Summary
Background
Accurate prediction of treatment response to neoadjuvant chemotherapy (NACT) in individual patients with locally advanced gastric cancer (LAGC) is essential for personalized medicine. We aimed to develop and validate a deep learning radiomics nomogram (DLRN) based on pretreatment contrast-enhanced computed tomography (CT) images and clinical features to predict the response to NACT in patients with LAGC.
Methods
719 patients with LAGC were retrospectively recruited from four Chinese hospitals between Dec 1st, 2014 and Nov 30th, 2020. The training cohort and internal validation cohort (IVC), comprising 243 and 103 patients, respectively, were randomly selected from center I; the external validation cohort1 (EVC1) comprised 207 patients from center II; and EVC2 comprised 166 patients from another two hospitals. Two imaging signatures, reflecting the phenotypes of the deep learning and handcrafted radiomics features, were constructed from the pretreatment portal venous-phase CT images. A four-step procedure, including reproducibility evaluation, the univariable analysis, the LASSO method, and the multivariable logistic regression analysis, was applied for feature selection and signature building. The integrated DLRN was then developed for the added value of the imaging signatures to independent clinicopathological factors for predicting the response to NACT. The prediction performance was assessed with respect to discrimination, calibration, and clinical usefulness. Kaplan-Meier survival curves based on the DLRN were used to estimate the disease-free survival (DFS) in the follow-up cohort (n = 300).
Findings
The DLRN showed satisfactory discrimination of good response to NACT and yielded the areas under the receiver operating curve (AUCs) of 0.829 (95% CI, 0.739–0.920), 0.804 (95% CI, 0.732–0.877), and 0.827 (95% CI, 0.755–0.900) in the internal and two external validation cohorts, respectively, with good calibration in all cohorts (p > 0.05). Furthermore, the DLRN performed significantly better than the clinical model (p < 0.001). Decision curve analysis confirmed that the DLRN was clinically useful. Besides, DLRN was significantly associated with the DFS of patients with LAGC (p < 0.05).
Interpretation
A deep learning-based radiomics nomogram exhibited a promising performance for predicting therapeutic response and clinical outcomes in patients with LAGC, which could provide valuable information for individualized treatment.
Keywords: Deep learning, Radiomics nomogram, Locally advanced gastric cancer, Neoadjuvant chemotherapy
Abbreviations: LAGC, locally advanced gastric cancer; NACT, neoadjuvant chemotherapy; CT, computed tomography; DLRN, deep learning radiomics nomogram; TRG, tumor regression grade; GR, good response; PR, poor response; ROI, regions of interest; LASSO, least absolute shrinkage and selection operator; ICC, interclass correlation coefficient; ROC, Receiver operating characteristic; NRI, Net reclassification index; IDI, integrated discrimination improvement; AIC, Akaike information criterion; DCA, decision curve analysis; DFS, disease free survival
Research in context.
Evidence before this study
We searched PubMed and Web of Science with the terms “(radiomics OR deep learning) AND (predict OR prediction) AND (response OR non-response) AND neoadjuvant chemotherapy AND gastric cancer” for papers published from database inception to Dec 31, 2021, with no language restrictions. We found seven original studies that applied radiomics analysis to predict response to neoadjuvant chemotherapy, displaying moderate performance with areas under the receiver operating curve of 0.70-0.75.
Added value of this study
To our knowledge, no report has investigated the potential benefits of combining deep learning and radiomics to enhance prediction performance. Our deep learning radiomics model, combining clinical factors with deep learning and radiomics features, outperformed the clinical model by discrimination and calibration in the large multicenter cohorts. Additionally, higher deep learning radiomics nomogram scores were significantly associated with improved disease-free survival, providing useful complementary information for individualized treatment.
Implications of all the available evidence
Our findings show that the deep learning radiomics model has a better predictive performance of therapeutic response and prognosis before treatment than the clinical model in patients with locally advanced gastric cancer, which could provide a novel tool for guiding neoadjuvant therapy and individualized treatment strategies. Future prospective multicenter studies with larger cohorts, ideally randomized controlled trials, are needed to validate our findings.
Alt-text: Unlabelled box
Introduction
Gastric cancer (GC) remains the fourth leading cause of cancer-related deaths worldwide and one of the most common malignancies in Asia.1 The majority of patients are diagnosed at an advanced stage with a poor prognosis, the 5-year overall survival rate is 30–40% after curative resection.2 Relapse-related death remains a challenge for curative treatment. Currently, several strategies have evolved to improve survival. Specifically, neoadjuvant chemotherapy (NACT) has demonstrated favorable results with improved R0 resection rate and prognosis in patients with local advanced GC (LAGC).3,4 Nevertheless, not all patients benefit from neoadjuvant therapies, due to the nature of tumor heterogeneity.5 Clinically, histopathological examination is still the gold standard method for response evaluation; however, it is only accessible after surgery, delaying the timely adjustment of therapy. Therefore, a reliable approach to early and individual predictive response is an urgent requisite for personalized treatment for patients with LAGC.
Several studies have proposed methods to identify good responders to NACT and found that some clinicopathological and molecular biomarkers, including tumor location, tumor differentiation, tumor-infiltrating lymphocytes (TILs), and death-associated protein-3 (DAP-3), are valuable for curative prediction.5, 6, 7, 8 However, none provide quantifiable risk measures, and the accuracy is limited. By contrast, some quantitative features reflective of tumor pathophysiological or metabolic characteristics, derived from different imaging modalities, such as low-dose computed tomography (CT) perfusion imaging and diffusion kurtosis imaging (DKI), had been shown to be associated with the response towards NACT.9, 10, 11, 12 However, these methods exhibit technical differences or may focus on the mean values, discarding a wealth of information on spatial tumor heterogeneity.
Recently, radiomics—converting medical images into mineable high-throughput quantitative features—has garnered increasing attention.13,14 Emerging evidence has confirmed that radiomics is an innovative strategy for tumor diagnosis and prediction of GC.15, 16, 17 Indeed, several radiomics models have been established to predict response to NACT in patients with LAGC.18, 19, 20 However, the clinical utility of these models is limited by the potential risks of overfitting due to small sample sizes and lack of independent validation. Moreover, the introduction of deep learning (DL) features enables radiomics to obtain intricate structures related to specific tasks, which showed excellent performance in tumor characterization and prognostic prediction in terms of esophageal, gastric, rectal and nasopharyngeal cancers.21, 22, 23, 24 To the best of our knowledge, to date, no study has included the association of deep learning radiomics to the therapeutic response prediction of NACT in patients with LAGC.
