Abstract
Background
Preeclampsia (PE) is a significant cause of maternal and neonatal morbidity and mortality worldwide. However, the pathogenesis of PE is unclear and reliable early diagnostic methods are still lacking. The purpose of this review is to summarize potential metabolic biomarkers and pathways of PE, which might facilitate risk prediction and clinical diagnosis, and obtain a better understanding of specific metabolic mechanisms of PE.
Methods
This review included human metabolomics studies related to PE in the PubMed, Google Scholar, and Web of Science databases from January 2000 to November 2021. The reported metabolic biomarkers were systematically examined and compared. Pathway analysis was conducted through the online software MetaboAnalyst 5.0.
Results
Forty-one human studies were included in this systematic review. Several metabolites, such as creatinine, glycine, L-isoleucine, and glucose and biomarkers with consistent trends (decanoylcarnitine, 3-hydroxyisovaleric acid, and octenoylcarnitine), were frequently reported. In addition, eight amino acid metabolism-related, three carbohydrate metabolism-related, one translation-related and one lipid metabolism-related pathways were identified. These biomarkers and pathways, closely related to renal dysfunction, insulin resistance, lipid metabolism disorder, activated inflammation, and impaired nitric oxide production, were very likely to contribute to the progression of PE.
Conclusion
This study summarized several metabolites and metabolic pathways, which may be associated with PE. These high-frequency differential metabolites are promising to be biomarkers of PE for early diagnosis, and the prominent metabolic pathway may provide new insights for the understanding of the pathogenesis of PE.
Keywords: gestational hypertension, metabolite, biomarker, prediction
Introduction
Preeclampsia (PE), a disorder of pregnancy associated with new-onset hypertension after 20 gestational weeks, is a progressive and heterogeneous disease involving multiple organ systems. Different classifications of PE based on onset of clinical symptoms [early-onset PE (EO-PE)<34 weeks and late-onset PE (LO-PE) ≥34 weeks; preterm PE <37 weeks and term PE ≥37 weeks] or severity of disease (severe and mild) had been proposed.1 PE complicates about 3–5% of all pregnancies and is one of the leading causes of maternal and perinatal morbidity and mortality.2 In addition, the adverse impact of PE may persist even after delivery, as evidence suggests that women who suffered from PE and their corresponding children tend to have an increased risk of cardiovascular disease later in life.3 Despite decades of research, the pathogenesis of this disease remains unclear. The only effective treatment for PE is timely termination of pregnancy, which often results in adverse neonatal outcomes (such as preterm birth), and the only helpful prevention comes from low-dose aspirin prophylaxis.4 Therefore, identification of possible predictive markers for PE is necessary for early identification and prevention of women at risk.
Over the course of pregnancy, mothers face a unique physiological challenge that requires complex metabolic adaptations to support fetal growth and development.5 Regarding the complexity and heterogeneity of PE, it is unlikely to find a single biomarker that could predict all sorts of PE disorders. As an emerging high-throughput technique, metabolomics is dedicated to simultaneously and quantitatively assessing all small molecule metabolites within specific biological samples, enabling the delineation of the metabolic phenotype and phenotypic perturbations in various obstetric diseases, including PE.6 Therefore, it has the potential to identify the metabolites and metabolic pathways related to PE, which may advance more comprehensive understanding of the pathophysiology of PE and facilitate its diagnosis and prevention.
In this study, we reviewed all metabolomics studies related to PE from January 2000 to November 2021 and comprehensively summarized and analyzed the information from these studies. The aim of this review was to identify metabolic biomarkers that may facilitate risk prediction, clinical diagnosis, and obtain an improved understanding of specific metabolic mechanisms of PE.
Methods
Literature Search
We obtained relevant publications by searching the PubMed, Google Scholar, and Web of Science databases from January 2000 to November 2021. The searching terms were (“metabolomics” or “metabonomics” or “metabolome” or “metabolic profiling” or “metabolic signature” or “metabolic biomarker” or “metabolic profile”) AND (“pre-eclampsia” [MeSH]). Two researchers searched the articles independently and a third researcher made a final decision in cases of disagreement.
Inclusion and Exclusion Criteria
The inclusion criteria were as follows: 1) metabolomics studies on PE, 2) full text available in English, and 3) studies clearly recording the positive or negative relationship between metabolite markers and PE.
We included all the relevant metabolomics studies on PE except 1) animal studies, 2) review articles, 3) studies evaluating drug effects, 4) results reporting unidentified metabolites, and 5) studies only reporting insignificant metabolites.
Information Extraction
After reading the full articles and supplementary materials, the following information was extracted from the final retained studies: first author, title, publication date, publication journal, study subjects (sample sizes, singleton/twins), diagnostic criteria of PE, analytical platforms, biological specimen, study design, subtypes of PE, gestational age of sample collection, and potential metabolic biomarkers with directions of change (increase or decrease in concentration). In addition, studies by the same first or corresponding author were judged whether there were overlaps in content.
Statistical Analysis
Frequencies on biological samples, sampling time, subtypes of PE, and significant metabolites along with their changing trends were counted and graphed. All the metabolites were matched with their unique Human Metabolome Database (HMDB) IDs to unify the metabolites with different terminologies but with the same chemical structure in different literature. Pathway analysis, including enrichment analysis and topology analysis, was performed using the MetaboAnalyst 5.0 online software (https://www.metaboanalyst.ca).7 The significant pathways were selected based on the criteria of the raw P <0.05.
Results
Study Characteristics
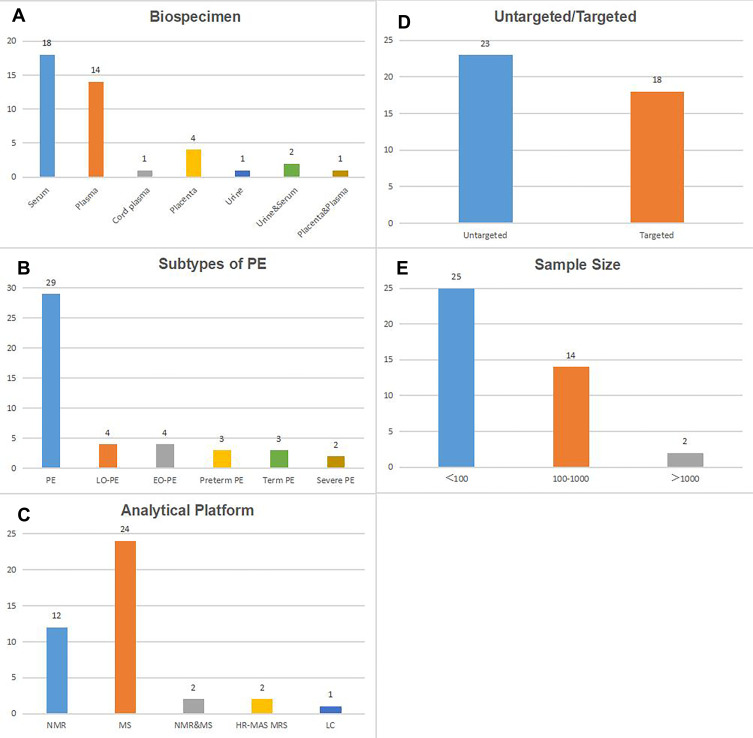
After literature searching and screening, a total of 41 studies8–48 were included. Characteristics of the 41 studies are presented in Table 1. Blood (serum, plasma, and cord plasma) is the mostly used biospecimen (in 33 studies), followed by placenta (in 4 studies) and urine (in 1 study). Meanwhile, 2 studies simultaneously collected urine and serum, and 1 study used both placenta and plasma (Figure 1A). These included articles involving different etiological subtypes of PE, including PE (in 29 studies), LO-PE (in 4 studies), EO-PE (in 4 studies), preterm PE (in 3 studies), term PE (in 3 studies) and severe PE (in 2 studies) (Figure 1B). Regarding the analytic platform, 12 studies used nuclear magnetic resonance (NMR), 24 used mass spectrometry (MS), 2 used both NMR and MS, 2 used high resolution magic angle spinning magnetic resonance spectroscopy (HR-MAS MRS), the remaining 1 used liquid chromatography (LC) (Figure 1C). Besides, 23 studies were untargeted and the remaining 18 studies were targeted (Figure 1D). The total sample sizes ranged from 8 to 1528, with 16 studies including more than 100 subjects (Figure 1E). The majority of the studies were performed among singleton pregnancies.
Table 1.
