Abstract
Glycans are ubiquitous and play important biological roles, yet chemical methods for probing their structure and function within cells remain limited. Strategies for studying other biomacromolecules, such as proteins, often exploit chemoselective reactions for covalent modification, capture, or imaging. Unlike amino acids that constitute proteins, glycan building blocks lack distinguishing reactivity because they are composed primarily of polyol isomers. Moreover, encoding glycan variants through genetic manipulation is complex. Therefore, we formulated a new, generalizable strategy for chemoselective glycan modification that directly takes advantage of cellular glycosyltransferases. Many of these enzymes are selective for the products they generate yet promiscuous in their donor preferences. Thus, we designed reagents with bioorthogonal handles that function as glycosyltransferase substrate surrogates. We validated the feasibility of this approach by synthesizing and testing probes of d-arabinofuranose (d-Araf), a monosaccharide found in bacteria and an essential component of the cell wall that protects mycobacteria, including Mycobacterium tuberculosis. The result is the first probe capable of selectively labeling arabinofuranose-containing glycans. Our studies serve as a platform for developing new chemoselective labeling agents for other privileged monosaccharides. This probe revealed an asymmetric distribution of d-Araf residues during mycobacterial cell growth and could be used to detect mycobacteria in THP1-derived macrophages.
Graphical Abstract
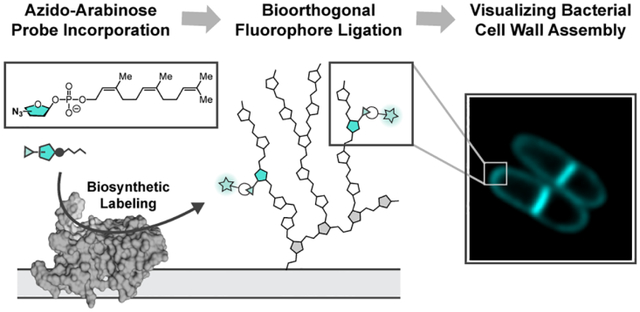
Monomer-selective bioconjugation reactions have transformed the study of biomolecules, affording molecular-level insight into structure, function, localization, and dynamics.1–3 Proteins exhibit significant functional group variation, and this diversity has been exploited in bioconjugation reactions. In contrast, glycans and their component sugars cannot easily be distinguished from one another based on complementary reactivity, as their structural diversity derives predominantly from the stereo- and constitutional isomerism of polyol monomers (Figure 1A). As a result, the systematic interrogation of glycan structure-function relationships at the bacterial cell surface is limited.
Figure 1.
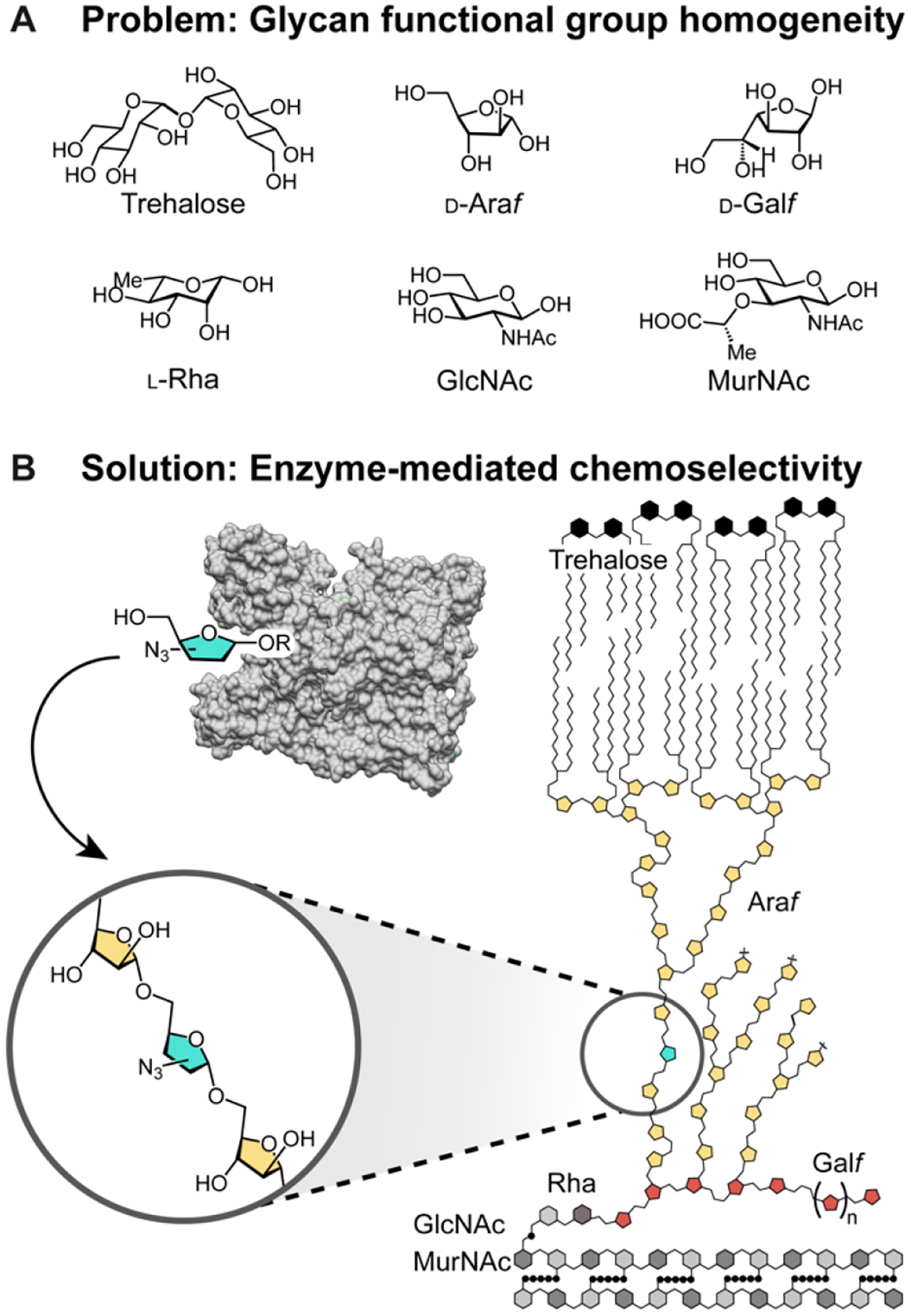
