Abstract
Cellular mechanosensing is pivotal for virtually all biological processes, and many molecular mechano-sensors and their way of function are being uncovered. In this work, we suggest that c-Src kinase acts as a direct mechano-sensor. c-Src is responsible for, among others, cell proliferation, and shows increased activity in stretched cells. In its native state, c-Src has little basal activity, because its kinase domain binds to an SH2 and SH3 domain. However, it is known that c-Src can bind to p130Cas, through which force can be transmitted to the membrane. Using molecular dynamics simulations, we show that force acting between the membrane-bound N-terminus of the SH3 domain and p130Cas induces partial SH3 unfolding, thereby impeding rebinding of the kinase domain onto SH2/SH3 and effectively enhancing kinase activity. Forces involved in this process are slightly lower or similar to the forces required to pull out c-Src from the membrane through the myristoyl linker, and key interactions involved in this anchoring are salt bridges between negative lipids and nearby basic residues in c-Src. Thus, c-Src appears to be a candidate for an intriguing mechanosensing mechanism of impaired kinase inhibition, which can be potentially tuned by membrane composition and other environmental factors.
Significance
The nonreceptor tyrosine kinase c-Src is involved in a vast diversity of biological processes such as cell adhesion and migration, cell cycle progression, and apoptosis. Because it is also implicated in mechanosensing pathways, we hypothesized that c-Src, while tethered to the cellular membrane and the adaptor protein p130Cas, can be directly activated by force. Our extensive force-probe molecular dynamics simulation reveal a novel tension-dependent regulatory mechanism at atomistic detail in which force impedes c-Src kinase deactivation. Our results suggest a new mode of c-Src regulation that integrates mechanical signals into the plethora of pathways controlled by this pivotal kinase.
Introduction
The tyrosine kinase c-Src controls a plethora of cellular processes, from cell growth to differentiation. It is a central regulatory hub that couples signals from cell surface receptors to the intracellular signaling machinery (1). c-Src can switch between active and inactive states, with multiple biochemical factors controlling the switching (2). Uncontrolled kinase activity can lead to excessive cell proliferation and cancerogenesis (3), rendering c-Src kinase an important drug target. In addition to biochemical stimuli, also mechanical force can activate c-Src (4). However, force is currently considered to cause c-Src activation only indirectly by acting on interaction partners or substrates of c-Src, such as actin-filament-associated proteins (5), integrins (6), or beta-catenin (7). Whether and how mechanical force affects c-Src dynamics and function is currently unknown.
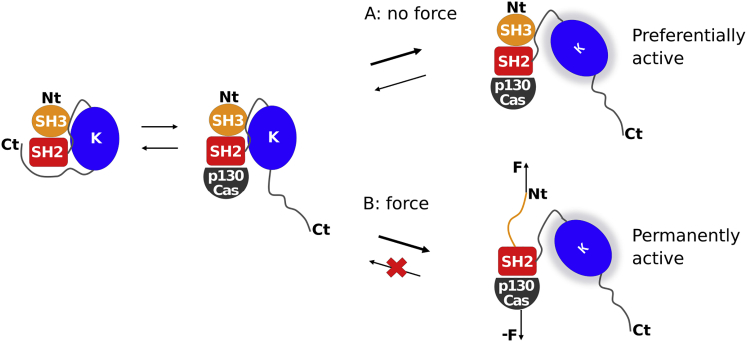
We here hypothesize, and test by molecular dynamics (MD) simulations, whether mechanical force acting on c-Src itself can enhance its kinase activity, as a new Src-mediated mode of force-sensing. Indeed, force has been identified as an important trigger of enzyme activation, with molecules such as titin and twitchin kinases in muscle (8,9) and focal adhesion kinase (FAK) at cell-matrix contact sites (10,11). A common theme of these force-sensing kinases is that the autoinhibitory domain of the kinase is pulled off from the kinase domain, leading to activation. In the case of c-Src, a different mechano-activation mechanism must be at play. c-Src consists of an N-terminal myristoylation site, followed by a disordered linker, the autoinhibitory SH3 and SH2 domains, a linker connecting SH2 to the kinase N-terminus, and the kinase domain (Fig. 1 A). A short C-terminal tail, if phosphorylated at Y527, folds back onto the SH2 domain, thereby clamping the SH3/SH2 domains to the kinase domain and autoinhibiting it by stabilizing the interface between the kinase and the SH2/SH3 domains. A peptide from the force-sensing p130Cas protein (12), on tyrosine phosphorylation, can displace the C-terminal tail of c-Src by binding though the phosphorylated tyrosine into the same pocket of SH2.
Figure 1.
The balance of Src kinase and p130Cas binding and the impact of external force on it. When p130Cas binds to Src kinase, it is preferentially active, with the Src C-terminal tail domain and p130Cas competitively binding to the SH2 domain. According to our hypothesis, however, external force is applied to the system at the N-terminus through the cellular membrane and at the C-terminus through the cytoskeleton. Rebinding of the kinase to the SH2-SH3 systems is prevented, making the protein permanently active. The N- and C-termini of the constructs are denoted as Nt and Ct, respectively. To see this figure in color, go online.
