Abstract
Drought is the main limiting factor of maize productivity, therefore improving drought tolerance in maize has potential practical importance. Cloning and functional verification of drought–tolerant genes is of great importance to understand molecular mechanisms under drought stress. Here, we employed a bioinformatic pipeline to identify 42 ZmHDZ drought responsive genes using previously reported maize transcriptomic datasets. The coding sequences, exon–intron structure and domain organization of all the 42 genes were identified. Phylogenetic analysis revealed evolutionary conservation of members of the ZmHDZ genes in maize. Several regulatory elements associated with drought tolerance were identified in the promoter regions of ZmHDZ genes, indicating the implication of these genes in plant response to drought stress. 42 ZmHDZ genes were distributed unevenly on 10 chromosomes, and 24 pairs of gene duplications were the segmental duplication. The expression of several ZmHDZ genes was upregulated under drought stress, and ZmHDZ9 overexpressing transgenic plants exhibited higher SOD and POD activities and higher accumulation of soluble proteins under drought stress which resulted in enhanced developed phenotype and improved resistance. The present study provides evidence for the evolutionary conservation of HD-ZIP transcription factors homologs in maize. The results further provide a comprehensive insight into the roles of ZmHDZ genes in regulating drought stress tolerance in maize.
Keywords: Maize, HD-ZIP, Drought stress, Overexpression, Genetic transformation
Introduction
Abiotic stresses such as salinity, drought and heat are major factors that influence the growth, development and yield of crop plants (Abou-Elwafa and Shehzad 2021). To cope with these adverse stresses, plants adopt complex responses at the physiological, metabolic, and molecular levels (Tan et al. 2017), accordingly, numerous defense responsive genes are transcriptionally activated or inhibited during this process (Sekhwal et al. 2015). The expression of stress-responsive genes is mainly controlled by specific transcription factors (TFs) (Hu et al. 2008; Rabara et al. 2014). TFs are proteins that bind to a specific DNA sequence to control the transcription rate of a gene. Based on the characteristics of the DNA-binding domain, TFs are classified into different families, including v-avian myeloblastosis viral oncogene homolog (MYB), NAC, WRKY, APETALA2/Ethylene Responsive Factor (AP2/ERF) and homeodomain-leucine zipper (HD-ZIP) (Du et al. 2014; Li et al. 2016, 2019b; Wang et al. 2020; Zhang et al. 2020; Zhao et al. 2018). HD-ZIP, which is exclusively found in plants, is a transcription factor composed of conserved HD and LZ motifs and plays pivotal roles in regulating plant development and in response to abiotic stresses (Ariel et al. 2007). Since the first plant HD-Zip gene KNOTTED1 was cloned from maize, HD-ZIP genes were identified and characterized in Arabidopsis (Ciarbelli et al. 2008), rice (Oryza sativa) (Meijer et al. 2000), tobacco (Nicotiana tabacum) (Li et al. 2019a), sunflower (Helianthus annuus) (Dezar et al. 2010), wheat (Triticum aestivum) (Yue et al. 2018) and soybean (Glycine max) (Belamkar et al. 2014; Valdés et al. 2012).
According to the encoded protein structure, physiological function and conserved domain, HD-Zip family members were classified into four categories: I–IV (Ariel et al. 2007). The HD-ZIP I genes are mostly implicated in regulating plant response to abiotic, environmental and biological stresses, optical signal reaction and organ development. In Arabidopsis, HOMEOBOX7 (AtHB7) and HOMEOBOX12 (AtHB12) play crucial roles in the primary response to drought by mediating the negative feedback effect on ABA signaling in plant response to drought (Ré et al. 2014). The overexpression of ZmHDZ1 in rice reduces plant tolerance to salinity stress, suggesting that ZmHDZ1 adversely regulates plant tolerance to salinity via ABA-mediated signal transduction pathways (Wang et al. 2017). The expression of the HD-ZIP II gene was regulated by optical signals and involved in organ development, hormone response and shade avoidance. Similar to A. thaliana and several other dicots, numerous rice drought-responsive genes including HOX6, HOX22 and HOX24, have been assigned to HD-ZIP groups I and II, and the expression of these genes was upregulated under drought conditions (Agalou et al. 2008). The overexpression of Hahb-4 in Arabidopsis resulted in shortened stems and internodes, tight catkins and improved drought tolerance (Dezar et al. 2005; Manavella et al. 2006). The genes belonging to HD-ZIP group III mainly control the formation of vascular tissue, polar transport and embryo development. Five HD-ZIP III genes were identified in Arabidopsis, among them, IFL1, AtHB9 and AtHB14 are related to the development of the apical meristem, vascular bundle and lateral tissues, which affect the development of root tip during the embryonic stage and control the formation of the root tip (Byrne 2006). The HD-ZIP genes of group IV are mostly implicated in root development and epithelial cell differentiation. The Arabidopsis HOMEOBOX10 (AtHB10) manipulates the transcription of specific genes in the epidermal cells and mostly functions in the hairy root, epidermal hair and seed coat (Dezar et al. 2010). Taken together, the overexpression of exogenous HD-ZIP genes could promote abiotic stress tolerance of transgenic plants.
Maize is an important food and industrial crop worldwide, and water shortage is a key factor in reducing maize yield, therefore improving drought tolerance in maize has potential practical significance (Su et al. 2021). Cloning and functional verification of drought–tolerance genes is of great importance to understand molecular mechanisms under drought stress. In this study, 42 maize HD-ZIP (ZmHDZ) genes associated with drought tolerance were identified from previous transcriptomic datasets (accession number PRJNA477643) generated under drought-stressed and rewatering treatments (Cao et al. 2019). The full-length genomic and coding sequences, exon–intron structure and domain organization of the 42 genes were identified. Moreover, phylogenetic and expression profile analyses of these genes were performed. A representative HD-ZIP gene, i.e., ZmHDZ9, was functionally characterized by overexpression in the A. thaliana Col-0 wild type (WT). ZmHDZ9 transgenic plants exhibited a higher drought–tolerant phenotype compared to the WT plants, indicating that ZmHDZ9 enhances drought tolerance in transgenic plants.
Materials and methods
Identification and bioinformatics analysis of HD-ZIP genes in maize
The Hidden Markov Model (HMM) profile of the HD domain (PF00046) obtained from Pfam database (http://pfam.sanger.ac.uk/) was employed as a query to identify all HD-containing sequences in maize by searching HD domain sequence against the maize genome database using BlastP program (p value = 0.001). The PFAM and SMART (http://smart.embl-heidelberg.de/) were implemented to identify homologs of the HD-ZIP domains-containing (PF02183) transcription factors in maize (ZmHDZs). The genomic sequence, molecular weight (MW), isoelectric point (PI) and localization information of ZmHDZ genes were redeemed from the EnsemblPlants database (http://plants.ensembl.org/index.html). The Gene Structure Display Server 2.0 was employed for the annotation of exon–intron structures of ZmHDZ genes. The un-rooted phylogenetic tree was generated using the Neighbor-Joining algorithm and the Dayhoff PAM matrix as in the MEGA6 software (http://www.megasoftware.net/). PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used for annotation of the promoter regions of ZmHDZ genes (2 kb upstream of the transcription start site (TSS)) for the identification of cis-elements. Seven cis-regulatory elements including the ABA-responsive element (ABRE), MYB binding site (MBS), SA-responsive elements (TCA), TC-rich, MeJA-responsive elements (CGTCA-motif), LTR and TATC-box were chosen for further analysis. A coexpression network map of genes was generated by the online software Cytoscape version 3.7.1 (https://cytoscape.org). The chromosomal location and duplication information of the identified 42 ZmHDZ genes were analyzed using TBtools software (Chen et al. 2020a).