Characteristics of the 41 Included Studies
| First Author (Year) | Country | S/Twin | Gestational Age | Outcome | Case | Control | Bio-Specimen | Analytic Platform | Targeted/Untargeted | Up-Regulated | Down-Regulated |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Kivelä (2021)8 | Finland | S | Predo:13,20,28w Radiel:13,23,35w (pooled across the 3 measurement points) |
PE | 43 (Predo) 11 (Radiel) |
254 (Predo) 292 (Radiel) |
Serum | NMR | Targeted | Extremely large VLDL; very large VLDL; large VLDL; medium VLDL; small VLDL; triglycerides in VLDL; total triglycerides; monounsaturated fatty acids; L-isoleucine; L-leucine | None |
| Jääskeläinen (2021)9 | Finland | S | 10–15w | PE | 46 | 53 | Serum | LC-MS | Untargeted | Monoacylglyceride 18:1; urea; glycerophosphocholine | None |
| 23–41w | PE | 57 | 14 | Serum | LC-MS | Untargeted | None | None | |||
| Harville (2021)10 | USA | S | 6+1–13+6w | PE | 18 | 109 | Serum | LC-MS and NMR | Untargeted | Bolasterone; cerasinone | L-Asparagine; N-dimethylglycine; trimethylamine |
| Tan (2020)11 | China | S | 26.6±1.7w(case) 26.1±1.4w(control) |
PE | 29 | 29 | Serum | UPLC-MS | Targeted | LysoPC(18:0); LysoPC(22:6); phytosphingosine; palmitic acid | Behenic acid; 14-methyl hexadecanoic acid; 1,25-dihydroxyvitamin D3-26,23-lactone |
| Sovio (2020)12 | UK | S | 36w | Term PE | 165 | 323 | Serum | UPLC-MS/MS | Untargeted | sFlt-1/PlGF; 4-hydroxyglutamate; C-glycosyltryptophan; 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | None |
| Lee (2020)13 | Korea | S | 16–24w | PE | 33 | 66 | Plasma | GC-TOF MS and LC-Orbitrap MS | Untargeted | 3-Phosphoglycerate; glutamate; palmitoleic acid; xanthine; LysoPE C16:1; LysoPE C17:0; LysoPE C20:0; PC C32:1; PC C32:2; PI C38:3; SM C30:1; SM C34:1 | Lyxose; propane-1,3-diol; LysoPC C19:0; LysoPC C22:1; OxPC C38:4 + 1O; OxPI C38:4 + 1O; PE C23:1e; PE C24:1e; PE C34:3e; PI C36:2; SM C28:1 |
| Within 3 d of delivery | PE | 13 | 21 | Plasma | GC-TOF MS and LC-Orbitrap MS | Untargeted | 3,6-Anhydro-d-galactose; erythritol; 2-deoxytetronic acid; isomaltose; myo-inositol; thymine; xylitol; FAHFA C18:0; OxPC C38:4 + 1O(1Cyc); OxPE C38:3 + 1O; OxPI C38:4 + 1O; PC C36:5e; PC C38:5; PC C40:7; PE C36:4; SM C30:1; SM C32:1; SM C33:2; SM C34:2; SM C38:1 | None | |||
| Kenny (2020)14 | UK | S | 15±1w | PE | 97 | 335 | Plasma | LC-MS/MS | Targeted | Dilinoleoyl-glycerol | 1-Heptadecanoyl-2-hydroxy-sn-glycero-3 phosphocholine; PlGF |
| Preterm PE | 23 | 335 | Plasma | LC-MS/MS | Targeted | Dilinoleoyl-glycerol; choline; 2-hydroxybutanoic acid; NG-monomethyl-l-arginine; decanoylcarnitine | PlGF | ||||
| Term PE | 74 | 335 | Plasma | LC-MS/MS | Targeted | L-Isoleucine; decanoylcarnitine | 1-Heptadecanoyl-2-hydroxy-sn-glycero-3 phosphocholine | ||||
| Hong (2020)15 | USA | S | Within 1–3 d after delivery | Preterm PE | 79 | 980 | Plasma | LC-MS | Untargeted | Symmetric dimethylarginine; C2 carnitine; xanthosine; xanthine; C34:1 DAG; C34:2 DAG; C36:3 DAG; C38:4 DAG; C22:5 LPC; C38:6 PC; C40:9 PC; C50:2 TAG; C52:3 TAG; creatine; creatinine; cystine; glutamine; homocitrulline; N-acetyltryptophan; N-acetylputrescine; N6,N6,N6-trimethyllysine; pyroglutamic acid; C3 carnitine; C5-DC carnitine; octanoylcarnitine; 4-acetamidobutanoate; imidazole propionate; 6.8-dihydroxypurine; uric acid; L-carnitine; C16:0 ceramide; C34:1 DAG/TAG fragment; C36:2 DAG; C38:5 DAG; C22:6 LPC; C22:6 LPE; C40:6 PC; C52:2 TAG; methylguanidine | Anthranilic acid; bilirubin; cytosine; 1-methylguanine; 7-methylguanine; cortisol; C14:0 CE; C18:2 CE; C16:0 CE; C18:0 CE; C18:3 CE; cortisone |
| Sander (2019)16 | UK | Mainly S | 35.9±2.8w(case) 36.7±3.2w(control) |
PE | 32 | 35 | Plasma | LC-MS | Untargeted | Taurodeoxycholic acid isomer; methionine sulfoxide; 3-hydroxyanthranilic acid; N1-methyl-pyridone-carboxamide; urocanic acid; phosphatidylinositol isomer(32:1); diacylglycerol isomer(32:2); diacylglycerol isomer(32:1) | Hydroxyhexacosanoic acid isomer |
| Liu (2019)17 | China | S | 34w | PE | 10 | 10 | Plasma | LC‐MS/MS | Targeted | 16-HDoHE; 8-HEPE; LTB4; 8-HETE | 18-HEPE; maresin; 14,15-DiHETE; 8,9-EEQ; 17-HDoHE |
| Lee (2019)18 | Korea | S | Prior to preterm delivery or at the time of cesarean delivery | PE | 14 | 9 | Plasma | GC-MS | Targeted | Cholesterol/desmosterol; cholesterol/7-dehydrocholesterol | Individual cholesterol esters/cholesterol; total cholesterol esters/cholesterol |
| Do Nascimento (2019)19 | Brazil | Not mentioned | Not mentioned | PE | 14 | 10 | Plasma | LC-HRMS | Targeted | None | L-Leucine; γ-aminobutyric acid; L-serine; L-isoleucine; L-methionine; L-valine; glycine; L-betaine; L-histidine; L-lysine; L-arginine; L-alanine |
| Austdal (2019)20 | Norway | S | Before delivery | PE | 62 | 65 | Serum | HR-MAS MRS | Targeted | Glycerophosphocholine; phosphocholine; aspartate; creatine | Ethanolamine; dihydroxyacetone; glutamate; taurine; L-isoleucine; L-valine; L-threonine; L-lysine; glucose; 3-hydroxybutyric acid; ascorbate; glycine |
| Yang (2018)21 | China | Not mentioned | Immediately after delivery | PE | 16 | 16 | Placenta | GC-MS | Targeted | Octadecanoic acid; 5,8,11-cis-eicosatrienoic acid; cis-11,14-eicosadienoic acid; 11,14,17-cis-eicosatrienoic acid; 9,12,15-cis-octadecatrienoic acid; 8,11,14-cis-eicosatrienoic acid; hexadecanoic acid; 7,10,13,16,19-cis-docosapentaenoic acid | Heneicosanoic acid; 11,14-eicosadienoic acid; 11-trans-octadecenoic acid; tridecanoic acid; heptanoic acid; nonanoic acid; decanoic acid |
| Powell (2018)22 | Australia | S | 26–41w | PE | 17 | 82 | Serum | NMR | Untargeted | Creatinine; acetyl groups | Acetic acid; 3-methylhistidine |
| Jääskeläinen (2018)23 | Finland | S | After delivery | PE | 42 | 53 | Cord plasma | LC-MS | Untargeted | Urea; creatine; creatinine; homocitrulline; guanidinopropionate; L-carnitine; trimethyllysine; γ-butyrobetaine; butyrylcarnitine; octenoylcarnitine; choline; 5-aminovaleric acid betaine; N-methyl-2-pyridone-5-carboxamide; acetylputrescine; 4-acetamidobutanoate; indolecarboxylic acid; indolelactic acid; phosphatidylcholines (PCs)[PC(16:1/18:1)PC(16:0/16:0)PC(16:0/18:1)] | 4-Pyridoxic acid |
| Zhou (2017)24 | China | S | Immediately after the cesarean section | Severe PE | 11 | 11 | Placenta | GC-MS | Targeted | 2-Phosphoenolpyruvate; β-citryl-L-glutamate; ornithine; L-threonine; glutamine; L-asparagine; L-tyrosine; L-lysine; methionine; cysteine; L-phenylalanine; L-leucine; L-isoleucine; glycine; palmitelaidate; DPA; adrenat; γ-linoleate; 9-heptadecenoate; DHA; dihomo-γ-linoleate; 11,14-eicosadienoate; arachidonate; cis-vaccenate; linoleate; trans-vaccenate; 11,14,17-eicosatrienoate; heptadecanoate; pentadecanoate; nonadecanoate; margarate; myristate; arachidate; behenate; stearate; 10,13-dimethyltetradecanoate; oxalate; cabamate | Citraconate; caprylate |
| Kelly (2017)25 | USA | S | 10–18w | PE | 47 | 62 | Plasma | LC-MS | Untargeted | 16-α-hydroxypregnenolone; ins-1-P-Cer(d18:1/22:0); C38:3 PC; 13’-carboxy-γ-tocopherol; C40:0 PC plasmalogen; dimethamine; quinoline; montecristin; terpinolene oxide; C34:2 PI; C50:1 TAG; C40:6 PC; C50:1 TAG; 1-(10-methylhexadecanyl)-2-(8-[3]-ladderane-octanyl)-sn-glycerophosphocholine; C16:3 CE; TAG source fragment; C54:1 TAG; C50:1 TAG; C37:3 PS; 13’-carboxy-γ-tocopherol; C32:1 PC; C18:0 LysoPE; C50:2 TAG; C38:5 DAG; C54:2 TAG; TAG source fragment; saccharin; cohibin A; chlormezanone; C32:1 PC; mabioside D; C36:4 DAG; artemoin A; quinoline; C16:0 LysoPC | Lutein; 3-hydroxy-10-apo-b, y-carotenal; GABA; N-phenylacetylphenylalanine; C40:1 PS; methylthioadenosine_isomer; C18:0 MAG |
| Chen (2017)26 | China | Not mentioned | 21.7–27.9w(case) 18.6–27.4w(control) |
PE | 20 | 20 | Serum | LC-Q-TOF-MS | Untargeted | Phytosphingosine; phenylethylamine; sphinganine; phosphocholine; indole; ubiquinone-1; L-phenylalanine; PI(22:0/22:0); PI(20:4(5Z,8Z,11Z,14Z)/0:0); PE(P-16:0/0:0); PE(18:1(9Z)/0:0); PC(14:0/0:0); PC(13:0/0:0); palmitoylcarnitine; palmitic amide; MG(0:0/18:2(9Z,12Z)/0:0); MG(0:0/16:0/0:0); linoleyl-L-carnitine; Cer(d18:1/26:1(17Z)); C17 sphinganine; C16 sphinganine | Palmitic acid; ribose; PS(18:2(9Z,12Z)/0:0); PS(20:2(11Z,14Z)/0:0) |
| Bahado-Singh(2017)27 | UK | S | Within 3 d of assessment | LO-PE | 59 | 115 | Serum | NMR | Untargeted | L-Carnitine; 2-hydroxybutyric acid; citric acid; glucose; L-leucine; lactate; L-valine; 3-hydroxybutyric acid; 3-hydroxyisovaleric acid; pyruvate; methylhistidine | Dimethylamine |