(A) Monosaccharides within glycans derive their identity from polyol stereo- and constitutional isomerism. (B) The core cell wall structure of mycobacteria and corynebacteria is comprised of six unique monomers. This structure, termed the mycolyl-arabinogalactan-peptidoglycan complex (mAGP) is a dense glycolipid matrix that protects cells from environmental stresses, including antibiotics. Biosynthetic incorporation directly leverages the activity of cellular glycosyltransferases for specific monosaccharides to introduce modifications into cell surface glycans.
In the absence of a viable chemical approach to site-selective glycan labeling, metabolic engineering has been used to modify and study cell surface glycans in eukaryotic systems.4–7 This method generally relies on the cellular uptake of non-natural monosaccharides, followed by extensive biosynthetic processing to nucleotide-sugar analogs. As nucleotide-sugars can serve as donors for cytosolic glycosyltransferases, these intermediates are incorporated into growing glycan chains that are subsequently exported to the cell surface. While effective in eukaryotes, probes can experience unintended fates leading to undesired labeling.8–9 The adaptation of metabolic incorporation to prokaryotes has been historically challenging.10–14 Mammals utilize 35 unique monosaccharide building blocks, while bacteria employ over 600.15–16 The structural diversity of bacterial monosaccharides and glycans necessitates a complex, and often poorly understood carbohydrate metabolism.17–18 As a result, site-specific modification of bacterial glycans is challenging.17
We assumed that unique sugar monomers could be best distinguished through direct enzyme recognition and modification to achieve selective glycan modification.19 We therefore sought to exploit existing catalysts with the requisite selectivity—the endogenous glycosyltransferases. These biocatalysts have an evolved selectivity for a specific small molecule substrate donor and a specific polysaccharide acceptor (Figure 1B).20–24
Here, we describe the development and application of a new approach to chemoselective glycan bioconjugation. This strategy, termed biosynthetic incorporation, leverages the activity of endogenous extracellular enzymes using substrate surrogates (Figure 2). This biocatalytic manifold can side-step challenges associated with small molecule-based glycan bioconjugation and metabolic engineering by exploiting the intrinsic selectivity of the target biocatalysts.
Figure 2.
