This study compares the performance of the BinaxNOW rapid antigen test versus reverse transcriptase polymerase chain reaction for community-based detection of infection with the Omicron variant of SARS-CoV-2 in San Francisco, California, during January 2022.
Visual Abstract. SARS-CoV-2 RT-PCR Versus BinaxNOW Rapid Antigen Test During Omicron Surge.

This study compares the performance of the BinaxNOW rapid antigen test versus reverse transcriptase polymerase chain reaction for community-based detection of infection with the Omicron variant of SARS-CoV-2 in San Francisco, California, during January 2022.
Abstract
Background:
SARS-CoV-2 rapid antigen tests are an important public health tool.
Objective:
To evaluate field performance of the BinaxNOW rapid antigen test (Abbott) compared with reverse transcriptase polymerase chain reaction (RT-PCR) for detecting infection with the Omicron variant of SARS-CoV-2.
Design:
Cross-sectional surveillance study.
Setting:
Free, walk-up, outdoor, urban community testing and vaccine site led by Unidos en Salud, serving a predominantly Latinx community highly impacted by COVID-19.
Participants:
Persons seeking COVID-19 testing in January 2022.
Measurements:
Simultaneous BinaxNOW and RT-PCR from nasal, cheek, and throat swabs, including cycle threshold (Ct) measures; a lower Ct value is a surrogate for higher amounts of virus.
Results:
Among 731 persons tested with nasal swabs, there were 296 (40.5%) positive results on RT-PCR; 98.9% were the Omicron variant. BinaxNOW detected 95.2% (95% CI, 91% to 98%) of persons who tested positive on RT-PCR with a Ct value below 30, 82.1% (CI, 77% to 87%) of those who tested positive on RT-PCR with a Ct value below 35, and 65.2% (CI, 60% to 71%) of all who were positive on RT-PCR. Among 75 persons with simultaneous nasal and cheek swabs, BinaxNOW using a cheek swab failed to detect 91% (20 of 22) of specimens that were positive on BinaxNOW with a nasal swab. Among persons with simultaneous nasal and throat swabs who were positive on RT-PCR with a Ct value below 30, 42 of 49 (85.7%) were detected by nasal BinaxNOW, 23 of 49 (46.9%) by throat BinaxNOW, and 44 of 49 (89.8%) by either.
Limitation:
Participants were a cross-sectional sample from a community-based sentinel surveillance site, precluding study of viral or symptom dynamics.
Conclusion:
BinaxNOW detected persons with high SARS-CoV-2 levels during the Omicron surge, enabling rapid responses to positive test results. Cheek or throat swabs should not replace nasal swabs. As currently recommended, high-risk persons with an initial negative BinaxNOW result should have repeated testing.
Primary Funding Source:
University of California, San Francisco.
SARS-CoV-2 rapid antigen tests are a valuable public health tool in the ongoing COVID-19 pandemic. They enable immediate identification of active SARS-CoV-2 infection with high viral levels, which can lead to faster isolation and curtail transmission chains (1–3). Antigen tests also enable the rapid diagnosis needed for initiation of time-sensitive outpatient COVID-19 therapies (4, 5). Many community testing programs, including school programs, use rapid tests; home antigen testing is increasingly becoming available in the United States and is widely available in some countries. Widespread use of rapid antigen tests is based on performance evaluations in the context of the ancestral, Alpha, or Delta variants (6, 7).
It is vital to evaluate performance of rapid antigen tests when new SARS-CoV-2 variants emerge. The U.S. Food and Drug Administration advised caution on rapid test use for detection of the Omicron variant in December 2021 (8). The Omicron variant has more than 50 mutations compared with ancestral lineages; the majority are in the spike protein, but 4 are in the nucleocapsid gene—the target gene in the BinaxNOW assay (Abbott). Although laboratory studies suggest that the performance of BinaxNOW should not be affected when used to detect the Omicron variant, field validation is needed (7). We sought to examine performance of the BinaxNOW rapid antigen test at our community-based site in the Mission District of San Francisco, California, a setting in which we have evaluated its performance with previous variants and routinely used it for walk-up low-barrier testing (9, 10).
Methods
This report includes data collected in January 2022 at our free, outdoor, walk-up testing and vaccine site situated in an outdoor parking lot in the heart of the Mission Cultural District of San Francisco. The site is led by Unidos en Salud, an academic (University of California, San Francisco [UCSF] and Chan Zuckerberg Biohub), community (San Francisco Latino Task Force for COVID-19), and public health (San Francisco Department of Public Health) collaboration that conducts SARS-CoV-2 surveillance and serves communities with the highest risk for COVID-19 (9–11). Unidos en Salud serves a San Francisco community with a large proportion of frontline workers, immigrants, and monolingual families living in multigenerational households, the majority of whom are Latinx.