The amino-terminal domain of p130Cas is attached to the actin cytoskeleton through FAK and talin, and cytoskeletal traction forces of an adherent cell subject p130Cas to stretching forces (12,13). Accordingly, force can propagate onto the Src SH2 domain from bound and tensed p130Cas on one side (12), and onto the SH3 domain through the N-terminal membrane-embedded myristoyl linker on the other side (Fig. 1). In the p130Cas-bound state, the autoinhibitory SH2/SH3 domains of c-Src are already detached from the kinase domain, i.e., the kinase domain is in an active state. To our knowledge, the kinase domain only transiently interacts with substrates such as p130Cas, but otherwise is not linked to force-bearing structures. Thus, c-Src kinase is subjected to pulling forces acting between the membrane it is anchored to and p130Cas. Our hypothesis is that force leads to SH2 and/or SH3 unfolding. Partial or full domain unfolding in turn renders the rebinding of the kinase domain unlikely, thereby shifting the equilibrium of c-Src further to the activated state. To test this hypothesis, we here examined the conformational changes in the SH2/SH3 domains under force, in presence and absence of the kinase domain, using MD simulations. In particular, we assessed two important questions: 1) Is the p130Cas-Src interaction strong enough and is the Src kinase sufficiently strongly anchored into the membrane to transmit large enough forces and thereby activate Src kinase? 2) If so, what are the molecular details of this activation?
Although the impact of a mechanical stimulus on c-Src dynamics and function has not been examined by simulations so far, a growing body of MD studies has shed light on the molecular mechanisms underlying c-Src function and regulation under different biochemical stimuli. MD simulations have indeed resolved the conformational transitions between active and inactive states and the effects of activation loop phosphorylation and autoinhibition by SH2/SH3 on these dynamics (14, 15, 16, 17, 18, 19). An additional and critical role of protonation state changes was recently revealed by MD simulations (20). In this study, to probe c-Src dynamics under pulling forces, we subjected an SH3/SH2 construct with a fragment of p130Cas bound to the SH2 domain to force-probe MD simulations. Our simulation setup thus aimed to mimic the effect of force sensed by p130Cas bound to the SH2 domain on the one side, and by the membrane-attached SH3 domain of c-Src on the other. This allowed the probing of the sequence of unfolding of the pulled SH3/SH2 inhibitory domain in the unclamped and active state, with the kinase domain detached. We observed a loss of the SH3-SH2 interface followed by SH3 unfolding, and obtained a highly similar unfolding process, albeit at higher forces, when the kinase domain is bound. Our results suggest that mechanical force propagates onto c-Src from the membrane and p130Cas to impede the rebinding of the kinase domain by detachment and unfolding of the autoinhibitory SH3 domain, thereby effectively enhancing c-Src activity. Our study puts forward c-Src as a potential intrinsic mechano-sensor, providing a new perspective on how cells sense forces at cell-matrix or cell-cell junctions.
Methods
Modeling and equilibrium simulations of the p130Cas-c-Src complex
The complex between c-Src and p130Cas was obtained from combining the coordinates from the structure of the Lck kinase SH2 domain in complex to the SH2-binding peptide of human p130Cas (positions 759 to 767, PDB accession code: 1x27 (21)) with the crystal structure of human c-Src (PDB accession code: 2src (22)).
The SH2 domain of the Lck kinase was first superimposed to the SH2 domain of c-Src (RMSD = 0.68 Å) and then deleted, leaving the p130Cas peptide bound to c-Src. The C-terminal autoinhibitory intrinsically disordered tail of c-Src, which competes with the binding site of the p130Cas peptide, was then deleted.
The p130Cas peptide (Nter-WMEDpYDpYVHLQG-Cter), which contains two phosphotyrosine (pY) residues, can dock onto the SH2 domain of c-SRC in two possible orientations, with its N-terminal oriented either toward the N-terminal or the C-terminal end of c-SRC. To assess the preferred orientation of the p130Cas peptide, we modeled both binding poses and performed equilibrium MD simulations for both complexes.
MD simulations were performed using Gromacs version 5.0.6 (23) (equilibrium simulations) and 2016 (force-probe simulations). After adding missing hydrogens and inserting the complexes in a cubic box with sides of approximately 9 nm containing approximately 73,000 atoms; the protein-containing box was solvated with TIP3P water molecules and Na+ and Cl− ions were added to achieve electroneutrality and reach a total ionic strength of 0.165 M. The Amber99sb∗-ILDN force field (24) was adopted to describe the interactions between the particles and the parameters for the Na and Cl ions were obtained from Joung and Cheatham (25), which reported a more accurate description of ionic behavior. The force field parameters for the noncanonical amino-acid phosphotyrosine and for adenosine triphosphate (ATP) bound to c-Src were obtained from the AMBER parameters database curated by the Bryce Group at Manchester University (26).
