Abstract
FmtA is a factor which affects the methicillin resistance level in methicillin-resistant Staphylococcus aureus. Since FmtA has two of three conserved motifs which are typically found in penicillin-binding proteins (PBPs) and β-lactamases, we investigated the penicillin-binding activity of recombinant FmtA and found no such activity. Immunoblotting analysis revealed that FmtA localizes in the membrane fraction. To investigate the function of FmtA, high-pressure liquid chromatography analysis of cell wall muropeptides was performed with an fmtA-inactivated mutant and its parent. The mutant showed a reduced cross-linking and partially reduced amidation of glutamate residues in the peptidoglycan of the mutant. The transcription of fmtA was dose dependently increased by the addition of β-lactam antibiotics, fosfomycin, and bacitracin, while its transcription was not changed by the addition of vancomycin or tetracycline. These results reveal that Fmt is a membrane-located, non-penicillin-binding protein and that mutation of fmtA affects the cell wall structure, although its precise function is still unknown.
Methicillin-resistant Staphylococcus aureus (MRSA) has an extra penicillin-binding protein, PBP 2′ or PBP 2A, which shows a low affinity for β-lactam antibiotics and which functions in the presence of otherwise inhibitory concentrations of β-lactam antibiotics (5, 14). The series of fem and aux factors, llm, fmt, and sigB were identified as factors which affect the methicillin resistance level (1, 3, 4, 11, 26). These genes are located on chromosomal DNA outside the mec element. Most of them were thought to be associated with peptidoglycan synthesis (6, 7, 13, 17, 18, 25) and to function in accordance with PBP 2′ in the presence of methicillin. However, many factors other than the fem series are considered to be involved in peptidoglycan synthesis (4). Investigation of these genes is important to understanding the variety of levels of resistance to methicillin in clinical S. aureus strains.
Recently, we found a novel gene, fmtA (we renamed fmt as fmtA), which affects the methicillin resistance level (9). Inactivation of fmtA resulted in reduction of the methicillin resistance level in MRSA; in particular, homogeneous resistance was converted to heterogeneous resistance. Complementation experiments revealed that fmtA alone restored the mutation, indicating that the reduction of methicillin resistance by the Tn551 insertion was not due to a polar effect on a downstream gene. Therefore, fmtA is thought to be responsible for the alteration in methicillin resistance. The putative protein, FmtA, has SXXK and S(Y)XN motifs which are typically found in β-lactamases and low-molecular-weight PBPs. In this study, we demonstrated that the FmtA mutation affects cell wall structure and that its expression is enhanced in the presence of β-lactam antibiotics.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains and plasmids used in this study are listed in Table 1. Staphylococcus and Escherichia were grown in either Trypticase soy broth or brain heart infusion broth (both from Beckton Dickinson Microbiology Systems, Cockeysville, Md.) and Luria-Bertani broth, respectively. When needed, erythromycin (30 μg/ml), chloramphenicol (10 μg/ml), or ampicillin (100 μg/ml) was added to the medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Origin or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| KSA8 | Homogeneous Mcr | 9 |
| COL | Homogeneous Mcr | 9 |
| RN4220 | 8325-4r− | R. Novick |
| RN4220-TS339C | Mutagenized fmtA::cat | This study |
| COL-TS339 | Mutagenized fmtA::Tn551 | 9 |
| COL-TS339C | fmtA::cat gene (transduction from RN4220-TS339C) | This study |
| KSA8 TS339 | fmtA::Tn551 (transduction from COL-TS339) | 9 |
| KSA8 TS339C | fmtA::cat gene (transduction from RN4220-TS339C) | This study |
| HK9605 | COL(pHK4147) | This study |
| HK9610 | COL(pHK4125) | This study |
| HK9710 | RN4220(pHK4080) | This study |
| E. coli | ||
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZM15 Tn10 (Tetr)] | 2 |
| HMS174 | recA1 hsdR Rifr | 16 |
| HK4172 | HMS174(pHK4171) | This study |
| Plasmids | ||
| pCL52.1 | Shuttle vector (thermosensitive); SPr in E. coli, Tcr in S. aureus | 19 |
| pLI50 | Shuttle vector; Ampr in E. coli, Cmr in S. aureus | 10 |
| pGEM T-easy | PCR cloning vector; Ampr in E. coli (Promega) | |
| pET28C | Expression vector (Novagen) | |
| pSL24 | Promoterless xylE gene for reporter system | 19 |
| pHK4033 | pUC19 with 3.6-kb HindIII fragment of KSA8 | This study |
| pHK4079 | pCL52.1 with 3.5-kb EcoRI-SalI fragment from pHK4033 | This study |
| pHK4080 | pCL52.1 with 3.5-kb EcoRI-SalI fragment plus cat gene | This study |
| pHK4130 | pGEM-T easy with 1.2-kb PCR fragment (fmtA) of KSA8 | This study |
| pHK7925 | pLI50 with 1.4-kb fmtA of KSA8 | This study |
| pHK4171 | pET28c with fmtA of KSA8 | This study |
| pHK4125 | pSL24 with fmtA promoter region (BamHI-HindIII fragment) | This study |
| pHK4147 | pSL24 with orf1 promoter region (BamHI-EcoRI fragment) | This study |
DNA manipulations.