Persons seeking testing provided demographic characteristics, symptoms and onset date, vaccination status, reason for testing, and informed consent. There was no age restriction. On all days, certified laboratory assistants collected bilateral anterior nasal swabs according to manufacturer instructions (12). A second anterior nasal swab was immediately collected for reverse transcriptase polymerase chain reaction (RT-PCR). Certified readers read BinaxNOW cards, and results were returned within an hour of testing using secure messaging in the Primary.Health platform. Three invalid results (out of 921 [0.3%]) were excluded from these analyses. Photographed cards were reread by a blinded trained expert, and these results were considered final for analyses. On 9 January 2022, we also collected simultaneous oral cheek swabs on 75 persons randomly selected from among those seeking testing that day and tested these for SARS-CoV-2 with BinaxNOW and RT-PCR. On 14 January 2022, we also collected simultaneous oral throat (tonsillar) swabs on 115 persons randomly selected from among those seeking testing that day and tested for SARS-CoV-2 with BinaxNOW and RT-PCR.
Bilingual (Spanish and English) staff from Unidos en Salud called persons diagnosed with COVID-19 to offer supportive services, including home deliveries of supplies, food, and care items, as previously described (11). Persons who were eligible for COVID-19 treatment were referred to their primary health provider or Zuckerberg San Francisco General Hospital.
As previously described (13), RT-PCR using probes specific to N and E genes was performed on the nasal and oral swabs collected in DNA/RNA Shield (Zymo Research) with an internal human positive control (RNase P). The assay limit of detection is 100 viral copies per milliliter; cycle threshold (Ct) values below 40 were considered positive. Cycle threshold is a surrogate measure of the amount of virus in a clinical specimen; lower values are associated with higher rates of positive viral culture results (14–16). Variant lineage was determined by full genome sequencing using the ARTIC Network V3 primers and the Illumina NovaSeq platform, followed by consensus genome assembly using the COVID module of the freely available CZ ID (formerly IDseq) pipeline, as described previously (17, 18). Complete genomes were deposited in GISAID (Accession EPI_ISL_882925, with virus name “hCoV-19/USA/CA-UCSF-JDxxxx/2022”).
Among persons with a positive RT-PCR result for SARS-CoV-2, both overall and among those with RT-PCR Ct values below 30 and below 35, we calculated the proportion testing positive on BinaxNOW. We also calculated the proportion of persons with a negative RT-PCR result who tested negative on BinaxNOW and the proportions with positive RT-PCR results and Ct values of 30 or higher and 35 or higher, respectively; 95% confidence intervals were calculated using the Clopper–Pearson method. We examined assay performance in strata defined by presence versus absence of symptoms at the time of testing, age, and vaccination status. Mean Ct values among symptomatic and asymptomatic persons and vaccinated and unvaccinated persons were compared using difference in means, with confidence intervals calculated using the t distribution. We performed analogous analyses among persons with simultaneous nares and cheek swabs and simultaneous nares and throat swabs.
Ethics Statement
The UCSF Committee on Human Research determined that the study met criteria for public health surveillance. All participants provided informed consent for dual testing.
Role of the Funding Source
The funders had no role in the design, conduct, or analysis of the study or the decision to submit the manuscript for publication.
