Abstract
Cadherins are a superfamily of adhesion proteins involved in a variety of biological processes that include the formation of intercellular contacts, the maintenance of tissue integrity, and the development of neuronal circuits. These transmembrane proteins are characterized by ectodomains composed of a variable number of extracellular cadherin (EC) repeats that are similar but not identical in sequence and fold. E-cadherin, along with desmoglein and desmocollin proteins, are three classical-type cadherins that have slightly curved ectodomains and engage in homophilic and heterophilic interactions through an exchange of conserved tryptophan residues in their N-terminal EC1 repeat. In contrast, clustered protocadherins are straighter than classical cadherins and interact through an antiparallel homophilic binding interface that involves overlapped EC1 to EC4 repeats. Here we present molecular dynamics simulations that model the adhesive domains of these cadherins using available crystal structures, with systems encompassing up to 2.8 million atoms. Simulations of complete classical cadherin ectodomain dimers predict a two-phased elastic response to force in which these complexes first softly unbend and then stiffen to unbind without unfolding. Simulated α, β, and γ clustered protocadherin homodimers lack a two-phased elastic response, are brittle and stiffer than classical cadherins and exhibit complex unbinding pathways that in some cases involve transient intermediates. We propose that these distinct mechanical responses are important for function, with classical cadherin ectodomains acting as molecular shock absorbers and with stiffer clustered protocadherin ectodomains facilitating overlap that favors binding specificity over mechanical resilience. Overall, our simulations provide insights into the molecular mechanics of single cadherin dimers relevant in the formation of cellular junctions essential for tissue function.
Significance
Multicellular organisms rely on cellular adhesion to survive, and this adhesion is mediated by diverse sets of proteins that include cadherins responsible for organ assembly and tissue integrity maintenance. As parts of cell-cell junctions in epithelial and cardiac tissues, classical cadherins experience forces and must be mechanically robust. In contrast, clustered protocadherins are responsible for neuronal connectivity and are exposed to more subtle mechanical stimuli. We used simulations to study the mechanics of isolated cadherin complexes and found that classical cadherins exhibit a two-phased elastic response that might prevent loss of adhesion during mild mechanical stress. Conversely, we predict that clustered protocadherin complexes are brittle. Our results suggest that each set of cadherins has evolved to adopt distinct mechanical properties.
Introduction
Cadherins are a large superfamily of glycoproteins that mediate cell-cell adhesion in a calcium (Ca2+)-dependent manner and whose members are involved in morphogenesis, tissue-integrity maintenance, and neuronal circuit development (1, 2, 3, 4, 5, 6, 7). The defining characteristics of the cadherin superfamily are their extracellular cadherin (EC) “repeats,” composed of approximately 100 amino acids of similar sequence and a Greek key fold, as well as their highly conserved amino acid motifs that form Ca2+-binding regions between EC repeats (3,8, 9, 10). Classical cadherin ectodomains have five EC repeats, while the clustered protocadherin (PCDH) ectodomains have six (Fig. 1 A and B). Other members of the cadherin superfamily have longer ectodomains with up to 34 EC repeats (6,11,12). Adhesive contacts across cell junctions (trans) are formed by interactions between these cadherin ectodomains protruding from opposing cells.
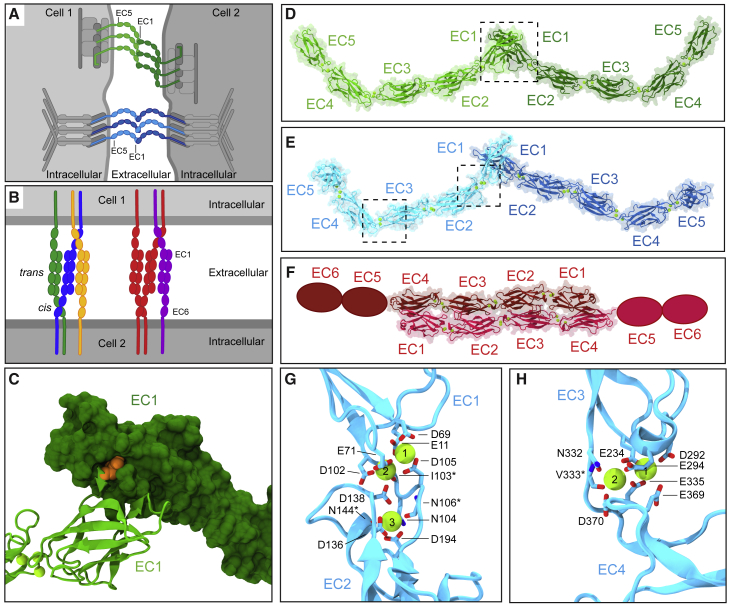
Figure 1.
Cadherin binding modes and their Ca2+-binding sites. (A) Schematics of epithelial cells and their intercellular contacts mediated by cadherins. Highlighted are the adherens junction (CDH1: greens) and the desmosome (DSGs and DSCs: blues). Proteins that connect cadherins to the cytoskeleton are shown in grays. (B) PCDH complexes at the interface of two neuronal surfaces showing trans and cis interactions. Colors denote different isoforms. (C) Detail of trans tryptophan exchange mechanism in classical cadherins. One monomer is shown in surface while another is shown in ribbon representation. Tryptophan residue at position two (Trp2) in one monomer is shown in orange. (D–F) Models of trans (D) CDH1 homodimer, (E) DSG-DSC heterodimer, and (F) PCDHβ6 homodimer complexes (missing EC5 and EC6 repeats are shown as ovals). Proteins are shown as ribbons with their molecular surfaces in transparent representation. Ca2+ ions are shown as green spheres. (G and H) Detail of the DSG2 EC1-2 and EC3-4 Ca2+-binding sites, respectively. Ca2+-coordinating residues are shown in stick representation and labeled. Backbone coordination is marked with an asterisk. Some backbone and side chain atoms are not shown for visualization purposes.
Multiple studies have revealed the details of the molecular complexes formed by various classical cadherins in epithelial adhesive structures, such as adherens junctions and desmosomes (Fig. 1 A) (13,14). A crystallographic model of the complete classical epithelial cadherin (CDH1) ectodomain (PDB: 3Q2V) shows both homophilic tip-to-tip trans interactions and cis (same cell) contacts in a crystal packing lattice that reveals a hypothetical adherens junction architecture (15). In this structure, the ectodomains adopt slightly curved conformations (Fig. 1 D) and the tip-to-tip trans interactions are mediated by a tryptophan (Trp2) exchange between the N-terminal EC1 repeats of dimeric complexes (8,15,16) (Fig. 1 C and D). Biophysical studies have shown the importance of Ca2+ in maintaining the stability and shape of the CDH1 ectodomain (17,18) as well as the relevance of CDH1 Trp2 residues in adhesion (19, 20, 21, 22), while single-molecule experiments have quantified the mechanical strength and lifetime of CDH1 homophilic bonds (23, 24, 25).
In parallel, all-atom molecular dynamics (MD) simulations of classical cadherin EC1/EC1 (26) and EC1-2/EC1-2 (24,27,28) complexes suggested that forced unbinding proceeds without the unfolding of EC repeats and that Ca2+ rigidifies EC linker regions. Simulations of the complete monomeric EC1-EC5 ectodomain of C-cadherin, a frog classical cadherin (16) have also predicted that its slightly bent shape is stable in the presence of bound Ca2+ and that the ectodomain can be straightened at a low force (29), resembling tertiary structure elasticity observed for ankyrin repeat stacks, tandem titin Ig domains, and other elongated proteins (30, 31, 32, 33, 34, 35, 36). Stretching after unbending resulted in mechanical unfolding at high forces, with predictions of Ca2+-dependent unfolding pathways and force peaks consistent with experimental results (29,31,37). Whether unbending and unbinding before unfolding occur in the same way for the complete CDH1 EC1-5/EC1-5 dimer, or for multiple dimers in an adherens junction, remains unexplored.
While adherens junctions are formed by homophilic CDH1 dimers, desmosomes are formed by heterophilic and homophilic complexes of desmoglein (DSG) and desmocollin (DSC) cadherin proteins (38, 39, 40, 41, 42, 43) (Fig. 1 A and E). The structures of several isoforms of DSG and DSC proteins have been solved and include those for DSG2-DSG2 (PDB: 5ERD) and DSC1-DSC1 (PDB: 5IRY) homodimer complexes that also interact tip-to-tip and exchange Trp2 residues between their N-terminal EC1 repeats (44). Unlike other classical cadherins, which coordinate three Ca2+ ions at each linker region between EC repeats (Fig. 1 G) (10), DSG coordinates only two Ca2+ ions between EC3 and EC4 (Fig. 1 H). Structurally, this results in a more pronounced bend in the overall ectodomain shape of DSG compared with DSC and CDH1 proteins in the crystal structures (44). There is extensive experimental evidence suggesting that desmosomes are composed of both homophilic and heterophilic complexes between DSC and DSG proteins (39,45, 46, 47, 48). To date, however, only structures for desmosomal proteins forming homophilic tip-to-tip complexes have been reported (44), and the corresponding structures do not suggest possible architectures for desmosomes. A recent study used MD simulations and low-resolution cryo-electron tomography maps to build an atomistic model of mouse liver DSG2-DSC2 desmosomes (49), but how single homophilic and heterophilic dimers unbind in response to force and how the mechanical strength varies across the complexes formed by different isoforms remains to be determined.
Unlike CDH1, DSGs, and DSCs, which are all members of the classical cadherins, the clustered PCDHs belong to a different subfamily involved in neuronal adhesion and self-recognition (50, 51, 52, 53). To ensure proper neuronal connectivity, axons and dendrites must make favorable connections to other neurons while avoiding redundant self-adhesion (Fig. 1 B) (54). The clustered PCDHs were named after the clustering of their genes into distinct groups consisting of variable and constant regions, which gives rise to the α, β, and γ subfamilies (51,52). The homodimeric structures of parts of the ectodomains of members from each subfamily (α, β, and γ) have been solved and reveal an extended antiparallel overlapping binding interface encompassing repeats EC1-4 (Fig. 1 F) (55, 56, 57, 58, 59, 60, 61). The α PCDHs are important in establishing and maturing neural circuitry during development, although their absence is nonlethal (62,63). Much less is known about β PCDHs, but they are expressed in the nervous system (53). Finally, the γ PCDHs have been identified as being vital for neuronal survival (64). Structures for representative members of each subfamily include PCDHα7 (PDB: 5DZV) (59), PCDHβ6 (PDB: 5DZX) (59), and PCDHγB3 (PDB: 5K8R) (58). Because the binding interface differs from those of classical cadherins, it is uncertain how clustered PCDHs respond to force, and how this difference would manifest in their unbinding pathways and function.
Here, we use all-atom steered MD (SMD) simulations (65, 66, 67, 68) to visualize and quantify the response of single cadherin trans dimers to the application of an external tensile force. This was achieved through simulations in which the C-terminal Cα atom of each monomer was pulled at various speeds (10, 1, and 0.5 or 0.1 nm/ns). Our simulations revealed a two-phased elastic response in which soft unbending over approximately 10-nm changes in end-to-end distances were observed before stiffening, leading to unbinding without unfolding for the classical cadherins. The clustered PCDHs, however, were straighter in isolation when compared with classical cadherins, and their complexes lacked the soft elastic response phase and instead exhibited complex unbinding pathways featuring intermediates. Additionally, we quantified the forces that these proteins can withstand before unbinding and found that clustered PCDHs unbind at higher forces than classical cadherin dimers when stretched at fast speeds. Overall, these results offer insights into the dynamics and mechanics of cadherin dimeric complexes with implications for their function as single units in adhesion sites during initial contact formation between cells. A companion article (69) reports on analyses of their response in junctions where multiple cadherin dimeric complexes work together to provide additional functionality. The combined work thus provides a molecular view of how cadherin-based cellular adhesion sites and junctions may function in vivo.
