Abstract
Introduction: Tumor extracellular vesicles (EVs) and their relevance to various processes of tumor growth have been vigorously investigated over the past decade. However, obtaining direct evidence of spontaneous EV transfer in vivo remains challenging. In our previous study, a single-guide RNA (sgRNA): Cas9 ribonucleoprotein complex, which can efficiently delete target genes, was delivered into recipient cells using an engineered EV. Aim: Applying this newly discovered exosomal bio-cargo to track the uptake and distribution of tumor EVs. Methods: Tumor cells of interest were engineered to express and release the sgRNA:Cas9 complex, and a reporter cell/system containing STOP-fluorescent protein (FP) elements was also generated. EV-delivered Cas9 proteins from donor cells were programmed by a pair of sgRNAs to completely delete a blockade sequence and, in turn, recuperated the expression of FP in recipient reporter cells. Thus, fluorescently illuminated cells indicate the uptake of EVs. To improve the efficiency and sensitivity of this tracking system in vivo, we optimized the sgRNA design, which could more efficiently trigger the expression of reporter proteins. Results: We demonstrated the EV-mediated crosstalk between tumor cells, and between tumor cells and normal cells in vitro. In vivo, we showed that intravenously administered EVs can be taken up by the liver. Moreover, we showed that EVs derived from melanoma xenografts in vivo preferentially target the brain and liver. This distribution resembles the manifestation of organotrophic metastasis of melanoma. Conclusion: This study provides an alternative tool to study the distribution and uptake of tumor EVs.
Keywords: CRISPR-Cas9, EV tracking, tumor EV distribution
Introduction
Extracellular vesicles (EVs) have been identified as alternative mediators of cell-to-cell communication. They can be secreted by all types of cells under both physiological and pathological conditions. EVs consist of a heterogeneous group of membrane vesicles with sizes varying from 50 nm to 1000 nm. EVs can be produced by either budding of the plasma membrane (formerly known as microvesicles) or derived from intraluminal vesicles that are packaged in multivesicle bodies (formerly known as exosomes). A wide range of bio-cargo can be packaged into EVs during their formation, including proteins, RNAs, DNAs, and lipids.1,2 In addition, cells of different types or under different physiological and pathological conditions have a specific profile of EV bio-cargo, indicating the selective sorting of EV bio-cargo. 3 Once EVs are taken up by recipient cells, bio-cargoes from donor cells are internalized into recipient cells and regulate multiple biological functions. Mounting evidence has shown that EVs are highly relevant to various physiological conditions and diseases.4–6 In particular, tumor-derived EVs have been shown to play important roles in many processes of tumorigenesis, including angiogenesis, 7 metastasis,8,9 immune escape, 10 and drug resistance. 11 Furthermore, tumor EVs can initiate the formation of a niche for metastasis. 12 Tumor EVs derived from different tissues bear distinct integrin expression patterns closely related to tumor organotropic metastasis. 13
Although the functional implications of EVs have been substantially described, tracking of EV transfer between cells, particularly intercellular transfer in vivo, remains a challenge. Furthermore, the depiction of the tissue-specific distribution of EVs, if any, can facilitate a better understanding of the functional role of tumor-derived EVs. To address this demand, we design an EV tracking system taking advantage of the CRISPR-Cas9 system. CRISPR-Cas9 is a versatile technology that can precisely and specifically edit genes of interest. RNA-guided DNA endonuclease Cas9 protein directed by customized single-guide RNAs (sgRNAs) cleaves the targeted genome and generates a double-strand break (DSB), which can be repaired by either non-homologous end-joining (NHEJ) or homology-directed repair (HDR). Genome repaired by error-prone NHEJ can be frequently introduced into insertion or deletion mutations, whereas HDR can lead to precise DNA modifications.14,15 We previously discovered that CRISPR-Cas9 components can be loaded into EVs in the form of sgRNA:Cas9 ribonucleoprotein. 16 In the recipient cells that contain the STOP-DsRed reporter system, we visualized genome editing of EV-delivered sgRNA:Cas9 ribonucleoprotein complex. 16 Thus, we speculate that this newly discovered bio-cargo can be utilized as a tool to visualize the uptake of EVs. Aiming to track the functional transfer of EVs from tumor cells, we engineered donor tumor cells that released a pair of sgRNAs and Cas9 proteins through EVs. sgRNA pairs direct the endonuclease Cas9 to generate two concomitant DSBs, leading to a complete deletion of the STOP sequence of the reporter protein (fluorescent protein [FP]) and subsequently illuminating the reporter cells or reporter animal models. Using this tracking system, we visualized the transfer of EVs between tumor cells and between tumor cells and normal cells in the neighborhood in vitro. In vivo, we revealed the transfer of EVs between tumor xenografts and other organs in tumor-bearing reporter mice. Due to the high efficiency of CRISPR-Cas9 for genome editing, 17 EV-delivered sgRNA:Cas9 ribonucleoprotein may serve as a promising tool for tracking EVs. Our study broadens the strategy for studying EV uptake, facilitating a better understanding of tumor-derived EVs.
Materials and Method
The reporting of this study conforms to ARRIVE 2.0 guidelines [1]. All animal care and methods aimed to minimize the suffering of experimental animals were performed following The Eighth Edition of the Guide for the Care and Use of Laboratory Animals [2] and were approved by the Institutional Review Board of Nanjing University (Nanjing, China). The approval number of this animal research was IACUC-2106007.