Therefore, we aimed to develop and validate a deep learning radiomics nomogram (DLRN) for early prediction of good response prior to the administration of NACT in a large-scale multicenter patient cohort. We also explored the ability of DLRN to predict prognosis in a subset of patients as the secondary goal of this study.
Methods
Patients
Ethics approvals were obtained from the Institutional Review Board of all participating centers, and the requirement for informed consent was waived due to the retrospective nature of this multicenter study.
A total of 719 patients with histologically confirmed GC at a locally advanced stage (cT2-4N0/+M0) who received NACT followed by gastrectomy from four independent hospitals in China were recruited. The detailed inclusion/exclusion criteria and enrolment process are illustrated in Appendix E1 and Fig. S1.
Subsequently, 346 patients with LAGC between Jan 1st, 2017 and Nov 30th, 2020 from center I were reviewed and randomly divided into a training cohort (TC, n = 243) and an internal validation cohort (IVC, n = 103) at a ratio of 7:3. Furthermore, 207 patients from center II (Yunnan Cancer Hospital) between Dec 31st, 2014 and Aug 31st, 2020 were collected as external validation cohort 1 (EVC1). We also curated an external validation cohort 2 (EVC2) comprising 166 patients from center III (The Sixth Affiliated Hospital of Sun Yat-sen University, n = 130) and center IV (Sichuan Provincial Cancer Hospital, n = 36) between Jan 1st,2015 and April 30th, 2020. The sample size estimation is shown in Appendix E2.
All baseline clinical characteristics, including age, sex, body mass index (BMI), tumor differentiation, CEA, CA199, and clinical T(cT) and N(cN) stages, according to the 8th AJCC TNM staging system,25 were retrieved from medical records (Table S1). In addition, a follow-up cohort (n = 300) was used for survival analysis in LAGC patients (Appendix E3).
Neoadjuvant chemotherapy protocols and response assessment
All enrolled patients received 2–4 cycles of NACT before surgery (i.e., SOX regimen: Oxaliplatin 130 mg/m2, intravenous drip, on day 1; and S-1 80 mg/m2, orally, on days 1–14; repeated every 21 days), and gastrectomy was performed within 2 weeks after NACT. The response to NACT was assessed by consensus of two subspecialty pathologists with 20 years and 10 years of experience in gastrointestinal cancer, respectively, who were blinded to the study data. The tumor regression grade (TRG) criteria were selected according to the recent National Comprehensive Cancer Network (NCCN) guideline (version 4, 2021).26 Briefly, the lack of any viable cancer cell in the primary lesion or lymph nodes was defined as complete response (TRG 0), while TRG 3 was defined as no evident tumor regression with extensive residual cancer. The presence of single cells or rare small groups of cancer cells was classified TRG 1, and evident tumor regression but more than single cells or rare small groups cells was defined as TRG 2. Furthermore, patients with TRGs 0-1 were considered good response (GR), while those with TRGs 2–3 were defined as poor response (PR) (Fig. S2).
CT examination and image preprocessing
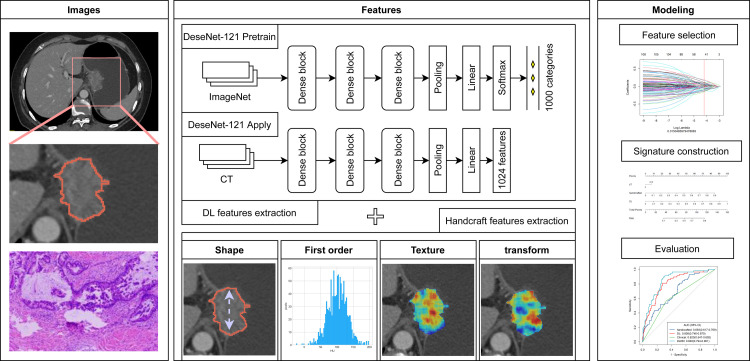
Figure 1 shows the workflow of this study. All patients underwent enhanced CT examination within two weeks before NACT. Portal venous-phase CT images were retrieved from Picture Archiving and Communication Systems (PACS) for further evaluation. Details regarding the CT image acquisition settings and image processing are shown in Appendix E4 and Table S2. Tumor regions of interest (ROIs) were manually delineated by reader 1 (K.W., with 8 years of experience in abdominal CT interpretation) on the central slice of CT images with the largest tumor, according to relevant literature,27 using the MITK software (version 2013.12.0; http://www.mitk.org/). After one month, 50 patients were randomly chosen and segmented again by readers 1 and 2 (Z.L., with 15 years of experience in abdominal CT interpretation) to evaluate the inter-/intra-observer reproducibility of radiomics features.
Figure 1.
Workflow of the study. Workflow of deep learning radiomics nomogram (DLRN) modeling for good response (GR) prediction in patients with locally advanced gastric cancer (LAGC). CT, computed tomography.
DL radiomics feature extraction
A total of 1125 handcrafted features and 1024 DL features were extracted from each ROI based on portal venous-phase CT images to quantify the tumor phenotype (Appendix E5). Herein, we adapted the DenseNet-121 architecture to extract DL features.28 The handcrafted features included shape, first order statistics, texture, and transformation features.
Deep learning and handcrafted signature building
As shown in Appendix E6, feature selection and signature building were performed in TC according to the following steps: (i) intra-/interclass correlation coefficients (ICCs) were calculated to explore the feature reproducibility; (ii) reservation of features with p < 0.01 using the univariable analysis, independent t-test or Mann–Whitney U test, as appropriate; (iii) the least absolute shrinkage and selection operator (LASSO) method was used to select the significant features; (iv) signature building with multivariable logistic regression analysis. Consequently, two types of signatures, handcrafted and DL, reflecting the different phenotypic characteristics of the tumors, were built as the predictors of GR status. The signatures combining the DL and handcrafted features were also constructed.
DLRN construction
In the TC, univariable analysis was carried out to select statistically significant clinicopathological variables (p < 0.05). Multivariable logistic regression was conducted to build the DLRN by combining the handcraft and DL signatures, as well as the significant clinicopathological factors. Additionally, a clinical model containing only clinicopathological variables was built for comparison.
Performance evaluation
The performances of all established models were measured using the receiver operator characteristic (ROC) analysis, and the area under the ROC curve (AUC) was calculated and compared among the cohorts using the DeLong test. Also, the predictive accuracy, sensitivity, and specificity were measured. The calibration curve was plotted to assess the calibration of all the models in the training and validation cohorts via bootstrapping with 1000 resamples, accompanied by the Hosmer-Lemeshow goodness-of-fit test.
