Abstract
The occurrence of mutations in the genes coding for gyrase (gyrA and gyrB) and topoisomerase IV (parE and parC) of Salmonella typhimurium experimental mutants selected in vitro and in vivo and of 138 nalidixic acid-resistant Salmonella field isolates was investigated. The sequencing of the quinolone resistance-determining region of these genes in highly fluoroquinolone-resistant mutants (MICs of 4 to 16 μg/ml) revealed the presence of gyrA mutations at codons corresponding to Gly-81 or Ser-83, some of which were associated with a mutation at Asp-87. No mutations were found in the gyrB, parC, and parE genes. An assay combining allele-specific PCR and restriction fragment length polymorphism was developed to rapidly screen mutations at codons 81, 83, and 87 of gyrA. The MICs of ciprofloxacin for the field isolates reached only 2 μg/ml, versus 16 μg/ml for some in vitro-selected mutants. The field isolates, like the mutants selected in vivo, had only a single gyrA mutation at codon 83 or 87. Single gyrA mutations were also found in highly resistant in vitro-selected mutants (MIC of ciprofloxacin, 8 μg/ml), which indicates that mechanisms other than the unique modification of the intracellular targets could participate in fluoroquinolone resistance in Salmonella spp. A comparison of experimental mutants selected in vitro, field strains, and mutants selected in vivo suggests that highly fluoroquinolone-resistant strains are counterselected in field conditions in the absence of selective pressure.
Human nontyphoidal Salmonella infections are increasingly frequent in developed countries (39), and the emergence of fluoroquinolone-resistant Salmonella strains is a serious matter of concern since this class of antibacterial agents constitutes a treatment of choice in cases of acute salmonellosis due to multiresistant strains (1, 8, 32, 37), whose prevalence is increasing (12, 41). Only a few cases of treatment failure due to fluoroquinolone resistance in Salmonella strains (including Salmonella typhi) have been reported (17, 30, 44), but there is evidence of an increasing incidence of strains that are resistant to nalidixic acid and strains that exhibit decreased susceptibility to the most recent fluoroquinolones used in human therapeutics (5–7, 14, 15, 19, 27, 34, 35). For example, 14% of the epidemic multiresistant Salmonella typhimurium strains of phage type DT104 isolated in 1997 in the United Kingdom were highly resistant to nalidixic acid and showed a decreased susceptibility to ciprofloxacin (MIC, 0.125 to 0.5 μg/ml) (41). Although the emergence of strains with low-level resistance to fluoroquinolones during hospital therapy has been described (27), a hypothesis of the selection and spread of such quinolone-resistant strains following the recent introduction of fluoroquinolones in veterinary therapy has been proposed (29).
In Salmonella spp., like in Escherichia coli and in many other gram-negative organisms (2, 10, 11, 14, 19, 21, 27, 40, 42), quinolone resistance is conferred by point mutations in the gyrA gene coding for the A subunit of gyrase, whose complex with DNA is the primary target of quinolones. Resistance mutations of gyrA have been clustered in a region of the gene product between amino acids 67 and 106, termed the quinolone resistance-determining region (QRDR) (45). Amino acid changes at Ser-83 (to Phe, Tyr, or Ala) or at Asp-87 (to Gly, Asn, or Tyr) are the most frequently observed in nalidixic acid-resistant strains. Double mutations at both residues 83 and 87 have been identified in fluoroquinolone-resistant clinical isolates of E. coli and Salmonella spp. Alterations at residue Gly-81 (to Ser or Cys) have also been identified in low-level quinolone-resistant spontaneous mutants of E. coli and S. typhimurium (33, 45). A QRDR was also identified in the gyrB gene of E. coli, where mutations can cause reductions in quinolone susceptibility but to a lesser extent than the common gyrA mutations (46). In S. typhimurium, one gyrB mutation associated with quinolone resistance was found (13), and in another study, complementation tests suggested a combination of gyrA and gyrB mutations in a highly resistant isolate (17). However, the contribution of gyrB mutations to quinolone resistance is still unclear. In gram-negative bacteria, topoisomerase IV, whose ParC and ParE subunits are homologous to GyrA and GyrB, respectively, is a secondary target for quinolones (20). Mutations in the genes parC and parE at positions equivalent to those identified in gyrA and gyrB participate in high-level resistance to quinolones (4, 18, 43).
The goals of this study were (i) to investigate the sequential selection, under experimentally controlled selective pressure conditions, of quinolone resistance mutations in the genes coding for the gyrase and topoisomerase IV of S. typhimurium and (ii) to evaluate the incidence of these mutations in field isolates. For that purpose, experimental quinolone-resistant mutants were obtained in vitro and in vivo under the selective pressure of enrofloxacin, a new generation fluoroquinolone used for animal therapy. As the systematic sequencing of the gyrase and topoisomerase IV genes of numerous isolates is not suitable, we used these experimental mutants to develop a rapid assay combining allele-specific PCR (22, 26) and restriction fragment length polymorphism (AS-PCR-RFLP) for the screening of point mutations responsible for all amino acid changes encoded by the gyrA gene at codons 83 and 87 and previously described amino acid changes at position 81. Then we used this assay to screen the gyrA mutations of all the nalidixic acid-resistant Salmonella strains of animal origin recently isolated from cattle or poultry in major production areas and obtained from the two French national networks of surveillance (5, 24, 25).
MATERIALS AND METHODS
Field isolates.
A total of 138 Salmonella strains resistant to nalidixic acid and isolated between 1985 and 1997 (116 in 1996 and 1997) were received from the two French national networks of surveillance, the SALMONELLA network and the RESABO network (5, 25). Various serotypes were represented: 71 S. typhimurium, 22 S. hadar, 13 S. kottbus, 11 S. newport, and 9 S. virchow strains, 6 Salmonella spp., and one strain each of S. anatum, S. montevideo, S. panama, S. regent, S. saintpaul, and S. senftenberg. The origins of the strains were documented and precautions were taken to avoid duplicate samples. These strains were collected from 29 different geographic regions; 74 strains were from poultry flocks (chickens, turkeys, and ducks), 56 were from bovine herds, and the remainder were from diverse origins (rabbit, horse, sheep, and farm environments).