| Bahado-Singh(2017)28 | UK | S | 11–14w | Term PE | 35 | 63 | Serum | DI-LC-MS/MS | Targeted | PC aa C40:6 | Glucose; putrescine |
| 11–14w | Term PE | 35 | 63 | Serum | 1H NMR | Targeted | None | Urea; dimethyl sulfone | |||
| 30–34w | Term PE | 35 | 63 | Serum | DI-LC-MS/MS | Targeted | None | Serotonin; t4-OH-pro; hexose | |||
| 30–34w | Term PE | 35 | 63 | Serum | 1H NMR | Targeted | Acetic acid; dimethyl sulfone | None | |||
| Koster (2015)29 | Netherlands | S | 8+0–13+6w | EO-PE | 68 | 500 | Serum | UPLC-MS/MS | Targeted | Hexanoylcarnitine; octenoylcarnitine; L-octanoylcarnitine; decenoylcarnitine; decanoylcarnitine; dodecenoylcarnitine; lauroylcarnitine; tetradecenoylcarnitine; hexadecenoylcarnitine | Isobutyrylcarnitine; linoleylcarnitine; stearoylcarnitine |
| LO-PE | 99 | 500 | Serum | UPLC-MS/MS | Targeted | Octenoylcarnitine; L-octanoylcarnitine; decenoylcarnitine; decanoylcarnitine; dodecenoylcarnitine | Linoleylcarnitine; palmitoylcarnitine; oleylcarnitine; stearoylcarnitine | ||||
| Bahado-Singh(2015)30 | UK | S | Within 3 d of assessment | EO-PE | 50 | 108 | Serum | NMR | Untargeted | 3-Hydroxyisovaleric acid; betaine; glucose; propylene glycol | Acetic acid; acetone; choline; formate; glycerol; glycine; isopropanol; L-phenylalanine; succinate |
| Austdal (2015)31 | Norway | S | After delivery | PE | 19 | 15 | Placenta | HR-MAS MRS | Untargeted | None | Glutamine; glutamate; taurine; L-valine; 3-hydroxybutyrate; ascorbate |
| Austdal (2015)32 | Norway | S | 11+0–13+6w | PE | 26 | 561 | Urine | 1H NMR | Untargeted | Creatinine; glycine; α-hydroxyisobutyrate; L-histidine; dimethylamine; 4-deoxythreonic acid | Hippurate; L-threonine; proline betaine; creatine; lactate |
| Serum | 1H NMR | Untargeted | Signals from triglycerides; 3-hydroxybutyric acid | Pyruvate; phosphatidylcholine; lactate | |||||||
| Moon (2014)33 | Korea | Not mentioned | 22.2–40.5w(case) 24.1–37.6w(control) |
PE | 30 | 30 | Serum | GC-MS | Targeted | Androsterone; etiocholanolone; 6β-OH-androstenedione; 11β-OH-androstenedione; pregnenolone; progesterone; 17α-hydroxypregnenolone; 17α-hydroxyprogesterone; 6β-hydroxycortisol; 7α-hydroxycholesterol; 7β-hydroxycholesterol; 4β-hydroxycholesterol; 20α-hydroxycholesterol; 24S-hydroxycholesterol; 27-hydroxycholesterol | 16α-OH-dehydroepiandrosterone; 16α-OH-androstendione; cholesterol |
| Kuc (2014)34 | Netherlands | S | 8+0–13+6w | EO-PE | 68 | 500 | Serum | UPLC-MS/MS | Targeted | None | Taurine; L-asparagine |
| LO-PE | 99 | 500 | Serum | UPLC-MS/MS | Targeted | None | Glycylglycine | ||||
| Korkes (2014)35 | Brazil | Not mentioned | Immediately after cesarean delivery | PE | 10 | 10 | Placenta | MALDI-MS | Untargeted | Glycerophosphoserines [GP03]-(PS); macrolide lactone polyketide [PK04] | Glycerophosphoethanolamines [GP02]-(PEt); glycerophosphoglycerols [GP04]-(PG); glycerophosphoinositols [GP06]-(PI); glycerophosphoinositol monophosphates [GP07]; diradylglycerols [GL02]-(DG); triradylglycerols [GL03]; acidic glycosphingolipids [SP06]- (GM3); steroid conjugates [ST05]; other acyl sugars [SL05]; flavonoids [PK12] |
| At the time of delivery | PE | 10 | 10 | Plasma | MALDI-MS | Untargeted | Glycerophosphocholines [GP01]-(PC); glycerophosphoserines [GP03]-(PS); glycerophosphates [GP10]-(PAc); phosphosphingolipids [SP03]-(SM); neutral glycosphingolipids [SP05]; steroid conjugates [ST05]; flavonoids [PK12] | Glycerophosphoglycerols [GP04]-(PG); sterols [ST01]; other acyl sugars [SL05]; glycerophosphoethanolamines [GP02]-(PEt) | |||
| Austdal (2014)36 | Norway | Not mentioned | 35.9(21.7–37.9) w (case) 35.2(18.6–37.4) w(control) |
PE | 10 | 10 | Urine | NMR | Untargeted | Isobutyrate; dimethylamine | Glycine; p-cresol sulfate; hippurate; L-histidine; L-asparagine; trigonelline; glucose |
| Serum | NMR | Untargeted | L-Histidine | None | |||||||
| Senyavina (2013)37 | Russia | Not mentioned | Not mentioned | PE | 6 | 25 | Serum | HPLC | Targeted | Xanthine; uric acid | Hypoxanthine |
| Diaz (2013)38 | Portugal | Not mentioned | 14–26w | Preterm PE | 9 | 84 | Urine | 1H NMR | Untargeted | N-methyl-2-pyridone-5-carboxamide; 3-methylhistidine; 4-deoxyerythronic acid; cis-aconitate; citric acid; glutamine; hippurate; indoxyl sulphate; N-methylnicotinamide; sucrose; U35 | 2-Ketoglutarate; 4-OH-hippurate; acetic acid; L-carnitine; creatinine; formate; fumarate; galactose; L-isoleucine; lactose; phenylacetyl-glutamine; p-cresol sulphate; scyllo-ino; succinate; trigonelline; L-tyrosine; U14; U4; correlates to trigonelline |
| Bahado-Singh(2013)39 | UK | S | 11+0–13+6w | LO-PE | 30 | 59 | Serum | NMR | Targeted | Glycerol; 1-methylhistidine; L-valine; L-carnitine; acetone; isopropanol; pyruvate; 3-hydroxyisovaleric acid; glucose; 2-hydroxybutyric acid; creatinine; creatine; citrate; 3-hydroxybutyric acid | Trimethylamine; acetamide; dimethylamine |
| Bahado-Singh(2012)40 | UK | S | 11+0–13+6w | EO-PE | 30 | 60 | Serum | NMR | Untargeted | 3-Hydroxyisovaleric acid; glucose; glutamine; propylene_glycol; pyruvate; trimethylamine | Acetic acid; L-alanine; choline; formate; glycerol; glycine; L-isoleucine; isopropanol; L-leucine; methionine; L-phenylalanine; serine; succinate; L-threonine |
| Odibo (2011)41 | USA | S | 11–14w | PE | 41 | 41 | Serum | LC-MS/MS | Targeted | C6OH (hydroxyhexanoylcarnitine); L-phenylalanine; glutamate; L-alanine | None |
| Kenny (2010)42 | Ireland | S | 15±1w | PE | 60 | 60 | Plasma | UPLC-MS | Untargeted | Monosaccharide(s); decanoylcarnitine; oleic acid; docosahexaenoic acid and/or docosatriynoic acid; γ-butyrolactone and/or axolan-3-one; 2-oxovaleric acid and/or axo-methylbutanoic acid; acetoacetic acid; hexadecenoyl-eicosatetraenoyl-sn-glycerol; di-(octadecadienoyl)-sn-glycerol; sphingosine 1-phosphate; sphinganine 1-phosphate; vitamin D3 derivatives | 5-Hydroxytryptophan; methylglutaric acid and/or adipic acid |
| Turner (2010)43 | UK | Not mentioned | Not mentioned | PE | 11 | 11 | Plasma | 1H NMR | Targeted | L-Histidine; L-tyrosine; L-phenylalanine | None |
| Kenny (2008)44 | UK | Not mentioned | 245(186–272) d(case) 243(186–282) d(control) |
PE | 20 | 20 | Plasma | UPLC-MS | Targeted | L-Alanine; 2-hydroxy-3-methyl-butanoic acid; 2-ethyl-3-hydroxypropionic acid; 2-oxoglutaric acid; glutamic acid; xylitol or ribitol; uric acid; creatinine | None |
| Turner (2007)45 | UK | Not mentioned | Not mentioned | PE | 11 | 11 | Plasma | 1H NMR | Untargeted | None | VLDL lipids; lactate; fucose; L-threonine; glycoprotein; L-proline; 3-hydroxybutyric acid |
| Jain (2014)46 | USA | Not mentioned | Immediately after delivery | PE | 3 | 5 | Placenta | ESI/MS | Untargeted | Free fatty acids | Plasmenyl phosphatidylethanolamine |
| McBride (2021)47 | UK | S | 26–28w | PE | 74 | 1454 | Plasma | NMR | Untargeted | None | None |
| He (2021)48 | USA | S | Not mentioned | Severe PE | 44 | 20 | Plasma | LC-MS/MS | Untargeted | None | LPC 15:0; LPC 20:5; PC35:1e; LPE 18:2; LPE 20:4; cholesterol ester 22:5; ceramide d30:1; LPC 16:0/PC 16:0e; LPC 16:1/PC 16:1e; LPC 18:2e/PC18:2e |
Abbreviation: S, singleton.
Figure 1.
Numbers of the included studies according to biospecimen (A), subtypes of PE (B), analytical platform (C), untargeted/targeted (D) and sample size (E).
High-Frequency Metabolic Biomarkers of PE
A total of 415 metabolic biomarkers were reported in the 41 included articles. Amino acids (eg, creatinine, glycine, L-isoleucine, creatine, L-phenylalanine, L-histidine, L-leucine, L-valine, and L-glutamine) are among the most frequently reported altered metabolites that were related with PE. Meanwhile, lipids8,18 and related metabolites, such as phosphatidylcholines,13,15,23,25,26,28,32,48 lysophosphatidylcholine,11,13,15,23,25,48 glycerophosphocholine,9,20,25,35 acylcarnitines,15 and sphingolipids13 were generally detected. Thirty-three metabolites were reported in ≥ 3 studies (Table 2). Creatinine, glycine, L-isoleucine, and glucose were the most reported metabolites (appeared in 7 articles). Decanoylcarnitine, 3-hydroxyisovaleric acid, xanthine, and octenoylcarnitine showed a consistent upward trend in PE patients versus controls, while succinate, formate, and taurine were consistently reported to be down-regulated in patients with PE across different biospecimens.