Routine DNA manipulations, digestion of DNA with restriction enzymes and shrimp alkaline phosphatase, DNA ligations, gel electrophoresis, Southern blotting of DNA and hybridization, and DNA sequencing were performed essentially as described previously (16). Restriction enzymes and shrimp alkaline phosphatase were purchased from Boehringer Mannheim Biochemica, Tokyo, Japan, and T4 DNA ligase was from New England BioLabs, Beverly, Mass. Hybridization was performed by means of a chemiluminescence procedure (ECL direct labelling kit or 3′-oligolabelling kit; Amersham Life Science, Bucks, United Kingdom). The DNA sequences of both strands were determined by the dideoxy chain termination method with the Autoread sequencing kit (Pharmacia Biotechnology, Tokyo, Japan). PCR reagents were from Boehringer Mannheim, and PCR was performed with the GeneAmp PCR System 2400 (Perkin-Elmer).
Construction of the cat insertional mutant.
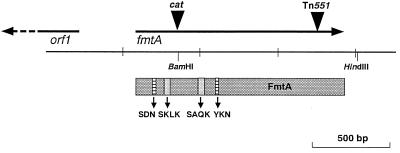
In COL-TS339, Tn551 was inserted at the C terminus of fmtA (Fig. 1), which raised the possibility that the FmtA truncated at the C terminus still possesses partial enzyme activity. Therefore, we constructed a mutant which possesses an inactivated fmtA gene by insertion of cat into its N-terminal region. A plasmid (pHK4080) containing the cat gene inserted at the N terminus of fmtA was first constructed. A 3.5-kb EcoRI-SalI fragment from pHK4033 was cloned into pCL52.1 (a thermosensitive plasmid; Table 1) to generate pHK4079. Then, the cat gene from Sau3AI-digested Escherichia coli-S. aureus shuttle vector pLI50 (10) was ligated into pHK4079 by using the BamHI site in the N-terminal region of fmtA (Fig. 1) to generate pHK4080. The recombinant plasmid was electroporated into RN4220 to generate strain HK9710. HK9710 was grown at 42°C in the presence of antibiotics (3 μg of tetracycline per ml and 10 μg of chloramphenicol per ml) to select strains with the plasmid integrated into the chromosomal DNA. A single colony was then incubated at 30°C in the presence of chloramphenicol for 24 to 48 h to allow excision of the integrated plasmid. Then, the strain that grew in the presence of chloramphenicol but not in the presence of tetracycline was isolated. The cat gene was transduced by phage 80 alpha to KSA8 and COL strains. The insertional inactivation of fmtA in the transductants by cat was confirmed by Southern hybridization. The MIC was determined by a microdilution method that has been described elsewhere (9).
FIG. 1.
Mapping of fmtA region of S. aureus KSA8. The arrows represent the open reading frames and the direction of transcription. The box represents the FmtA protein and contains the four motifs which are typically found in PBPs and β-lactamases.
Muropeptide analysis.
Strains KSA8, KSA8-TS339, and KSA8-TS339C were further investigated because the reduction of the resistance level in KSA8-TS339 was the most remarkable among the strains tested. Preparation of murein and reduction of muropeptide were as described elsewhere (15, 18). Digested muropeptides were fractionated by reverse-phase high-pressure liquid chromatography (RP-HPLC) as described elsewhere (15, 18).
