANSFERABLE PLASMIDS
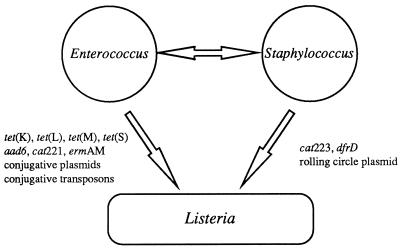
Multiple antibiotic resistance in L. monocytogenes which appeared to be transferable to Enterococcus, Streptococcus, and Staphylococcus was reported for three clinical strains (Table 1), two isolated in France (2, 35, 38) and one in Switzerland (21). The first strain, BM4210, was isolated in 1988 at the Hospital of Dijon from an 84-year-old patient with meningoencephalitis (35) and was resistant to chloramphenicol (MIC, 64 μg/ml), erythromycin (MIC, 2,048 μg/ml), streptomycin (MIC, 256 μg/ml), and tetracycline (MIC, 128 μg/ml)-minocycline (MIC, 8 μg/ml) (Tables 1 and 2). The resistance determinants were carried by plasmid pIP811, which was more stable in the genera Enterococcus and Streptococcus than in Listeria and Staphylococcus (35). Transfer by conjugation of pIP811 was obtained at higher frequencies between strains of L. monocytogenes and/or Enterococcus-Streptococcus than among Staphylococcus strains (35). Moreover, pIP811 exhibited high structural similarity with pAMβ1, the prototype of broad-host-range resistance plasmids from Enterococcus-Streptococcus (17, 35). The genes conferring resistance to chloramphenicol, erythromycin, and streptomycin were homologous to cat221 (encoding a chloramphenicol acetyltransferase), ermAM (encoding an rRNA methylase), and aad6 (encoding a 6-N-aminoglycoside nucleotidyltransferase) (Table 3), which are common in Enterococcus and Streptococcus (35) (Fig. 1). Taken together, these observations strongly suggest that emergence of multiple antibiotic resistance in L. monocytogenes BM4210 results from acquisition under natural conditions of a plasmid originating from Enterococcus-Streptococcus (35).
TABLE 1.
Resistance phenotypes in Listeria spp.
| Resistance phenotypea | Country of originb (no. of strains) | Reference(s) |
|---|---|---|
| L. monocytogenes | ||
| Humans | ||
| Cm Cl Gm Sm To | Greece | 46 |
| Cm Em Sm Tc-Mc | France (1) | 35 |
| Cm Em Tc-Mc | France (1) | 38 |
| Switzerland (1) | 21 | |
| Tc | UK (33) | 30, 36 |
| France (3) | 42 | |
| Sm | ND (1) | 44 |
| Italy (2) | 14 | |
| Switzerland (2) | 9 | |
| France (1) | 9 | |
| Em | UK (1) | 30 |
| Cp | France | 42 |
| Food and environment | ||
| Tp | France (1) | 9 |
| Tc-Mc | France (31) | 9 |
| Italy (5) | 9 | |
| Czechoslovakia (1) | 9 | |
| Km Sm Su | Italy (2) | 14 |
| Km Rif Sm Su | Italy (1) | 14 |
| Cm Em Km Rif Sm Su | Italy (1) | 14 |
| L. innocua (food and environment) | ||
| Tc | Canada (2) | 44 |
| Italy (1) | 14, 15 | |
| Spain (8) | 18 | |
| Tc-Mc | Italy (16) | 9 |
| Sweden (4) | 9 | |
| Spain (1) | 9 | |
| France (1) | 9 | |
| Em Tc | Italy (1) | 14, 15 |
| Sm Tc-Mc | Italy (1) | 9 |
| Em | Italy (1) | 14 |
| Sm Su | Italy (2) | 14 |
| Km Sm Su | Italy (1) | 14 |
| L. welshimeri (food) | ||
| Tc-Mc | Italy (2) | 9 |
Cm, chloramphenicol; Cl, clindamycin; Cp, ciprofloxacin; Em, erythromycin; Gm, gentamicin; Km, kanamycin; Mc, minocycline; Rif, rifampin; Sm, streptomycin; Su, sulfonamide; Tc, tetracycline; To, tobramycin; Tp, trimethoprim.
UK, United Kingdom; ND, non determined.
TABLE 2.