Selection of experimental quinolone-resistant mutants. (i) In vitro selection.
Spontaneous quinolone-resistant mutants were obtained by stepwise selection from two susceptible S. typhimurium strains: BN82, isolated from a sick calf, and BN18, isolated from a septicemic pigeon. At the first step, 100 μl of overnight cultures of the susceptible strains, grown in a brain heart infusion medium, were plated on Mueller-Hinton agar medium supplemented with enrofloxacin (0.1 μg/ml; Bayer AG, Leverkusen, Germany). Resistant colonies of each strain were collected, one of which was restained for the next step. Further steps with increasing concentrations of enrofloxacin in the Mueller-Hinton agar were conducted similarly. Seven and eight selection steps were achieved with the initial strains, BN82 and BN18, respectively, up to a final selecting concentration of enrofloxacin of 75 μg/ml. MICs were determined after two subcultures in liquid and solid media without drugs.
(ii) In vivo selection.
Two groups of 20 chickens hatched under sterile conditions were reared in two germfree isolators. On day 6, the axenic birds of both groups were given about 106 CFU of strain BN82 or strain BN18 per os. During this 2-month experiment, the birds had free access to sterile feed and water. Four 5-day treatments of enrofloxacin (Baytril; Bayer AG) added to the drinking water were administered to the two groups of birds; these treatments alternated with periods of 10 days without treatment. Doses of enrofloxacin added to the drinking water were successively 1/8, 1/4, 1/2, and 1/1 of the recommended therapeutic dose (6.25, 12.5, 25, and 50 μg/ml, respectively). Samples of feces were taken weekly from each bird, diluted in sterile water, and plated on Mueller-Hinton agar supplemented with 0.5, 1, 2, 4, 6, and 8 μg of enrofloxacin per ml. After an overnight culture, resistant colonies were collected.
Susceptibility tests.
For all the strains, antibiotic susceptibilities were first evaluated by the disk diffusion method with disks containing nalidixic acid (30 μg; Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France), flumequine (30 μg; Sanofi Diagnostics Pasteur), enrofloxacin (5 μg, Difco Laboratories, Detroit, Mich.), and ciprofloxacin (5 μg, Sanofi Diagnostics Pasteur). For all the field isolates, all the in vitro-selected mutants and for the in vivo-selected mutants exhibiting the highest resistance levels MICs of nalidixic acid (Sigma, Steinheim, Germany), flumequine (Sigma), enrofloxacin, and ciprofloxacin (Bayer AG) were determined by the agar doubling dilution method with solid Mueller-Hinton medium and inocula of 104 CFU per spot. The MIC was defined as the lowest concentration of the drug that completely inhibited visible growth after incubation for 18 h at 37°C. The following MIC breakpoints (c and C, in micrograms per milliliter), defined by the Comité de l’Antibiogramme de la Société Française de Microbiologie (CASFM) (9), were used to classify strains as susceptible (MIC ≤ c), intermediate (c < MIC ≤ C), or resistant (MIC > C): nalidixic acid, 8 and 16; flumequine, 4 and 8; and ciprofloxacin, 1 and 2. For enrofloxacin, Bayer’s recommendations were 0.5 and 2.
Amplification and sequencing of QRDR regions of gyrA, gyrB, parC, and parE genes.
The sequences of all the primers used in the PCR amplifications of gyrA (STGYRA1 and STGYRA12), gyrB (STGYRB5 and STGYRB6), parC (STPARC1 and STPARC2), and parE (STPARE1 and STPARE2) are given in Table 1. Template DNA was prepared as follows. Strains were grown overnight at 37°C with shaking in 5 ml of brain heart infusion. Amounts of 1.5 ml of culture were pelleted, and cells were boiled in 200 μl of H2O. After centrifugation, the supernatants were kept at −20°C. PCR was performed in a total volume of 25 μl, which contained 5 μl of supernatant, 25 pmol of each primer, 200 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, and 0.5 U of Taq polymerase (Promega, Madison, Wis.). After an initial denaturation step of 3 min at 94°C, amplification was performed over 30 cycles, each one consisting of 1 min at 94°C, 1 min at hybridization temperature (55°C for gyrA, 58°C for gyrB, and 52°C for parC and parE), and 1 min at 72°C, with a final extension step of 10 min at 72°C. Primers and free nucleotides were removed with a Qiaquick spin PCR purification kit (Qiagen S.A., Courtaboeuf, France). Sequences were determined by the method of Sanger et al. (36) in an automatic DNA sequencer (Perkin-Elmer Applied Biosystem 373A) with primers STGYRA1, STGYRB7, STPARC1, and STPARE1 for gyrA, gyrB, parC, and parE fragments, respectively (Table 1). The sequences of the QRDRs were determined between amino acids 54 and 171 of GyrA, 397 and 520 of GyrB, 12 and 130 of ParC, and 421 and 524 of ParE.
TABLE 1.
Primers used for PCR amplifications and sequencing
| Primer | Sequence |
|---|---|
| STGYRA1 | 5′-TGTCCGAGATGGCCTGAAGC-3′ |
| STGYRA12 | 5′-CGTTGATGACTTCCGTCAG-3′ |
| STGYRB5 | 5′-AAGCGCGATGGCAAAGAAG-3′ |
| STGYRB6 | 5′-AACGGTCTGCTCATCAGAAAGG-3′ |
| STGYRB7 | 5′-GAAATGACCCGCCGTAAAGG-3′ |
| STPARC1 | 5′-ATGAGCGATATGGCAGAGCG-3′ |
| STPARC2 | 5′-TGACCGAGTTCGCTTAACAG-3′ |
| STPARE1 | 5′-GACCGAGCTGTTCCTTGTGG-3′ |
| STPARE2 | 5′-GCGTAACTGCATCGGGTTCA-3′ |
| STGYRA-HinfI/87 | 5′-ATGTAACGCAGCGAGAATGGCTGCGCCATACGAACGATGGAG-3′ |
| AS-81 | 5′-GGTAAATACCATCCCCACG-3′ |
Detection of gyrA mutations by AS-PCR-RFLP assay.