Recombinant FmtA protein.
To produce recombinant FmtA protein, the DNA fragment corresponding to signal peptide-processed FmtA was amplified by PCR with two primers and was cloned in-frame downstream from the His tag (six His residues) sequence in the pET28c expression vector (Novagen, Madison, Wis.) to generate pHK4171. The presence of the plasmid was verified by sequencing. The plasmid was electroporated into E. coli HMS174, and the transformant (strain HK4172) was used to purify the recombinant protein, as described elsewhere (21). Recombinant FmtA was used to prepare rabbit anti-FmtA antiserum by a method described elsewhere (21).
PBP assay.
The recombinant FmtA protein was used for PBP assay. The recombinant protein (0.5 μg; 10 pmol) was dialyzed against 50 mM Tris-HCl (pH 7.5) and was incubated with various concentrations of [3H]benzylpenicillin (20, 50, and 100 pmol; 10 to 30 Ci/mmol; Amersham International, Bucks, United Kingdom) at 37°C for 15 min, and then the reaction was stopped by the addition of the sample loading buffer. Samples were applied to the well and were then electrophoresed in a 10% polyacrylamide gel. PBP activity was detected by fluorography.
Fractionation of proteins from S. aureus cells.
S. aureus cells growing to the late exponential phase were collected by centrifugation at 10,000 × g. After washing of the cells with phosphate-buffered saline (PBS), the cells were suspended in PBS containing phenylmethylsulfonyl fluoride (1 mM) and were digested with lysostaphin (final concentration, 100 μg/ml) for 30 min at 37°C. Then, the supernatants obtained after centrifugation at 10,000 × g were used as the whole-cell lysate fraction. Various fractions of S. aureus cells were prepared as described elsewhere (20).
Construction of fmtA promoter fusion.
The putative fmtA promoter region was ligated into BamHI and HindIII sites within the xylE transcriptional fusion vector pSL24 (Table 1). The DNA fragment containing the promoter region was amplified with two oligonucleotide primers (5′-AATAAGCTTACACACGCATGTATAACTAGT-3′ and 5′-ACAGGATCCAGAACCAATGCTAGAAGGATC-3′) generating BamHI or HindIII sites from KSA8 chromosomal DNA for direct cloning into the upstream region of the promoterless xylE reporter gene of pSL24 to generate pHK4125. The putative orf1 promoter region, which was oriented in the transcriptional direction opposite that of fmtA, was ligated into BamHI and EcoRI sites in pSL24. orf1 is located 450 bp upstream of fmtA (Fig. 1), and its function is unknown. The DNA fragment was amplified with two oligonucleotide primers (5′-AATGAATTCACACACGCATGTATAACTAGT-3′ and 5′-ACAGGATCCAGAACCAATGCTAGAAGGATC-3′), generating BamHI or EcoRI sites, and was then cloned into pSL24 to generate pHK4147. Both PCR fragments were verified by sequencing. The recombinant plasmids were electroporated into RN4220 and were then transduced to strain COL by using phage 80 alpha.
Catechol 2,3-dideoxygenase assays.
An S. aureus COL strain with pHK4125 (strain HK9610) or pHK4147 (strain HK9605) was grown to an optical density at 660 nm (OD660) of 1.0 with or without antibiotics, and quantitative assays were performed spectrophotometrically as described by Zukowski et al. (27). One unit is defined as an increase of 12 units at OD375 in 1 min. Specific activity is defined as milliunits per milligram of protein. This experiment was repeated three times, and then the mean and standard deviation (SD) were calculated. Significance was compared by the unpaired t test.
Chemicals and reagents.
Oxacillin, methicillin, bacitracin, and vancomycin were from Sigma Chemical Co., St. Louis, Mo. Erythromycin, chloramphenicol, and fosfomycin were from Wako Chemicals (Tokyo, Japan). Cefoxitin and tetracycline were from Daiichi Seiyaku (Tokyo, Japan) and Lederle Japan (Tokyo, Japan), respectively.
RESULTS
cat insertional mutant.
