Abstract
Foxtail millet (Setaria italica) is rich in nutrients and extremely beneficial to human health. We identified and comprehensively analyzed 89 MADS-box genes in the foxtail millet genome. According to the classification of MADS-box genes in Arabidopsis thaliana and rice, the SiMADS-box genes were divided into M-type (37) and MIKC-type (52). During evolution, the differentiation of MIKC-type MADS-box genes occurred before that of monocotyledons and dicotyledons. The SiMADS-box gene structure has undergone much differentiation, and the number of introns in the MIKC-type subfamily is much greater than that in the M-type subfamily. Analysis of gene duplication events revealed that MIKC-type MADS-box gene segmental duplication accounted for the vast majority of gene duplication events, and MIKC-type MADS-box genes played a major role in the amplification of SiMADS-box genes. Collinearity analysis showed highest collinearity between foxtail millet and maize MADS-box genes. Analysis of tissue-specific expression showed that SiMADS-box genes are highly expressed throughout the grain-filling process. Expression analysis of SiMADS-box genes under eight different abiotic stresses revealed many stress-tolerant genes, with induced expression of SiMADS33 and SiMADS78 under various stresses warranting further attention. Further, some SiMADS-box proteins may interact under external stress. This study provides insights for MADS-box gene mining and molecular breeding of foxtail millet in the future.
Subject terms: Evolution, Genetics, Plant sciences
Introduction
The MADS-box genes are divided into two categories: ARG80/SRF genes in animals and fungi, also known as M-type genes in plants; and MEF2 genes in animals and yeast, known as MIKC-type genes in plants1. The MADS genes of M-type are divided into three types: Mα, Mβ, and Mγ. And MIKC-type MADS genes are divided into two major categories, namely MIKC* and MIKCC. MIKCC is further divided into 14 subfamily in rice: GLO-like, DEF-like, GGM13-like, OsMADS32-like, AGL12-like, STMADS11-like, AG-like, AGL17-like, AGL15-like, FLC-like, TM3-like, AGL6-like, AGL2-like and SQUA-like1. Serving as transcription factors, MADS-box genes are widely distributed in eukaryotes (plants, animals and fungi)2. Evolutionarily speaking, replication of the original MADS-box gene occurred before the differentiation of animals and plants3, indicating that the MADS-box gene family is relatively ancient. There are many reports on the functions of MADS-box genes, which regulate many growth processes, such as flower organ development4, embryo development5, fruit development6,7 and vegetative organ development8. For example, the discovery of the ABC genetic model explained how the combined functions of three types of genes (A, B and C) determine the characteristics of four floral organs4. The activity of organ-recognition genes B and C was found to be closely related to three MADS-box genes (SEP1–3)9. In recent years, MADS-box genes have also been thoroughly studied in other plants. Several key MADS-box genes of moso bamboo (Phyllostachys edulis) are involved in inflorescence development, and ectopic overexpression of PeMADS5 in Arabidopsis thaliana leads to early flowering and abnormal flower phenotypes10. The MADS-box gene of pineapple is closely related to flower density, and some MADS-box genes are involved in CAM photosynthesis and are regulated by a biological clock11. Similarly, many MADS-box genes involved in floral organ and fruit development have also been found in tomato12. MADS-box genes even play an important role in tuber plant dormancy13. Thus, MADS-box genes have many different roles.
In around 6000 BC in northern China, foxtail millet (Setaria italica) was domesticated from Setaria viridis14 and, together with Panicum miliaceum, became the main food crop at that time15,16. Foxtail millet is rich in nutrients such as calcium, dietary fiber, polyphenols, fat and protein17,18, which promote human health. Moreover, foxtail millet is a drought-resistant plant, which has a certain reference to the stress adaptation of crops and serves as a model C4 plant for studies. MADS-box genes have scarcely been studied in C4 plants. However, whole-genome sequencing of a foxtail millet species16,19 enabled us to conduct an in-depth analysis of the MADS-box gene family in this plant.
Analyses of gene or protein families in foxtail millet include WRKY20, ZIP21, TPS22, SSPs23, and WD4024, but studies of the MADS-box gene family in this plant are incomplete. Moreover, how SiMADS-box genes participate in the growth and development of foxtail millet and in its response to stress is not clear. Therefore, in-depth research on the MADS-box gene family in millet is still needed, as it will provide great assistance for gene mining and molecular breeding. We identified 89 MADS-box genes from the whole genome of foxtail millet, and they were classified into subfamilies according to the MADS-box gene family classification in A. thaliana25 and rice1: 37 genes belonged to the M-type and 52 to the MIKC-type. We analyzed their gene structure, exon/intron distribution, conserved motifs, molecular weight (Mw), isoelectric point (pI) and subcellular localization, and gene duplication events were revealed. Homology between SiMADS-box genes and MADS-box genes of other species was assessed and a phylogenetic tree was constructed. Finally, the tissue-specific expression of 12 SiMADS-box genes and their responses to different abiotic stresses are discussed.
Results
Identification of MADS-box genes in foxtail millet
Two BLAST methods were used to identify 89 MADS-box genes in foxtail millet. The genes were named SiMADS1–SiMADS89 according to their chromosomal positions. Their genetic characteristics, including coding sequence (CDS) length, protein molecular weight, pI and subcellular localization, were determined. Among the 89 SiMADS-box proteins, SiMADS7 and SiMADS74 were the smallest with only 60 amino acids, and the largest one (SiMADS21) had 483 amino acids. The molecular weight of the proteins ranged from 6.78 to 53.18 kDa, and pI ranged from 4.41 (SiMADS21) to 11.39 (SiMADS35). Predicted subcellular localization revealed 1 in the endoplasmic reticulum, 11 in the mitochondria, 19 in the chloroplast, 14 in the cytoplasm, and 44 in the nucleus (Table S1).
Phylogenetic analysis and classification of the SiMADS-box genes
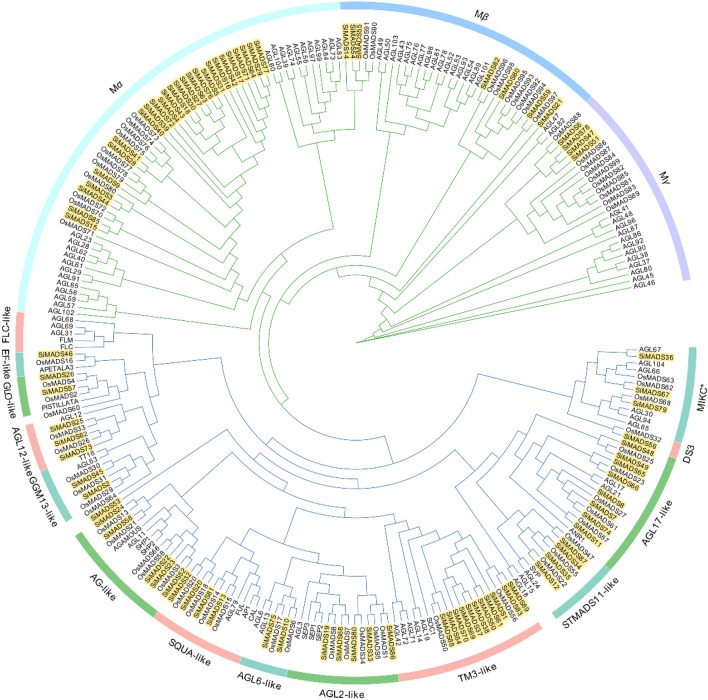
The phylogenetic relationships of the 89 SiMADS-box proteins was studied by multiple sequence alignment. MADS-box gene classification of A. thaliana and rice was referenced. The foxtail millet MADS-box phylogenetic tree classified the SiMADS-box genes into two subgroups: 37 M-type genes and 52 MIKC-type genes. The SiMADS-box genes were similar to those of the monocotyledon rice, and could be further divided into 17 groups: 14 in the MIKC-type gene subfamily, and 3 in the M-type gene subfamily (Fig. 1). Interestingly, the FLC-like subfamily did not contain SiMADS-box genes, in contrast to both Arabidopsis25 and rice1. We also investigated the number of MADS-box genes in maize26, sorghum26, rice1, Arabidopsis25 and Brassica rapa27. The numbers and proportions of MADS-box genes in each subfamily are shown in Table 1. C4 plants (foxtail millet, maize, sorghum) had similar proportions of MIKC-type MADS-box genes as C3 plants. However, there were significant differences between the M-type genes in C4 vs. C3 plants. The proportion of Mα subfamily members in C4 plants was much higher than in C3 plants. On the other hand, the proportion of Mβ and Mγ subfamily members in C4 plants was much lower than in C3 plants. These results suggest that after the divergence of C3 and C4 plants, perhaps more members of the Mα subfamily were retained by the C4 plants.
