Abstract
IQGAP1 is a multidomain scaffold protein that coordinates the direction and impact of multiple signaling pathways by scaffolding its various binding partners. However, the spatial and temporal resolution of IQGAP1 scaffolding remains unclear. Here, we use fluorescence imaging and correlation methods that allow for real-time live-cell changes in IQGAP1 localization and complex formation during signaling. We find that IQGAP1 and PIPKIγ interact on both the plasma membrane and in cytosol. Epidermal growth factor (EGF) stimulation, which can initiate cytoskeletal changes, drives the movement of the cytosolic pool toward the plasma membrane to promote cytoskeletal changes. We also observe that a significant population of cytosolic IQGAP1-PIPKIγ complexes localize to early endosomes, and in some instances form aggregated clusters which become highly mobile upon EGF stimulation. Our imaging studies show that PIPKIγ and PI3K bind simultaneously to IQGAP1, which may accelerate conversion of PI4P to PI(3,4,5)P3 that is required for cytoskeletal changes. Additionally, we find that IQGAP1 is responsible for PIPKIγ association with two proteins associated with cytoskeletal changes, talin and Cdc42, during EGF stimulation. These results directly show that IQGAP1 provides a physical link between phosphoinositides (through PIPKIγ), focal adhesion formation (through talin), and cytoskeletal reorganization (through Cdc42) upon EGF stimulation. Taken together, our results support the importance of IQGAP1 in regulating cell migration by linking phosphoinositide lipid signaling with cytoskeletal reorganization.
Significance
IQGAP1 is scaffold protein that can bind to many different proteins simultaneously to organize several cell signaling pathways by scaffolding. We use advanced imaging methods to visualize IQGAP1 interactions in cells, specifically with phosphoinositide kinases, that had previously not been detailed. We see that IQGAP1 facilitates several cellular events upon epidermal growth factor (EGF) stimulation. These effects include the movement of the cytosolic IQGAP1-PIPKIγ complexes toward the plasma membrane, the simultaneous binding of PIPKIγ and PI3K to IQGAP1 that may promote the production of PI(3,4,5)P3, and PIPKIγ’s associations with talin and Cdc42—two proteins associated with cell migration. Overall, we show that IQGAP1 links phosphoinositides and cytoskeletal reorganization in response to EGF, highlighting the role of IQGAP1 in cell movement.
Introduction
Phosphoinositides are signaling lipids that are located on the cytoplasmic leaflet of a cell's plasma and internal membranes. The adaptable phosphorylation of the hydroxyl groups at the inositol ring positions 3, 4, and 5 gives rise to seven phosphoinositide species. Phosphoinositides, in particular phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2) and phosphatidylinositol-3,4,5-trisphosphate (PI(3,4,5)P3) (1), play pivotal roles in the regulation of a wide variety of cellular processes as secondary messengers of cell signaling pathways (2,3). Spatiotemporal regulation of phosphoinositide-mediated biological processes is achieved by adjustments of the phosphorylation states of phosphoinositides by specific kinases and phosphatases in a time- and space-dependent manner (4). The majority of PI(4,5)P2 is generated from phosphatidylinositol-4-phosphate (PI(4)P) by type I phosphatidylinositol phosphate kinases (PIPKIs) (5). PI(4,5)P2 is in turn phosphorylated to PI(3,4,5)P3 by class I phosphatidylinositol-3-OH kinase (PI3K) (6), which is the key step in the activation of the PI3K/Akt signaling pathway promoting metabolism, proliferation, and angiogenesis (7,8).
Cell migration is a dynamic process that involves the cytoskeleton and focal adhesions, which in turn require coordination between phosphoinositides and cytoskeletal proteins. Irregular and enhanced cell migration and motility is considered a hallmark of several diseases that include cancer (9,10). Although the overall cellular concentration of PI(4,5)P2 remains relatively constant, it accumulates at a protrusive structure characterized by intense actin polymerization essential for cell polarity in migrating cells known as a leading edges (11,12). Changes in local PI(4,5)P2 concentrations are controlled by redistribution and activation of PI(4,5)P2 generating enzymes such as PIPKIs (13) among others. Additionally, the comparatively much smaller PI(3,4,5)P3 pools generated by PI3K activity localize to the leading edges (14,15). We focus specifically on PIPKIγ whose expression is closely linked to proliferation and cancer progression. PI(4,5)P2 levels are comparatively much higher than those of PI(3,4,5)P3, which is primarily produced in response to receptor-mediated signaling events. These receptors generally comprise receptor tyrosine kinases such as the epidermal growth factor receptor (EGFR), whose agonist EGF is a well-studied activator of the PI3K/Akt pathway (8).
Spatiotemporal coordination and organization of cell signals, such as those involved in phosphoinositide signaling, are often provided by scaffolding proteins that form heterogeneous complexes with several different partner proteins that may also have specific cellular localization and may be generated at specific time points (16). One such scaffolding protein is IQGAP1 (IQ-domain containing Ras GTPase Activating Protein 1), which is a large multidomain protein (195 kDa). Through scaffolding, IQGAP1 is known to affect key cellular functions, including proliferation, migration, cell adhesion, motility, and metabolism (17,18). IQGAP1 has been thought to form distinct scaffolds in terms of cell type and intracellular localization. However, the spatiotemporal dynamics of these protein complex formations remain unknown.
Recent studies indicate IQGAP1's propensity to scaffold the components of the PI3K/Akt signaling pathway that has been linked to cell migration (19,20). IQGAP1 associates with multiple PIPKIs, including the PIPKIα and PIPKIγ isoforms (21), as well as PI3K isoforms. IQGAP1 and PIPKI isoforms localize to specific sites on intracellular membranes and the plasma membrane that include the leading edge of migrating cells, cell-cell contact sites, and focal adhesions where they share many binding partners. In addition to these protein targets, IQGAP1 also binds PI(4,5)P2 and PI(3,4,5)P3 at distinct sites as demonstrated by in vitro studies (19,21). It has been hypothesized that PIPKI binding to IQGAP1 causes a conformational change that is a prerequisite for interactions with additional partners such as PI(4,5)P2 (17). IQGAP1 is also known to bind to EGFR (22). The activation of the PI3K/Akt pathway in response to EGF is affected by IQGAP1-PIPKI interactions (23) and, therefore, may play a mediating role in the maintenance of PI(3,4,5)P3 levels (19).
IQGAP1, PIPKIs, PI(4,5)P2, and PI(3,4,5)P3 are known to interact with several proteins that regulate cytoskeletal reorganization and focal adhesion formation. These cytoskeletal regulatory proteins include small GTPases such as Cdc42, a major initiator of the formation of the leading edge of migrating cells in the case of IQGAP1 and PI(4,5)P2 (24, 25, 26). The polarized distributions of PI(4,5)P2 and PI(3,4,5)P3 ensure that actin polymerization occurs at the leading edge of migrating cells by differentially binding to actin regulatory and focal adhesion proteins (11,27, 28, 29). A recent study showed that IQGAP1 binds to actin and regulates actin polymerization by organizing filaments into thin bundles, suppressing barbed end growth, and inhibiting filament disassembly (30). The PIPKI isoforms mediate PI(4,5)P2's associations with actin regulatory and focal adhesion proteins (13), and it is therefore likely that IQGAP1 provides scaffolding between phosphoinositide signaling and the cytoskeleton.
Since IQGAP1 lacks catalytic activity (31), it participates in cellular signaling primarily as a scaffold (18). However, IQGAP1's interactions, including those with phosphoinositide kinases, have only been studied by biochemical methods using recombinant purified proteins, and its ability to coordinate signaling pathways in living cells is not well understood. Here, we characterize the direct physical interactions between IQGAP1 and different phosphoinositide kinases by using an array of biophysical fluorescence imaging techniques that allow us to detect protein-protein interactions both spatially and temporally. The techniques and the rationale for using them are described in materials and methods.
While IQGAP1 has been reported to enhance cell migration upon EGF stimulation by linking phosphoinositide lipid signaling with cytoskeletal reorganization, its ability to simultaneously bind and coordinate these pathways in space and time have not been elucidated using either biochemical techniques or confocal imaging. Here, we have used fluorescence imaging and correlation methods to investigate and monitor distinct IQGAP1complexes that include PIPK1γ, PI3K, talin, and Cdc42 in different cellular compartments with high resolution in the absence and presence of EGF stimulation. We observe IQGAP1-PIPKIγ complexes not only at the plasma membrane but also on early endosomes and cytosolic aggregated clusters. We see that IQGAP1 scaffolds PIPKIγ and PI3K that possibly promotes the production of PI(3,4,5)P3. We also see that IQGAP1 scaffolds PIPKIγ′s EGF-mediated association with talin and Cdc42. These results directly show that IQGAP1 provides a physical link between phosphoinositides (through PIPKIγ), focal adhesion formation (through talin), and cytoskeletal reorganization (through Cdc42) upon EGF stimulation.
Materials and methods
Materials
HeLa, HepG2A, and NIH/3T3 cells were obtained from the American Type Culture Collection (Manassas, VA, USA). Untagged eGFP and mCherry plasmids were obtained from Clontech (Mountain View, CA, USA). dsRed-PIPKIγ was a kind gift from Dr. Richard Anderson (University of Wisconsin, Madison, WI, USA). PI4K plasmids, eGFP-PH Btk1, eGFP-PH-PLCδ1, and mCherry-PH-SidM were a kind gift from Dr. Tamas Balla (NIH, Bethesda, MD, USA), PI3K 110 plasmids were a kind gift from Dr. Jonathan Backer (Albert Einstein School of Medicine, Bronx, NY, USA) All other plasmids were obtained through AddGene (Boston, MA, USA) from the following investigators: eGFP-IQGAP1 (Dr. David Sacks), eGFP- PIPKIγ (Dr. Pietro de Camilli), eYFP-p85 PI3K (Dr. Lewis Cantley), mCherry-PH-PLCδ1(Dr. Narasimhan Gautam), mCherry-PH-Akt1(Dr. Moritoshi Sato), mCherry-PH-Akt2 (Dr. Ivan Yudishkin), eGFP-Cdc42 (Dr. Gary Bokoch), mCherry-talin (Dr. Michael Davidson). On-Target Plus SmartPool IQGAP1-siRNA (GE Dharmacon, Lafayette, CO, USA) was used to knock down IQGAP1 while the scrambled small interfering RNA (siRNA) was obtained from Ambion (Thermo Fisher Scientific, Waltham, MA, USA).
Cell culture
HeLa, HepG2A, and NIH 3T3 cells were incubated in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific) supplemented with 5% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA, USA) at 37°C with 5% CO2. Transfection of plasmids and siRNA was performed using Lipofectamine 3000 (Thermo Fisher Scientific) in antibiotic-free medium as per the manufacturer's instructions. For treatment with agonist, cells were cultured in a serum-free high-glucose DMEM for 2 h, following which they were supplemented with EGF (100 ng/mL), isoproterenol (10 μM), or LY2942001 (1 μM) and incubated for at least 1 h.