Results
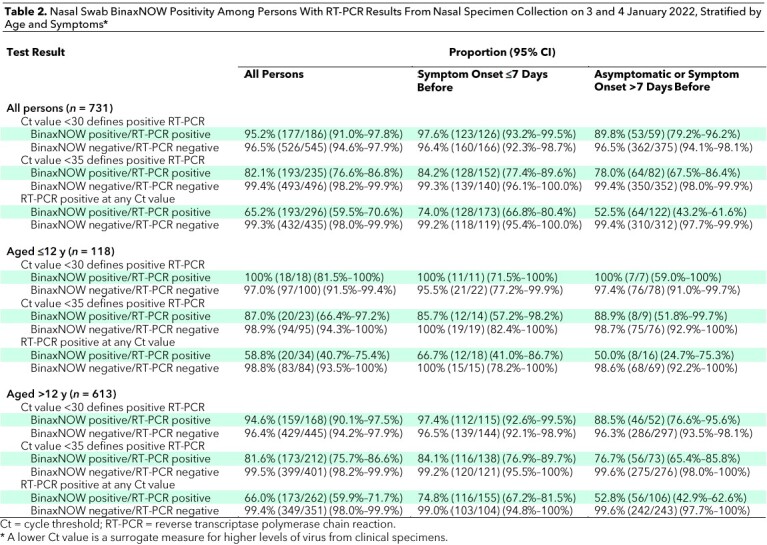
Testing for SARS-CoV-2 with RT-PCR or BinaxNOW was performed on 731 samples on 3 and 4 January 2022 at our community testing site. Self-identified race/ethnicity among participants included Latinx (77.4%), Central or South American Indian (6.8%), White (5.2%), Asian (3.1%), and Black (1.8%). Among participants, 46.0% identified as male, 53.6% identified as female, and 0.4% identified as other; 16.1% reported age 12 years or younger (Table 1). Of the 731 participants, 310 (42.7%) reported symptoms. The most common symptoms were cough (62.3%), sore throat (46.1%), and congestion (44.2%), and the least common symptom was anosmia (2.9%). Primary motivations reported for testing were clearance for work (26.4%), known or suspected exposure (23.1%), school requirement (17.7%), and symptoms (16.3%). Among the 731 persons, 89 (12.2%) reported being unvaccinated, 134 (18.3%) reported having started but not completed their primary vaccine series, 267 (36.5%) reported having completed their primary vaccine series, 166 (22.7%) reported having completed the primary series plus a booster, and 75 (10.3%) either declined to report vaccine status or reported receipt of a non-mRNA vaccine.
Table 1.
Baseline Demographic Characteristics of Participants (Tested 3 and 4 January 2022), Stratified by Nasal Swab RT-PCR and BinaxNOW Result*
Overall, 296 of 731 (40.5%) persons tested were positive for SARS-CoV-2 on RT-PCR. Full genome sequencing was attempted on all 182 samples that were positive on RT-PCR with a Ct value below 30. Of these, variant lineages could be determined for 178 genomes (97.8%) with no assignment ambiguity (Pangolin v3.1.19, lineage version 2022-01-20); 176 (98.9%) were Omicron (BA.1 or BA.1.1), and the remaining 2 were Delta (AY.44).
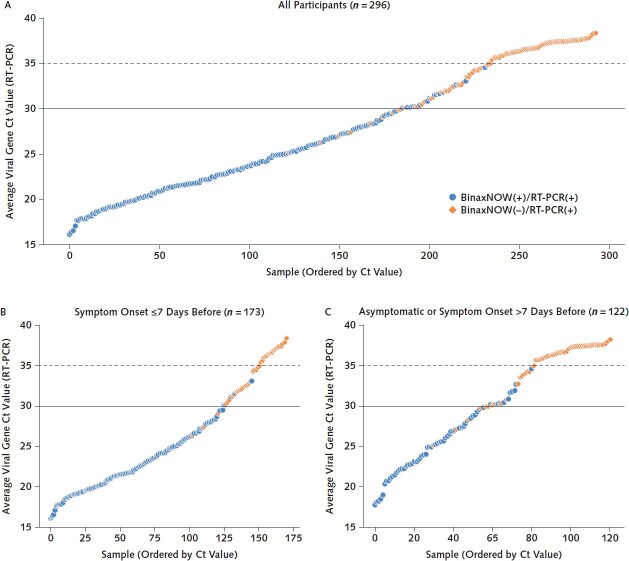
Similar to performance with other variants, the BinaxNOW assay maintained the highest performance among persons who were positive on RT-PCR with low Ct values. There were 177 persons who were positive on BinaxNOW among the 186 who were positive on RT-PCR with a Ct value below 30. There were no positive BinaxNOW results among the 61 persons who were positive on RT-PCR with a Ct value above 35 (Figure 1). BinaxNOW detected infection in 95.2% (95% CI, 91% to 98%) and 82.1% (CI, 77% to 87%) of persons who tested positive on RT-PCR with Ct values below 30 and below 35, respectively; 65.2% (CI, 60% to 71%) of all persons who tested positive on RT-PCR (regardless of Ct value) tested positive on BinaxNOW. BinaxNOW performance was similar among persons aged 12 years or younger and those older than 12 years (Table 2).
Figure 1. RT-PCR Ct values and BinaxNOW rapid antigen test results among all participants with positive RT-PCR results tested on 3 and 4 January 2022 (A) and stratified according to COVID-19 symptoms (B and C).