Figure 1.
Unrooted phylogenetic tree showing relationships among MADS-box proteins of Setaria italica, rice and Arabidopsis thaliana. The phylogenetic tree was derived using the ML method in MEGA X. The tree shows the 17 phylogenetic subfamilies. MADS-box proteins from rice are marked with the prefix ‘Os’.
Table 1.
The number and proportion of each type of MADS-box genes in foxtail millet, maize, sorghum, rice, Arabidopsis and Brassica rapa.
| Total | MIKCC | MIKC* | Mα | Mβ | Mγ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | Percentage | Number | Percentage | Number | Percentage | Number | Percentage | Number | Percentage | ||
| S. italica | 89 | 49 | 55.06 | 3 | 3.37 | 26 | 29.21 | 7 | 7.87 | 4 | 4.49 |
| Maize | 75 | 39 | 52.00 | 4 | 5.33 | 27 | 36.00 | 3 | 4.00 | 2 | 2.67 |
| Sorghum | 65 | 33 | 50.77 | 2 | 3.08 | 26 | 40.00 | 2 | 3.08 | 2 | 3.08 |
| Rice | 75 | 39 | 52.00 | 4 | 5.33 | 13 | 17.33 | 9 | 12.00 | 10 | 13.33 |
| Arabidopsis | 108 | 39 | 36.11 | 8 | 7.41 | 25 | 23.15 | 20 | 18.52 | 16 | 14.81 |
| B. rapa | 167 | 89 | 53.29 | 16 | 9.58 | 11 | 6.59 | 29 | 17.37 | 22 | 13.17 |
Therefore, members of the Mα subfamily may have played a major role in the evolution of C4 plants.
Gene structure, motif composition, and protein-interaction predictions for the SiMADS-box gene family
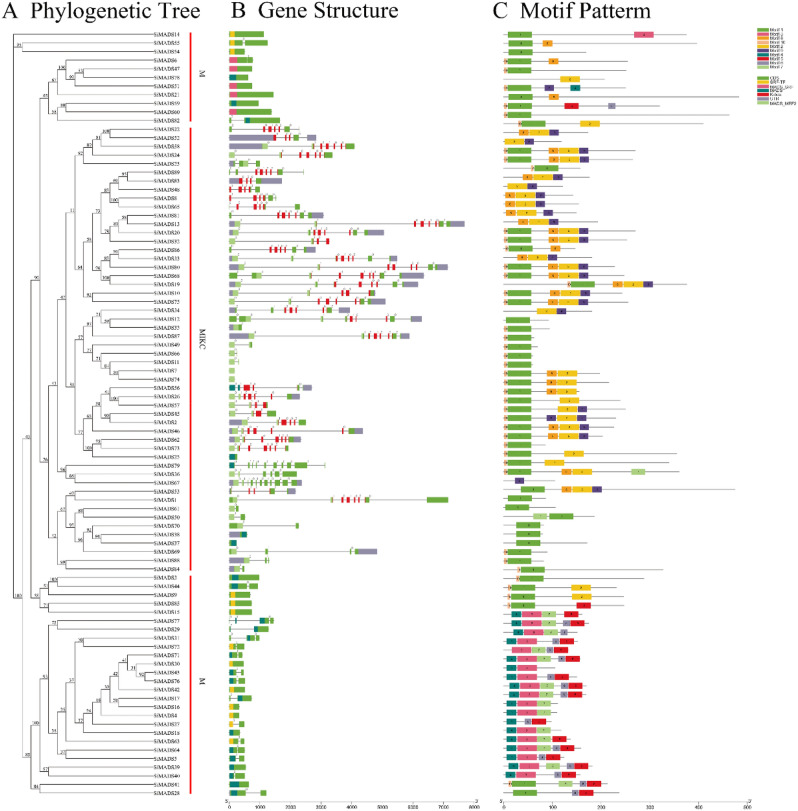
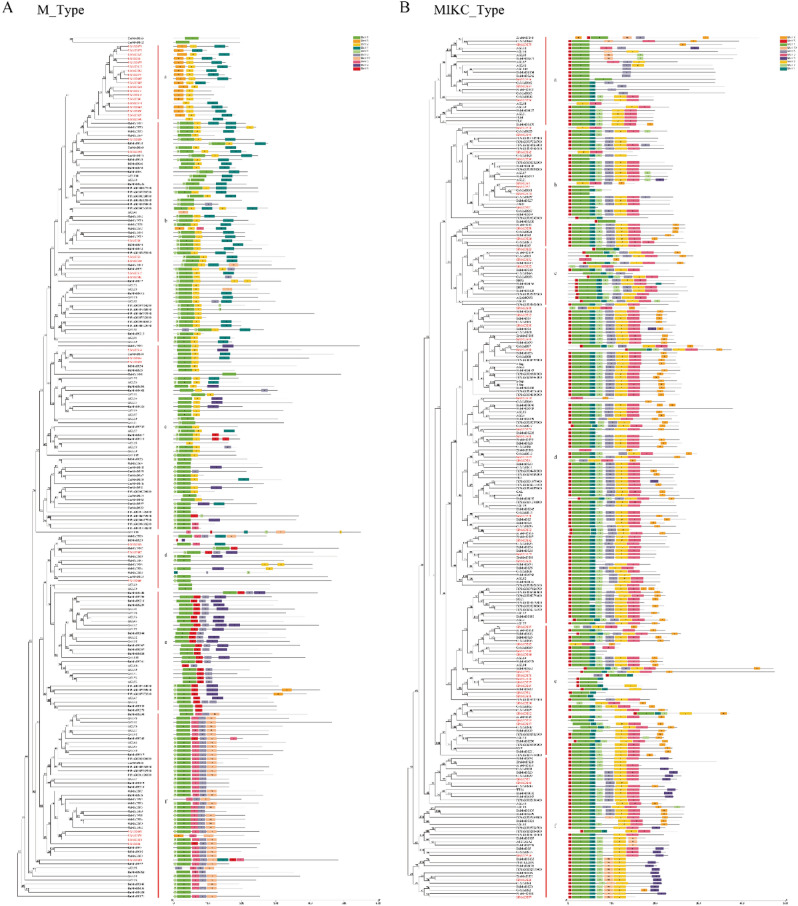
Introns are ubiquitous in eukaryotes, which is an important feature different from prokaryotes. In higher organisms, introns have been reported to regulate gene expression at multiple levels. The main function of introns is to generate different exon combinations through differential splicing to translate different proteins, which improves the complexity of the proteome28,29. There are two arguments about the origin of introns, namely the hypotheses of early introns and late introns, which are still inconclusive at present. However, more researches tend to the early intron hypothesis, that is, there are a large number of introns in relatively old ancestors, which means that the loss of a large number of introns may be common in eukaryotic evolution, and the acquisition of a large number of introns may be rare30–33. The latest research also shows that introns themselves have important functions independent of their coding genes. Introns can mediate the cell's response to starvation34, and it can also regulate the growth rate under stress conditions and improve the adaptability of yeast35. Therefore, introns are very important to organisms, so we also analyzed the differences in introns of two types of foxtail millet MADS genes. Exon and intron structures of SiMADS-box genes were obtained by comparing their CDSs with the corresponding genomic DNA sequences. In general, the exon/intron structures of different members of the same subfamily should be similar. As shown in Fig. 2, the introns of SiMADS-box genes ranged from 0 to 10. The mean number of introns in the MIKC-type subfamily was about 4.3, and that in the M-type subfamily was about 0.6, suggests that M-type gene has undergone great differentiation. In the M-type subfamily, the lowest average number of intron, 0.25, was found for the Mγ branch. Among the 37 M-type genes, 19 (51.35%) had no intron structure. This may be further evidence for significantly different rates of evolution for the M-type branches. However, the average number of introns for the MIKC* branch of the MIKC-type subfamily was largest, as high as 8. The foxtail millet MADS-box genes were found to contain mainly the MADS, MADS-SRF, MADS-MEF2 and K-box domains.
Figure 2.
Phylogenetic relationships, gene structure and architecture of the conserved protein motifs in 89 genes from S. italica. (A) The phylogenetic tree was constructed based on the full-length sequences of S. italica MADS-box proteins. (B) Exon–intron structure of S. italica MADS-box genes. Lines represent introns, boxes represent exons, and domains are color-coded. Number indicates the phase of the corresponding intron. (C) Amino acid motifs in the SiMADS-box proteins (1–10) are represented by colored boxes. Black lines indicate relative protein lengths. Sequence information for each motif is provided in Table S2.