Imaging
Cells plated on MatTek chambers (MatTek, Ashland, MA, USA) were fixed using 3.7% formaldehyde and permeabilized with 0.2% nonyl phenoxypolyethoxylethanol in phosphate-buffered saline (PBS) for 5 min and then blocked in PBS containing 1% bovine serum albumin for 1 h. If appropriate, cells were then incubated with the primary antibody diluted to 1:1000 for 1.5 h at 37°C, followed by incubation with Alexa-labeled secondary antibody for 0.5 h at the same temperature. Cells were washed with Tris-buffered saline after the incubations. Images of the cells were obtained using a Zeiss LSM 510 confocal microscope. Live-cell images were taken on the same microscope while the cells were incubated in a custom-built chamber at 37°C and 5% CO2. The fluorescence images were obtained with a Nikon (Minato, Tokyo, Japan) Eclipse Ti inverted microscope with a TIRF Illuminator and a Nikon 60× CFI Apo TIRF oil objective. For excitation sources, coherent 488 nm, 561 nm, and 647 nm sapphire lasers were used. Images were captured with an Andor3iXon CCD camera (Belfast, UK). The images were analyzed using Zeiss software and/or ImageJ (32) (National Institutes of Health, Bethesda, MD, USA).
Fluorescence biophysics techniques
Fluorescence lifetime imaging (FLIM) in conjugation with Förster resonance energy transfer (FRET) allows for visualization and quantification of protein-protein interactions in live cells. This method utilizes the decrease of the donor's fluorescence lifetime due to transfer to an acceptor as a means to generate an image that highlights the cellular location(s) where the protein-protein interaction occurs. Given the large size of the tagged proteins, any observed changes in donor lifetime in the presence of the proteins tagged with an acceptor group can be attributed to changes due to direct physical protein-protein interactions. We also utilize techniques based on fluorescence fluctuation spectroscopy, which observes subtle changes in the intensities and movement of fluorescently labeled proteins in conjugation with fluorescent microscopy. One such technique is fluorescence correlation spectroscopy (FCS), which was used in this study to characterize the differences in the mobility of micropopulations of proteins in small focal areas across a living cell at both basal and stimulated states. The movement of the fluorescently tagged proteins inside the excited focal volume generates a signal that is subsequently autocorrelated to quantify physical quantities such as diffusion coefficients and concentrations (33, 34, 35). An extension of FCS is fluorescence cross-correlation spectroscopy (FCCS) whereby we observe concerted protein movements. In this case, the proteins are labeled with two spectrally distinct fluorophores that enable the correlative analysis of the dynamic behavior of the two proteins. Interacting proteins show synchronous diffusion through the focal volume (36,37), which can be related to the degree of protein-protein interaction. Additionally, we performed number and brightness (N&B) studies to study changes in protein localization and oligomerization state across the whole cell unlike in the case of FCS and FCCS, which are limited to small focal areas.
Fluorescence lifetime imaging microscopy
FLIM is a method to measure FRET between labeled proteins in cells. FRET reduces the fluorescence lifetime of a donor molecule due to transfer of energy to an acceptor. Because of its steep distance dependence in the low-nanometer range, FRET transfer requires a direct physical interaction between the proteins to which the probes are attached. If the donor is excited with intensity modulated light, the phase of the acceptor fluorescence will be shifted. The fluorescence lifetime can be calculated from these phase shifts. We can determine the phase and modulation for each individual pixel of the image and plot the phase shifts and modulation decreases on a phasor plot. The phasor plot is a combined graphical representation of all the raw FLIM data in a vector space. The phasor space is constructed by using two phasor vectors (G, S), where each component is represented as shown in the following equation:
| (1) |
mx,y and φx,y are the modulation ratio and the phase delay measured for a particular modulation frequency (ω) at a pixel location (x,y). For a single lifetime population, the values from all of the pixels will fall on the phasor arc with longer lifetimes displayed to the left and shorter lifetimes to the right. Since FRET shortens donor lifetimes, FRET will manifest itself by moving the (g(ω),s(ω)) data point to the right. For a mixed population of donor molecules, some of which undergoing FRET, the resulting phasor points localize inside the phasor arc (38). Distributions inside the phasor arc therefore indicate FRET transfer and, hence, association of the two labeled proteins. The lifetime of an individual pixel can be calculated by
| (2) |
For our experiments, we used green fluorescent donors (such as enhanced green fluorescent protein (eGFP)) or yellow fluorescent donor eYFP, with red fluorescent proteins such as mCherry and dsRed acting as the FRET acceptors.
FLIM was performed by acquiring images of live cells plated on MatTek chambers using a 2-photon MaiTai laser (Spectra-Physics, Santa Clara, CA, USA) (excitation 850 nm at 80 MHz) and a Nikon inverted confocal microscope in an ISS Alba System (Champaign, IL, USA).The Images were analyzed using ISS VistaVision and ImageJ software packages. Atto 425 fluorescent dye (t = 3.61 ns) was used to calibrate the sample lifetimes.
Fluorescence correlation spectroscopy
FCS measurements of cells expressing fluorescent fusion protein were done on a 2-photon MaiTai laser (Spectra-Physics) (excitation 930 nm at 80 MHz) and a Nikon inverted confocal microscope in an ISS Alba System. The beam waist, ω0, and focal volume were calibrated with various concentrations of Alexa 488 dye (D = 425 μm2/s). Measurements of each cell were taken over 30 s and repeated more than four times. For each cell, the traces were averaged to obtain the final autocorrelation function to be used for fitting. The power of the excitation laser was adjusted such that there was sufficient signal-to-noise ratio and minimal photobleaching.
| (3) |
where τ is the correlation time, τD is the average time a particle spends in the confocal volume, N is the average number of molecules in the confocal volume, and S is the structural parameter. S was set to 100 (quasi-infinite) for two-dimensional diffusion. The diffusion coefficient, D, is calculated from the τD of a molecule using the Einstein relation for diffusion r2 = 4D × τD, where r is the radius of the observation volume.
The concentrations are derived from fitting the experimental data using the 2-photon cross-correlation fitting model that was provided to us by ISS, who manufactured our system (39).
This model presumes that:
-
•
Ca and Da (concentration and diffusion coefficient of acceptor) supposed to be in Ch1 will exist in Ch1 and cross-correlation channel;
-
•
Cd and Dd (concentration and diffusion coefficient of donor) supposed to be in Ch2 will exist in Ch2 and cross-correlation channel;
-
•
Cad and Dad (concentration and diffusion coefficient of bounded donor and acceptor) will exist in Ch1, Ch2, and cross-correlation channel;
-
•
w0 and z0 denote lateral radius and axial radius of the focal area, respectively.
Simply,
Ch1 fits with equation
| (4) |
Ch2 fits with equation
| (5) |
Cross correlation fits with equation
| (6) |
Number and brightness
N&B analysis is a powerful tool that has been used previously to quantify graphically the aggregation state of diffusing proteins in living cells (40, 41, 42, 43). N&B analysis can determine of the number (N) of diffusing particles within a given focal area and the intrinsic brightness (B) of each particle represented by pixels in an image and provides a map of brightness for every pixel using the following equation:
| (7) |
where I denotes the intensity of the signal, σ2 is the variance of the signal, σ02 is the readout noise variance of the detection electronics, and offset is the detector offset. The analysis has been described in detail earlier (40). Higher variance in fluorescence is associated with higher-order oligomers. Therefore, the brightness-versus-intensity map can be used to determine the size of the aggregate at a given location as the brightness B can directly relate to the dimensions of the fluorescent molecules (41).
The number of particles can be derived by the formula
| (8) |
We prefer to use the B value because it accounts for the immobile fraction as it assigns them a value of 1. The approximate value of oligomeric state can be calculated using the following calculation:
| (9) |
We used untagged eGFP, YFP, and mCherry as controls to calculate the baseline monomeric values. Specifically for eGFP, the baseline B value was about 1.05, which was comparable with previous studies. Based on this observation and adjusting for offset, we chose to use the percentage of pixels with B values above 1.5 to quantify the oligomeric aggregates in the cells.
N&B studies were performed by acquiring images of live cells plated on MatTek chambers (MatTek, MA, USA) using a 2-photon MaiTai laser (Spectra-Physics) and a Nikon inverted confocal microscope in an ISS Alba System. The images were analyzed using ISS VistaVision and SimFCS 4 (Irvine, CA, USA) software packages. The analysis has been described in more detail elsewhere (42,43).
Coimmunoprecipitation
HeLa cells were lysed with 500 μL of buffer containing 150 mM NaCl, 20 mM HEPES, 2 mM MgCl2, 5 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, and cOmplete protease inhibitor cocktail tablet (Roche/Millipore Sigma, St. Louis, MO, USA). After preclearing non-specifically binding proteins by incubating lysate with 20 μL of Protein A beads (Thermo Fisher Scientific), the lysate was incubated with 5 μL of anti-IQGAP1 antibody (Abcam, Boston, MA, USA) overnight at 4°C after the removal of beads. Subsequently an additional 20 μL of Protein A beads was added to the mixture, which was then gently rotated for 4 h at 4°C. The unbound proteins were separated from the beads, which were then washed twice with the lysis buffer. The bound proteins were then eluted from the beads in sample buffer at 95°C for 3 min and were analyzed using SDS-PAGE and Western blotting.
Mass spectroscopy
The protein sample obtained from coimmunoprecipitation was run on an SDS-PAGE gel and stained using Coomassie blue. The pieces of gel containing the protein were cut into 1 × 1 mm pieces and placed in 1.5 mL tubes with 1 mL of water for 30 min. This sample was sent to a University of Massachusetts Medical School Mass Spectroscopy Facility (Dr. John Leszyk) to undergo mass spectroscopy as described previously (44). Raw data files were peak processed with Proteome Discoverer (version 2.1, Thermo Fisher Scientific) before database searching with Mascot Server (version 2.5) against the Uniprot database. Search results were then loaded into the Scaffold Viewer (Proteome Software, Portland, OR, USA) for peptide/protein validation and label-free quantitation. The data were further analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (45) (National Institutes of Health).
Statistics
All data were analyzed using Sigmaplot (SysStat, San Jose, CA, USA) using Student's t-test substituted by the Mann-Whitney rank sum test if the data were not normally distributed. A difference between two groups was considered statistically significant only with a p-value below 0.05.