Average viral Ct values of all persons with positive RT-PCR and/or BinaxNOW results (n = 296) are plotted in ascending order of Ct value. Each point represents 1 person. Blue circles represent persons whose samples were positive on both BinaxNOW and RT-PCR. Orange diamonds represent persons with positive results on RT-PCR but negative results on BinaxNOW. Ct = cycle threshold; RT-PCR = reverse transcriptase polymerase chain reaction.
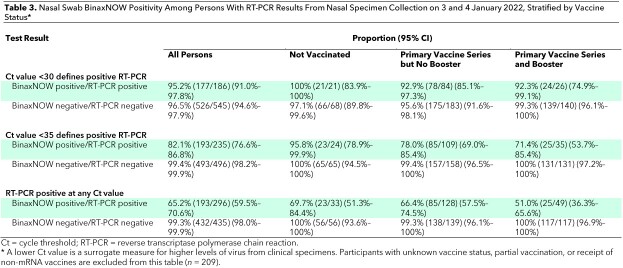
Table 2.
Nasal Swab BinaxNOW Positivity Among Persons With RT-PCR Results From Nasal Specimen Collection on 3 and 4 January 2022, Stratified by Age and Symptoms*
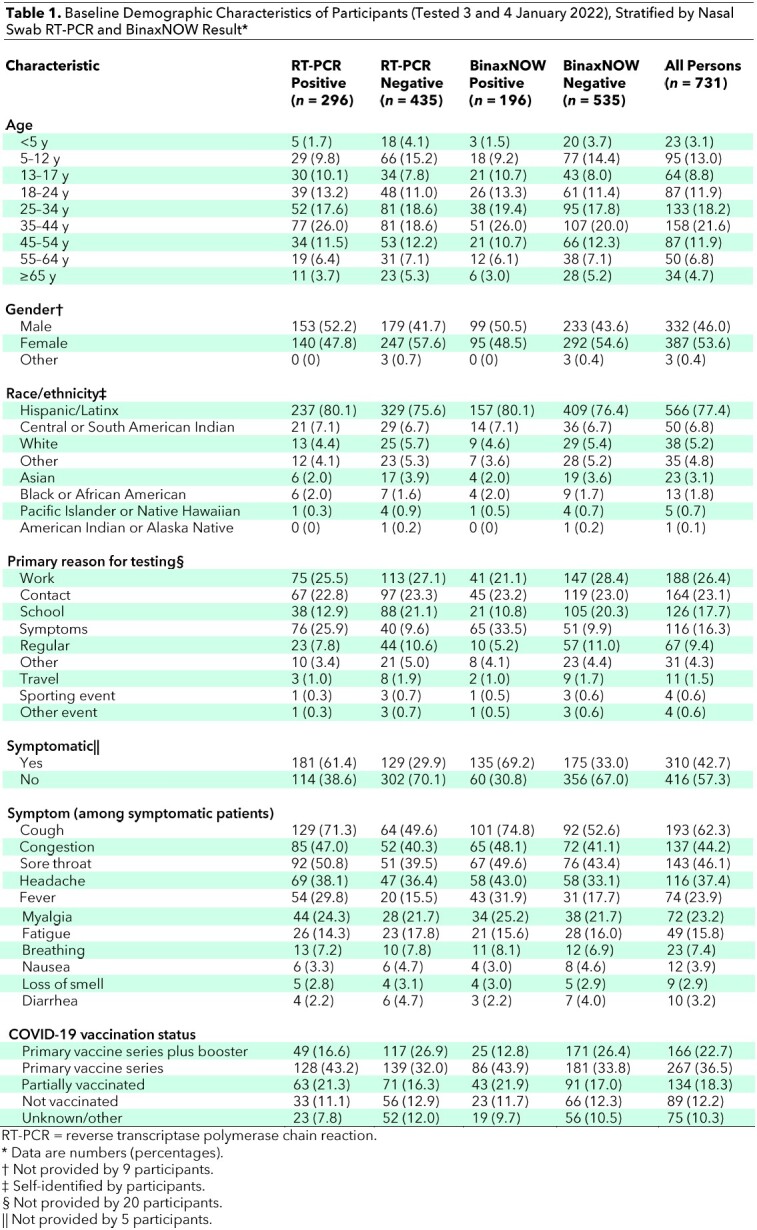
Among persons who were positive on RT-PCR with a Ct value below 30, BinaxNOW detected SARS-CoV-2 in 53 of 59 (89.8% [CI, 79% to 96%]) who were asymptomatic and 123 of 126 (97.6% [CI, 93% to 100%]) who reported symptoms (Figure 1 and Table 2). The mean Ct value was higher among asymptomatic (30.1 [SD, 6.1]) than symptomatic (25.9 [SD, 6.2]) persons (difference in means, 4.2 [CI, 2.8 to 5.6]) (Figure 2, A). The Ct values increased with the number of days from onset of symptoms, as previously described (Figure 2, B) (19). The mean Ct value was similar among persons who had received at least 1 vaccine dose (27.8 [SD, 6.4]) versus unvaccinated persons (27.8 [SD, 6.6]) (difference in means, 0.04 [CI, −75.5 to 75.6]). BinaxNOW detection of infection among persons with a Ct value below 30 who had received a booster, had completed the primary series only, and were unvaccinated was 92.3% (CI, 75% to 99%), 92.9% (CI, 85% to 97%), and 100% (CI, 84% to 100%), respectively (Table 3). Of the 435 persons with a negative RT-PCR result, 432 (99.3% [CI, 98.0% to 99.9%]) were also negative on BinaxNOW.