MICs of antibiotics against resistant Listeria spp.
| Species | MIC (μg/ml)a
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CL | CM | EM | GM | KM | RIF | SM | TO | SU | TC | MC | TP | |
| L. monocytogenes | >2 | 8–>16 | 8 | >8 | 64–>256 | 32–64 | 32–64 | 8 | >1,024 | 64–128 | >4 | 1,024 |
| 64 | >32 | 256 | 8 | |||||||||
| 2,048 | ||||||||||||
| L. innocua | —b | 64–>256 | 64 | — | 128 | >1,024 | 64–>256 | 4–32 | — | |||
| 512 | ||||||||||||
| L. welshimeri | — | — | — | — | — | — | 128 | 4–16 | — | |||
CL, clindamycin; CM, chloramphenicol; EM, erythromycin; GM, gentamicin; KM, kanamycin; MC, minocycline; RIF, rifampin; SM, streptomycin; SU, sulfonamide; TO, tobramycin; TC, tetracycline; TP, trimethoprim.
—, no resistance.
TABLE 3.
Distribution of antibiotic resistance genes in Listeria spp.
| Resistance phenotypea | Gene(s) [no. of strains] | Reference(s) |
|---|---|---|
| L. monocytogenes | ||
| Humans | ||
| Cm Em Sm Tc-Mc | cat221, ermAM, aad6, tet(S) [1] | 21, 35, 38 |
| Tc-Mc | tet(M)/int-Tn [24] | 36 |
| Tc | tet(L) [1] | 36 |
| Food and environment | ||
| Em | erm(C) [1] | 40 |
| Tc-Mc | tet(M)/int-Tn [36] | 9 |
| tet(M) [1] | 9 | |
| L. innocua (food) | ||
| Tc | tet(K) [1] | 15 |
| tet(M) [9] | 15 | |
| tet(K) + tet(M) [2] | 15 | |
| Tc-Mc | tet(M) [14] | 9 |
| tet(M)/int-Tn [5] | 9 | |
| tet(S) [3] | 9 | |
| Sm Tc-Mc | aad6, tet(M) [1] | 9 |
| L. welshimeri (food) | ||
| Tc-Mc | tet(M)/int-Tn [1] | 9 |
| tet(S) [1] | 9 |
Cm, chloramphenicol; Em, erythromycin; Km, kanamycin; Mc, minocycline; Rif, rifampin; Sm, streptomycin; Su, sulfonamide; Tc, tetracycline; Tp, trimethoprim.
FIG. 1.
Schematic representation of the origin of antibiotic resistance determinants and their genetic basis in Listeria spp.
The tetracycline-minocycline resistance determinant was characterized as a novel class of genes named tet(S), similar to tet(M) and tet(O), which confer resistance by ribosomal protection (6) (Table 3). The tet(S) gene, initially detected in strain BM4210, was later found in the two other multiresistant L. monocytogenes clinical isolates (6) (Fig. 1). The gene was also detected, alone or associated with tet(M) and/or tet(L), in 22 clinical isolates of E. faecalis resistant to tetracycline-minocycline (7) (Fig. 1). Transfer of tet(S) from eight strains of E. faecalis harboring this gene alone, to E. faecalis and/or to L. monocytogenes, was obtained by conjugation, confirming the enterococcal origin of tet(S) (7). Further analysis of five clinical strains of E. faecalis harboring tet(S) showed that transfer of the gene between strains of E. faecalis required the presence in the donor of a conjugative plasmid. Transfer occurred from chromosome to chromosome and resulted in the acquisition by the recipient of a ca. 40-kb tet(S) element associated with the exchange of large pieces of the chromosome (19).
The two L. monocytogenes strains isolated subsequently in Bordeaux (2, 38) and Zurich (21) were resistant to chloramphenicol, erythromycin, and tetracycline-minocycline but not to streptomycin, in contrast to BM4210 (Table 1). The strain from Bordeaux harbored plasmid pUBX1, which carried the resistance genes and was self-transferable by conjugation to L. monocytogenes, E. faecalis, S. agalactiae, and Staphylococcus aureus (2). Resistance in the strain from Zurich was due to the presence of plasmid pWDB100, which was self-transferable to L. monocytogenes, Enterococcus, and Staphylococcus epidermidis. Comparative analysis of the restriction endonuclease-generated profiles of pIP811, pUBX1, and pWDB100 DNAs showed that the plasmids were indistinguishable (2, 21). Thus, the plasmids detected in strains of L. monocytogenes isolated from patients hospitalized in distinct geographic areas are closely related. The three plasmids also showed significant similarity to pIP501, the broad-host-range plasmid from S. agalactiae (13, 21). These results are consistent with the hypothesis that Enterococcus-Streptococcus spp. represent the origin of multiple antibiotic resistance in L. monocytogenes (2, 7, 21, 35). In a more recent study (22), plasmid pWDB100/m (2,679 bp) was detected in S. epidermidis transconjugants that had acquired the multiresistance plasmid pWDB100. Plasmid pWDB100/m conferred resistance to chloramphenicol and corresponded to an excised derivative of pWDB100 (Fig. 1). The gene encoding a chloramphenicol acetyltransferase belonged to the family of cat determinants of the pC223 family of plasmids that replicate using a single-stranded intermediate (22).