Table 2.
High-Frequency Metabolic Biomarkers of PE
| No. | Metabolites | Hits | Up | Down |
|---|---|---|---|---|
| 1 | Creatinine | 7 | 6 plasma15c 44 serum22 39b cord plasma23 urine32 | 1 urine38c |
| 2 | Glucose | 7 | 4 serum27b 30a 39b 40a | 3 serum20 28b urine36 |
| 3 | L-Isoleucine | 7 | 3 serum8 plasma14d placenta24e | 4 plasma19 serum 20 40a urine38c |
| 4 | Glycine | 7 | 2 placenta24e urine32 | 5 plasma19 serum 20 30a 40a urine36 |
| 5 | L-Phenylalanine | 6 | 4 placenta24e serum26 41 plasma43 | 2 serum30a 40a |
| 6 | Decanoylcarnitine* | 5 | 5 plasma 14cd serum29ab 42 | |
| 7 | L-Carnitine | 5 | 4 plasma15c cord-plasma23 serum27b 39b | 1 urine38c |
| 8 | Creatine | 5 | 4 plasma15c serum20 39b cord-plasma23 | 1 urine32 |
| 9 | L-Histidine | 5 | 3 urine32 serum36 plasma43 | 2 plasma19 urine36 |
| 10 | L-Leucine | 5 | 3 serum8 27b placenta24e | 2 plasma19 serum40a |
| 11 | 3-Hydroxybutyric acid | 5 | 3 serum27b 32 39b | 2 serum20 plasma45 |
| 12 | L-Valine | 5 | 2 serum27b 39b | 3 serum20 plasma19 placenta31 |
| 13 | L-Threonine | 5 | 1 placenta24e | 4 serum20 40a urine32 plasma45 |
| 14 | Acetic acid** | 5 | 1 serum28b | 4 serum22 30a 40a urine38c |
| 15 | 3-Hydroxyisovaleric acid** | 4 | 4 serum27b 30a 39b 40a | |
| 16 | Pyruvate** | 4 | 3 serum27b 39b 40a | 1 serum32 |
| 17 | Glutamine | 4 | 3 plasma15c placenta24e serum40a | 1 placenta31 |
| 18 | Dimethylamine** | 4 | 2 urine32 36 | 2 serum27b 39b |
| 19 | Choline | 4 | 2 plasma14d cord-plasma23 | 2 serum30a 40a |
| 20 | Alanine | 4 | 2 serum41 plasma44 | 2 plasma19 serum40a |
| 21 | Glutamate | 4 | 2 plasma13 serum41 | 2 serum20 placenta 31 |
| 22 | Lactate** | 4 | 1 serum27b | 1 serum32 urine32 plasma45 |
| 23 | Asparagine | 4 | 1 placenta24e | 1 serum10 34a urine36 |
| 24 | Xanthine | 3 | 3 plasma13 15c serum37 | |
| 25 | Octenoylcarnitine* | 3 | 3 cord-plasma23 serum29ab | |
| 26 | Urea | 3 | 2 serum9 cord-plasma23 | 1 serum28b |
| 27 | Trimethylamine | 3 | 1 serum40a | 2 serum10 39b |
| 28 | Glycerol** | 3 | 1 serum39b | 2 serum30a 40a |
| 29 | Hippurate** | 3 | 1 urine38c | 2 urine32 36 |
| 30 | Isopropanol** | 3 | 1 serum39b | 2 serum30a 40a |
| 31 | Succinate** | 3 | 3 serum30a 40a urine38c | |
| 32 | Formate** | 3 | 3 serum30a 40a urine38c | |
| 33 | Taurine | 3 | 3 serum20 34a placenta31 |
Notes: *Identified by LC-MS only; **Identified by NMR only; aEO-PE; bLO-PE; cPreterm PE; dTerm PE; eSevere PE.
Among the 33 high-frequency metabolites, 12 metabolites were just identified by single type of technology. For instance, decanoylcarnitine and octenoylcarnitine were identified by LC-MS, and other ten metabolites (acetic acid, 3-hydroxyisovaleric acid, pyruvate, dimethylamine, lactate, glycerol, hippurate, isopropanol, succinate, and formate) were found by NMR.
Potential of Metabolic Marker Panels to Predict and Classify PE
Several studies have assessed the potential of metabolic biomarker panels to predict and diagnose PE (Table 3). Lee et al13 used a panel of five metabolites (SM C28:1, SM C30:1, LysoPC C19:0, LysoPE C20:0, and propane-1,3-diol) to predict PE, which exhibited an area under the receiver operating curve (AUC) of 0.868 in a mid-trimester cohort and 0.858 in an at-delivery cohort; the authors also evaluated a panel of another three metabolites (SM C30:1, oxidized PE C38:3, and isomaltose), yielding an AUC of 0.972 in the at-delivery cohort. Powell et al22 identified a panel of four metabolite parameters (3-methylhistidine, creatinine, acetyl groups, and acetate) that could predict PE, which resulted in an AUC of 0.938 and 0.936 in discovery and validation sets, respectively. Odibo et al41 used four significant metabolites (hydroxyhexanoylcarnitine, phenylalanine, glutamate, and alanine), achieving an AUC value of 0.824 to predict PE and 0.846 to predict EO-PE; they also assessed the predictive ability of three significant amino acids (phenylalanine, glutamate, and alanine), which got an AUC of 0.818. Kenny et al42 used a panel of 14 metabolites to predict PE, reporting an AUC value of 0.940 in discovery group. Liu et al,17 Zhou et al,24 and Chen et al26 all calculated AUC of single metabolites, resulting AUC values ranging from 0.795 to 0.935.
Table 3.
The Potential of Metabolic Markers for the Prediction of PE
| Author | Comparison Groups | Bio-Specimen | Sensitivity | Specificity | AUC | Potential Biomarkers | |
|---|---|---|---|---|---|---|---|
| Sovio12 | Preterm PE | 12w | - | - | 0.887 | PGAPE, PAPPA, PlGF, 4-hydroxyglutamate | |
| Preterm PE | 20w | - | - | 0.911 | PGAPE, sFlt1/PlGF, 4-hydroxyglutamate | ||
| Preterm PE | 28w | - | - | 0.926 | PGAPE, sFlt1/PlGF, 4-hydroxyglutamate, C-glycosyltryptophan | ||
| Lee13 | PE | Mid-trimester cohort | 0.751 | 0.830 | 0.868 | SM C28:1, SM C30:1, LysoPC C19:0, LysoPE C20:0, propane-1,3-diol | |
| PE | At-delivery cohort | 0.693 | 0.846 | 0.858 | SM C28:1, SM C30:1, LysoPC C19:0, LysoPE C20:0, propane-1,3-diol | ||
| 0.786 | 0.714 | 0.844 | SM C30:1 | ||||
| 0.838 | 0.799 | 0.904 | SM C30:1, oxidized PE C38:3 | ||||
| 0.923 | 0.857 | 0.972 | SM C30:1, oxidized PE C38:3, isomaltose | ||||
| Kenny14 | Preterm PE | 15±1w | 0.650 | 0.830 | 0.780 | PlGF, dilinoleoyl-glycerol, 1-heptadecanoyl-2-hydroxy-sn-glycero-3 phosphocholine | |
| Liu17 | PE | 34w | - | - | 0.895 | 14,15-DiHETE | |
| - | - | 0.830 | 16-HDoHE | ||||
| - | - | 0.820 | LTB4 | ||||
| - | - | 0.795 | 8,9-EEQ | ||||
| Powell22 | PE | 26–41w | 0.870 0.600 |
0.917 0.955 |
0.938(discovery) 0.936(validation) |
3-Methylhistidine, creatinine, acetyl groups, acetate | |
| Zhou24 | Severe PE | Immediately after the cesarean section | 0.870 | 0.830 | 0.930 | Arachidonate | |
| 0.930 | 0.830 | 0.930 | Docosapentaenoate | ||||
| 0.860 | 0.890 | 0.920 | γ-Linoleate | ||||
| 0.930 | 0.890 | 0.930 | Myristate | ||||
| 0.860 | 0.830 | 0.920 | Dihomo-γ-linoleate | ||||
| Chen26 | PE | 21.7–27.9w(PE) 18.6–27.4w(control) |
- | - | 0.935 | PC(14:0/00) | |
| - | - | 0.928 | Proline betaine | ||||
| - | - | 0.923 | Proline | ||||
| Bahado-Singh27 | LO-PE | Within 3 d of assessment | 0.304 | 0.804 | 0.629(validation) | Carnitine, pyruvate, acetone | |
| 0.391 | 0.804 | 0.722(validation) | UtPI, pyruvate, carnitine, glycerol | ||||
| 0.348 | 0.826 | 0.734(validation) | Maternal weight, UtPI, pyruvate, carnitine | ||||
| Bahado-Singh28 | Term PE | First trimester | 0.727 | 0.574 | 0.701 | Putrescine, urea, carnitine | |
| Term PE | Third trimester | 0.742 | 0.723 | 0.761 | Methylhistidine, serotonin, citrate, hexose, propylene glycol | ||
| Term PE | Integrated first and third trimester | 0.816 | 0.710 | 0.817 | Urea (1st), SM C18:1 (1st), citrate (3rd), hexose (3rd) | ||
| Koster29 | EO-PE | 8+0–13+6w | - | - | 0.878(training) 0.784(validation) |
MC, MAP, PAPPA, PlGF, taurine, stearoylcarnitine | |
| LO-PE | 8+0–13+6w | - | - | 0.833(training) 0.700(validation) |
MC, MAP, PAPPA, PlGF, stearoylcarnitine | ||
| Bahado-Singh30 | EO-PE | Within 3 d of assessment | 0.825 0.750 |
0.823 0.744 |
0.896(discovery) 0.835(validation) |
2-Hydroxybutyrate,3-hydroxyisovalerate, acetone, citrate, glycerol | |
| 0.908 0.900 |