Figure 2. SARS-CoV-2 Ct values by symptom status, days since symptom onset, and vaccination status among persons with positive RT-PCR results tested on 3 and 4 January 2022.
The figure shows RT-PCR Ct values and BinaxNOW rapid antigen test results among symptomatic and asymptomatic participants (A) and stratified by days since symptom onset among symptomatic participants (B) and by vaccination status (completed primary mRNA vaccine series plus booster, completed primary mRNA vaccine series without booster, incomplete mRNA primary vaccine series, no vaccine, and unknown vaccination status or self-reported receipt of non-mRNA vaccine) (C). Each point represents 1 person. Blue circles represent persons whose samples were positive on both BinaxNOW and RT-PCR. Orange diamonds represent persons with positive results on RT-PCR but negative results on BinaxNOW. Box plots show the first quartile, median, and third quartile in the shaded regions. Ct = cycle threshold; RT-PCR = reverse transcriptase polymerase chain reaction.
Table 3.
Nasal Swab BinaxNOW Positivity Among Persons With RT-PCR Results From Nasal Specimen Collection on 3 and 4 January 2022, Stratified by Vaccine Status*
Simultaneous nares and oral cheek specimens were collected from an additional 75 persons on 9 January 2022. Of these, 46 of 75 (61%) were positive on RT-PCR from the nares, 22 of which had a Ct value below 30 (Appendix Table). Among the 46 nares specimens that were positive on RT-PCR, 22 were positive on BinaxNOW with the nares specimen, whereas only 2 (4.3% [CI, 0% to 15%]) were positive on BinaxNOW with the oral cheek specimen. Thirteen of the 46 nares specimens that were positive on RT-PCR were also positive on RT-PCR with the oral cheek specimen. There were no specimens that were positive on RT-PCR from the oral cheek collection and negative on RT-PCR from the nares.
Appendix Table.
BinaxNOW Detection With RT-PCR Referent From Nasal or Cheek Specimen Collection on 9 January 2022*

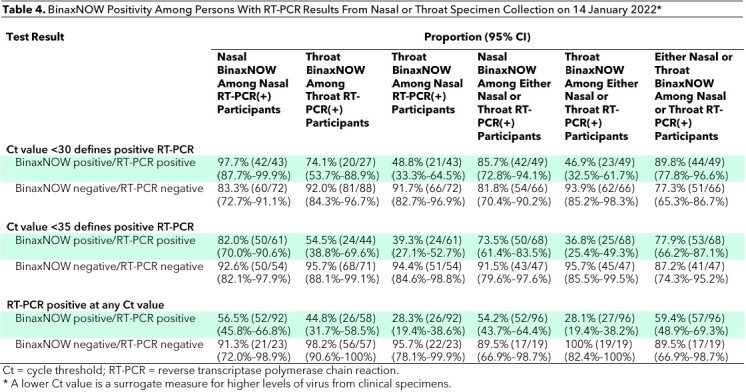
Simultaneous nares and oral throat specimens were collected from an additional 115 persons on 14 January 2022. Of these, 92 were positive on RT-PCR from the nares. Among persons with nares specimens that were positive on RT-PCR with a Ct value below 30 (n = 43) and below 35 (n = 61), BinaxNOW detected SARS-CoV-2 in 97.7% (CI, 87.7% to 99.9%) and 82.0% (CI, 70.0% to 90.6%), respectively, using the nasal swab alone and in 74.1% (CI, 53.7% to 88.9%) and 54.5% (CI, 38.8% to 69.6%), respectively, using the throat swab alone (Table 4). For persons with a positive RT-PCR result with a Ct value below 30 from either a nasal or throat specimen, BinaxNOW detected SARS-CoV-2 in 85.7% (CI, 72.8% to 94.1%) using a nasal swab alone, 46.9% (CI, 32.5% to 61.7%) using a throat swab alone, and 89.8% (CI, 77.8% to 96.6%) from either the nose or the throat (Figure 2).