RNA extraction and RT-qPCR
Trizol reagent (TaKaRa, Dalian, China) was used to extract total RNA from leaves according to the manufacturer’s procedure. After DNase treatment, the integrity and purity of isolated RNA were analyzed using spectrophotometry and electrophoresis on1% agarose gel. cDNA was synthesized using about 2 μg of RNA using the First Strand cDNA Synthesis SuperMixKit (YEASEN) for RT-qPCR assay.
RT-qPCR analysis was conducted according to the procedure of TB Green PremixExTaq™ II (TaKaRa, Dalian, China). The 18S RNA was chosen as a reference gene. The 2−∆∆CT method was employed to estimate the relative expression (Livak and Schmittgen 2001). RT-qPCR experiments were conducted using three biological and three technical replicates.
Subcellular localization
The full-length cDNA sequence of ZmHDZ9 was ligated into the pSAT1-cCFP-C and pSAT1-nVenus-C vectors (Biovector NTCC Inc., Beijing, China) to produced ZmNF-YC2-GFP, ZmAP2-GFP and ZMM4-GFP fusion proteins. The recombinant plasmids were introduced into the maize protoplast following the procedure described by Gomez-Cano et al. (2019). After overnight incubation at 24 °C in the dark, a laser scanning confocal microscope (excited at 488 nm for GFP) was employed for the detection of GFP signals in the protoplast (Carl Zeiss LSM710, Jena, Germany).
The complete coding region of ZmHDZ9 was amplified and ligated into the pMDC83-GFP vector to generate a fusion protein. The recombinant vector was transferred by electroporation into Agrobacterium tumefaciens EHA105 competent cells and transiently expressed in 4-weeks old N. Benthamiana leaves following the infiltration procedure described by Yang et al. (2000). The laser scanning fluorescence microscope (Carl Zeiss LSM710) was employed to assay the GFP-associated fluorescence of fusion constructs after 48 h.
Vector construction and generation of transgenic plants
The complete ZmHDZ9 cDNA sequence was amplified. After restriction enzyme digestion, the PCR product was ligated into the corresponding restriction sites of the binary vector pFGC5941 derived by the CaMV 35S promoter. The intactness of the binary vectors to ZmHDZ9 cDNA sequence was verified by enzymatic restriction digestion and PCR assay, followed by sequencing. The construct (pFGC5941-ZmHDZ9) was transferred by electroporation into Agrobacterium tumefaciens LBA 4404 competent cells and transformed into A. thalianaCol-0 wild-type plants using the floral dip approach (Clough and Bent 1998). The integration of the transgene in the genomes of T1 plants was verified by PCR assay (Abou-Elwafa et al. 2010). Two independent homozygous lines, OE1 and OE2, were chosen for further analysis.
Drought stress assays of ZmHDZ9 transgenic Arabidopsis plants
After surface sterilization, seeds of OE1 and OE2 lines were stored for 15 d at 4 °C. Sterilized seeds were cultivated on 1/2 MS medium and grown in a climate chamber (light intensity of 100 μmol m−2 s−1 of photosynthetically active radiation for 14 h at 22 °C). Four-leaf old seedlings were transplanted into a 3:1 soil and vermiculite mixture. Drought stress was applied to 9–10 leaves-old seedlings by withholding water for 12 days.
Measurement of physiological and biochemical parameters
Twelve days after water withholding, leaves were sampled for measuring physiological parameters. Leaves from four Arabidopsis seedlings were randomly sampled from each treatment. Chlorophyll and soluble protein contents in the leaves were measured referencing the guidance of physiological experiments (Zhang 1992). Chlorophyll content was analyzed using the HPLC procedure (Montefiori et al. 2009). The activity of superoxide dismutase (SOD) and peroxidase (POD) was assayed using analytical kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) following the procedure described by Zhang et al. (2012).
Coexpression and statistical analysis
According to the provided target gene expression in each sample, Pearson correlation analysis between the 42 ZmHDZ genes based on their expression levels measured as Fragments per kilobase of transcript per million mapped fragments (FPKM) was performed to obtain correlation coefficients and p value significance between genes. Finally, the filtered network into Cytoscape version 3.7.1 (https://cytoscape.org) was employed to create the coexpression network map. Student’s t-test was employed to statistically analyze the differences in experimental data between groups. p values of ≤ 0.05 or 0.01 indicate significant or highly significant respectively, differences between sample groups. All experimental data were presented as averages of three replicates.
Results
Identification of ZmHDZ genes under drought-stressed and rewatering conditions
Forty-two HD-ZIP genes, designated as ZmHDZ1 to ZmHDZ42, implicated in drought tolerance regulation redeemed from the previously generated transcriptomic datasets (Cao et al. 2019), were screened using the online tools Pfam and SMART (Table 1). The 42 ZmHDZ genes encode polypeptides with 176 to 894 amino acids. The estimated molecular weight of ZmHDZ proteins ranged from 17.82 (ZmHDZ2) and 96.87 (ZmHDZ27) kDa, and the isoelectric point (PI) ranged from 4.36 (ZmHDZ1) to 10.15 (ZmHDZ17).
Table 1.