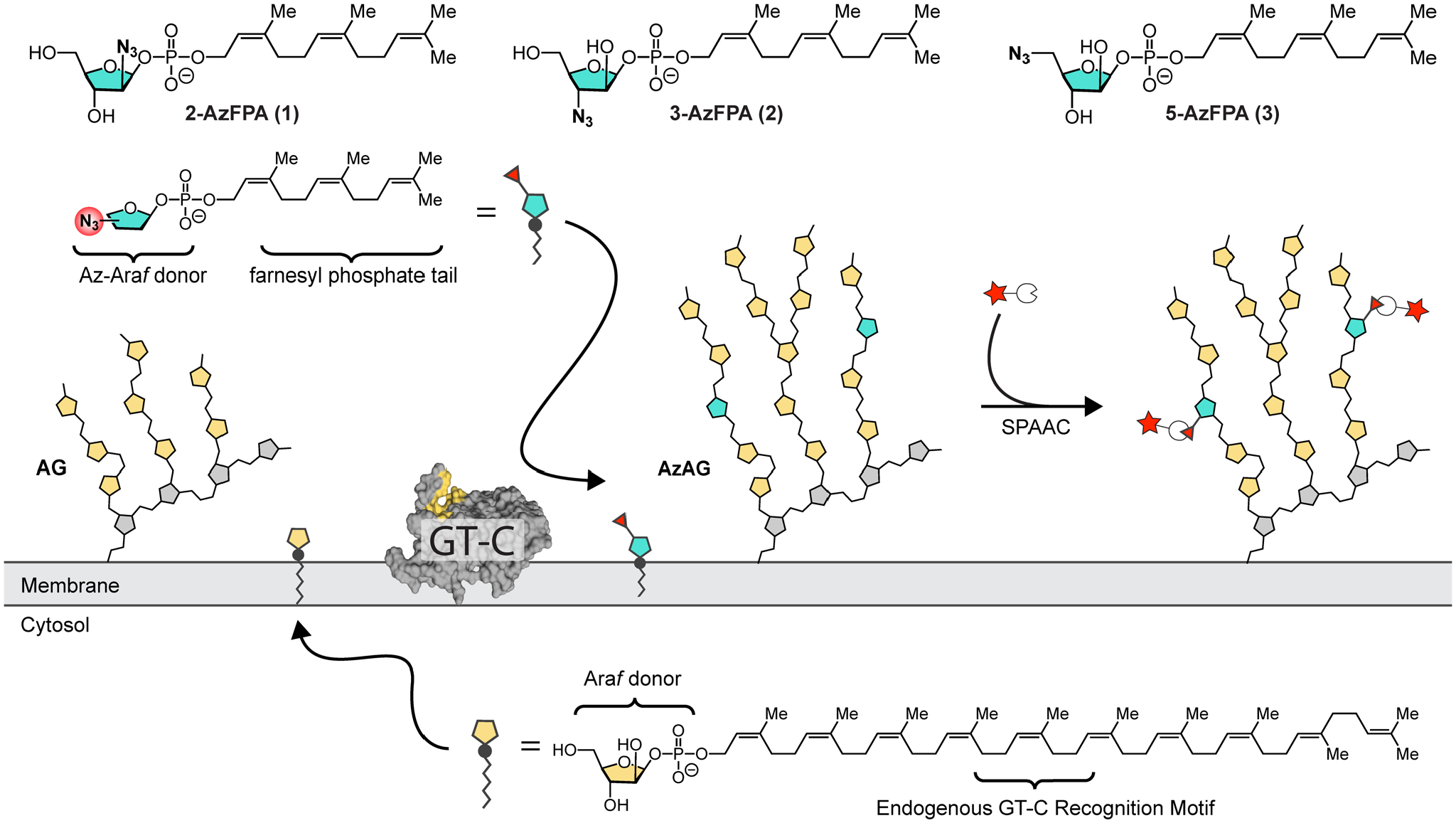
To harness the catalytic activity of arabinofuranosyltransferase GT-Cs for bioconjugation, an azide-modified substrate surrogate was designed based on structural homology to the endogenous d-Araf donor (DPA). Three azide regioisomers were produced (1–3). Exogenous delivery of AzFPA was designed to result in substrate incorporation, which could subsequently be detected and quantified using SPAAC-mediated fluorophore conjugation.
We assessed the feasibility of our chemoselective labeling strategy by targeting d-arabinofuranose (d-Araf). This arabinose isomer is not found in humans but is present in microbes. d-Araf is an essential component of the cell wall of the order Mycobacteriales.25–26 Although many constituents of this order are benign, Mycobacterium tuberculosis (Mtb), Corynebacterium diphtheriae, and Mycobacterium leprae are notorious human pathogens.27–29 These organisms utilize d-Araf for the construction of the core glycolipid component of their cell wall, the mycolyl-arabinogalactan-peptidoglycan complex (mAGP).30 Within the mAGP, the arabinan is thought to help maintain the structural integrity of the cell envelope. Indeed, mAGP biosynthesis is the target of the front-line antituberculosis drug ethambutol.31
d-Araf residues are an excellent test of our strategy. First, no methods to selectively visualize d-Araf are known. Second, the biosynthesis of the activated sugar proceeds through late-stage epimerization of the C2 hydroxyl from the ribose-phospholipid to the corresponding arabinose-phospholipid donor rather than other sources of free arabinose.32 Consequently, metabolic engineering approaches are unlikely to result in specific d-Araf labeling.33
In mycobacteria, integral membrane glycosyltransferases (GT-Cs) mediate d-Araf incorporation into cell-surface glycans.20–24 In contrast to the nucleotide-sugar substrates of cytosolic glycosyltransferases, these GT-Cs are transmembrane enzymes that recognize polyprenyl phosphate-linked sugar donors.34 Our group previously used the non-natural glycolipid donor (Z,Z)-farnesyl phosphoryl-β-d-arabinofuranose (FPA) as a surrogate for the extended (C55) endogenous arabinofuranose donor decaprenyl phosphoryl-β-d-arabinofuranose (DPA) in C. glutamicum and M. smegmatis.35 This lipid substitution facilitates exogenous reagent delivery, as extended polyprenyl glycosides are poorly soluble and form micelles.23, 36 Here, we employ this non-natural (Z,Z)-farnesyl phosphoryl donor scaffold as a vehicle to introduce azide-modified d-Araf derivatives into the mAGP.