Materials and methods
Simulated systems
Nine molecular systems for simulation were built in VMD with the psfgen, solvate, and autoionize plugins (70) (Tables 1 and S1). Five of these included classical cadherin trans dimers built using the following three crystallographic structures: Mus musculus (mm) CDH1 EC1-5 (PDB: 3Q2V) (15) with residues 1–536 (UNP residues 157–692); Homo sapiens (hs) DSG2 EC1-5 (PDB: 5ERD) (44) with residues 1 to 553 (UNP residues 50–602); and hs DSC1 EC1-5 (PDB: 5IRY) (44) with residues 1–539 (UNP residues 135–673). The DSG2 and DSC1 proteins were selected for simulation because these desmosomal cadherins had complete structures with the highest resolution and because they are expected to form heterophilic complexes in vitro (44) and in vivo (45). The first two molecular systems (linear and diagonal) included CDH1 EC1-5 in two different orientations within the simulation box, which allowed us to simulate two different in vivo conditions. The next three systems consisted of the desmosomal DSG2 and DSC1 homodimers and the DSG2-DSC1 heterodimer. To create the DSG2-DSC1 heterodimer, the first six Cα atoms of a DSC1 protomer were aligned with the first six Cα atoms of one of the protomers in the DSG2 homodimer structure. This DSG2 protomer was then replaced with the aligned DSC1 protomer to create the heterodimeric trans complex. A 20-ps vacuum equilibration with constraints placed on all but residues two through six of both DSG2 and DSC1 was performed, which allowed the Trp2 residues of each monomer to fully insert into the hydrophobic pocket of the opposing monomer. Coordinates of the DSG2-DSC1 heterodimer are available upon request.
Table 1.
Overview of MD simulations
| System | Label | tsim (ns) | Slowest speed (nm/ns) | Size (#atoms) | Size (nm3) |
|---|---|---|---|---|---|
| Linear CDH1 | S1a-f | 158.1 | 0.5 | 321,547 | 54.9 × 7.5 × 8.2 |
| Diagonal CDH1 | S2a-e | 151.3 | 0.5 | 2,868,694 | 43.9 × 22.7 × 29.4 |
| DSG2-DSG2 | S3a-d | 219.2 | 0.1 | 429,545 | 59.9 × 9.0 × 8.3 |
| DSG2-DSC1 | S4a-d | 251.0 | 0.1 | 558,418 | 61.2 × 9.6 × 9.9 |
| DSC1-DSC1 | S5a-d | 207.0 | 0.1 | 365,669 | 60.9 × 8.7 × 7.3 |
| PCDHα7 | S6a-d | 172.5 | 0.1 | 500,917 | 54.0 × 10.0 × 9.6 |
| PCDHβ6 | S7a-d | 221.6 | 0.1 | 394,722 | 43.4 × 10.0 × 9.5 |
| PCDHγB3 | S8a-d | 234.1 | 0.1 | 335,584 | 50.0 × 8.4 × 8.4 |
| PCDHβ6-A | S9a-b | 121.2 | – | 102,143 | 22.2 × 8.2 × 6.3 |
Labels indicate the system and protein used. Initial size of the systems is indicated in the last column.
The final four molecular systems for simulation included clustered PCDHs and were built using structures for mm PCDHα7 EC1-5 (PDB: 5DZV) (59) with residues 1–528 (UNP residues 30–557), mm PCDHβ6 EC1-4 (PDB: 5DZX) (59) with residues 3–416 (UNP residues 31–444), and hs PCDHγB3 EC1-4 (PDB: 5K8R) (58) with residues 1–414 (UNP residues 31–444). Three of them were built using homodimeric complexes, while the last one was built as a monomer (monomer A of mm PCDHβ6 EC1-4).
Missing residues in one monomer of the CDH1 structure, which had multiple monomers in the asymmetric unit, were added by using the same residues from the other monomer after spatial superposition. The PCDHα7 structure had residues 494–500 in monomer A and residues 157–159 and 498–500 in monomer B missing, which were added by building a model of the protein in SWISS-MODEL (71) and by copying only the missing residues back into the original crystallographic structure. Water molecules and Ca2+ ions in the crystal structures were incorporated into the final systems, but final protein models had sugars, alternative conformations, and crystallizing reagents removed. Hydrogen atoms were added to protein structures with the psfgen plugin in VMD. Residues Glu and Asp were assigned a negative charge, while Lys and Arg residues were assigned a positive charge. Histidine residues were assumed to be neutral and their protonation states were chosen to form hydrogen bonds with surrounding residues. Termini (N- and C-) were assumed to be charged. Water molecules (TIP3P) and randomly placed ions were used to solvate and ionize the systems at a concentration of 150 mM NaCl. Sizes of the systems can be found in Table 1.
Simulations
MD simulations using explicit solvent (72, 73, 74, 75, 76, 77, 78, 79, 80) were performed with NAMD 2.11 and 2.12 (81) using the CHARMM36 (82) force field for proteins with the CMAP backbone correction (83). A cutoff of 12 Å with a switching distance of 10 Å was used for van der Waals interactions, and a pair list was generated for atoms within 13.5 Å that was updated every 40 fs. To compute long-range electrostatic forces, the Particle Mesh Ewald method (84) with a grid point density of more than 1 Å−3 was used. A uniform integration time step of 2 fs for evaluation of bonded and nonbonded interactions was used together with the SHAKE algorithm (85). Langevin dynamics was utilized to enforce constant temperature (T = 300°K) when indicated, with a damping coefficient of γ = 0.1 ps−1 unless otherwise stated. Constant number, pressure, and temperature simulations (NpT) at p = 1 atmosphere were conducted using the hybrid Nosé-Hoover Langevin piston method with a 200 fs decay period and a 50 fs damping time constant (81). Simulations with constraints on Cα atoms used a harmonic spring constant of kr = 1 kcal mol−1 Å−2. All systems were minimized for 5000 steps and were equilibrated with backbone constraints for 200 ps, followed by a free, un-constrained equilibration of 20 ns, except for the diagonal CDH1 system, for which the C-terminal Cα atoms remained constrained and the PCDHβ6 system for which the free equilibration was performed for 19.1 ns.
Constant velocity stretching simulations used the SMD method and the NAMD Tcl forces interface. SMD simulations (66, 67, 68,86, 87, 88) were performed by attaching Cα atoms of C-terminal residues to independent virtual springs of stiffness ks = 1 kcal mol−1 Å−2. The stretching direction was set along the x-axis matching the vector connecting terminal regions of the protein, with protein ends free to move in y and z directions unless otherwise stated (Tables 1 and S1). The free ends of the springs were moved away from the protein in opposite directions at a constant velocity. For each system, SMD simulations were performed at 10, 1, and either 0.5 or 0.1 nm/ns. Applied forces were computed using the extension of the virtual springs and values of these forces as well as position coordinates of the C-terminal Cα atoms were saved every 40 fs, while the coordinates of the whole system were saved every 1 ps.
Simulation analysis procedures and tools
Plotted forces are those applied to one of the C-terminal atoms from a dimer pair. All applied forces were calculated from SMD spring extensions in the x direction, unless harmonic constraints were applied in the y and z directions, in which case the total magnitude of the force applied, including constraints, was reported. Stiffness was computed through linear regression fits of force-distance plots. Maximum force peaks for each protein C-terminal end of the dimer pair were computed from 50-ps running averages to eliminate local fluctuations. Reported force peaks are averages computed from values for each protein C-terminal end. End-to-end distances for complexes were computed as the magnitude of the distance between the C-terminal Cα atoms in a dimer, unless otherwise stated. Distances between residues were computed between listed atoms, unless otherwise specified. To calculate the orientation of successive EC repeats during unbinding, the principal axes of the leading repeat were first aligned to the x-, y-, and z-axes using the Orient plugin in VMD. The principal axes of the following repeat were then calculated, and the x and y coordinates of the third principal axes of the second EC repeat were plotted, thus providing information about their relative orientation. This process was repeated for EC1-EC2, EC2-EC3, EC3-EC4, and EC4-EC5 (when applicable) on structures saved before simulation, after the monomer had been straightened, and after the dimers had been unbound. To compare shapes of cadherin ectodomains, conformations from equilibrium trajectories taken every 50 ps for one of the monomers were aligned to the initial conformation of the simulated system (aligned along x) based on Cα atoms. Projections of each Cα atom coordinate in the xy and xz were plotted along with their averages. Plots were prepared in Xmgrace. Molecular images were created in the molecular graphics program VMD (70).
Results
To visualize, quantify, and compare the response of cadherin ectodomain dimers to external forces, we built eight molecular systems for simulation as models representing initial isolated encounter complexes that may lead to the formation of cellular junctions. Below we describe results from equilibrium and SMD simulations for each of these systems including adherens junction cadherins (two mm CDH1 systems), desmosomal cadherins (hs DSG2-DSG2, DSG2-DSC1, and DSC1-DSC1 systems), and clustered PCDHs (mm PCDHα7, mm PCDHβ6, and hs PCDHγB3 systems; species omitted for clarity in text below). A ninth system with a monomeric fragment of PCDHβ6 was also built to explore the shape of clustered PCDHs. Combined, these systems and simulations may serve as predictive models for single-molecule force spectroscopy experiments.
Adherens junction cadherins exhibit a two-phased response to force before unbinding
We constructed two different CDH1 systems that include two ectodomains of this protein forming a homodimer and that differ in the way putative intracellular cytoskeletal attachments at each side are considered. The first is a linear CDH1 system in which the ectodomains were free to move and rotate when force was applied to C-terminal ends in the direction of the vector between C-terminal Cα atoms (Fig. 2 A). This represents a system in which no attachment to the cytoskeleton is considered and in which both proteins are already aligned for stretching in a linear fashion along the axis that joins their C-terminal ends. In the second, a diagonal CDH1 system had harmonic constraints applied to the C-terminal Cα atoms to restrict their movement in the plane perpendicular to the stretching direction and thereby mimic attachment to the underlying cytoskeleton during SMD (Fig. 2 B). In this system, monomers were positioned in a slanted orientation expected for dimers in an adherens junction (15). Both systems were equilibrated for 20 ns (simulations S1a and S2a; Tables 1 and S1) and subsequently stretched. Monomers retained the curved shape during equilibrations (Fig. S1), while stretching at all speeds proceeded in two phases in which the protein complex first unbends before subsequent unbinding without unfolding of EC repeats (Fig. 2 A and B; Video S1). As expected, Ca2+ ions at linker regions and C-terminal disulfide bonds prevented EC unfolding.
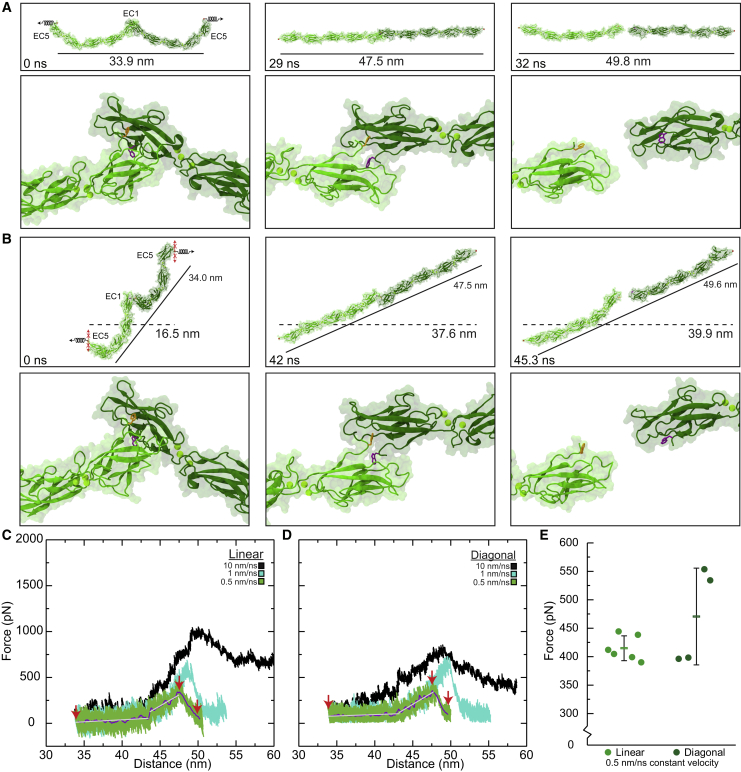
Figure 2.