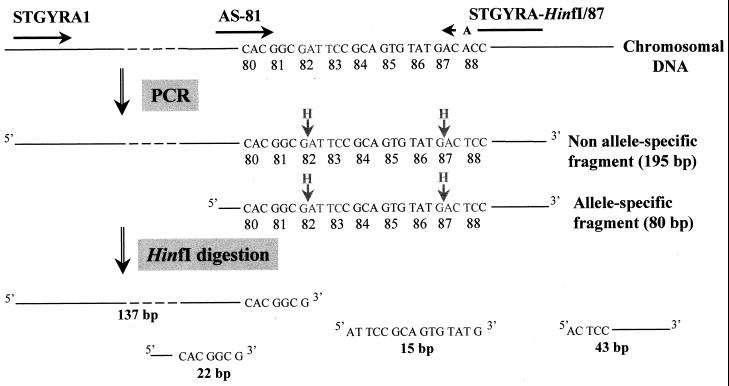
We developed the AS-PCR-RFLP assay to detect common mutations related to quinolone resistance at codons 81, 83, and 87 of the gyrA gene (Fig. 1). A PCR amplification was performed with three primers. The forward primer STGYRA1 and the reverse primer STGYRA-HinfI/87 (Table 1) were expected to produce a 195-bp fragment with a HinfI restriction site at the codon corresponding to Ser-83. As previously described for E. coli (28), the reverse primer STGYRA-HinfI/87, whose sequence is different by one base from the gene sequence, introduced an artificial HinfI cleavage site including the Asp-87 codon according to the primer-specified restriction site modification method (16). A second allele-specific forward primer, AS-81, whose 3′-terminal nucleotide corresponds to the first nucleotide of codon 81, permitted the amplification of an 80-bp fragment only in the absence of this nucleotide, a fragment that also contained both the natural and the artificial HinfI cleavage sites. PCR was performed as described for the amplification of the sequenced fragments, with 25 pmol of primers STGYRA1 and AS-81, 50 pmol of primer STGYRA-HinfI/87, and a hybridization temperature of 57°C.
FIG. 1.
Schematic diagram of AS-PCR-RFLP assay of a strain with no mutation at codon 81, 83, or 87.
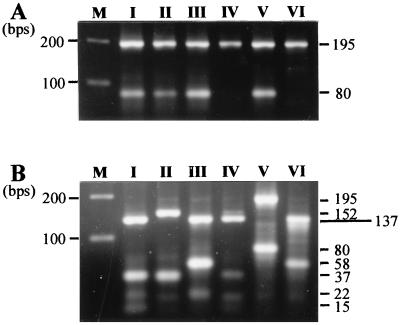
Restriction enzyme digestions were performed in a 7-μl mixture containing 5 μl of the PCR product and 5 U of HinfI. Both digested and undigested PCR products were resolved in a single 3% (wt/vol) agarose gel in 1× Tris-borate-EDTA at 120 V for 2 to 3 h. A 100-bp DNA ladder was used as a molecular marker. Table 2 presents the theoretical fragments generated by AS-PCR-RFLP for six representative strains whose gyrA QRDR sequencing revealed the absence of mutation (strain BN82) or the presence of different combinations of mutations. The corresponding profiles are shown in Fig. 2. These strains were used as controls for the identification of the gyrA genotypes of the tested strains.
TABLE 2.
MICs for strains, nucleotide changes in the gyrA QRDR, inferred amino acid substitutions, and corresponding AS-PCR-RFLP assay results for six representative strains
| Strains | MIC (μg/ml)a
|
gyrA sequencing results (amino acid) at codonb:
|
AS-PCR-RFLP assay results
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calculated fragment sizes (bp)
|
Pattern | |||||||||||||||
| Nal | Flu | Enr | Cip | 80 | 81 | 82 | 83 | 84 | 85 | 86 | 87 | 88 | Before HinfI digestion | After HinfI digestion | ||
| BN82 | 4 | 1 | 0.06 | 0.03 | CAC (His) | GGC (Gly) | GAT (Asp) | TCC (Ser) | GCA (Ala) | GTG (Val) | TAT (Tyr) | GAC (Asp) | ACC (Thr) | 195, 80 | 137, 43, 22, 15 | I |
| BN99 | >1,024 | 32 | 2 | 0.5 | --- | --- | --- | -T- (Phe) | --- | --- | --- | --- | --- | 195, 80 | 152, 43,c 37c | II |
| BNJ18 | >1,024 | 64 | 4 | 0.5 | --- | --- | --- | --- | --- | --- | --- | A-- (Asn) | --- | 195, 80 | 137, 58, 22 | III |
| BN18/42 | >1,024 | 256 | 8 | 2 | --- | T-- (Cys) | --- | --- | --- | --- | --- | --- | --- | 195 | 137, 43, 15 | IV |
| BN82/61 | >1,024 | 256 | 32 | 8 | --- | --- | --- | -T- (Phe) | --- | --- | --- | -G- (Gly) | --- | 195, 80 | 195, 80 | V |
| BN18/61 | >1,024 | 512 | 32 | 4 | --- | T-- (Cys) | --- | --- | --- | --- | --- | -G- (Gly) | 195 | 137, 58 | VI | |
Nal, nalidixic acid; Flu, flumequine; Enr, enrofloxacin; Cip, ciprofloxacin.
Dashes indicate identity to BN82.
43- and 37-bp fragments were not separated at current electrophoresis times.
FIG. 2.
AS-PCR-RFLP patterns of six representative strains before (A) and after (B) HinfI digestion. See Table 2 for theoretical fragment sizes according to the gyrA sequence. Lane M, molecular weight marker.
Detection of parC mutations at codon 80.