The endogenous monosaccharide donor and polysaccharide acceptors for cell wall arabinosylation have been well-characterized, yet few 3D structures for the target arabinosyltransferase enzymes have been reported.37 We reasoned that the efficiency of incorporation could vary between different isomers. Accordingly, we focused on preparing and evaluating all three possible regioisomeric azido-(Z,Z)-farnesyl phosphoryl-β-d-arabinofuranose (AzFPA) derivatives. The AzFPA regioisomers were synthesized from commercially available arabinofuranose and ribofuranose monomers. The key azide functionality was installed through nucleophilic displacement or nucleophilic epoxide opening.38 We appended the (Z,Z)-farnesyl recognition motif and then removed the protecting groups to afford the target compounds (Scheme SI-1). The synthetic routes were optimized to provide access to the desired substrate surrogates for microbiological studies.
The collection of AzFPA isomers was evaluated for incorporation in C. glutamicum and M. smegmatis using fluorescent labeling and flow cytometry (Figure 3). Bacteria were cultured to mid-logarithmic phase with each AzFPA isomer, washed to remove any unassociated probe, and treated with AF647-conjugated dibenzocyclooctyne (DBCO) to install the fluorophore via a strain-promoted azide-alkyne click reaction (SPAAC).39 At the indicated dose, a minimal effect on bacterial viability was observed due to probe treatment (Figure SI-1).
Figure 3.

(A) Flow cytometry analysis of AzFPA (250 μM) labeled C. glutamicum and M. smegmatis treated with DBCO-AF647. Mean fluorescence intensity (MFI) was calculated using the geometric mean and plotted relative to a dye-only control. Error bars denote the standard error of the mean of three replicate experiments. (B) Fluorescence emission (633 nm) from isolated mAGP from AzFPA (250 μM) labeled C. glutamicum and M. smegmatis reacted with DBCO-AF647. Error bars denote the standard error of the mean of two replicate experiments.
Analysis of the fixed samples by flow cytometry revealed that cells treated with 2-AzFPA (1) and 5-AzFPA (3) could be labeled through a strained azide-alkyne click cycloaddition with a fluorophore.39 The 2-azido isomer exhibited the brightest staining in M. smegmatis, while the 5-azido derivative led to more C. glutamicum labeling (Figure 3A). The 3-AzFPA (2) derivative afforded minimal staining of either species. The fluorescence observed with compounds 1 and 3 was not the result of non-specific staining, as Escherichia coli, an organism lacking arabinose-containing glycans, exhibited no signal (Figure SI-2).
The observed selectivity is consistent with recently disclosed structural data of the M. smegmatis arabinofuranosyltransferase EmbA bound to the endogenous arabinose donor DPA, determined by cryoelectron microscopy.37 This structure indicates key hydrogen-bonding contacts occur at the C-3 hydroxyl group of DPA in the catalytic pocket. Thus, the poor incorporation of the 3-AzFPA (2) regioisomer could result from disrupted enzyme-substrate complementarity. A critical finding from the experiments above is that each species prefers a different substrate isomer. This observation highlights the value of testing different isomers. We expect such preferences could enable the selective functionalization of distinct glycans within mixed microbial communities.
To evaluate whether AzFPA was found in the mAGP, we isolated this polysaccharide. We applied a standard procedure to cells exposed to AzFPA and then the strained alkyne dye AF647.35, 40 The fluorescence emission of each isolated polysaccharide fraction was used as a measure of the degree of incorporation (Figure 3B). The trends in fluorescence for mAGP modification were consistent with the relative cellular fluorescence observed by flow cytometry. The reduced fluorescence intensity across mAGP samples could be attributed to the relatively harsh conditions employed in the isolation protocol.
We next tested our probes in confocal fluorescence microscopy to visualize the localization of the mAGP within live cells. As before, the bacteria were cultured in the presence of AzFPA and stained with AF647-DBCO. The relative incorporation trends determined by flow cytometry were mirrored by the intensities of fluorescence observed by microscopy; the 5-and 2-AzFPA probes afforded the most pronounced signal in C. glutamicum and M. smegmatis, respectively (See Supporting Information). Together, these data indicate that the AzFPA probes are competent substrates for relevant glycosyltransferases and can be selectively incorporated into cell-surface glycans.
Having validated the utility of our platform for fluorescence-based applications, we used 2-AzFPA (1) to visualize cell wall biosynthesis. Live-cell confocal imaging of M. smegmatis revealed brighter staining at the poles and septum of dividing cells (Figure 4A). The spatial localization of the probe was quantified across cell length. The resultant fluorescence intensity plot was consistent with that obtained using other cell wall probes in M. smegmatis, namely, 7-hydroxycoumarin-3-carboxylic acid-3- amino-D-alanine (HADA), a fluorescent d-alanine analog that is incorporated into nascent peptidoglycan and a mycolic acid probe, quencher-trehalose-fluorophore (QTF).41–42
Figure 4.