Cells, Reagents, Antibodies, and Animals
Human adenocarcinomic alveolar basal epithelial cells A549, human bronchial epithelial cells (HBE) and HEK 293 T were purchased from the Institute of Biochemistry and Cell Biology at Shanghai Institute for Biological Science Academy of Science (Shanghai, China). The mouse melanoma cell line B16-F10 was purchased from the American Type Culture Collection. A549, HEK 293 T, and B16-F10 cells were maintained in Dulbecco's modified Eagle's medium (Gibco, 11965092, CA, USA) supplemented with 10% fatal bovine serum (FBS; Gibco, 10099141), 100 units/ml penicillin (Gibco, 15140148), and 100 μg/ml streptomycin (Gibco, 15140163). All EV donor or recipient cells were cultured using EV-depleted FBS, in which EVs were removed from FBS by ultracentrifugation at 100,000 × g for 18 h. STR profiling and detection of mycoplasma contamination were performed to authenticate all cell lines. Antibodies against SpCas9 (#14697, 1:1000) and β-actin (#8457, 1:1000) were purchased from Cell Signalling Technology (MA, USA). Antibody against CD63 (sc-5275, 1:500) was purchased from Santa Cruz Biotechnology (TX, USA). Antibody against CD9 (ab92726, 1:1000) was purchased from Abcam (Cambridge, UK). Antibody against Flag (MA1-91878, 1:1000) was purchased from Thermo Fisher Scientific (MA, USA). Antibodies against Alix (12422-1-AP, 1:1000), TSG101 (14497-1-AP, 1:1000), and ITGβ5 (28543-1-AP, 1:1000) were purchased from Proteintech (IL, USA). Male B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J mice aged 6 to 8 weeks were purchased from The Jackson Laboratory (ME, USA). C57BL/6J mice aged 6 to 8 weeks were purchased from the Model Animal Research Centre of Nanjing University (Nanjing, China). Mice were weighed and allocated randomly into either control or treatment groups. Mice were housed under specific pathogen-free and 12 h light/12 h dark conditions at Nanjing University. They were free access to pellet food and sterile water. Mice in each group were treated randomly to control confounders. The sex of animals has no influence on the results. Mice were anesthetized before conducting in vivo imaging analysis by injecting with pentobarbital (50 mg/kg, i.p.), which was approved by the Institutional Review Board of Nanjing University. Mice were sacrificed by cervical dislocation.
Confocal Imaging Analysis
Reporter cells were seeded onto glass coverslips in 12-well plates, fixed with 4% paraformaldehyde (PFA), and stained with Hoechst 33342 (Thermo Fisher Scientific, 62249) at room temperature for 15 min. Mouse tissues were dissected and fixed in 4% PFA (Thermo Fisher Scientific, R37814) at 4 °C overnight, followed by a thorough rinse in phosphate buffer saline (PBS) (3 times, 10 min per rinse). Thereafter, tissues were placed in 30% sucrose at 4 °C overnight until the tissue sank to the bottom and were completely submerged in an optimal cutting temperature (OCT) compound and frozen in dry ice. Embedded tissues were moved to a pre-cooled cryomicrotome (Leica, Germany) and cut into 5 to 10 μm sections. Sections were carefully retrieved onto glass slides and stained with Hoechst 33342 (Thermo Fisher Scientific, 62249). Subsequently, the slides were rinsed in PBS three times and stained with Prolong Diamond Antifade 10 (Thermo Fisher Scientific, p36961). Slides were imaged using a confocal imaging system (TCS SP8-MaiTai M, Leica, Wetzler, Germany) and imaged using the TCS SP8 software (Leica). The results were analyzed in three independent experiments. The counts of fluorescent signals were analyzed using Image J software, and the percentage of conversion efficiency (positive counts/Dapi counts) was calculated.
Guide RNA Design
Guide RNAs targeting STOP sequences were designed using the CRISPR Design Tool (http://zlab.bio/guide-design-resources). All guide RNAs were synthesized by Genescript (Nanjing, China). The guide RNA sequences are listed in the Supplemental Material.
DNA Constructs
The synthesized oligonucleotides corresponding to sgRNA pairs were annealed to form cDNAs. Each of the sgRNA pairs was cloned into a lentiCRISPRv2 vector (Addgene) downstream of the U6 promoter using the BsmBI restriction site, and the U6 promoter sgRNA2 element was amplified by Polymerase Chain Reaction (PCR) from one of the paired plasmids and cloned into the other plasmid at the EcoRI restriction site. The mCherry/GFP nanobody (Nb) sequence was synthesized from Genescript (Nanjing) and cloned into the lentiCRISPRv2 vector. mCherry or green fluorescent protein (GFP) was amplified by PCR using primers for flanking and cloned into the psin-puromycin vector (Addgene) between MluI and SpeI restriction sites; CD63 was amplified by PCR, and the amplicons were cloned into psin-mCherry/GFP vector. The STOP element sequence was synthesized by GenScript (Nanjing). GFP was amplified by PCR from the pcDNA6.2-GFP plasmid (Thermo Fisher Scientific). The STOP element and GFP were finally cloned into the psin-puromycin vector using MluI and SpeI restriction sites. CAG-STOP-tdTomato was cloned from Rosa tdTomato mice and cloned into psin-puromycin between the MluI and SpeI restriction sites. Plasmid sequences are listed in the Supplemental Material.
Lentivirus Construction and Generation of Stable Cell Lines
To prepare lentivirus particles, the plasmid of interest, psPAX2 and pMD2G at a molar ratio of 10:5:1 were co-transfected into HEK 293 T cells with Lipofectamine 2000 (Thermo Fisher Scientific, 11668019). A549 cells or B16-F10 cells were seeded in 6-well plates and incubated with CRISPR-Cas9 lentivirus and CD63-mCherry/GFP lentivirus sequentially supplemented with 4 μg/ml polybrene. Thereafter, the cells were cultured as a polyclonal population and kept under selection using 1 μg/ml puromycin (Santa Cruz Biotechnology, sc-205821). Reporter A549, HBE or HEK 293 T cells were incubated with STOP-GFP or STOP-tdTomato lentivirus.
Transfection of Cells With Plasmids
Donor cells were seeded into 75-cm2 flasks. When cells reached a 70% confluence, plasmid 4 expressing sgRNA and Cas9 protein and plasmid 2 expressing CD63-GFP fused protein were co-transfected using Lipofectamine 2000 (Thermo Fisher Scientific, 11668019) according to the manufacturer's instruction. Six hours later, the spent Opti-MEM was replaced with fresh Dulbecco's modified Eagle's medium (DMEM) culture medium. After 24 h, the culture medium was collected and prepared for EV isolation.
Injection of Plasmids into Mice 20
Plasmids were transformed into Escherichia coli DH5α competent cells (Tsingke, TSC01, Beijing, China), which were then cultured in Luria-Bertani (LB) medium in a shaking incubator at 37 °C overnight. Plasmids were extracted and purified using EndoFree Maxi Plasmid Kit V2 (Tiangen, DP120, Beijing, China) according to the manufacturer's instructions. The purified plasmids were dissolved in PBS and administered to mice (5 mg/kg) via regular tail vein injection for 3 days (1 injection per day).