Forced unbinding of trans CDH1 dimers in silico. (A) Snapshots of CDH1 unbinding at a stretching speed of 0.5 nm/ns (simulation S1d; Table 1). Stretched C-terminal Cα atoms are shown as red spheres. Springs in first panel indicate position and direction of forces applied along the vector joining the C-terminal Cα atoms (linear system) of the two monomers. The time and end-to-end distance between the C-terminal atoms are indicated in the top panels. Lower panels show the loss of the Trp2 exchange between CDH1 protomers. Trp2 residues are shown as orange and purple sticks for each monomer. (B) Snapshots of CDH1 unbinding at a stretching speed of 0.5 nm/ns (simulation S2d) shown as in (A). Force was applied with constraints (red Xs) to prevent motion perpendicular to the stretching direction (diagonal system). This mimics attachment to the cytoskeleton. All simulations showed straightening before unbinding without unfolding. Solid lines indicate end-to-end distance (between C-terminal Cα atoms) and dashes lines indicate membrane-to-membrane separation. (C) Force vs. end-to-end distance plot for linear constant velocity stretching of the CDH1 dimer at 10 nm/ns (S1b, black), 1 nm/ns (S1c, cyan), and 0.5 nm/ns (S1d, green; 1 ns running average shown in purple; gray lines are linear fits used to determine elasticity). Red arrowheads indicate time points in (A). (D) Force vs. end-to-distance plot for the diagonal constant velocity stretching of the CDH1 dimer shown as in (C) for simulations S2b-d. Red arrowheads indicate time points in (B). Forces in (C) and (D) are shown as monitored for one of the monomers. (E) Average magnitude peak force for simulations S1d-f (linear) and S2d-e (diagonal). Dots are force peaks from individual monomers. The bar represents the average from all protomers within a system (error bars are standard deviations).
Stretching of the CDH1 trans dimer at 0.5 nm/ns (simulation S1d, Table S1, 0–33.4 ns) causes soft unbending of the inherent curvature of CDH1 monomers, followed by stiff phase, prior to unbinding of the trans interaction. Monomers begin to re-bend immediately after unbinding. Proteins are depicted in ribbon representation (greens), while water molecules and solute ions are not shown for clarity.
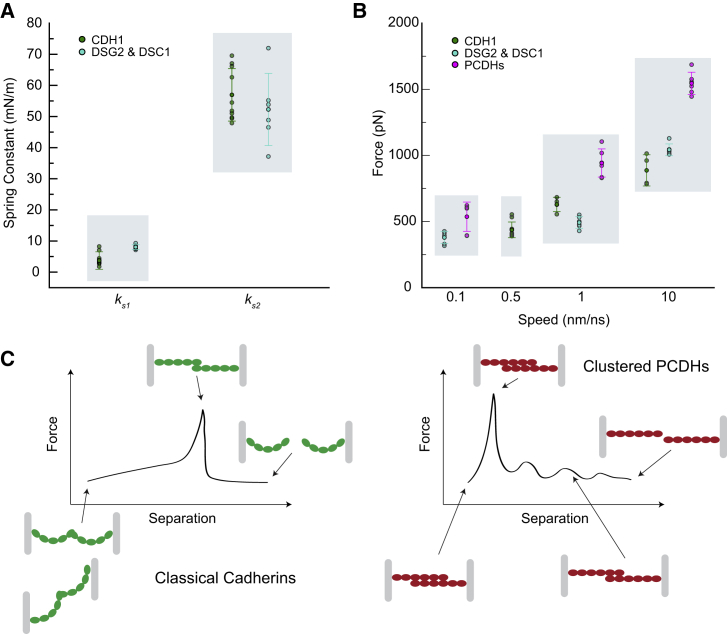
A quantitative characterization of the two phases of CDH1's mechanical response by spring constants associated to unbending and by force peaks associated to unbinding was dependent on the loading rate as expected (34,74,89,90). At the slowest stretching speed used for the CDH1 systems (simulations S1d–f and S2d–e at 0.5 nm/ns, with simulation repeats starting from different states obtained from the equilibrations), both the linear and diagonal systems displayed an initial unbending of CDH1 monomers at small forces of approximately 80 to approximately 100 pN with extensions of approximately 10 nm (Fig. 2 C and D). The unbending of monomers was associated with soft elastic responses with spring constants of ks1l ∼ 3.8 ± 3.6 mN/m (average over three repeats and considering both monomers) and ks1d ∼ 3.4 ± 0.4 mN/m (average over two repeats and considering both monomers) in the linear and diagonal systems, respectively (Fig. 2 C and D). As the proteins unbend, a second phase was observed with stiffer associated spring constants (ks2l ∼ 55.0 ± 7.9 mN/m for linear, ks2d ∼ 60.0 ± 9.4 mN/m for diagonal) over approximately 4-nm extensions (Fig. 2 C and D). While stiffness and changes in end-to-end distances (between the C-terminal ends) were similar for both the linear and diagonal systems before rupture, the separation between hypothetical membrane planes would be drastically different. For the linear system, the protein dimer had already been aligned along the stretching axis, so an initial separation of approximately 16.5 nm increases to approximately 33.9 nm upon rotation, and then through the two phases up to approximately 47.5 nm before dimer rupture with a total plane separation increase of approximately 31 nm. For the diagonal system, the separation between planes started at approximately 16.5 nm and increased to approximately 37.6 nm before dimer rupture, with an increase in separation of planes of approximately 21 nm before rupture, suggesting that this diagonal arrangement would result in smaller increases in separation between the two membranes before rupture.
Analyses of simulation trajectories show that the CDH1 tandem EC repeats straightened and twisted, as quantified by computing inter-repeat orientations (Fig. S2 A and B). Some tandem EC pairs straightened and twisted more than others, e.g., CDH1 EC4-5, compared with EC2-3 in both the linear and the diagonal systems (Fig. S2 A and B). The less flexible EC2-3 linker might facilitate proper CDH1 cis binding mediated by EC1 and EC2 while the more flexible EC4-5 linker may prevent trans-bond rupture during mild mechanical stress, either from regular cellular activities or external stimuli.
We also monitored the peak force necessary to separate dimers at the slowest stretching speed tested (0.5 nm/ns), which differed depending on the system and starting state. For instance, the first stretching simulation for the linear system had a force peak of Fp ∼ 408.4 pN ± 5.0 pN (average over two sides), whereas the diagonal system had a magnitude force peak of Fp ∼ 397.2 pN ± 1.5 pN (average over two sides). Data from triplicate repeats for the linear system (simulations S1d-f; Fp ∼ 414.8 pN ± 21.8 pN) and duplicate repeats for the diagonal system (simulations S2d-e; Fp ∼ 470.6 pN ± 85.5 pN) indicate that the difference in average force needed to separate CDH1 dimers was not statistically significant when comparing the two stretching approaches (Fig. 2 E; p = 0.15).
An inspection of the SMD trajectories and forces reveals that the rupture of the CDH1 dimer correlated with the extraction of Trp2 residues from their binding pockets for both the linear and diagonal systems, as reported by an increase in distance between the Trp2 Hε atom from one monomer and the backbone Asp90 O atom from the other while unbinding forces peaked (Figs. 2 A, B, S3 A, B, and S4; Video S2). Residues involved in unbinding are highly conserved (Fig. S5 A; Table S2), and despite the different stretching geometries, unbinding pathways were similar. After unbinding, each CDH1 monomer quickly began to rebend, as indicated by a decrease in the end-to-end distance between N- and C-termini within each monomer and by a partial recovery of inter-repeat orientations (Figs. 2 A, B, S2, S3 C, and D), thus suggesting that the curved shape of CDH1 is preferred in equilibrium.
Focused view of the stretching of the trans CDH1 dimer at 0.5 nm/ns (simulation S1d, Table S1, 0–33.4 ns). Dislodging of Trp2 (orange) from the hydrophobic pocket of the binding partner is observed. One monomer is shown in surface representation, while the other is shown in ribbon.
Overall, our stretching simulations of entire CDH1 ectodomain dimers show how these complexes unbend softly first, then stiffen before unbinding at extensions of approximately 14 nm, with unbinding pathways involving extraction of Trp2 residues from their binding pockets and rupture of EC1-EC1 contacts, regardless of the geometric arrangement used to apply forces. The end-to-end distance for a linear trans dimer of CDH1 is approximately 37 nm, which is much larger than the distance between cells at the adherens junction, which typically range between 15 and 25 nm (91). For CDH1 dimers to fit within an adherens junction, there would need to be a change in monomer shape or a change in orientation. The approximately 30° tilt of CDH1, with respect to a hypothetical cell plane created by the C-termini of homodimers within the crystal packing lattice, creates an end-to-end distance of approximately 19 nm (15). This tilted orientation, as seen in the diagonal simulations, does not affect the strength of trans dimers and allows for proper cis dimerization. Therefore, the diagonal system creates a hypothetical cell-cell distance closer to that observed in tissue that does not affect its tensile adhesive properties.
Desmosomal cadherins exhibit a stiffer initial response to force before unbinding
To determine whether the response of desmosomal cadherins to force is similar to what we observed for CDH1 in simulations, we built three different systems containing desmosomal cadherins that included a DSG2 homodimer, a DSG2-DSC1 heterodimer, and a DSC1 homodimer. The DSG2 and DSC1 homodimers were taken directly from crystal structures (44), while the DSG2-DSC1 heterodimer was constructed from the existing structures because high-resolution experiment-based models of heterodimers are not available (see Materials and Methods). All three systems were equilibrated for 20 ns (simulations S3a, S4a, and S5a; Tables 1 and S1) with protein monomers within the complexes maintaining a curved shape (Fig. S1). Subsequent stretching SMD simulations showed that all three systems exhibited a similar two-phased response to force at all three pulling speeds used (10, 1, and 0.1 nm/ns; Fig. 3 D; Video S3). Because the prevailing experimentally derived model of desmosomal structure shows dimers forming linear trans contacts between cells (49,92), stretching was performed in a linear fashion as opposed to the diagonal configuration. The initial unbending, in which each monomer lost its curvature, was followed by further unbending and subsequent unbinding through the loss of the Trp2 exchange as well as of a network of interactions at the EC1-EC1 interface (Figs. 3 A–C, S6 A, and S7; Video S4). Additionally, unbinding was observed without unfolding of protein secondary structure in all simulations, with Ca2+ ions at linker regions and EC5 disulfide bonds preventing unraveling of β strands.
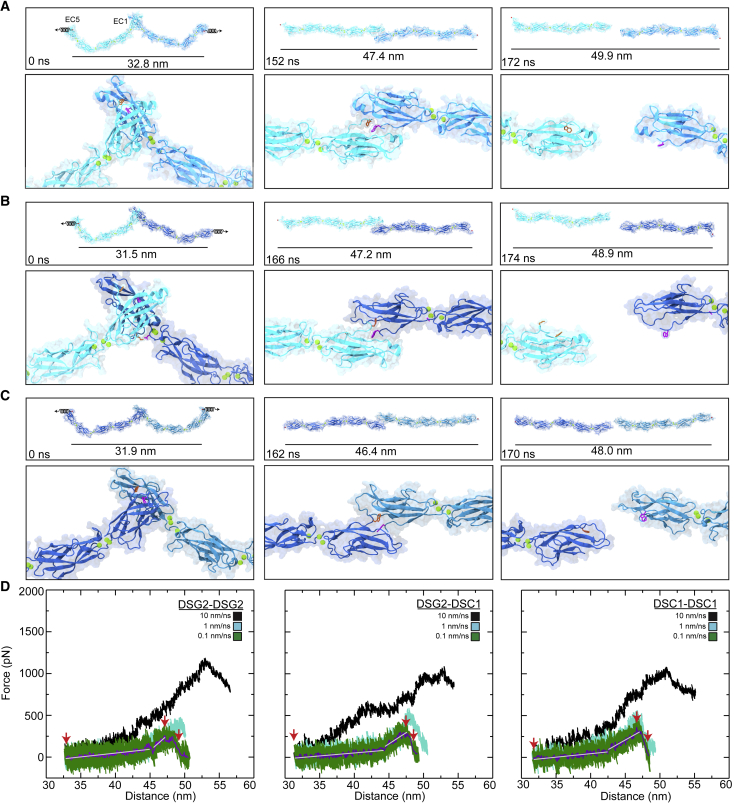
Figure 3.