Molecular information of 42 ZmHDZ genes identified in the maize genome
| Gene name | ID gene identifier | Chromosome | Protein size (aa) | Molecular weight (kDa) | Theoretical IP |
|---|---|---|---|---|---|
| ZmHDZ1 | GRMZM2G021339 | 4 | 339 | 37 | 4.36 |
| ZmHDZ2 | GRMZM2G122076 | 4 | 272 | 17.82 | 6.55 |
| ZmHDZ3 | GRMZM2G003304 | 1 | 270 | 29.99 | 4.75 |
| ZmHDZ4 | GRMZM2G351330 | 2 | 261 | 29.38 | 4.76 |
| ZmHDZ5 | GRMZM2G178741 | 9 | 344 | 37.57 | 6.5 |
| ZmHDZ6 | GRMZM2G117164 | 5 | 235 | 26.41 | 4.77 |
| ZmHDZ7 | GRMZM2G002915 | 2 | 283 | 31.22 | 5.12 |
| ZmHDZ8 | GRMZM2G056600 | 7 | 261 | 28.46 | 4.5 |
| ZmHDZ9 | GRMZM2G041462 | 7 | 239 | 26.24 | 5.22 |
| ZmHDZ10 | GRMZM2G041127 | 2 | 305 | 33.04 | 4.53 |
| ZmHDZ11 | GRMZM2G139963 | 1 | 344 | 37.6 | 6.75 |
| ZmHDZ12 | GRMZM2G034113 | 2 | 244 | 26.79 | 5.22 |
| ZmHDZ13 | GRMZM2G132367 | 1 | 326 | 35.53 | 4.85 |
| ZmHDZ14 | GRMZM2G134260 | 9 | 293 | 31.1 | 9.65 |
| ZmHDZ15 | GRMZM2G366130 | 2 | 325 | 34.52 | 7.59 |
| ZmHDZ16 | GRMZM2G068672 | 6 | 261 | 27.37 | 9.73 |
| ZmHDZ17 | GRMZM2G307397 | 5 | 247 | 26.54 | 10.15 |
| ZmHDZ18 | GRMZM2G148074 | 1 | 321 | 34.83 | 8.62 |
| ZmHDZ19 | GRMZM2G047715 | 4 | 331 | 35.54 | 5.33 |
| ZmHDZ20 | GRMZM2G477415 | 5 | 221 | 23.8 | 9.3 |
| ZmHDZ21 | GRMZM2G106276 | 9 | 272 | 28.03 | 9.7 |
| ZmHDZ22 | GRMZM2G105834 | 9 | 296 | 31.56 | 9.38 |
| ZmHDZ23 | GRMZM2G131476 | 1 | 262 | 28.24 | 8.14 |
| ZmHDZ24 | GRMZM2G127537 | 7 | 333 | 36.02 | 7.28 |
| ZmHDZ25 | GRMZM2G044752 | 2 | 277 | 25.05 | 8.92 |
| ZmHDZ26 | GRMZM2G126808 | 1 | 292 | 30.96 | 8.63 |
| ZmHDZ27 | GRMZM2G003509 | 1 | 894 | 96.87 | 7.21 |
| ZmHDZ28 | GRMZM2G178102 | 3 | 865 | 93.94 | 6.98 |
| ZmHDZ29 | GRMZM2G469551 | 1 | 872 | 96.35 | 6.08 |
| ZmHDZ30 | GRMZM2G001289 | 2 | 794 | 85.19 | 5.55 |
| ZmHDZ31 | GRMZM2G004957 | 10 | 797 | 85.65 | 6.38 |
| ZmHDZ32 | GRMZM2G118063 | 10 | 719 | 76.66 | 4.78 |
| ZmHDZ33 | GRMZM2G122897 | 10 | 844 | 91.06 | 5.91 |
| ZmHDZ34 | GRMZM2G126646 | 4 | 698 | 76.17 | 6.62 |
| ZmHDZ35 | GRMZM2G145690 | 5 | 692 | 75.81 | 6.58 |
| ZmHDZ36 | GRMZM2G396527 | 10 | 268 | 29.29 | 5.22 |
| ZmHDZ37 | GRMZM2G008286 | 8 | 176 | 19.6 | 7.48 |
| ZmHDZ38 | GRMZM2G004334 | 6 | 687 | 74.91 | 6.04 |
| ZmHDZ39 | GRMZM5G803812 | 5 | 348 | 37.53 | 6.45 |
| ZmHDZ40 | GRMZM2G023291 | 1 | 856 | 93.17 | 6.58 |
| ZmHDZ41 | GRMZM2G119999 | 1 | 305 | 33.34 | 4.57 |
| ZmHDZ42 | AC233899.1_FG004 | 9 | 370 | 39.87 | 7.99 |
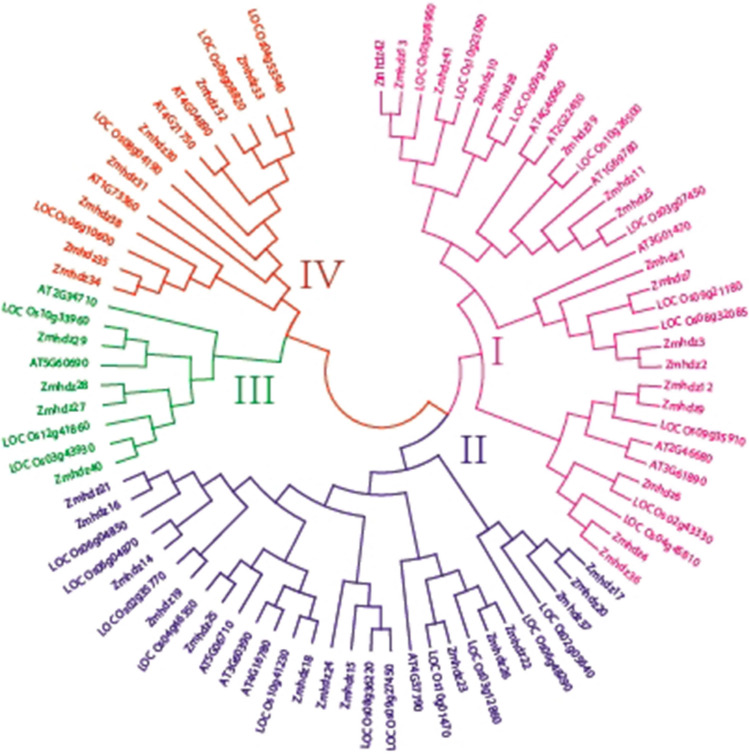
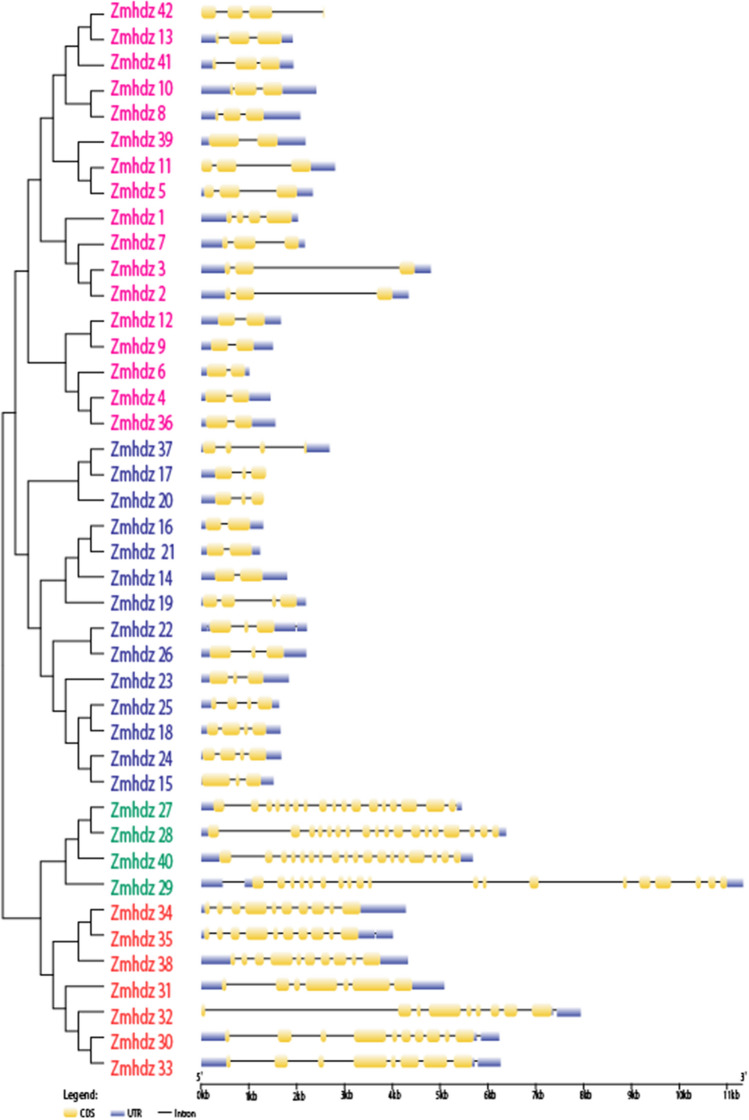
The evolutionary relationship of HD-ZIP genes in maize, Arabidopsis and rice was analyzed by phylogenetic analysis. A phylogenetic tree comprising 42 maize, 15 Arabidopsis and 28 rice HD-ZIP protein sequences was constructed (Fig. 1). Phylogenetic analysis classified ZmHDZ proteins into four groups, each of which contains Arabidopsis and rice members, indicating that HD-ZIP genes in maize, Arabidopsis and rice have a parallel evolution relationship. Seventeen ZmHDZ genes were assigned to group I, 14 ZmHDZ genes were assigned to group II, 7 ZmHDZ genes were assigned to group III, and 4 ZmHDZ genes were assigned to group IV. Furthermore, most of the ZmHDZ genes in the same evolutionary subgroup shared a similar exon–intron structure in terms of intron/exon number and position. The number of exons exhibits substantial variations among the 42 ZmHDZ genes, with most of the ZmHDZ genes having 2–4 exons, whereas only four of them have 17–19 exons (Fig. 2).