The online MEME program was used to analyze the motifs of the 89 SiMADS-box proteins, and a structural diagram of these proteins was constructed. We identified 10 conserved motifs (Table S2). The motif composition of SiMADS-box proteins in the same subfamily was similar, and differed in different subfamilies. As can be seen from Fig. 2, the motif order for the MIKC-type subfamily was mainly 10–1–8–2–9, and these motifs could be conserved. The conserved motif order for the Mα subfamily was suggested to be 4–3–7–6–5, where the differences in the conserved motif patterns might be related to the proteins' specific functions, but this requires further elucidation.
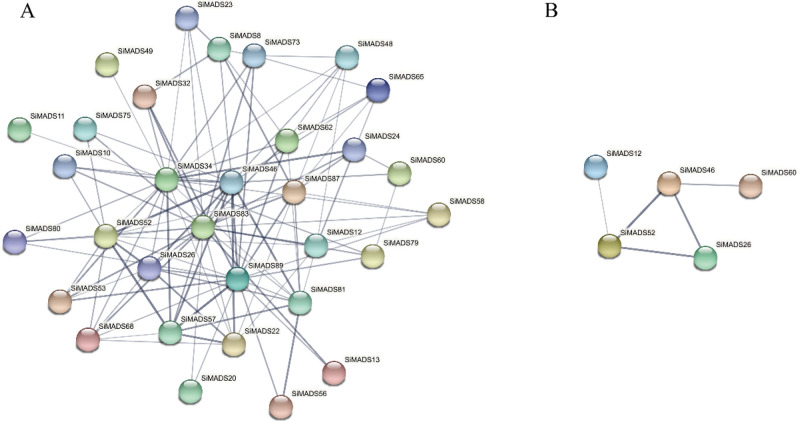
These results showed that motif sequences patterns in a same subfamily are basically similar, which may also indicate that these proteins have similar functions or participate together in some pathways. We therefore turned to STRING36 to predict interactions among the 89 SiMADS-box proteins. The results are shown in Fig. 3. Among the 89 SiMAD-box proteins, 32 were predicted to interact with each other. Among these putatively interacting proteins, 31 were MIKC-type, and only 1 (SiMADS60) was M-type. The higher number of interactions predicted for the MIKC-type MADS-box proteins might be related to their function.
Figure 3.
Predicted interactions between foxtail millet MADS-box proteins. (A) Prediction of the protein–protein interaction network among 89 SiMADS. (B) Prediction of the protein–protein interaction network among 5 SiMADS in stress treatments.
Chromosomal distribution and synteny analysis of the SiMADS-box genes
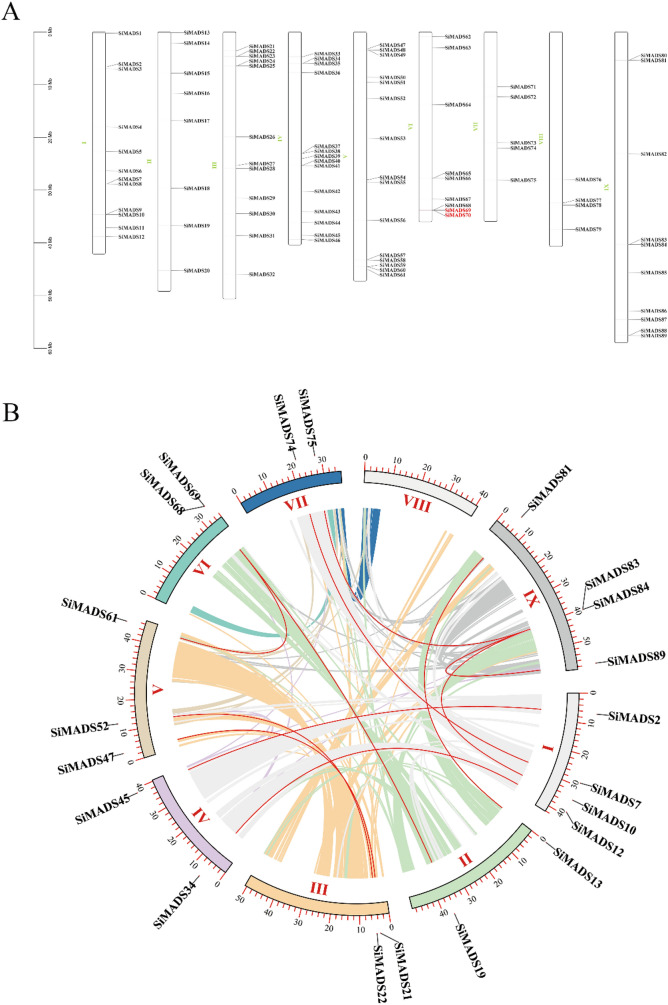
The chromosomal positions of the SiMADS-box genes are shown in Fig. 4A. Chromosome VIII had the least number of MADS-box genes (4), and chromosome V, the most (15). Referring to the closely related genes in A. thaliana falling into each other in 200 kB as tandem duplicates37, only one pair of tandem duplicates (SiMADS69 and SiMADS70) were found among the foxtail millet MADS-box genes (Fig. 4A). The number of linked genes in linkage group (LG) III/V was higher than that in other linkage groups, whereas the distribution of SiMADS-box genes was largest in LG IX (5). There were 11 pairs of segment duplicates among SiMADS-box genes (Fig. 4B, Table S3), many more than the number of tandem duplicates. Meanwhile, only one of the 11 pairs of segment duplicates belonged to M-type MADS-box gene, while the rest belonged to MIKC MADS-box gene (Table S3, Table 1). This may further explain why this is the largest branch of the foxtail millet MADS-box gene family, with the highest number of genes (49) and the highest proportion (55.06%) in each subfamily. Moreover, the proportion of this subfamily was higher than that in other species, even sorghum and corn, which are also C4 plants (Table 1). At the same time, the analysis of MADS-box gene structure also found that the MIKCC branch, which has a large number of introns. These results suggest that some SiMADS-box genes may have been generated by gene-replication events, which may have been the main driving force for SiMADS-box gene evolution.
Figure 4.
The chromosomal distribution and synteny blocks of the S. italica MADS-box genes. (A). Schematic representation of the chromosomal distribution of the S. italica MADS-box genes. Vertical bars represent the chromosomes of S. italica. The chromosome number is indicated to the left of each chromosome. The scale on the left represents chromosome length. (B). Schematic representation of the chromosomal distribution and interchromosomal relationships of S. italica MADS-box genes. Colored lines indicate all synteny blocks in the S. italica genome and red lines indicate duplicated MADS-box gene pairs. Chromosome number is indicated at the bottom of each chromosome.
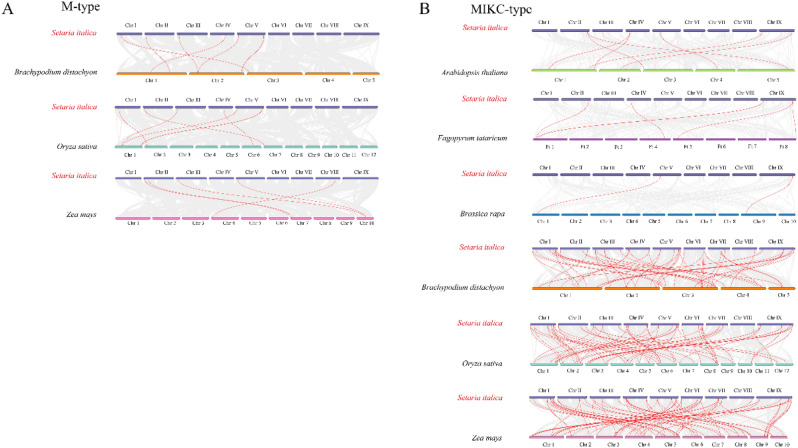
Collinearity analysis was conducted between M-type SiMADS-box genes and other plants (A. thaliana, Brassica rapa, Fagopyrum tataricum, Brachypodium distachyon, rice, and maize) (Fig. 5). The three dicotyledonous plants showed no collinearity with M-type SiMADS-box genes. There were only a few collinear genes between the M-type genes in foxtail millet, and those in B. distachyon, rice, and maize (Fig. 5, Table S4). Analysis of the collinearity map between MIKC-type SiMADS-box genes and those of other plants revealed highest collinearity with maize (71), followed by rice (62), B. distachyon (53), A. thaliana (7), F. tataricum (6) and B. rapa (2). Further analysis of these collinear genes revealed that seven MIKC-type SiMADS-box genes (SiMADS20, SiMADS36, SiMADS52, SiMADS80, SiMADS81, SiMADS86, and SiMADS87) exist in both monocotyledons and dicotyledons. Among these seven genes, only two (SiMADS52 and SiMADS81) were included in the genes with segmental duplications. In addition, two MIKC-type genes (SiMADS35, SiMADS66) and two M-type genes (SiMADS15, SiMADS78) showed collinearity only with C4 plants (maize). In general, foxtail millet exhibited the highest collinearity with maize, suggesting that these C4 plants may have a close genetic relationship. In addition, for both M-type and MIKC-type SiMADS-box genes, the number of genes with collinearity to the monocots was much higher than that with collinearity to the dicots.