Net reclassification index (NRI) and integrated discrimination improvement (IDI) were calculated to compare the performance between DLRN and the clinical model. The confusion matrix of DLRN was also depicted. Additionally, stratified analyses were performed on all patients’ clinicopathological characteristics and CT protocol. The clinical usefulness of the models was evaluated with decision curve analysis (DCA) by quantifying the net benefit at various threshold probabilities. Besides, the association between DLRN score and disease-free survival (DFS) was evaluated in the follow-up cohort using Kaplan-Meier curve.
Statistical analysis
The differences in the clinicopathological characteristics between patients in different groups or cohorts were compared using the independent t-test or Mann–Whitney U test for continuous variables, and Fisher's exact test or chi-Squared test for categorical variables, as appropriate. The DFS probabilities were evaluated by Kaplan-Meier survival analysis and the log-rank test. The optimal cutoff value was determined using the maximally selected rank statistics method, and patients were classified into high-risk or low-risk groups. The univariable and multivariable analyses with Cox proportional hazards regression, with backward stepwise elimination and Akaike information criteria (AIC), were used to construct the models for DFS prediction. The proportional hazards assumption of models was verified by examining the scaled Schoenfeld residual test. All statistical analyses were performed using R software version (version 3.6.3, http://www.R-project.org, Appendix E7). A two-sided p < 0.05 was considered statistically significant.
Role of the funding source
The funding source had no involvement in study design, data collection, data analysis, or manuscript preparation or approval. All authors had full access to all the data and approved the final manuscript for submission.
Results
Baseline information
The baseline characteristics of all 719 patients with LAGC are summarized in Tables 1 and S1. Patients in the four cohorts were balanced for the efficacy of NACT, with the GR rate of 23.9% and 23.3% for TC and IVC, and 17.9% and 24.7% for EVC1 and EVC2, respectively. No significant difference was detected in the age, BMI, sex, differentiation, pre-NACT CEA, CA199 and tumor location, as well as cN stage between the GR and PR groups in all four cohorts (p > 0.05). Moreover, cT stage showed significant differences between the two groups in TC and EVC2 (p < 0.05), and was then constructed for the clinical model.
Table 1.
Clinicopathological characteristics of patients with LAGC in the training and validation cohorts.
| Characteristics | Training cohort |
Internal Validation cohort |
External Validation cohort 1 |
External Validation cohort 2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GR (n = 58) | PR (n = 185) | P value | GR (n = 24) | PR (n = 79) | P value | GR (n = 37) | PR (n = 170) | P value | GR (n = 41) | PR (n = 125) | P value | |
| Age(y), median (IQR) | 61.0(54.3–66.0) | 60.0(53.0–65.0) | 0.581 | 61.5(52.5–65.3) | 60.0(52.5–64.0) | 0.821 | 58.0(51.0–66.0) | 55.0(48.0–63.0) | 0.115 | 61.0(53.0–67.0) | 60.0(51.0–64.0) | 0.409 |
| BMI, median (IQR) | 22.9(20.6–25.0) | 22.8(20.4–24.8) | 0.989 | 21.9(20.1–24.7) | 22.3(20.4–25.2) | 0.676 | 21.6(20.3–22.5) | 21.6(20.0–24.0) | 0.657 | 22.4(19.9–23.2) | 22.5(20.2–24.1) | 0.401 |
| Sex, No. (%) | 1.000 | 0.728 | 0.070 | 0.955 | ||||||||
| Female | 13(22.4%) | 40(21.6%) | 2(8.3%) | 11(13.9%) | 6(16.2%) | 56(32.9%) | 9(22.0%) | 30(24.0%) | ||||
| Male | 45(77.6%) | 145(78.4%) | 22(91.7%) | 68(86.1%) | 31(83.8%) | 114(67.1%) | 32(78.0%) | 95(76.0%) | ||||
| Differentiation (%) | 0.164 | 0.106 | 0.440 | 1.000 | ||||||||
| well | 1(1.7%) | 6(3.2%) | 3(12.5%) | 2(2.5%) | 0(0.0%) | 4(2.3%) | 2(4.9%) | 5(4.0%) | ||||
| Moderately | 9(15.5%) | 49(26.5%) | 8(33.3%) | 23(29.1%) | 9(24.3%) | 28(16.5%) | 11(26.8%) | 33(26.4%) | ||||
| Poorly | 48(82.8%) | 130(70.3%) | 13(54.2%) | 54(68.4%) | 28(75.7%) | 138(81.2%) | 28(68.3%) | 87(69.6%) | ||||
| Pre-NACT CEA (%) | 0.060 | 0.290 | 1.000 | 1.000 | ||||||||
| ≤5(Normal) | 41(70.7%) | 103(55.7%) | 11(45.8%) | 48(60.8%) | 18(48.6%) | 81(47.6%) | 21(51.2%) | 66(52.8%) | ||||
| >5(Abnormal) | 17(29.3%) | 82(44.3%) | 13(54.2%) | 31(39.2%) | 19(51.4%) | 89(52.4%) | 20(48.8%) | 59(47.2%) | ||||
| Pre-NACT CA199(%) | 0.325 | 0.074 | 0.716 | 0.873 | ||||||||
| ≤20(Normal) | 47(81.0%) | 136(73.5%) | 16(66.7%) | 67(84.8%) | 28(75.7%) | 136(80.0%) | 32(78.0%) | 94(75.2%) | ||||
| >20(Abnormal) | 11(19.0%) | 49(26.5%) | 8(33.3%) | 12(15.2%) | 9(24.3%) | 34(20.0%) | 9(22.0%) | 31(25.0%) | ||||
| Locations, No. (%) | 0.185 | 0.162 | 0.457 | 0.093 | ||||||||
| Cardia | 26(44.8%) | 84(45.4%) | 10(41.7%) | 44(55.7%) | 7(18.9%) | 17(10.0%) | 14(34.1%) | 41(32.8%) | ||||
| Gastric body | 11(19.0%) | 55(29.7%) | 5(20.8%) | 18(22.8%) | 11(29.7%) | 54(31.8%) | 6(14.6%) | 40(32.0%) | ||||
| Gastric antrum | 18(31.0%) | 35(18.9%) | 9(37.5%) | 13(16.5%) | 19(51.4%) | 97(57.1%) | 20(48.8%) | 40(32.0%) | ||||
| Whole stomach | 3(5.2%) | 11(5.9%) | 0(0.00%) | 4(5.1%) | 0(0.00%) | 2(1.2%) | 1(2.4%) | 4(3.20%) | ||||
| Clinical T stage (%) | 0.001* | 0.200 | 0.472 | 0.005* | ||||||||
| T2 | 0(0.0%) | 1(0.5%) | 2(8.3%) | 1(1.3%) | 0(0.0%) | 8(4.71%) | 4(9.8%) | 5(4.0%) | ||||
| T3 | 33(56.7%) | 60(32.4%) | 8(33.3%) | 29(36.7%) | 12(32.4%) | 40(23.5%) | 29(70.7%) | 64(51.2%) | ||||
| T4a | 25(43.1%) | 109(58.9%) | 13(54.2%) | 48(60.8%) | 20(54.1%) | 101(59.4%) | 4(9.8%) | 44(35.2%) | ||||
| T4b | 0(0.0%) | 15(8.1%) | 1(1.3%) | 1(4.2%) | 5(13.5%) | 21(12.4%) | 4(9.8%) | 12(9.6%) | ||||
| Clinical N stage (%) | 0.057 | 0.509 | 0.081 | 0.937 | ||||||||
| N0 | 11(19.0%) | 17(9.2%) | 4(16.7%) | 11(13.9%) | 4(10.8%) | 23(13.5%) | 3(7.3%) | 7(5.6%) | ||||
| N1 | 21(36.2%) | 51(27.6%) | 10(41.7%) | 22(27.8%) | 14(37.8%) | 30(17.6%) | 15(36.6%) | 42(33.6%) | ||||
| N2 | 16(27.6%) | 66(35.7%) | 6(25.0%) | 24(30.4%) | 9(24.3%) | 52(30.6%) | 16(39.0%) | 53(42.4%) | ||||
| N3 | 10(17.2%) | 51(27.6%) | 4(16.7%) | 22(27.8%) | 10(27.0%) | 65(38.2%) | 7(17.1%) | 23(18.4%) | ||||
Abbreviations: BMI, body mass index; NACT, neoadjuvant chemotherapy; CEA, carcinoembryonic antigen;.