Fig. 1.
Phylogenetic analysis of HD-ZIP genes in maize, Arabidopsis and rice. An un-rooted phylogenetic tree was generated using the Neighbor-Joining algorithm and the Dayhoff PAM matrix employed in the MEGA6 software with 1000 bootstrap replicates. Different colors represent different subgroups
Fig. 2.
Structural analysis of 42 ZmHDZ genes. Yellow boxes indicate exons, blue boxes indicate the UTRs. Exon and intron sizes can be determined using the horizontal line at the bottom
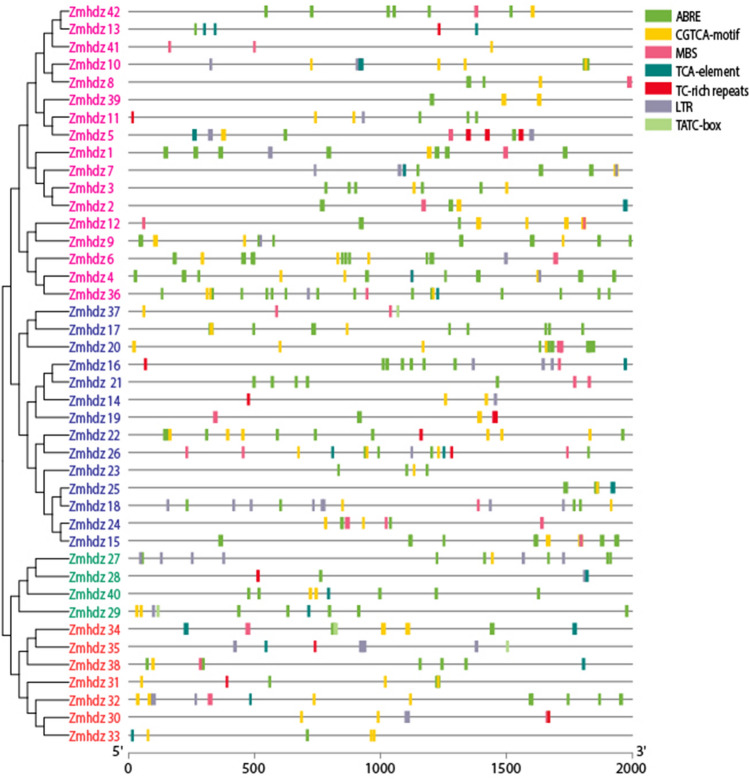
Stress-related Cis-elements analysis in the promoters of ZmHDZ genes
To further reveal the structural characteristics and potential regulatory mechanisms of ZmHDZ genes, the MEME SUITE online tool was employed to annotate the promoter region of ZmHDZ (2 kb upstream TSS) to identify the stress-related cis-elements, such as ABRE, MBS, TCA, TC-rich, CGTCA, LTR and TATC-box. The annotation revealed the clustering of the ABRE, CGTCA, MBS, TC-rich and LTR cis-regulatory elements in the promoter regions of most of the ZmHDZ genes (Fig. 3). For instance, the ABRE was identified in 36 ZmHDZ genes; among them, ZmHDZ36 contained 14 ABRE, followed by eight in each of ZmHDZ4, ZmHDZ17and ZmHDZ6, and six ABREs in ZmHDZ9. MBS, which is involved in drought stress response, was identified in 20 ZmHDZ genes. Three MBS elements were identified in each of ZmHDZ24 and ZmHDZ26, two were detected in each of ZmHDZ12, ZmHDZ21, ZmHDZ37 and ZmHDZ42. The CGTCA-motif, which is a MeJA-responsive element involved in the methyl jasmonate inducibility, was found in 38 ZmHDZ gene promoter regions, such as ZmHDZ9, ZmHDZ32, ZmHDZ36 andZmHDZ6. Six CGTCA elements were enriched in ZmHDZ22, indicating the implication of most of ZmHDZ genes in regulating tolerance to abiotic stresses.
Fig. 3.
Distribution of the major identified stress associated cis-elements in the promoter region (− 2 kb upstream TSS) of ZmHDZ genes. ABRE, CGTCA-motif, TCA-element, and TATC-box: hormone-responsive element; MBS: dehydration-responsive element; TC-rich repeats: stress-responsive element, LTR: low-temperature response. Different colors represent different cis-regulatory elements
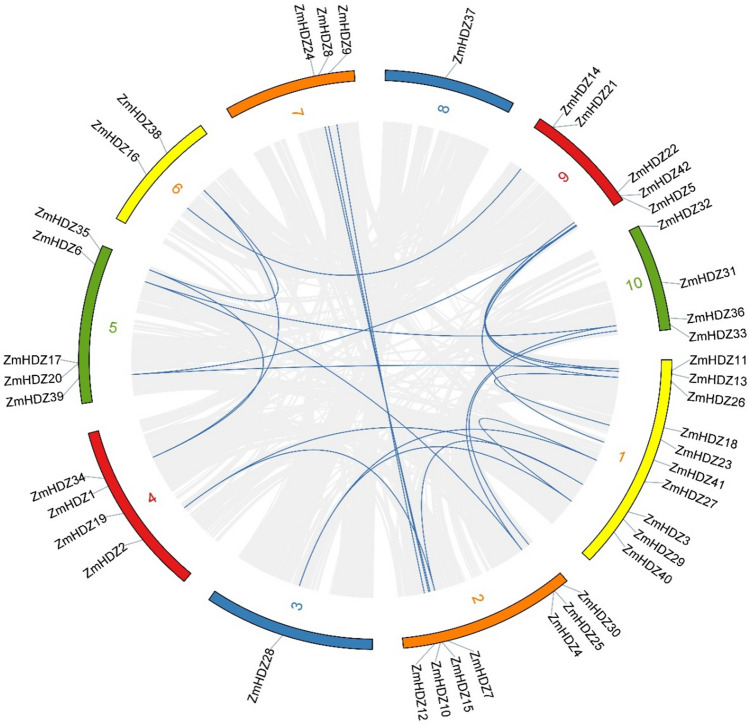
Chromosomal distribution and gene duplication of ZmHDZ genes
The identified 42 ZmHDZ genes were unevenly distributed on the 10 maize chromosomes. The highest number of ZmHDZ genes (10 genes) was mapped on chromosome 1, whereas only a single gene was mapped on each of chromosomes 3 and 8. A total of 24 pairs of duplicated genes were identified, all of which were fragment repeats, such as ZmHDZ12/ZmHDZ9, ZmHDZ13/ZmHDZ42, and ZmHDZ16/ZmHDZ14. Eleven of the 42 ZmHDZ genes participated in gene replication and promoted the diversity of the ZmHDZ gene family, suggesting that some ZmHDZ genes might promote the evolution of the ZmHDZ gene family through segmental duplication (Fig. 4).
Fig. 4.