Results
IQGAP1 associates with PIPKIγ at plasma membrane and in cytosol
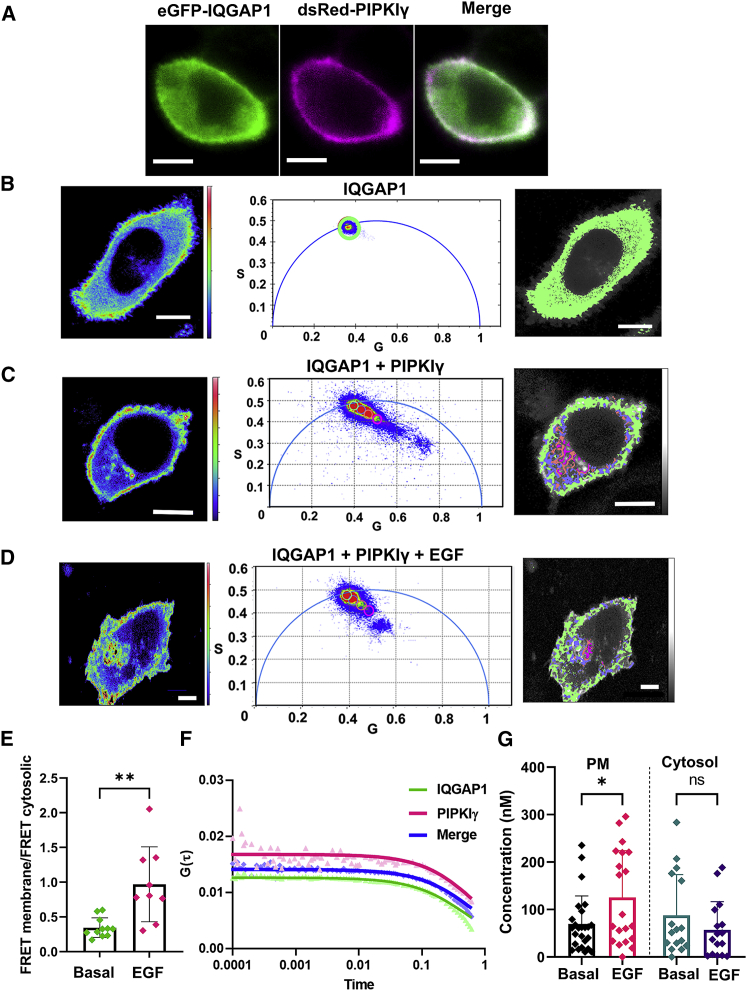
Fluorescence confocal microscopy images of HeLa cells expressing both eGFP-IQGAP1 and dsRed-PIPKIγ show a high degree of colocalization between the two proteins at the plasma membrane (Fig. 1 A), corroborating previous studies (21). Fluorescence lifetime imaging microscopy (FLIM) of these cells expressing eGFP-IQGAP1 generates an intensity profile image showing that IQGAP1 is distributed across the cytosol and plasma membrane region but is absent in the nucleus (Fig. 1 B, left). A phasor analysis (Fig. 1 B, center) provides a graphical representation of fluorescence lifetime distributions that allows the observer to distinguish and separate individual lifetime populations within an FLIM image. In the phasor plot, the individual lifetimes from each pixel in the FLIM image are plotted as the sine versus the cosine of the phase angle between the exciting and emitted light (multiplied in each case by the amplitude modulation ratio). Since phasor representation does not require fitting, the raw lifetime data without any modifications are seen (for a detailed description of the phasor analysis of FLIM images, see (46)). In HeLa cells expressing only eGFP-IQGAP1, eGFP lifetimes exhibit a largely homogeneous population as indicated by a single circular spot on the arc of the phasor plot (Fig. 1 B, center), analogous to cells expressing only free eGFP (Fig. S1 A). The pixels constituting this spot represent a uniform lifetime across the entire cell (Fig. 1 B, right).
Figure 1.
IQGAP1 strongly associates with PIPKIγ. (A) A representative image showing colocalization between eGFP-IQGAP1 and dsRed-PIPKIγ in HeLa cells. (B) A false-color image of a representative cell (left) shows that eGFP-IQGAP1 expressed in HeLa cells has a broad distribution across the cell except the nucleus. Plotting the raw lifetime values of each pixel in the image as a phasor plot (described in materials and methods, center) shows that eGFP-IQGAP1 has a single fluorescent lifetime indicated by a homogenous population on the phasor arc. The pixels included in the green circle of the phasor plot (lifetime center = 2.56 ns) are false-colored green and overlaid on a grayscale image of the cell (right), leading to the conclusion that the fluorescence lifetime is similar throughout the cell. The green pixels completely cover the gray of the cell. (C) When coexpressed with dsRed-PIPKIγ, the representative cell image (left) shows that IQGAP1 is primarily localized to the membrane. The phasor plots (center) show distinct eGFP-IQGAP1 lifetime populations both on the arc and inside the arc due to reduced lifetimes caused by FRET. The distinct regions on the phasor plots are highlighted by a green circle indicating higher (non-FRET) lifetimes (lifetime center = 2.55 ns, top) or by blue, orange, and magenta circles indicating shortened lifetimes (blue lifetime center = 2.1 ns, orange lifetime center = 1.9 ns, magenta lifetime center = 1.7 ns). The pixels underlying these circles are false colored and overlaid on grayscale cell images (right). (D) After stimulation with 100 ng/mL EGF, we still see a comet-like projection in cells expressing both eGFP-IQGAP1 and dsRed-PIPKIγ in the phasor plots (center), showing distinct eGFP-IQGAP1 lifetime populations both on the arc and inside the arc due to reduced lifetimes caused by FRET. The distinct regions on the phasor plots from the representative image (left) are highlighted as described above (right). n ≥ 10 in at least two individual experiments. (E) The ratios of FRET pixels in the membrane versus FRET pixels in the cytosol were quantified per image from the experiments described above in basal and EGF treatment groups. (F) Representative correlation curves from HeLa cells expressing eGFP-IQGAP1 and dsRed-PIPKIγ simultaneously. (G) Concentrations of the IQGAP1-PIPKIγ complexes were derived from the cross-correlated curves of plasma-membrane-localized focal areas on HeLa cells expressing eGFP-IQGAP1 and dsRed-PIPKIγ both at a basal state and after being stimulated with EGF (100 ng/mL for 1 h). φ denotes fluorescence lifetime in figure legends. n ≥ 10 in at least two individual experiments; siRNA treatment was used for the control groups. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001; ns, not significant. Error bars denote standard deviation. Scale bars, 10 μm. To see this figure in color, go online.
When we coexpress eGFP-IQGAP1 and dsRed-labeled PIPKIγ, eGFP-IQGAP1 localizes primarily toward the plasma membrane (Fig. 1, A and C). The average eGFP-IQGAP1 lifetime decreases by ∼10% due to FRET to dsRed-PIPKIγ compared with cells that express only eGFP (Fig. S1 B). This decrease in fluorescence lifetime is significantly greater than the ∼1%–3% decrease in lifetime typically associated with nonspecific binding and indicates an interaction between IQGAP1 and PIPKIγ (Fig. S2, A and B). In phasor plots, the eGFP/dsRed FRET results in a shift of the eGFP lifetimes inside the arc, giving the data a comet-like appearance. The points inside the phasor arc correspond to a reduced lifetime due to FRET. The phasor representation of the data allows us to distinguish between distinct FRET (i.e., inside the arc) and non-FRET (on the arc) pixel subpopulations (Fig. 1 C, center). When these lifetimes (identified in the phasor plot by four circles) are visualized in the whole-cell image (Fig. 1 C, right), we can identify the lower lifetime FRET populations corresponding to high IQGAP1/PIPK1γ complexes. Surprisingly, we see these FRET populations not only at the plasma membrane but also across the cytoplasm, indicating previously unknown subcellular locations of IQGAP1-PIPKIγ interactions (Fig. 1 C, right).
We then stimulated HeLa cells expressing fluorescently tagged proteins with EGF to observe its effects on IQGAP1-PIPKIγ interactions. Under these conditions, we see the comet-like appearances inside the phasor arc endure (Fig. 1 D, left). Additionally, FRET and non-FRET pixel subpopulations remain across the cytosol and on the plasma membrane (Fig. 1 D, right). While there is no difference in the overall (global) lifetime decrease with stimulation (Fig. S2 B), analysis of membrane-localized and cytosolic FRET reveals that the relative proportion of FRET between IQGAP1 and PIPKIγ on the plasma membrane increases upon EGF stimulation (Fig. 1 E).
To better understand IQGAP1-PIPKIγ complexes at the plasma membrane, we monitored their correlated movement by FCCS. These measurements show three correlation curves from fluctuations of eGFP-IQGAP1 particles, dsRed-PIPKIγ particles, and the cross correlation, which synchronizes with the shape of the other two curves only when there is a physical interaction between the two proteins (Fig. 1 F). Deriving the concentrations of the IQGAP1-PIPKIγ complexes after fitting the cross-correlation curves revealed a statistically significant concentration increase of the plasma membrane resident IQGAP1-PIPK1γ complexes upon EGF stimulation (Fig. 1 G). Thus, the interactions between IQGAP1 and PIPKIγ on plasma membrane significantly increase when stimulated with EGF. A similar interaction between IQGAP1 and PIPKIγ is also seen in NIH 3T3 fibroblasts and the liver cancer cell line HepG2 (Fig. S2 C).