EV Isolation via Ultracentrifugation
The cell culture medium was harvested and centrifuged at 300 × g for 5 min to remove dead cells. The supernatant was centrifuged at 3000 × g for 30 min to remove the cell debris, the supernatant was centrifuged at 10,000 × g for 30 min to remove large vesicles, and the supernatant was centrifuged at 110,000 × g for 70 min to collect EVs. Finally, the EVs were resuspended in a convenient volume of PBS. Isolated EVs were lysed using RIPA buffer (Beyotime, P0013C, Shanghai, China) on ice. Lysates were centrifuged and the supernatants were collected. Protein concentrations of EVs were determined using Bicinchoninic Acid protein assay kit (Thermo Fisher Scientific, 23235). EVs were quantified by protein level. Same amount of EVs was used for characterization, PCR, and western blot analysis. EVs were stored at −80 °C
Animal Experiments
Pilot experiments were performed to determine the sample size. Five male mice were intravenously injected with plasmids (5 mg/kg body weight). Thereafter, the livers were collected and cryosectioned. EVs (50 μg) were injected intravenously (5 or 3 injections) or intraperitoneally (5 injections) into the mice (n = 5) within 10 days. Then, the livers, lungs, kidneys, and spleens were collected and cryosectioned. Melanoma cells (1 × 106) were implanted subcutaneously. After 21 days, tissues including the livers, brains, lungs, kidneys, and spleens were collected and prepared for confocal imaging analysis.
Samples from all animal samples were analyzed.
RNA Immunoprecipitation (IP) Assays
EVs (60 μg) were lysed as described previously. 16 Lysates were incubated with anti-Flag antibody or IgG and then bound to Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology, sc-2003). Finally, immunoprecipitants were eluted from beads using lysis buffer and treated with Radio Immunoprecipitation Assay (RIPA) for western blot analysis or TRIzol reagent (TaKaRa, T9108, Dalian, China) for RNA analysis.
qRT-PCR
Total RNA was prepared from cells or EVs using TRIzol reagent (TaKaRa, T9108). cDNA was synthesized using AMV reverse transcriptase (TaKaRa, 2621), and quantitative Real-time PCR (qRT-PCR) was performed using ABI7300 (Applied Biosystems). The mRNA levels were normalized to 18 s rRNA (ΔCt). The data are shown as 2^−ΔCt. The levels of sgRNA were calculated based on the standard curve, which was generated by amplifying a series of synthetic sgRNAs with known concentrations. The levels of sgRNA in the cells were normalized to the expression of 18 s rRNA.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism version 8.0. The technical and biological triplicates of each experiment were performed. Comparisons were performed using a one-way analysis of variance. Results are expressed as mean ± standard error of the mean.
Results
The Experimental Design of EV Tracking System Based on EV-Delivered CRISPR-Cas9 Components
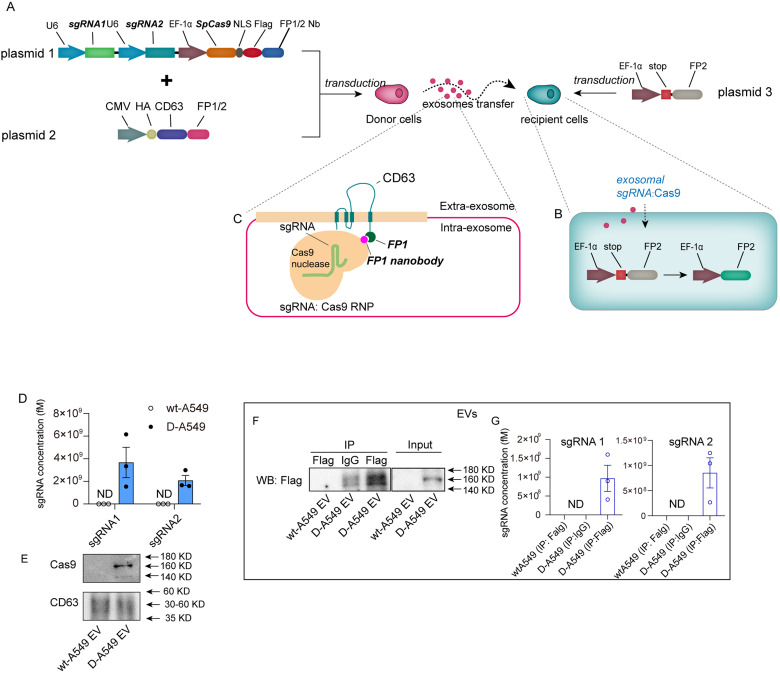
To perform EV tracking strategy, this study generated a donor cell line that expressed a pair of sgRNAs and Cas9 proteins by transfecting plasmid 1, and a reporter cell line that harbored the STOP-FP element introduced by plasmid 3 (Figure 1A). The STOP sequence blocks the expression of FP by causing a frameshift mutation. 16 sgRNA pairs expressed in donor cells were designed to delete the entire STOP sequence, resulting in the recuperation of FP expression (Figure 1B). Thus, the reporter cells are marked with a fluorescent signal once they take up the exosomal sgRNAs:Cas9 derived from donor cells. As previously reported, 16 Cas 9 protein was fused with a Nb of FP, which could capture CD63-FP due to its affinity, boosting the loading efficiency of Cas9 protein (Figure 1C). The FP used in donor cells is different from that used in recipient reporter systems.
Figure 1.
The design and characterization of EV tracking system. (A) Schematic of EV tracking system. This system consisted of donor A549 cells that expressed sgRNAs and Cas9 proteins and recipient cells that contained STOP-FP elements. (B) Schematic of the recuperation of FP expression in reporter cells. (C) Schematic of engineered EVs. FP Nb and its affinitive Nb were fused with exosomal membrane protein CD63 and Cas9 proteins, respectively. Cas9 proteins were captured by EVs through the bond of FP-FP Nb, selectively being sorted into EVs. (D) qRT-PCR analysis of sgRNA pair levels in EVs derived from wild-type A549 or donor A549 cells (n = 3 in each group). (E) Western blot analysis of Cas9 protein levels in EVs derived from wild-type A549 cells or donor A549 cells (n = 3 in each group). (F) RNA immunoprecipitation (IP) assay of exosomal Cas9 and sgRNAs derived from wild-type A549 cells or donor A549 cells. Cas9 proteins were tagged with Flag. Lysates of EVs were blotted using Flag antibody (input) or immunoprecipitated with Flag and blotted using Flag antibody (IP). IP with anti-IgG served as control. (G) sgRNAs analysis of immunoprecipitants following IP assay in (F). Abbreviations: EV: extracellulaer vesicle; FP: fluorescent protein; Nb: nanobody; sgRNAs: single-guide RNAs.