We obtained a mutant, COL-TS339C, from COL by inactivating fmtA by the insertion of cat (Fig. 1). The oxacillin MIC for this mutant was reduced (64 μg/ml) compared with that for the parent (512 μg/ml). The MIC was similar to that for COL-TS339 (64 μg/ml), which carries a Tn551 insertion at the C terminus of fmtA. To eliminate the possibility of introducing another mutation during construction, we transduced the cat gene from COL-TS339C into COL. The oxacillin MIC was reduced for all transductants, as was observed for COL-TS339C. The cat gene was also transduced into another MRSA strain, strain KSA8, and the transductant was designated KSA8-TS339C. The oxacillin MIC for KSA8-TS339C was also reduced. The MICs for KSA8 and KSA8-TS339C were 512 and 16 μg/ml, respectively. The MIC of oxacillin for each cat insertional mutant was identical to that for the respective Tn551 insertional mutant.
Muropeptide analysis of mutants.
The muropeptide profiles of KSA8, KSA8-TS339, and KSA8-TS339C were established by RP-HPLC. The profile of KSA8 was similar to those of other wild-type strains described earlier (15, 18). As for strain BB270, the highest peak was observed in the dimeric fraction, and large amounts of oligomers were observed in the profile for strain KSA8, consistent with the typical high degree of cross-linking of staphylococcal peptidoglycan. The two fmtA-inactivated mutants had similar muropeptide profiles. Compared to the wild type, both mutants showed slightly reduced amounts of oligomeric muropeptides, as can especially be seen from the increase in the amounts of monomers relative to the amounts of all other fractions and the clear decrease in large-molecular-mass oligomers that eluted at longer retention times (Table 2). A detailed resolution of the monomer fractions was also observed (data not shown). All peaks were identified by comparison with those for standard samples on the basis of mass spectrometry and/or amino acid analysis (15, 18). In KSA8-TS339 and KSA8-TS339C, the amount of the major monomeric component (M4, a monomer pentapeptide with five glycine peak) relative to those of M9 (peak 7; glutamic acid instead of glutamine in M4) and D5 (dimer from M4 and M9) was reduced compared to that for the parent strain; while these changes were not drastic, they were most pronounced in KSA8-TS339C (fmtA::cat).
TABLE 2.
Cell wall composition from RP-HPLC data
| Strain | % Mono-mers | % Dimers | % Trimers | % Oligomers | % Cross-linking | Ratio M4/M9 |
|---|---|---|---|---|---|---|
| KSA8 | 5.4 | 8.5 | 7.7 | 78.4 | 80 | 3.5 |
| KSA8-TS339 | 7.9 | 11.7 | 11.0 | 69.4 | 75.5 | 3.1 |
| KSA8-TS339C | 8.2 | 11.7 | 10.4 | 69.7 | 74.9 | 1.7 |
Penicillin-binding activity of recombinant FmtA.
When the membrane fraction of COL was used for the penicillin-binding assay, we failed to detect a band corresponding to FmtA (data not shown). Since FmtA lacks one of three motifs which are typically found in β-lactamases and PBPs, FmtA may have a very weak affinity of binding to penicillin. We therefore assessed the penicillin-binding activity of purified recombinant FmtA (rFmtA) with a six-residue His tag at the N terminus. rFmtA (0.5 μg) was incubated with [3H]benzylpenicillin and was subjected to polyacrylamide gel electrophoresis followed by fluorography. We did not detect any radioactive band (data not shown), suggesting that FmtA lacks covalent penicillin-binding affinity.
Identification and localization of FmtA by immunoblotting.
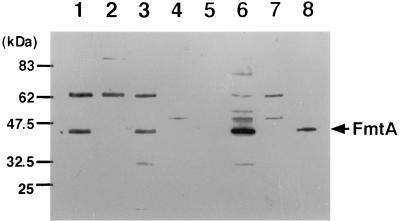
Using anti-FmtA serum, we found an immunoreactive band in whole-cell lysates of COL with a molecular mass that corresponded to the molecular mass of FmtA calculated from the DNA sequence; this band was missing from whole-cell lysate of COL-TS339C (Fig. 2). A 65-kDa band of unknown origin was observed in all strains. To investigate the cellular localization of FmtA, we performed Western blotting analysis with various fractions of COL cells. Figure 2 shows that FmtA was predominantly found in the membrane fraction but not in supernatant, cell wall, or cytoplasmic fraction.