Table 4.
BinaxNOW Positivity Among Persons With RT-PCR Results From Nasal or Throat Specimen Collection on 14 January 2022*
Discussion
Test positivity for infection with the SARS-CoV-2 Omicron variant was extremely high—40% prevalence on RT-PCR—at a walk-up community testing site amidst the COVID-19 Omicron surge in San Francisco in January 2022. This cross-sectional analysis confirms that the ability of the BinaxNOW rapid antigen test to detect infection with the Omicron variant, particularly at higher virus levels, is similar to its ability to detect prior variants (9, 10, 19). The BinaxNOW assay rapidly identifies persons with the highest virus levels, who are likely to pose the greatest risk for transmission at the time of the test (14). A positive rapid test result thus enables immediate public health and personal action for isolation, disease mitigation, and clinical care, in a disease process where chains of transmission need to be broken and therapies are time-sensitive. With the increasing availability of this test in the United States, this information can inform optimal emerging public health strategies that hinge on rapid diagnosis and treatment.
Our data support the recommendation for repeated rapid antigen testing for persons at risk for COVID-19 who have an initial negative BinaxNOW result. Persons who have low levels of virus detectable on RT-PCR but not antigen testing may be at either the upswing or the downswing of the viral dynamic curve for SARS-CoV-2 (20). In the setting of an acute surge, many persons are likely to have been recently infected, and thus those initially testing negative using BinaxNOW may subsequently develop higher viral loads associated with greater infectiousness and may become detectable on repeated testing 1 to 2 days later. Persons at high risk for COVID-19 with a single negative test result should thus wear masks and undergo repeated rapid or RT-PCR testing regardless of symptoms, especially during the type of COVID-19 surge that was ongoing during sample collection in this study (21).
BinaxNOW detected at least 90% of all RT-PCR–positive cases with high viral loads (Ct value <30), regardless of symptoms or vaccine status. Cycle threshold values were higher among asymptomatic versus symptomatic persons but were similar by vaccine status, the latter of which is consistent with prior observations in pre-Omicron variants (22–24).
Omicron putatively has a shorter incubation period than prior variants. Symptoms may be intense in the upper respiratory tract, and the virus appears to be less pathogenic in the lower airway compared with prior variants (25). One small study with 5 positive health care workers suggested detection is faster from oral versus nasal swabs (26). Although we only examined 75 paired nasal and oral cheek swabs in a cross-sectional analysis, our data suggest that a simple oral cheek swab does not increase detection of SARS-CoV-2. Even with the prominent clinical feature of pharyngitis in persons with the Omicron variant, our data from 115 paired nasal and throat swabs also argue against replacing nasal swabs with throat swabs for diagnosis. At the time of presentation to a community-based testing site, we saw only a small (<5%) increase in detection of COVID-19 with addition of a throat swab to a nasal swab. Although these findings do not directly contradict the potential for earlier positivity via throat swab, they do suggest that in a high-throughput community testing site, the complexity and cost of such an approach would need to be formally evaluated before adoption.
The ongoing presence of a walk-up COVID-19 community testing and surveillance site permitted the rapid collection and analyses of these data. The Unidos en Salud collaboration, established in April 2020 near the start of the pandemic, provides a model of how a community, academic, and public health partnership can provide testing and vaccine services to a disproportionately affected community while also generating data to inform both community members and the wider scientific discourse (1, 9–11, 27–30). More than half the clients seeking services at this site self-reported not having insurance coverage or not being able to identify a primary provider where they might otherwise access health services (1). Easy access to rapid testing at a site located in the heart of the community, with friendly community staffing and accessible hours (including weekends), serves a vital role in supporting the frontline workers disproportionately exposed to infection, who are often working multiple jobs to support the ongoing functioning of the economy.