Schematic representations for chromosomal allocation and ZmHDZ genes relationships. Grey lines reveal collinearity between maize genomes, and the blue lines represent repeated ZmHDZ gene pairs
Expression patterns of ZmHDZ genes under drought-stressed and rewatering conditions
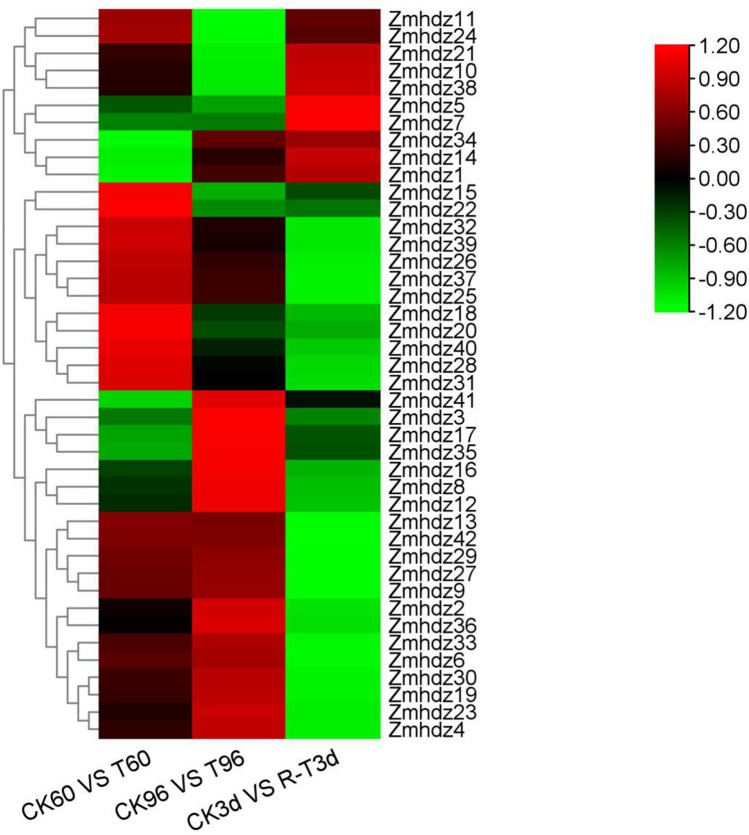
To explore the potential role of ZmHDZ genes, the expression profiles of the 42 ZmHDZ genes were analyzed (Fig. 5). The results showed that the 42 genes exhibited different expression profiles. The expression levels of 13 ZmHDZ genes, such as ZmHDZ4, ZmHDZ6 and ZmHDZ9, ZmHDZ13 and ZmHDZ29, were upregulated at 60 and 96 h under drought stress induction and downregulated after rewatering, indicating that they were strongly induced by drought and may play pivotal roles in regulating plant tolerance to drought stress. The expression of ZmHDZ21, ZmHDZ34 and ZmHDZ41 was downregulated at 60 and 96 h under drought stress induction but was upregulated after rewatering. The expression of the remaining 26 genes, such as ZmHDZ38, ZmHDZ3, ZmHDZ17, exhibited irregular expression patterns.
Fig. 5.
Expression profiles of the 42 ZmHDZ genes under drought and rewatering conditions. Drought stress is induced at three-leaf stage for 60 h (T60) and 96 h (T96) by 20% PEG6000 and rewatered after 3d denoted as T3d. CK60, CK96, and CK3d indicate the control non-transgenic plants exposed to water deficit for 60 and 96 h, and 3 d after rewatering, respectively. The ratio between the expression levels of CK60 and the T60 is expressed as log2(FC) of CK60 versus T60. Similarly, log2(FC) of CK96 versus the T96 and log2(FC) of CK3d versus the T3d were calculated
Coexpression network analysis of ZmHDZ genes
A coexpression map based on gene expression similarity was constructed to identify the biological function and networks between the 42 ZmHDZ genes. In many cases, gene coexpression may imply the presence of a functional linkage between genes. As shown in Fig. 6, the nodes represent genes, and genes with similar profiles connected and formed a network. ZmHDZ4, − 6, − 9, − 14, − 27, − 32, and − 40 exhibited more connection with other genes, indicating that they are key regulatory genes.
Fig. 6.
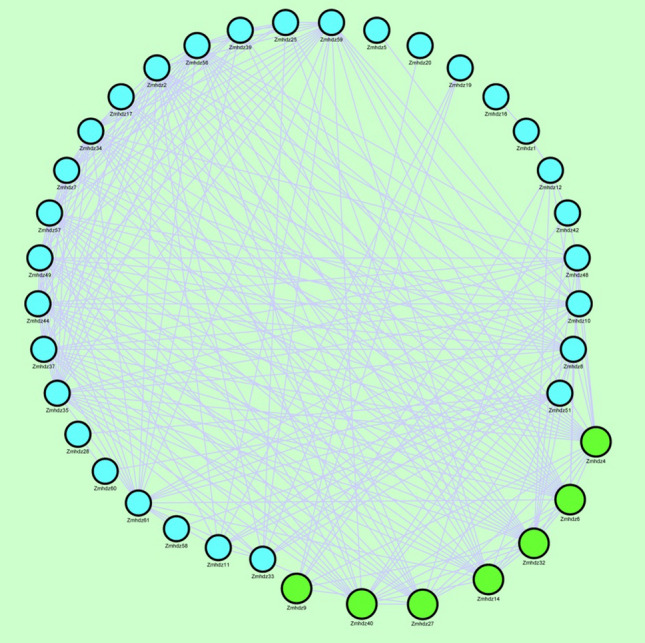
Coexpression network map of ZmHDZ genes. The straight line represents the regulatory relationship of gene existence. The larger the green circle, the stronger association of the gene with other genes
Subcellular localization of ZmHDZ9
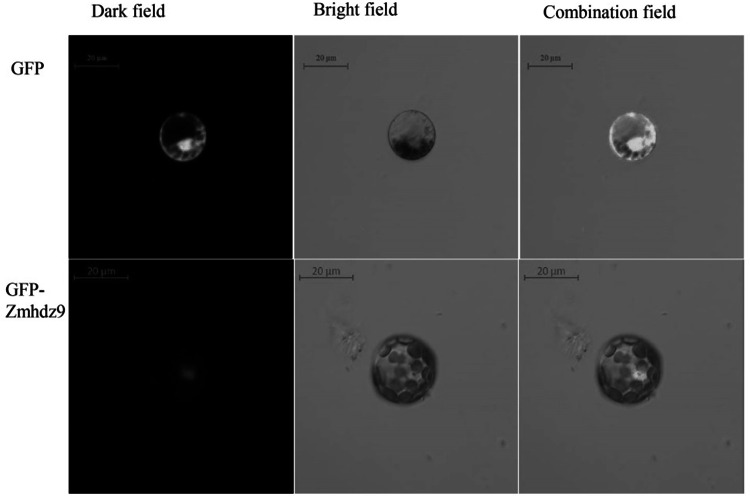
ZmHDZ9, a core gene upregulated under drought stress, was selected as a candidate gene for further functional analysis. Subcellular localization of ZmHDZ9 protein was detected. The recombinant vectors pMDC83-ZmHDZ9-GFP and pMDC83-GFP were constructed and transferred into tobacco (N. benthamian) leaves. The GFP control was evenly observed either in the nucleus and the cytoplasm, while pMDC83-ZmHDZ9-GFP fusion protein was exclusively observed in the nucleus, indicating that ZmHDZ9 protein is localized in the nucleus (Fig. 7).