FIG. 2.
Western blots of fractions from S. aureus COL. The fractionation method is described in Materials and Methods. The samples were prepared from COL (lanes 1 and 4 to 7), COL-TS339C (lane 2), and COL-TS339 with pHK7925 (lane 3) and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in 10% polyacrylamide gels and were then subjected to Western blotting. Anti-FmtA serum was used for primary serum. The FmtA band is marked with an asterisk. Lanes: 1 to 3, whole-cell lysate; 4, culture supernatant; 5, cell wall extract; 6, membrane extract; 7, cytoplasmic extract; 8, recombinant FmtA.
Transcriptional fusion studies. (i) Effect of oxacillin.
In the presence of 1 μg of oxacillin per ml, fmtA promoter activity was similar to that without oxacillin (Table 3). The activity was slightly enhanced by the addition of 10 μg of oxacillin per ml and was increased 2.5 times by the addition of 100 μg of oxacillin per ml (one-fourth the MIC). On the contrary, the orf1 promoter activities in the presence of oxacillin, even 100 μg of oxacillin per ml, were quite similar to those without oxacillin (Table 3).
TABLE 3.
XylE activity in the presence of β-lactam antibiotics
| Oxacillin concn (μg/ml) | XylE activity (mU/mg)
|
|
|---|---|---|
| fmtA | orf1 | |
| None | 14.8 ± 1.4a | 20.9 ± 0.9 |
| 1 | 17.0 ± 1.4 | 21.8 ± 1.3 |
| 10 | 22.6 ± 2.3 | 21.1 ± 1.6 |
| 100 | 42.8 ± 3.5 | 18.2 ± 0.6 |
Mean ± SD.
(ii) Effects of various antibiotics.
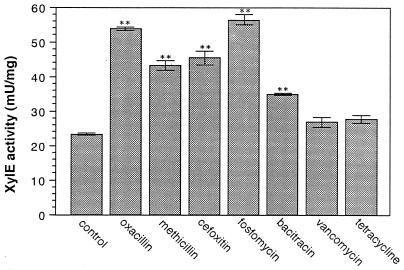
An increase in XylE activity was observed by the addition of one-fourth the MICs of oxacillin (100 μg/ml), methicillin (200 μg/ml), cefoxitin (64 μg/ml), fosfomycin (8 μg/ml), and bacitracin (32 μg/ml), while the activity in the presence of one-fourth the MICs of vancomycin (0.5 μg/ml) and tetracycline (32 μg/ml) was similar to that of the control (Fig. 3). In particular, after the addition of β-lactams and fosfomycin, the activity was two to three times higher than that of the control.
FIG. 3.
XylE activity of fmtA promoter in the presence of antibiotics. The preparation of samples for the XylE assay and the XylE assay are described in Materials and Methods. The experiment was repeated three times. Bars represent 1 SD. ∗∗, P < 0.01.
DISCUSSION
RP-HPLC analysis of muropeptide is a powerful tool for investigation of cell wall structure, although some important parameters like glycan chain length variations, secondary wall polymer properties, or degree of O acetylation are not usually analyzed (15, 18). For strain KSA8-TS339, there were slight increases in the amounts of the nonamidated monomers, dimers, and trimers, related to the slight decrease in the degree of cross-linking, compared with the amounts of these oligomers for the parent strain, strain KSA8. Thus, the HPLC profile of KSA8-TS339 resembled that of a glnR mutant, as reported previously (17), although the alteration of the RP-HPLC profile for the glnR mutant was more drastic than that observed for the fmtA mutant. The same effects were found in the muropeptide profile of KSA8-TS339C, but the effects were more exaggerated. This might be related to the fact that FmtA inactivation occurred at the C terminus of the protein for strain KSA8-TS339, while the cat interposition took place close to the N terminus in strain KSA8-TS339C (Fig. 1). glnR codes for a glutamine synthetase repressor (6), and inactivation of glnR resulted in an increase in the amounts of nonamidated monomers, dimers, and trimers and a reduced level of cell wall cross-linking (17). FmtA does not have homology with glutamine synthetase or its associated enzymes. It was reported that S. aureus grown in the presence of penicillin decreased the amidation of glutamate residues and the degree of cross-linking, indicating that penicillin affects the structure of the stem pentapeptide other than its cross-linking (12, 24). Penicillin does not inhibit glutamine synthetase directly, so that the blocking of amidation in glutamate residues should be caused by the indirect effect(s) of penicillin. Therefore, the inhibition of amidation in glutamate residues by fmtA inactivation may be an indirect effect, and the precise function of FmtA remains to be elucidated.