Limitations of our study included the use of self-reported vaccine status, which may have led to misclassification, and the use of a cross-sectional sample. Persons who were asymptomatic at the time of testing may have subsequently developed symptoms, and our analyses did not assess the evolution of Ct values or RT-PCR and BinaxNOW test positivity (overall or by sample site) over time. Trained technicians collected the samples and read the BinaxNOW cards. Although results could vary when these actions are performed by nontechnicians, such as in the home setting, the overall finding of similar detection of the Omicron variant compared with prior variants by this rapid antigen test remains applicable. The performance of BinaxNOW observed in this study was in the setting of high SARS-CoV-2 prevalence, resulting in a high positive predictive value. In settings with low disease prevalence, RT-PCR should be performed to confirm a positive BinaxNOW result, particularly in persons without symptoms or exposures (9).
In conclusion, BinaxNOW detected persons with high SARS-CoV-2 levels during the Omicron surge in a community walk-up setting, enabling rapid responses to positive test results. BinaxNOW is one important component of the diagnostic armamentarium for SARS-CoV-2. Cheek or throat swabs should not replace nasal swabs. As currently recommended, repeated testing should be done for high-risk persons with an initial negative BinaxNOW test result.
Footnotes
This article was published at Annals.org on 15 March 2022.
* Drs. DeRisi and Havlir contributed equally to this work.
References
- 1. Rubio LA , Peng J , Rojas S , et al; CLIAHUB Consortium. The COVID-19 symptom to isolation cascade in a Latinx community: a call to action. Open Forum Infect Dis. 2021;8:ofab023. [PMID: ] doi: 10.1093/ofid/ofab023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mina MJ , Parker R , Larremore DB . Rethinking Covid-19 test sensitivity—a strategy for containment. N Engl J Med. 2020;383:e120. [PMID: ] doi: 10.1056/NEJMp2025631 [DOI] [PubMed] [Google Scholar]
- 3. Larremore DB , Wilder B , Lester E , et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv. 2021;7. [PMID: ] doi: 10.1126/sciadv.abd5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jayk Bernal A, Gomes da Silva MM, Musungaie DB, et al; MOVe-OUT Study Group.. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N Engl J Med. 2022;386:509-20. [PMID: ] doi: 10.1056/NEJMoa2116044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O'Shaughnessy JA; U.S. Food and Drug Administration. Letter to Pfizer, Inc. (Karen Baker) on Emergency Use Authorization 105. 22 December 2021. Accessed at www.fda.gov/media/155049/download on 7 March 2022.
- 6. Centers for Disease Control and Prevention. Interim Guidance for Antigen Tests for SARS-CoV-2. Updated 9 September 2021. Accessed at www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html on 8 January 2022.
- 7. Regan J , Flynn JP , Choudhary MC , et al. Detection of the Omicron variant virus with the Abbott BinaxNow SARS-CoV-2 rapid antigen assay. Open Forum Infect Dis. 2022;9:ofac022. [PMID: ] doi: 10.1093/ofid/ofac022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. U.S. Food and Drug Administration. Coronavirus (COVID-19) Update: December 28, 2021 [press release]. Accessed at www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-december-28-2021 on 7 March 2022.
- 9. Pilarowski G , Marquez C , Rubio L , et al. Field performance and public health response using the BinaxNOW™ rapid severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigen detection assay during community-based testing. Clin Infect Dis. 2021;73:e3098-e3101. [PMID: ] doi: 10.1093/cid/ciaa1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pilarowski G , Lebel P , Sunshine S , et al. Performance characteristics of a rapid severe acute respiratory syndrome coronavirus 2 antigen detection assay at a public plaza testing site in San Francisco. J Infect Dis. 2021;223:1139-44. [PMID: ] doi: 10.1093/infdis/jiaa802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kerkhoff AD , Sachdev D , Mizany S , et al. Evaluation of a novel community-based COVID-19 ‘Test-to-Care' model for low-income populations. PLoS One. 2020;15:e0239400. [PMID: ] doi: 10.1371/journal.pone.0239400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abbott. BinaxNOW COVID-19 Ag Card (PN 195-000) – Instructions for Use. Accessed at www.fda.gov/media/141570/download on 7 March 2022.