TETRACYCLINE RESISTANCE MEDIATED BY CONJUGATIVE PLASMIDS AND TRANSPOSONS IN LISTERIA SPP.
Tetracycline resistance is the most frequent resistance trait in L. monocytogenes isolated from humans (Table 1). As already mentioned, the three self-transferable multiresistant plasmids from L. monocytogenes isolated from humans carried the tet(S) gene (21, 35, 38). In a study performed in the United Kingdom with 1,288 clinical strains isolated between 1967 and 1981, 1983 and 1986, and 1987 and 1990, 33 isolates were found to be resistant to tetracycline (30, 36). Twenty-five strains were selected for further analysis (36), and 24 were cross resistant to tetracycline (MIC, 64 to 128 μg/ml) and minocycline (MIC > 4 μg/ml). These strains harbored tet(M) associated with int-Tn, the gene encoding the integrase involved in the mobility of the Tn1545-Tn916 family of conjugative transposons from Enterococcus-Streptococcus (36, 37) (Fig. 1). Cotransfer by conjugation of tet(M) and int-Tn among L. monocytogenes strains and from L. monocytogenes to E. faecalis was detected in 7 of 12 strains at frequencies similar to those obtained with the prototype element Tn1545. One strain was resistant to tetracycline alone (MIC, 64 μg/ml) and harbored tet(L) carried by plasmid pIP813 (5 kb) (36) (Tables 1 to 3) (Fig. 1). This small plasmid was able to replicate in E. faecalis and Escherichia coli and could belong to the family of plasmids which possess a broad host range including gram-positive and gram-negative bacteria and replicate using a single-stranded intermediate. The tet(L) gene codes for a protein which promotes active efflux of tetracycline from the bacteria, whereas tet(M) directs the synthesis of a cytoplasmic protein that protects the ribosome from inhibition by the antibiotic (45). These tetracycline resistance determinants are common in Enterococcus-Streptococcus (7, 45). These observations indicate that two types of movable genetic elements, conjugative plasmids and transposons originating from Enterococcus-Streptococcus, are responsible for the emergence of resistance to tetracycline in L. monocytogenes isolated in clinical situations (36). More recently, three strains resistant to tetracycline were detected among 685 strains from human sources collected in France in 1994 and 1995 (42).
The incidence of tetracycline resistance is increasing in strains of Listeria spp. isolated from food and environmental sources (Table 1). The first two strains of L. innocua resistant to tetracycline were identified in a study (44) on the susceptibilities to nine antibiotics of 26 strains isolated from milk in Canada (Table 1). Subsequently, 11 tetracycline-resistant strains (MIC, 128 to >256 μg/ml) were detected among 85 strains of L. innocua isolated from meat and cheese in Italy in 1991 (14, 15). One of them was also resistant to erythromycin. Genetic study of tetracycline resistance was performed for the 11 isolates and for 1 strain isolated subsequently from cheese (15); tet(M) was found in 9 strains, tet(K) was found in 1 strain, and the two genes were associated in two strains (Tables 1 to 3) (Fig. 1). Like tet(L), tet(K) codes for a protein conferring resistance to tetracycline by efflux (45). In contrast to tet(L), tet(K) is widely spread in Staphylococcus spp. but more rarely found in Enterococcus-Streptococcus (45). The tet(M) gene could be transferred by conjugation from one L. innocua strain to L. monocytogenes, L. ivanovii, and E. faecalis (15). Another study (18) on the susceptibilities to nine antibiotics of 32 strains of L. innocua, 28 strains of L. monocytogenes, and 13 strains of L. seeligeri isolated in Spain from food and the environment found 8 L. innocua strains resistant to tetracycline (MIC, 64 μg/ml) (Tables 1 and 2). In the most recent study to date performed on 1,040 strains of Listeria spp. isolated from food and environmental sources and collected worldwide (9), 61 strains were resistant to tetracycline (MIC, 64 to 128 μg/ml) and minocycline (MIC, 4 to 32 μg/ml): 37 L. monocytogenes strains, 22 L. innocua strains, and 2 L. welshimeri strains; one strain of L. innocua was resistant to tetracycline-minocycline and to streptomycin (Tables 1 and 2). Among the 61 strains, the tet(S) gene was detected in 3 L. innocua strains and 1 L. welshimeri strain and tet(M) was identified in 57 strains of Listeria spp. together with int-Tn in 42 isolates (Tables 1 and 3). In the L. innocua and L. welshimeri species, tet(S) was located in the chromosome, whereas in L. monocytogenes BM4210, it was carried by plasmid pIP811. The strain of L. innocua resistant to tetracycline-minocycline and streptomycin harbored tet(M) (9) (Table 1). These results confirm the notion of easy exchange of tetracycline resistance genes under natural conditions between the genera Enterococcus-Streptococcus and Listeria and between different species of Listeria (7, 9, 15, 36).