Forced unbinding of desmosomal trans dimers in silico. (A) Snapshots of the DSG2 dimer unbinding at a stretching speed of 0.1 nm/ns (Simulation S3d; Table 1). Stretched C-terminal Cα atoms are shown as red spheres. Springs in first panel indicate the position and direction of the forces applied along the vector joining the C-terminal Cα atoms of the two monomers. The time and end-to-end distance between the C-terminal atoms are indicated in the top panels. The lower panels show the loss of the Trp2 exchange between DSG2 protomers. The Trp2 residues are shown as orange and purple sticks for each monomer. (B) Snapshots of DSG2-DSC1 unbinding at a stretching speed of 0.1 nm/ns (simulation S4d) shown as in (A). A salt bridge formed between DSC1 Asp101 and DSG2 Lys17 is shown in purple and orange sticks. (C) Snapshots of DSC1 dimer unbinding at a stretching speed of 0.1 nm/ns (simulation S5d) shown as in (A). All simulations showed straightening before unbinding without unfolding. (D) Force vs. end-to-end distance plots for constant velocity stretching of the three simulation systems at 10 nm/ns (S3b, S4b, S5b, black), 1 nm/ns (S3c, S4c, S5c, cyan), and 0.1 nm/ns (S3d, S4d, S5d, green; the 1-ns running averages are shown in purple; the gray lines are the linear fits used to determine elasticity). Red arrowheads indicate time points in (A), (B), and (C) respectively. Force is shown as monitored for one of the monomers for all plots.
Stretching of the DSG2-DSG2 trans dimer at 0.1 nm/ns (simulation S3d, Table S1) results in unbending of the inherent curvature of the DSG2 monomers, followed by a stiff phase, prior to unbinding of the trans interaction. Monomers begin to re-bend immediately after unbinding. System depicted as in Video S1. Similar trajectories were observed for the DSG2-DSC1 heterodimer and the DSC1-DSC1 homodimer.
Focused view of the stretching of the trans DSG2 homodimer at 0.1 nm/ns (simulation S3d, Table S1). Dislodging of the Trp2 (orange) from the hydrophobic pocket of the binding partner is observed. One monomer is shown in surface representation, while the other is shown in ribbon. Similar trajectories were observed for the DSG2-DSC1 heterodimer and the DSC1-DSC1 homodimer.
At the slowest stretching speed (0.1 nm/ns), the mechanical responses of the DSG2 and DSC1 homodimers and the DSG2-DSC1 heterodimer were similar. At the beginning of each simulation, the end-to-end distances were 32.8 nm, 31.5 nm, and 31.9 nm for the DSG2 homodimer, the DSG2-DSC1 heterodimer, and the DSC1 homodimer systems respectively, while at the force peak the systems were stretched to 47.4 nm, 47.2 nm, and 46.4 nm (Fig. 3 A–C), resulting in extensions that were greater than 10 nm. Analyses of forces (Fig. 3 D, gray lines) revealed soft unbending for the DSG2 homodimer, the DSG2-DSC1 heterodimer, and the DSC1 homodimer with spring constants of ksa1 ∼ 8.4 mN/m, ksb1 ∼ 7.7 mN/m, and ksc1 ∼ 8.0 mN/m, respectively (values obtained considering extension of both monomers in each case). The spring constants associated with the straightened phase (approximately 2–5 nm) were ksa2 ∼ 54.6 mN/m, ksb2 ∼ 54.5 mN/m, and ksc2 ∼ 47.7 mN/m, respectively. Overall, the initial, soft phase responses were stiffer than what we observed for CDH1, while spring constants for the second, stiffer, phase were similar.
Interestingly, force peaks at all three pulling speeds were comparable among the different desmosomal systems (Fig. 3 D). At the slowest stretching speed (0.1 nm/ns), the average force peaks from both monomers for the DSG2 homodimer, the DSG2-DSC1 heterodimer, and the DSC1 homodimer systems were Fp ∼ 323.4 pN ± 6.2 pN, Fp ∼ 395.7 pN ± 0.7 pN, and Fp ∼ 419.8 pN ± 6.3 pN, respectively (averages over two sides). As observed for CDH1, a loss of the Trp2 exchange correlated with the rupture force peak for all systems (Fig. S6 A), indicating that this is one crucial interaction in desmosomal trans dimers. However, several other interactions involving conserved residues at the EC1-EC1 interface also persisted until unbinding occurred in all three systems (Figs. S5 B and C, S6 A, and S7), suggesting these other interactions, in combination with the Trp2 exchange, contribute to the force peaks. After unbinding, we observed quick rebending in all systems in each of the separate monomers, as monitored by the end-to-end distance between N- and C-termini within each monomer (Fig. S6 B) and by a partial recovery of inter-repeat orientations (Figs. S8–S10). As with the CDH1 system, curvature in each monomer seems to be an inherent property of classical cadherins.
In addition to Trp2, there are other residues that formed transient interactions during equilibrium and stretching simulations that may have functional consequences. In the DSG2 homodimer system, the rupture and reformation of a salt bridge involving Arg97 in one monomer and residues Glu30 and Glu31 in the other resulted in a small dip and subsequent increase in the force response (Fig. S6 A, first panel). Similar interactions were not seen in either the DSG2-DSC1 or DSC1 homodimer systems as the residues at these positions differ in DSC1, which suggests the combination of DSG and DSC monomers in the desmosome results in distinct mechanical responses. Additionally, a salt bridge formed during equilibration of the DSG2-DSC1 system between DSG2 Lys17 and DSC1 Asp101. This interaction was one among several that was predicted to be responsible for the heterophilic interaction specificity observed in experiments (44). While this interaction broke well before the force peak (Fig. S6 A, second panel), it did persist during equilibration and may have functional consequences for specificity that do not relate to force response. For instance, this interaction introduced significant twisting between DSC1 and DSG2 as compared with the DSG2-DSG2 system. This twisting could potentially influence the way desmosomal cadherins interact with one another in the desmosome, and thus influence the structure of the desmosome itself.
In summary, the mechanical response of desmosomal cadherins predicted by simulations was similar to what we observed in simulations of CDH1, but with a stiffer first phase of extension and with similar unbinding force peaks despite simulations being carried out at a slower stretching speed (0.1 nm/ns compared with 0.5 nm/ns), which should have resulted in decreased force peaks (74,90,93,94). Specific interactions among conserved residues might explain this behavior (Figs. S5 B, C, and S6 A). These results suggest that desmosomal cadherin dimers, especially those formed by DSC1, might be more resistant to external mechanical stimuli than those formed by CDH1.
Clustered PCDHs lack a two-phased response to force
The next three molecular systems simulated involved PCDH homodimers, including those formed by PCDHα7 EC1-5, a representative of the α clustered PCDH subfamily; by PCDHβ6 EC1-4, representing the β subfamily; and by PCDHγB3 EC1-4, a γ subfamily representative. All homodimers comprise a large antiparallel EC1-4 interface that was maintained during initial equilibrium simulations lasting approximately 20 ns, with some fluctuations in contacts in longer equilibrations (95). Subsequent constant velocity SMD simulations on each of these clustered PCDH systems at three different stretching speeds of 10 nm/ns, 1 nm/ns, and 0.1 nm/ns revealed a response that was distinct to that observed for CDH1 and the desmosomal cadherins. This response was generally characterized by EC repeats slipping past each other as some salt bridge interactions ruptured and others transiently formed resulting in short-lived binding intermediates (Video S5). Forces monitored throughout the SMD simulations did not show evidence for a two-phased response with soft unbending of monomers, but rather exhibited only a stiff phase that led to a main force peak followed by smaller force peaks for intermediates when these were present. At the fastest stretching speed, one of the PCDHγB3 EC1-4 monomers unfolded before unbinding, but this was not observed in any of the other systems at the stretching speeds tested in our simulations, as described below and despite the absence of disulfide bonds in terminal EC repeats for clustered PCDHs. This suggests that Ca2+ ions strengthen linker regions to prevent unfolding in most cases.
Stretching of the PCDHβ6 trans homodimer at 0.1 nm/ns (simulation S7d, Table S1, 0–180 ns) results in rupture of the EC1-EC4 interface and the formation of transient intermediates before complete unbinding of the complex. System depicted as in Video S1.
At the beginning of the PCDHα7 dimer simulation at the slowest stretching speed (0.1 nm/ns; simulation S6d), the end-to-end distance between the C-terminal Cα atoms of the complex increased little, from 28.6 nm to 30.4 nm, while the applied force increased rapidly to a peak value of Fp ∼ 394.1 ± 1.6 pN (average over two sides; Figs. 4 A, D, left panel, and S11 A–C, left panel). The force vs. end-to-end distance plot lacks the first soft-phase observed for classical cadherin and shows a broad semiplateaued peak with values slowly diminishing as two salt bridge interactions, one between Glu91 in one monomer and Lys373 in the other, and the other between Arg348 in one monomer and Asp41 in the other, broke one after another (Figs. S11 A, left panel, and S12 A). Interestingly, residue charge is conserved at positions 91, 348, and 373, but not at position 41 (Fig. S5 D). Eventually, the monomers separated completely from each other when the end-to-end distance was 41.8 nm (Fig. 4 A). No drastic changes in monomer lengths and orientations between EC repeats were observed during this forced unbinding (Figs. S11 B, left panel, and S13). The increase in end-to-end distance seen during this unbinding was a result of EC repeats slipping past one another, rather than from the unbending of each monomer as observed for the classical cadherins.
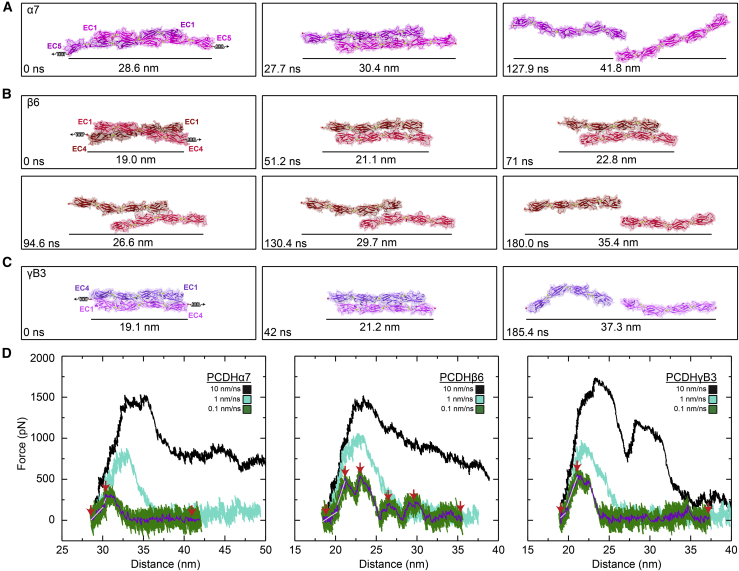
Figure 4.