Figure 5.
Synteny analysis of MADS-box genes between S. italica and other plant species. (A) Synteny analysis of the M-type MADS-box genes between S. italica and three representative plant species (B. distachyon, Oryza sativa, Zea mays). (B) Synteny analysis of the MIKC-type MADS-box genes between S. italica and six representative plant species (A. thaliana, F. tataricum, B. rapa, B. distachyon, O. sativa, Z. mays). Gray lines in the background indicate the collinear blocks within S. italica and other plant genomes, while red lines highlight the syntenic MADS gene pairs.
Evolutionary analysis of MADS-box proteins from foxtail millet and several other species
We investigated the evolutionary relationship of M-type MADS-box proteins in the monocotyledons rice (31 genes), B. distachyon (17 genes) and maize (17 genes), and the dicotyledons A. thaliana (55 genes), F. tataricum (26 genes) and Brassica napus (30 genes) (Fig. 6A, Table S5). According to the phylogenetic tree, the M-type MADS-box proteins could be divided into six subfamilies, labeled a–f. The motifs of the M-type MADS-box proteins were analyzed by online MEME analysis. Motifs 7, 1 and 2 were conserved and distributed almost alternately in the whole subfamily. However, there were large differences among subfamilies. Subfamily a had the largest distribution of SiMADS-box members (19); their conserved motif order was 8–2–4, but they lacked motif 7–1. The main order of the b and c subfamily motifs was 7–1–2–4. The motifs of subfamily d were mainly 7–1, and subfamily e did not contain any SiMADS-box members. The main order of the motif of subfamily f was 7–1–3–6–10.
Figure 6.
Phylogenetic relationship and motif composition of MADS-box proteins from S. italica with six different plant species (A. thaliana, F. tataricum, B. napus, B. distachyon, O. sativa, Z. mays). (A) Evolutionary relationship between M-type MADS-box proteins and motif composition. (B) Evolutionary relationship between MIKC-type MADS-box proteins and motif composition. Different-colored boxes represent different motifs and their positions in each MADS-box protein sequence. Sequence information for each motif is provided in Table S2.
To explore the evolutionary relationship between MIKC-type MADS-box proteins of foxtail millet and the monocotyledonous plants rice (38 genes), B. distachyon (29 genes) and maize (16 genes), and dicotyledonous plants A. thaliana (43 genes), (38 genes), and B. napus (33 genes), we constructed a phylogenetic tree (Fig. 6B). There were six subfamilies (a–f) in the MIKC-type MADS-box family. Compared to the M-type MADS-box proteins, the motif distribution of the MIKC subfamily was more conservative. Its motif appeared most frequently in the order 5–1–4–7–6–2–3, and it was distributed in almost all subfamilies. However, there were also differences in some subfamilies, in particular subfamily f where motif 6 seemed to be replaced by 10. In addition, there was a new motif 9, which may confer unique physiological functions on this subfamily.
Expression patterns of the SiMADS-box genes in different foxtail millet tissues
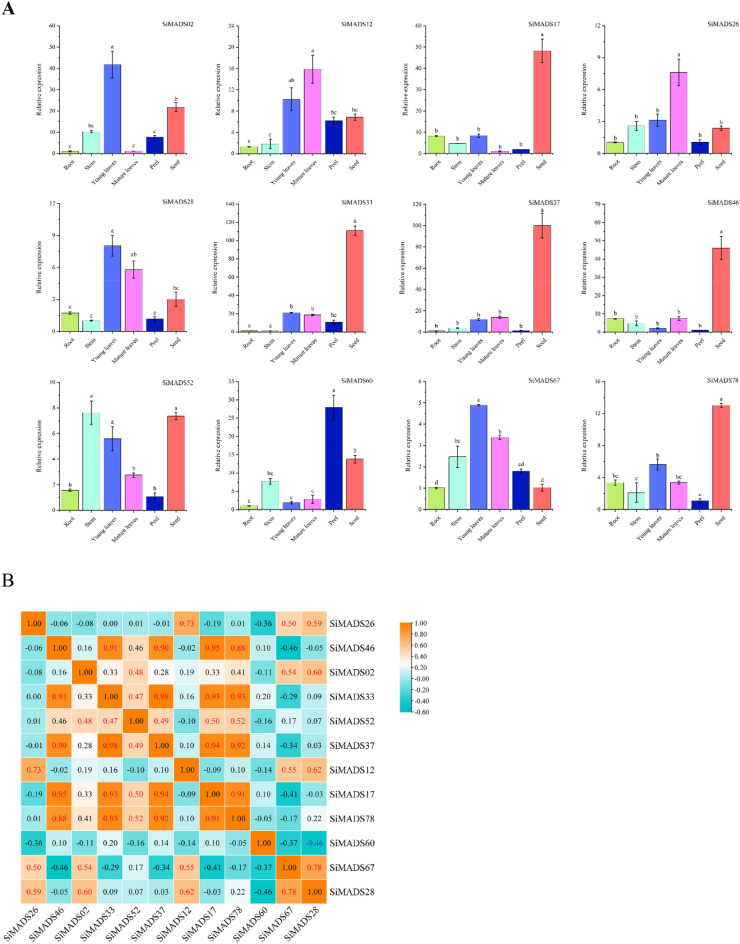
To study the physiological function of the SiMADS-box genes, the spatiotemporal expression of some members of the gene family was detected by qRT-PCR. Accumulation of eight MIKC-type and four M-type SiMADS-box gene transcripts in roots, stems, leaves (young leaves/mature leaves), peel and fruit in the middle stage of grain-filling was detected. Each of these genes belonged to a different subfamily. Since the function of MADS-box genes in floral organs is relatively clear, this study focused on the role of MADS-box genes in other tissues. These genes were expressed in all organs, but some were predominantly expressed in only a few tissues (Fig. 7A). Among them, SiMADS17 and SiMADS46 were highly expressed in roots, and SiMADS02, SiMADS52 and SiMADS60 were highly expressed in stems. In addition to SiMADS46 and SiMADS60, other genes were highly expressed in young leaves. Six genes (SiMADS12, SiMADS26, SiMADS28, SiMADS33, SiMADS37, SiMADS46) were highly expressed in mature leaves. Four genes (SiMADS02, SiMADS12, SiMADS33, SiMADS60) were highly expressed in the peel. All genes except SiMADS67 were highly expressed in the fruit, and in particular, the relative expression values of MIKC-type genes SiMADS33 and SiMADS37 were extremely high. These results, exhibiting differential expression patterns of SiMADS-box genes in different tissues of foxtail millet, indicated that the SiMADS-box genes have multiple functions in foxtail millet growth and development.
Figure 7.
Tissue-specific expression of 12 S. italica MADS-box genes and their correlation with expression patterns in the middle stage of grain filling. (A) Expression patterns of 12 S. italica MADS-box genes in the root, stem, young leaves, mature leaves, peel and seed organs were examined by qRT-PCR. Error bars were obtained from three measurements. Lowercase letter above the bar indicates significant difference (α = 0.05, LSD) among treatments. (B) Positive number: positively correlated; negative number: negatively correlated. Red numbers indicate a significant correlation at the 0.05 level.
We also examined the correlation between SiMADS-box gene expression patterns in foxtail millet root, stem, leaf, peel and fruit; most of the genes were positively correlated (Fig. 7B). Expression of the M-type gene SiMADS28 and MIKC-type gene SiMADS67 was significantly positively correlated, and their expression was also significantly positively correlated with MIKC-type genes SiMADS2, SiMADS12 and SiMADS26. These results indicated similar functions for M-type and MIKC-type SiMADS-box genes during plant growth and development. For example, expression of the M-type genes SiMADS17 and SiMADS78 showed a significant positive correlation, and their expression was also significantly positively correlated with MIKC-type genes SiMADS33, SiMADS37, SiMADS46 and SiMADS52, which were strongly expressed in fruit.