TRIMETHOPRIM RESISTANCE MEDIATED BY AN RC PLASMID IN L. MONOCYTOGENES
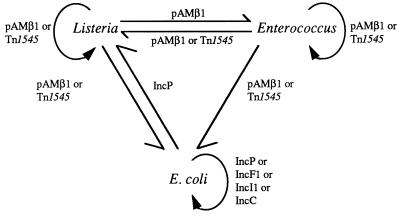
L. monocytogenes BM4293, resistant to high levels of trimethoprim (MIC, 1024 μg/ml), was detected in France among 1,040 strains of Listeria spp. from various sources (9) (Tables 1 and 2). This strain was susceptible to sulfamethoxazole, but due to trimethoprim resistance the synergism usually observed between the two antibiotics was abolished. Since resistance to trimethoprim alone cannot be detected in vitro by using the disk of co-trimoxazole, it is thus advisable to study independently the two components of the combination (1, 9, 28). The trimethoprim resistance gene was identical to dfrD from Staphylococcus haemolyticus, which encodes S2DHFR, of the second class of high-level trimethoprim-resistant dihydrofolate reductases identified in gram-positive bacteria (5). This determinant was carried by plasmid pIP823 (3,712 bp), which possesses all the features of the pUB110/pC194 plasmid family which replicates by the rolling-circle (RC) mechanism (8) (Fig. 1). Plasmid pIP823 had an exceptionally broad host range of replication since it was maintained stably in L. monocytogenes, E. faecalis, S. aureus, and B. subtilis but also in E. coli (8). Conjugative mobilization of pIP823 was obtained by self-transferable plasmids between L. monocytogenes and E. faecalis, between L. monocytogenes and E. coli, and between strains of E. coli and by the streptococcal conjugative transposon Tn1545 between strains of L. monocytogenes, from E. faecalis to L. monocytogenes and E. faecalis, and from L. monocytogenes and E. faecalis to E. coli (8) (Fig. 2). The high degree of similarity between the RC plasmids indicates easy horizontal transfer of these replicons between gram-positive cocci and bacilli, including Listeria. It was proposed that trimethoprim resistance in L. monocytogenes could originate in staphylococci.
FIG. 2.
Saga of plasmid pIP823. Arrows represent direction and extent of mobilization of plasmid pIP823. Circular arrows indicate intrageneric (Listeria or Enterococcus) or intraspecies (E. coli) transfer. Linear arrows indicate intergeneric transfer. The genetic elements (plasmids or transposons) able to mobilize pIP823 in trans are indicated.
RESISTANCE TO OTHER ANTIBIOTICS IN LISTERIA SPP.
Resistance to streptomycin was observed in several clinical strains of L. monocytogenes (Tables 1 and 2). A strain resistant to streptomycin (MIC, 30 μg/ml) was detected among 195 Listeria spp. isolated from various sources (human samples, food, and environment) collected worldwide (44). Analysis of susceptibilities to 11 antibiotics of 54 clinical L. monocytogenes strains isolated in Italy between 1987 and 1991 revealed the presence of two strains resistant to streptomycin (MIC, 64 μg/ml) (14). The already mentioned strain of L. innocua resistant to tetracycline-minocycline and to streptomycin (MIC, 512 μg/ml) produced a 6-N-streptomycin adenylyltransferase encoded by aad6 (9), a gene largely spread in Enterococcus-Streptococcus (Tables 1 to 3). More recently, three L. monocytogenes strains resistant to low levels of streptomycin (MIC, 32 to 64 μg/ml) were identified among 60 strains from human samples isolated worldwide (9). In these strains, no streptomycin-modifying activity was detected, suggesting that low-level resistance could be due to ribosomal mutations (9).