The Generation of Parts in Tracking System
In order to investigate the transfer of tumor-derived EVs using our tactic, human adenocarcinoma alveolar basal epithelial cells A549 were selected as EV donor cells. To unlock the fluorescent signal in reporter cells, the entire STOP sequence must be removed by a pair of sgRNAs expressed simultaneously. To express the sgRNA pairs more efficiently and economically, we constructed a single plasmid that expressed sgRNA pairs under tandem U6 promoters (Supplementary Figure S1A). Lentivirus-expressing sgRNA pairs were transduced into A549 cells (D-A549). The percentages of transduced cells were shown in Supplementary Figure S1B. The levels of sgRNA1, sgRNA2, and Cas9 proteins were significantly detected in the donor cells (Supplementary Figure S1C-S1E). To boost the sorting of Cas9 into EVs, mCherry Nb and mCherry were fused to Cas9 protein and CD63, respectively, as shown in Figure 1A. Nanoparticle tracking analysis (NTA) showed that the diameters of EVs from D-A549 cells were not altered compared to those from wild-type A549 (wt-A549) cells (Supplementary Figure S1F). Transmission electron microscopy (TEM) analysis showed that EVs from D-A549 cells had a typical EV morphology (Supplementary Figure S1G). Western blots analysis showed that the membranal proteins (CD9 and CD63) and proteins involved in EV biogenesis (ALIX and TSG101) were not altered by the transfection in D-A549 cells (Supplementary Figure S1H). Considerable levels of sgRNA pairs and Cas9 proteins were detected in EVs from D-A549 cells but not in wt-A549 cells (Figure 1D and E). sgRNA concentrations in EVs were calculated using a standard curve (Supplementary Figure S1H). RNA-immunoprecipitation assay confirmed that sgRNA pairs and Cas9 proteins were associated as a functional ribonucleoprotein complex in EVs, as reported previously 16 (Figure 1F and G). These data indicate that the donor cells were successfully established.
The Transfer of Tumor-Derived EVs In Vitro
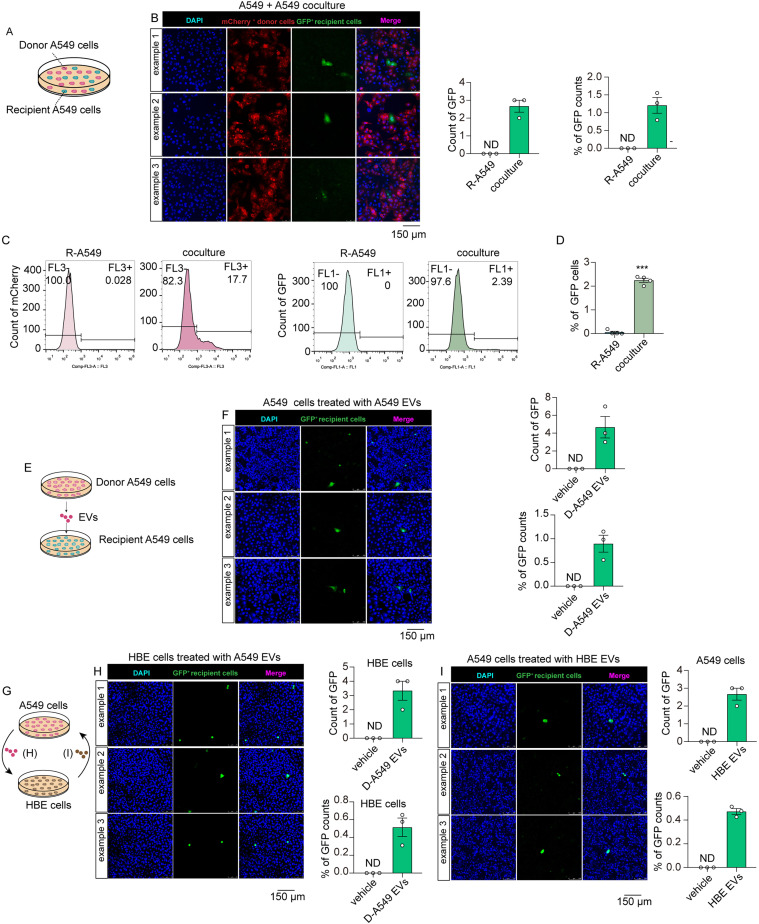
Given the demonstration that cancer cells cooperate to facilitate tumor growth, we investigated whether EVs can be transferred between cancer cells. We generated the recipient A549 (R-A549) cells that expressed the stop-GFP element. D-A549 and R-A549 were cocultured for 4 days (Figure 2A). Thereafter, the co-cultured cells were subjected to fluorescence analysis using a confocal microscope. The result showed substantial expression of mCherry proteins, indicating the existence of donor cells (Figure 2B). As anticipated, GFP-marked reporter cells appeared discernibly in the co-cultured cells, indicating the uptake of donor cell-derived sgRNA:Cas9 by recipient cells (Figure 2B). Additionally, flow cytometry analysis confirmed the expression of GFP in recipient cells, showing a significant color switch in coculture system (Figure 2C and D). To investigate whether this bio-cargo transfer is mediated via EVs, we directly incubated EVs isolated from D-A549 cells with R-A549 cells (Figure 2E). As anticipated, discernible GFP signals could be detected in R-A549 cells, indicating an EV-mediated communication between tumor cells (Figure 2F). Next, we investigated whether this system could indicate the EV-mediated communication between tumorous and non-tumorous cells. HBE were selected as non-tumorous cell model. As shown in Figure 2G, HBE cells were incubated with EVs derived from D-A549 cells. Significant GFP signals were detected in HBE cells, indicating that the tumor-derived EVs could be internalized by normal cells (Figure 2H). To investigate whether EVs derived from HBE cells can be taken up by tumor cells, HBE cells were engineered into CRISPR-Cas9 donor cells, and their EVs were incubated with R-A549 cells (Figure 2G). The result showed that tumor cells could also take up the EVs from normal cells (Figure 2I). In summary, the EV-delivered CRISPR-Cas9 system combined with a matched reporter system can be utilized to track EVs. In addition, this system proves the EV-mediated communications between tumor cells, and between tumor cells and normal cells.