(A) Localization analysis of M. smegmatis grown with 2-AzFPA (250 μM) and labeled with AF647 (500 μM). Each line denotes an individual cell (n=10). (B) Confocal fluorescence microscopy images of C. glutamicum grown with 5-AzFPA (250 μM) for 2 or 5 hours. (C) Confocal fluorescence microscopy images of C. glutamicum grown with 5-AzFPA (250 μM) and HADA (500 μM) for 2 hours. (Scale bars: 3 μm).
To better understand arabinogalactan biosynthesis, we visualized 5-AzFPA (3) incorporation in C. glutamicum (Figure 4B). Because the incorporation of our probes is contingent upon glycosyltransferase activity, we expected to observe brighter staining in areas where cell wall biosynthesis and remodeling are most active. Like M. smegmatis, C. glutamicum grows asymmetrically, with peptidoglycan biosynthesis occurring most rapidly at the old pole and slower at the new pole and septal plane.43 Exposure of cells to 5-AzFPA (3) over two doubling times afforded pronounced polar and septal staining. As cells underwent multiple division cycles after five doubling times, the staining became distributed across the cell envelope. The ability of cells to continue dividing in the presence of the probes and the morphology of the bacteria indicate that no major deleterious changes to the cell envelope occur.
The asymmetry in the fluorescence pattern was similar to that observed previously for the peptidoglycan.42 Because the peptidoglycan serves as the base cell wall structure to which the arabinogalactan is conjugated, we anticipated that arabinogalactan and peptidoglycan assembly would coincide. To test this hypothesis, we incubated C. glutamicum with 5-AzFPA (3) and HADA to visualize the mAGP and peptidoglycan simultaneously.42 The fluorescent signals co-localized (Figure 4C). These data support our hypothesis and indicate that our probe can be used in concert with established tools to explore cell envelope assembly.
To assess the utility of our probes in a more complex environment, we examined their efficacy for visualizing bacteria in a phagocytic cell. Mtb infection initiates from aerosol particles that enter the lungs of a host.44 The bacteria then recruit macrophages to the lung that phagocytose the invading pathogen.45 To examine whether labeled bacteria could be detected, we pre-stained M. smegmatis with 2-AzFPA (1) then exposed them to THP1-derived macrophages (Figure 5A). Bacteria were taken up by the phagocytic cells, and the fluorescent signal was stable (Figure 5B). These data indicate that our probes can be used to visualize d-Araf residues in more complex systems, such as infection models.
Figure 5.
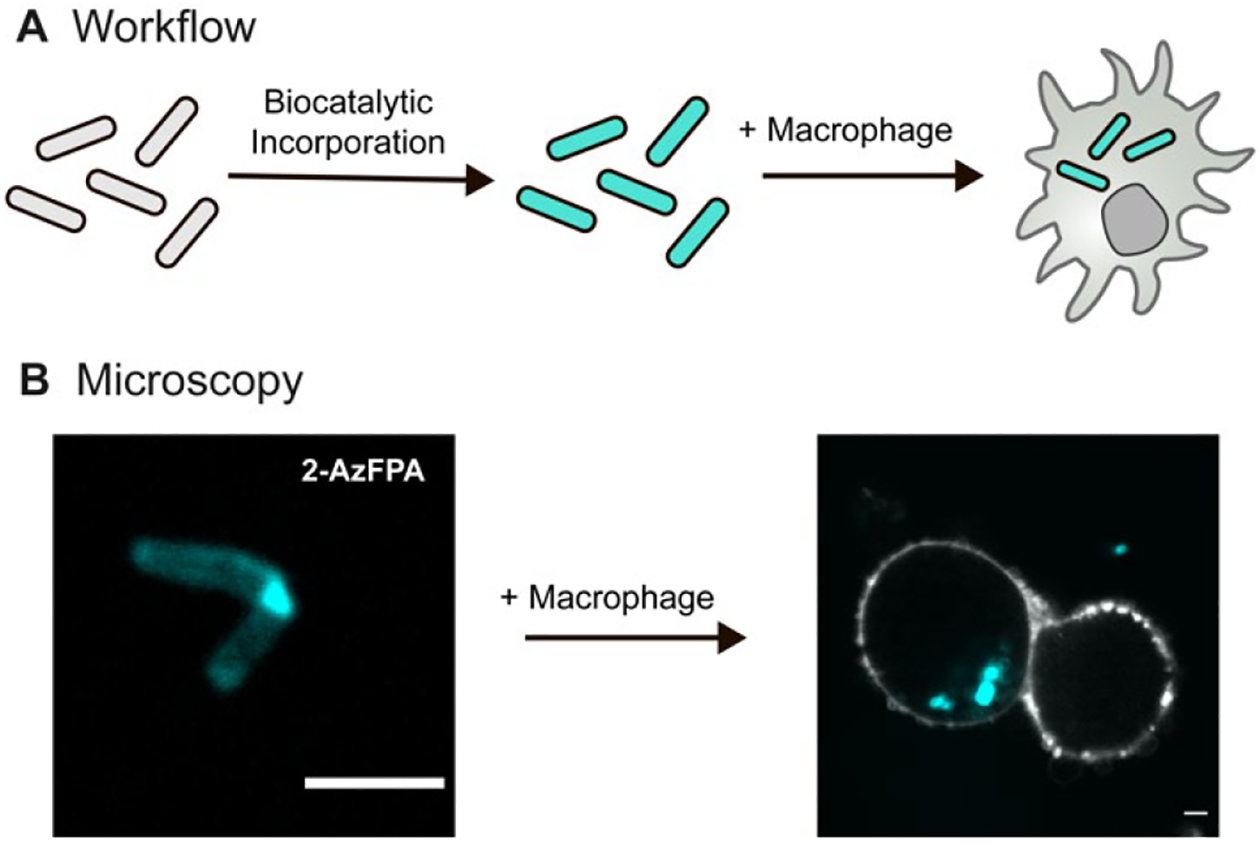
(A) Schematic of the macrophage uptake workflow. First M. smegmatis cells are exposed to AzFPA and then AF647. The resulting labeled cells were mixed with THP1-derived macrophages. (B) Confocal fluorescence microscopy images of labeled M. smegmatis (cyan) that had been taken up by THP1-derived macrophages (MOI: 10:1). Fluorophore-conjugated (405 nm) Wheat germ agglutinin was used to stain the plasma membrane (white) (Scale bars: 3 μm).