A strain resistant to erythromycin (MIC > 32 μg/ml) was detected in a study on antibiotic susceptibilities of 621 clinical strains isolated in the United Kingdom between 1987 and 1990 (30) (Tables 1 and 2) but was not studied further.
In a study in Italy in 1991 on 98 L. monocytogenes isolates from food, two strains were resistant to streptomycin, sulfamethoxazole, and kanamycin, one strain was resistant to streptomycin, sulfamethoxazole, kanamycin, and rifampin, and one strain was resistant to the latter four antibiotics as well as to erythromycin and chloramphenicol (14) (Table 1). Among 85 strains of L. innocua from food-borne sources, the same study detected one strain resistant to erythromycin, one strain resistant to erythromycin and tetracycline (already mentioned), two strains resistant to streptomycin and sulfamethoxazole, and one strain resistant to these two antibiotics and to kanamycin (Tables 1 and 2). Among these strains an L. monocytogenes strain and an L. innocua strain resistant to erythromycin carried the ermC gene which was transferable by conjugation to Listeria and E. faecalis without association with a conjugative plasmid (40) (Table 3).
In a recent study of 685 strains collected in France from human sources in 1994 and 1995, one strain was resistant to ciprofloxacin (42).
A multiresistant strain of L. monocytogenes was isolated in Greece from a neonate who developed meningitis 21 days after birth. The strain was resistant to gentamicin (MIC > 8 μg/ml), streptomycin (MIC > 1,000 μg/ml), chloramphenicol (MIC > 16 μg/ml), and clindamycin (MIC > 2 μg/ml) and was moderately susceptible to tobramycin (MIC = 8 μg/ml) (46).
No cases of resistance to penicillins, gentamicin, or vancomycin have been reported for strains of Listeria spp. originating from humans, food, or the environment (9, 14, 22, 44).
CONCLUSIONS
Listeriosis is an emerging food-borne disease, and numerous outbreaks that occurred during the last 10 years could be linked to contaminated food (16, 20, 25, 34, 41, 43). With the exception of tetracycline resistance, the proportion of Listeria spp. from clinical, food-borne, or environmental sources resistant to antibiotics remains low. Nevertheless, emergence of multiresistant strains casts doubt on the notion, well established until now, of Listeria as a bacterial genus uniformly susceptible to antibiotics. Since the detection in 1988 of the first clinical strain of L. monocytogenes multiresistant to antibiotics (35), other strains of Listeria spp. from various sources resistant to one or more antibiotics have been reported (2, 9, 14, 15, 18, 21, 30, 36, 38, 44). Multiple resistance in L. monocytogenes was linked to the presence of a self-transferable plasmid that was proposed to originate in Enterococcus-Streptococcus (35). Resistance to tetracycline results from the acquisition of genes carried by self-transferable plasmids or by conjugative transposons also originating in Enterococcus-Streptococcus (7, 9, 15, 36). Tetracycline use in therapy and as a supplement in animal food probably favored the dissemination of tetracycline resistance determinants among the various species of Listeria. The recently detected food-borne strain of L. monocytogenes resistant to trimethoprim is of particular interest, since the trimethoprim-sulfamethoxazole combination is used in the treatment of listeriosis, especially in patients allergic to the penicillins (9). Thus, emergence and dissemination of this resistance trait in clinical strains of L. monocytogenes could have important therapeutic consequences. Recently, a clinical isolate of L. monocytogenes resistant to multiple antibiotics including gentamicin was reported (46). Resistance to other antibiotics such as streptomycin, erythromycin, kanamycin, sulfamethoxazole, or rifampin has also been observed (9, 14, 30, 44). These examples clearly demonstrate that Listeria spp. are capable of acquiring antibiotic resistance genes from foreign sources.