0.908 0.884 |
0.956(discovery) 0.916(validation) |
3-Hydroxyisovalerate, arginine, glycerol, UtPI | ||||
| Austdal32 | PE | 11+0–13+6w | 0.192 | - | 0.694 | Hippurate/creatinine | |
| 0.423 | - | 0.778 | Hippurate/creatinine, MAP, maternal age | ||||
| 0.538 | - | 0.807 | Hippurate/creatinine, MAP, UtPI, maternal age | ||||
| Bahado-Singh39 | LO-PE | 11+0–13+6w | 0.600 | 0.966 | 0.885(primary dataset) | Methylhistidine, glycerol, maternal weight, maternal race, acetoacetate | |
| 0.767 | 1.000 | 0.960(primary dataset) | Valine, maternal weight, maternal race, pyruvate, 3-hydroxybutyrate, 1-methylhistidine, glycerol, trimethylamine, maternal medical disorder | ||||
| 0.400 | 0.941 | 0.790(expanded dataset) | Glycerol | ||||
| 0.400 | 0.950 | 0.796(expanded dataset) | Glycerol, maternal weight | ||||
| 0.567 | 0.950 | 0.783(expanded dataset) | Glycerol, 1-methylhistidine | ||||
| Bahado-Singh40 | EO-PE | 11–13w | 0.759 | 0.951 | 0.904 | Citrate, glycerol, hydroxyisovalerate, methionine | |
| 0.826 | 0.984 | 0.980 | Acetate, glycerol, hydroxyisovalerate, UtPI, fetal CRL | ||||
| 0.500 | 0.950 | 0.840 | Glutamine, pyruvate, propylene glycol, trimethylamine, hydroxy butyrate, maternal weight, maternal medical disorders | ||||
| 0.600 | 0.970 | 0.940 | Pyruvate, propylene glycol, trimethylamine, 3-hydroxisovalerate, UtPI | ||||
| Odibo41 | PE | 11–14w | - | - | 0.824 | Hydroxyhexanoylcarnitine, phenylalanine, glutamate, alanine | |
| EO-PE | - | - | 0.846 | Hydroxyhexanoylcarnitine, phenylalanine, glutamate, alanine | |||
| PE | - | - | 0.818 | Phenylalanine, glutamate, alanine | |||
| Kenny42 | PE | 15±1w | - | - | 0.940(discover) 0.920(validation) |
5-Hydroxytryptophan, monosaccharide(s), decanoylcarnitine, methylglutaric acid and/or adipic acid, oleic acid, docosahexaenoic acid and/or docosatriynoic acid, γ-butyrolactone and/or oxolan-3-one, 2-oxovaleric acid and/or oxo-methylbutanoic acid, acetoacetic acid, hexadecenoyl-eicosatetraenoyl-sn-glycerol, di-(octadecadienoyl)-sn-glycerol, sphingosine 1-phosphate, sphinganine 1-phosphate, vitamin D3 derivatives | |
| McBride47 | PE | 22–28w | - | - | 0.620(training and testing) 0.620(validation) 0.710(training and testing) 0.630(validation) 0.680(training and testing) 0.660(validation) |
Risk factor (BMI, age, parity, smoking) Risk factor (BMI, age, parity, smoking) Metabolite (154 metabolites) Metabolite (154 metabolites) Combined (BMI, parity, 26 metabolites) Combined (BMI, parity, 26 metabolites) |
|
Abbreviations: AUC, area under the curve; PGAPE, the predicted gestational age of pre-eclampsia; PAPPA, pregnancy-associated plasma protein A; PLGF, placenta growth factor; sFlt1, soluble fms-like tyrosine kinase 1; UtPI, uterine artery pulsatility index; MC, maternal characteristics; MAP, mean arterial pressure; fetal CRL, fetal crown-rump length; BMI, body mass index.
As several studies27,29,30,32,39,40,47 reported, the distinguish performance had been remarkably improved after incorporating metabolites into maternal characteristics (eg, weight, age, and race), medical parameters (eg, mean arterial pressure and uterine artery plasticity index) or previously established biomarkers (eg, pregnancy-associated plasma protein A and placenta growth factor). In addition, Sovio et al12 and Kenny et al14 tried to build models for prediction of preterm-PE. Both of them found that metabolite markers are effectively complemented to existing biomarkers, for example, soluble fms-like tyrosine kinase receptor 1 or placenta growth factor. Besides, Bahado-Singh and coworkers28 combined first and third trimester metabolites with maternal characteristics to predict term-PE.
Metabolic Pathways for PE
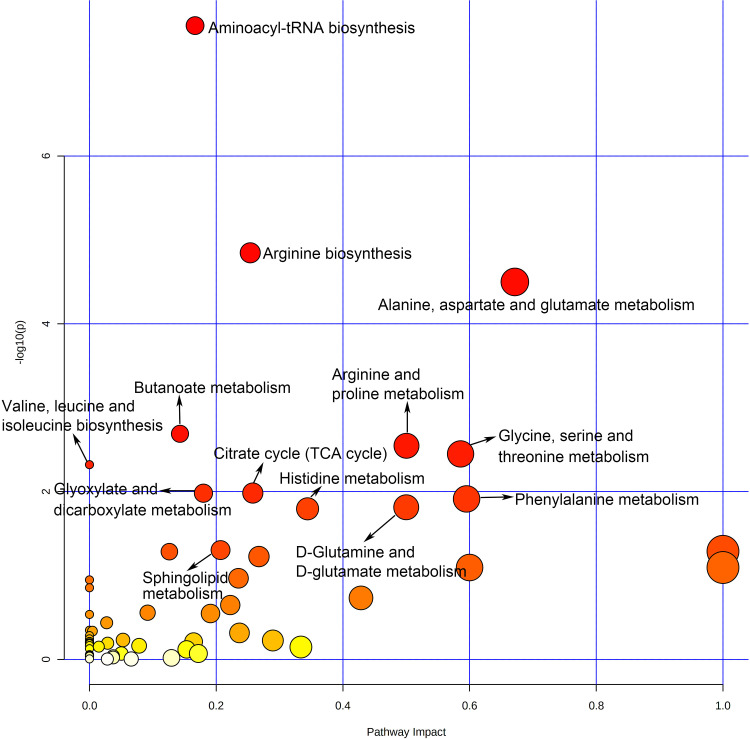
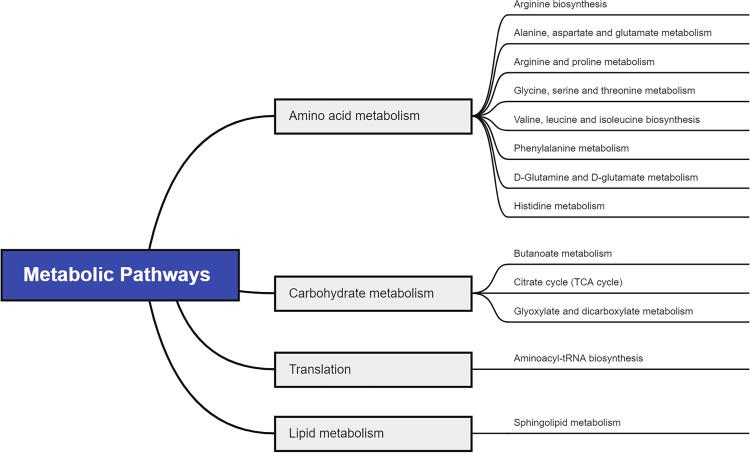
To figure out the potential metabolic pathways involved in the development of PE, 415 potential metabolite markers were imported to MetaboAnalyst for enrichment analysis and topological analysis. Figure 2 shows the results of the analysis. In comparison of patients with PE and normal controls, thirteen metabolic pathways were significantly enriched (Raw P<0.05, Supplementary Table S1). There are eight amino acid metabolism-related pathways, including arginine biosynthesis; alanine, aspartate and glutamate metabolism; arginine and proline metabolism; glycine, serine and threonine metabolism; valine, leucine and isoleucine biosynthesis; phenylalanine metabolism; D-glutamine and D-glutamate metabolism and histidine metabolism. Besides, three carbohydrate metabolism-related pathways were reported: butanoate metabolism; citrate cycle; and glyoxylate and dicarboxylate metabolism. Aminoacyl-tRNA biosynthesis was a translation-related pathway and sphingolipid metabolism was a lipid metabolism-related pathway (Figure 3).
Figure 2.
Overview of pathway analysis of PE.
Figure 3.
Pathway analysis for significant metabolites of PE.
Discussion
In this study, 41 metabolomics studies on PE were comprehensively reviewed and analyzed to identify valuable metabolic biomarkers and pathways involved in the pathologies of PE. Thirty-three high-frequency metabolites (reported in ≥3 studies) were found. In addition, several metabolic pathways involved with PE were identified, particularly alanine, aspartate and glutamate metabolism with a high impact.
Reliability of Metabolic Biomarkers/Metabolite Panels and Their Clinical Implications
Metabolomics studies have identified more than 30 metabolic biomarkers related to PE. Several metabolic biomarkers were repeatedly identified across these studies. Notably, the levels of some metabolic biomarkers, eg, decanoylcarnitine, 3-hydroxyisovaleric acid, xanthine, octenoylcarnitine, succinate, formate, and taurine, were consistently increased or decreased across these studies. The consistency increased the reliability of these metabolites as PE biomarkers. However, several metabolites were inconsistent across studies, for instance, creatinine, glycine, L-isoleucine, and glucose. The disparities may partly be explained by study differences in gestational age when biospecimens were collected, maternal characteristics (such as age, obesity, ethnic origin, and sort and severity of diseases), analytical platforms, and bioinformatics and biostatistical tools.
Seventeen metabolomics studies12–14,17,22,24,26–30,32,39–42,47 used metabolic biomarker panels to discriminate patients with PE or subtypes of PE, yielding AUC values of 0.620–0.980 (Table 2). Also, more effective clinical models were achieved by incorporating metabolic biomarkers with traditional predictive biomarkers or clinical parameters. The results appeared promising. However, the related studies generally had small sample size and limited validation. Also, untargeted metabolomics only acquired semiquantitative metabolite concentrations. Further studies, such as independent external cohorts with large sample size, are needed for obtaining reliable conclusions. Moreover, before metabolite panels are applied to clinical practice, additional robust quantitative analyses of these metabolite panels are necessary.