The presence of mutations at codon 80 (Ser) of the parC gene was investigated by HaeII digestion of the parC PCR fragment. Any base substitution at codon 80 (AGC) leads to a disruption of the HaeII restriction site except the substitution AGC→GGC. Therefore, any substitution at codon 80 can be detected except for the one encoding Ser80-Gly.
RESULTS
Selection and characterization of experimental quinolone-resistant mutants. (i) In vitro selection.
Two lines of quinolone-resistant mutants derived from the susceptible S. typhimurium strains BN82 and BN18 were obtained through a stepwise selection with enrofloxacin (Table 3). After seven or eight selection steps, two highly resistant mutants were obtained; the MICs (32 and 64 μg/ml) of enrofloxacin for these strains were 512- and 1,024-fold higher than those for the corresponding susceptible strains. The QRDRs of the genes gyrA, gyrB, parC, and parE of these mutants were amplified and sequenced. No sequence alterations were found in the QRDRs of the genes gyrB, parC, and parE, but mutations were detected in the QRDR of the gyrA gene. One mutant, BN18/82, had a single base mutation giving rise to a Gly81-Cys substitution, whereas another mutant, BN82/71, had two mutations conferring the amino acid substitutions Ser83-Phe and Asp87-Gly.
TABLE 3.
Quinolone resistance phenotypes and mutations of the multistep spontaneous mutants selected with increasing concentrations of enrofloxacin
| Strain | No. of selection steps (selecting concn of enrofloxacin [μg/ml]) | MIC (μg/ml)a
|
Codon no. of mutation(s) in:
|
|||||
|---|---|---|---|---|---|---|---|---|
| Nal | Flu | Enr | Cip | gyrAb | parCd | gyrB and parE | ||
| BN18 | 0 | 8 | 1 | 0.06 | 0.03 | Nonec | Nonec | Nonec |
| BN18/11e | 1 (0.1) | 16 | 4 | 0.5 | 0.06 | None | None | NDf |
| BN18/21 | 2 (0.5) | 32 | 8 | 1 | 0.125 | None | None | ND |
| BN18/22e | 2 (1) | 256 | 32 | 2 | 0.5 | 81 | None | ND |
| BN18/32e | 3 (6) | >1,024 | 128 | 8 | 1 | 81 | None | ND |
| BN18/41e | 4 (10) | >1,024 | 256 | 8 | 2 | 81 | None | ND |
| BN18/51e | 5 (15) | >1,024 | 512 | 16 | 2 | 81 | None | ND |
| BN18/62e | 6 (30) | >1,024 | 512 | 16 | 2 | 81 | None | ND |
| BN18/61 | 6 (30) | >1,024 | 512 | 32 | 4 | 81, 87c | Nonec | Nonec |
| BN18/71e | 7 (50) | >1,024 | 512 | 32 | 8 | 81 | None | ND |
| BN18/82 | 8 (75) | >1,024 | 512 | 32 | 8 | 81c | Nonec | Nonec |
| BN82 | 0 | 4 | 1 | 0.06 | 0.03 | Nonec | Nonec | Nonec |
| BN82/11e | 1 (0.1) | 16 | 2 | 0.25 | 0.06 | None | None | ND |
| BN82/22 | 2 (0.5) | 128 | 16 | 2 | 0.25 | None | None | ND |
| BN82/21e | 2 (1) | 1,024 | 32 | 4 | 0.5 | 83 | None | ND |
| BN82/31e | 3 (6) | >1,024 | 128 | 8 | 2 | 83 | None | ND |
| BN82/41e | 4 (10) | >1,024 | 256 | 16 | 2 | 83 | None | ND |
| BN82/52 | 5 (25) | >1,024 | 256 | 16 | 4 | 83 | None | ND |
| BN82/51e | 5 (20) | >1,024 | 256 | 32 | 4 | 83, 87 | None | ND |
| BN82/61e | 6 (50) | >1,024 | 256 | 32 | 8 | 83, 87c | Nonec | ND |
| BN82/71 | 7 (75) | >1,024 | 1024 | 64 | 16 | 83, 87c | Nonec | Nonec |
Nal, nalidixic acid; Flu, flumequine; Enr, enrofloxacin; Cip, ciprofloxacin.
AS-PCR-RFLP assay results.
Sequencing results.
HaeII digestion of the parC gene QRDR.
Isolate retained for the next selection step.
ND, not done.
To determine the order in which mutations were acquired during the selection steps, we developed the AS-PCR-RFLP assay (Fig. 1) to detect mutations at codons 81, 83, and 87 (Fig. 2). The first selection step did not result in any substitutions in the gene studied. For the second-step mutants, single Gly81-Cys and Ser83-Phe mutations were detected in the selection lines BN18 and BN82, respectively. They were associated, respectively, with 16- and 64-fold increases of the MICs of nalidixic acid. The MICs of enrofloxacin were also increased, and only the isolate BN82/21, mutated at Ser-83, was resistant (MIC ≥ 4 μg/ml). Both mutations were associated with an eightfold increase of the MICs of ciprofloxacin (from 0.06 to 0.5 μg/ml), but the isolates remained susceptible, according to the criteria of the CASFM.
Further selection steps resulted in an increase of the quinolone MICs. One isolate of step 6 from the BN18 selection line (BN18/61) that was not retained for additional selection steps had a second substitution, Asp87-Gly, but required only twofold more enrofloxacin or ciprofloxacin for inhibition than the previous-step mutant and a single mutant of the same step. Moreover, other isolates obtained in the same selection line after seven or eight steps exhibited higher resistance levels only with the single substitution Gly81-Cys. In the BN82 selection line, the second mutation, Asp87-Gly, also conferred only twofold increases of the MICs of enrofloxacin and ciprofloxacin. Nevertheless, this twofold MIC increase conferred resistance to ciprofloxacin, according to the CASFM criteria.
(ii) In vivo selection.