Figure 2.
The transfer of EVs between tumor cells and between tumor and normal cells. (A) Schematic of A549 coculture system. (B) Confocal imaging analysis of fluorescent expression in donor A549 cells and recipient A549 cells (n = 3 in each group). Objective magnification: 20×. (C) Flow cytometry analysis of the tdTomato and GFP cells in coculture system (n = 4 in each group). (D) The percentage of GFP cells following the flow cytometry analysis in C (n = 4 in each group). (E) Schematic of A549 cells incubated with A549 EVs. (F) Confocal imaging analysis of fluorescent expression in recipient A549 cells treated with vehicle or EVs derived from D-A549 cells (n = 3 in each group). Objective magnification: 20×. (G) Schematic of HBE cells incubated with A549 EVs and vice versa. EVs from donor A549 cells were incubated with normal HBE cells; in another way, HBE cells were engineered into CRISPR-Cas9 donor cells and their EVs were incubated with A549 cells. (H) Confocal imaging analysis of fluorescent expression in recipient HBE cells treated with vehicle or EVs from D-A549 cells (n = 3 in each group). Objective magnification: 20×. (I) Confocal imaging analysis of fluorescent expression in recipient A549 cells treated with vehicle or EVs from D-HBE cells (n = 3 in each group). Objective magnification: 20×. Abbreviation: EVs: extracellular vesicles; HBE: human bronchial epithelial cell.
The Study of EV Uptake in Living Mice Using EV-Delivered CRISPR-Cas9 Components
Next, we investigated whether this system could be used to study EV uptake in living mice. A mouse model that ubiquitously expressed STOP-tdTomato was selected as the reporter mouse (tdTomato reporter mouse). The STOP sequence from the tdTomato reporter mouse consists of three repeated STOP elements in a string, as shown in Supplementary Figure S3A. To completely delete the STOP sequence, we devised two strategies for sgRNA design: (1) sgRNA pair targeting the bilateral flanks of the intact STOP sequence (tdsgRNAs3STOP) (Supplementary Figure S2A) or (2) sgRNA pair targeting the bilateral flanks of each single STOP element (tdsgRNAs1STOP) (Supplementary Figure S2B). sgRNA pairs were synthesized and cloned into vectors as described in Supplementary Figure S1A to generate plasmids 4 and 5 (Supplementary Figure S2E). A reporter cell harboring STOP-tdTomato elements was generated to assess the efficiency of sgRNA pairs in vitro (Supplementary Figure S2C). PCR analysis showed clear detection of the STOP-tdTomato sequence in the reporter cells (Supplementary Figure S2D). The plasmids expressing tdsgRNAs3STOP or tdsgRNAs1STOP were transfected into tdTomato reporter cells (Supplementary Figure S2E). Confocal imaging analysis showed a discernible fluorescent signal of tdTomato in cells transfected with tdsgRNAs3STOP or tdsgRNAs1STOP, indicating the successful deletion of STOP elements by both types of sgRNA design (Supplementary Figure S2F and S2G). We observed a more significant fluorescent signal exhibited by the cells treated with tdsgRNAs3STOP (Supplementary Figure S2H). Therefore, tdsgRNA3STOP pairs were selected for further studies. Thereafter, to determine whether these sgRNA pairs were effective in vivo, plasmid 4 was administered intravenously into mice (Supplementary Figure S3A). Vehicle plasmids were administered as control. After 24 h, the livers were collected to assess sgRNA efficiency in vivo. As anticipated, the levels of Cas9 mRNA and sgRNA pairs could be detected in the livers of mice injected with both types of CRISPR-Cas9 vectors Supplementary (Figure S3B and S3C), indicating the successful expression of both plasmids in vivo. The confocal images revealed that the livers of reporter mice exhibited significant expression of tdTomato (Supplementary Figure S3D).
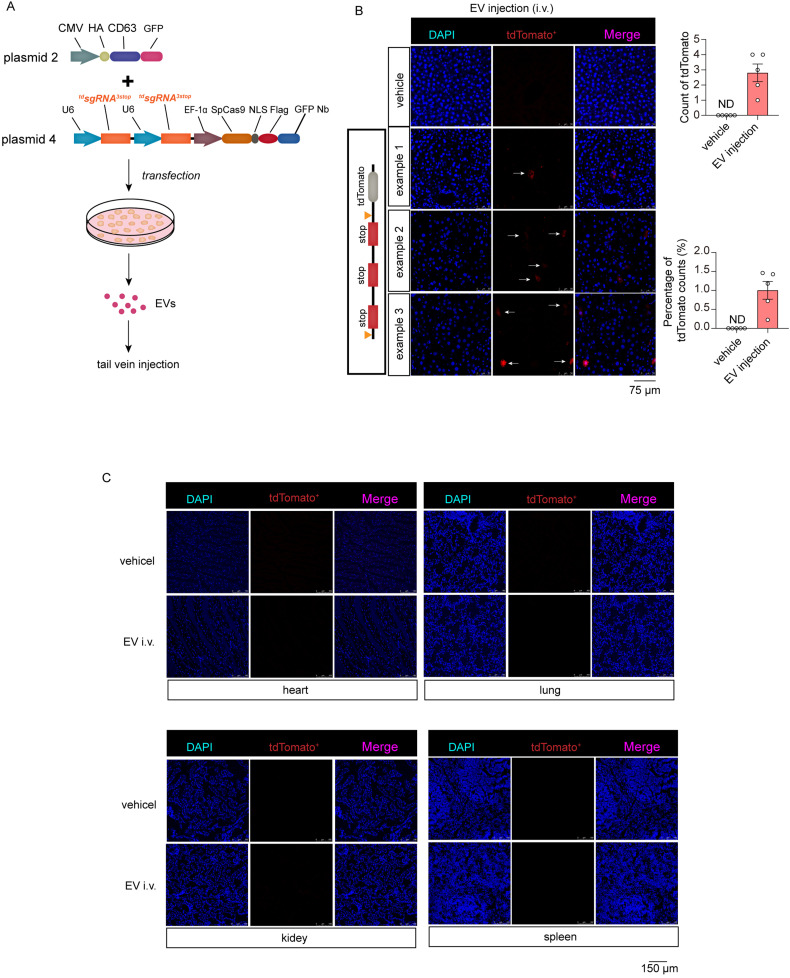