Expression patterns of MADS-box genes in foxtail millet during fruit development
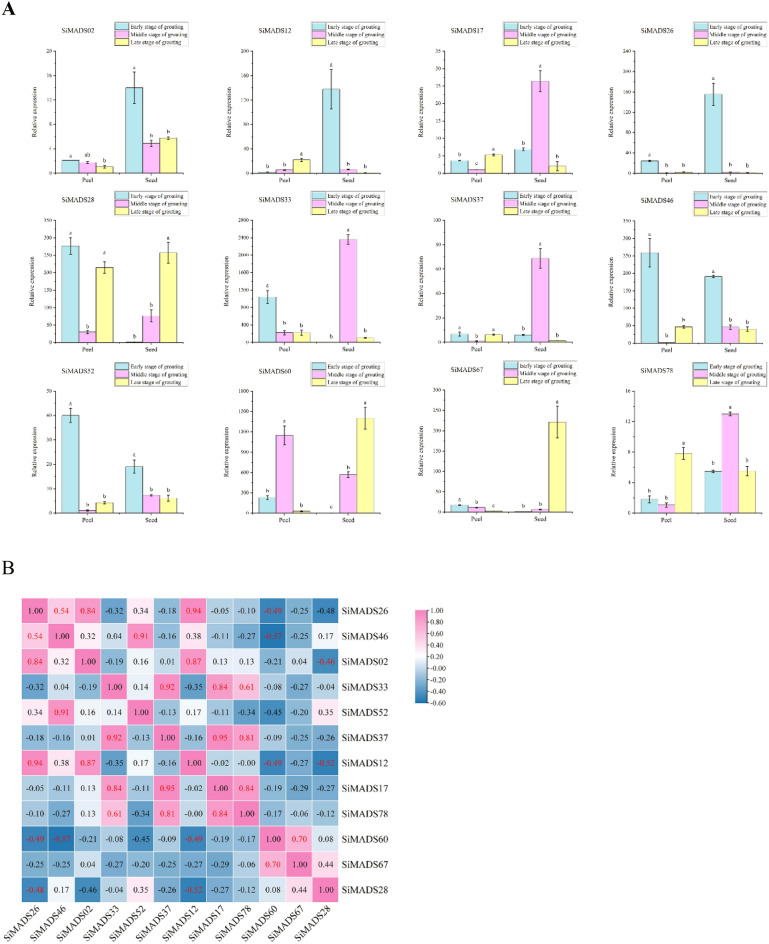
Foxtail millet fruit are rich in calcium, dietary fiber, polyphenols, fats, proteins and other nutrients17,18. Tissue-specific expression of SiMADS-box genes was also found to be high in fruit. Therefore, we determined the expression level of SiMADS-box genes in peel and fruit before, during and after grain-filling (Fig. 8A). Two genes (SiMADS02 and SiMADS17) showed very low expression in the peel. Five genes (SiMADS28, SiMADS33, SiMADS46, SiMADS52, SiMADS60) were highly expressed in the peel, mainly at the early filling stage. M-type SiMADS60 also exhibited high expression in the middle filling stage, and M-type SiMADS28 in the late filling stage.
Figure 8.
Expression pattern and correlation of 12 S. italica MADS-box genes during fruit development. (A) qRT-PCR was used to detect the expression patterns of 12 S. italica MADS-box genes in peel and fruit before, during and after grain filling. Error bars were obtained from three measurements. Lowercase letter above the bar indicates significant difference (α = 0.05, LSD) among treatments. (B) Positive number: positively correlated; negative number: negatively correlated. Red numbers indicate a significant correlation at the 0.05 level.
Compared to the peel, all genes were highly expressed in the fruit, eight genes in particular (SiMADS12, SiMADS26, SiMADS28, SiMADS33, SiMADS37, SiMADS46, SiMADS60, SiMADS67). Among these genes, there were differences in expression for those belonging to the MIKC-type and M-type. MIKC-type genes were mainly highly expressed in the early filling stage (SiMADS12, SiMADS26, SiMADS46) and middle filling stage (SiMADS33, SiMADS37), whereas M-type genes SiMADS28 and SiMADS60 were mainly highly expressed in the late filling stage.
We also studied the correlation between SiMADS-box gene-expression patterns in peel and fruit at different filling stages and found both positive and negative correlations (Fig. 8B). For example, the MIKC-type SiMADS12 and SiMADS26, which were highly expressed in the early stage of fruit-filling, were significantly negatively correlated with the M-type SiMADS28 and SiMADS60, which were highly expressed in the late stage of grain-filling. This also revealed that there were differences in expression of SiMADS-box genes of the MIKC-type and M-type. There were also some similarities, for example, a significant positive correlation between M-type SiMADS17 and SiMADS78 expression, which were also significantly positively correlated with expression of the MIKC-type genes SiMADS33 and SiMADS37, which were highly expressed in fruit at the middle stage of filling.
Expression patterns of SiMADS-box genes in response to different abiotic stresses
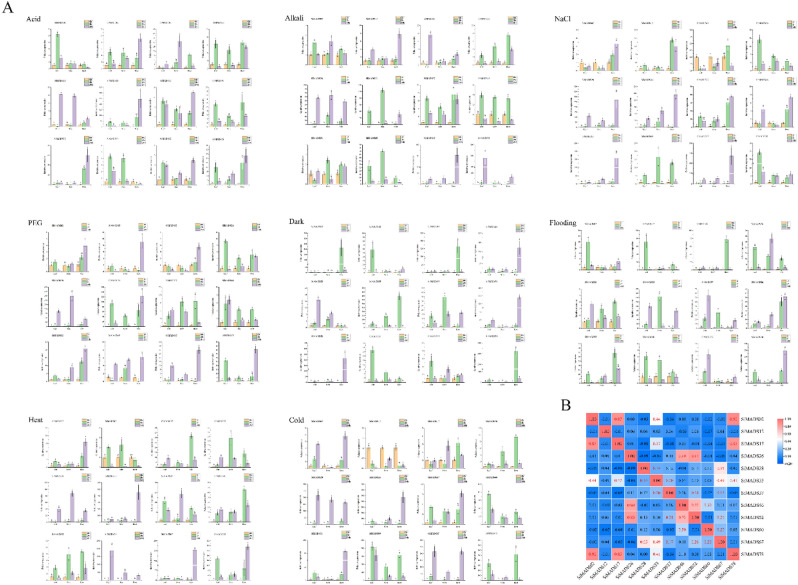
To determine whether the expression of SiMADS-box genes is affected by different abiotic stresses, we analyzed the expression of 12 SiMADS-box genes under eight kinds of abiotic stress: acid, alkali, NaCl, polyethylene glycol (PEG), flooding, dark, heat and cold. We used qRT-PCR to analyze the expression patterns of these 12 genes in leaves, stems and roots under the different treatments. Some SiMADS-box genes were significantly induced, whereas others were suppressed (Fig. 9A). For example, SiMADS33 and SiMADS78 were significantly induced under several abiotic stresses (acid, alkali, dark, heat and cold treatments). In addition, some genes showed different patterns under different treatments, such as SiMADS28, the expression of which was significantly upregulated under alkali, salt and PEG treatments, but changed little under flooding. SiMADS60 expression was significantly upregulated in both heat and cold treatments, but its expression also did not change significantly under flooding. SiMADS33 and SiMADS67 tended to be highly expressed in roots, SiMADS78 tended to be expressed in roots and leaves, and expression of SiMADS28, SiMADS37 and SiMADS60 was significantly upregulated in roots, stems and leaves. In general, the expression levels of genes that tended to be expressed in roots peaked after 24 h of stress, whereas the expression levels of those that tended to be expressed in stems and leaves could be detected after 2 h of stress. A correlation between SiMADS-box gene-expression patterns under stress was also observed (Fig. 9B). Most SiMADS-box genes were positively correlated. For example, there was a significant positive correlation between the expressions of SiMADS33 and SiMADS28, SiMADS67 and SiMADS78, and between SiMADS02 and SiMADS17 (P < 0.05).
Figure 9.
Gene expression of 12 S. italica MADS-box genes in plants subjected to abiotic stresses (acid, alkali, NaCl, PEG, dark, flooding, heat, cold) at the seedling stage. (A) qRT-PCR was used to detect the expression patterns of 12 S. italica MADS-box genes in roots, stems and leaves at different times. Error bars were obtained from three measurements. Lowercase letter above the bar indicates significant difference (α = 0.05, LSD) among treatments. (B) Positive number: positively correlated; negative number: negatively correlated. Red numbers indicate a significant correlation at the 0.05 level.