NOTE: Chi-squared or Fisher's exact tests, were used to compare the differences in categorical variables, whereas student t or Mann-Whitney U test was used to compare the differences in continuous variables, as appropriate. *P < 0.05.
Radiomics and DL signatures validation
Finally, 10 and 18 features were selected to build the radiomics signature and DL signature, respectively. The detailed process and the selected features for the construction of the Rad-score are described in Appendix E8 and Table S3. As shown in Tables 2 and S4, the DL-based signature achieved better predictive performance than the corresponding handcrafted signatures, with AUCs of 0.808 (95% CI, 0.746–0.870) and 0.806 (95% CI, 0.705–0.907) in the TC and IVC, respectively. Then, the moderate performances of the DL-based signature were validated, with AUCs of 0.720 (95% CI, 0.631–0.808) and 0.734 (95% CI, 0.642–0.827) in the EVC1 and EVC2, respectively, although slightly lower than the corresponding handcrafted signatures. Furthermore, the signature combining both handcrafted features and DL features achieved an improved performance than either of them alone.
Performance and validation of DLRN
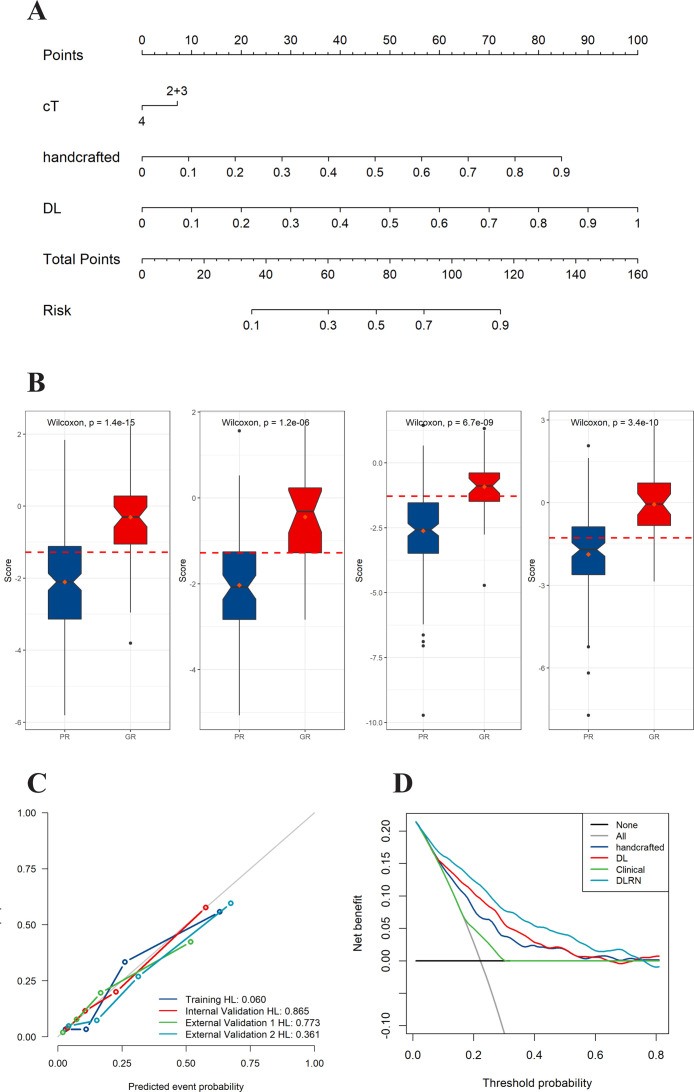
In the TC, the handcraft-based signature, DL-based signature, and cT stages were independent factors for GR prediction using backward stepwise multivariable analysis with the lowest AIC criteria (Table S5 and Fig. S3), and were then combined into DLRN (Figure 2A). These selected imaging features are weakly correlated or uncorrelated (Fig. S4).
Figure 2.
Deep learning radiomics nomogram (DLRN) and its performance. (A) DLRN with the handcrafted and deep learning signatures and clinical T stage. (B) Box plots showing patterns of correlation between therapeutic response and DLRN score for in the TC, IVC, EVC1, and EVC2, respectively. (C) Calibration curves of DLRN in all the four cohorts. (D) Decision curve analysis for DLRN, deep learning signature, handcrafted signature, and clinical model.