The solvated and neutralized complex was first energy minimized using a steep descent algorithm, with an energy minimization step of 0.1 nm and a force tolerance of 1 kJ mol−1, to resolve any particle-particle clash. Solvent was subsequently equilibrated in two steps. In a first step, which was run considering an NVT ensemble, the complex was positionally restrained using a force constant of 1000 kJ mol−1 nm−1 in x, y, and z dimensions while the solvent was allowed to relax for a total time of 100 ns using a timestep of 0.002 ps. In this step, particles were assigned random velocities following a Maxwell-Boltzmann distribution at the temperature of the simulation, which was 298.15 K. The temperature was kept constant using Bussi’s V-rescale thermostat (27) with coupling constant of 0.1 ps, with solvent and protein particles coupled separately. Newton’s equation of motion was integrated using a velocity Verlet algorithm, and setting a cutoff of 1 nm to compute Lennard-Jones and Coulomb interactions between nonbonded particles. Coulomb interactions beyond the cutoff were computed in the Fourier space using the Particle Mesh Ewald (PME) summation method and using a grid spacing of 0.16 nm and a four-order polynomial for spline interpolation. Bonds were treated using harmonic potential energy functions, with parameters from the adopted AMBER force fields and covalent bonds involving hydrogens constrained at every step using holonomic constraints. After this NVT step, a 1-ns-long NPT step was performed using a Parrinello-Rahman barostat (28) to isotropically couple pressure, with the pressure for protein and solvent particles separately coupled every 2.0 ps to a reference pressure of 1 bar. Subsequent to the NVT and NPT, the positional restraint of nonsolvent particles was lifted, and the system was equilibrated for a total of 300 ns. The dynamics of the p130Cas peptide were monitored in the two modeled conformations, to understand which of the orientations was the most stable when bound to c-Src. Further simulations were performed with the more stable construct, in which the direction of the p130Cas peptide was aligned with the c-Src C-terminal peptide in the SH2 binding grove. We visualized three-dimensional structures using VMD (29) and contact maps using CONAN (30).
For the force-probe simulations, we subjected the construct to an external force by pulling apart the N-terminal of the SH3 domain and the two phosphorylated tyrosines of p130Cas with equal and opposite velocities. The system with the kinase domain present included approximately 196,000 atoms in a box of approximately 20 × 10 × 10 nm, whereas without the kinase domain present included approximately 127,000 atoms in a box of approximately 20 × 8 × 8 nm. Five velocities were chosen: 0.01, 0.05, 0.1, 0.5, and 1 nm/ns, and the force was applied through an umbrella potential with a force constant of 500 kJ/mol/nm2. One of the 15 unfolding trajectories, performed at 0.5 m/s, showed unbinding of the p130Cas-Src interaction. We consider this a very unlikely scenario caused most likely by the bias introduced by high-speed force-probe MD simulations in which external forces close to the application point are artificially higher because of high loading rates and friction (31). We excluded this trajectory from further analysis, but note that p130Cas detachment is an alternative pathway under force, terminating the force-propagation onto c-Src. Determining the exact likelihood of this minority pathway is, however, outside the scope of this work.
Random conformations of a Src N-terminal peptide (2-GSNKSKPKDASQRRRSLE-19) were generated with Modeller (version 9.20 (32)). These were N-terminally myristoylated, C-terminally capped with N-methylamid (NME) and the myristoyl moiety embedded in a POPC membrane with 10% PIP2 in the upper (intracellular) leaflet as used previously (33, 34, 35) using Charmm-Gui (36). The membrane-peptide complex was inserted in a triclinic box with approximate dimensions of 6.5 × 6.5 × 10 nm, solvated with TIP3P water and NaCl added to reach neutrality and a concentration of 0.15 M, leading to a system with approximately 44,000 atoms in total. A set of simulations with only a myristoylated and NME-capped glycine residue in a pure POPC bilayer was simulated with the same conditions as listed below, albeit with a smaller system size of 5 × 5 × 10.5 nm corresponding to approximately 26,000 atoms.
Gromacs version 2018.5 was used for the simulations involving a membrane along with the CHARMM36 force field (March 2019 version (37)). The system was energy minimized using the steepest descent algorithm with a step size of 0.1 nm and a force tolerance of 1000 kJ/mol. Equations of motion were integrated using the leap-frog integrator with a time step of 0.002 ps. Van-der-Waals interactions were smoothly switched to zero between 1.0 and 1.2 nm using the force-switch method. Long-range coulomb interactions beyond 1.2 nm were treated with the PME method. All bonds involving hydrogen atoms were constrained using the LINCS algorithm. During two equilibration steps in the NVT and NPT ensemble, peptide backbone heavy atoms were position restrained with a force constant of 1000 kJ/mol/nm and side chain heavy atoms with 500 kJ/mol/nm. Lipids were restrained to the membrane plane with a force constant of 1000 kJ/mol/nm. The temperature was adjusted to 300 K during a 100 ps simulation in the NVT ensemble using the v-rescale thermostat with a coupling constant of 0.5 ps. This was followed by 1.5 ns in the NPT ensemble in which the pressure was equilibrated to 1 bar using semi-isotropic pressure coupling with the Berendsen barostat with a coupling constant of 5 ps. After equilibration, the barostat was switched to the Parinello-Rahman barostat.