Discussion
Characteristics of SiMADS-box genes
We analyzed 89 SiMADS-box proteins with molecular weight ranging from 6.78 to 53.18 kDa and pI ranging from 4.41 to 11.39. Several alkaline residue-rich fragments in the MADS domain contain nuclear localization signals38, so the MADS-box proteins are thought to be located in the nucleus39. Many MADS-box genes have been subcellularly localized, and indeed, most of them are located in the nucleus, such as AGL1540, AGL2441, AGL6142, AGL8043 in A. thaliana and OsMADS22, OsMADS47, OsMADS5044 in rice. In this study, most of the SiMADS-box genes were predicted to be located in the nucleus, as expected. The SiMADS-box genes were divided into M-type (37) and MIKC-type (52). It is interesting that no SiMADS-box genes were included in the FLC-like subfamily, which is associated with vernalization45. This may be because foxtail millet does not have a vernalization requirement, so this branch may have been lost in this species. Gene-structure analysis showed a great difference in the average number of introns between the two subfamilies, with the average number of introns in the MIKC-type subfamily being much larger than that in the M-type subfamily. This is similar to the distribution of introns in rice1 and A. thaliana25, and indicates that the two types of SiMADS-box genes have different evolutionary paths, possibly due to their different tendencies to lose or gain introns during differentiation25. We speculate that in the evolution of foxtail millet MADS genes, under the pressure of natural selection, M-type MADS genes may tend to lose a large number of introns. As a result, the number of introns in the M-type MADS gene is very small or no intron, which suggests that the M-type MADS gene may have played a role in the evolution of foxtail millet. As for why the M-type and MIKC_type MADS genes produced different evolutionary paths, it is currently unclear, which is also worthy of in-depth research in the future. In addition, the MIKC and M subfamilies have their own unique conserved motif sequences, implying that their functions may also differ. A phylogenetic tree, constructed with MADS genes of other species, showed that the motif distribution in the MIKC subfamily was more conserved than that in the M-type MADS genes. In the M-type phylogenetic tree, the subfamily not only included the most SiMADS-box members (19), but it also had a unique conserved motif (motif 8), but lacked motif 7–1. In the MIKC-type phylogenetic tree, subfamily f was relatively unique, and new motifs 9 and 10 appeared. The unique motifs in these subgroups may endow the subfamily with special physiological functions. On the other hand, similar amino acid structures in each subfamily suggest similar physiological functions.
Evolution of SiMADS-box genes
Most of the duplications of foxtail millet genes stemmed from whole genome duplication (WGD) events that are common to all Gramineae19. We analyzed the duplication events in foxtail millet MADS-box genes and found only one pair of tandem repeats (SiMADS69 and SiMADS70) in the MIKC-type genes, but 11 pairs of duplicated segment genes. Among the SiMADS-box genes with segmental duplications, only 1 pair belonged to the M-type, and the other 10 pairs belonged to the MIKC-type. Among these repetitive events, the TM3-like subfamily had three pairs. This is similar to the situation of segmental duplications in rice1, that is, MIKC-type genes account for the vast majority of the segmental duplications. Therefore, some SiMADS-box genes may have been generated by gene-replication events, in which the amplification of MIKC-type genes is the main force driving the amplification of the number of SiMADS-box genes, and the number of MIKC-type genes (52) is thus much larger than that of the M-type genes (37). In addition, collinearity analysis with other species showed the highest collinearity for both M-type and MIKC-type genes with maize, and collinearity with monocotyledons was much higher than that with dicotyledons. Some of the MIKC genes showed collinearity with both monocots and dicots. Therefore, we speculate that the differentiation of the MIKC subfamily occurred earlier than that of monocotyledons and dicotyledons. In addition, the genes may be so fundamental to the different plants that they cannot be lost. However, these genes included only two duplicated genes, suggesting that the other nine duplicated genes were formed after the differentiation of monocotyledons and dicotyledons. We found four genes that were only collinear with maize, and these genes may be characteristic of C4 plants. These results were expected, because WGD of foxtail millet occurred before the separation of sorghum and maize19. Foxtail millet, as a monocotyledonous C4 crop46,47, is more closely related to maize and monocotyledons. Naturally, it has the most collinear genes with maize, which is also a C4 crop, and should have more collinear genes with other monocotyledons than dicotyledons.
Temporal and spatial expression of SiMADS-box genes
The expression of MADS-box genes has been investigated in the tissues of different species, for example, in root, leaf and inflorescence of A. thaliana25, and in roots, stems, leaves and flower organs of Brachypodium distachyon48. In rice, most MADS-box genes are specifically expressed in the panicle and seed1. In this study, we found expression of SiMADS-box genes in various organs and to varying degrees. Most of the genes were highly expressed in fruit, especially MIKC-type SiMADS33 and SiMADS37. The correlation of genes' expression in different tissues was also explored. Expression of M-type and MIKC-type genes was more positively correlated in certain tissues, indicating their similar functions in these tissue parts.
Furthermore, the expression of SiMADS-box genes in the peel and fruit before, during and after grain-filling was discussed. Different genes were involved in the whole process of fruit development during grain-filling, that is, genes were highly expressed before, during or after grain filling. However, M-type genes SiMADS28 and SiMADS60 tended to be highly expressed in the late filling period, whereas MIKC-type genes tended to be highly expressed in the early filling period (SiMADS12, SiMADS26, SiMADS46) and middle filling period (SiMADS33, SiMADS37). There were not only significant positive correlations, but also significant negative correlations between MIKC-type and M-type genes in the early, middle and late stages of grain-filling. These results indicate that the functions of MIKC-type and M-type genes in the process of grain-filling can be similar or different. Predicted protein interactions indicated that SiMADS46 protein interacts with SiMADS26, SiMADS52 and SiMADS60 proteins (Fig. 3B). The correlation analysis results of the relative expression levels of 12 genes also showed significant correlations between SiMADS46 and SiMADS26, SiMADS52 and SiMADS60. These results also confirm the reliability of the predicted protein-interaction results, and suggest that these proteins may indeed interact.
Previous studies have found that OsMADS22 (LOC_Os02g52340) and OsMADS55 (LOC_Os06g11330) are highly expressed in the stem, significantly inhibiting stem elongation in coordination with negative regulation of brassinosteroid content49,50. However, the homologous gene SiMADS12 was not highly expressed in the stem. OsMADS29 (LOC_Os02g07430) plays an important role in the development of rice seeds51, and its homologous gene SiMADS2 was also highly expressed at the early stage of seed filling, suggesting that SiMADS2 may also play a crucial role in the development of foxtail millet seeds.
Response of SiMADS-box genes to abiotic stress
Studies have been more focused on exploring the role of MADS-box genes in floral organ development, and less on exploring their response to various abiotic stresses48,52 However, some studies have found that the response of M-type MADS-box genes to abiotic stress is very important48. We found some genes to be significantly induced under stress, such as SiMADS33 and SiMADS78 under acid, alkali, dark, heat and cold treatments. SiMADS33 and SiMADS78 may be key genes in stress tolerance, warranting further study. Some genes showed different expression patterns under different treatments, such as SiMADS28 which was significantly upregulated under alkali, salt and PEG treatments, but changed little under flooding. Some genes may be greatly affected by temperature stress, such as the significant increase of SiMADS60 expression under cold and heat stress treatments. Expression tendencies also differed, for example, SiMADS33 and SiMADS67 tended to be highly expressed in roots, whereas SiMADS78 tended to be expressed in roots and leaves. The response time of the different genes to the stresses also differed. It was frequently found that those genes which were highly expressed in stems and leaves could respond to stress quickly, with significant changes in expression detected after 2 h of the stress. However, the response time of genes that were highly expressed in roots was relatively slow, reaching peak expression after 24 h of stress. The expression of SiMADS-box genes under the stress treatments was mostly positively correlated, suggesting interactions between genes, so as to jointly deal with the adverse effects of the abiotic stress on the plants. Protein-interaction prediction showed that the three MIKC proteins (SiMADS26, SiMADS46, SiMADS52) interacted, and that SiMADS46 also interacted with the M-type protein SiMADS60 (Fig. 3B). In addition, correlation analysis of the relative expression of the 12 genes under eight abiotic stresses also showed a significant positive correlation among SiMADS26, SiMADS46, and SiMADS52. SiMADS46 and SiMADS60 were also significantly positively correlated. Therefore, we speculate that there is interaction between these proteins under external environmental stress, which makes the plant better through the stress period.
In addition, other genes may also have important biological functions, such as SiMADS52, which is highly expressed in stems. Its collinearly related gene AGAMOUS (AT4G18960) regulates sepal senescence by promoting the production of jasmonic acid53. In addition, AGAMOUS-like15 (AGL15) and AGL1854,55 can delay sepal senescence and anther ablation in A. thaliana, thereby controlling the time of flower senescence. The homologous gene OsMADS58 (LOC_Os05g11414) plays a key role in regulating flower meristem decisions, and OsMADS3 (LOC_Os01g10504) plays an important role in regulating stamen characteristics56. This suggests that SiMADS52 may also have a similar function, warranting further study. Previous studies have found that OsMADS2 (LOC_Os01g66030) and OsMADS4 (LOC_Os05g34940) play an important role in style and stamen development57, and their homolog SiMADS26 may play a similar major role in this development.