EGF-induced changes to PIPKIγ localization are partly mediated by IQGAP1
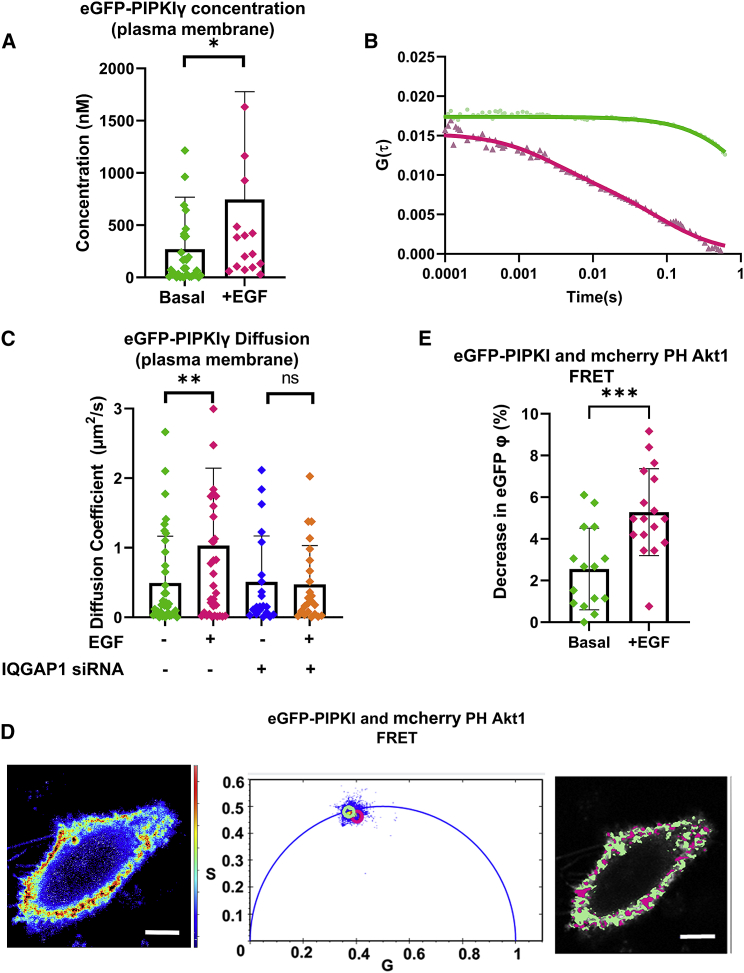
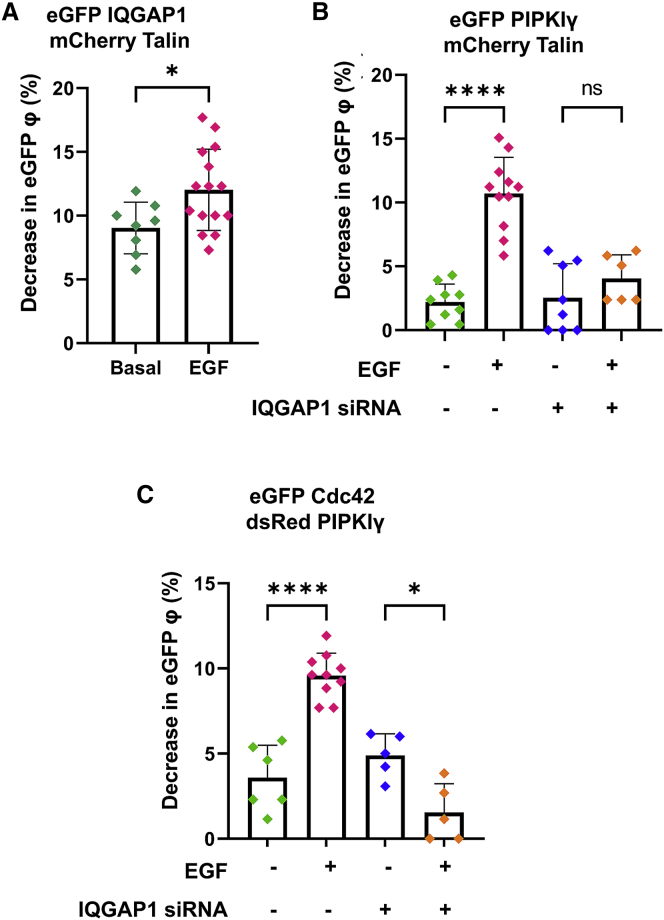
We used FCS to quantify the concentrations and diffusion coefficients of eGFP-PIPKIγ populations localized at the plasma membrane. The concentrations of eGFP-PIPKIγ particles can be derived from the FCS correlation curves after fitting the raw data from plasma-membrane-localized focal areas (Fig. 2 A; representative curves, Fig. 2 B). The data reveal a statistically significant increase in the plasma membrane concentrations of PIPKIγ upon EGF stimulation. Moreover, the diffusion coefficients of eGFP-PIPKIγ particles also show a statistically significant increase in their values. An siRNA-mediated knockdown of IQGAP1 (Fig. S3 A) abrogates any EGF-mediated changes in PIPKI diffusion and concentrations. These results suggest that IQGAP1 is partly responsible for alterations to PIPKI particle behavior induced by EGF (Fig. 2 C).
Figure 2.
PIPKI moves to plasma membrane in response to EGF. (A) Concentrations of eGFP-PIPKIγ particles derived from FCS correlation curves in plasma-membrane-localized focal areas on cells both at a basal state and after EGF stimulation (100 ng/mL). (B) Diffusion coefficients of eGFP-PIPKIγ particles derived from FCS correlation curves in plasma-membrane-localized focal areas on cells both at a basal state and after EGF stimulation (100 ng/mL) in the presence and absence of IQGAP1 expression. (C) FCS correlation curves from cells expressing eGFP-PIPKIγ that represent the range of diffusion coefficients (D) from our imaging data. The lighter-shade dots represent the raw data while the darker-colored curve represents the fitted curve. Here, the green curve represents a low diffusion curve with a D of 0.004 μm2/s while the red curve represents a D of 1.2 μm2/s. (D) Images of a representative HeLa cell coexpressing mCherry-PH-Akt1 and eGFP-PIPKIγ (left), where the pixels of distinct regions on phasor plots (center) are highlighted to show the distinct regions. The distinct regions on the phasor plots are represented by a green circle indicating higher (non-FRET) lifetimes (lifetime center = 2.55 ns) or by magenta or yellow circles indicating shortened lifetimes (magenta lifetime center = 2.00 ns). The pixels underlying these circles are false colored and overlaid on grayscale cell images (right). (E) Coexpression of mCherry-PH-Akt1 and eGFP-PIPKIγ in HeLa cells results in decreased eGFP IQGAP1 lifetimes due to FRET only when stimulated by EGF (100 ng/mL). φ denotes fluorescence lifetime in figure legends. Scrambled (nonspecific) siRNA treatment was used for the control groups. n ≥ 10 measured in at least two independent experiments; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001; ns, not significant. Error bars denote standard deviation. Scale bars, 10 μm. To see this figure in color, go online.
IQGAP1 is known to bind to both PI(3,4,5)P3 and PI(4,5)P2. Fluorescently labeled pleckstrin homology (PH) domains have been shown to accurately report the cellular distribution of PI(4,5)P2 and/or PI(3,4,5)P3 pools (47,48). The PH domain of PLCδ1 binds specifically to PI(4,5)P2 (49, 50, 51) while the PH domains of Akt1, Btk1 (50), and Akt2 (52) bind PI(3,4,5)P3. For the eGFP-mCherry FRET pair, the distance at which half of the excitation energy of eGFP is transferred to mCherry is about 5.4 nm (53). Here, we observe the interactions of IQGAP1 and PIPKIγ, respectively, with fluorescently tagged PH domains using FLIM-FRET. Note that the bound PH domain occludes PI(4,5)P2 from other binding partners, although it is not expected to substantially interfere with EGF signaling (47,49, 50, 51). Using these sensors, we observe FRET between eGFP-PIPKIγ and mCherry-PH-Akt1 upon EGF stimulation. This result suggests that PIPKIγ, which produces PI(4,5)P2 from PI(4)P, is colocalized with the newly generated pools of PI(3,4,5)P3, presumably through IQGAP1, corroborating our earlier results (Fig. 2, D and E).
Focusing on the plasma membrane (Fig. 2 D), we find a high level of FRET between eGFP-PIPKIγ and mCherry-PH- PLCδ1 in the basal state that is not affected by EGF signaling. We also observe FRET in cells that express both eGFP-IQGAP1 and mCherry-PH-PLCδ1, confirming earlier results (21) that indicate an interaction between IQGAP1 and PI(4,5)P2. When the FRET and non-FRET populations are visualized separately, this interaction seems to be localized to plasma membrane and the nuclear regions, corroborating the predominant cellular localization of PI(4,5)P2. This interaction is not affected by EGF or the PI3K antagonist, LY294002. However, EGF stimulation enhances the interaction between eGFP-IQGAP1 and mCherry-PH-Akt1, which binds PI(3,4,5)P3, within an hour of stimulation, while treatment with LY294002 instead of EGF results in a loss of the FRET signal (Fig. S3 B). Taken together, these results suggest that PIPKIγ (which produces PI(4,5)P2) remains bound to IQGAP1 as PI3K produces PI(3,4,5)P3.
PIPKIγ and IQGAP1 interact in the cytosol
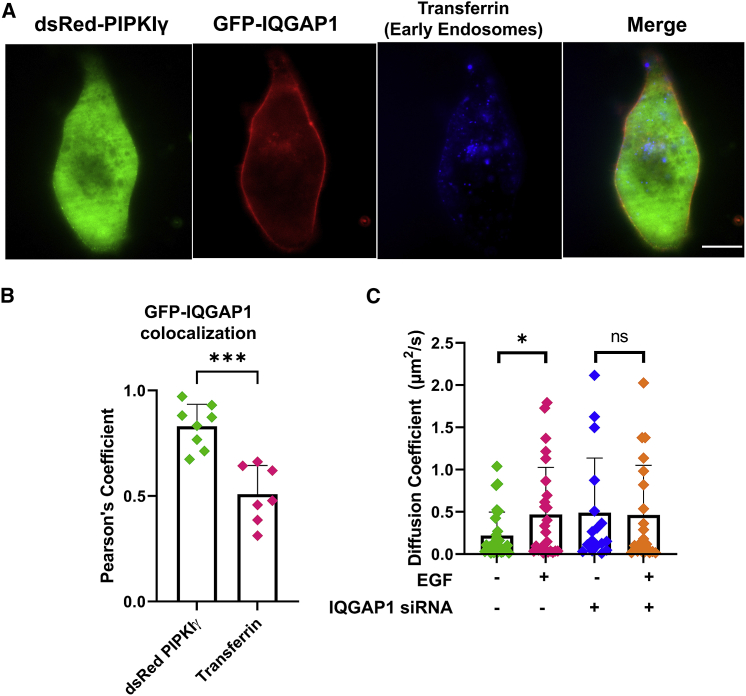
PIPKIγ populations outside the plasma membrane have not yet been studied in detail. We first characterized its cellular localization by confocal imaging microscopy using fluorescent markers such as transferrin or LysoTracker that act as a sensor for early endosomes and lysosomes, respectively. We then found that cytosolic populations of IQGAP1 and PIPKIγ colocalize with transferrin, which predominantly localizes in early endosomes (Fig. 3 A), but these populations localize to a much lower extent to LysoTracker (Fig. S4 A). Quantification using Pearson's coefficient based on these immunofluorescence studies confirms that eGFP-IQGAP1 strongly colocalizes with dsRed-PIPKIγ. While eGFP-IQGAP1's colocalization with transferrin is statistically significant when compared with the negative control (Fig. 3 B), this interaction is significantly smaller than the interaction of IQGAP1 and PIPKIγ. These data suggests that cytosolic IQGAP1 and PIPKIγ interactions are localized to early endosomes as well as other cellular sites.
Figure 3.
PIPKI and IQGAP1 interact in the cytosol. (A) A representative image showing colocalization between eGFP-IQGAP1, dsRed-PIPKIγ, and transferrin that acts as a marker for early endosomes in HeLa cells. (B) Colocalization quantified using Pearson's coefficient based on immunofluorescence studies show that eGFP-IQGAP1 strongly colocalizes with dsRed-PIPKIγ significantly higher than its colocalization with transferrin (positive control = 0.92 ± 0.02, negative control = 0.004 ± 0.007). (C) Diffusion coefficients of eGFP-PIPKIγ particles derived from FCS correlation curves in cytosol-localized focal areas on cells both at a basal state and after EGF stimulation (100 ng/mL) in the presence and absence of IQGAP1 expression. Scrambled (nonspecific) siRNA treatment was used for the control groups. n ≥ 5 measured in at least two independent experiments; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001; ns, not significant. Error bars denote standard deviation. Scale bar, 10 μm. To see this figure in color, go online.