Thereafter, tdsgRNA3STOP pairs and Cas9 proteins were loaded into EVs to investigate whether they could be used to study the uptake of EVs in living mice. Donor cells were transfected with plasmid 4 expressing tdsgRNAs3STOP and Cas9 proteins (Figure 3A). Plasmids expressing CD63-GFP fused proteins (plasmid 2) were co-transfected to boost the loading of Cas9 proteins into EVs (Figure 3A). EVs were isolated from the medium of cells transfected with vehicle or CRISPR-Cas9 plasmids. Mice were injected with EVs for 5 times or 3 times, respectively. Mice that received 5 injections of CRISPR-Cas9 EVs showed discernible tdTomato fluorescent signals in the liver, while mice received 3 injections only showed dim fluorescent signals in the liver (Figure 3B and Supplementary Figure S3E). However, we could not observe any expression of FP in other organs, such as the heart, lung, kidney, and spleen (Figure 3C). This indicates EVs injected through tail vein were primarily taken up by liver. Given that contaminating transfection artifact may account for some effects of EVs, the cells in liver might be illuminated by contaminating transfection plasmids not the EVs. To rule out this, we performed a control experiment using a reporter plasmid (GFP-expressing plasmid). Vehicle EVs isolated from GFP plasmid-transfected cells and GFP plasmids were injected into mice via tail vein respectively. We speculate uncontaminated EVs cannot illuminate liver with GFP signal. As anticipated, mice injected with GFP plasmids showed significant GFP signals in livers, whereas mice injected with EVs had no GFP signals detected in livers (Supplementary Figure S3F). This result indicates EVs are hardly contaminated with plasmids. Next, we investigated if the system could track EVs administrated intraperitoneally. Unfortunately, we could not observe any fluorescent signals in livers (Supplementary Figure S3G), indicating that EVs injected intraperitoneally are not efficiently taken up by liver. Taken together, these results indicate an uptake of intravenously injected-EVs by liver in vivo.
Figure 3.
The study of EV uptake in living mice using EV-delivered CRISPR-Cas9 components. (A) Schematic of generating the EVs loaded with tdsgRNAs3STOP and Cas9 proteins. (B) Confocal image analysis of tdTomato in livers of mice injected with vehicle EVs, or EVs loaded with tdsgRNAs3STOP and Cas9 proteins (n = 5 for each group). Mice were received 5 injections (50 μg EV each time). Objective magnification: 40×. (C). Confocal image analysis of tdTomato in hearts, lungs, kidneys, and spleens of mice injected with vehicle EVs, or EVs loaded with tdsgRNAs3STOP and Cas9 proteins (n = 5 for each group). Mice were received 5 injections (50 μg EV each time). Objective magnification: 20×. Abbreviations: EVs: extracellular vesicles; sgRNAs: single-guide RNAs.
The Systemic Transfer of EVs Derived from Melanoma Cells
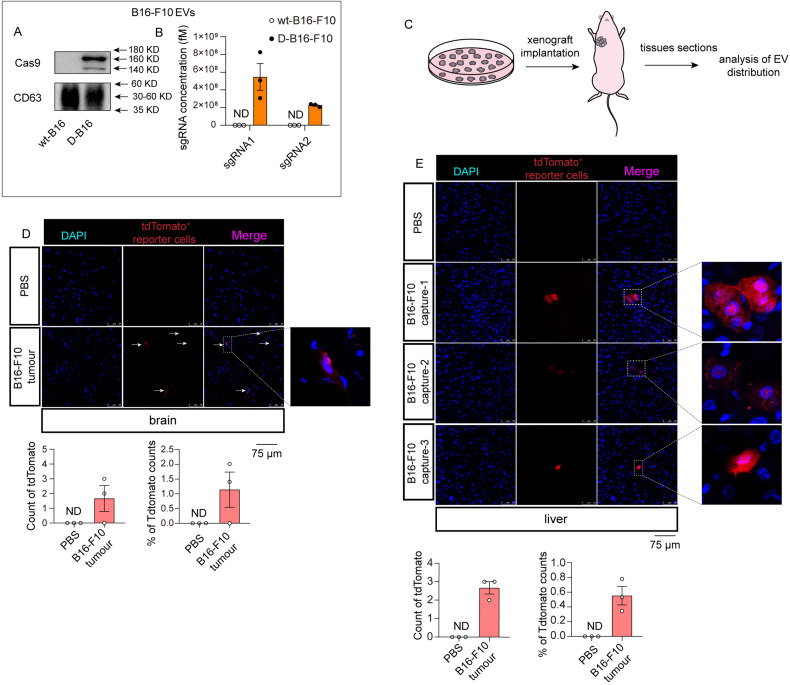
Melanoma has a high metastatic and invasive ability. 21 In light of the functional implication of tumor EVs in initiating distant metastasis by forming a pre-metastasis niche,12,13 we sought to investigate whether melanoma-derived EVs can be taken up by other organs in vivo. The melanoma cell line B16-F10 was selected and transduced with lentivirus expressing tdsgRNAs3STOP and Cas9 proteins to generate donor cells (D-B16-F10) (Supplementary Figure S4A). Considerable levels of Cas9 and sgRNAs were detected in both D-B16-F10 cells (Supplementary Figure S4B-S4D) and their EVs (Figure 4A and B). To investigate EV transfer under natural pathological conditions, D-B16-F10 cells were subcutaneously implanted into mice (Figure 4C). PBS was injected into mice as control. After 21 days, tissues, including the brain and liver, were collected and sectioned. Confocal imaging analysis showed that the fluorescent tdTomato signal could be detected in the liver and brain (Figure 4D and E), indicating that this system can study the uptake of naturally released EVs in vivo. Notably, tdTomato signals were only detected in the brain and liver, although not in other tissues such as the heart, kidney, lung, and spleen (Supplementary Figure S4E). Reportedly, integrins determine the tropism of tumor-derived exosomes. Especially, liver tropism of exosomes requires ITGβ5. 13 Western blot analysis revealed that ITGβ5 was abundantly expressed in B16 EVs (Supplementary Figure S4F). This result indicates the liver-specific transfer of B16 EVs may be also mediated by ITGβ5. These results indicate that melanoma-derived EVs are primarily taken up by the liver and brain, suggesting the target preference of tumor-derived EVs.