Metabolic Dysregulations Implicated in PE Pathogenic Mechanisms
The pathological process of PE involves a series of biological changes and is incompletely elucidated.49 Until now the diagnosis of PE has been focused on blood pressure and urinary protein levels with low specificity or sensitivity. Therefore, it is necessary to identify novel biomarkers to improve diagnosis and deepen the understanding of the pathogenesis of PE. Herein, we describe some important metabolites that may play a role in the etiology and early identification of PE patients. Amino acids (for example, creatinine, glycine, L-isoleucine, creatine, L-phenylalanine, L-histidine, L-leucine, L-valine, and L-glutamine) are the most frequently reported altered metabolites related with PE. According to our analysis, renal dysfunction, insulin resistance, lipid metabolism disorder, inflammatory activation and impaired NO production were interestingly concerned with the pathophysiology of PE.
Renal Dysfunction
Renal dysfunction is seen in asymptomatic women in whom PE will eventually develop, and proteinuria and decreased renal function are classic hallmarks of PE.50 The levels of related low molecular weight substances, such as creatinine,15,22,32,38,39,44 lactate,27,32,45 urea,9,23,28 and creatine,15,20,23,32,39 could be used to monitor renal function and to assist the diagnosis of PE.
Insulin Resistance
Insulin resistance is a clear risk factor for PE.51 Elevated insulin derived from insulin resistant may cause increases in sympathetic nervous system activity and renal sodium retention, which could increase blood pressure.52 Glucose and several metabolites (eg, L-valine, and L-leucine) that can influence or reflect insulin release or insulin sensitivity were frequently reported in the included studies.
Metabolomics studies showed an upward trend in glucose in PE.27,30,39,40 Insulin is a hypoglycemic hormone, promoting blood sugar into cells for use as energy.53 Insulin resistance refers to impaired insulin sensitivity, and therefore a high blood glucose level could be explained.
L-isoleucine, L-leucine, and L-valine are essential branched chain amino acids (BCAAs). High levels of BCAAs can activate the mammalian target of rapamycin complex 1 that phosphorylates insulin receptor substrate 1, leading to insulin resistance.54 In addition, a BCAA dysmetabolism model proposed that the accumulation of mitotoxic metabolites promotes pancreatic β-cell mitochondrial dysfunction.55 Hence, the BCAAs are particularly sensitive to insulin resistant conditions and/or insulin deficiency.56 Moreover, as a typical partial-degradation product of BCAAs, 3-hydroxybutyric acid is synthesized in the liver from acetyl-CoA and can be used as an energy source when blood glucose is low. Blood level of 3-hydroxybutyric acid can serve as a biomarker of insulin resistance and impaired glucose tolerance.57
Glutamate, glutamine, L-asparagine, L-alanine, and D-alanine are derived from intermediates of central metabolism, mostly the citric acid cycle, in one or two steps. A recent study performed high-throughput metabolite profiling in two large and well characterized community-based cohorts, observed a significant association of insulin resistance with glutamine and glutamate and found that administration of glutamine leads to both impaired glucose tolerance and decreased blood pressure.58
Lipid Metabolism Disorder
Excessive lipid in maternal serum may induce endothelial dysfunction secondary to oxidative stress. Lipid dysfunction may start early in pregnancies destined for PE development,59 suggesting that related metabolomics may be used to predict the onset of PE. It was found that dietary acetic acid could reduce serum total cholesterol and triacylglycerol by inhibition of lipogenesis in liver and increment in faecal bile acid excretion in rats fed with cholesterol-rich diet.60 Meanwhile, acetic acid was suggested to exhibit anti-hypertension effects via reducing the activity of renin-angiotensin system.61
Furthermore, fatty acids are necessary for proper pregnancy progression and fetal growth and development.62 Carnitine, acylcarnitines (ie, decanoylcarnitine and octenoylcarnitine) play key role in transporting acyl-groups (including fatty acids) from the cytoplasm into the mitochondria so that they can be broken down to produce energy, which is known as β-oxidation.63 It was also concluded that incomplete muscle fatty acid β-oxidation causes acylcarnitine accumulation and associated oxidative stress, raising the possibility that these metabolites play a role in muscle insulin resistance.64
Glycerol is a three-carbon alcohol metabolite that constitutes the backbone to which fatty acids and lipids are bound to form triglycerides and phospholipids. The metabolism of stored fats leads to the release of glycerol and fatty acids into the bloodstream. The glycerol component can be converted into glucose by the liver and provides energy for cellular metabolism. Pyruvate is a key intermediate in several metabolic pathways throughout the cell. Particularly, pyruvate can be made from glucose through glycolysis, converted back to carbohydrates (such as glucose) via gluconeogenesis, or to fatty acids through a reaction with acetyl-CoA. Therefore, altered glucose, pyruvate and glycerol levels are accompanied by perturbations of lipid metabolism.
Activated Inflammation
Inflammation and related oxidative stress can induce endothelial dysfunction, which are the features of PE.65 Taurine is a vaso-protective and anti-hypertensive agent.66 It can be synthesized by the body from cysteine when vitamin B6 is present. Decreases in placental taurine levels could impair trophoblastic invasion and spiral artery remodeling67,68 and produce relative fetal intrauterine hypoxia, which may contribute to PE.69
Women with PE also showed altered choline and glycine, and hippurate, which may be related to increased oxidative stress. Choline and glycine are connected through the metabolic pathway of homocysteine, which increase the risk of PE.70 Glycine is a precursor to glutathione, a tripeptide important for protection against oxidative stress.71 Histidine is a precursor for histamine. L-histidine has anti-oxidant, anti-inflammatory and anti-secretory properties, mainly due to its singlet oxygen and hydroxyl radical scavenging characteristics. In many recent nonclinical studies, L-histidine has been shown to have significant potential as an anti-inflammatory therapeutic agent in a range of diseases.72
Impaired NO Production
Nitric oxide (NO) plays an important role in regulating various cardiovascular function. NO stimulates soluble guanylate cyclase to produce cGMP, which activates cGMP dependent Protein Kinase G (PKG) and subsequently results in smooth muscle cell relaxation, reduction of vascular resistance and eventually increase of blood perfusion in tissues.73 Metabolomics studies on PE found changed levels of dimethylamine and L-phenylalanine. These metabolites were suggested to affect NO production in many ways.
Dimethylamine may have dietary origins, but is also derived from asymmetric dimethylarginine, a biomarker of increased renal disorder74 and cardiovascular risk.75,76 Asymmetric dimethylarginine is an endogenous inhibitor of NO synthases that attenuates NO production by inhibition of nitric oxide synthase (NOS) activity. Asymmetric dimethylarginine also enhances NOS uncoupling to produce reactive oxygen species such as superoxide anion and peroxynitrite, which could further reduce the cardiovascular NO bioavailability.75 Besides, tetrahydrobiopterin (BH4) has been reported to be a powerful oxidant of oxygen.77 L-phenylalanine, as the essential amino acid, may increase BH4-dependent NO production, thus helps restore vascular function and decrease blood pressure in rats.78
Methodological Considerations of Metabolomics Studies on PE
In metabolomics research, NMR and MS are common analytical techniques, and many high-frequent reported metabolites were identified by either NMR or MS. Different methods have their strengths and weaknesses. NMR is quantitative and requires minimal sample preparation making it more reproducible among different studies, while MS can offer a combined sensitivity and selectivity platform for metabolomics study.79 The chosen method may influence the reporting of results to an extent. Unfortunately, only two included articles in this review employed both methods. More studies combining the two may provide the greatest opportunity to identify accurate and sensitive metabolic biomarkers of PE.
Limitations of Current Metabolomics Studies on PE
Several limitations of the existing metabolomics studies on PE should be noted. First, the biological samples are limited to blood samples in the majority of studies. Other types of samples (eg, urine and placenta) should be analyzed to allow researchers to obtain a more comprehensive understanding of metabolic signatures. Second, many studies had relatively small sample sizes, limiting statistical power and the generalizability of their research results. Third, the identification and analysis of metabolites are mostly based on public databases (eg, Human Metabolome Database), therefore some unidentified metabolites may be ignored. Fourth, when researchers used blood or other biofluids, profiling of metabolites may be greatly dynamic and influenced by diet, immune status, lifestyle, and environmental factors.80 Unfortunately, the effect of sample collection on results is often overlooked in metabolomics studies. Finally, insufficient validations exist for many studies. Multiple independent validations, including external cohorts and animal and cell experiments, are necessary before translating the results into clinical practice. Future studies addressing these limitations may be performed to complement, validate, and expand current findings. In addition, the integration of metabolomics with other omics platforms (eg, genomics, transcriptomics, and proteomics) will enable researchers to obtain a global perspective of the complexity of PE.