Four different peptide-starting conformations were sampled for 500 ns. Force-probe simulations were started from frames at 300, 400, and 500 ns after extending the box to a length of 19 nm in the z-direction (increasing the number of atoms to approximately 75,000). We applied an umbrella potential with a force constant of 500 kJ/mol/nm2 to pull the peptide NME cap heavy atoms with reference to the lipids of the membrane upper leaflet within a cylinder of 2-nm radius at constant velocities of 1, 0.5, 0.1, 0.05, and 0.01 nm/ns. We visualized the frames using PyMOL (Schrödinger L & DeLano W, 2020, PyMOL; http://www.pymol.org/pymol).
Results
Both p130Cas as well as membrane-anchoring have been previously established to be part of the mechanotransduction pathways at focal adhesion sites. Thus, they are prime candidates for c-Src tethering at focal adhesions, and have been the points of force application in this study. p130Cas binding to the SH2 domain results in the displacement of the Src C-terminal tail from the same binding site and in Src kinase activation, with the SH3-SH2 domains at most only loosely interacting with the kinase domain (Fig. 1 B). In this state, force propagates onto the SH2-SH3 domains from the bound p130Cas and the membrane-embedded myristoylated N-terminus of c-Src, while the kinase is not part of the force-propagation pathway. In the first set of MD simulations, we therefore focused on the force response of p130Cas-bound SH3/SH2 domains in absence of the kinase domain.
Currently, a structure comprising the SH2 domain and a p130Cas peptide is not available, and we modeled a p130Cas-c-Src complex based on the autoinhibited structure of c-Src (see methods). In principle, the p130Cas peptide with its phosphorylated tyrosines can be oriented similarly to the C-terminal peptide of c-Src when bound to SH2 (N-terminal end of the peptide oriented toward the N-terminal end of c-Src), or in the opposite direction. We modeled both orientations and assessed the peptide’s binding stability in a particular orientation by monitoring its dynamics in the bound state. In equilibrium MD simulations, the root mean-square fluctuation profiles of p130Cas showed a consistently lower root mean-square fluctuation and a higher number of contacts when the C-terminal end of the peptide oriented along the C-terminal end of c-Src (Fig. S1). This pose also features significantly more overall contacts between p130Cas and c-Src (Fig. S2), suggesting that this binding mode would be preferred within the p130Cas-c-Src complex (Fig. S3). The structural homology with the c-Src peptide, the higher structural stability, and the stronger intermolecular interaction together strongly suggest this binding mode to be the more stable and preferred conformation of the p130Cas-c-Src complex, and we chose this conformation for all force-probe simulations where the p130Cas fragment is considered.
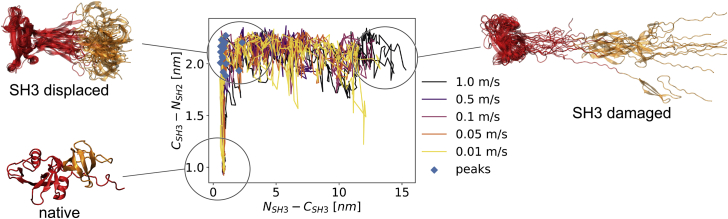
We examined the response of the p130Cas-bound SH2-SH3 domain of Src kinase to a pulling force by force-probe MD simulations (38,39). We extended the protein under mechanical force by a total of 15 nm. When examining the unfolding trajectories, we find the SH2-SH3 domains to extend in two steps, an initial detachment from one another followed by SH3 partial unfolding (Fig. 2). The initial stretching of the protein is characterized by a displacement of the SH3 domain by about 1.2 nm as well as small extensions within both domains. The point of major rupture (maximal pulling force, blue dots in Fig. 2) shows an intact, but displaced SH3 domain, with the interdomain linker connecting SH2 and SH3 stretched. At this point, most trajectories exhibit a force peak followed by the unfolding of the SH3 domain. The final poses at 15 nm show that the SH3 domain is largely damaged, with only three of its six beta strands still folded. The two processes, initial stretching and SH3 unfolding, occur consecutively in all trajectories (Fig 2).
Figure 2.
Autoinhibitory SH3-SH2 domains of c-Src first detach and then unfold from the SH3 domain. Shown is the evolution of the end-to-end distance of the SH3-SH2 linker (y axis, measuring detachment) and the end-to-end distance of the SH3 domain (x axis, measuring SH3 unfolding) across all 15 pulling trajectories (color coded by velocity). The two processes, detachment from one another and SH3 unfolding, occur consecutively, and the transition between them involves the force peak in most trajectories (blue dots, snapshot “SH3 displaced”). This holds true both in the trajectories with the kinase domain removed (shown here) and when it is present (see Fig. S4B for comparison). To see this figure in color, go online.