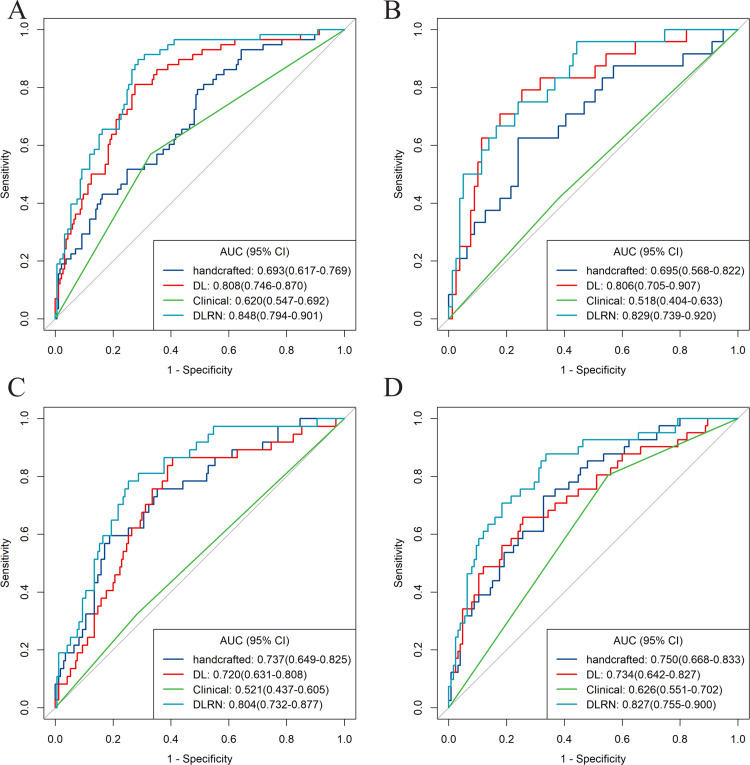
As shown in Figure 2B, there were significant differences in the DLRN scores between the GR and PR groups in all cohorts (all p < 0.001). The DLRN showed a good performance for GR prediction in the TC with an AUC of 0.848 (95% CI, 0.794–0.901), which was further confirmed in all the validation cohorts, with AUCs larger than 0.800 (Figure 3). The stratified analyses revealed that the performance of DLRN was not affected by age, sex, BMI, tumor location, the version of CT, type of CT contrast agent, contrast agent concentration, contrast agent infused rate, and slice thickness (Fig. S5). Furthermore, DLRN showed significantly higher AUCs than the clinical model in all four cohorts, which also outperformed handcrafted signature in TC, IVC, and EVC2, as well as the DL signature in EVC1 and EVC1 (p < 0.05) (Table 2 and Fig. S6).
Figure 3.
Receiver operating characteristic (ROC) curves of the four models. ROC curves of DLRN, deep learning signature, handcrafted signature, and clinical model, for predicting good responder (GR) in the (A) training cohort, (B) internal validation cohort, (C) external validation cohort 1, and (D) external validation cohort 2, respectively.
Table 2.
Performance of models.
| models | C-index (95%CI) |
||||
|---|---|---|---|---|---|
| training cohort | Internal Validation cohort | External Validation cohort 1 | External Validation cohort 2 | AIC | |
| Handcrafted signature | 0.693(0.617–0.769) | 0.695(0.568–0.822) | 0.737(0.649–0.825) | 0.750(0.668–0.833) | 698.91 |
| DL signature | 0.808(0.746–0.870) | 0.806(0.705–0.907) | 0.720(0.631–0.808) | 0.734(0.642–0.827) | 682.42 |
| Clinical model | 0.620(0.547–0.692) | 0.518(0.404–0.633) | 0.521(0.437–0.605) | 0.626(0.551–0.702) | 750.23 |
| DLRN | 0.848(0.794–0.901) | 0.829(0.739–0.920) | 0.804(0.732–0.877) | 0.827(0.755–0.900) | 585.65 |
| NRI (95%CI) | P values | ||||
| DLRN vs Clinical | 0.461(0.326–0.596) | 0.473(0.264–0.682) | 0.270(0.103–0.437) | 0.508(0.356–0.660) | <0.001 |
| IDI (95%CI) | P values | ||||
| DLRN vs Clinical | 0.240(0.180–0.301) | 0.245(0.152–0.339) | 0.191(0.121–0.261) | 0.247(0.168–0.326) | <0.001 |
Abbreviations: DL, deep learning; DLRS, deep learning radiomics signature; DLRS, deep learning radiomics nomogram.
The NRI and IDI analysis revealed that the integration of image signatures into the DLRN performed satisfactorily in all cohorts, indicating improved classification accuracy for GR prediction than the clinical model. Furthermore, the AIC values of the DLRN were lower than the corresponding clinical models or the two image signatures (Table 2). The calibration curves of the DLRN demonstrated that model-predicted GR was well-calibrated with the actual observation in all cohorts (p > 0.05) (Figure 2C). Additionally, the DCA graphically indicated that the DLRN provided a large net benefit than other models over the relevant threshold range in the whole cohorts (Figure 2D).
Preoperative predictors of survival
We also evaluated the prognostic value of DLRN in the 300 LAGC patients with follow-up data. The median follow-up period was 19 (range, 2–55) months. The LR, DM, or deaths from any cause occurred in 128 (42.7%) patients at a median follow-up period of 12 (range, 2–46) months.
Schoenfeld individual tests showed that the cox model fitted the proportional hazard assumption requirement (p > 0.05). We identified the optimal DLRN score to predict DFS as -0.313, and all patients were divided into high-risk and low-risk groups accordingly. The Kaplan–Meier curves demonstrated that higher DLRN scores were significantly associated with a better DFS (hazard ratio [HR], 0.886; 95% CI, 0.789–0.996, log-rank test, p = 0.003) (Figure 4A). Table 3 summarizes the results of univariable and multivariable Cox regression analysis of the predictors of DFS in the follow-up cohort, indicating that DLRN is an independent prognostic factor of DFS (HR, 0.500; 95% CI 0.283–0.886, p = 0.018), as well as diffuse tumors, poor differentiation and the cN3 stage before NCRT (Figure 4B). The final Cox regression model yielded a C-index of 0.693 (95% CI, 0.588–0.781).
Figure 4.
Kaplan-Meier curves and forest plot of Disease-free survival (DFS) on the follow-up LAGC cohort. (A) Kaplan–Meier curves of DFS between the groups with low and high DLRN scores in the follow-up cohort. (B) Forest plot illustrating multivariable Cox regression analyses for DFS in the follow-up cohort.
Table 3.