It has been proposed that Enterococcus-Streptococcus constitute a reservoir of resistance genes not only for other gram-positive bacteria but also for gram-negative bacilli (10, 47). It has been further suggested that, in humans and animals, the digestive tract was the privileged site for acquisition, by Listeria spp., of conjugative plasmids and transposons from Enterococcus-Streptococcus (11). The observations on antibiotic resistance in Listeria spp. reported in this review clearly support these hypotheses. Strains of E. faecalis harboring conjugative plasmids or transposons encoding β-lactamases and enzymes that confer high-level resistance to gentamicin have been described (39). Acquisition by L. monocytogenes of resistance to these antibiotics would represent a major therapeutic problem in clinical settings. It can be anticipated that the current favorable situation of quasiuniform susceptibility of Listeria spp. to the antibiotics used in clinical practice will deteriorate under the selective pressure exerted by mis- or overuse of the drugs. Antibiotics have also often been used indiscriminately in animal breeding. Since the food origin of human infections is now recognized, it is thus important to test for antibiotic resistance in food-borne and clinical isolates of Listeria and to reconsider critically the use of antibiotics as supplements in animal feed.
ACKNOWLEDGMENTS
We are grateful to G. Gerbaud for reading the manuscript. P.C. thanks Milton H. Saier for laboratory hospitality.
This work was supported in part by a Bristol-Myers Squibb Unrestricted Biomedical Research Grant in Infectious Disease.
REFERENCES
- 1.Amyes S G B, Towner K J. Trimethoprim resistance; epidemiology and molecular aspects. J Med Microbiol. 1990;31:1–19. doi: 10.1099/00222615-31-1-1. [DOI] [PubMed] [Google Scholar]
- 2.Arpin C, Carlier C, Courvalin P, Quentin C. Analysis of an antibiotic resistance plasmid from a clinical isolate of Listeria monocytogenes, abstr. 26/C3. 12 Réunion Interdisc. Paris, France: Chimiothér. Anti-Infect.; 1992. [Google Scholar]
- 3.Biavasco F, Giovanetti E, Miele A, Vignaroli C, Facinelli B, Varaldo P E. In vitro conjugative transfer of VanA vancomycin resistance between enterococci and Listeriae of different species. Eur J Clin Microbiol Infect Dis. 1996;15:50–59. doi: 10.1007/BF01586185. [DOI] [PubMed] [Google Scholar]
- 4.Boisivon A, Guiomar C, Carbon C. In vitro bactericidal activity of amoxicillin, gentamicin, rifampicin, ciprofloxacin and trimethoprim-sulfamethoxazole alone or in combination against Listeria monocytogenes. Eur J Clin Microbiol Infect Dis. 1990;9:206–209. doi: 10.1007/BF01963839. [DOI] [PubMed] [Google Scholar]
- 5.Charpentier E, Courvalin P. Emergence of the trimethoprim resistance gene dfrD in Listeria monocytogenes BM4293. Antimicrob Agents Chemother. 1997;41:1134–1136. doi: 10.1128/aac.41.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charpentier E, Gerbaud G, Courvalin P. Characterization of a new class of tetracycline-resistance gene tet(S) in Listeria monocytogenes BM4210. Gene. 1993;131:27–34. doi: 10.1016/0378-1119(93)90665-p. [DOI] [PubMed] [Google Scholar]
- 7.Charpentier E, Gerbaud G, Courvalin P. Presence of the Listeria tetracycline resistance gene tet(S) in Enterococcus faecalis. Antimicrob Agents Chemother. 1994;38:2330–2335. doi: 10.1128/aac.38.10.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charpentier E, Gerbaud G, Courvalin P. Conjugative mobilization of the rolling-circle plasmid pIP823 from Listeria monocytogenes BM4293 among gram-positive and gram-negative bacteria. J Bacteriol. 1999;181:3368–3374. doi: 10.1128/jb.181.11.3368-3374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charpentier E, Gerbaud G, Jacquet C, Rocourt J, Courvalin P. Incidence of antibiotic resistance in Listeria spp. J Infect Dis. 1995;172:277–281. doi: 10.1093/infdis/172.1.277. [DOI] [PubMed] [Google Scholar]