We used FCS to study the levels and mobility of PIPKIγ in the cytosol. We find that in the basal state eGFP-PIPKIγ is characterized by significantly lower diffusion coefficients in the cytosol (Fig. 3 C), where the mobility is significantly lower in the cytosol (0.29 μm2/s) than in the plasma membrane (0.77 μm2/s). Surprisingly, EGF stimulation resulted in an increase in the diffusion coefficients of these PIPKIγ particles. Like the results with the membrane-localized PIPKIγ particles in Fig. 2 B, we see that there is no EGF-mediated response in the absence of IQGAP1 expression, suggesting that IQGAP1 plays a key role in mediating the large change in PIPKIγ diffusion in response to EGF stimulation (Fig. 3 C; representative curves, Fig. 2 B).
IQGAP1 mediates PIPKIγ colocalization with PI3K
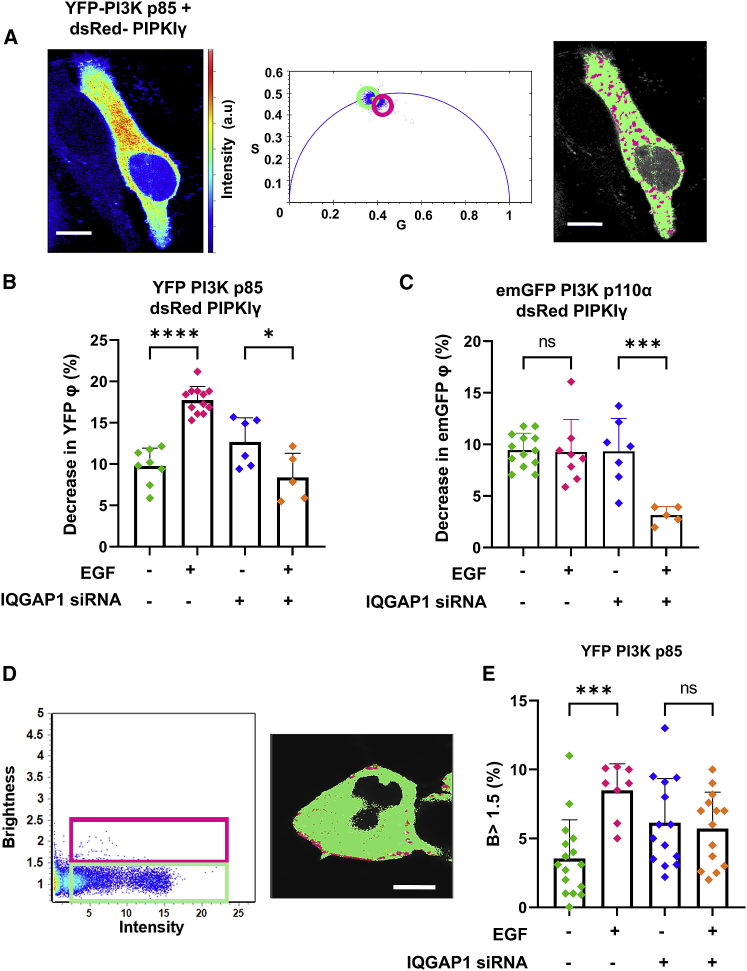
IQGAP1 has been shown to associate with PI3K subunits (p85, p110β, and p110α) (19), which we have corroborated in pull-down studies and immunofluorescence experiments by quantifying a positive colocalization (Pearson’s coefficient 0.48 ± 0.16, Fig. S4 B). A previous study showed that PIPKIα coimmunoprecipitated with the PI3K p110α subunit in the presence of IQGAP1 despite a lack of direct physical PIPKIγ-PI3K binding, suggesting that IQGAP1 scaffolds these kinases together (12). Here, we test the ability of IQGAP1 to scaffold PIP3K and PIPKIγ by observing FRET between dsRed-PIPKIγ with both emGFP-PI3K p110α and eYFP-PI3K p85, indicating an association between PIPKIγ and PI3K in HeLa cells (Fig. 4 A). The interaction between PI3K-p110α and PIPKIγ is not affected by EGF stimulation (not shown). In contrast, the interaction between the PI3K p85 subunit and PIPKIγ increases upon stimulation with EGF as demonstrated by a statistically significant decrease in lifetime (Fig. 4 B). A heterogeneous lifetime population is seen on the phasor plot (Fig. 4 A, center). Lifetime imaging shows that FRET occurs at the plasma membrane as well as some intracellular structures (Fig. 4 A, right). Because the two kinases do not bind to each other, we hypothesize that this interaction is due to scaffolding mediated by IQGAP1.
Figure 4.
IQGAP1 mediates an association between PIPKIγ and PI3K. (A) A representative cell image shows the eYFP fluorescence of HeLa cells expressing both dsRed-PIPKIγ and eYFP-PI3K p85 (left). eYFP lifetimes for some pixels are inside the arc due to reduced lifetime caused by FRET (center). The distinct regions on the phasor plots are highlighted by a green circle indicating higher (non-FRET) lifetimes (lifetime center = 2.55 ns) and a magenta circle indicating shortened lifetimes (lifetime center = 2.00 ns). The pixels underlying these circles are false colored and overlaid on grayscale cell images (right). (B) EGF treatment decreases the fluorescence lifetime of HeLa cells expressing eYFP-PI3K p85-dsRed-PIPKIγ, indicating that EGF enhances the interactions of PIPKIγ with p85. Furthermore, suppressing IQGAP1 levels with siRNA decreased the PIPKIγ-p85 interactions. (C) EGF treatment does not decrease the fluorescence lifetime of HeLa cells expressing eGFP PI3K p110α-dsRed-PIPKIγ, while suppressing IQGAP1 levels with siRNA decreased PIPKIγ-p110α interactions in response to EGF. (D) A brightness-versus-intensity plot, where individual pixels of HeLa cell images under stimulation by EGF (100 ng/mL for 1 h) expressing eYFP-PI3K p85 are highlighted either in a green box (B < 1.5) or a magenta box (B > 1.5) (left). The distribution of these highlighted pixels can be seen overlaid (right) on a representative cell. This EGF-stimulated cell shows localization of IQGAP1 clusters at the plasma membrane. (E) There is a statistically significant increase in the number of pixels having higher B values (magenta pixels, B > 1.5) that represent the oligomeric species or clusters of eYFP-PI3K p85 after the cells are stimulated with EGF compared with basal levels both in the presence and absence of IQGAP1. φ denotes fluorescence lifetime in figure legends. Scrambled (nonspecific) siRNA treatment was used for the control groups. n ≥ 5 measured in at least two independent experiments. In both cases, lifetimes of the control and IQGAP1 siRNA groups are significantly different from eYFP-p85 alone and from emGFP-p110α alone (p < 0.001). ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001; ns, not significant. Error bars denote standard deviation. Scale bars, 10 μm. To see this figure in color, go online.
We tested the idea that IQGAP simultaneously scaffolds PIPKIγ and PI3K by measuring their association in cells treated with siRNA (IQGAP1). We find changes in interactions between PIPKIγ and PI3K subunits p85 and p110α upon EGF stimulation, unlike the control groups in these IQGAP1 knockdown cells (Fig. 4, B and C). Our results show the dynamic effect of IQGAP1 on PIPKI-PI3K interactions where IQGAP1's scaffolding of PIPKIγ and PI3K is essential for a robust response to EGF that involves the generation of PI(3,4,5)P3.
PI3K-p85 dimerization has been reported earlier (54), and we wondered whether IQGAP1 could assemble multiple PI3K-p85 subunits in an EGF-dependent manner. To this end, we performed N&B studies (Fig. 4 E). We calculated the percentage of pixels that correspond to a B value of above 1.5 as a representative of oligomeric species after background correction and calibration (40). We find that EGF stimulation promotes PI3K-p85 complex formation only when IQGAP1 is present. In the absence of IQGAP1 p85 dimerization occurs, but it is not sensitive to EGF stimulation. Thus, IQGAP1 helps to promote EGF signals by sequestering multiple p85 subunits.
IQGAP1 mediates EGF-induced PIPKIγ interactions with talin and Cdc42
IQGAP1 has been found to form complexes with Cdc42, guanine nucleotide exchange factor FGD6, the Rho GTPase-activating protein ARHGAP10, filamin, and talin close to focal adhesions (55). Talin-PI(4,5)P2 mediates the activation and the function of integrins (56), including the formation of talin-integrin clusters (57). eGFP-IQGAP1 strongly binds to mCherry-talin in HeLa cells when quantified by FLIM-FRET, and this interaction is slightly enhanced by EGF stimulation (Fig. 5 A). PIPKIγ is the only PIPKI isoform known to bind to talin (58). Using FLIM-FRET, we see that eGFP-PIPKIγ interacts with mCherry-talin only when the cells are stimulated by EGF. This EGF-mediated increase in FRET (decrease in donor lifetime) is completely abrogated when IQGAP1 is knocked down, indicating that the PIPKIγ-talin interaction is scaffolded by IQGAP1 (Fig. 5 B).
Figure 5.
IQGAP1 interacts with talin and mediates PIPKIγ interactions with talin and Cdc42. (A) There is a decrease in eGFP fluorescence lifetime of due to FRET between eGFP-IQGAP1 and mCherry-talin. The interaction between IQGAP1 and talin is further enhanced by EGF stimulation (100 ng/mL). (B) eGFP lifetime also decreases due to FRET in HeLa cells expressing both eGFP-PIPKIγ and mCherry-talin only when stimulated with EGF (100 ng/mL). These EGF-mediated interactions are abrogated by IQGAP1 downregulation, indicating that IQGAP1 is required for PIPKIγ and talin interactions. (C) A similar effect is seen in HeLa cells expressing both eGFP-Cdc42 and dsRed-PIPKIγ where FRET is seen only upon EGF (100 ng/mL) stimulation but not with IQGAP1. n ≥ 5 measured in at least two independent experiments. φ denotes fluorescence lifetime in figure legends. Scrambled (nonspecific) siRNA treatment was used for the control groups. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001; ns, not significant. Error bars denote standard deviation. To see this figure in color, go online.
Cdc42 is a major initiator for the formation of the leading edge of a moving cell. Cdc42 and the other small GTPases RhoA and Rac1 are known to stimulate PIPKIγ activity and elevate the cellular levels of PI(4,5)P2 (59). However, Cdc42 is not known to physically associate with any PIPKI isoforms, unlike Rac1 and RhoA (60). Interestingly, our studies reveal that mCherry-PIPKIγ exhibits FRET with eGFP-Cdc42 only when the cells are stimulated by EGF. However, this EGF-mediated interaction is eliminated when IQGAP1 is knocked down using siRNA, indicating that a PIPKIγ-Cdc42 interaction is promoted by IQGAP1 when cells are stimulated with EGF (Fig. 5 C). These results directly show that IQGAP1 provides a physical link between phosphoinositides (through PIPKIγ), focal adhesion formation (through talin), and cytoskeletal reorganization (through Cdc42) upon EGF stimulation.