Uni- and multivariable cox regression analysis of predictors of disease-free survival.
| Univariable Analysis |
Multivariable Analysis |
|||
|---|---|---|---|---|
| Characteristics | Hazard ratio (95% CI) | P Value | Hazard ratio (95% CI) | P Value |
| Age | 0.999(0.981–1.017) | 0.924 | ||
| BMI | 0.971(0.921–1.022) | 0.267 | ||
| Sex (female vs. male) | 0.784(0.518–1.185) | 0.248 | ||
| Tumor differentiation | ||||
| Well and moderate | Ref | Ref | ||
| Poor | 1.809(1.182–2.769) | 0.006 | 1.740(1.129–2.680) | 0.012* |
| Pre-CRT CEA (≤5 vs. >5) | 1.235 (0.873–1.748) | 0.234 | ||
| Pre-CRT CA199(≤20 vs. >20) | 1.385(0.939–2.044) | 0.101 | ||
| Location (%) | ||||
| Cardia | Ref | Ref | ||
| Corpus | 1.473(0.960–2.261) | 0.076 | 1.132(0.731–1.753) | 0.579 |
| Antrum | 1.170(0.736–1.861) | 0.506 | 1.256(0.786–2.010) | 0.340 |
| Diffuse | 2.768(1.507–5.083) | 0.001 | 2.830(1.524–5.254) | 0.001* |
| Clinical T stage | ||||
| T2 and T3 | Ref | |||
| T4a | 1.298(0.888–1.897) | 0.177 | ||
| T4b | 2.025(1.013–4.048) | 0.046 | ||
| Clinical N stage | ||||
| 0 | Ref | Ref | ||
| 1 | 1.070(0.544–2.105) | 0.844 | 0.870(0.437–1.732) | 0.692 |
| 2 | 1.196(0.620–2.306) | 0.593 | 0.871(0.444–1.710) | 0.688 |
| 3 | 2.893(1.544–5.422) | 0.001 | 2.378(1.250–4.524) | 0.008* |
| DLRN (low vs. high) | 0.446(0.256–0.778) | 0.004 | 0.500(0.283–0.886) | 0.018* |
Discussion
In the present study, we developed a non-invasive imaging signature by DL and radiomics analysis based on pretreatment venous-phase CT images, and independently validated its ability to predict response to NACT in large, multicenter cohorts. Furthermore, the imaging signature incorporated into the DLRN model exhibited improved performance in response prediction compared to the clinical models. Notably, the DLRN was significantly associated with DFS, providing useful complementary information about prognosis in patients with LAGC.
LAGC is clinically heterogeneous, necessitating accurate treatment response and prognosis prediction for the selection of appropriate treatment. Accumulating evidence has been reported to predict the response to NACT in patients with LAGC. Some texture features, such as entropy and delta gray-level cooccurrence matrix (GLCM) contrast, were found to be able to identify responders to NACT.29,30 In terms of radiomics, previous studies have focused on the application of CT-based features prior to NACT administration, displaying moderate performance with AUCs 0.70–0.75.18, 19, 20,31 Moreover, the detection radiomics model constructed by the baseline and restaging CT could be utilized for the early detection tasks of pathological downstaging (pDS), with excellent AUCs close to 0.90 in the internal validation cohort.20 Nevertheless, the optimistic result was mainly attributed to restage CT, which provided post-treatment tumor information and could not achieve an earlier prediction to guide NACT administration. However, these findings were of limited clinical relevance, due to the relatively small sample size and lack of validation in multicenter cohorts. Relatively, the lower AUC of 0.679 in the external validation cohort for the radiomics signature proposed in a larger population (323 cases) was more convincing, indicating that the radiomics features should be combined with auxiliary features to improve the prediction performance.32 Intriguingly, all the ten selected features used in our radiomics signature were transformation factors, especially the Laplacian of Gaussian (LoG) and wavelet-based features, providing more detailed information about tumoral heterogeneity.
A DL method, based on the DenseNet-121 architecture, was applied for DL feature extraction in this study. It was worth noting that, different from handcraft features, the DL method did not require delineated delineation, which not only reduces contour variability of different manual segmentation, but also enhances efficiency. Moreover, DL provides in-depth information included specific task in neural nets’ hidden layers without predefined features. The features captured by DL algorithm could predict lymph node metastasis, occult peritoneal metastasis, or survival outcomes for resectable GCs.15,24,33 The DL signature in our study presented a promising performance in GR prediction with AUCs larger than 0.72, similar to a previous study predicting the treatment response in esophageal squamous cell carcinoma by DL features.23 Similarly, the DL prediction model outperformed the handcrafted signature and the clinical model in discrimination ability in most of cohorts. All these indicated that DL offers a wealth of information reflecting tumor spatial heterogeneity and tumor microenvironment related to tumor chemosensitivity.
Furthermore, the prediction ability of DLRN was far better than that of the clinical model in all cohorts (p < 0.05). Previous studies have pointed out that various clinical or molecular risk factors are associated with neoadjuvant response. However, these metrics were not consistent across all studies. The distribution of cT, incorporated into our clinical model, was significantly different in some rather than all cohorts. Specifically, the AUC of the clinical model was only 0.518 in IVC, which was significantly lower than that of other models, including DLRN, handcrafted, and DL signatures. Tumor differentiation or no clinical factors were associated with treatment response in other studies.19,32 Additionally, the clinical factors only reflect specific aspects of the tumor. Patients with the same above features exhibited differential responses. This may be the reason for the ill-performance of the clinical model on different patient distribution. DLRN mines high-dimensional imaging features, followed by comprehensive quantification of intratumor heterogeneity, thereby improving the performance. In the present study, DLRN was constructed with 10 radiomics features, 18 DL features, and one clinical factor. These selected imaging features were not redundant but complementary, as shown in the heatmaps that these features are weakly correlated or uncorrelated. Moreover, the lowest AIC, and improved NRI and IDI also confirmed that the enhanced discrimination performance of DLRN was indeed due to feature integration instead of model overfitting.
Another interesting finding was that our DLRN is significantly associated with the DFS of patients with LAGC. Several studies proved that LAGC patients with different NACT treatment efficacy had varied prognosis.34,35 However, improved TRG after NACT might not necessarily mean prolonged survival. In the current study, compared to patients at high risk of GR, those at low risk of GR had greatly diminished the likelihood of favorable long-term outcomes even after NACT followed by curative resection. Patients with higher DLRN scores were significantly associated with a better DFS. In the multivariable Cox regression model, DLRN was an independent prognostic factor of DFS. Since the outcomes in low DLRN scores were not satisfactory, timely alternative curative-intent treatment approaches should be offered to avoid unnecessary toxicity and improve their survival outcomes. Accordingly, it is feasible to guide the treatment plan and implement personalized treatment based on our DLRN model.