- 10.Courvalin P. Transfer of antibiotic resistance genes between gram-positive and gram-negative bacteria. Antimicrob Agents Chemother. 1994;38:1447–1451. doi: 10.1128/aac.38.7.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doucet-Populaire F, Trieu-Cuot P, Dosbaa I, Andremont A, Courvalin P. Inducible transfer of conjugative transposon Tn1545 from Enterococcus faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. Antimicrob Agents Chemother. 1991;35:185–187. doi: 10.1128/aac.35.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espaze E P, Reynaud A E. Antibiotic susceptibilities of Listeria: in vitro studies. Infection. 1988;16:S160–S164. doi: 10.1007/BF01639741. [DOI] [PubMed] [Google Scholar]
- 13.Evans R P, Macrina F L. Streptococcal R plasmid pIP501: endonuclease site map, resistance determinant location, and construction of novel derivatives. J Bacteriol. 1983;154:1347–1355. doi: 10.1128/jb.154.3.1347-1355.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Facinelli B, Giovanetti E, Varaldo P E, Casolari P, Fabio U. Antibiotic resistance in foodborne listeria. Lancet. 1991;338:1272. doi: 10.1016/0140-6736(91)92138-r. [DOI] [PubMed] [Google Scholar]
- 15.Facinelli B, Roberts M C, Giovanetti E, Casolari C, Fabio U, Varaldo P E. Genetic basis of tetracycline resistance in food-borne isolates of Listeria innocua. Appl Environ Microbiol. 1993;59:614–616. doi: 10.1128/aem.59.2.614-616.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farber J M, Peterkin P I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flamm R K, Hinrichs D J, Thomashow M F. Introduction of pAMβ1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect Immun. 1984;44:157–161. doi: 10.1128/iai.44.1.157-161.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franco Abuin C M, Quinto Fernandez E J, Fente Sampayo C, Rodriguez Otero J L, Dominguez Rodriguez L, Cepeda Saez A. Susceptibilities of Listeria species isolated from food to nine antimicrobial agents. Antimicrob Agents Chemother. 1994;38:1655–1657. doi: 10.1128/aac.38.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francois B, Charles M, Courvalin P. Conjugative transfer of tet(S) between strains of Enterococcus faecalis is associated with the exchange of large fragments of chromosomal DNA. Microbiology. 1997;143:2145–2154. doi: 10.1099/00221287-143-7-2145. [DOI] [PubMed] [Google Scholar]
- 20.Gellin B G, Broome C V. Listeriosis. JAMA. 1989;261:1313–1320. [PubMed] [Google Scholar]
- 21.Hadorn K, Hächler H, Schaffner A, Kayser F H. Genetic characterization of plasmid-encoded multiple antibiotic resistance in a strain of Listeria monocytogenes causing endocarditis. Eur J Clin Microbiol Infect Dis. 1993;12:928–937. doi: 10.1007/BF01992167. [DOI] [PubMed] [Google Scholar]
- 22.Hadorn K, Kayser F H, Hächler H. Miniplasmid derived from Listeria monocytogenes multiresistance plasmid pWDB100 upon conjugal transfer into Staphylococcus epidermidis carries chloramphenicol resistance gene identical with staphylococcal gene. System Appl Microbiol. 1994;17:492–500. [Google Scholar]
- 23.Hof H. Therapeutic activities of antibiotics in listeriosis. Infection. 1991;19(Suppl. 4):229–233. doi: 10.1007/BF01644039. [DOI] [PubMed] [Google Scholar]
- 24.Hof H, Nichterlein T, Kretschmar M. Management of listeriosis. Clin Microbiol Rev. 1997;10:345–357. doi: 10.1128/cmr.10.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacquet C, Miegeville A-F, Catimel B, Huynh G, Courtieu A L, Rocourt J. La listériose humaine en France en 1991, 1992 et 1993. Bilan à partir des souches adressées aux centres nationaux de référence. Bull Epidém Hebdom. 1994;28:123–125. [Google Scholar]
- 26.Jones E M, MacGowan A P. Antimicrobial chemotherapy of human infection due to Listeria monocytogenes. Eur J Clin Microbiol Infect Dis. 1995;14:165–175. doi: 10.1007/BF02310351. [DOI] [PubMed] [Google Scholar]
- 27.Lorber B. Listeriosis. Clin Infect Dis. 1997;24:1–11. doi: 10.1093/clinids/24.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Lyon B R, Tennent J M, May J W, Skurray R A. Trimethoprim resistance encoded on a Staphylococcus aureus gentamicin resistance plasmid; cloning and transposon mutagenesis. FEMS Microbiol Lett. 1986;33:189–192. [Google Scholar]