Our findings highlight the use of synthetic glycolipid donors for selective modification of cell surface glycans. Using a suite of tools for in cellulo d-Araf functionalization, we identified probes that afford species-selective glycan modification. These findings indicate that biosynthetic incorporation can be exploited to selectively modify the glycans of different species—even when these glycans are constructed from identical building blocks. The disclosed AzFPA reagents will enable new studies, including the facile purification of glycans, the visualization of polysaccharide trafficking, biosynthesis, and remodeling, and the identification of new protein-carbohydrate interactions at the cell interface. We anticipate that our findings will serve as a foundation for further expanding the biosynthetic incorporation platform to other monosaccharide components of complex glycans.
Supplementary Material
ACKNOWLEDGMENT
The authors thank H. L. Hodges, R. L. McPherson, C. M. Jarvis, C. R. Isabella and K. I. Taylor for helpful scientific discussions as well as R. L. McPherson, S. D. Brucks and S. M. Smelyansky for their assistance in reviewing the manuscript. The authors thank the NIH-NIAID (Al-126592 to L.L.K.), the NIH Common Fund (UO1GM125288 to L.L.K.), the NIH (R01A1022553 and R01AR073252 to B.D.B.) NIH-NIGMS (F32 GM142288) to D.E.K.), and the NSERC (PGSD Fellowship for V.M.M.) for financial support.
Funding Sources
This research was supported by the National Institute of Allergy and Infectious Disease (R01 Al-126592), the NIH Common Fund (U01GM125288) and the NIH (R01A1022553 and R01AR073252).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website. Experimental procedures, characterization data, supporting figures and schemes (pdf).
The authors declare no competing financial interest.
REFERENCES
- 1.Boutureira O; Bernardes GJ, Advances in chemical protein modification. Chem Rev 2015, 115, 2174–95. [DOI] [PubMed] [Google Scholar]
- 2.Gunnoo SB; Madder A, Bioconjugation - using selective chemistry to enhance the properties of proteins and peptides as therapeutics and carriers. Org Biomol Chem 2016, 14, 8002–8013. [DOI] [PubMed] [Google Scholar]
- 3.Roy S; Cha JN; Goodwin AP, Nongenetic Bioconjugation Strategies for Modifying Cell Membranes and Membrane Proteins: A Review. Bioconjug Chem 2020, 31, 2465–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilormini PA; Batt AR; Pratt MR; Biot C, Asking more from metabolic oligosaccharide engineering. Chem Sci 2018, 9, 7585–7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cioce A; Bineva-Todd G; Agbay AJ; Choi J; Wood TM; Debets MF; Browne WM; Douglas HL; Roustan C; Tastan O; Kjaer S; Bush JT; Bertozzi CR; Schumann B, Metabolic Engineering Optimizes Bioorthogonal Glycan Labeling in Living Cells. 2021. ChemRxiv. DOI: 10.26434/chemrxiv.13514365.v1. (accessed 2021-09-07) [DOI] [Google Scholar]
- 6.Dube DH; Bertozzi CR, Metabolic oligosaccharide engineering as a tool for glycobiology. Curr Opin Chem Biol 2003, 7, 616–625. [DOI] [PubMed] [Google Scholar]
- 7.Kayser H; Ats C; Lehmann J; Reutter W, New amino sugar analogues are incorporated at different rates into glycoproteins of mouse organs. Experientia 1993, 49, 885–887. [DOI] [PubMed] [Google Scholar]
- 8.Boyce M; Carrico IS; Ganguli AS; Yu SH; Hangauer MJ; Hubbard SC; Kohler JJ; Bertozzi CR, Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc Natl Acad Sci U S A 2011, 108, 3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin W; Qin K; Fan X; Peng L; Hong W; Zhu Y; Lv P; Du Y; Huang R; Han M; Cheng B; Liu Y; Zhou W; Wang C; Chen X, Artificial Cysteine S-Glycosylation Induced by Per-O-Acetylated Unnatural Monosaccharides during Metabolic Glycan Labeling. Angew Chem Int Ed 2018, 57, 1817–1820. [DOI] [PubMed] [Google Scholar]
- 10.Andolina G; Wei R; Liu H; Zhang Q; Yang X; Cao H; Chen S; Yan A; David Li X; Li X, Metabolic Labeling of Pseudaminic Acid-Containing Glycans on Bacterial Surfaces. ACS Chem Biol 2018, 13, 3030–3037. [DOI] [PubMed] [Google Scholar]