PI(4,5)P2 and PIPKIγ have been found to play a major role in cell migration, especially by modulating actin reorganization and focal adhesion (61,62), in conjugation with IQGAP1 (19,21,55). A proteomics screen of proteins that were pulled down with endogenous IQGAP1 in HeLa cells revealed that IQGAP1 binds to several proteins that are known to be involved in these roles (Fig. S5). Interestingly, almost all of these proteins have already been found to interact with PI(4,5)P2, PIPKIγ, or PI(3,4,5)P3 (58,63,64).
Discussion
Scaffold proteins play a critical role in fine-tuning cellular signaling pathways by spatially and temporally organizing signaling components. These proteins regulate signaling pathways through a variety of mechanisms, including tethering, localizing to a specific organelle, coordinating positive and negative feedback signals, and sequestering various components. Understanding how scaffolds organize proteins requires quantification of protein-protein interactions, which have been historically characterized by biochemical techniques such as Western blots using both recombinant proteins and whole-cell lysates, and have been visualized using imaging techniques such as immunofluorescence. While these techniques are able to suggest interactions, verification of physical association, as well as their spatial and temporal dynamics, needs to be carried out in live cells, especially in the case of scaffold protein interactions. Scaffold proteins are typically large and tend to complex with many different proteins, making it difficult to characterize limited and temporary protein-protein interactions. Here, we use a variety of advanced imaging techniques to visualize the physical interactions that occur between these proteins in live cells.
The benefits of live-cell measurements are demonstrated by our observations of IQGAP1-PIPKIγ interactions where we have used a combination of FLIM-FRET and FCCS to observe these interactions at both cell-wide and local levels, and how these change with EGF stimulation. Because of the primary plasma membrane localization of these proteins, it had been assumed that their interactions occur exclusively at the plasma membrane. Using FLIM-FRET, we find that this is not the case. In fact, we see IQGAP1-PIPKIγ complexes in the cytosol. We observe increased membrane localization of PIPKIγ in response to EGF stimulation. Based on the results with the PI(3,4,5)P3 sensor PH-Akt1, we find that this plasma membrane population of PIPKIγ as well as IQGAP1 may be targeted to or create the pools of newly generated PI(3,4,5)P3. FCS measurements show that this PIPKIγ species diffuses faster in response to EGF stimulation only in the presence of IQGAP1. These results support a model in which IQGAP1 scaffolds PIPKIγ to phosphoinositides in response to stimuli, suggesting its potential to regulate the phosphoinositide signaling pathway.
Our data show that IQGAP1 promotes association of PI3K subunits and PIPKIγ, especially after stimulation with EGF. These kinases do not bind to each other in the absence of IQGAP1, indicating that IQGAP1 scaffolding enables their proximity. These results support the hypothesis that IQGAP1 scaffolds PIPKIγ and PI3K to the incomplete phosphorylated phosphoinositides in response to EGF stimulation, hence promoting the production of PI(3,4,5)P3. We find that that the brightness values B in the IQGAP1 siRNA groups are significantly higher than those of control unstimulated cells but are significantly lower than control cells stimulated with EGF. It is possible that the absence of higher IQGAP1 levels may allow for some homo-oligomerization of p85 molecules due to proximity, which may account for the higher B values when compared with control cells. This effect could also be caused by a slight increase in EGFR phosphorylation upon the knockdown of IQGAP1. These results highlight the lack of a strong EGF response in the absence of IQGAP1. This EGF response is statistically larger than the increase caused by the downregulation of IQGAP1, suggesting that IQGAP1 is necessary for a robust response in response to EGF activation. The PI3K signaling cascade further results in the activation of Akt by PDK1, which in turn regulates several downstream partners (65). Several of these downstream components of the PI3K/Akt pathway are also known to bind to IQGAP1 (66), including Akt1. Hence, our results support the existence of a complex assembled by IQGAP1-PIPKIγ that integrates the PI3K/Akt signaling pathways at the site of PI(3,4,5)P3 generation.
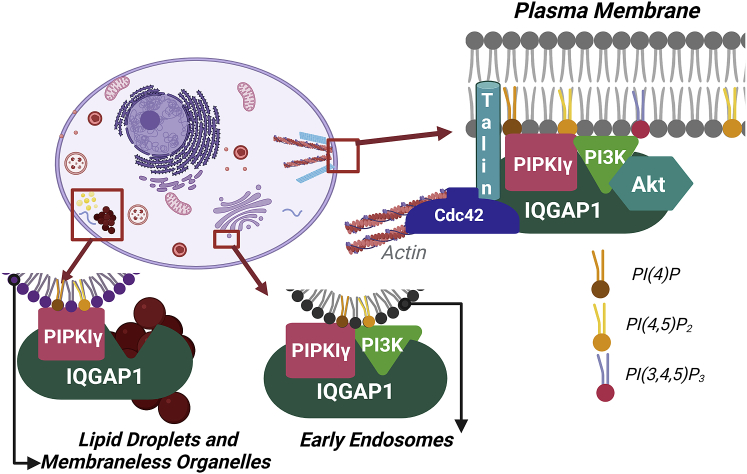
We were especially interested in the cytosolic population of PIPKIγ. Although less prevalent than the plasma-membrane-based population, it nevertheless exhibits unique characteristics. On testing a variety of organelle-specific markers, the only one that colocalizes with PIPKIγ is transferrin, which is a marker for early endosomes. Thus, we see that cytosolic IQGAP1 and PIPKIγ potentially interact at endosomes. Interestingly, IQGAP1 is also known to localize to Rab11-associated recycling endosomes to a lesser extent than early endosomes or lysosomes (67). A recent study describes the generation of PI(3,4,5)P3 by PI3K, suggesting an endosomal localization of phosphoinositide kinases along the endosomal membrane and IQGAP1 scaffolding at that location (68). We also see that cytosolic PIPKIγ populations are marked by extremely low diffusion coefficients (<0.5 μm2/s). Like the plasma-membrane-localized population, EGF stimulation only affects diffusion in the presence of IQGAP1. It is also possible that these cytosolic IQGAP1-PIPKIγ complexes are in phase-separated liquid-liquid domains, consistent with the low diffusion coefficients (69).
The spatiotemporal organization of biological molecules and reactions is essential for the working of the cell. This spatiotemporal organization might occur through a process called liquid-liquid phase separation (LLPS). Biomolecules such as proteins or nucleic acids condense into a dense phase that coexists with a dilute phase during LLPS (70). Phase separation into liquid droplet-like structures allows for rapid, reversible condensation of specific proteins and nucleic acid molecules into discrete assemblies that dynamically exchange biomolecules with the surrounding cytosol. Some environmental conditions such as temperature and pH are able to promote LLPS, providing cells with an opportunity to respond to external stimuli. A second step of LLPS involves the diffusion of specific proteins or other molecules into the phase without considerably contributing to the stability of the phase. This process can increase local concentrations of molecules by severalfold (71). LLPS eventually results in the formation of membrane-less organelles that include nucleoli, stress granules, P granules, and Cajal bodies, which are involved in multiple cellular processes including gene regulation, ribosome function, and regulation of signal transduction.
The ability of proteins to undergo LLPS is influenced by electrostatic interactions between different subdomains, the amino acid composition, post-translational modifications (e.g., phosphorylation and acetylation), extension of the polypeptide chain of the protein, and/or interactions with other proteins (72). Recent studies show that multivalent interactions between scaffold proteins drive LLPS (71,73,74). Therefore, it is very likely that IQGAP1 is involved in LLPS through its versatile scaffolding of its many binding partners based on cell type and stimuli (71).
Additionally, cells are known to store energy in the form of neutral lipids packaged into micrometer-sized organelles named lipid droplets (LDs). LDs consist of a core of triglycerides and sterol esters surrounded by a phospholipid monolayer. They emerge from the endoplasmic reticulum. While LDs have been studied for a long time, their biogenesis has been poorly understood (75). Both PI(4,5)P2 and its precursor PI(4)P have been identified to be found on LD surface. Therefore, it is likely that PIPKIs localize to LDs. A recent study identified that LD formation is driven by LLPS (76), and another study found that stress granules and LDs are formed using the same signaling pathways (77). More studies have established that dysregulated LLPS (78) including accumulation of LDs (79) and stress granules (80) are a prominent characteristic of cancer cells. These results suggest that IQGAP1 facilitates PIPKIγ diffusion and, in turn, its association with cytosolic supramolecular protein complexes that might be phase separated. We speculate that IQGAP1 may corral PIPKIγ in these domains in the absence of stimulation to regulate the levels of PI(4,5)P2, thereby fine-tuning the phosphoinositide signaling pathway. In the context of the recent LLPS studies, the cytosolic IQGAP1-PIPKIγ populations might be involved in connecting the protein and nucleic acid-dominated membrane-less organelles and lipid-rich LDs during LLPS by forming the nexus of protein-lipid interactions.
Cdc42 initiates the formation of the leading edge of a moving cell, which is a site of intense actin polymerization and cell polarity. Cdc42 and other small GTPases, RhoA and Rac1, stimulate PIPKIγ activity and elevate the cellular level of PI(4,5)P2 (59). However, unlike Rac1 and RhoA, Cdc42 is not known to physically associate with any PIPKIγ isoforms (60). Therefore, the proximity between PIPKIγ and Cdc42 is probably mediated by IQGAP1 scaffolding. Cdc42 is required for PI(4,5)P2 -induced actin polymerization (81), and Cdc42 and PI(4,5)P2 together mediate actin nucleation by activating WASP, which in turn activates actin regulatory proteins (64). We note that several proteins involved in actin nucleation and actin polymerization also bind to IQGAP1 and PI3K (Fig. S5). Using N&B experiments, we also observe that IQGAP1 promotes the formation of new actin clusters in HeLa cells, especially along the plasma membrane, in response to EGF stimulation (Fig. S6). These results indicate that IQGAP1 promotes directed actin polymerization by coordinating with Cdc42 and phosphoinositide signaling, thus corroborating earlier studies that identified the role of IQGAP1-Cdc42 in cell migration (82, 83, 84, 85, 86). We also demonstrate that IQGAP1 plays a role in mediating PIPKIγ binding to the PI(4,5)P2-binding focal adhesion protein talin. PIPKIγ targets IQGAP1 to the plasma membrane and therefore to phosphoinositides (21). PIPKIγ is the only PIPKIγ isoform known to bind to talin (58). As this PIPKIγ-talin binding is regulated by phosphorylation (87), it is possible that the kinases involved in this process are also scaffolded by IQGAP1. The interaction between PIPK1 and talin has been shown to spatially modulate PI(4,5)P2 levels near focal adhesions (58).