- 13. Crawford ED , Acosta I , Ahyong V , et al. Rapid deployment of SARS-CoV-2 testing: the CLIAHUB. PLoS Pathog. 2020;16:e1008966. [PMID: ] doi: 10.1371/journal.ppat.1008966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jefferson T , Spencer EA , Brassey J , et al. Viral cultures for coronavirus disease 2019 infectivity assessment: a systematic review. Clin Infect Dis. 2021;73:e3884-e3899. [PMID: ] doi: 10.1093/cid/ciaa1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Singanayagam A , Patel M , Charlett A , et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25. [PMID: ] doi: 10.2807/1560-7917.ES.2020.25.32.2001483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Riemersma KK, Grogan BE, Kita-Yarbro A, et al. Shedding of infectious SARS-CoV-2 despite vaccination. medRxiv. Preprint posted online 6 November 2021. doi: 10.1101/2021.07.31.21261387 [DOI]
- 17. Martinez D, Miyasaki T, Chow E. UCSF CAT COVID-19 Tailed 275bp v3 ARTIC protocol v1 V.1. 12 April 2021. Accessed at www.protocols.io/view/ucsf-cat-covid-19-tailed-275bp-v3-artic-protocol-v-bsfknbkw on 10 February 2022.
- 18. Peng J , Liu J , Mann SA , et al; IDseq Team. Estimation of secondary household attack rates for emergent spike L452R severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants detected by genomic surveillance at a community-based testing site in San Francisco. Clin Infect Dis. 2022;74:32-9. [PMID: ] doi: 10.1093/cid/ciab283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pollock NR , Jacobs JR , Tran K , et al. Performance and implementation evaluation of the Abbott BinaxNOW rapid antigen test in a high-throughput drive-through community testing site in Massachusetts. J Clin Microbiol. 2021;59. [PMID: ] doi: 10.1128/JCM.00083-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kissler SM , Fauver JR , Mack C , et al. Viral dynamics of acute SARS-CoV-2 infection and applications to diagnostic and public health strategies. PLoS Biol. 2021;19:e3001333. [PMID: ] doi: 10.1371/journal.pbio.3001333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mina MJ , Peto TE , García-Fiñana M , et al. Clarifying the evidence on SARS-CoV-2 antigen rapid tests in public health responses to COVID-19. Lancet. 2021;397:1425-7. [PMID: ] doi: 10.1016/S0140-6736(21)00425-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Acharya CB, Schrom J, Mitchell AM, et al. No significant difference in viral load between vaccinated and unvaccinated, asymptomatic and symptomatic groups infected with SARS-CoV-2 Delta variant. medRxiv. Preprint posted online 29 September 2021. doi: 10.1101/2021.09.28.21264262 [DOI]
- 23. Kissler SM , Fauver JR , Mack C , et al. Viral dynamics of SARS-CoV-2 variants in vaccinated and unvaccinated persons [Letter]. N Engl J Med. 2021;385:2489-91. [PMID: ] doi: 10.1056/NEJMc2102507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Puhach O, Adea K, Hulo N, et al. Infectious viral load in unvaccinated and vaccinated patients infected with SARS-CoV-2 WT, Delta and Omicron. medRxiv. Preprint posted online 11 January 2022. doi: 10.1101/2022.01.10.22269010 [DOI] [PubMed]
- 25. Meng B, Ferreira IATM, Abdullahi A, et al. SARS-CoV-2 Omicron spike mediated immune escape, infectivity and cell-cell fusion. bioRxiv. Preprint posted online 22 December 2021. doi: 10.1101/2021.12.17.473248 [DOI]
- 26. Adamson B, Sikka R, Wyllie AL, et al. Discordant SARS-CoV-2 PCR and rapid antigen test results when infectious: a December 2021 occupational case series. medRxiv. Preprint posted online 5 January 2022. doi: 10.1101/2022.01.04.22268770 [DOI]
- 27. Chamie G , Marquez C , Crawford E , et al; CLIAhub Consortium. Community transmission of severe acute respiratory syndrome coronavirus 2 disproportionately affects the Latinx population during shelter-in-place in San Francisco. Clin Infect Dis. 2021;73:S127-S135. [PMID: ] doi: 10.1093/cid/ciaa1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laurie MT , Liu J , Sunshine S , et al. SARS-CoV-2 variant exposures elicit antibody responses with differential cross-neutralization of established and emerging strains including Delta and Omicron. J Infect Dis. 2022. [PMID: ] doi: 10.1093/infdis/jiab635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marquez C , Kerkhoff AD , Naso J , et al. A multi-component, community-based strategy to facilitate COVID-19 vaccine uptake among Latinx populations: from theory to practice. PLoS One. 2021;16:e0257111. [PMID: ] doi: 10.1371/journal.pone.0257111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fields J, Gutierrez JR, Marquez C, et al. Community-academic partnerships to address Covid-19 inequities: lessons from the San Francisco Bay Area. NEJM Catalyst. 23 June 2021. doi: 10.1056/CAT.21.0135 [DOI]