Conclusion
In summary, this study presents a systematic review and analysis of metabolomics research into different aspects of PE. A series of small-molecule metabolites, including various amino acids, have been reported to be associated with PE. Those metabolites consistently reported to be associated with PE, including decanoylcarnitine, 3-hydroxyisovaleric acid, and taurine, warrant particular attention. Metabolic pathways, such as alanine, aspartate and glutamate metabolism, are suggested to be involved in the development and progression of PE. In the future, the construction of diagnostic or predictive models for PE may be achieved by combining these potential metabolite markers themselves or combining these metabolite markers with clinical parameters. Furthermore, the identification of interventional or therapeutic targets based on the underlying metabolic mechanisms is worthy of exploration.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lisonkova S, Joseph KS. Incidence of preeclampsia: risk factors and outcomes associated with early- versus late-onset disease. Am J Obstet Gynecol. 2013;209(6):544e541–544 e512. doi: 10.1016/j.ajog.2013.08.019 [DOI] [PubMed] [Google Scholar]
- 2.Abalos E, Cuesta C, Grosso AL, Chou D, Say L. Global and regional estimates of preeclampsia and eclampsia: a systematic review. Eur J Obstet Gynecol Reprod Biol. 2013;170(1):1–7. doi: 10.1016/j.ejogrb.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 3.Pittara T, Vyrides A, Lamnisos D, Giannakou K. Preeclampsia and long-term health outcomes for mother and infant: an umbrella review. BJOG. 2021;128:1421–1430. doi: 10.1111/1471-0528.16683 [DOI] [PubMed] [Google Scholar]
- 4.Burton GJ, Redman CW, Roberts JM, Moffett A. Pre-eclampsia: pathophysiology and clinical implications. BMJ. 2019;366:l2381. doi: 10.1136/bmj.l2381 [DOI] [PubMed] [Google Scholar]
- 5.Liang L, Rasmussen MH, Piening B, et al. Metabolic dynamics and prediction of gestational age and time to delivery in pregnant women. Cell. 2020;181(7):1680–1692 e1615. doi: 10.1016/j.cell.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nobakht BF. Application of metabolomics to preeclampsia diagnosis. Syst Biol Reprod Med. 2018;64(5):324–339. doi: 10.1080/19396368.2018.1482968 [DOI] [PubMed] [Google Scholar]
- 7.Pang Z, Chong J, Zhou G, et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49(W1):W388–W396. doi: 10.1093/nar/gkab382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kivelä J, Sormunen-Harju H, Girchenko PV, et al. Longitudinal metabolic profiling of maternal obesity, gestational diabetes and hypertensive pregnancy disorders. J Clin Endocrinol Metab. 2021;106:e4372–e4388. doi: 10.1210/clinem/dgab475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jääskeläinen T, Kärkkäinen O, Jokkala J, et al. A non-targeted LC-MS metabolic profiling of pregnancy: longitudinal evidence from Healthy and pre-eclamptic pregnancies. Metabolomics. 2021;17(2):20. doi: 10.1007/s11306-020-01752-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harville EW, Li YY, Pan K, McRitchie S, Pathmasiri W, Sumner S. Untargeted analysis of first trimester serum to reveal biomarkers of pregnancy complications: a case-control discovery phase study. Sci Rep. 2021;11(1):3468. doi: 10.1038/s41598-021-82804-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan B, Ma Y, Zhang L, Li N, Zhang J. The application of metabolomics analysis in the research of gestational diabetes mellitus and preeclampsia. J Obstet Gynaecol Res. 2020;46(8):1310–1318. doi: 10.1111/jog.14304 [DOI] [PubMed] [Google Scholar]
- 12.Sovio U, McBride N, Wood AM, et al. 4-Hydroxyglutamate is a novel predictor of pre-eclampsia. Int J Epidemiol. 2020;49(1):301–311. doi: 10.1093/ije/dyz098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SM, Kang Y, Lee EM, et al. Metabolomic biomarkers in midtrimester maternal plasma can accurately predict the development of preeclampsia. Sci Rep. 2020;10(1):16142. doi: 10.1038/s41598-020-72852-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenny LC, Thomas G, Poston L, et al. Prediction of preeclampsia risk in first time pregnant women: metabolite biomarkers for a clinical test. PLoS One. 2020;15(12):e0244369. doi: 10.1371/journal.pone.0244369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong X, Zhang B, Liang L, et al. Postpartum plasma metabolomic profile among women with preeclampsia and preterm delivery: implications for long-term health. BMC Med. 2020;18(1). doi: 10.1186/s12916-020-01741-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sander KN, Kim DH, Ortori CA, et al. Untargeted analysis of plasma samples from pre-eclamptic women reveals polar and apolar changes in the metabolome. Metabolomics. 2019;15(12):157. doi: 10.1007/s11306-019-1600-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Zu L, Cai W, et al. Metabolomics revealed decreased level of omega-3 PUFA-derived protective eicosanoids in pregnant women with pre-eclampsia. Clin Exp Pharmacol Physiol. 2019;46(8):705–710. doi: 10.1111/1440-1681.13095 [DOI] [PubMed] [Google Scholar]
- 18.Lee SM, Moon JY, Lim BY, et al. Increased biosynthesis and accumulation of cholesterol in maternal plasma, but not amniotic fluid in pre-eclampsia. Sci Rep. 2019;9(1):1550. doi: 10.1038/s41598-018-37757-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Do Nascimento E, Menezes HC, Resende RR, Goulart VAM, Cardeal ZL. Determination of amino acids in plasma samples of preeclampsia patients by liquid chromatography coupled to high-resolution mass spectrometry. J Braz Chem Soc. 2019;30(10):2136–2143. [Google Scholar]
- 20.Austdal M, Silva GB, Bowe S, et al. Metabolomics identifies placental dysfunction and confirms flt-1 (FMS-like tyrosine kinase receptor 1) biomarker specificity. Hypertension. 2019;74(5):1136–1143. doi: 10.1161/HYPERTENSIONAHA.119.13184 [DOI] [PubMed] [Google Scholar]
- 21.Yang X, Xu P, Zhang F, et al. AMPK hyper-activation alters fatty acids metabolism and impairs invasiveness of trophoblasts in preeclampsia. Cell Physiol Biochem. 2018;49(2):578–594. doi: 10.1159/000492995 [DOI] [PubMed] [Google Scholar]
- 22.Powell KL, Carrozzi A, Stephens AS, et al. Utility of metabolic profiling of serum in the diagnosis of pregnancy complications. Placenta. 2018;66:65–73. doi: 10.1016/j.placenta.2018.04.005 [DOI] [PubMed] [Google Scholar]
- 23.Jääskeläinen T, Kärkkäinen O, Jokkala J, et al. A non-targeted LC-MS profiling reveals elevated levels of carnitine precursors and trimethylated compounds in the cord plasma of pre-eclamptic infants. Sci Rep. 2018;8(1):14616. doi: 10.1038/s41598-018-32804-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X, Han T-L, Chen H, Baker PN, Qi H, Zhang H. Impaired mitochondrial fusion, autophagy, biogenesis and dysregulated lipid metabolism is associated with preeclampsia. Exp Cell Res. 2017;359(1):195–204. doi: 10.1016/j.yexcr.2017.07.029 [DOI] [PubMed] [Google Scholar]
- 25.Kelly RS, Croteau-Chonka DC, Dahlin A, et al. Integration of metabolomic and transcriptomic networks in pregnant women reveals biological pathways and predictive signatures associated with preeclampsia. Metabolomics. 2017;13(1). doi: 10.1007/s11306-016-1149-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen T, He P, Tan Y, Xu D. Biomarker identification and pathway analysis of preeclampsia based on serum metabolomics. Biochem Biophys Res Commun. 2017;485(1):119–125. doi: 10.1016/j.bbrc.2017.02.032 [DOI] [PubMed] [Google Scholar]
- 27.Bahado-Singh RO, Syngelaki A, Mandal R, et al. Metabolomic determination of pathogenesis of late-onset preeclampsia. J Maternal Fetal Neonatal Med. 2017;30(6):658–664. doi: 10.1080/14767058.2016.1185411 [DOI] [PubMed] [Google Scholar]
- 28.Bahado-Singh R, Poon LC, Yilmaz A, et al. Integrated proteomic and metabolomic prediction of term preeclampsia. Sci Rep. 2017;7(1):16189. doi: 10.1038/s41598-017-15882-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koster MPH, Vreeken RJ, Harms AC, et al. First-trimester serum acylcarnitine levels to predict preeclampsia: a metabolomics approach. Dis Markers. 2015;2015:1–8. doi: 10.1155/2015/857108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bahado-Singh RO, Syngelaki A, Akolekar R, et al. Validation of metabolomic models for prediction of early-onset preeclampsia. Am J Obstet Gynecol. 2015;213(4):530.e531–530.e510. doi: 10.1016/j.ajog.2015.06.044 [DOI] [PubMed] [Google Scholar]
- 31.Austdal M, Thomsen LCV, Tangeras LH, et al. Metabolic profiles of placenta in preeclampsia using HR-MAS MRS metabolomics. Placenta. 2015;36(12):1455–1462. doi: 10.1016/j.placenta.2015.10.019 [DOI] [PubMed] [Google Scholar]
- 32.Austdal M, Tangeras LH, Skrastad RB, et al. First trimester urine and serum metabolomics for prediction of preeclampsia and gestational hypertension: a prospective screening study. Int J Mol Sci. 2015;16(9):21520–21538. doi: 10.3390/ijms160921520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moon JY, Moon MH, Kim KT, et al. Cytochrome P450-mediated metabolic alterations in preeclampsia evaluated by quantitative steroid signatures. J Steroid Biochem Mol Biol. 2014;139:182–191. doi: 10.1016/j.jsbmb.2013.02.014 [DOI] [PubMed] [Google Scholar]
- 34.Kuc S, Koster MPH, Pennings JLA, et al. Metabolomics profiling for identification of novel potential markers in early prediction of preeclampsia. PLoS One. 2014;9(5):e98540. doi: 10.1371/journal.pone.0098540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korkes HA, Sass N, Moron AF, et al. Lipidomic assessment of plasma and placenta of women with early-onset preeclampsia. PLoS One. 2014;9(10):e110747. doi: 10.1371/journal.pone.0110747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Austdal M, Skrastad RB, Gundersen AS, Austgulen R, Iversen A-C, Bathen TF. Metabolomic biomarkers in serum and urine in women with preeclampsia. PLoS One. 2014;9(3):e91923. doi: 10.1371/journal.pone.0091923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senyavina NV, Khaustova SA, Grebennik TK, Pavlovich SV. Analysis of purine metabolites in maternal serum for evaluating the risk of gestosis. Bull Exp Biol Med. 2013;155(5):682–684. doi: 10.1007/s10517-013-2225-y [DOI] [PubMed] [Google Scholar]