Forced unbinding of clustered PCDH trans homodimers in silico. (A) Snapshots of the PCDHα7 EC1-5 dimer unbinding at a stretching speed of 0.1 nm/ns (Simulation S6d; Table 1). Stretched C-terminal Cα atoms are shown as red spheres. Springs in first panel indicate position and direction of forces applied along the vector joining the C-terminal Cα atoms of the two monomers. Time and end-to-end distance between C-terminal atoms is indicated in each panel. (B) Snapshots of PCDHβ6 EC1-4 dimer unbinding at a stretching speed of 0.1 nm/ns (simulation S7d) shown as in (A). (C) Snapshots of PCDHγB3 EC1-4 dimer unbinding at a stretching speed of 0.1 nm/ns (simulation S8d) shown as in (A) and (B). (D) Force vs. end-to-end distance plots for constant velocity stretching of the three simulation systems at 10 nm/ns (S6b, S7b, S8b, black), 1 nm/ns (S6c, S7c, S8c, cyan), and 0.1 nm/ns (S6d, S7d, S8d, green; 1 ns running averages shown in purple; gray lines are linear fits used to determine elasticity). Red arrowheads indicate time points in (A), (B), and (C). Force is shown as monitored for one of the monomers for all plots.
The unbinding pathway for the PCDHβ6 dimer was more complex, with transient intermediates associated with multiple force peaks at the slowest stretching speed of 0.1 nm/ns (Fig. 4 B; simulation S7d). The end-to-end distance between the C-terminal Cα atoms of the complex again increased little from 19.0 nm to 21.1 nm as the first force peak was reached at Fp ∼ 552.6 ± 41.8 (Figs. 4 D, middle panel, and S11 C, middle panel). Three subsequent force peaks were evident, with forces reaching Fp ∼ 603.3 ± 5.6, Fp ∼ 273.4 ± 21.1, and Fp ∼ 330.5 ± 17.0 at end-to-end distances of 22.8 nm, 26.6 nm, and 29.7 nm, respectively (Fig. 4 B and D, middle panel). The multiple force peaks observed during unbinding of the PCDHβ6 dimer were associated with different salt bridge interactions that formed and ruptured during the trajectory. The initial salt bridge interaction between Glu289 in one monomer and Arg157 in the other ruptured as another salt bridge interaction between Arg157 in one monomer and Glu213 in the other formed (Fig. S12 B). This new interaction broke when Glu165 from one monomer formed a salt bridge with Arg4 from the other. This salt bridge also broke as a new salt bridge between Glu165 and Arg4 of swapped monomers formed (Video S6). Eventually this last salt bridge interaction ruptured as the monomers separated from one another (Fig. S11 A, middle panel). Again, residue charge is conserved for some of the residues forming salt bridges, but not all (Fig. S5 E). The monomers were completely separated from each other when the end-to-end distance was 35.4 nm (Fig. 4 B). The inter-repeat orientations were more stable in one monomer than the other, likely owing to rearrangements throughout the trajectory (Fig. S14). The change in lengths for the monomers was negligible in response to force when compared with changes seen for the CDH1 and desmosomal systems (Fig. S11 B, middle panel) as the increase in end-to-end distance seen during unbinding was again due to EC repeats slipping past one another.
Close up view of the transient interaction that forms between Arg4 (A) and Glu165 (B) during stretching of the PCDHβ6 trans homodimer at 0.1 nm/ns (simulation S7d, Table S1, 0–180 ns). Chain B is shown in bright pink while chain A is shown in dark red color.
For the third system, which included the PCDHγB3 dimer, the stretching simulation at 0.1 nm/ns revealed an increase in end-to-end distance of the dimer from 19.1 nm to 21.2 nm when the force reached a peak at Fp ∼ 612.6 ± 13.5 pN (Figs. 4 C, D, right panel, and S11 C, right panel). Two salt bridge interactions, the first one between Lys340 in one monomer and Glu77 in the other (Fig. S12 C), and the second one between Glu125 in one monomer and Lys292 in the other, ruptured back-to-back as force peaked and fell during unbinding (Fig. S11 A, right panel). A second small force spike (Fp ∼ 234.4 ± 5.3 pN) was observed at an end-to-end distance of 33.5 nm when a new salt bridge interaction, between Arg4 in one monomer and Glu77 in the other, formed transiently and then ruptured (Fig. S11 A, right panel). All residue involved in these interactions were highly conserved within the species analyzed (Fig. S5 F). The monomers were completely separated when the end-to-end distance reached 37.3 nm (Fig. 4 C). Changes in lengths and inter-repeat orientation for each of the monomers in response to application of force to the complex were again minor (Figs. S11 B, right panel, and S15). Predicted glycosylation sites in PCDHγB3 are not at residues forming contacts at any point during the unbinding trajectory and hence glycosylation is not expected to interfere with the unbinding pathway and intermediates (Video S7). Similar to what we observed for PCDHα7 and PCDHβ6, the increase in end-to-end of distance for the PCDHγB3 trans dimer during unbinding was caused by EC repeats slipping past one another.
Location of glycosylation sites in the PCDHγB3 trans homodimer during forced stretching at 0.1 nm/ns (simulation S8d, Table S1, 0–185.4 ns). Protein is shown in gray ribbon representation, residues that form interactions with each other are shown as magenta spheres, while glycosylation sites are shown as cyan spheres. Glycosylation is not expected to interfere with unbinding pathway.
Overall, our simulations of clustered PCDHs revealed a mechanical response in which stiff dimers break without an unbending soft phase, but with force quickly climbing over approximately 2 nm extensions to reach peak maxima that are generally larger than what we observed for classical cadherins. Intermediates were observed in some cases as the separation between the monomers ends increased over approximately 10 nm and the monomers passed each other keeping their rather straight conformations. An additional approximately 100-ns long control simulation of the PCDHβ6 EC1-4 monomer (simulations S9a-b) confirmed that this fragment is straighter than what was observed for CDH1, DSG2, and DSC1 over approximately 20 ns (Fig. S1), suggesting that the brittle response of clustered PCDH complexes ultimately originates from the straighter conformations of their monomers.
Discussion
Multimodular proteins, such as cadherins, are known to be involved in various mechanical processes in vivo (96,97). The simulations presented here offer a unique and comparative view of the forced unbinding trajectories of adhesive complexes formed by three cadherin subtypes, including CDH1, the desmosomal cadherins, and the clustered PCDHs. The dimeric complexes analyzed already display evident structural differences that are reflected in their mechanical responses. The classical cadherins CDH1, DSG2, and DSC1 all have bent ectodomains that remain in this conformation throughout equilibrium simulations (Fig. S1) and that interact tip-to-tip through contacts mediated by their EC1 repeats. We predict that soft unbending (<10 mN/m over approximately 10 nm extensions; Fig. 5 A and C) in response to force precedes unbinding characterized by the extraction of swapped Trp2 residues and rupture of various EC1-EC1 contacts. In contrast, our simulations predict that clustered PCDHs are straighter (Fig. S1), that their response to force lacks a soft unbending phase, and that the PCDHα7, PCDHβ6, and PCDHγB3 overlapped dimers minimally stretch (approximately 2 nm) before unbinding forces peak at generally larger values than those observed for classical cadherins (Fig. 5 B and C), with subsequent formation of transient, weaker interactions as monomers pass each other before complete separation. These results, which pertain to single dimers stretched at fast speeds, should help in interpreting experimental results, including those from bulk equilibrium and from single-molecule force spectroscopy measurements. In addition, our results should help in advancing our understanding of the assembly of larger complexes that form cellular structures involved in adhesion and signaling.
Figure 5.
Predicted elasticity for classical cadherins and clustered PCDHs. (A) Summary of spring constants for the soft (ks1) and stiff (ks2) phases predicted in simulations of classical cadherins. (B) Summary of unbinding force peaks for classical cadherins and clustered PCDHs at different stretching speeds as predicted by simulations. Averages are shown along with error bars (standard deviations) (C) Illustration of unbending and unbinding stages for classical cadherins (left) and clustered PCDHs (right).
Dissociation constants measured in equilibrium for the protein fragments simulated here generally range between approximately 2 μM and 100 μM (Supplementary Discussion and Table S3) (44,56,59,60,98), with little correlation between binding mechanism, buried surface area, and experimentally measured bond strengths in near-equilibrium conditions. It is intriguing that the dissociation constants for classical cadherins that form small EC1-EC1 contacts with little buried surface area (<1000 Å2) are not drastically different from those measured for PCDH complexes with significantly larger interfaces (>1500 Å2, Table S3). This indicates that the nature and details of the contacts, including the exchange of Trp2 for classical cadherins and specific salt bridges and hydrogen bonds for all complexes, are more relevant in determining the affinity of the bond in equilibrium. Transient variations in contacts and buried surface area for large interfaces such as those of clustered PCDHs (95) might also explain why binding affinities are not as strong as expected. Alternatively, because force is known to restructure the energy landscape of protein-protein interactions (99), the atomistic differences in contact interfaces and buried surface area among cadherin complexes might be more relevant upon force application and under nonequilibrium conditions.
While there have been several studies exploring the mechanical unbinding strength of classical cadherin bonds using single-molecule force spectroscopy experiments (19,20,23,100, 101, 102, 103, 104), the strength of the clustered PCDH bonds has not been experimentally probed, and a direct comparison with results from our simulations is difficult. A fairly linear increase in force associated with unbending of classical cadherin ectodomains as predicted by our simulations is likely to be buried in experimental force profiles within the phase attributed to stretching of linkers used to attach cadherin to surfaces. Unbinding force peaks from experiments are typically obtained at stretching speeds that range from 1 μm/s to 20 μm/s (10−6 to 10−5 nm/ns), while our simulations are carried out at stretching speeds that are as slow as 0.1 nm/ns, which results in expected larger unbinding forces (88,93,105,106). In addition, our simulations suggest that at fast stretching speeds specificity observed in near equilibrium conditions might be overridden and less relevant than interface size, as clustered PCDHs seem to generally unbind at higher forces than classical cadherins at the fastest speeds used (Fig. 5 B), a prediction that could be tested using high-speed single-molecule force spectroscopy (90,94).
Relevant mechanical stimuli for cadherins in vivo are expected to be diverse (107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119). As cells divide and tissues develop, cell-cell contacts will be experiencing tension. Similarly, epithelial and cardiac tissues are subject to constant stress from routine physiological stretching and shearing forces, as well as from external forces, such as cuts and abrasions. Although there is little information on the magnitude of the forces that cadherins may experience in vivo (120, 121, 122), the spatial and time scales of certain physiological events can serve to analyze cadherin responses in the context of our findings. For instance, we expect slow processes during tissue development (minutes to hours) and faster events in cardiac tissue where cardiomyocyte adhesions can move substrates at more than 1 μm/s (123) and sarcomere lengths can change at speeds of approximately 2 μm/s (124). Yet, these are stretching speeds that are orders of magnitude slower than what we have used in our simulations, and thus near-equilibrium conditions may better represent the response of cadherins in these contexts. In contrast, cadherins in tissues exposed to the catastrophic impact of a bullet (>1000 nm/ns) (125) or to bruising by an external object (>1 nm/ns) (126) might be stretched at the speeds we have used in our simulations (0.1 nm/ns), where rupture of the cadherin bond might prevent cell damage.
Regardless of the type of stimulus and stretching speeds used, we do expect that the first, soft stretching phase of classical cadherins associated with unbending will be part of their mechanical response. This soft response is observed in other protein systems, such as ankyrin repeat stacks and tandem titin Ig domains (30, 31, 32, 33, 34, 35, 36), and might be important in the context of cell oscillations or small-scale tissue stretching (127, 128, 129, 130, 131), where classical cadherin bonds would act as molecular shock absorbers and be mechanically robust, while clustered PCDH bonds might break instead. Ectodomain bending and unbending might also preclude trans contacts that go beyond EC1 (tip-to-tip) in classical cadherins, while the more rigid and straighter ectodomains of clustered PCDHs could facilitate the antiparallel overlap observed in structures. Looking at our simulated clustered PCDH unbinding trajectories in reverse as a model for possible bond formation suggests that to have antiparallel EC1-4 overlap these ectodomains should maintain their straight conformation, as observed here in equilibrium simulations of PCDHβ6. Interestingly, transient intermediates observed for clustered PCDH proteins during simulated unbinding may help drive trans bond assembly, especially in the context of cellular fluctuations that may facilitate a ratchet-like mechanism (132) of binding for rigid ectodomains. Softer ectodomains, like those of classical cadherins, might just bend and preserve EC1 contacts, rather than favoring overlap. In turn, the larger interface achieved through overlap by PCDH proteins permits greater variation in the number and type of residues involved in the binding interface, a key determinant of strict homophilic specificity observed for clustered PCDH proteins.