Notably, the present study has some limitations. Firstly, although the multicenter sample size was larger, inherent bias was inevitable due to the retrospective nature and different distribution. Thus, further well-designed prospective studies are warranted to validate the generalizability and clinical applicability of our DLRN model. Secondly, we delineated the tumor on the largest slice with two-dimensional(2D) rather than the entire tumor(3D), which could not be representative of the entire tumor, and some radiomics features might be affected from 2D vs 3D. Hence, 3D analysis of the whole tumor deserved further investigation. Thirdly, for the survival analysis, the relative short-term follow-up part of a subset patients might increase the risk of type II error, with more censored data. A large number of patients in a multicenter study with prolonged follow-up time, and more unperceived clinicopathological factors that simultaneously impact the prognosis of LAGC after NACT, should be investigated further. Lastly, the biological significance of DLRN, especially DL features, still needs comprehensive elucidation. Thus, future investigations integrating imaging and molecular or gene data may provide insight into more micro information and their relationship.
In summary, we developed and validated a CT-based model using deep learning and radiomics analysis for the early prediction of response prior to NACT for patients with LAGC. The proposed DLRN, incorporating imaging signature and clinical factors, exhibited a promising performance for predicting response and clinical outcomes, and provide valuable information for individualized treatment in patients with LAGC. However, future prospective studies are required to confirm the clinical utility of our DLRN model.
Contributors
YC and XY were responsible for conception and design. YC, JZ, KW, LW, and ZS provided statistical analysis. All authors were involved in drafting and technical support in deep learning methods. JR assisted in statistical analysis. YC, JZ, and YL were involved in drafting the manuscript, and XM, XY, and XG were involved in reviewing the manuscript. YC and XY had full access to all the data. YC, ZL, XM, and XY verified the underlying data. All authors approved the final manuscript for submission.
Data sharing statement
Due to the privacy of patients, the data related to patients cannot be available for public access but can be obtained from the corresponding author on reasonable request approved by the institutional review board of all enrolled centers.
Declaration of interests
JR is an employee of GE Healthcare. YC received funding from National Natural Science Foundation of China (No. 82001789), China Postdoctoral Science Foundation (No. 2021M700897), and Project of Shanxi Provincial Health Commission (No. 2021XM51 and 2019058). ZL received funding from National Natural Science Foundation of China (No. 82001986), and Applied Basic Research Projects of Yunnan Province, China, Outstanding Youth Foundation (202101AW070001). YL received funding from National Natural Science Foundation of China (No. 82002702), and Youth Project of Natural Science Foundation of Hunan Science (No. 2020JJ5905). XY received funding from National Natural Science Foundation of China (No. 82171923), and Project of Shanxi Provincial Health Commission (No. 2020064). XG received funding from National Natural Science Foundation of China (No. 81871439). All other authors declare no competing interests.
Funding
This study was supported by the National Natural Science Foundation of China (No. 82001789, 82171923, 82001986, 81871439, and 82002702), the China Postdoctoral Science Foundation (No. 2021M700897), the Project of Shanxi Provincial Health Commission (No. 2021XM51, 2020064, and 2019058), the Applied Basic Research Projects of Yunnan Province, China, Outstanding Youth Foundation (202101AW070001), and the Youth Project of Natural Science Foundation of Hunan Science (No. 2020JJ5905).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.eclinm.2022.101348.
Contributor Information
Xiaochun Meng, Email: mengxch3@mail.sysu.edu.cn.
Xiaotang Yang, Email: yangxt210@126.com.
Xin Gao, Email: xingaosan@163.com.
Appendix. Supplementary materials
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Machlowska J., Baj J., Sitarz M., Maciejewski R., Sitarz R. Gastric cancer: epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21114012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Batran S.E., Homann N., Pauligk C., et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): a randomised, phase 2/3 trial. Lancet. 2019;393:1948–1957. doi: 10.1016/S0140-6736(18)32557-1. [DOI] [PubMed] [Google Scholar]
- 4.Fazio N., Biffi R., Maibach R., et al. Preoperative versus postoperative docetaxel-cisplatin-fluorouracil (TCF) chemotherapy in locally advanced resectable gastric carcinoma: 10-year follow-up of the SAKK 43/99 phase III trial. Ann Oncol. 2016;27:668–673. doi: 10.1093/annonc/mdv620. [DOI] [PubMed] [Google Scholar]
- 5.Lorenzen S., Blank S., Lordick F., Siewert J.R., Ott K. Prediction of response and prognosis by a score including only pretherapeutic parameters in 410 neoadjuvant treated gastric cancer patients. Ann Surg Oncol. 2012;19:2119–2127. doi: 10.1245/s10434-012-2254-1. [DOI] [PubMed] [Google Scholar]
- 6.Jiang L., Ma Z., Ye X., Kang W., Yu J. Clinicopathological factors affecting the effect of neoadjuvant chemotherapy in patients with gastric cancer. World J Surg Oncol. 2021;19:44. doi: 10.1186/s12957-021-02157-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zurlo I.V., Schino M., Strippoli A., et al. Predictive value of NLR, TILs (CD4+/CD8+) and PD-L1 expression for prognosis and response to preoperative chemotherapy in gastric cancer. Cancer Immunol Immunother. 2021 doi: 10.1007/s00262-021-02960-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia Y., Ye L., Ji K., et al. Death-associated protein-3, DAP-3, correlates with preoperative chemotherapy effectiveness and prognosis of gastric cancer patients following perioperative chemotherapy and radical gastrectomy. Br J Cancer. 2014;110:421–429. doi: 10.1038/bjc.2013.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider P.M., Eshmuminov D., Rordorf T., et al. (18)FDG-PET-CT identifies histopathological non-responders after neoadjuvant chemotherapy in locally advanced gastric and cardia cancer: cohort study. BMC Cancer. 2018;18:548. doi: 10.1186/s12885-018-4477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Z., Cheng X., Ge Y., Shao L., Xuan Y., Yan G. An application study of low-dose computed tomography perfusion imaging for evaluation of the efficacy of neoadjuvant chemotherapy for advanced gastric adenocarcinoma. Gastric Cancer. 2018;21:413–420. doi: 10.1007/s10120-017-0763-0. [DOI] [PubMed] [Google Scholar]