Fig. 7.
ZmHDZ9 subcellular localization analysis. Fusion proteins were transiently expressed under the control of the CaMV35S promoter in tobacco leaves and detected using a laser scanning confocal microscope. The green color indicates GFP signals. Scale Bars = 20 µm
Overexpression of ZmHDZ9 improves drought tolerance in transgenic Arabidopsis plants
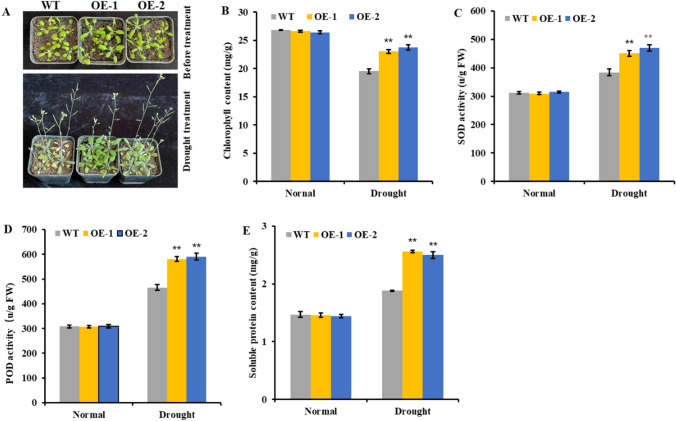
Overexpression analysis of ZmHDZ9 in A. thaliana wild-type (WT) plants was employed to dissect the function of ZmHDZ9 in regulating drought tolerance. Two independent homozygous transgenic lines, OE1 and OE2, were chosen for analysis. There were no visible morphological variations between the transgenic and WT plants. However, 12 days after water withholding, the leaves of the WT plants appeared yellow and wilted, whereas transgenic plants exhibited vigorous, even though the tip section of the leaves became yellow, indicating that ZmHDZ9 transgenic plants exhibited drought stress-tolerant phenotype (Fig. 8A).
Fig. 8.
Overexpression of ZmHDZ9 enhanced drought tolerances in transgenic Arabidopsis plants. A Phenotypes of WT and ZmHDZ9-OE seedlings after 12 days water withholding B chlorophyll content, C SOD activity, D POD activity, E Soluble protein content. * and ** indicate P ≤ 0.05 or 0.01, respectively. Values are presented as averages of three biological replicates ± SD
The related physiological and biochemical parameters were also assayed. No significant differences were observed in the leaf contents of chlorophyll and soluble protein, as well as the SOD and POD activities between the WT and transgenic plants under non-stressed conditions. After 12 days of water withholding, leaf chlorophyll and soluble protein contents and the SOD and POD activities of the two transgenic lines overexpressing ZmHDZ9 were significantly (p < 0.01) higher than those of the WT plants, indicating that the overexpression of ZmHDZ9 could withstand drought stress (Fig. 8B–E).
Discussion
HD-ZIP transcription factors play pivotal roles in regulating plant growth, development and response to various abiotic and biotic stresses. However, comprehensive studies to dissect the roles of these TFs have less been reported in maize. In this study, employing the online tools Pfam and SMART, 42 ZmHDZ drought-responsive genes were identified from the previously reported maize transcriptomic datasets, and further bioinformatics and expression profile analysis was conducted. Phylogenetic analysis showed that the 42 ZmHDZ genes were allocated into four groups, which was consistent with the classification of HD-Zip genes in rice and Arabidopsis (Fig. 1), and genes in the same subgroup exhibited similar gene exon–intron structure. Analyzing the type and number of cis-regulatory elements in the gene promoter region is helpful to explore its function and regulation mechanism (Lechner et al. 2011). In this study, several cis-regulatory elements including, ABRE, MBS, TCA, TC-rich, CGTCA, LTR and TATC-boxes were observed in the promoter regions of the 42 ZmHDZ genes, among which, ABRE, CGTCA, MBS, TC-rich and LTR were observed in most of the ZmHDZ genes. In addition, several ZmHDZ genes contain both ABRE and MBS regulatory elements. ABRE is an ABA-responsive regulatory element with a conserved, 8-bp long (PyACGTGG/TC) cis-element and an ACGT core sequence. However, a single ABRE copy is not sufficient for ABA-responsive gene expression induction. To function as an active cis-regulatory element, ABRE requires other ABRE copies or another specific cis-regulatory element in proximity, which is designated as the coupling element (Fujita et al. 2011; Nakashima et al. 2009). MBS is an MYB binding site required for MYB transcriptional activation of drought inducible genes (Kaur et al. 2017). Furthermore, ABRE and TCA elements have been reported as key cis-regulatory elements that play crucial roles in regulating plant response to various stresses (Li et al. 2020), indicating that ZmHDZ is a positive regulator of drought stress in maize. The 42 ZmHDZ genes were unevenly distributed on the 10 chromosomes of maize. Collinearity analysis of the 42 ZmHDZ genes showed that 31 ZmHDZ genes (73.8%) were in a replication relationship, for a total of 24 gene pairs, all of which were segmental repeats. Tandem and segmental duplications are the driving forces for gene family expansion and evolution. The protein sequence and gene structure of two genes in one gene pair were highly similar, which could ensure the stability of maize evolution.
Under drought and recovery treatments, 42 ZmHDZ genes exhibited different expression profiles. The expression profiles of several ZmHDZ genes, such as ZmHDZ4, ZmHDZ6, ZmHDZ9 and ZmHDZ13, were upregulated under drought stress and downregulated after rewatering, whereas other genes exhibited irregular expression profiles, indicating that different genes are involved in different regulatory pathways and have different functions. Coexpression analysis is a powerful approach to deduce the transcriptional regulation of genes. ZmHDZ 4, − 6, − 9, − 14, − 27, − 32 and − 40 exhibited more connection with other ZmHDZ genes, indicating that those genes are likely to be more implicated in drought tolerance in maize.
Transgenic expression of HD-ZIP genes was found to promote plant ability to tolerate abiotic stresses (Zhu et al. 2018). The ectopic overexpression of AtHDG11 significantly promoted drought and salt tolerance in tall fescue (Festuca arundinacea Schreb.) (Cao et al. 2009). Moreover, the contents of proline, abscisic acid (ABA), and soluble sugar, as well as POD and SOD activities, were highly elevated in transgenic rice plants overexpressing AtEDT1/HDG11under drought stress (Yu et al. 2013). In rice, the overexpression of ZmHDZ4 improves drought tolerance and enhances plant sensitivity to induced abscisic acid (Wu et al. 2016). To further dissect the molecular function of ZmHDZ genes, ZmHDZ9, which was positively upregulated under drought stress, was selected for functional analysis by overexpression in the A. thaliana Col-0 wild-type strain. ZmHDZ9 transgenic Arabidopsis plants showed much better tolerance than Col-0 WT plants under drought stress. Thus, it is suggested that ZmHDZ9 has a similar biological function with its Arabidopsis homolog, AtHB7, which was reported to be implicated in plant response to drought stress (Lee and Chun 1998), and the overexpression of AtHB7 enhances the development and regeneration of transgenic tomato plants (Pehlivan 2019). Chlorophyll is an essential component of the photosynthetic system in plants (Abou-Elwafa and Amein 2016; Chen et al. 2020b). In the study, the results showed that chlorophyll content was drastically reduced in both the ZmHDZ9 transgenic and Arabidopsis WT plants in response to drought stress. However, the reduction was much higher in the WT plants. Thus, it is conceivable that ZmHDZ9 transgenic plants exhibited an enhanced developed phenotype and improved tolerance to drought conditions. Drought exacerbates the accumulation of reactive oxygen species (ROS) in plants, causing oxidative stress. POD and SOD are both important antioxidant enzymes in plants that maintain the balance of free radicals by eliminating harmful free radicals in plants (Lobo et al. 2010; Sharma et al. 2012). The observed higher SOD and POD activities in ZmHDZ9 transgenic plants compared to the WT plants indicate that the overexpression of ZmHDZ9 could protect cells from oxidative damage induced by drought stress. Soluble proteins are important osmotic regulatory substances and nutrients. The accumulation of soluble proteins in plant cells can improve cells’ ability to retain water and protect cell membranes; thus, they are frequently used as indicators of tolerance ability (Jiang et al. 2020; Zhang et al. 2021). The higher abundance of soluble protein in ZmHDZ9 transgenic plants compared to the WT plants under water deficit conditions indicates that the overexpression of ZmHDZ9 improves drought tolerance in transgenic Arabidopsis plants.