IQGAP1 forms protein complexes that regulate cytoskeletal dynamics in several different organisms (88). Our results indicate that IQGAP1 mediates important cell processes by acting as an interface between PIPKIγ, PI(4,5)P2, actin, focal adhesion proteins, and Cdc42. By corralling the PI(4,5)P2 and PI(3,4,5)P3 machinery to the regulators of the cytoskeletal elements, IQGAP1 may coordinate cytoskeletal rearrangements in response to EGF stimulation. Spatiotemporal coordination allows protein complexes generated by IQGAP1 scaffolding to mediate cell motility, cytoskeletal reorganization, actin polymerization, leading-edge formation, and focal adhesions (for a model, see Fig. 6).
Figure 6.
A proposed model of IQGAP1-PIPKIγ interactions in various cellular localizations, created with BioRender.com To see this figure in color, go online.
While the full extent of IQGAP1's activities in healthy conditions remain unclear, IQGAP1 is upregulated in several different types of cancers (89), including hepatocellular cancer (90,91), prostate cancer (92), glioma (93), melanoma, and breast cancer (94). In most studies, perturbing IQGAP1 levels by upregulation or downregulation has a corresponding effect on cell migration in normal and cancer cell lines (92, 93, 94, 95). IQGAP1 knockout cells have significantly lower ability to metastasize in in vivo models (96). In the same vein, mutation and overexpression of the EGFR family have been closely associated with many types of cancers, including breast cancer, lung cancer, head and neck cancer, ovarian cancer, colorectal cancer, gastric cancer, glioma, melanoma, and medulloblastoma. Meanwhile, dysregulation of the PI3K/Akt signaling pathway has been linked to enhanced cell migration and motility in various diseases such as cancer (9,10). It was also observed that IQGAP1 promotes cell migration in an interdependent manner with PIPKIγ. In various cancer cells, overexpression of either protein led to increased cell migration that was eliminated by knocking down the other protein (21). In this context, our studies provide novel insights into IQGAP1 scaffolding, highlighting the possibility of IQGAP1-EGF-phosphinositide signaling as a target for anticancer therapy.
Author contributions
All authors conceived and planned the experiments. V.S.Y. carried out the experiments. All authors contributed to analysis and the interpretation of the results. V.S.Y. took the lead in writing the manuscript with input from all authors. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
Acknowledgments
We would like to acknowledge Drs. Richard Anderson (University of Wisconsin, Madison), Jonathan Backer (Albert Einstein School of Medicine), and Tamas Balla (NIH) for helpful discussion. We want to thank Dr. John Leszyk for his assistance with mass spectrometry, and Yemi Osayame, Christopher Dallarosa, and Dr. Osama Garwain for their assistance. This work has been funded by NSF CHE 1904886 (A.G.) and NIH R01GM 68737 (S.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Editor: Ilya Levental.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.01.018.
Supporting material
References
- 1.Berridge M.J., Irvine R.F. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature. 1984;312:315–321. doi: 10.1038/312315a0. [DOI] [PubMed] [Google Scholar]
- 2.Prestwich G.D. Phosphoinositide signaling; from affinity probes to pharmaceutical targets. Chem. Biol. 2004;11:619–637. doi: 10.1016/j.chembiol.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 3.Balla T., Szentpetery Z., Kim Y.J. Phosphoinositide signaling: new tools and insights. Physiology (Bethesda) 2009;24:231–244. doi: 10.1152/physiol.00014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunney T.D., Katan M. Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat. Rev. Cancer. 2010;10:342–352. doi: 10.1038/nrc2842. [DOI] [PubMed] [Google Scholar]
- 5.Park S.J., Itoh T., Takenawa T. Phosphatidylinositol 4-phosphate 5-kinase type I is regulated through phosphorylation response by extracellular stimuli. J. Biol. Chem. 2001;276:4781–4787. doi: 10.1074/jbc.M010177200. [DOI] [PubMed] [Google Scholar]
- 6.Rameh L.E., Cantley L.C. The role of phosphoinositide 3-kinase lipid products in cell function. J. Biol. Chem. 1999;274:8347–8350. doi: 10.1074/jbc.274.13.8347. [DOI] [PubMed] [Google Scholar]
- 7.Osaki M., Oshimura M., Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis. 2004;9:667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- 8.Wee P., Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers (Basel) 2017;9:52. doi: 10.3390/cancers9050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue G., Hemmings B.A. PKB/Akt-dependent regulation of cell motility. J. Natl. Cancer Inst. 2013;105:393–404. doi: 10.1093/jnci/djs648. [DOI] [PubMed] [Google Scholar]
- 10.Saxena N.K., Sharma D., et al. Anania F.A. Concomitant activation of the JAK/STAT, PI3K/AKT, and ERK signaling is involved in leptin-mediated promotion of invasion and migration of hepatocellular carcinoma cells. Cancer Res. 2007;67:2497–2507. doi: 10.1158/0008-5472.CAN-06-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thapa N., Anderson R.A. PIP2 signaling, an integrator of cell polarity and vesicle trafficking in directionally migrating cells. Cell Adh Migr. 2012;6:409–412. doi: 10.4161/cam.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma V.P., DesMarais V., et al. Narang A. Immunostaining evidence for PI(4,5)P2 localization at the leading edge of chemoattractant-stimulated HL-60 cells. J. Leukoc. Biol. 2008;84:440–447. doi: 10.1189/jlb.0907636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi S., Thapa N., et al. Anderson R.A. PIP kinases define PI4,5P(2)signaling specificity by association with effectors. Biochim. Biophys. Acta. 2015;1851:711–723. doi: 10.1016/j.bbalip.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y.E., Iijima M., et al. Devreotes P. Receptor-mediated regulation of PI3Ks confines PI(3,4,5)P3 to the leading edge of chemotaxing cells. Mol. Biol. Cell. 2003;14:1913–1922. doi: 10.1091/mbc.E02-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plantard L., Arjonen A., et al. Stromblad S. PtdIns(3,4,5)P(3) is a regulator of myosin-X localization and filopodia formation. J. Cell Sci. 2010;123:3525–3534. doi: 10.1242/jcs.069609. [DOI] [PubMed] [Google Scholar]
- 16.Good M.C., Zalatan J.G., Lim W.A. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi S., Anderson R.A. IQGAP1 is a phosphoinositide effector and kinase scaffold. Adv. Biol. Regul. 2016;60:29–35. doi: 10.1016/j.jbior.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith J.M., Hedman A.C., Sacks D.B. IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol. 2015;25:171–184. doi: 10.1016/j.tcb.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi S., Hedman A.C., et al. Anderson R.A. Agonist-stimulated phosphatidylinositol-3,4,5-trisphosphate generation by scaffolded phosphoinositide kinases. Nat. Cell Biol. 2016;18:1324–1335. doi: 10.1038/ncb3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziemba B.P., Falke J.J. A PKC-MARCKS-PI3K regulatory module links Ca2+ and PIP3 signals at the leading edge of polarized macrophages. PLoS One. 2018;13:e0196678. doi: 10.1371/journal.pone.0196678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi S., Thapa N., et al. Anderson R.A. IQGAP1 is a novel phosphatidylinositol 4,5 bisphosphate effector in regulation of directional cell migration. EMBO J. 2013;32:2617–2630. doi: 10.1038/emboj.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNulty D.E., Li Z., et al. Annan R.S. MAPK scaffold IQGAP1 binds the EGF receptor and modulates its activation. J. Biol. Chem. 2011;286:15010–15021. doi: 10.1074/jbc.M111.227694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen M., Choi S., et al. Anderson R.A. The specificity of EGF-stimulated IQGAP1 scaffold towards the PI3K-Akt pathway is defined by the IQ3 motif. Sci. Rep. 2019;9:9126. doi: 10.1038/s41598-019-45671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heo W.D., Inoue T., et al. Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raucher D., Stauffer T., et al. Meyer T. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell. 2000;100:221–228. doi: 10.1016/s0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- 26.Logan M.R., Mandato C.A. Regulation of the actin cytoskeleton by PIP2 in cytokinesis. Biol. Cell. 2006;98:377–388. doi: 10.1042/BC20050081. [DOI] [PubMed] [Google Scholar]
- 27.Tapon N., Hall A. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr. Opin. Cell Biol. 1997;9:86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- 28.Cai X., Lietha D., et al. Schaller M.D. Spatial and temporal regulation of focal adhesion kinase activity in living cells. Mol. Cell Biol. 2008;28:201–214. doi: 10.1128/MCB.01324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Insall R.H., Weiner O.D. PIP3, PIP2, and cell movement—similar messages, different meanings? Dev. Cell. 2001;1:743–747. doi: 10.1016/s1534-5807(01)00086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoeprich G.J., Sinclair A.N., et al. Goode B.L. Single-molecule imaging of IQGAP1 regulating actin filament dynamics. Mol. Biol. Cell. 2021;33 doi: 10.1091/mbc.E21-04-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurella V.B., Richard J.M., et al. Worthylake D.K. Crystal structure of the GTPase-activating protein-related domain from IQGAP1. J. Biol. Chem. 2009;284:14857–14865. doi: 10.1074/jbc.M808974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golebiewska U., Nyako M., et al. McLaughlin S. Diffusion coefficient of fluorescent phosphatidylinositol 4,5-bisphosphate in the plasma membrane of cells. Mol. Biol. Cell. 2008;19:1663–1669. doi: 10.1091/mbc.E07-12-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiantia S., Ries J., Schwille P. Fluorescence correlation spectroscopy in membrane structure elucidation. Biochim. Biophys. Acta. 2009;1788:225–233. doi: 10.1016/j.bbamem.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Schwille P. Fluorescence correlation spectroscopy and its potential for intracellular applications. Cell Biochem. Biophys. 2001;34:383–408. doi: 10.1385/CBB:34:3:383. [DOI] [PubMed] [Google Scholar]
- 36.Krieger J.W., Singh A.P., et al. Wohland T. Imaging fluorescence (cross-) correlation spectroscopy in live cells and organisms. Nat. Protoc. 2015;10:1948–1974. doi: 10.1038/nprot.2015.100. [DOI] [PubMed] [Google Scholar]
- 37.Bacia K., Schwille P. Practical guidelines for dual-color fluorescence cross-correlation spectroscopy. Nat. Protoc. 2007;2:2842–2856. doi: 10.1038/nprot.2007.410. [DOI] [PubMed] [Google Scholar]
- 38.Digman M.A., Caiolfa V.R., et al. Gratton E. The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 2008;94:L14–L16. doi: 10.1529/biophysj.107.120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen Y. ISS; 2020. VistaVision FFS Processing and Analysis. [Google Scholar]