Materials and methods
Plant materials, growth conditions, and abiotic stress treatments in foxtail millet
The test material in this study was the foxtail millet Setaria italica cv. Yugu 1, typical to northern China, and planted in a greenhouse. We obtained samples of roots, stems, leaves, peels and fruit in the middle grain-filling stage, and of peels and fruit in the early and late filling stages. All samples were taken from five plants under the same growing conditions, quickly frozen in liquid nitrogen, and stored at − 80 °C. The expression levels of 12 SiMADS-box genes were detected. In addition, we carried out stress treatments on foxtail millet plants at the seedling stage (28 days), including salt (5% NaCl), acid (0.1 mol/L HCl), alkali (0.2 M NaOH), darkness (complete shading), flooding (whole plant), drought (10% PEG6000), heat (40 °C), and cold (4 °C). Five repeats were carried out for each stress treatment, and samples were collected at 0, 2 and 24 h for qRT-PCR analysis.
Total RNA extraction, cDNA reverse transcription and qRT-PCR analysis
Total RNA was extracted by RNA extraction kit (TaKaRa Bio). The qRT-PCR primers (Table S6) were designed using Primer 5.0 software. In this experiment, the actin gene (Si001873mg)58 was used as an internal control (http://www.pantherdb.org/genes/gene.do?acc=SETIT%7CEnsemblGenome%3DSETIT_001873mg%7CUniProtKB%3DK3XJ00). Standard qRT-PCR with SYBR Premix Ex Taq II (TaKaRa Bio) was repeated at least three times on a CFX96 Real-Time PCR System (Bio-Rad). The qRT-PCR data were analyzed by the 2−(ΔΔCt) method.
Genome-wide Identification of MADS-box genes in foxtail millet
The Setaria italica genome was downloaded from EnsemblPlants (https://plants.ensembl.org/info/website/ftp/index.html). The MADS-box protein sequences of A. thaliana (https://www.Arabidopsis.org/) and rice (http://Rice.plantbiology.msu.edu/) were downloaded separately. First, the whole foxtail millet genome was aligned with AtMADS and OsMADS protein sequences (score value ≥ 100 and e-value ≤ 1e−10), yielding the candidate MADS-box genes. Second, the hidden Markov model (HMM) of the M-type SRF-TF domain (PF00319), MIKC-type SRF-TF domain (PF00319) and MIKC-type K-box domain (PF01486) was downloaded from the Pfam database (http://pfam.xfam.org/)59, and HMMER 3.0 software (with default parameters) (http://HMMER.org/)60 was used to search for MADS-box proteins. The obtained sequences were further verified by SMART tool (http://SMART.emblheidelberg.de/)61 to identify a putative MADS domain. Finally, the sequences of 89 MADS-box proteins were obtained. Then their length, molecular weight and pI were determined on the ExPasy website (https://web.expasy.org/compute_pi/). Subcellular localization of MADS-box proteins was predicted by WoLFPSORT (https://wolfpsort.hgc.jp/)62.
Phylogenetic analysis and classification of the SiMADS-box gene family
According to the classification of MADS-box genes in rice and A. thaliana and the SiMADS-box domain, the identified SiMADS-box genes were divided into groups. Phylogenetic trees were constructed using protein sequences from A. thaliana, O. sativa, Brassica napus, Zea mays, Fagopyrum tataricum, and Brachypodium distachyon) downloaded from the UniProt database (https://www.UniProt.org). We used MUSCLE sequence alignment for the protein sequences, and then constructed the ML phylogenetic tree with IQ-tree wrapper, and bootstrap number set to 1000.
Chromosomal distribution and gene duplication of SiMADS-box genes
We use Circos to obtain information on the genes' physical location in the foxtail millet genome and localized all SiMADS-box genes to the chromosomes. The Multiple Collinearity Scan toolkit X (MCScanX) was used with default parameters to scan the collinearity of SiMADS-box genes and then analyze gene-duplication events. We used a Double Synteny Plotter to analyze the homology of SiMADS-box genes among species63.
Gene structure, conserved motif analysis and protein-interaction prediction
The structural map of SiMADS-box genes was constructed by sequence alignment between the CDS and the corresponding genomic DNA sequence. The online MEME tool (http://meme-suite.org/tools/meme)64 was used to analyze the full-length conserved motifs of the SiMADS-box family of proteins, and the maximum conservative motif search value was set to 10. SiMADS-box protein interactions were predicted using STRING (https://string-preview.org/).
Statistical analysis
Analysis of variance (ANOVA) was performed with JMP6.0 software (SAS Institute), and least significant difference (LSD) was used for comparisons at the 0.05 and 0.01 levels. The histogram was drawn with OriginPro2019b software (OriginLab).
Ethics approval and consent to participate
The foxtail millet accession (Yugu 1) was supplied by Professor Jianping Cheng of Guizhou University. These plant materials are widely used all over the world and no permits are required for the collection of plant samples. This article does not contain any studies with human participants or animals performed by the authors. The methods were carried out in accordance with the relevant guidelines and regulations. We confirm that all experimental protocols were approved by Guizhou University.
Supplementary Information
Acknowledgements
We thank all of the colleagues in our laboratory for providing useful discussions and technical assistance. We are very grateful to the editor and reviewers for critically evaluating the manuscript and providing constructive comments for its improvement.
Author contributions
Methodology, D.L.; Investigation, D.L.; Formal analysis, D.L., A.H., G.X., H.Y., L.F., X.W., L.L. and D.X.; Writing—original draft, D.L. and Y.F.; Supervision, J.C.; Writing—review & editing, J.R. and J.Y. All authors read and approved the final manuscript.
Funding
This research was supported by the National Science Foundation of China (31560578, Cheng JP, http://www.nsfc.gov.cn), Sichuan International Science and Technology Cooperation and Exchange Research and Development Project (2018HH0116, Yan J, http://kjt.sc.gov.cn), Guizhou Science and Technology Support Project (No. 20201Y125). Funds were used for the design of the study, collection, analysis, data interpretation, and writing the manuscript, as well as for the open access payment.
Data availability
Information on the entire Setaria italica genome sequence was from the Ensembl Genomes website (http://ensemblgenomes.org/). The Setaria italica materials (Yugu 1) used in the experiments were supplied by Prof. Jianping Cheng of Guizhou University. The datasets supporting the conclusions of this article are included in the article and its Supplementary Material.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jun Yan, Email: yanjun62@qq.com.
Jianping Cheng, Email: chengjianping63@qq.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-07103-9.