- 11.Clark EL; Emmadi M; Krupp KL; Podilapu AR; Helble JD; Kulkarni SS; Dube DH, Development of Rare Bacterial Monosaccharide Analogs for Metabolic Glycan Labeling in Pathogenic Bacteria. ACS Chemical Biology 2016, 11, 3365–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demeester KE; Liang H; Jensen MR; Jones ZS; Ambrosio EAD; Scinto SL; Zhou J; Grimes CL; D’Ambrosio EA; Scinto SL; Zhou J; Grimes CL, Synthesis of Functionalized N-Acetyl Muramic Acids to Probe Bacterial Cell Wall Recycling and Biosynthesis. J Amer Chem Soc 2018, 140, 9458–9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang H; DeMeester KE; Hou CW; Parent MA; Caplan JL; Grimes CL, Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications. Nat Commun 2017, 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mas Pons J; Dumont A; Sautejeau G; Fugier E; Baron A; Dukan S; Vauzeilles B, Identification of Living Legionella pneumophila Using Species-Specific Metabolic Lipopolysaccharide Labeling. Angew Chem Int Ed 2014, 53, 1275–1278. [DOI] [PubMed] [Google Scholar]
- 15.Herget S; Toukach PV; Ranzinger R; Hull WE; Knirel YA; von der Lieth C-W, Statistical analysis of the Bacterial Carbohydrate Structure Data Base (BCSDB): Characteristics and diversity of bacterial carbohydrates in comparison with mammalian glycans. BMC Struct Biol 2008, 8, 35–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imperiali B, Bacterial carbohydrate diversity - a Brave New World. Curr Opin Chem Biol 2019, 53, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolbe K; Möckl L; Sohst V; Brandenburg J; Engel R; Malm S; Bräuchle C; Holst O; Lindhorst TK; Reiling N, Azido Pentoses: A New Tool To Efficiently Label Mycobacterium tuberculosis Clinical Isolates. ChemBioChem 2017, 18, 1172–1176. [DOI] [PubMed] [Google Scholar]
- 18.Fraenkel DG; Vinopal RT, Carbohydrate Metabolism in Bacteria. Annu Rev Microbiology 1973, 27, 69–100. [Google Scholar]
- 19.Tommasone S; Allabush F; Tagger YK; Norman J; Kopf M; Tucker JHR; Mendes PM, The challenges of glycan recognition with natural and artificial receptors. Chem Soc Rev 2019, 48, 5488–5505. [DOI] [PubMed] [Google Scholar]
- 20.Alderwick LJ; Seidel M; Sahm H; Besra GS; Eggeling L, Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J Biol Chem 2006, 281, 15653–61. [DOI] [PubMed] [Google Scholar]
- 21.Seidel M; Alderwick LJ; Birch HL; Sahm H; Eggeling L; Besra GS, Identification of a novel arabinofuranosyltransferase AftB involved in a terminal step of cell wall arabinan biosynthesis in Corynebacterianeae, such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J Biol Chem 2007, 282, 14729–40. [DOI] [PubMed] [Google Scholar]
- 22.Skovierova H; Larrouy-Maumus G; Zhang J; Kaur D; Barilone N; Kordulakova J; Gilleron M; Guadagnini S; Belanova M; Prevost MC; Gicquel B; Puzo G; Chatterjee D; Brennan PJ; Nigou J; Jackson M, AftD, a novel essential arabinofuranosyltransferase from mycobacteria. Glycobiology 2009, 19, 1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J; Angala SK; Pramanik PK; Li K; Crick DC; Liav A; Jozwiak A; Swiezewska E; Jackson M; Chatterjee D, Reconstitution of functional mycobacterial arabinosyltransferase AftC proteoliposome and assessment of decaprenylphosphorylarabinose analogues as arabinofuranosyl donors. ACS Chem Biol 2011, 6, 819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jankute M; Alderwick LJ; Moorey AR; Joe M; Gurcha SS; Eggeling L; Lowary TL; Dell A; Pang PC; Yang T; Haslam S; Besra GS, The singular Corynebacterium glutamicum Emb arabinofuranosyltransferase polymerises the alpha(1-->5) arabinan backbone in the early stages of cell wall arabinan biosynthesis. Cell Surf 2018, 2, 38–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotake T; Yamanashi Y; Imaizumi C; Tsumuraya Y, Metabolism of L-arabinose in plants. J Plant Res 2016, 129, 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolucka BA, Biosynthesis of D-arabinose in mycobacteria – a novel bacterial pathway with implications for antimycobacterial therapy. FEBS J 2008, 275, 2691–2711. [DOI] [PubMed] [Google Scholar]
- 27.Organization, W. H., Global Tuberculosis Report 2019. World Health Organization; 2019. [Google Scholar]