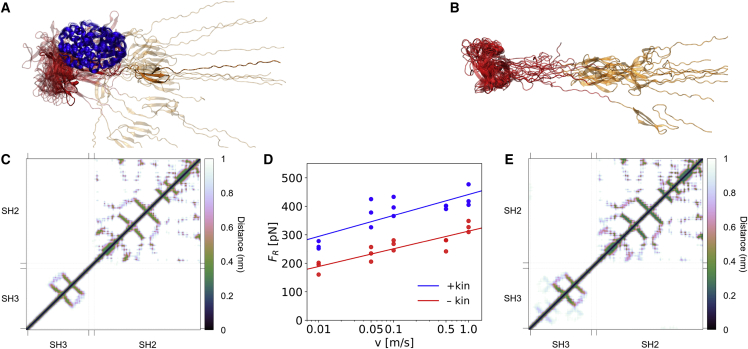
In the force-probe MD simulations described above, we considered the kinase domain to be fully detached from the autoinhibitory domains. However, the kinase domain is still present and can, in principle, albeit with low probability, rebind to the SH2/SH3 domains, as p130Cas and the Src kinase’s own C-terminal tail compete with each other for the same SH2 binding site (Fig. S3). To examine the force response of Src in the hypothetical p130Cas-bound yet inhibited state, we repeated these simulations with the entire kinase domain present. We again find that the unfolding happens through the same mechanism: first, the SH3 domain loses contact to the SH2 domain, and then, the SH3 domain starts unfolding (Figs. 3 and S4). The kinase domain, being not directly part of the force transduction pathway, remains stable (Fig. S5). The observed force profiles for both systems, with and without kinase and across all loading rates, are largely similar, with a large peak at the extension of 7 nm followed by a forceless stretching (Fig. S6). Thus, the presence of the kinase domain does not alter the force-induced unfolding mechanism of SH3-SH2. The dependence of the first peak size (rupture force) on the logarithm of pulling speed is close to linear, fitting the Bell model (Fig. 3 D), and shows highly similar slopes for the two systems. This conforms to the observed similarity between all unfolding trajectories (Fig. S4). However, because the kinase domain indirectly stabilizes the SH2-SH3 domains, we find an increase of the required rupture force by about 120 pN across all pulling velocities when the kinase domain is present.
Figure 3.
The unfolding pathways of SH2-SH3 domains of Src kinase are unaffected by the presence of the kinase domain. The final frames (top) and contact maps (bottom) of the forced unfoldings of Src kinase (A and C) and Src kinase without the kinase domain (B and E) are very similar: the SH2 domain remains fully folded and only three β-strands of the SH3 domain are present. The unfolding forces with the kinase domain present are, however, consistently higher than if it is removed (D), at each pulling velocity. The average force difference between the two constructs is 120 pN across all velocities. The final frames are defined at an extension of 15 nm. The contact maps between (C) and (E) are based on heavy atom-heavy atom distances between the SH3 and SH2 domains. To see this figure in color, go online.
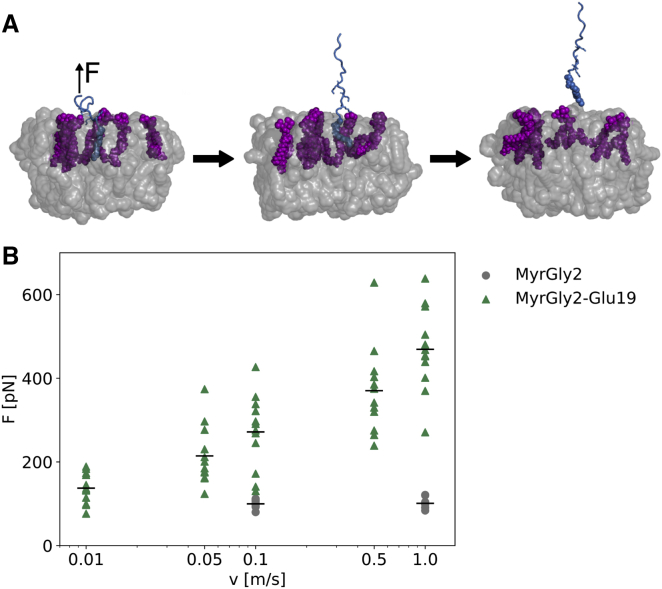
Our data indicate that force-induced unfolding of the SH2-SH3 autoinhibitory domains is a plausible mechanism that can impede their rebinding to the kinase domain. A prerequisite of this scenario is that the protein remains membrane bound to maintain force propagation. To test this, we asked whether the Src membrane anchorage is strong enough to withstand the forces required for SH2-SH3 domain rupture. We myristoylated a peptide consisting of the first 19 amino acids of the Src N-terminal peptide (Myr-G2SNKSKPKDASQRRRSLE19), embedded the myristoyl moiety in a lipid bilayer made out of POPC lipids with 10% PIP2 lipids, and subjected it to force-probe MD simulations.