In vivo selection experiments were conducted with the same two susceptible strains in two groups of animals. Resistant isolates in the feces of birds monocontaminated with the susceptible strains were collected and then treated with increasing doses of enrofloxacin. The occurrence of mutations at codons 81, 83, and 87 of the gyrA gene of these isolates was investigated by the AS-PCR-RFLP assay. Cumulative data obtained from the two lines of selection are presented in Table 4. One single treatment with enrofloxacin at 6.25 μg/ml was sufficient to select isolates highly resistant to nalidixic acid (MIC > 1,024 μg/ml) and resistant to enrofloxacin (MIC = 4 μg/ml). The mutants from both groups each had a mutation at codon 83 except for one isolate that had a mutation at codon 87. Sequencing of the QRDR of the gyrA gene of this isolate revealed an Asp87-Asn substitution. After the second treatment, mutants with higher resistance levels (MICs of enrofloxacin and ciprofloxacin, 16 and 4 μg/ml, respectively) were isolated. However, they did not reach the resistance levels of in vitro-selected mutants, and no additional mutations were detected at gyrA codons screened by the AS-PCR-RFLP assay or at codon 80 of parC. No further increases of the fluoroquinolone MICs were observed with the isolates collected following the third and the fourth treatments. Ten days after the fourth treatment, isolates for which enrofloxacin MICs were 16 μg/ml (4 μg/ml for ciprofloxacin) were still detected in the feces, but at a drastically reduced frequency (data not shown).
TABLE 4.
Quinolone resistance phenotypes and mutations of all the isolates selected in vivo in chickens treated with four successive doses of enrofloxacin
| Time of isolation (dose [μg/ml]) | No. of isolates | MIC for the most resistant isolates (μg/ml)a
|
No. of isolates with gyrA mutation(s) at indicated codon/totalb
|
No. of isolates with parC mutations at codon 80 (Ser)c | |||||
|---|---|---|---|---|---|---|---|---|---|
| Nal | Flu | Enr | Cip | 81 (Gly) | 83 (Ser) | 87 (Asp) | |||
| Before treatment | 5 | 32 | 8 | 1 | 0.125 | None | None | None | ND |
| After treatment: | |||||||||
| 1 (6.25) | 27 | >1,024 | 128 | 4 | 2 | None | 26/27 | 1/27 | None |
| 2 (12.5) | 29 | >1,024 | 512 | 16 | 4 | None | 29/29 | None | None |
| 3 (25) | 20 | >1,024 | 512 | 16 | 4 | None | 20/20 | None | None |
| 4 (50) | 11 | >1,024 | 512 | 16 | 4 | None | 11/11 | None | None |
Nal, nalidixic acid; Flu, flumequine; Enr, enrofloxacin; Cip, ciprofloxacin.
AS-PCR-RFLP assay results.
HaeII digestion of the parC gene QRDR was performed for the most resistant isolates obtained after each treatment. ND, not done.
Characterization of nalidixic acid-resistant field strains.
A total of 138 nalidixic acid-resistant Salmonella field isolates of various serotypes were screened by the AS-PCR-RFLP assay for mutations at codons 81, 83, and 87 of the gyrA gene (Table 5). Only six strains were resistant to enrofloxacin, and none of them was resistant to ciprofloxacin. All the isolates had a mutation at either codon 83 or codon 87, but none of them had a double mutation like those in experimental mutants selected in vitro. HaeII digestion of the parC PCR fragments of all the isolates did not reveal any mutations at codon 80. Mutations at codons 83 and 87 of gyrA were not equally distributed among the different serotypes: all the S. newport and S. virchow strains and 52 out of the 71 S. typhimurium strains were mutated at codon 83, whereas most of the S. hadar and S. kottbus strains were mutated at codon 87 (21 out of 22 and 10 out of 13, respectively).
TABLE 5.
Fluoroquinolone resistance phenotype and mutations of 138 nalidixic acid-resistant veterinary Salmonella isolates of various serotypes
| MIC (μg/ml)a
|
No. of isolates | No. of isolates (%) with gyrA mutations at codonb:
|
No. of isolates with parC mutations at codon 80 (Ser)c | |||||
|---|---|---|---|---|---|---|---|---|
| Nal | Flu | Enr | Cip | 81 (Gly) | 83 (Ser) | 87 (Asp) | ||
| 128—512 | 8—16 | 0.5—1 | 0.125—0.5 | 132 | None | 74 (56) | 58 (44) | None |
| 512—1,024 | 16—32 | 4 | 1—2 | 5 | None | 5 | None | None |
| 1,024 | 64 | 4 | 2 | 1 | None | 1 | None | None |
Nal, nalidixic acid; Flu, flumequine; Enr, enrofloxacin; Cip, ciprofloxacin.
AS-PCR-RFLP assay results.
HaeII digestion of the parC gene QRDR.
DISCUSSION
Among the mechanisms of quinolone resistance, alterations of the gyrA gene have a major role in the resistance of gram-negative bacteria such as E. coli (11), Neisseria gonorrhoeae (2), or Klebsiella pneumoniae (10). Topoisomerase IV appears to be a secondary target of quinolone in these bacteria, and alterations in the parC gene are often found to be associated with single or double gyrA mutations in strains exhibiting high resistance levels (4, 18, 43). Classically, these amino acid substitutions are encoded by the parC gene at positions homologous to the quinolone-resistance-conferring substitutions encoded by gyrA. But in clinical human and veterinary isolates of Salmonella spp., only gyrA mutations were clearly found to be involved in quinolone resistance (31, 34). Our study also supports the hypothesis of a nonimplication of parC mutations in the quinolone resistance of Salmonella spp.; no mutations were found at codon 80 of the parC genes of 138 nalidixic acid-resistant field isolates, some of which exhibited a phenotype of intermediate resistance to ciprofloxacin (MIC = 2 μg/ml). Moreover, complete sequencing of the QRDRs of the genes gyrA, gyrB, parC, and parE of experimental in vitro- and in vivo-selected mutants exhibiting high-level resistances to ciprofloxacin (MICs of up to 16 μg/ml) revealed the presence of alterations only in the gyrA gene.