- 28.Sharma NC; Efstratiou A; Mokrousov I; Mutreja A; Das B; Ramamurthy T, Diphtheria. Nat Rev Dis Primers 2019, 5, 81. [DOI] [PubMed] [Google Scholar]
- 29.Pinheiro RO; de Souza Salles J; Sarno EN; Sampaio EP, Mycobacterium leprae-host-cell interactions and genetic determinants in leprosy: an overview. Future Microbiol 2011, 6, 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alderwick LJ; Harrison J; Lloyd GS; Birch HL, The Mycobacterial Cell Wall--Peptidoglycan and Arabinogalactan. Cold Spring Harb Perspect Med 2015, 5, a021113, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takayama K; Kilburn JO, Inhibition of synthesis of arabinogalactan by ethambutol in Mycobacterium smegmatis. Antimicrob Agents Chemother 1989, 33, 1493–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikušová K; Huang H; Yagi T; Holsters M; Vereecke D; D’Haeze W; Scherman MS; Brennan PJ; McNeil MR; Crick DC, Decaprenylphosphoryl Arabinofuranose, the Donor of the d-Arabinofuranosyl Residues of Mycobacterial Arabinan, Is Formed via a Two-Step Epimerization of Decaprenylphosphoryl Ribose. J. Bacteriol 2005, 187, 8020–8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolbe K; Mockl L; Sohst V; Brandenburg J; Engel R; Malm S; Brauchle C; Holst O; Lindhorst TK; Reiling N, Azido Pentoses: A New Tool To Efficiently Label Mycobacterium tuberculosis Clinical Isolates. Chembiochem 2017, 18, 1172–1176. [DOI] [PubMed] [Google Scholar]
- 34.Lairson LL; Henrissat B; Davies GJ; Withers SG, Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem 2008, 77, 521–55. [DOI] [PubMed] [Google Scholar]
- 35.Calabretta PJ; Hodges HL; Kraft MB; Marando VM; Kiessling LL, Bacterial Cell Wall Modification with a Glycolipid Substrate. J Am Chem Soc 2019, 141, 9262–9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angala SK; Joe M; McNeil MR; Liav A; Lowary TL; Jackson M, Use of Synthetic Glycolipids to Probe the Number and Position of Arabinan Chains on Mycobacterial Arabinogalactan. ACS Chem Biol 2021, 16, 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L; Zhao Y; Gao Y; Wu L; Gao R; Zhang Q; Wang Y; Wu C; Wang M; Zhu Y; Zhang B; Bi L; Zhang L; Yang H; Guddat L; Xu W; Wang Q; Li J; Besra GS; Rao Z, Structures of cell wall arabinosyltransferases with ethambutol. Science 2020, 368, 1211–1219. [DOI] [PubMed] [Google Scholar]
- 38.Kraft MB; Martinez Farias MA; Kiessling LL, Synthesis of lipid-linked arabinofuranose donors for glycosyltransferases. J Org Chem 2013, 78, 2128–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agard NJ; Prescher JA; Bertozzi CR, A Strain-Promoted [3+2] Azide-Alkyne Cycloaddition for Covalent Modification of Biomolecules in Living Systems. J Am Chem Soc 2004, 126, 15046–15047. [DOI] [PubMed] [Google Scholar]
- 40.Besra GS; Khoo K; McNeil MR; Dell A; Morris HR; Brennan PJ, A New Interpretation of the Structure of the Mycolyl-Arabinogalactan Complex of Mycobacterium tuberculosis As Revealed through Characterization of Oligoglycosylalditol Fragments by Fast-Aton Bombardment Mass Spectrometry and 1H Nuclear Magnetic Resonance Spectroscopy. Biochemistry 1995, 34, 4257–4266. [DOI] [PubMed] [Google Scholar]
- 41.Hodges HL; Brown RA; Crooks JA; Weibel DB; Kiessling LL, Imaging mycobacterial growth and division with a fluorogenic probe. Proc Natl Acad Sci USA 2018, 115, 5271–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuru E; Hughes HV; Brown PJ; Hall E; Tekkam S; Cava F; de Pedro MA; Brun YV; VanNieuwenhze MS, In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl 2012, 51, 12519–12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kieser KJ; Rubin EJ, How sisters grow apart: mycobacterial growth and division. Nat Rev Microbiol 2014, 12, 550–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cambier CJ; Falkow S; Ramakrishnan L, Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell 2014, 159, 1497–509. [DOI] [PubMed] [Google Scholar]
- 45.Queval CJ; Brosch R; Simeone R, The Macrophage: A Disputed Fortress in the Battle against Mycobacterium tuberculosis. Front Microbiol 2017, 8, 2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