- 29.MacGowan A P, Holt H A, Reeves D S. In-vitro synergy testing of nine antimicrobial combinations against Listeria monocytogenes. J Antimicrob Chemother. 1990;25:561–566. doi: 10.1093/jac/25.4.561. [DOI] [PubMed] [Google Scholar]
- 30.MacGowan A P, Reeves D S, McLauchlin J. Antibiotic resistance in Listeria monocytogenes. Lancet. 1990;336:513–514. [PubMed] [Google Scholar]
- 31.McLauchlin J, Audurier A, Taylor A G. Treatment failure and recurrent human listeriosis. J Antimicrob Chemother. 1991;27:851–857. doi: 10.1093/jac/27.6.851. [DOI] [PubMed] [Google Scholar]
- 32.Nunez-Poulsen P, Carvajal A, Lester A, Andreasen J. In vitro susceptibility of Listeria monocytogenes isolated from human blood and cerebrospinal fluid. APMIS. 1988;96:223–228. [PubMed] [Google Scholar]
- 33.Pérez-Diaz J C, Vicente M F, Baquero F. Plasmids in Listeria. Plasmid. 1982;8:112–118. doi: 10.1016/0147-619x(82)90049-x. [DOI] [PubMed] [Google Scholar]
- 34.Pinner R W, Schuchat A, Swaminathan B, Hayes P S, Deaver K A, Weaver R E, Plikaytis B D, Reeves M, Broome C V, Wenger J D the Listeria Study Group. Role of foods in sporadic listeriosis. II. Microbiologic and epidemiologic investigation. JAMA. 1992;267:2046–2050. [PubMed] [Google Scholar]
- 35.Poyart-Salmeron C, Carlier C, Trieu-Cuot P, Courtieu A-L, Courvalin P. Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet. 1990;335:1422–1426. doi: 10.1016/0140-6736(90)91447-i. [DOI] [PubMed] [Google Scholar]
- 36.Poyart-Salmeron C, Trieu-Cuot P, Carlier C, MacGowan A, McLauchlin J, Courvalin P. Genetic basis of tetracycline resistance in clinical isolates of Listeria monocytogenes. Antimicrob Agents Chemother. 1992;36:463–466. doi: 10.1128/aac.36.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poyart-Salmeron C, Trieu-Cuot P, Carlier C, Courvalin P. Molecular characterization of two proteins involved in the excision of the pneumococcal transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 1989;8:2425–2433. doi: 10.1002/j.1460-2075.1989.tb08373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quentin C, Thibaut M C, Horovitz J, Bebear C. Multiresistant strain of Listeria monocytogenes in septic abortion. Lancet. 1990;336:375. doi: 10.1016/0140-6736(90)91916-x. [DOI] [PubMed] [Google Scholar]
- 39.Rice L B, Eliopoulos G M, Wennersten C, Goldmann D, Jacoby G A, Moellering R C. Chromosomally mediated β-lactamase production and gentamicin resistance in Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:272–276. doi: 10.1128/aac.35.2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts M C, Facinelli B, Giovanetti E, Varaldo P E. Transferable erythromycin resistance in Listeria spp. isolated from food. Appl Environ Microbiol. 1996;62:269–270. doi: 10.1128/aem.62.1.269-270.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocourt J. Listeria et listériose humaine. Ann Inst Pasteur. 1990;1:25–30. [Google Scholar]
- 42.Rocourt J, Saint Cloment C, Brouille F, Charpentier E, Gerbaud G, Courvalin P. Antibiotic susceptibility of Listeria monocytogenes strains isolated from humans in France, abstr. 125/C10. 16 Réunion Interdisc. Paris, France: Chimiothér. Anti-Infect.; 1996. [Google Scholar]
- 43.Schuchat A, Swaminathan B, Broome C V. Epidemiology of human listeriosis. Clin Microbiol Rev. 1991;4:169–183. doi: 10.1128/cmr.4.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slade P J, Collins-Thompson D L. Listeria, plasmids, antibiotic resistance, and food. Lancet. 1990;336:1004. doi: 10.1016/0140-6736(90)92464-s. [DOI] [PubMed] [Google Scholar]
- 45.Speer B S, Shoemaker N B, Salyers A A. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin Microbiol Rev. 1992;5:387–399. doi: 10.1128/cmr.5.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsakaris A, Papa A, Douboyas J, Antoniadis A. Neonatal meningitidis due to multi-resistant Listeria monocytogenes. J Antimicrob Chemother. 1997;39:553–554. doi: 10.1093/jac/39.4.553. [DOI] [PubMed] [Google Scholar]
- 47.Vicente M F, Baquero F, Pérez-Diaz J C. Conjugative acquisition and expression of antibiotic resistance determinants in Listeria spp. J Antimicrob Chemother. 1988;21:309–318. doi: 10.1093/jac/21.3.309. [DOI] [PubMed] [Google Scholar]