- 38.Diaz SO, Barros AS, Goodfellow BJ, et al. Second trimester maternal urine for the diagnosis of trisomy 21 and prediction of poor pregnancy outcomes. J Proteome Res. 2013;12(6):2946–2957. doi: 10.1021/pr4002355 [DOI] [PubMed] [Google Scholar]
- 39.Bahado-Singh RO, Akolekar R, Mandal R, et al. First-trimester metabolomic detection of late-onset preeclampsia. Am J Obstet Gynecol. 2013;208(1):58.e51–57. doi: 10.1016/j.ajog.2012.11.003 [DOI] [PubMed] [Google Scholar]
- 40.Bahado-Singh RO, Akolekar R, Mandal R, et al. Metabolomics and first-trimester prediction of early-onset preeclampsia. J Maternal Fetal Neonatal Med. 2012;25(10):1840–1847. doi: 10.3109/14767058.2012.680254 [DOI] [PubMed] [Google Scholar]
- 41.Odibo AO, Goetzinger KR, Odibo L, et al. First-trimester prediction of preeclampsia using metabolomic biomarkers: a discovery phase study. Prenat Diagn. 2011;31(10):990–994. doi: 10.1002/pd.2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kenny LC, Broadhurst DI, Dunn W, et al. Robust early pregnancy prediction of later preeclampsia using metabolomic biomarkers. Hypertension. 2010;56(4):741–749. doi: 10.1161/HYPERTENSIONAHA.110.157297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner E, Brewster JA, Simpson NAB, Walker JJ, Fisher J. Aromatic amino acid biomarkers of preeclampsia - A nuclear magnetic resonance investigation. Hypertens Pregnancy. 2008;27(3):225–235. doi: 10.1080/10641950801955725 [DOI] [PubMed] [Google Scholar]
- 44.Kenny LC, Broadhurst D, Brown M, et al. Detection and identification of novel metabolomic biomarkers in preeclampsia. Reprod Sci. 2008;15(6):591–597. doi: 10.1177/1933719108316908 [DOI] [PubMed] [Google Scholar]
- 45.Turner E, Brewster JA, Simpson NA, Walker JJ, Fisher J. Plasma from women with preeclampsia has a low lipid and ketone body content–a nuclear magnetic resonance study. Hypertens Pregnancy. 2007;26(3):329–342. doi: 10.1080/10641950701436073 [DOI] [PubMed] [Google Scholar]
- 46.Jain S, Jayasimhulu K, Clark JF. Metabolomic analysis of molecular species of phospholipids from normotensive and preeclamptic human placenta electrospray ionization mass spectrometry. Front Biosci. 2004;9:3167–3175. doi: 10.2741/1470 [DOI] [PubMed] [Google Scholar]
- 47.McBride N, Yousefi P, Sovio U, et al. Do mass spectrometry-derived metabolomics improve the prediction of pregnancy-related disorders? Findings from a UK birth cohort with independent validation. Metabolites. 2021;11(8). doi: 10.3390/metabo11080530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He B, Liu Y, Maurya MR, et al. The maternal blood lipidome is indicative of the pathogenesis of severe preeclampsia. J Lipid Res. 2021;62:100118. doi: 10.1016/j.jlr.2021.100118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rana S, Lemoine E, Granger JP, Karumanchi SA. Preeclampsia: pathophysiology, challenges, and perspectives. Circ Res. 2019;124(7):1094–1112. doi: 10.1161/CIRCRESAHA.118.313276 [DOI] [PubMed] [Google Scholar]
- 50.Jeyabalan A, Conrad KP. Renal function during normal pregnancy and preeclampsia. Front Biosci. 2007;12(1):2425–2437. doi: 10.2741/2244 [DOI] [PubMed] [Google Scholar]
- 51.Scioscia M, Gumaa K, Rademacher TW. The link between insulin resistance and preeclampsia: new perspectives. J Reprod Immunol. 2009;82(2):100–105. doi: 10.1016/j.jri.2009.04.009 [DOI] [PubMed] [Google Scholar]
- 52.Da silva AA, Do Carmo JM, Li X, Wang Z, Mouton AJ, Hall JE. Role of hyperinsulinemia and insulin resistance in hypertension: metabolic syndrome revisited. Can J Cardiol. 2020;36(5):671–682. doi: 10.1016/j.cjca.2020.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qaid MM, Abdelrahman MM. Role of insulin and other related hormones in energy metabolism—a review. Cogent Food Agric. 2016;2(1):1267691. doi: 10.1080/23311932.2016.1267691 [DOI] [Google Scholar]
- 54.Yoon MS. The emerging role of branched-chain amino acids in insulin resistance and metabolism. Nutrients. 2016;8(7):405. doi: 10.3390/nu8070405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. 2014;10(12):723–736. doi: 10.1038/nrendo.2014.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu J, Xie G, Jia W, Jia WJ. Insulin resistance and the metabolism of branched-chain amino acids. Front Med. 2013;7(1):53–59. doi: 10.1007/s11684-013-0255-5 [DOI] [PubMed] [Google Scholar]
- 57.Zhang Q, Ford LA, Goodman KD, et al. LC-MS/MS method for quantitation of seven biomarkers in human plasma for the assessment of insulin resistance and impaired glucose tolerance. J Chromatogr B. 2016;1038:101–108. doi: 10.1016/j.jchromb.2016.10.025 [DOI] [PubMed] [Google Scholar]
- 58.Cheng S, Rhee EP, Larson MG, et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125(18):2222–2231. doi: 10.1161/CIRCULATIONAHA.111.067827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Demirci O, Tuğrul AS, Dolgun N, Sözen H, Eren S. Serum lipids level assessed in early pregnancy and risk of pre-eclampsia. J Obstet Gynaecol Res. 2011;37(10):1427–1432. doi: 10.1111/j.1447-0756.2011.01562.x [DOI] [PubMed] [Google Scholar]
- 60.Fushimi T, Suruga K, Oshima Y, Fukiharu M, Tsukamoto Y, Goda T. Dietary acetic acid reduces serum cholesterol and triacylglycerols in rats fed a cholesterol-rich diet. Br J Nutr. 2006;95(5):916–924. doi: 10.1079/BJN20061740 [DOI] [PubMed] [Google Scholar]
- 61.Kondo S, Tayama K, Tsukamoto Y, Ikeda K, Yamori Y. Antihypertensive effects of acetic acid and vinegar on spontaneously hypertensive rats. Biosci Biotechnol Biochem. 2001;65(12):2690–2694. doi: 10.1271/bbb.65.2690 [DOI] [PubMed] [Google Scholar]
- 62.Godhamgaonkar AA, Wadhwani NS, Joshi SR. Exploring the role of LC-PUFA metabolism in pregnancy complications. Prostaglandins Leukot Essent Fatty Acids. 2020;163:102203. doi: 10.1016/j.plefa.2020.102203 [DOI] [PubMed] [Google Scholar]
- 63.Grube M, Meyer Zu Schwabedissen H, Draber K, et al. Expression, localization, and function of the carnitine transporter octn2 (slc22a5) in human placenta. Drug Metabol Disposit. 2005;33(1):31–37. doi: 10.1124/dmd.104.001560 [DOI] [PubMed] [Google Scholar]
- 64.Aguer C, McCoin CS, Knotts TA, et al. Acylcarnitines: potential implications for skeletal muscle insulin resistance. FASEB J. 2015;29(1):336–345. doi: 10.1096/fj.14-255901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sánchez-Aranguren LC, Prada CE, Riaño-Medina CE, Lopez M. Endothelial dysfunction and preeclampsia: role of oxidative stress. Front Physiol. 2014;5:372. doi: 10.3389/fphys.2014.00372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wójcik OP, Koenig KL, Zeleniuch-Jacquotte A, Costa M, Chen Y. The potential protective effects of taurine on coronary heart disease. Atherosclerosis. 2010;208(1):19–25. doi: 10.1016/j.atherosclerosis.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Desforges M, Parsons L, Westwood M, Sibley CP, Greenwood SL. Taurine transport in human placental trophoblast is important for regulation of cell differentiation and survival. Cell Death Dis. 2013;4(3):e559. doi: 10.1038/cddis.2013.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Desforges M, Ditchfield A, Hirst CR, et al. Reduced placental taurine transporter (TauT) activity in pregnancies complicated by pre-eclampsia and maternal obesity. Adv Exp Med Biol. 2013;776:81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reuvekamp A, Velsing-Aarts FV, Poulina IE, Capello JJ, Duits AJ. Selective deficit of angiogenic growth factors characterises pregnancies complicated by pre-eclampsia. Br J Obstet Gynaecol. 1999;106(10):1019–1022. doi: 10.1111/j.1471-0528.1999.tb08107.x [DOI] [PubMed] [Google Scholar]
- 70.Braekke K, Ueland PM, Harsem NK, Karlsen A, Blomhoff R, Staff AC. Homocysteine, cysteine, and related metabolites in maternal and fetal plasma in preeclampsia. Pediatr Res. 2007;62(3):319–324. doi: 10.1203/PDR.0b013e318123fba2 [DOI] [PubMed] [Google Scholar]
- 71.Friesen RW, Novak EM, Hasman D, Innis SM. Relationship of dimethylglycine, choline, and betaine with oxoproline in plasma of pregnant women and their newborn infants. J Nutr. 2007;137(12):2641–2646. doi: 10.1093/jn/137.12.2641 [DOI] [PubMed] [Google Scholar]
- 72.Wade AM, Tucker HN. Antioxidant characteristics of L-histidine. J Nutr Biochem. 1998;9(6):308–315. [Google Scholar]
- 73.Carnicer R, Crabtree MJ, Sivakumaran V, Casadei B, Kass DA. Nitric oxide synthases in heart failure. Antioxid Redox Signal. 2013;18(9):1078–1099. doi: 10.1089/ars.2012.4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Teerlink T, Hennekes MW, Mulder C, Brulez HF. Determination of dimethylamine in biological samples by high-performance liquid chromatography. J Chromatogr B. 1997;691(2):269–276. doi: 10.1016/S0378-4347(96)00476-8 [DOI] [PubMed] [Google Scholar]
- 75.Tsikas D, Thum T, Becker T, et al. Accurate quantification of dimethylamine (DMA) in human urine by gas chromatography-mass spectrometry as pentafluorobenzamide derivative: evaluation of the relationship between DMA and its precursor asymmetric dimethylarginine (ADMA) in health and disease. J Chromatogr B. 2007;851(1–2):229–239. doi: 10.1016/j.jchromb.2006.09.015 [DOI] [PubMed] [Google Scholar]
- 76.Böger RH. Asymmetric dimethylarginine, an endogenous inhibitor of nitric oxide synthase, explains the “L-arginine paradox” and acts as a novel cardiovascular risk factor. J Nutr. 2004;134(10):2842S–2847S; discussion 2853S. doi: 10.1093/jn/134.10.2842S [DOI] [PubMed] [Google Scholar]
- 77.Laursen JB, Somers M, Kurz S, et al. Endothelial regulation of vasomotion in apoE-deficient mice: implications for interactions between peroxynitrite and tetrahydrobiopterin. Circulation. 2001;103(9):1282–1288. doi: 10.1161/01.CIR.103.9.1282 [DOI] [PubMed] [Google Scholar]
- 78.Wang Z, Cheng C, Yang X, Zhang C. L-phenylalanine attenuates high salt-induced hypertension in Dahl SS rats through activation of GCH1-BH4. PLoS One. 2021;16(4):e025012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Emwas AH. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol Biol. 2015;1277:161–193. [DOI] [PubMed] [Google Scholar]
- 80.Souza RT, Mayrink U, Leite DF, et al. Metabolomics applied to maternal and perinatal health: a review of new frontiers with a translation potential. Clinics. 2019;74. doi: 10.6061/clinics/2019/e894 [DOI] [PMC free article] [PubMed] [Google Scholar]