Our observations suggest that each set of cadherins has evolved to adopt various features (13,133, 134, 135), including mechanical properties suitable for their roles in vivo. How the curvature and shape have evolved and are sequence-encoded in classical cadherins and clustered PCDHs is unknown, but is easy to speculate that both the length of loops that form linker regions and the nature and size of their residues are relevant for bending, especially in subtle cases without Ca2+-free linker regions (136,137). Intriguingly, the clearest indicator of bending in classical cadherin linker regions that bind two or three Ca2+ ions seems to be the presence of a glutamate residue in the DXNDN motif, which is conserved in all linker regions of clustered PCDHs and modified to DXNEN at the EC3-4 linker of classical cadherins (16). The longer glutamate sidechain may contribute degrees of freedom that, along with other linker-specific features, facilitate bending without compromising Ca2+ binding. A disease-causing mutation in DSG2 (D105E) that transforms the DXNDN motif into DXNEN at the EC1-2 linker (138) confirms that a subtle change to this motif can alter function, perhaps through shape changes that alter binding properties (139). This hypothesis might be tested in future simulations that also explore how the response of single trans dimers changes and determines the properties of larger complexes present in adhesive contacts, including those with mixtures of classical and nonclassical cadherins (140, 141, 142, 143). Elucidating the molecular basis of the mechanics of cadherin ectodomains, alone and in complexes, may serve to further understand their function and to also rationally design modular proteins with desired mechanical properties (144,145).
Author contributions
B.L.N. prepared and simulated CDH1 systems. C.N. prepared and simulated systems with desmosomal cadherins. S.W. and R.A.S. prepared and simulated clustered PCDH systems. M.S. trained co-authors and supervised research. B.L.N., C.N., S.W., and M.S. designed research and wrote and edited the manuscript with feedback from R.A.S.
Acknowledgments
This work was supported by the Ohio State University and by the Human Frontier Science Program (RGP0056/2018). Simulations were performed using the NCSA-Blue Waters (GLCPC), TACC-Stampede, PSC-Bridges (XSEDE MCB140226), OSC-Owens, and OSC-Pitzer (PAS1037 and PAA0217) supercomputers. B.L.N. was supported by an OSU/NIH cellular, molecular biochemical sciences program training grant fellowship (T32GM086252), and by an OSU presidential fellowship. C.N. was supported by an OSU/NIH molecular biophysics training grant (TG32GM118291). R.A.-S. was a Pelotonia fellow. M.S. was an Alfred P. Sloan fellow (FR-2015-6794).
Editor: Alexander Dunn.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.02.007.
Supporting citations
Reference (146) appears in the supporting material.
Supporting material
References
- 1.Takeichi M. Functional correlation between cell adhesive properties and some cell surface proteins. J. Cell Biol. 1977;75:464–474. doi: 10.1083/jcb.75.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kemler R., Babinet C., et al. Jacob F. Surface antigen in early differentiation. Proc. Natl. Acad. Sci. U S A. 1977;74:4449–4452. doi: 10.1073/pnas.74.10.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brasch J., Harrison O.J., et al. Shapiro L. Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol. 2012;22:299–310. doi: 10.1016/j.tcb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaidel-Bar R. Cadherin adhesome at a glance. J. Cell Sci. 2013;126:373–378. doi: 10.1242/jcs.111559. [DOI] [PubMed] [Google Scholar]
- 5.Weiner J.A., Jontes J.D. Protocadherins, not prototypical: a complex tale of their interactions, expression, and functions. Front. Mol. Neurosci. 2013;6:4. doi: 10.3389/fnmol.2013.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sotomayor M., Gaudet R., Corey D.P. Sorting out a promiscuous superfamily: towards cadherin connectomics. Trends Cell Biol. 2014;24:524–536. doi: 10.1016/j.tcb.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canzio D., Maniatis T. The generation of a protocadherin cell-surface recognition code for neural circuit assembly. Curr. Opin. Neurobiol. 2019;59:213–220. doi: 10.1016/j.conb.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro L., Fannon A.M., et al. Hendrickson W.A. Structural basis of cell-cell adhesion by cadherins. Nature. 1995;374:327–337. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- 9.Overduin M., Harvey T.S., et al. Ikura M. Solution structure of the epithelial cadherin domain responsible for selective cell adhesion. Science. 1995;267:386–389. doi: 10.1126/science.7824937. [DOI] [PubMed] [Google Scholar]
- 10.Nagar B., Overduin M., et al. Rini J.M. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- 11.Hirano S., Takeichi M. Cadherins in brain morphogenesis and wiring. Physiol. Rev. 2012;92:597–634. doi: 10.1152/physrev.00014.2011. [DOI] [PubMed] [Google Scholar]
- 12.Sadeqzadeh E., de Bock C.E., Thorne R.F. Sleeping giants: emerging roles for the fat cadherins in health and disease. Med. Res. Rev. 2014;34:190–221. doi: 10.1002/med.21286. [DOI] [PubMed] [Google Scholar]
- 13.Rübsam M., Broussard J.A., Niessen C.M., et al. Adherens junctions and desmosomes coordinate mechanics and signaling to orchestrate tissue morphogenesis and function: an evolutionary perspective. Cold Spring Harb. Perspect. Biol. 2018;10:a029207. doi: 10.1101/cshperspect.a029207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mège R.M., Ishiyama N. Integration of cadherin adhesion and cytoskeleton at adherens junctions. Cold Spring Harb. Perspect. Biol. 2017;9:a028738. doi: 10.1101/cshperspect.a028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison O.J., Jin X., et al. Honig B. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 2011;19:244–256. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boggon T.J., Murray J., et al. Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 17.Pokutta S., Herrenknecht K., et al. Engel J. Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding. Eur. J. Biochem. 1994;223:1019–1026. doi: 10.1111/j.1432-1033.1994.tb19080.x. [DOI] [PubMed] [Google Scholar]
- 18.Prasad A., Pedigo S. Calcium-dependent stability studies of domains 1 and 2 of epithelial cadherin. Biochemistry. 2005;44:13692–13701. doi: 10.1021/bi0510274. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Sivasankar S., et al. Chu S. Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc. Natl. Acad. Sci. U S A. 2009;106:109–114. doi: 10.1073/pnas.0811350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sivasankar S., Zhang Y., et al. Chu S. Characterizing the initial encounter complex in cadherin adhesion. Structure. 2009;17:1075–1081. doi: 10.1016/j.str.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison O.J., Bahna F., et al. Shapiro L. Two-step adhesive binding by classical cadherins. Nat. Struct. Mol. Biol. 2010;17:348–357. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciatto C., Bahna F., et al. Shapiro L. T-cadherin structures reveal a novel adhesive binding mechanism. Nat. Struct. Mol. Biol. 2010;17:339–347. doi: 10.1038/nsmb.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rakshit S., Zhang Y., et al. Sivasankar S. Ideal, catch, and slip bonds in cadherin adhesion. Proc. Natl. Acad. Sci. U S A. 2012;109:18815–18820. doi: 10.1073/pnas.1208349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manibog K., Li H., et al. Sivasankar S. Resolving the molecular mechanism of cadherin catch bond formation. Nat. Commun. 2014;5:3941. doi: 10.1038/ncomms4941. [DOI] [PubMed] [Google Scholar]
- 25.Manibog K., Sankar K., et al. Sivasankar S. Molecular determinants of cadherin ideal bond formation: conformation-dependent unbinding on a multidimensional landscape. Proc. Natl. Acad. Sci. U S A. 2016;113:E5711–E5720. doi: 10.1073/pnas.1604012113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayas M.V., Schulten K., Leckband D. Forced dissociation of the strand dimer interface between C-cadherin ectodomains. Mech. Chem. Biosyst. 2004;1:101–111. [PubMed] [Google Scholar]
- 27.Cailliez F., Lavery R. Cadherin mechanics and complexation: the importance of calcium binding. Biophys. J. 2005;89:3895–3903. doi: 10.1529/biophysj.105.067322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cailliez F., Lavery R. Dynamics and stability of E-cadherin dimers. Biophys. J. 2006;91:3964–3971. doi: 10.1529/biophysj.106.087213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sotomayor M., Schulten K. The allosteric role of the Ca2+ switch in adhesion and elasticity of C-cadherin. Biophys. J. 2008;94:4621–4633. doi: 10.1529/biophysj.107.125591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard J., Bechstedt S. Hypothesis: a helix of ankyrin repeats of the NOMPC-TRP ion channel is the gating spring of mechanoreceptors. Curr. Biol. 2004;14:R224–R226. doi: 10.1016/j.cub.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 31.Sotomayor M., Corey D.P., Schulten K. In search of the hair-cell gating spring: elastic properties of ankyrin and cadherin repeats. Structure. 2005;13:669–682. doi: 10.1016/j.str.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Lee G., Abdi K., et al. Marszalek P.E. Nanospring behaviour of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 33.Lee E.H., Hsin J., et al. Schulten K. Tertiary and secondary structure elasticity of a six-Ig titin chain. Biophys. J. 2010;98:1085–1095. doi: 10.1016/j.bpj.2009.12.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsin J., Strümpfer J., et al. Schulten K. Molecular origin of the hierarchical elasticity of titin: simulation, experiment, and theory. Annu. Rev. Biophys. 2011;40:187–203. doi: 10.1146/annurev-biophys-072110-125325. [DOI] [PubMed] [Google Scholar]
- 35.Kappel C., Zachariae U., et al. Grubmüller H. An unusual hydrophobic core confers extreme flexibility to HEAT repeat proteins. Biophys. J. 2010;99:1596–1603. doi: 10.1016/j.bpj.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Argudo D., Capponi S., et al. Grabe M. A multiscale model of mechanotransduction by the ankyrin chains of the NOMPC channel. J. Gen. Physiol. 2019;151:316–327. doi: 10.1085/jgp.201812266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oroz J., Valbuena A., et al. Carrión-Vázquez M. Nanomechanics of the cadherin ectodomain: “canalization” by Ca2+ binding results in a new mechanical element. J. Biol. Chem. 2011;286:9405–9418. doi: 10.1074/jbc.M110.170399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green K.J., Simpson C.L. Desmosomes : new perspectives on a classic. J. Invest. Dermatol. 2007;127:2499–2515. doi: 10.1038/sj.jid.5701015. [DOI] [PubMed] [Google Scholar]
- 39.Delva E., Tucker D.K., Kowalczyk A.P. The desmosome. Cold Spring Harb. Perspect. Biol. 2009;1:a002543. doi: 10.1101/cshperspect.a002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson J.L., Najor N.A., Green K.J. Desmosomes : regulators of cellular signaling and adhesion in epidermal health and disease. Cold Spring Harb. Perspect. Med. 2014;4:a015297. doi: 10.1101/cshperspect.a015297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujiwara M., Nagatomo A., et al. Suzuki S.T. Desmocollin-2 alone forms functional desmosomal plaques, with the plaque formation requiring the juxtamembrane region and plakophilins. J. Biochem. 2015;158:339–353. doi: 10.1093/jb/mvv048. [DOI] [PubMed] [Google Scholar]
- 42.Arnemann J., Sullivan K.H., et al. Buxton R.S. Stratification-related expression of isoforms of the desmosomal cadherins in human epidermis. J. Cell Sci. 1993;104:741–750. doi: 10.1242/jcs.104.3.741. [DOI] [PubMed] [Google Scholar]