- 40.Planes N., Digman M.A., et al. Caballero-George C. Number and brightness analysis to study spatio-temporal distribution of the angiotensin II AT1 and the endothelin-1 ETA receptors: influence of ligand binding. Biochim. Biophys. Acta Gen. Subj. 2019;1863:917–924. doi: 10.1016/j.bbagen.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Digman M.A., Dalal R., et al. Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys. J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ossato G., Digman M.A., et al. Gratton E. A two-step path to inclusion formation of huntingtin peptides revealed by number and brightness analysis. Biophys. J. 2010;98:3078–3085. doi: 10.1016/j.bpj.2010.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plotegher N., Gratton E., Bubacco L. Number and Brightness analysis of alpha-synuclein oligomerization and the associated mitochondrial morphology alterations in live cells. Biochim. Biophys. Acta. 2014;1840:2014–2024. doi: 10.1016/j.bbagen.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith T.C., Fridy P.C., et al. Luna E.J. Supervillin binding to myosin II and synergism with anillin are required for cytokinesis. Mol. Biol. Cell. 2013;24:3603–3619. doi: 10.1091/mbc.E12-10-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 46.Sun Y., Day R.N., Periasamy A. Investigating protein-protein interactions in living cells using fluorescence lifetime imaging microscopy. Nat. Protoc. 2011;6:1324–1340. doi: 10.1038/nprot.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szentpetery Z., Balla A., et al. Balla T. Live cell imaging with protein domains capable of recognizing phosphatidylinositol 4,5-bisphosphate; a comparative study. BMC Cell Biol. 2009;10:67. doi: 10.1186/1471-2121-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varnai P., Balla T. Live cell imaging of phosphoinositide dynamics with fluorescent protein domains. Biochim. Biophys. Acta. 2006;1761:957–967. doi: 10.1016/j.bbalip.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 49.Lemmon M.A., Ferguson K.M., et al. Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. U S A. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varnai P., Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia P., Gupta R., et al. Rebecchi M.J. The pleckstrin homology domain of phospholipase C-delta 1 binds with high affinity to phosphatidylinositol 4,5-bisphosphate in bilayer membranes. Biochemistry. 1995;34:16228–16234. doi: 10.1021/bi00049a039. [DOI] [PubMed] [Google Scholar]
- 52.Ebner M., Lucic I., et al. Yudushkin I. PI(3,4,5)P3 engagement restricts Akt activity to cellular membranes. Mol. Cell. 2017;65:416–431.e6. doi: 10.1016/j.molcel.2016.12.028. [DOI] [PubMed] [Google Scholar]
- 53.Lam A.J., St-Pierre F., et al. Lin M.Z. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LoPiccolo J., Kim S.J., et al. Backer J.M. Assembly and molecular architecture of the phosphoinositide 3-kinase p85alpha homodimer. J. Biol. Chem. 2015;290:30390–30405. doi: 10.1074/jbc.M115.689604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steenblock C., Heckel T., et al. Hoflack B. The Cdc42 guanine nucleotide exchange factor FGD6 coordinates cell polarity and endosomal membrane recycling in osteoclasts. J. Biol. Chem. 2014;289:18347–18359. doi: 10.1074/jbc.M113.504894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orlowski A., Kukkurainen S., et al. Rog T. PIP2 and talin join forces to activate integrin. J. Phys. Chem. B. 2015;119:12381–12389. doi: 10.1021/acs.jpcb.5b06457. [DOI] [PubMed] [Google Scholar]
- 57.Lawson C., Schlaepfer D.D. Integrin adhesions: who's on first? What's on second? Connections between FAK and talin. Cell Adh. Migr. 2012;6:302–306. doi: 10.4161/cam.20488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ling K., Doughman R.L., et al. Anderson R.A. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- 59.Weernink P.A., Meletiadis K., et al. Jakobs K.H. Activation of type I phosphatidylinositol 4-phosphate 5-kinase isoforms by the Rho GTPases, RhoA, Rac1, and Cdc42. J. Biol. Chem. 2004;279:7840–7849. doi: 10.1074/jbc.M312737200. [DOI] [PubMed] [Google Scholar]
- 60.Santarius M., Lee C.H., Anderson R.A. Supervised membrane swimming: small G-protein lifeguards regulate PIPK signalling and monitor intracellular PtdIns(4,5)P2 pools. Biochem. J. 2006;398:1–13. doi: 10.1042/BJ20060565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roach A.N., Wang Z., et al. Du G. Phosphatidic acid regulation of PIPKI is critical for actin cytoskeletal reorganization. J. Lipid Res. 2012;53:2598–2609. doi: 10.1194/jlr.M028597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Senju Y., Kalimeri M., et al. Lappalainen P. Mechanistic principles underlying regulation of the actin cytoskeleton by phosphoinositides. Proc. Natl. Acad. Sci. U S A. 2017;114:E8977–E8986. doi: 10.1073/pnas.1705032114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nader G.P., Ezratty E.J., Gundersen G.G. FAK, talin and PIPKIgamma regulate endocytosed integrin activation to polarize focal adhesion assembly. Nat. Cell Biol. 2016;18:491–503. doi: 10.1038/ncb3333. [DOI] [PubMed] [Google Scholar]
- 64.Higgs H.N., Pollard T.D. Activation by Cdc42 and PIP(2) of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J. Cell Biol. 2000;150:1311–1320. doi: 10.1083/jcb.150.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manning B.D., Cantley L.C. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi S., Anderson R.A. And Akt-ion! IQGAP1 in control of signaling pathways. EMBO J. 2017;36:967–969. doi: 10.15252/embj.201796827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Samson E.B., Tsao D.S., et al. Diehl M.R. The coordinating role of IQGAP1 in the regulation of local, endosome-specific actin networks. Biol. Open. 2017;6:785–799. doi: 10.1242/bio.022624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thapa N., Chen M., et al. Anderson R.A. Phosphatidylinositol-3-OH kinase signalling is spatially organized at endosomal compartments by microtubule-associated protein 4. Nat. Cell Biol. 2020;22:1357–1370. doi: 10.1038/s41556-020-00596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murthy A.C., Dignon G.L., et al. Fawzi N.L. Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. Nat. Struct. Mol. Biol. 2019;26:637–648. doi: 10.1038/s41594-019-0250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alberti S., Gladfelter A., Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pudewell S., Wittich C., et al. Ahmadian M.R. Accessory proteins of the RAS-MAPK pathway: moving from the side line to the front line. Commun. Biol. 2021;4:696. doi: 10.1038/s42003-021-02149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kanaan N.M., Hamel C., et al. Combs B. Liquid-liquid phase separation induces pathogenic tau conformations in vitro. Nat. Commun. 2020;11:2809. doi: 10.1038/s41467-020-16580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hong K., Song D., Jung Y. Behavior control of membrane-less protein liquid condensates with metal ion-induced phase separation. Nat. Commun. 2020;11:5554. doi: 10.1038/s41467-020-19391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lyon A.S., Peeples W.B., Rosen M.K. A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 2021;22:215–235. doi: 10.1038/s41580-020-00303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Skotland T., Sagini K., et al. Llorente A. An emerging focus on lipids in extracellular vesicles. Adv. Drug Deliv. Rev. 2020;159:308–321. doi: 10.1016/j.addr.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 76.Zoni V., Khaddaj R., et al. Vanni S. Pre-existing bilayer stresses modulate triglyceride accumulation in the ER versus lipid droplets. eLife. 2021;10:e62886. doi: 10.7554/eLife.62886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amen T., Kaganovich D. Small molecule screen reveals joint regulation of stress granule formation and lipid droplet biogenesis. Front. Cell Dev. Biol. 2020;8:606111. doi: 10.3389/fcell.2020.606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang S., Fagman J.B., et al. Liu B. Protein phase separation and its role in tumorigenesis. eLife. 2020;9:e60264. doi: 10.7554/eLife.60264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Z., Liu H., Luo X. Lipid droplet and its implication in cancer progression. Am. J. Cancer Res. 2020;10:4112–4122. [PMC free article] [PubMed] [Google Scholar]
- 80.Lu J., Qian J., et al. Zhang W. Emerging roles of liquid-liquid phase separation in cancer: from protein aggregation to immune-associated signaling. Front. Cell Dev. Biol. 2021;9:631486. doi: 10.3389/fcell.2021.631486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen F., Ma L., et al. Alt F.W. Cdc42 is required for PIP(2)-induced actin polymerization and early development but not for cell viability. Curr. Biol. 2000;10:758–765. doi: 10.1016/s0960-9822(00)00571-6. [DOI] [PubMed] [Google Scholar]
- 82.Watanabe T., Wang S., et al. Kaibuchi K. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev. Cell. 2004;7:871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 83.Fukata M., Watanabe T., et al. Kaibuchi K. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 2002;109:873–885. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- 84.Bashour A.M., Fullerton A.T., et al. Bloom G.S. IQGAP1, a Rac- and Cdc42-binding protein, directly binds and cross-links microfilaments. J. Cell Biol. 1997;137:1555–1566. doi: 10.1083/jcb.137.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nouri K., Timson D.J., Ahmadian M.R. New model for the interaction of IQGAP1 with CDC42 and RAC1. Small GTPases. 2020;11:16–22. doi: 10.1080/21541248.2017.1321169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brandt D.T., Marion S., et al. Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J. Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee S.Y., Voronov S., et al. De Camilli P. Regulation of the interaction between PIPKI gamma and talin by proline-directed protein kinases. J. Cell Biol. 2005;168:789–799. doi: 10.1083/jcb.200409028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hedman A.C., Smith J.M., Sacks D.B. The biology of IQGAP proteins: beyond the cytoskeleton. EMBO Rep. 2015;16:427–446. doi: 10.15252/embr.201439834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.White C.D., Brown M.D., Sacks D.B. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009;583:1817–1824. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.White C.D., Khurana H., et al. Schmidt V.A. IQGAP1 and IQGAP2 are reciprocally altered in hepatocellular carcinoma. BMC Gastroenterol. 2010;10:125. doi: 10.1186/1471-230X-10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schmidt V.A., Chiariello C.S., et al. Bahou W.F. Development of hepatocellular carcinoma in Iqgap2-deficient mice is IQGAP1 dependent. Mol. Cell Biol. 2008;28:1489–1502. doi: 10.1128/MCB.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moon H., Ruelcke J.E., et al. Hill M.M. Diet-induced hypercholesterolemia promotes androgen-independent prostate cancer metastasis via IQGAP1 and caveolin-1. Oncotarget. 2015;6:7438–7453. doi: 10.18632/oncotarget.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Diao B., Liu Y., et al. Xu G.Z. IQGAP1siRNA inhibits proliferation and metastasis of U251 and U373 glioma cell lines. Mol. Med. Rep. 2017;15:2074–2082. doi: 10.3892/mmr.2017.6257. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 94.Casteel D.E., Turner S., et al. Pilz R.B. Rho isoform-specific interaction with IQGAP1 promotes breast cancer cell proliferation and migration. J. Biol. Chem. 2012;287:38367–38378. doi: 10.1074/jbc.M112.377499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang X.X., Wang K., et al. Wang X.H. Targeted knockdown of IQGAP1 inhibits the progression of esophageal squamous cell carcinoma in vitro and in vivo. PLoS One. 2014;9:e96501. doi: 10.1371/journal.pone.0096501. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 96.Hebert J.D., Tian C., et al. Hynes R.O. The scaffold protein IQGAP1 is crucial for extravasation and metastasis. Sci. Rep. 2020;10:2439. doi: 10.1038/s41598-020-59438-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.