Three classes could be distinguished among the experimental in vitro- and in vivo-selected isolates and the field isolates on the basis of the quinolone MICs and gyrA mutations. First, isolates with no mutations in the QRDRs of the gyrase genes were resistant to nalidixic acid and exhibited slightly reduced susceptibilities to fluoroquinolones. Second, isolates with one gyrA mutation exhibited phenotypes from a low to a very high fluoroquinolone resistance level. Last, double gyrA mutants, selected exclusively in vitro, were highly fluoroquinolone resistant. We confirmed the essential role of the first gyrA mutation in the acquisition of very high-level resistance to nalidixic acid and in the decrease of susceptibility to fluoroquinolones. However, most of the increases of the MICs during the selection experiments did not correlate with the acquisition of gyrA mutations. The implication of the second mutation at position 87 remains unclear, but it may not be essential for the acquisition of a high resistance level in Salmonella spp., since we have demonstrated that high resistance levels could be reached with only a single mutation in the QRDR of gyrA. We cannot rule out the presence of mutations outside of the sequenced region, but these results suggest that other mechanisms are responsible for high-level quinolone resistance in Salmonella spp. These mechanisms could include a decrease in permeability to quinolones through modifications of the outer membrane proteins or an active efflux mechanism. Decreased accumulation of enrofloxacin and ciprofloxacin has recently been observed in nalidixic acid-resistant Salmonella isolates (31).
The Ser83-Phe substitution identified in the mutants derived from strain BN82 in vitro was frequently reported in previous studies (6, 14, 27, 35, 44). Other substitutions at this position (Ser83-Tyr and Ser83-Ala) have been described and are also associated with nalidixic acid resistance (19, 33). The single substitutions Asp87-Gly, Asp87-Tyr, and Asp87-Asn are also currently present in Salmonella strains resistant to nalidixic acid (6, 19, 34, 44). We identified a novel Gly81-Cys replacement that conferred an eightfold increase in the MIC of ciprofloxacin for the mutants selected in vitro. This substitution was previously observed in E. coli (45) and Mycobacterium tuberculosis (38). A Gly81-Ser substitution has been observed in spontaneous mutants of S. typhimurium, conferring resistance to nalidixic acid only when associated with an Ala67-Pro substitution (33). However, no alterations were found at codon 81 in the in vivo-selected isolates and in the field strains of various serotypes. We also obtained double gyrA mutants, with a Asp87-Gly substitution associated with either a Ser83-Phe or a Gly81-Cys substitution, in vitro. This second mutation at codon 87 conferred only a twofold increase in the MIC of ciprofloxacin in the two cases. Such double mutations at codons 83 and 87 have also been found in clinical isolates of S. typhimurium var. Copenhagen exhibiting very high-level resistance (MIC of ciprofloxacin, 32 μg/ml), but complementation tests revealed that a gyrB mutation was probably also involved (19). Another similar double mutation was detected in an S. typhi isolate from India that exhibited only a slight decrease in susceptibility to fluoroquinolones (MIC of ciprofloxacin, 0.256 μg/ml) (6). In our studies, single resistance mutations in the field isolates suggest that mutations at codon 83 confer a higher resistance level than mutations at codon 87. However, isolates with mutations at the same position exhibited a wide range of resistance levels, which suggests the presence of additional resistance mechanisms.
We could not select mutants in vivo that reached the level of resistance of the in vitro-selected mutants. This is obviously not due to a limited bioavailability of fluoroquinolone in the gut (e.g., through tight binding to fecal material), since under similar conditions, fully resistant E. coli could be selected (unpublished results). These highly fluoroquinolone-resistant mutants also exhibited drastically altered growth on solid media (smaller colony size) compared to the field isolates, the mutants, selected in vivo, and the corresponding susceptible strains (unpublished results). These results suggest that the mechanisms leading to a high level of resistance could be deleterious and could therefore be counterselected under in vivo conditions. Nucleoid partitioning defects have already been reported in S. typhimurium strains mutated genes that code for topoisomerase IV (23). Moreover, in previous studies on mutations in quinolone-resistant Salmonella, as in our studies, no mutations in the parC gene were reported (31, 35). This situation is different from those of other gram-negative bacteria in which the participation of parC mutations in fluoroquinolone resistance has been widely documented (2, 10, 11, 18, 43). From our works on fluoroquinolone-resistant E. coli from avian origin, we could illustrate the cumulation of mutations in the gyrA and parC genes and the role of parC mutations in high-level resistance (unpublished results). parC mutations leading to high fluoroquinolone resistance in E. coli and other gram-negative bacteria may have a prohibitive cost to the fitness of Salmonella spp. A recent study has demonstrated that most Salmonella mutants resistant to streptomycin, rifampin, or nalidixic acid lost their virulence in mice but could rapidly recover it by accumulating various compensatory mutations without concomitant loss of resistance (3). Studies with animal models should be conducted to investigate if compensatory mutations could also restore the fitness in growth-deficient, highly fluoroquinolone-resistant strains.
In many countries, salmonellae are the leading cause of food-borne disease, and the increasing incidence of multiresistant strains (especially in the S. typhimurium serovar of phage type DT104) represents a risk to public health. The hypothesis that the introduction of fluoroquinolones into veterinary therapy could have contributed to the emergence of fluoroquinolone resistances in bacterial pathogens responsible for human infections, like E. coli, Campylobacter spp., or Salmonella spp., has been raised (29). Only a few cases of salmonellosis treatment failure due to fluoroquinolone resistance have been reported up to now, but the increasing number of strains resistant to nalidixic acid is a matter of concern. Animals may constitute a reservoir where nalidixic acid-resistant strains with initial mutations in gyrA could persist and acquire additional resistance mechanisms upon further exposure to fluoroquinolones. Our study suggests that the mechanisms leading to high fluoroquinolone resistance alter the viability of Salmonella and are thus counterselected in field conditions. However, it may be that the higher the incidence of nalidixic acid-resistant strains, the higher the risk of selecting strains combining fluoroquinolone resistance and normal growth capacities following the acquisition of compensatory mutations. Thus, monitoring the incidence of gyrA mutations associated with quinolone resistance appears to be important.