- 43.Lowndes M., Rakshit S., et al. Nelson W.J. Different roles of cadherins in the assembly and structural integrity of the desmosome complex. J. Cell Sci. 2014;127:2339–2350. doi: 10.1242/jcs.146316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrison O.J., Brasch J., et al. Shapiro L. Structural basis of adhesive binding by desmocollins and desmogleins. Proc. Natl. Acad. Sci. U S A. 2016;113:7160–7165. doi: 10.1073/pnas.1606272113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chitaev N.A., Troyanovsky S.M. Direct Ca2+-dependent heterophilic interaction between desmosomal cadherins, desmoglein and desmocollin, contributes to cell-cell adhesion. J. Cell Biol. 1997;138:193–201. doi: 10.1083/jcb.138.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Getsios S., Amargo E.V., et al. Green K.J. Coordinated expression of desmoglein 1 and desmocollin 1 regulates intercellular adhesion. Differentiation. 2004;72:419–433. doi: 10.1111/j.1432-0436.2004.07208008.x. [DOI] [PubMed] [Google Scholar]
- 47.Waschke J., Bruggeman P., et al. Drenckhahn D. Pemphigus foliaceus IgG causes dissociation of desmoglein 1-containing junctions without blocking desmoglein 1 transinteraction. J. Clin. Invest. 2005;115:3157–3165. doi: 10.1172/JCI23475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garrod D., Chidgey M. Desmosome structure, composition and function. Biochim. Biophys. Acta. 2008;1778:572–587. doi: 10.1016/j.bbamem.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 49.Sikora M., Ermel U.H., et al. Frangakis A.S. Desmosome architecture derived from molecular dynamics simulations and cryo-electron tomography. Proc. Natl. Acad. Sci. U S A. 2020;117:27132–27140. doi: 10.1073/pnas.2004563117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sano K., Tanihara H., et al. Suzuki S. Protocadherins: a large family of cadherin-related molecules in central nervous system. EMBO J. 1993;12:2249–2256. doi: 10.1002/j.1460-2075.1993.tb05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Q., Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 52.Yagi T. Clustered protocadherin family. Dev. Growth Differ. 2008;50:131–141. doi: 10.1111/j.1440-169X.2008.00991.x. [DOI] [PubMed] [Google Scholar]
- 53.Mountoufaris G., Canzio D., et al. Maniatis T. Writing, reading, and translating the clustered protocadherin cell surface recognition code for neural circuit assembly. Annu. Rev. Cell Dev. Biol. 2018;34:471–493. doi: 10.1146/annurev-cellbio-100616-060701. [DOI] [PubMed] [Google Scholar]
- 54.Lawrence Zipursky S., Grueber W.B. The molecular basis of self-avoidance. Annu. Rev. Neurosci. 2013;36:547–568. doi: 10.1146/annurev-neuro-062111-150414. [DOI] [PubMed] [Google Scholar]
- 55.Schreiner D., Weiner J.A. Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proc. Natl. Acad. Sci. U S A. 2010;107:14893–14898. doi: 10.1073/pnas.1004526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rubinstein R., Thu C.A., et al. Honig B. Molecular logic of neuronal self-recognition through protocadherin domain interactions. Cell. 2015;163:629–642. doi: 10.1016/j.cell.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicoludis J.M., Lau S.-Y., et al. Gaudet R. Structure and sequence analyses of clustered protocadherins reveal antiparallel interactions that mediate homophilic specificity. Structure. 2015;23:2087–2098. doi: 10.1016/j.str.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicoludis J.M., Vogt B.E., et al. Gaudet R. Antiparallel protocadherin homodimers use distinct affinity- and specificity-mediating regions in cadherin repeats 1-4. Elife. 2016;5:e18449. doi: 10.7554/eLife.18449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goodman K.M., Rubinstein R., et al. Shapiro L. Structural basis of diverse homophilic recognition by clustered α- and β-protocadherins. Neuron. 2016;90:709–723. doi: 10.1016/j.neuron.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodman K.M., Rubinstein R., et al. Shapiro L. γ-Protocadherin structural diversity and functional implications. Elife. 2016;5:e20930. doi: 10.7554/eLife.20930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brasch J., Goodman K.M., et al. Shapiro L. Visualization of clustered protocadherin neuronal self-recognition complexes. Nature. 2019;569:280–283. doi: 10.1038/s41586-019-1089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirayama T., Yagi T. The role and expression of the protocadherin-alpha clusters in the CNS. Curr. Opin. Neurobiol. 2006;16:336–342. doi: 10.1016/j.conb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 63.Hasegawa S., Hamada S., Yagi T., et al. The protocadherin-alpha family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Mol. Cell. Neurosci. 2008;38:66–79. doi: 10.1016/j.mcn.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 64.Lefebvre J.L., Zhang Y., et al. Sanes J.R. γ-Protocadherins regulate neuronal survival but are dispensable for circuit formation in retina. Development. 2008;135:4141–4151. doi: 10.1242/dev.027912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Izrailev S., Stepaniants S., Isralewitz B. Computational Molecular Dynamics: Challenges, Methods, Ideas. Springer; 1998. Steered molecular dynamics; pp. 39–65. [Google Scholar]
- 66.Grubmüller H. Force probe molecular dynamics simulations. Methods Mol. Biol. 2005;305:493–515. doi: 10.1007/978-1-59259-912-7_23. [DOI] [PubMed] [Google Scholar]
- 67.Sotomayor M., Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 68.Franz F., Daday C., Gräter F. Advances in molecular simulations of protein mechanical properties and function. Curr. Opin. Struct. Biol. 2020;61:132–138. doi: 10.1016/j.sbi.2019.12.015. [DOI] [PubMed] [Google Scholar]
- 69.Neel B.L., Nisler C.R., et al. Sotomayor M. Collective mechanical responses of cadherin-based adhesive junctions as predicted by simulations. Biophys. J. 2022 doi: 10.1016/j.bpj.2022.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 71.Waterhouse A., Bertoni M., et al. Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karplus M., Petsko G.A. Molecular dynamics simulations in biology. Nature. 1990;347:631–639. doi: 10.1038/347631a0. [DOI] [PubMed] [Google Scholar]
- 73.Daggett V., Levitt M. Realistic simulations of native-protein dynamics in solution and beyond. Annu. Rev. Biophys. Biomol. Struct. 1993;22:353–380. doi: 10.1146/annurev.bb.22.060193.002033. [DOI] [PubMed] [Google Scholar]
- 74.Lee E.H., Hsin J., et al. Schulten K. Discovery through the computational microscope. Structure. 2009;17:1295–1306. doi: 10.1016/j.str.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dror R.O., Dirks R.M., et al. Shaw D.E. Biomolecular simulation: a computational microscope for molecular biology. Annu. Rev. Biophys. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- 76.Smith J.C., Roux B. Eppur si muove! The 2013 Nobel Prize in Chemistry. Structure. 2013;21:2102–2105. doi: 10.1016/j.str.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 77.Singharoy A., Maffeo C., et al. Schulten K. Atoms to phenotypes: molecular design principles of cellular energy metabolism. Cell. 2019;179:1098–1111.e23. doi: 10.1016/j.cell.2019.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanbonmatsu K.Y., Tung C.-S. High performance computing in biology: multimillion atom simulations of nanoscale systems. J. Struct. Biol. 2007;157:470–480. doi: 10.1016/j.jsb.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jorgensen W.L., Chandrasekhar J., et al. Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 80.Jagger B.R., Kochanek S.E., et al. Mulholland A.J. Multiscale simulation approaches to modeling drug-protein binding. Curr. Opin. Struct. Biol. 2020;61:213–221. doi: 10.1016/j.sbi.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 81.Phillips J.C., Braun R., et al. Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang J., MacKerell A.D.J. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2014;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Buck M., Bouguet-Bonnet S., et al. MacKerell A.D. Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys. J. 2006;90:L36–L38. doi: 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089. [Google Scholar]
- 85.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 86.Isralewitz B., Baudry J., et al. Schulten K. Steered molecular dynamics investigations of protein function. J. Mol. Graph. Model. 2001;19:13–25. doi: 10.1016/s1093-3263(00)00133-9. [DOI] [PubMed] [Google Scholar]
- 87.Gräter F., Shen J., et al. Grubmüller H. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys. J. 2005;88:790–804. doi: 10.1529/biophysj.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sheridan S., Gräter F., Daday C. How fast is too fast in force-probe molecular dynamics simulations? J. Phys. Chem. B. 2019;123:3658–3664. doi: 10.1021/acs.jpcb.9b01251. [DOI] [PubMed] [Google Scholar]
- 89.Evans E.A., Calderwood D.A. Forces and bond dynamics in cell adhesion. Science. 2007;316:1148–1153. doi: 10.1126/science.1137592. [DOI] [PubMed] [Google Scholar]
- 90.Rico F., Gonzalez L., et al. Scheuring S. High-speed force spectroscopy unfolds titin at the velocity of molecular dynamics simulations. Science. 2013;342:741–743. doi: 10.1126/science.1239764. [DOI] [PubMed] [Google Scholar]
- 91.Miyaguchi K. Ultrastructure of the zonula adherens revealed by rapid-freeze deep-etching. J. Struct. Biol. 2000;132:169–178. doi: 10.1006/jsbi.2000.4244. [DOI] [PubMed] [Google Scholar]
- 92.Al-Amoudi A., Castano-Diez D., et al. Frangakis A.S. The three-dimensional molecular structure of the desmosomal plaque. Proc. Natl. Acad. Sci. U S A. 2011;108:6480–6485. doi: 10.1073/pnas.1019469108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Evans E., Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rico F., Russek A., et al. Scheuring S. Heterogeneous and rate-dependent streptavidin–biotin unbinding revealed by high-speed force spectroscopy and atomistic simulations. Proc. Natl. Acad. Sci. U S A. 2019;116:6594–6601. doi: 10.1073/pnas.1816909116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nicoludis J.M., Green A.G., et al. Gaudet R. Interaction specificity of clustered protocadherins inferred from sequence covariation and structural analysis. Proc. Natl. Acad. Sci. U S A. 2019;116:17825–17830. doi: 10.1073/pnas.1821063116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vogel V. Mechanotransduction involving multimodular proteins: converting force into biochemical signals. Annu. Rev. Biophys. Biomol. Struct. 2006;35:459–488. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- 97.Jaiganesh A., Narui Y., et al. Sotomayor M. Beyond cell-cell adhesion: sensational cadherins for hearing and balance. Cold Spring Harb. Perspect. Biol. 2017;10:a029280. doi: 10.1101/cshperspect.a029280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Katsamba P., Carroll K., et al. Honig B.H. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc. Natl. Acad. Sci. U S A. 2009;106:11594–11599. doi: 10.1073/pnas.0905349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen Y., Radford S.E., Brockwell D.J. Force-induced remodelling of proteins and their complexes. Curr. Opin. Struct. Biol. 2015;30:89–99. doi: 10.1016/j.sbi.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baumgartner W., Hinterdorfer P., et al. Drenckhahn D. Cadherin interaction probed by atomic force microscopy. Proc. Natl. Acad. Sci. U S A. 2000;97:4005–4010. doi: 10.1073/pnas.070052697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Panorchan P., Thompson M.S., et al. Wirtz D. Single-molecule analysis of cadherin-mediated cell-cell adhesion. J. Cell Sci. 2006;119:66–74. doi: 10.1242/jcs.02719. [DOI] [PubMed] [Google Scholar]