Figure 4.
The investigation into systematic transfer of EVs derived from melanoma cells. (A) Western blot analysis of Cas9 protein levels in EVs derived from wild-type melanoma cells or donor melanoma cells (n = 3 in each group). (B) qRT-PCR analysis of sgRNA pair levels in EVs derived from wild-type melanoma cells or donor melanoma cells (n = 3 in each group). (C) Schematic of melanoma xenograft implantation and EV distribution analysis. (D-E) Confocal analysis of tdTomato in brains (D) and livers (E) of normal mice (PBS) or mice bearing melanoma xenografts (n = 3 for each group). Objective magnification: 40×. Abbreviations: EVs: extracellular vesicles; sgRNAs: single-guide RNAs
Discussion
The tactic of obtaining direct evidence of EV uptake is challenging. This study devised an EV tracking system using CRISPR-Cas9 delivered by EVs and visualized the uptake and tissue distribution of EVs in vitro and in vivo.
Chemical fluorescent dyes have been utilized to track EVs, including lipophilic dyes (such as DiR, DiD, and DiI, 3 ) PKH26, Cy7, and luminagen (DPA-SCP). 22 Although this strategy can be conveniently performed, it can only trace artificially administered EVs, although not the ones naturally released in vivo. Therefore, generating a parent cell expressing a reporter protein provides another strategy for studying EV release and uptake in vivo. 23 However, this method cannot be easily used for studying EV transfer among closely contacted cells because the parent cells and recipient cells that have taken up EVs exhibit undistinguishable reporter signals. Therefore, bio-cargoes loaded in EVs combined with a matched report system are used as a more promising tool to track EVs in vivo. One of the prominent designs is an EV tracking system based on the Cre-LoxP system. 24 Donor cell-derived exosomal Cre can be taken up into recipient cells and induce a color switch from DsRed to GFP in recipient cells.
Inspired by our previous study, we speculate that CRISPR-Cas9 in EVs may serve as a more efficient tool for studying EV transfer. By co-transfecting plasmids that expressed CD63-mCherry protein, Cas9 fused with mCherry Nb could be selectively caught and packaged into EVs rather than a random release, ensuring sufficient loading of CRISPR-Cas9 elements for tracking analysis. 16 In parallel, we optimized the sgRNA design and expression to delete the STOP sequence in recipient cells efficiently. Introducing concomitant DSBs using a pair of sgRNAs can cause genomic deletions.17,25,26 The target interval of sgRNA pairs can range from approximately a hundred bases to megabases.26,27 This study found that sgRNA pairs distant from each other could cause a more efficient deletion of the genome in recipient cells. Additionally, to more practically express sgRNA pairs, we engineered a single plasmid to simultaneously express Cas9 and sgRNAs transcribed by tandem U6 promoters. These will ensure the functionality and sensitivity of the tracking system.
There is a caveat in the method of tracking EV administrated through tail vein (Figure 3). Kevin Dooley et al. have demonstrated that transient DNA-transfection reagent (such as plasmids) cannot be separated from crude EV preparation using differential ultracentrifugation, even after a serial of media changes or nuclease treatment. 28 This raises the concern that the contaminated plasmids in EVs preparation may account for the expression of fluorescent signal in liver of mice received EVs injection. To rule out this, we performed a control experiment, in which we show that mice injected with EVs isolated from GFP plasmid-transfected cells have no GFP signal in liver, while direct injection of GFP plasmids causes significant GFP in liver (Supplementary Figure S3F). This somehow negates the concern that contaminated plasmids transfer at a functional level but cannot preclude the possibility that plasmid transfer is occurring. Therefore, to more precisely evaluate the EV function, EVs should be purified using density gradient centrifugation, such as discontinuous iodixanol gradient centrifugation. 28 Usinga stable transduction cell line is maybe another way to preclude plasmid contamination in EVs pellets. In Figure 2 and Figure 4, EVs were isolated from donor cells that can stably express CRISPR-Cas9. These results demonstrate that fluorescent signals reflect the transfer of EVs, and our system can be used to track EV transfer in vivo and in vitro.
Melanoma brain metastases and melanoma liver metastases are responsible for the high mortality of melanoma.29,30 EVs released by melanoma xenografts can be transferred to the liver and brain, resembling the manifestation of melanoma metastasis. Tumor cell-derived EVs have been demonstrated to facilitate metastasis. The bio-information contained in EVs can rewire a permissive microenvironment for metastasis.12,31 Thus, our data prompted us to speculate that the organ preference of melanoma EVs may be associated with its organotrope metastasis.
Even though we tracked the transfer of natural released EVs from tumor cells, this EV-tracking system is still not able to unveil the transfer of EVs naturally released by normal organs. It needs engineered donor organs/tissues, also its utilization is associated with a match reporter system. Therefore, to extend the use of this system into tracking EVs released from normal organs, a more complicated reporter mouse model, in which Cas9 protein and sgRNA are expressed in the organs of interest, are needed. Nevertheless, this EV-tracking system still serves as an alternative tactic to facilitate the study of cancer-derived EVs and provide insight into the roles of EVs in tumorigenesis. In addition, this method also can be a laboratory tool to assess the ability and organotropism of metastasis of cancer, guiding clinical intervention.
Conclusion
We devise an EV-tracking system using CRISPR-Cas9 system. EV-delivered Cas9 proteins and sgRNAs can be taken up by recipient cells, in which they rescue the expression of FPs. Thus, the fluorescently illuminated cells indicate the uptake of EVs. Using this system, we show the transfer of EVs between lung cancer A549 cells, and between A549 and normal bronchial epithelial cells. Additionally, we reveal that uptake of EVs administrated through intravenously injection. Furthermore, we successfully observed the transfer of naturally released melanoma EVs and the tissue tropism transfer of these EVs. Given the high advantages of CRISPR-Cas9 system, hopefully this system has a high sensitivity and accuracy for EV-tracking. This strategy can facilitate the study regarding release and transfer of cancer EVs but is not yet to be utilized for tracking the EVs released from normal organs. More technological improvements are needed for further utilization of this EV-tracking strategy.