In conclusion, a bioinformatic pipeline was efficiently implemented to identify 42 ZmHDZ drought-responsive genes using previously generated maize transcriptomic datasets. Phylogenetic analysis revealed the first evidence for evolutionary conservation of components of the HD-ZIP TFs in maize. Several regulatory elements associated with drought-inducible genes were identified in the promoter regions of ZmHDZ genes, indicating the implication of these genes in regulating drought tolerance in plants. The expression of several ZmHDZ genes was elevated under drought conditions. Transgenic plants overexpressing ZmHDZ9 exhibited higher activities of SOD and POD and the accumulation of more soluble proteins in the plant cells under drought stress, which resulted in an enhanced developed phenotype and improved resistance. The present survey reveals the first evidence for evolutionary conservation of HD-ZIP transcription factor homologs in maize. In addition, our results provide a comprehensive insight into the roles of ZmHDZ genes in regulating drought–tolerance in maize.
Acknowledgements
Funding was provided by the National Natural Science Foundation of China (No. 31471452).
Declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiao Qiu, GuoRui Wang and Salah Fatouh Abou-Elwafa have contributed equally to this work.
Contributor Information
XiaoKang Guan, Email: guanxk06@henau.edu.cn.
Li Wei, Email: weili-wtc@126.com.
References
- Abou-Elwafa SF, Amein KA. Genetic diversity and potential high temperature tolerance in barley (Hordeum vulgare) World J Agric Res. 2016;4:1–8. [Google Scholar]
- Abou-Elwafa SF, Shehzad T. Genetic diversity, GWAS and prediction for drought and terminal heat stress tolerance in bread wheat (Triticum aestivum L.) Genet Resour Crop Evol. 2021;68:711–728. [Google Scholar]
- Abou-Elwafa SF, Büttner B, Chia T, Schulze-Buxloh G, Hohmann U, Mutasa-Göttgens E, Jung C, Müller AE. Conservation and divergence of autonomous pathway genes in the flowering regulatory network of Beta vulgaris. J Exp Bot. 2010;62:3359–3374. doi: 10.1093/jxb/erq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agalou A, Purwantomo S, Övernäs E, Johannesson H, Zhu X, Estiati A, de Kam RJ, Engström P, Slamet-Loedin IH, Zhu Z, Wang M, Xiong L, Meijer AH, Ouwerkerk PBF. A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol Biol. 2008;66:87–103. doi: 10.1007/s11103-007-9255-7. [DOI] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci. 2007;12:419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Belamkar V, Weeks NT, Bharti AK, Farmer AD, Graham MA, Cannon SB. Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genomics. 2014;15:950. doi: 10.1186/1471-2164-15-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME. Shoot meristem function and leaf polarity: the role of class III HD-ZIP Genes. PLOS Genet. 2006;2:e89. doi: 10.1371/journal.pgen.0020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y-J, Wei Q, Liao Y, Song H-L, Li X, Xiang C-B, Kuai B-K. Ectopic overexpression of AtHDG11 in tall fescue resulted in enhanced tolerance to drought and salt stress. Plant Cell Rep. 2009;28:579–588. doi: 10.1007/s00299-008-0659-x. [DOI] [PubMed] [Google Scholar]
- Cao L, Lu X, Zhang P, Wang G, Wei L, Wang T. Systematic analysis of differentially expressed maize ZmbZIP genes between drought and rewatering transcriptome reveals bZIP family members involved in abiotic stress responses. Int J Mol Sci. 2019;20:4103. doi: 10.3390/ijms20174103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Chen J, Cao F, Li H, Shan S, Tao Z, Lei T, Liu Y, Xiao Z, Zou Y, Huang M, Abou-Elwafa SF. Genotypic variation in the grain photosynthetic contribution to grain filling in rice. J Plant Physiol. 2020;253:153269. doi: 10.1016/j.jplph.2020.153269. [DOI] [PubMed] [Google Scholar]
- Ciarbelli AR, Ciolfi A, Salvucci S, Ruzza V, Possenti M, Carabelli M, Fruscalzo A, Sessa G, Morelli G, Ruberti I. The Arabidopsis homeodomain-leucine zipper II gene family: diversity and redundancy. Plant Mol Biol. 2008;68:465–478. doi: 10.1007/s11103-008-9383-8. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dezar CA, Gago GM, González DH, Chan RL. Hahb-4, a sunflower homeobox-leucine zipper gene, is a developmental regulator and confers drought tolerance to Arabidopsis thaliana plants. Transgenic Res. 2005;14:429–440. doi: 10.1007/s11248-005-5076-0. [DOI] [PubMed] [Google Scholar]
- Dezar CA, Giacomelli JI, Manavella PA, Ré DA, Alves-Ferreira M, Baldwin IT, Bonaventure G, Chan RL. HAHB10, a sunflower HD-Zip II transcription factor, participates in the induction of flowering and in the control of phytohormone-mediated responses to biotic stress. J Exp Bot. 2010;62:1061–1076. doi: 10.1093/jxb/erq339. [DOI] [PubMed] [Google Scholar]
- Du H, Huang M, Zhang Z, Cheng S. Genome-wide analysis of the AP2/ERF gene family in maize waterlogging stress response. Euphytica. 2014;198:115–126. [Google Scholar]
- Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124:509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- Gomez-Cano L, Yang F, Grotewold E. Isolation and efficient maize protoplast transformation. Bio-Protoc. 2019;9:e3346. [Google Scholar]
- Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol. 2008;67:169–181. doi: 10.1007/s11103-008-9309-5. [DOI] [PubMed] [Google Scholar]
- Jiang D, Lu B, Liu L, Duan W, Chen L, Li J, Zhang K, Sun H, Zhang Y, Dong H, Li C, Bai Z. Exogenous melatonin improves salt stress adaptation of cotton seedlings by regulating active oxygen metabolism. PeerJ. 2020;8:e10486–e10486. doi: 10.7717/peerj.10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A, Pati PK, Pati AM, Nagpal AK. In-silico analysis of cis-acting regulatory elements of pathogenesis-related proteins of Arabidopsis thaliana and Oryza sativa. PLoS ONE. 2017;12:e0184523. doi: 10.1371/journal.pone.0184523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner E, Leonhardt N, Eisler H, Parmentier Y, Alioua M, Jacquet H, Leung J, Genschik P. MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev Cell. 2011;21:1116–1128. doi: 10.1016/j.devcel.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Lee Y-H, Chun J-Y. A new homeodomain-leucine zipper gene from Arabidopsis thaliana induced by water stress and abscisic acid treatment. Plant Mol Biol. 1998;37:377–384. doi: 10.1023/a:1006084305012. [DOI] [PubMed] [Google Scholar]
- Li M, Qiao Y, Li Y, Shi Z, Zhang N, Bi C, Guo J. A R2R3-MYB transcription factor gene in common wheat (namely TaMYBsm1) involved in enhancement of drought tolerance in transgenic Arabidopsis. J Plant Res. 2016;129:1097–1107. doi: 10.1007/s10265-016-0857-5. [DOI] [PubMed] [Google Scholar]