ACKNOWLEDGMENTS
This work was supported by grants from the Conseil Régional de la Région Centre.
We thank C. Mouline and G. Flaujac for their technical assistance and A. Cloeckaert and J. P. Lafont for critical reading of the manuscript.
REFERENCES
- 1.Barnass S, Franklin J, Tabaqchali S. The successful treatment of multiresistant non-enteric salmonellosis with seven day oral ciprofloxacin. J Antimicrob Chemother. 1990;25:299–300. doi: 10.1093/jac/25.2.299. [DOI] [PubMed] [Google Scholar]
- 2.Belland R J, Morrison S G, Ison C, Huang W M. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol Microbiol. 1994;14:371–380. doi: 10.1111/j.1365-2958.1994.tb01297.x. [DOI] [PubMed] [Google Scholar]
- 3.Björkman J, Hughes D, Andersson D I. Virulence of antibiotic-resistant Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:3949–3953. doi: 10.1073/pnas.95.7.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breines D M, Ouabdesselam S, Ng E Y, Tankovic J, Shah S, Soussy C J, Hooper D C. Quinolone resistance locus nfxD of Escherichia coli is a mutant allele of the parE gene encoding a subunit of topoisomerase IV. Antimicrob Agents Chemother. 1997;41:175–179. doi: 10.1128/aac.41.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brisabois A, Cazin I, Breuil J, Collatz E. Surveillance of antibiotic resistance in Salmonella. Eurosurveillance. 1997;2:19–20. doi: 10.2807/esm.02.03.00181-en. [DOI] [PubMed] [Google Scholar]
- 6.Brown J C, Shanahan P M A, Jesudason M V, Thomson C J, Amyes S G B. Mutations responsible for reduced susceptibility to 4-quinolones in clinical isolates of multiresistant Salmonella typhi in India. J Antimicrob Chemother. 1996;37:891–900. doi: 10.1093/jac/37.5.891. [DOI] [PubMed] [Google Scholar]
- 7.Brown J C, Thomson C J, Amyes S G B. Mutations of the gyrA gene of clinical isolates of Salmonella typhimurium and three other Salmonella species leading to decreased susceptibility to 4-quinolone drugs. J Antimicrob Chemother. 1996;37:351–356. doi: 10.1093/jac/37.2.351. [DOI] [PubMed] [Google Scholar]
- 8.Cherubin C E, Eng R H K. Quinolones for the treatment of infections due to Salmonella. Rev Infect Dis. 1991;13:343–344. doi: 10.1093/clinids/13.2.343. [DOI] [PubMed] [Google Scholar]
- 9.Comité de l’Antibiogramme de la Société Française de Microbiologie. Zone sizes and MIC breakpoints for non-fastidious organisms. Clin Microbiol Infect. 1996;2(Suppl. 1):S46–S49. [PubMed] [Google Scholar]
- 10.Deguchi T, Fukuoka A, Yasuda M, Nakano M, Ozeki S, Kanematsu E, Nishino Y, Ishihara S, Ban Y, Kawada Y. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1997;41:699–701. doi: 10.1128/aac.41.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett M J, Jin Y F, Ricci V, Piddock L J V. Contribution of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother. 1996;40:2380–2386. doi: 10.1128/aac.40.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher I S T. Salmonella enteritidis and S. typhimurium in Western Europe for 1993-1995: a surveillance report from Salm-Net. Eurosurveillance. 1997;2:4–6. doi: 10.2807/esm.02.01.00191-en. [DOI] [PubMed] [Google Scholar]
- 13.Gensberg K, Jin Y F, Piddock L J V. A novel gyrB mutation in a fluoroquinolone-resistant clinical isolate of Salmonella typhimurium. FEMS Microbiol Lett. 1995;132:57–60. doi: 10.1111/j.1574-6968.1995.tb07810.x. [DOI] [PubMed] [Google Scholar]
- 14.Griggs D J, Gensberg K, Piddock L J V. Mutations in gyrA gene of quinolone-resistant salmonella serotypes isolated from humans and animals. Antimicrob Agents Chemother. 1996;40:1009–1013. doi: 10.1128/aac.40.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griggs D J, Hall M C, Jin Y F, Piddock L J V. Quinolone resistance in veterinary isolates of Salmonella. J Antimicrob Chemother. 1994;33:1173–1189. doi: 10.1093/jac/33.6.1173. [DOI] [PubMed] [Google Scholar]
- 16.Haliassos A, Chomel J C, Tesson L, Baudis M, Kruh J, Kaplan J C, Kitzis A. Modification of enzymatically amplified DNA for the detection of point mutations. Nucleic Acids Res. 1989;17:3606. doi: 10.1093/nar/17.9.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heisig P. High level fluoroquinolone resistance in a Salmonella typhimurium isolate due to alterations in both gyrA and gyrB genes. J Antimicrob Chemother. 1993;32:367–377. doi: 10.1093/jac/32.3.367. [DOI] [PubMed] [Google Scholar]
- 18.Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;40:879–885. doi: 10.1128/aac.40.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heisig P, Kratz B, Halle E, Graser Y, Altwegg M, Rabsch W, Faber J P. Identification of DNA gyrase A mutations in ciprofloxacin-resistant isolates of Salmonella typhimurium from men and cattle in Germany. Microb Drug Resist. 1995;1:211–218. doi: 10.1089/mdr.1995.1.211. [DOI] [PubMed] [Google Scholar]
- 20.Khodursky A B, Zechiedrich E L, Cozzarelli N R. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kureishi A, Diver J M, Beckthold B, Schollaardt T, Bryan L E. Cloning and nucleotide sequence of Pseudomonas aeruginosa DNA gyrase gyrA gene from strain PAO1 and quinolone-resistant clinical isolates. Antimicrob Agents Chemother. 1994;38:1944–1952. doi: 10.1128/aac.38.9.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwow S, Kellogg D E, McKinney N, Spasic D, Goda L, Levenson C, Sninsky J J. Effects of primer-template mismatches on the polymerase chain reaction: human deficiency virus type 1 model studies. Nucleic Acids Res. 1990;18:999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luttinger A L, Springer A L, Schmid M B. A cluster of genes that affects nucleoid segregation in Salmonella typhimurium. New Biol. 1991;3:687–697. [PubMed] [Google Scholar]