Supplemental Material
Supplemental material, sj-docx-1-tct-10.1177_15330338221085370 for In Vivo Visualized Tracking of Tumor-Derived Extracellular Vesicles Using CRISPR-Cas9 System by Yangyang Ye, Qian Shi, Ting Yang, Fei Xie, Xiang Zhang, Bin Xu, Jingwen Fang, Jiangning Chen, Yujing Zhang and Jing Li in Technology in Cancer Research & Treatment
Abbreviations
- DSBAL
double-strand break
- EVsAL
extracellular vesicles
- FPAL
fluorescent protein
- HBEAL
Human bronchial epithelial cell
- HDRAL
homology-directed repair
- NbAL
nanobody
- NHEJAL
non-homologous end-joining
- NTAAL
nanoparticle tracking analysis
- OCTAL
optimal cutting temperature
- PFAAL
paraformaldehyde
- sgRNAAL
single-guide RNA.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This study was supported by grants from the National Natural Science Foundation of China (grant numbers 31741066, 31771666, and 31972912) and Fundamental Research Funds for the Central Universities (grant numbers 020814380168, 020814380094, 020814380087, and 020814380137).
Ethical Approval: The reporting of this study conforms to ARRIVE 2.0 guidelines [1]. All animal care and methods aimed to minimize the suffering of experimental animals were performed following The Eighth Edition of the Guide for the Care and Use of Laboratory Animals [2] and were approved by the Institutional Review Board of Nanjing University (Nanjing, China). The approval number of this animal research was IACUC-2106007.
Author Contributions: Yangyang Ye, Qian Shi, Ting Yang, Fei Xie, Xiang Zhang, Bin Xu, and Jingwen Fang acquired data. Jing Li conceived and designed the study. Jing Li and Yujing Zhang obtained fundings. Jing Li, Yujing Zhang, and Jiangning Chen supervised the study. All authors approved the manuscript.
ORCID iD: Jing Li https://orcid.org/0000-0002-3000-1974
Supplemental Material: Supplemental material for this article is available online.
References
- 1.van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213‐228. [DOI] [PubMed] [Google Scholar]
- 2.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569‐579. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39(1):133‐144. [DOI] [PubMed] [Google Scholar]
- 4.Shah R, Patel T, Freedman JE. Circulating extracellular vesicles in human disease. N Engl J Med. 2018;379(22):2180‐2181. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Liang H, Zhang J, et al. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22(3):125‐132. [DOI] [PubMed] [Google Scholar]
- 6.Huang-Doran I, Zhang CY, Vidal-Puig A. Extracellular vesicles: novel mediators of cell communication in metabolic disease. Trends Endocrinol Metab. 2017;28(1):3‐18. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Zhang Y, Liu Y, et al. Microvesicle-mediated transfer of microRNA-150 from monocytes to endothelial cells promotes angiogenesis. J Biol Chem. 2013;288(32):23586‐23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Zhang S, Yao J, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527(7576):100‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salimi L, Akbari A, Jabbari N, et al. Synergies in exosomes and autophagy pathways for cellular homeostasis and metastasis of tumor cells. Cell Biosci. 2020;10(64):1‐18. 10.1186/s13578-020-00426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin Y, Cai X, Chen X, et al. Tumor-secreted miR-214 induces regulatory T cells: a major link between immune evasion and tumor growth. Cell Res. 2014;24(10):1164‐1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santos P, Almeida F. Role of exosomal miRNAs and the tumor microenvironment in drug resistance. Cells. 2020;9(6):1450‐1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa-Silva B, Aiello N, Ocean A, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17(6):816‐826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoshino A, Costa-Silva B, Shen T, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329‐335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ran FA, Cong L, Yan W, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimasu H, Ran FA, Hsu P, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935‐949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye Y, Zhang X, Xie F, et al. An engineered exosome for delivering sgRNA:Cas9 ribonucleoprotein complex and genome editing in recipient cells. Biomater Sci. 2020;8(10):2966‐2976. [DOI] [PubMed] [Google Scholar]
- 17.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819‐823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Percie du Sert N, Hurst V, Ahluwalia A, et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. BMJ Open Sci. 2020;4(1):e100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Worlein JM, Baker K, Bloomsmith M, et al. The eighth edition of the guide for the care and Use of laboratory animals (2011); implications for behavioral management. Am J Primatol. 2011;73(1):98‐98. [Google Scholar]
- 20.Fu Z, Zhang X, Zhou X, et al. In vivo self-assembled small RNAs as a new generation of RNAi therapeutics. Cell Res. 2021;31(6):631‐648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rastrelli M, Tropea S, Rossi C, et al. Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo. 2014;28(6):1005‐1011. [PubMed] [Google Scholar]
- 22.Betzer O, Barnoy E, Sadan T, et al. Advances in imaging strategies for in vivo tracking of exosomes. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2020;12(2):e1594. [DOI] [PubMed] [Google Scholar]
- 23.van der Vos KE, Abels E, Zhang X, et al. Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro Oncol. 2016;18(1):58‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zomer A, Maynard C, Verweij F, et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161(5):1046‐1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, Wang H, Shivalila C, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370‐1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maddalo D, Manchado E, Concepcion C, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516(7531):423‐427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vidigal JA, Ventura A. Rapid and efficient one-step generation of paired gRNA CRISPR-Cas9 libraries. Nat Commun. 2015;6(8083):1‐7. 0.1038/ncomms9083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dooley K, McConnell R, Xu K, et al. A versatile platform for generating engineered extracellular vesicles with defined therapeutic properties. Mol Ther. 2021;29(5):1729‐1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merola JP, Ocen J, Kumar S, et al. Survival in melanoma brain metastases in the era of novel systemic therapies. Neurooncol Adv. 2020;2(1):p. vdaa144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bakalian S, Marshall J, Logan P, et al. Molecular pathways mediating liver metastasis in patients with uveal melanoma. Clin Cancer Res. 2008;14(4):951‐956. [DOI] [PubMed] [Google Scholar]
- 31.Fong MY, Zhou W, Liu L, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17(2):183‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tct-10.1177_15330338221085370 for In Vivo Visualized Tracking of Tumor-Derived Extracellular Vesicles Using CRISPR-Cas9 System by Yangyang Ye, Qian Shi, Ting Yang, Fei Xie, Xiang Zhang, Bin Xu, Jingwen Fang, Jiangning Chen, Yujing Zhang and Jing Li in Technology in Cancer Research & Treatment