- Li Y, Bai B, Wen F, Zhao M, Xia Q, Yang D-H, Wang G. Genome-wide identification and expression analysis of HD-ZIP I gene subfamily in Nicotiana tabacum. Genes (Basel) 2019;10:575. doi: 10.3390/genes10080575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xiong H, Cuo D, Wu X, Duan R. Genome-wide characterization and expression profiling of the relation of the HD-Zip gene family to abiotic stress in barley (Hordeum vulgare L.) Plant Physiol Biochem. 2019;141:250–258. doi: 10.1016/j.plaphy.2019.05.026. [DOI] [PubMed] [Google Scholar]
- Li R, Zhu F, Duan D. Function analysis and stress-mediated cis-element identification in the promoter region of VqMYB15. Plant Signal Behav. 2020;15:1773664. doi: 10.1080/15592324.2020.1773664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods (San Diego, Calif) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lobo V, Patil A, Phatak A, Chandra N. Free radicals, antioxidants and functional foods: impact on human health. Pharmacogn Rev. 2010;4:118–126. doi: 10.4103/0973-7847.70902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella PA, Arce AL, Dezar CA, Bitton F, Renou J-P, Crespi M, Chan RL. Cross-talk between ethylene and drought signalling pathways is mediated by the sunflower Hahb-4 transcription factor. Plant J. 2006;48:125–137. doi: 10.1111/j.1365-313X.2006.02865.x. [DOI] [PubMed] [Google Scholar]
- Meijer AH, de Kam RJ, d'Erfurth I, Shen W, Hoge JHC. HD-Zip proteins of families I and II from rice: interactions and functional properties. Mol Gen Genet MGG. 2000;263:12–21. doi: 10.1007/pl00008671. [DOI] [PubMed] [Google Scholar]
- Montefiori M, McGhie TK, Hallett IC, Costa G. Changes in pigments and plastid ultrastructure during ripening of green-fleshed and yellow-fleshed kiwifruit. Sci Hortic. 2009;119:377–387. [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehlivan N. Stochasticity in transcriptional expression of a negative regulator of Arabidopsis ABA network. 3 Biotech. 2019;9:15. doi: 10.1007/s13205-018-1542-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabara RC, Tripathi P, Rushton PJ. The potential of transcription factor-based genetic engineering in improving crop tolerance to drought. OMICS. 2014;18:601–614. doi: 10.1089/omi.2013.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ré DA, Capella M, Bonaventure G, Chan RL. Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biol. 2014;14:150. doi: 10.1186/1471-2229-14-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhwal MK, Swami AK, Sharma V, Sarin R. Identification of drought-induced transcription factors in Sorghum bicolor using GO term semantic similarity. Cell Mol Biol Lett. 2015;20:1–23. doi: 10.2478/s11658-014-0223-3. [DOI] [PubMed] [Google Scholar]
- Sharma P, Jha AB, Dubey RS, Pessarakli M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J Botany. 2012;2012:217037. [Google Scholar]
- Su H, Liang J, Abou-Elwafa SF, Cheng H, Dou D, Ren Z, Xie J, Chen Z, Gao F, Ku L, Chen Y. ZmCCT regulates photoperiod-dependent flowering and response to stresses in maize. BMC Plant Biol. 2021;21:453. doi: 10.1186/s12870-021-03231-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Li M, Yang Y, Sun X, Wang N, Liang B, Ma F. Overexpression of MpCYS4, a phytocystatin gene from Malus prunifolia (Willd.) Borkh., enhances stomatal closure to confer drought tolerance in transgenic Arabidopsis and apple. Front Plant Sci. 2017;8:33. doi: 10.3389/fpls.2017.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés AE, Övernäs E, Johansson H, Rada-Iglesias A, Engström P. The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol Biol. 2012;80:405–418. doi: 10.1007/s11103-012-9956-4. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zha K, Chai W, Wang Y, Liu B, Jiang H, Cheng B, Zhao Y. Functional analysis of the HD-Zip I gene ZmHDZ1 in ABA-mediated salt tolerance in rice. J Plant Biol. 2017;60:207–214. [Google Scholar]
- Wang G, Yuan Z, Zhang P, Liu Z, Wang T, Wei L. Genome-wide analysis of NAC transcription factor family in maize under drought stress and rewatering. Physiol Mol Biol Plants. 2020;26:705–717. doi: 10.1007/s12298-020-00770-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Zhou W, Gong X, Cheng B. Expression of ZmHDZ4, a maize homeodomain-leucine zipper I gene, confers tolerance to drought stress in transgenic rice. Plant Mol Biol Report. 2016;34:845–853. [Google Scholar]
- Yang Y, Li R, Qi M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J Cell Mol Biol. 2000;22:543–551. doi: 10.1046/j.1365-313x.2000.00760.x. [DOI] [PubMed] [Google Scholar]
- Yu L, Chen X, Wang Z, Wang S, Wang Y, Zhu Q, Li S, Xiang C. Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 2013;162:1378–1391. doi: 10.1104/pp.113.217596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue H, Shu D, Wang M, Xing G, Zhan H, Du X, Song W, Nie X. Genome-wide identification and expression analysis of the HD-Zip gene family in wheat (Triticum aestivum L.) Genes (basel) 2018;9:70. doi: 10.3390/genes9020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL. The guidance of plant physiological experiment. Beijing: Higher Education Press; 1992. [Google Scholar]
- Zhang L, Zhao G, Xia C, Jia J, Liu X, Kong X. A wheat R2R3-MYB gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis. J Exp Bot. 2012;63:5873–5885. doi: 10.1093/jxb/ers237. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Chen J, Liang C, Liu F, Hou X, Zou X. Genome-wide identification and characterization of the bHLH transcription factor family in pepper (Capsicum annuum L.) Front Genet. 2020;11:1156. doi: 10.3389/fgene.2020.570156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tariq A, Zeng F, Chai X, Graciano C. Involvement of soluble proteins in growth and metabolic adjustments of drought-stressed Calligonum mongolicum seedlings under nitrogen addition. Plant Biol. 2021;23:32–43. doi: 10.1111/plb.13190. [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhang X, Guo R, Wang Y, Guo C, Li Z, Chen Z, Gao H, Wang X. Over-expression of a grape WRKY transcription factor gene, VlWRKY48, in Arabidopsis thaliana increases disease resistance and drought stress tolerance. Plant Cell Tissue Organ Cult (PCTOC) 2018;132:359–370. [Google Scholar]
- Zhu DX, Peng KP, Zhang MK, Jia Y, Wang JJ. Cloning and salt resistance function identification of GmHDL57 gene from Glycine max. Acta Agron Sin. 2018;44:1347–1356. [Google Scholar]