- 102.Schinner C., Erber B.M., et al. Waschke J. Stabilization of desmoglein-2 binding rescues arrhythmia in arrhythmogenic cardiomyopathy. JCI Insight. 2020;5:e130141. doi: 10.1172/jci.insight.130141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schinner C., Olivares-Florez S., et al. Waschke J. The inotropic agent digitoxin strengthens desmosomal adhesion in cardiac myocytes in an ERK1/2-dependent manner. Basic Res. Cardiol. 2020;115:46. doi: 10.1007/s00395-020-0805-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shafraz O., Rübsam M., et al. Sivasankar S. E-cadherin binds to desmoglein to facilitate desmosome assembly. Elife. 2018;7:e37629. doi: 10.7554/eLife.37629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Izrailev S., Stepaniants S., et al. Schulten K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys. J. 1997;72:1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dudko O.K., Hummer G., Szabo A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc. Natl. Acad. Sci. U S A. 2008;105:15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Leckband D. Force as a probe of membrane protein structure and function. Curr. Opin. Struct. Biol. 2001;11:433–439. doi: 10.1016/s0959-440x(00)00229-3. [DOI] [PubMed] [Google Scholar]
- 108.Ingber D.E. Cellular mechanotransduction: putting all the pieces together again. FASEB J. 2006;20:811–827. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- 109.Charras G., Yap A.S. Tensile forces and mechanotransduction at cell-cell junctions. Curr. Biol. 2018;28:R445–R457. doi: 10.1016/j.cub.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 110.Broussard J.A., Jaiganesh A., et al. Green K.J. Scaling up single-cell mechanics to multicellular tissues - the role of the intermediate filament-desmosome network. J. Cell Sci. 2020;133:jcs228031. doi: 10.1242/jcs.228031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arslan F.N., Eckert J., et al. Heisenberg C.-P. Holding it together: when cadherin meets cadherin. Biophys. J. 2021;120:4182–4192. doi: 10.1016/j.bpj.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schwartz M.A., DeSimone D.W. Cell adhesion receptors in mechanotransduction. Curr. Opin. Cell Biol. 2008;20:551–556. doi: 10.1016/j.ceb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leckband D.E., le Duc Q., et al. de Rooij J. Mechanotransduction at cadherin-mediated adhesions. Curr. Opin. Cell Biol. 2011;23:523–530. doi: 10.1016/j.ceb.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 114.Leckband D.E., de Rooij J. Cadherin adhesion and mechanotransduction. Annu. Rev. Cell Dev. Biol. 2014;30:291–315. doi: 10.1146/annurev-cellbio-100913-013212. [DOI] [PubMed] [Google Scholar]
- 115.Pruitt B.L., Dunn A.R., et al. Nelson W.J. Mechano-transduction: from molecules to tissues. PLoS Biol. 2014;12:e1001996. doi: 10.1371/journal.pbio.1001996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Katta S., Krieg M., Goodman M.B. Feeling force: physical and physiological principles enabling sensory mechanotransduction. Annu. Rev. Cell Dev. Biol. 2015;31:347–371. doi: 10.1146/annurev-cellbio-100913-013426. [DOI] [PubMed] [Google Scholar]
- 117.Lecuit T., Yap A.S. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat. Cell Biol. 2015;17:533–539. doi: 10.1038/ncb3136. [DOI] [PubMed] [Google Scholar]
- 118.Hoffman B.D., Yap A.S. Towards a dynamic understanding of cadherin-based mechanobiology. Trends Cell Biol. 2015;25:803–814. doi: 10.1016/j.tcb.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 119.Ladoux B., Nelson W.J., et al. Mège R.M. The mechanotransduction machinery at work at adherens junctions. Integr. Biol. (Camb). 2015;7:1109–1119. doi: 10.1039/c5ib00070j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tzima E., Irani-Tehrani M., et al. Schwartz M.A. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- 121.Borghi N., Sorokina M., et al. Dunn A.R. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc. Natl. Acad. Sci. U S A. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sim J.Y., Moeller J., et al. Pruitt B.L. Spatial distribution of cell–cell and cell–ECM adhesions regulates force balance while main-taining E-cadherin molecular tension in cell pairs. Mol. Biol. Cell. 2015;26:2456–2465. doi: 10.1091/mbc.E14-12-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pandey P., Hawkes W., et al. Iskratsch T. Cardiomyocytes sense matrix rigidity through a combination of muscle and non-muscle myosin contractions. Dev. Cell. 2018;44:326–336.e3. doi: 10.1016/j.devcel.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Peterson P., Kalda M., Vendelin M. Real-time determination of sarcomere length of a single cardiomyocyte during contraction. Am. J. Physiol. Physiol. 2013;304:C519–C531. doi: 10.1152/ajpcell.00032.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stefanopoulos P.K., Pinialidis D.E., et al. Filippakis K.N. Wound ballistics 101: the mechanisms of soft tissue wounding by bullets. Eur. J. Trauma Emerg. Surg. 2017;43:579–586. doi: 10.1007/s00068-015-0581-1. [DOI] [PubMed] [Google Scholar]
- 126.Sugiura R., Fujikawa T., et al. Nishimoto T. 2019 19th International Conference on Control, Automation and Systems (ICCAS) IEEE; 2019. Soft tissue bruise injury by blunt impact in human-robot interaction - difference of tolerance between chest and extremities; pp. 792–797. [Google Scholar]
- 127.Sanyour H., Childs J., et al. Hong Z. Spontaneous oscillation in cell adhesion and stiffness measured using atomic force microscopy. Sci. Rep. 2018;8:2899. doi: 10.1038/s41598-018-21253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kasas S., Ruggeri F.S., et al. Longo G. Detecting nanoscale vibrations as signature of life. Proc. Natl. Acad. Sci. U S A. 2015;112:378–381. doi: 10.1073/pnas.1415348112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nelson S.L., Proctor D.T., et al. Sutherland G.R. Vibrational profiling of brain tumors and cells. Theranostics. 2017;7:2417–2430. doi: 10.7150/thno.19172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Peyret G., Mueller R., et al. Ladoux B. Sustained oscillations of epithelial cell sheets. Biophys. J. 2019;117:464–478. doi: 10.1016/j.bpj.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pelling A.E., Sehati S., et al. Gimzewski J.K. Local nanomechanical motion of the cell wall of Saccharomyces cerevisiae. Science. 2004;305:1147–1150. doi: 10.1126/science.1097640. [DOI] [PubMed] [Google Scholar]
- 132.Kosztin I., Schulten K. Fluctuation-driven molecular transport through an asymmetric membrane channel. Phys. Rev. Lett. 2004;93:238102. doi: 10.1103/PhysRevLett.93.238102. [DOI] [PubMed] [Google Scholar]
- 133.Oda H., Takeichi M. Evolution: structural and functional diversity of cadherin at the adherens junction. J. Cell Biol. 2011;193:1137–1146. doi: 10.1083/jcb.201008173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gul I.S., Hulpiau P., et al. van Roy F. Evolution and diversity of cadherins and catenins. Exp. Cell Res. 2017;358:3–9. doi: 10.1016/j.yexcr.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 135.Green K.J., Roth-Carter Q., et al. Nichols S.A. Tracing the evolutionary origin of desmosomes. Curr. Biol. 2020;30:R535–R543. doi: 10.1016/j.cub.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jin X., a Walker M., et al. Shapiro L. Crystal structures of Drosophila N-cadherin ectodomain regions reveal a widely used class of Ca2+-free interdomain linkers. Proc. Natl. Acad. Sci. U S A. 2012;109:E127–E134. doi: 10.1073/pnas.1117538108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Araya-Secchi R., Neel B.L., Sotomayor M. An elastic element in the protocadherin-15 tip link of the inner ear. Nat. Commun. 2016;7:13458. doi: 10.1038/ncomms13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Syrris P., Ward D., et al. McKenna W.J. Desmoglein-2 mutations in arrhythmogenic right ventricular cardiomyopathy: a genotype-phenotype characterization of familial disease. Eur. Heart J. 2007;28:581–588. doi: 10.1093/eurheartj/ehl380. [DOI] [PubMed] [Google Scholar]
- 139.Dieding M., Debus J.D., et al. Anselmetti D. Arrhythmogenic cardiomyopathy related DSG2 mutations affect desmosomal cadherin binding kinetics. Sci. Rep. 2017;7:13791. doi: 10.1038/s41598-017-13737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Biswas S., Emond M.R., Jontes J.D. Protocadherin-19 and N-cadherin interact to control cell movements during anterior neurulation. J. Cell Biol. 2010;191:1029–1041. doi: 10.1083/jcb.201007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cooper S.R., Jontes J.D., Sotomayor M. Structural determinants of adhesion by Protocadherin-19 and implications for its role in epilepsy. Elife. 2016;5:e18529. doi: 10.7554/eLife.18529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tsai T.Y.-C., Sikora M., et al. Megason S.G. An adhesion code ensures robust pattern formation during tissue morphogenesis. Science. 2020;370:113–116. doi: 10.1126/science.aba6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hoshina N., Johnson-Venkatesh E.M., et al. Umemori H. Female-specific synaptic dysfunction and cognitive impairment in a mouse model of PCDH19 disorder. Science. 2021;372:eaaz3893. doi: 10.1126/science.aaz3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Carrion-Vazquez M., Oberhauser A.F., et al. Fernandez J.M. Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog. Biophys. Mol. Biol. 2000;74:63–91. doi: 10.1016/s0079-6107(00)00017-1. [DOI] [PubMed] [Google Scholar]
- 145.Li H., Cao Y. Protein mechanics: from single molecules to functional biomaterials. Acc. Chem. Res. 2010;43:1331–1341. doi: 10.1021/ar100057a. [DOI] [PubMed] [Google Scholar]
- 146.Marcozzi C., Burdett I.D., et al. Magee A.I. Coexpression of both types of desmosomal cadherin and plakoglobin confers strong intercellular adhesion. J. Cell Sci. 1998;111:495–509. doi: 10.1242/jcs.111.4.495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Stretching of the CDH1 trans dimer at 0.5 nm/ns (simulation S1d, Table S1, 0–33.4 ns) causes soft unbending of the inherent curvature of CDH1 monomers, followed by stiff phase, prior to unbinding of the trans interaction. Monomers begin to re-bend immediately after unbinding. Proteins are depicted in ribbon representation (greens), while water molecules and solute ions are not shown for clarity.
Focused view of the stretching of the trans CDH1 dimer at 0.5 nm/ns (simulation S1d, Table S1, 0–33.4 ns). Dislodging of Trp2 (orange) from the hydrophobic pocket of the binding partner is observed. One monomer is shown in surface representation, while the other is shown in ribbon.
Stretching of the DSG2-DSG2 trans dimer at 0.1 nm/ns (simulation S3d, Table S1) results in unbending of the inherent curvature of the DSG2 monomers, followed by a stiff phase, prior to unbinding of the trans interaction. Monomers begin to re-bend immediately after unbinding. System depicted as in Video S1. Similar trajectories were observed for the DSG2-DSC1 heterodimer and the DSC1-DSC1 homodimer.
Focused view of the stretching of the trans DSG2 homodimer at 0.1 nm/ns (simulation S3d, Table S1). Dislodging of the Trp2 (orange) from the hydrophobic pocket of the binding partner is observed. One monomer is shown in surface representation, while the other is shown in ribbon. Similar trajectories were observed for the DSG2-DSC1 heterodimer and the DSC1-DSC1 homodimer.
Stretching of the PCDHβ6 trans homodimer at 0.1 nm/ns (simulation S7d, Table S1, 0–180 ns) results in rupture of the EC1-EC4 interface and the formation of transient intermediates before complete unbinding of the complex. System depicted as in Video S1.
Close up view of the transient interaction that forms between Arg4 (A) and Glu165 (B) during stretching of the PCDHβ6 trans homodimer at 0.1 nm/ns (simulation S7d, Table S1, 0–180 ns). Chain B is shown in bright pink while chain A is shown in dark red color.
Location of glycosylation sites in the PCDHγB3 trans homodimer during forced stretching at 0.1 nm/ns (simulation S8d, Table S1, 0–185.4 ns). Protein is shown in gray ribbon representation, residues that form interactions with each other are shown as magenta spheres, while glycosylation sites are shown as cyan spheres. Glycosylation is not expected to interfere with unbinding pathway.