- 24.Martel J L, Dellac B, Bonnier M, Martin S, Thiese I. Proceedings of the Salmonella and Salmonellosis Meeting. Ploufragan, France: Zoopôle; 1997. Quinolone susceptibility of 192 Salmonella strains, isolated in France from bovine digestive pathology during 1995; pp. 443–447. [Google Scholar]
- 25.Martel J L, Chaslus-Dancla E, Coudert M, Poumarat F, Lafont J P. Survey of antimicrobial resistance in bacterial isolates from diseased cattle in France. Microb Drug Resist. 1995;1:273–283. doi: 10.1089/mdr.1995.1.273. [DOI] [PubMed] [Google Scholar]
- 26.Newton C R, Graham A, Heptinstall L E, Powell S J, Summers C, Kalsheker N, Smith J C, Markham A F. Analysis of any point mutation in DNA. The amplification refractory mutation system. Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouabdesselam S, Tankovic J, Soussy C J. Quinolone resistance mutations in the gyrA gene of clinical isolates of Salmonella. Microb Drug Resist. 1996;2:299–302. doi: 10.1089/mdr.1996.2.299. [DOI] [PubMed] [Google Scholar]
- 28.Ozeki S, Deguchi T, Yasuda M, Nakano M, Kawamura T, Nishino Y, Kawada Y. Development of a rapid assay for detecting gyrA mutations in Escherichia coli and determination of incidence of gyrA mutations in clinical strains isolated from patients with complicated urinary tract infections. J Clin Microbiol. 1997;35:2315–2319. doi: 10.1128/jcm.35.9.2315-2319.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piddock L J V. Does the use of antimicrobial agents in veterinary medicine and animal husbandry select antibiotic resistant bacteria that infect man and compromise antimicrobial chemotherapy? J Antimicrob Chemother. 1996;38:1–3. doi: 10.1093/jac/38.1.1. [DOI] [PubMed] [Google Scholar]
- 30.Piddock L J V, Griggs D J, Hall M C, Jin Y F. Ciprofloxacin resistance in clinical isolates of Salmonella typhimurium obtained from two patients. Antimicrob Agents Chemother. 1993;37:662–666. doi: 10.1128/aac.37.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piddock L J V, Ricci V, McLaren I, Griggs D J. Role of mutations in the gyrA and parC genes of nalidixic-acid-resistant Salmonella serotypes isolated from animals in the United Kingdom. J Antimicrob Chemother. 1998;41:635–641. doi: 10.1093/jac/41.6.635. [DOI] [PubMed] [Google Scholar]
- 32.Reid T M S. The treatment of non-typhi salmonellosis. J Antimicrob Chemother. 1992;29:4–8. doi: 10.1093/jac/29.1.4. [DOI] [PubMed] [Google Scholar]
- 33.Reyna F, Huesca M, Gonzalez V, Fuchs L Y. Salmonella typhimurium gyrA mutations associated with fluoroquinolone resistance. Antimicrob Agents Chemother. 1995;39:1621–1623. doi: 10.1128/aac.39.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ridley A, Threlfall E J. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella typhimurium DT 104. Microb Drug Resist. 1998;4:113–118. doi: 10.1089/mdr.1998.4.113. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz J, Castro D, Goñi P, Santamaria J A, Borrego J J, Vila J. Analysis of the mechanism of quinolone resistance in nalidixic acid-resistant clinical isolates of Salmonella serotype Typhimurium. J Med Microbiol. 1997;46:623–628. doi: 10.1099/00222615-46-7-623. [DOI] [PubMed] [Google Scholar]
- 36.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah P M. Use of quinolone for the treatment of patients with bacteremia. Rev Infect Dis. 1989;11(Suppl. 5):S1156–S1159. doi: 10.1093/clinids/11.supplement_5.s1156. [DOI] [PubMed] [Google Scholar]
- 38.Takiff H E, Salazar L, Guerrero C, Philipp W, Huang W M, Kreiswirth B, Cole S T, Jacobs W R, Jr, Telenti A. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother. 1994;38:773–780. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tauxe R V. Epidemiology of salmonella. Oral communication presented at session 94 of the 92nd General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1992. [Google Scholar]
- 40.Taylor D E, Chau A S-S. Cloning and nucleotide sequence of the gyrA gene from Campylobacter fetus subsp. fetus ATCC 27374 and characterization of ciprofloxacin-resistant laboratory and clinical isolates. Antimicrob Agents Chemother. 1997;41:665–671. doi: 10.1128/aac.41.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Threlfall E J, Ward L R, Rowe B. Increasing incidence of resistance to trimethoprime and ciprofloxacin in epidemic Salmonella typhimurium DT 104. Eurosurveillance. 1997;2:81–83. doi: 10.2807/esm.02.11.00187-en. [DOI] [PubMed] [Google Scholar]
- 42.Vila J, Ruiz J, Marco F, Barcelo A, Goñi P, Giralt E, De Anta T J. Association between double mutation in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and MICs. Antimicrob Agents Chemother. 1994;38:2477–2479. doi: 10.1128/aac.38.10.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vila J, Ruiz J, Goñi P, De Anta M T J. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40:491–493. doi: 10.1128/aac.40.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wain J, Hoa N T T, Chinh N T, Vinh H, Everett M J, Diep T S, Day N P J, Solomon T, White N J, Piddock L J V, Parry C M. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin Infect Dis. 1997;25:1404–1410. doi: 10.1086/516128. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida H, Bogaki M, Nakamura M, Yamanaka L M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother. 1991;35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]