While the peptide is stretched during the first part of the simulations, all positively charged residues except K5, corresponding to the positively charged residue closest to Myr, unbind from the PIP2 lipids. K5 and Myr simultaneously detach from the membrane (Fig. 4). Overall, the membrane detachment force for the myristoylated peptide falls into the same range of forces or is slightly higher than those required for SH3-SH3 stretching and unfolding (compare Fig. 3 D). The higher stability is more evident for higher pulling velocities. To exclude the possibility that these observations are an artifact of using different force fields for protein and membrane simulations, we repeated the force-probe MD simulations of Src using the Charmm36 force field, which yielded similar rupture forces as obtained with the Amber99sb∗-ILDN force field (Figs. S7 and 3 D). The results imply that both membrane detachment and Src rupture can in principle happen under force. Interestingly, force-probe MD simulations of a myristoylated and capped glycine residue in a pure POPC membrane result in threefold lower membrane detachment forces compared with the longer Src peptide. This strong reduction in the resistance against membrane detachment demonstrates the importance of electrostatic interactions between positively charged residues in the Src linker and negatively charged PIP2 lipids, as previously reported (40).
Figure 4.
Membrane detachment forces are similar to Src rupture forces and depend on electrostatic interactions. During the force-probe simulations (frames shown in A), the peptide (blue) is stretched before the PIP2 (purple)-interacting lysine residues (stick representation) and myristoyl moiety (VDW representation) detach from the membrane (gray: POPC lipids). Comparing the membrane detachment forces (B) of the myristoylated N-terminal Gly2-Glu19 peptide (green triangles) including basic residues interacting with negatively charged PIP2 lipids to a myristoylated glycine residue (gray circles) reveals approximately threefold higher forces. Black bars denote the means. To see this figure in color, go online.
In the p130Cas bound state, the kinase domain is already mostly dissociated from the inhibitory domain. In the hypothetical case of the kinase domain remaining bound, we observe that the extent of unfolding in the autoinhibitory domains, starting from the SH3 domain, drastically modifies and decreases the native contacts of these domains with the kinase domain. We performed additional analyses and simulations to address the question of how the interaction between the kinase and the SH3/SH2 domains is affected by the p130Cas-enabled mechanical unfolding pathway. We do not detect a significant weakening of the SH3/SH2-kinase interface, as measured by the rupture forces, when pulling the SH3/SH2 domains off the kinase domain in additional force-probe MD simulations in a vertical, nonphysiological direction (Fig. S8). This is likely because the forces for mechanically induced dissociation do not capture the extent of autoinhibition of the kinase domain by the partially unfolded SH3/SH2 domains. Indeed, the observed rupture forces do not correlate with the contact area between the kinase domain and the rest of the protein (Fig. S9).
Discussion
Our study proposes a possible scenario for how c-Src is activated by force: deactivation by kinase rebinding is impeded by partial unfolding of the autoinhibitory domains at medium forces (Fig. 1). These forces are in fact quite similar to the ones found for FAK unfolding and activation (11), about 250 pN (without the kinase domain) at 0.1 m/s pulling speed. Given the fact that the two pTyr of p130Cas and the Src-C-terminal tail have very similar binding affinity and thus permanently compete with one another for the SH2 binding site, the SH3 domain unfolding will significantly shift the equilibrium toward the activated state (Fig. 1).
Our simulations also show that the SH3 domain is weaker than the SH2 and unfolds first. The weaker mechanical stability of SH3 compared with SH2 is in line with the previous experimental finding from H/D exchange experiments that SH3 is more dynamic and unfolds more easily than SH2 (41). As a consequence, p130Cas stays bound to the more stable and intact SH2 domain and force continues acting on the inhibitory domain, while the SH3 domain is displaced and eventually unfolded. In fact, SH3 is required for autoinhibition, as shown by experiments of an SH3-deletion mutant, which shows activity even if the C-terminal Tyr527 is phosphorylated (42).
p130Cas is stretched under cytoskeletal traction forces (12), indicating that it does not detach from the cytoskeleton under force. Our findings extend this observation to the linkage of the central domain of p130Cas to Src kinase: force does not compromise anchoring of Src kinase to FAs through p130Cas, and at the same time reduces Src kinase inhibition through unfolding of its inhibitory SH3 domain. More specifically, our simulations predict force to continue to act even when the protein is already opened and stretched, i.e., for at least 15 nm of stretching.
c-Src conformations with partially unfolded autoinhibitory domains and a bound kinase domain showed no measurable decrease in the stability of the SH3/SH2-kinase interface in additional force-probe MD simulations. However, the question of kinase binding to the pulled SH3/SH2 domains is of thermodynamic nature, and rupture forces from nonequilibrium simulations are a less adequate measure. In addition, the kinase will eventually bind in a very different mode when the SH3 domain is effectively absent due to forced unfolding. The reduced rebinding in this case is corroborated by the finding that an SH3-deletion mutant shows high kinase activity. Quantitatively assessing the shift in the equilibrium between activated/dissociated and deactivated/bound states of c-Src as a function of force would require simulations or docking protocols for the rebinding of the kinase domain to partially unfolded SH3/SH2 conformations, which are the subject of future investigations.