- 11.Fu J., Tang L., Li Z.Y., et al. Diffusion kurtosis imaging in the prediction of poor responses of locally advanced gastric cancer to neoadjuvant chemotherapy. Eur J Radiol. 2020;128 doi: 10.1016/j.ejrad.2020.108974. [DOI] [PubMed] [Google Scholar]
- 12.Gao X., Zhang Y., Yuan F., et al. Locally advanced gastric cancer: total iodine uptake to predict the response of primary lesion to neoadjuvant chemotherapy. J Cancer Res Clin Oncol. 2018;144:2207–2218. doi: 10.1007/s00432-018-2728-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambin P., Rios-Velazquez E., Leijenaar R., et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambin P., Leijenaar R.T.H., Deist T.M., et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 15.Dong D., Tang L., Li Z.Y., et al. Development and validation of an individualized nomogram to identify occult peritoneal metastasis in patients with advanced gastric cancer. Ann Oncol. 2019;30:431–438. doi: 10.1093/annonc/mdz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y., Wang H., Wu J., et al. Noninvasive imaging evaluation of tumor immune microenvironment to predict outcomes in gastric cancer. Ann Oncol. 2020;31:760–768. doi: 10.1016/j.annonc.2020.03.295. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Y., Yuan Q., Lv W., et al. Radiomic signature of (18)F fluorodeoxyglucose PET/CT for prediction of gastric cancer survival and chemotherapeutic benefits. Theranostics. 2018;8:5915–5928. doi: 10.7150/thno.28018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun K.Y., Hu H.T., Chen S.L., et al. CT-based radiomics scores predict response to neoadjuvant chemotherapy and survival in patients with gastric cancer. BMC Cancer. 2020;20:468. doi: 10.1186/s12885-020-06970-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y., Wei K., Liu D., et al. A machine learning model for predicting a major response to neoadjuvant chemotherapy in advanced gastric cancer. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.675458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Q., Sun Z., Li X., et al. Advanced gastric cancer: CT radiomics prediction and early detection of downstaging with neoadjuvant chemotherapy. Eur Radiol. 2021;31:8765–8774. doi: 10.1007/s00330-021-07962-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X., Zhang D., Liu Z., et al. Deep learning radiomics-based prediction of distant metastasis in patients with locally advanced rectal cancer after neoadjuvant chemoradiotherapy: a multicentre study. EBioMedicine. 2021;69 doi: 10.1016/j.ebiom.2021.103442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng H., Dong D., Fang M.J., et al. Prognostic value of deep learning PET/CT-based radiomics: potential role for future individual induction chemotherapy in advanced nasopharyngeal carcinoma. Clin Cancer Res. 2019;25:4271–4279. doi: 10.1158/1078-0432.CCR-18-3065. [DOI] [PubMed] [Google Scholar]
- 23.Hu Y., Xie C., Yang H., et al. Computed tomography-based deep-learning prediction of neoadjuvant chemoradiotherapy treatment response in esophageal squamous cell carcinoma. Radiother Oncol. 2021;154:6–13. doi: 10.1016/j.radonc.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 24.Dong D., Fang M.J., Tang L., et al. Deep learning radiomic nomogram can predict the number of lymph node metastasis in locally advanced gastric cancer: an international multicenter study. Ann Oncol. 2020;31:912–920. doi: 10.1016/j.annonc.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Sano T., Coit D.G., Kim H.H., et al. Proposal of a new stage grouping of gastric cancer for TNM classification: international Gastric Cancer Association staging project. Gastric Cancer. 2017;20:217–225. doi: 10.1007/s10120-016-0601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ajani J.A., Bentrem D.J., Besh S., et al. Gastric cancer, version 2.2013: featured updates to the NCCN Guidelines. J Natl Compr Cancer Netw. 2013;11:531–546. doi: 10.6004/jnccn.2013.0070. https://www.nccn.org/professionals/physician_gls/pdf/gastric.pdf (NCCN National Comprehensive Cancer Network. NCCN Clinical Practice 415 Guidelines in Oncology: Gastric Cancer. Fort Washington, PA: National 416 Comprehensive Cancer Network, Version 4 (2021). 417. [2021/09/03]) [DOI] [PubMed] [Google Scholar]
- 27.Meng L., Dong D., Chen X., et al. 2D and 3D CT radiomic features performance comparison in characterization of gastric cancer: a multi-center study. IEEE J Biomed Health Inform. 2021;25:755–763. doi: 10.1109/JBHI.2020.3002805. [DOI] [PubMed] [Google Scholar]
- 28.Kermany D.S., Goldbaum M., Cai W., et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122–1131. doi: 10.1016/j.cell.2018.02.010. e1129. [DOI] [PubMed] [Google Scholar]
- 29.Giganti F., Marra P., Ambrosi A., et al. Pre-treatment MDCT-based texture analysis for therapy response prediction in gastric cancer: comparison with tumour regression grade at final histology. Eur J Radiol. 2017;90:129–137. doi: 10.1016/j.ejrad.2017.02.043. [DOI] [PubMed] [Google Scholar]
- 30.Mazzei M.A., Di Giacomo L., Bagnacci G., et al. Delta-radiomics and response to neoadjuvant treatment in locally advanced gastric cancer-a multicenter study of GIRCG (Italian Research Group for Gastric Cancer) Quant Imaging Med Surg. 2021;11:2376–2387. doi: 10.21037/qims-20-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z., Zhang D., Dai Y., et al. Computed tomography-based radiomics for prediction of neoadjuvant chemotherapy outcomes in locally advanced gastric cancer: a pilot study. Chin J Cancer Res. 2018;30:406–414. doi: 10.21147/j.issn.1000-9604.2018.04.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W., Peng Y., Feng X., et al. Development and validation of a computed tomography-based radiomics signature to predict response to neoadjuvant chemotherapy for locally advanced gastric cancer. JAMA Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.21143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang Y., Jin C., Yu H., et al. Development and validation of a deep learning CT signature to predict survival and chemotherapy benefit in gastric cancer: a multicenter, retrospective study. Ann Surg. 2020 doi: 10.1097/SLA.0000000000003778. [DOI] [PubMed] [Google Scholar]
- 34.Aoyama T., Nishikawa K., Fujitani K., et al. Early results of a randomized two-by-two factorial phase II trial comparing neoadjuvant chemotherapy with two and four courses of cisplatin/S-1 and docetaxel/cisplatin/S-1 as neoadjuvant chemotherapy for locally advanced gastric cancer. Ann Oncol. 2017;28:1876–1881. doi: 10.1093/annonc/mdx236. [DOI] [PubMed] [Google Scholar]
- 35.Ychou M., Boige V., Pignon J.P., et al. Perioperative chemotherapy compared with surgery alone for resectable gastroesophageal adenocarcinoma: an FNCLCC and FFCD multicenter phase III trial. J Clin Oncol. 2011;29:1715–1721. doi: 10.1200/JCO.2010.33.0597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.