Analysis of the fmtA promoter demonstrated that the transcriptional level of fmtA increased when the cells were exposed to β-lactam antibiotics, especially high concentrations of β-lactam antibiotics (Table 3). Immunoblotting analysis also confirmed that the amount of FmtA protein was enhanced by β-lactam antibiotics (data not shown). It has been reported that the expression of PBP 2′ and β-lactamases is regulated by mecR1-mecI and blaR1-blaI, respectively (8, 22, 23). These genes are located just upstream of mecA or blaZ and induce the expression of mecA or blaZ products in the presence of β-lactam antibiotics. There is no corresponding region like mecR1-mecI or blaR1-blaI upstream of fmtA. Transcription of fmtA was not induced with a low concentration (1 μg/ml) of oxacillin, so FmtA may be involved in the expression of high-level methicillin resistance. Enhancement of FmtA expression was also found in the presence of certain non-β-lactam cell wall inhibitors, especially fosfomycin, so this enhancement is not specific for β-lactam antibiotics but is specific for at least several cell wall synthesis inhibitors. These results imply that the induction of fmtA transcripts is mediated by an unknown system(s) other than mecR1-mecI or blaR1-blaI. Further biochemical research will be needed to elucidate the function of FmtA and the mechanism of its regulation.
ACKNOWLEDGMENTS
We thank Chia Y. Lee, Kansas Medical Center of Kansas University, for technical advice. Technical support by Karsten Servan during HPLC muropeptide analysis and advice by Kerstin Ehlert are gratefully acknowledged.
This work was supported by a grant-in-aid for Encouragement of Young Scientists (grant 10770119) from the Ministry of Education, Science, Sports and Culture of Japan and Health Sciences Research Grants for Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Berger-Bächi B, Strässle A, Gustafson J E, Kayser F H. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:1367–1373. doi: 10.1128/aac.36.7.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 3.de Lencastre H, de Jonge B L M, Matthews P R, Tomasz A. Molecular aspects of methicillin resistance in Staphylococcus aureus. J Antimicrob Chemother. 1994;33:7–24. doi: 10.1093/jac/33.1.7. [DOI] [PubMed] [Google Scholar]
- 4.de Lencastre H, Tomasz A. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2590–2598. doi: 10.1128/aac.38.11.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontana R. Penicillin-binding proteins and the intrinsic resistance to beta-lactams in gram-positive cocci. J Antimicrob Chemother. 1985;16:412–416. doi: 10.1093/jac/16.4.412. [DOI] [PubMed] [Google Scholar]
- 6.Gustafson J, Strässle A, Hächler H, Kayser F H, Berger-Bächi B. The femC locus of Staphylococcus aureus required for methicillin resistance includes the glutamine synthetase operon. J Bacteriol. 1994;176:1460–1467. doi: 10.1128/jb.176.5.1460-1467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henze Y, Sidow T, Wecke J, Labischinski H, Berger-Bächi B. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J Bacteriol. 1993;175:1612–1620. doi: 10.1128/jb.175.6.1612-1620.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiramatsu K, Suzuki E, Takayama H, Katayama T, Yokota T. Role of penicillinase plasmids in the stability of the mecA gene in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1990;34:600–604. doi: 10.1128/aac.34.4.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komatsuzawa H, Sugai M, Ohta K, Fujiwara T, Nakashima S, Suzuki J, Lee C Y, Suginaka H. Cloning and characterization of the fmt gene which affects the methicillin resistance level and autolysis in the presence of Triton X-100 in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:2355–2361. doi: 10.1128/aac.41.11.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C Y, Buranen S L, Ye Z-H. Construction of single-copy integration vector for Staphylococcus aureus. Gene. 1991;103:101–105. doi: 10.1016/0378-1119(91)90399-v. [DOI] [PubMed] [Google Scholar]