References
- 1.Arora R, et al. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 2007;8:242. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao HB, et al. Genome-wide identification and analysis of the MADS-box gene family and its potential role in fruit development and ripening in red bayberry (Morellarubra) Gene. 2019;717:144045. doi: 10.1016/j.gene.2019.144045. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez-Buylla ER, et al. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5328–5333. doi: 10.1073/pnas.97.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coen ES, Meyerowitz EM. The war of the whorls: Genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 5.Heck GR, Perry SE, Fernandez N. AGL15, a MADS domain protein expressed in developing embryos. Plant Cell. 1995;7:1271. doi: 10.1105/tpc.7.8.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liljegren SJ, et al. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;404:766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- 7.Gu Q, Ferrándiz C, Yanofsky MF, Martienssen R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development. 1998;125:1509–1517. doi: 10.1242/dev.125.8.1509. [DOI] [PubMed] [Google Scholar]
- 8.Ferrandiz C, Liljegren SJ, Yanofsky MF. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science. 2000;289:436–438. doi: 10.1126/science.289.5478.436. [DOI] [PubMed] [Google Scholar]
- 9.Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C function floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Tang D, Lin X, Ding M, Tong Z. Genome-wide identification of MADS-box family genes in moso bamboo (Phyllostachys edulis) and a functional analysis of PeMADS5 in flowering. BMC Plant Biol. 2018;18:176. doi: 10.1186/s12870-018-1394-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Fatima M, Zhou P, Ma Q, Ming R. Analysis of MADS-box genes revealed modified flowering gene network and diurnal expression in pineapple. BMC Genomics. 2020;21:8. doi: 10.1186/s12864-019-6421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, et al. Genome-wide analysis of the MADS-box transcription factor family in Solanumlycopersicum. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20122961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang S, et al. Functional annotation and identification of MADS-box transcription factors related to tuber dormancy in Helianthustuberosus L. 3 Biotech. 2019;9:378. doi: 10.1007/s13205-019-1897-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mamidi S, et al. A genome resource for green millet Setaria viridis enables discovery of agronomically valuable loci. Nat. Biotechnol. 2020;38:1203–1210. doi: 10.1038/s41587-020-0681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bettinger RL, Barton L, Morgan C. The origins of food production in north China: A different kind of agricultural revolution. Evol. Anthropol. Issues News Rev. 2010;19:9–21. doi: 10.1002/evan.20236. [DOI] [Google Scholar]
- 16.Bennetzen JL, et al. Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 2012;30:555–561. doi: 10.1038/nbt.2196. [DOI] [PubMed] [Google Scholar]
- 17.Devi PB, Vijayabharathi R, Sathyabama S, Malleshi NG, Priyadarisini VB. Health benefits of finger millet (Eleusinecoracana L.) polyphenols and dietary fiber: A review. J. Food Sci. Technol. 2014;51:1021–1040. doi: 10.1007/s13197-011-0584-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verma S, Srivastava S, Tiwari N. Comparative study on nutritional and sensory quality of barnyard and foxtail millet food products with traditional rice products. J. Food Sci. Technol. 2015;52:5147–5155. doi: 10.1007/s13197-014-1617-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang G, et al. Genome sequence of foxtail millet (Setariaitalica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 2012;30:549–554. doi: 10.1038/nbt.2195. [DOI] [PubMed] [Google Scholar]
- 20.Muthamilarasan M, et al. Global analysis of WRKY transcription factor superfamily in Setaria identifies potential candidates involved in abiotic stress signaling. Front. Plant Sci. 2015;6:910. doi: 10.3389/fpls.2015.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alagarasan G, Dubey M, Aswathy KS, Chandel G. Genome wide identification of orthologous ZIP genes associated with zinc and iron translocation in Setariaitalica. Front. Plant Sci. 2017;8:775. doi: 10.3389/fpls.2017.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karunanithi PS, et al. The foxtail millet (Setariaitalica) terpene synthase gene family. Plant J. 2020;103:781–800. doi: 10.1111/tpj.14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaur VS, Sood S, Tiwari S, Kumar A. Genome-wide identification and characterization of seed storage proteins (SSPs) of foxtail millet (Setariaitalica (L.) P. Beauv.) 3 Biotech. 2018;8:415. doi: 10.1007/s13205-018-1431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra AK, Muthamilarasan M, Khan Y, Parida SK, Prasad M. Genome-wide investigation and expression analyses of WD40 protein family in the model plant foxtail millet (Setariaitalica L.) PLoS One. 2014;9:e86852. doi: 10.1371/journal.pone.0086852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parenicova L, et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell. 2003;15:1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y, et al. Whole-genome survey and characterization of MADS-box gene family in maize and sorghum. Plant Cell Tissue Organ Cult. (PCTOC) 2010;105:159–173. doi: 10.1007/s11240-010-9848-8. [DOI] [Google Scholar]
- 27.Saha G, et al. Genome-wide identification and characterization of MADS-box family genes related to organ development and stress resistance in Brassicarapa. BMC Genomics. 2015;16:178. doi: 10.1186/s12864-015-1349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kriventseva EV, et al. Increase of functional diversity by alternative splicing. Trends Genet. 2003;19:124–128. doi: 10.1016/S0168-9525(03)00016-7. [DOI] [PubMed] [Google Scholar]
- 29.Stetefeld J, Ruegg MA. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem. Sci. 2005;30:515–521. doi: 10.1016/j.tibs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Shabalina SA, et al. Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol. Biol. Evol. 2010;27:1745–1749. doi: 10.1093/molbev/msq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy SW, Gilbert W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 32.Chorev M, Carmel L. The function of introns. Front. Genet. 2012;3:55. doi: 10.3389/fgene.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jo BS, Choi SS. Introns: The functional benefits of introns in genomes. Genomics Inform. 2015;13:112. doi: 10.5808/GI.2015.13.4.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parenteau J, et al. Introns are mediators of cell response to starvation. Nature. 2019;565:612–617. doi: 10.1038/s41586-018-0859-7. [DOI] [PubMed] [Google Scholar]
- 35.Morgan JT, Fink GR, Bartel DP. Excised linear introns regulate growth in yeast. Nature. 2019;565:606–611. doi: 10.1038/s41586-018-0828-1(. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szklarczyk D, et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holub EB. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001;2:516–527. doi: 10.1038/35080508. [DOI] [PubMed] [Google Scholar]
- 38.Immink RG, Gadella TW, Jr, Ferrario S, Busscher M, Angenent GC. Analysis of MADS box protein–protein interactions in living plant cells. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2416–2421. doi: 10.1073/pnas.042677699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gramzow L, Theissen G. A hitchhiker's guide to the MADS world of plants. Genome Biol. 2010;11:214. doi: 10.1186/gb-2010-11-6-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perry SE, Lehti MD, Fernandez DE. The MADS-domain protein AGAMOUS-like 15 accumulates in embryonic tissues with diverse origins. Plant Physiol. 1999;120:121–130. doi: 10.1104/pp.120.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujita H, et al. An Arabidopsis MADS-Box protein, AGL24, is specifically bound to and phosphorylated by meristematic receptor-like kinase (MRLK) Plant Cell Physiol. 2003;44:735–742. doi: 10.1093/pcp/pcg092. [DOI] [PubMed] [Google Scholar]
- 42.Bemer M, Wolters-Arts M, Grossniklaus U, Angenent GC. The MADS domain protein DIANA acts together with AGAMOUS-LIKE80 to specify the central cell in Arabidopsis ovules. Plant Cell. 2008;20:2088–2101. doi: 10.1105/tpc.108.058958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Portereiko MF, et al. AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell. 2006;18:1862–1872. doi: 10.1105/tpc.106.040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S, Jeong DH, An G. A possible working mechanism for rice SVP-group MADS-box proteins as negative regulators of brassinosteroid responses. Plant Signal. Behav. 2008;3:471–474. doi: 10.4161/psb.3.7.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becker A. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003;29:464–489. doi: 10.1016/s1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 46.Daniel, K. Y. T. & Jeffrey, S. A. in Photosynthesis Ch. Chapter 12, (2013).
- 47.Doust AN, Kellogg EA, Devos KM, Bennetzen JL. Foxtail millet: A sequence-driven grass model system. Plant Physiol. 2009;149:137–141. doi: 10.1104/pp.108.129627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei B, et al. Genome-wide analysis of the MADS-box gene family in Brachypodiumdistachyon. PLoS One. 2014;9:e84781. doi: 10.1371/journal.pone.0084781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li N, Wang Y, Lu J, Liu C. Genome-wide identification and characterization of the ALOG domain genes in rice. Int. J. Genomics. 2019;2019:2146391. doi: 10.1155/2019/2146391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S, Choi SC, An G. Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J. 2008;54:93–105. doi: 10.1111/j.1365-313X.2008.03406.x. [DOI] [PubMed] [Google Scholar]
- 51.Yang X, et al. Live and let die—The B(sister) MADS-box gene OsMADS29 controls the degeneration of cells in maternal tissues during seed development of rice (Oryzasativa) PLoS One. 2012;7:e51435. doi: 10.1371/journal.pone.0051435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jia J, et al. MADS-box family genes in sheepgrass and their involvement in abiotic stress responses. BMC Plant Biol. 2018;18:42. doi: 10.1186/s12870-018-1259-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jibran R, Tahir J, Cooney J, Hunter DA, Dijkwel PP. Arabidopsis AGAMOUS regulates sepal senescence by driving jasmonate production. Front. Plant Sci. 2017;8:2101. doi: 10.3389/fpls.2017.02101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adamczyk BJ, Lehti-Shiu MD, Fernandez DE. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 2007;50:1007–1019. doi: 10.1111/j.1365-313X.2007.03105.x. [DOI] [PubMed] [Google Scholar]
- 55.Fernandez DE, et al. The embryo MADS domain factor AGL15 acts postembryonically: Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell. 2000;12:183–198. doi: 10.1105/tpc.12.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamaguchi T, et al. Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryzasativa. Plant Cell. 2006;18:15–28. doi: 10.1105/tpc.105.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao SG, Ohmori S, Kimizu M, Yoshida H. Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development. Plant Cell Physiol. 2008;49:853–857. doi: 10.1093/pcp/pcn050. [DOI] [PubMed] [Google Scholar]
- 58.Min DH, et al. Characteristics of PP2C gene family in foxtail millet (Setariaitalica) Acta Agron. Sin. 2013;39:2135. doi: 10.3724/SP.J.1006.2013.02135. [DOI] [Google Scholar]
- 59.El-Gebali S, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horton P, et al. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007;35:W585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen C, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 64.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Information on the entire Setaria italica genome sequence was from the Ensembl Genomes website (http://ensemblgenomes.org/). The Setaria italica materials (Yugu 1) used in the experiments were supplied by Prof. Jianping Cheng of Guizhou University. The datasets supporting the conclusions of this article are included in the article and its Supplementary Material.