Because the high pulling velocities applied in MD simulations are known to cause higher than physiological rupture forces, we compare our results with previously published AFM studies of similar proteins. Pulling the myristoylated linker of recoverin out of a DPPC membrane required approximately 48 pN (43). We expect the necessary force for Src to be higher due to the electrostatic contributions of the linker residues adjacent to the myristoylation site. In the case of FAK, the autoinhibitory interface was released at around 25 pN followed by domains unfolding at forces below 50 pN (11), suggesting that the Src rupture force could lie within this range. We therefore expect the Src rupture force to be similar or below the membrane detachment force at experimental pulling speeds. Indeed, in the lowest velocities covered in our force-probe MD simulations, we find SH3 unfolding and myristoyl-membrane dissociation to require a similar mean force. This indicates that both scenarios can happen under force. Our results also underline the importance of electrostatic interactions for stable Src membrane binding. We thus hypothesize that the delicate balance between myristoyl unbinding and SH3 unfolding can be regulated by local lipid composition, protein or lipid phosphorylation, or salt concentration to favor either membrane detachment or force-enhanced Src activation.
We conclude that mechanical force activates Src kinase by unfolding the SH3 domain and thereby impeding rebinding of the autoinhibitory SH2-SH3 domains. Single-molecule stretching experiments previously proved highly valuable to dissect the mechanism of other mechanokinases, such as FAK and titin kinase (9,11). Similar experiments on the c-Src autoinhibitory domains in absence and presence of the kinase domain can test our predictions. Further exploration of the precise pathways affected by Src mechanosensing will be required to put this finding into cellular context.
Author contributions
C.D. designed and performed the research, analyzed the data, and wrote the manuscript. S.d.B. and D.M. performed research and analyzed data, S.d.B. wrote the manuscript. F.G. designed the research and wrote the manuscript.
Acknowledgments
We acknowledge funding through the Deutsche Forschungsgemeinschaft under Germany's Excellence Strategy – 2082/1–390761711, the Klaus Tschira Foundation, the state of Baden-Württemberg through bwHPC, as well as the DFG through grant INST 35/1134-1 FUGG. S.d.B. thanks the Carl Zeiss Foundation for financial support.
Editor: Alexandr Kornev.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.01.028.
Supporting material
References
- 1.Parsons S.J., Parsons J.T. Src family kinases, key regulators of signal transduction. Oncogene. 2004;23:7906–7909. doi: 10.1038/sj.onc.1208160. [DOI] [PubMed] [Google Scholar]
- 2.Chong Y.-P., Ia K.K., et al. Cheng H.-C. Endogenous and synthetic inhibitors of the Src-family protein tyrosine kinases. Biochim. Biophys. Acta. 2005;1754:210–220. doi: 10.1016/j.bbapap.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 3.Blume-Jensen P., Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Botvinick E.L., et al. Chien S. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 5.Han B., Bai X.-H., et al. Liu M. Conversion of mechanical force into biochemical signaling. J. Biol. Chem. 2004;279:54793–54801. doi: 10.1074/jbc.M406880200. [DOI] [PubMed] [Google Scholar]
- 6.Arias-Salgado E.G., Lizano S., et al. Shattil S.J. Src kinase activation by direct interaction with the integrin cytoplasmic domain. Proc. Natl. Acad. Sci. U S A. 2003;100:13298–13302. doi: 10.1073/pnas.2336149100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Röper J.-C., Mitrossilis D., et al. Farge E. The major β-catenin/E-cadherin junctional binding site is a primary molecular mechano-transductor of differentiation in vivo. eLife. 2018;7:e33381. doi: 10.7554/eLife.33381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gräter F., Shen J., et al. Grubmüller H. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys. J. 2005;88:790–804. doi: 10.1529/biophysj.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puchner E.M., Alexandrovich A., et al. Gautel M. Mechanoenzymatics of titin kinase. Proc. Natl. Acad. Sci. U S A. 2008;105:13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J., Aponte-Santamaría C., et al. Gräter F. Mechanism of focal adhesion kinase mechanosensing. PLoS Comput. Biol. 2015;11:e1004593. doi: 10.1371/journal.pcbi.1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer M.S., Baumann F., et al. Lietha D. Structural and mechanistic insights into mechanoactivation of focal adhesion kinase. Proc. Natl. Acad. Sci. U S A. 2019;116:6766–6774. doi: 10.1073/pnas.1820567116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawada Y., Tamada M., et al. Sheetz M.P. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Defilippi P., Stefano P.D., Cabodi S. p130Cas: a versatile scaffold in signaling networks. Trends Cell Biol. 2006;16:257–263. doi: 10.1016/j.tcb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Meng Y., Pond M.P., Roux B. Tyrosine kinase activation and conformational flexibility: lessons from Src-family tyrosine kinases. Acc. Chem. Res. 2017;50:1193–1201. doi: 10.1021/acs.accounts.7b00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fajer M., Meng Y., Roux B. The activation of c-Src tyrosine kinase: conformational transition pathway and free energy landscape. J. Phys. Chem. B. 2017;121:3352–3363. doi: 10.1021/acs.jpcb.6b08409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng Y., Shukla D., et al. Roux B. Transition path theory analysis of c-Src kinase activation. Proc. Natl. Acad. Sci. U S A. 2016;113:9193–9198. doi: 10.1073/pnas.1602790113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla D., Meng Y., et al. Pande V.S. Activation pathway of Src kinase reveals intermediate states as targets for drug design. Nat. Commun. 2014;5:3397. doi: 10.1038/ncomms4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng Y., Roux B. Locking the active conformation of c-Src kinase through the phosphorylation of the activation loop. J. Mol. Biol. 2014;426:423–435. doi: 10.1016/j.jmb.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon H.J., Lee S., et al. Wu S. Network approach of the conformational change of c-Src, a tyrosine kinase, by molecular dynamics simulation. Sci. Rep. 2018;8:5673. doi: 10.1038/s41598-018-23964-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai C.-C., Yue Z., Shen J. How electrostatic coupling enables conformational plasticity in a tyrosine kinase. J. Am. Chem. Soc. 2019;141:15092–15101. doi: 10.1021/jacs.9b06064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nasertorabi F., Tars K., et al. Ely K.R. Molecular basis for regulation of Src by the docking protein p130Cas. J. Mol. Recognit. 2006;19:30–38. doi: 10.1002/jmr.755. [DOI] [PubMed] [Google Scholar]
- 22.Xu W., Doshi A., et al. Harrison S.C. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol. Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 23.Van Der Spoel D., Lindahl E., et al. Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 24.Lindorff-Larsen K., Piana S., et al. Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joung I.S., Cheatham T.E. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.AMBER parameter database (Bryce Group: computational biophysics and drug design - University of Manchester). http://research.bmh.manchester.ac.uk/bryce/amber.
- 27.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 28.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 29.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 30.Mercadante D., Gräter F., Daday C. CONAN: a tool to decode dynamical information from molecular interaction maps. Biophys. J. 2018;114:1267–1273. doi: 10.1016/j.bpj.2018.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheridan S., Gräter F., Daday C. How fast is too fast in force-probe molecular dynamics simulations? J. Phys. Chem. B. 2019;123:3658–3664. doi: 10.1021/acs.jpcb.9b01251. [DOI] [PubMed] [Google Scholar]
- 32.Šali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 33.Cai X., Lietha D., et al. Schaller M.D. Spatial and temporal regulation of focal adhesion kinase activity in living cells. Mol. Cell. Biol. 2008 doi: 10.1128/MCB.01324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng J., Mertz B. Novel phosphotidylinositol 4,5-bisphosphate binding sites on focal adhesion kinase. PLoS One. 2015;10:e0132833. doi: 10.1371/journal.pone.0132833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Senju Y., Kalimeri M., et al. Lappalainen P. Mechanistic principles underlying regulation of the actin cytoskeleton by phosphoinositides. Proc. Natl. Acad. Sci. U S A. 2017;114:E8977–E8986. doi: 10.1073/pnas.1705032114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jo S., Kim T., et al. Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 37.Huang J., MacKerell A.D. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park S., Schulten K. Calculating potentials of mean force from steered molecular dynamics simulations. J. Chem. Phys. 2004;120:5946–5961. doi: 10.1063/1.1651473. [DOI] [PubMed] [Google Scholar]
- 39.Grubmüller H. In: Protein-Ligand Interactions: Methods and Applications. Ulrich Nienhaus G., editor. Humana Press; 2005. Force probe molecular dynamics simulations; pp. 493–515. [DOI] [Google Scholar]
- 40.Sigal C.T., Zhou W., et al. Resh M.D. Amino-terminal basic residues of Src mediate membrane binding through electrostatic interaction with acidic phospholipids. Proc. Natl. Acad. Sci. U S A. 1994;91:12253–12257. doi: 10.1073/pnas.91.25.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hochrein J.M., Lerner E.C., et al. Engen J.R. An examination of dynamics crosstalk between SH2 and SH3 domains by hydrogen/deuterium exchange and mass spectrometry. Protein Sci. 2006;15:65–73. doi: 10.1110/ps.051782206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okada M., Howell B.W., et al. Cooper J.A. Deletion of the SH3 domain of Src interferes with regulation by the phosphorylated carboxyl-terminal tyrosine. J. Biol. Chem. 1993;268:18070–18075. [PubMed] [Google Scholar]
- 43.Desmeules P., Grandbois M., et al. Salesse C. Measurement of membrane binding between recoverin, a calcium-myristoyl switch protein, and lipid bilayers by AFM-based force spectroscopy. Biophys. J. 2002;82:3343–3350. doi: 10.1016/S0006-3495(02)75674-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.