- 11.Maki H, Yamaguchi T, Murakami K. Cloning and characterization of a gene affecting the methicillin resistance level and the autolysis rate in Staphylococcus aureus. J Bacteriol. 1994;176:4993–5000. doi: 10.1128/jb.176.16.4993-5000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakel M, Ghuysen J-M, Kandler O. Wall peptidoglycan in Aerococcus viridans strains 201 Evans and ATCC 11563 and in Gaffkya homari ATCC 10400. Biochemistry. 1971;10:2170–2175. doi: 10.1021/bi00787a033. [DOI] [PubMed] [Google Scholar]
- 13.Ornelas-Soares A, de Lencastre H, de Jonge B L M, Tomasz A. Reduced methicillin resistance in a new Staphylococcus aureus transposon mutant that incorporates muramyl dipeptides into the cell wall peptidoglycan. J Biol Chem. 1994;269:27246–27250. [PubMed] [Google Scholar]
- 14.Reynolds P E, Fuller C. Methicillin resistant strains of Staphylococcus aureus; presence of an identical additional penicillin-binding protein in all strains examined. FEMS Microbiol Lett. 1986;33:251–254. [Google Scholar]
- 15.Roos M, Pittenauer E, Schmid E, Beyer M, Reinike B, Allmaier G, Labischinski H. Improved high-performance liquid chromatographic separation of peptidoglycan isolated from various Staphylococcus aureus strains for mass spectrometric characterization. J Chromatogr B. 1998;705:183–192. doi: 10.1016/s0378-4347(97)00506-9. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Stranden A, Roos M, Berger-Bächi B. Glutamine synthetase and heteroresistance in methicillin-resistant Staphylococcus aureus. Microb Drug Resist. 1996;2:201–207. doi: 10.1089/mdr.1996.2.201. [DOI] [PubMed] [Google Scholar]
- 18.Stranden A M, Ehlert K, Labischinski H, Berger-Bächi B. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J Bacteriol. 1997;179:9–16. doi: 10.1128/jb.179.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subrata S, Jiwen S, Lee C Y. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J Bacteriol. 1997;179:1614–1621. doi: 10.1128/jb.179.5.1614-1621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugai M, Akiyama T, Komatsuzawa H, Miyake Y, Suginaka H. Characterization of sodium dodecyl sulfate-stable Staphylococcus aureus bacteriolytic enzymes by polyacrylamide gel electrophoresis. J Bacteriol. 1990;172:6494–6498. doi: 10.1128/jb.172.11.6494-6498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugai M, Kawamoto T, Pérès S Y, Ueno Y, Komatsuzawa H, Fujiwara T, Kurihara H, Suginaka H, Oswald E. The cell cycle-specific growth-inhibitory factor produced by Actinobacillus actinomycetemcomitans is a cytolethal distending toxin. Infect Immun. 1998;66:5008–5019. doi: 10.1128/iai.66.10.5008-5019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki E, Kuwahara-Arai K, Richardson J F, Hiramatsu K. Distribution of mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicrob Agents Chemother. 1993;37:1219–1226. doi: 10.1128/aac.37.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tesch W, Ryffel C, Strässle A, Kayser F H, Berger-Bächi B. Evidence of a novel staphylococcal mec-encoded element (mecR) controlling expression of penicillin-binding protein 2′. Antimicrob Agents Chemother. 1990;34:1703–1706. doi: 10.1128/aac.34.9.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tipper D J, Strominger J L. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-d-alanine. Proc Natl Acad Sci USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S, de Lencastre H, Sali A, Tomasz A. A phosphoglucomutase-like gene essential for the optimal expression of methicillin resistance in Staphylococcus aureus: molecular cloning and DNA sequencing. Microb Drug Resist. 1996;2:277–286. doi: 10.1089/mdr.1996.2.277. [DOI] [PubMed] [Google Scholar]
- 26.Wu S, de Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zukowski M M, Gaffney D F, Speck D, Kauffmann M, Findeli A, Wisecup A, Lecocq J P. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci USA. 1983;80:1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]